Abstract
Histone chaperones serve a pivotal role in maintaining human physiological processes. They interact with histones in a stable manner, ensuring the accurate and efficient execution of DNA replication, repair and transcription. Retinoblastoma binding protein (RBBP)4 and RBBP7 represent a crucial pair of histone chaperones, which not only govern the molecular behavior of histones H3 and H4, but also participate in the functions of several protein complexes, such as polycomb repressive complex 2 and nucleosome remodeling and deacetylase, thereby regulating the cell cycle, histone modifications, DNA damage and cell fate. A strong association has been indicated between RBBP4/7 and some major human diseases, such as cancer, age-related memory loss and infectious diseases. The present review assesses the molecular mechanisms of RBBP4/7 in regulating cellular biological processes, and focuses on the variations in RBBP4/7 expression and their potential mechanisms in various human diseases, thus providing new insights for their diagnosis and treatment.
Keywords: RBBP4, RBBP7, biological function, human diseases, mechanism
1. Introduction
Histone chaperones serve a pivotal role in histone metabolism, facilitating histone binding to DNA during processes such as DNA replication and repair (1). Originally, Laskey et al (2) revealed that nucleophosmin in Xenopus eggs can bind histones and promote nucleosome formation independently of ATP. These two properties remain the only shared, and thus defining, characteristics of histone chaperones. Currently, histone chaperones are deemed to aid histone transfer but remain separate from the final histone-DNA complex (3). Extensive research has been conducted on the relationship between histone chaperones and human diseases (3-5). Their abnormal expression patterns, not only in tumors, but also in cardiovascular diseases, autoimmune disorders and neurodegenerative diseases, highlight their prognostic value in various human diseases. Given their role in chromatin dynamics and disease contexts, they present potential therapeutic targets and diagnostic markers in various human diseases.
Retinoblastoma binding protein (RBBP)4 and RBBP7 are recognized as histone chaperones, both of which belong to the WD40 family (6). The WD40 domain is a distinct protein motif that typically adopts a β-propeller architecture, mediating protein-protein interactions (PPIs). RBBP4/7 typically exist in complexes, such as the nucleosome remodeling and deacetylase (NuRD) complex (7) and polycomb repressive complex 2 (PRC2) (8), to regulate chromatin remodeling and gene expression through the interactions of their WD40 repeats with the H4 α1 helix and H3 tail (9,10). Dysregulation of RBBP4/7 may disrupt the normal chromatin remodeling process, resulting in altered gene expression patterns that contribute to the initiation and progression of cancer and other human diseases (11). Notably, alterations in the expression levels of RBBP4/7 have been observed in different types of human disease, such as esophageal squamous cell carcinoma (ESCC) and colorectal cancer (CRC). These changes are closely related to clinicopathological features (12-15).
The present review summarizes the pivotal role of RBBP4/7 in regulating cell fate, assessing their expression, functions, clinical features and associated mechanisms in human diseases.
2. Molecular structure of RBBP4 and RBBP7
RBBP4, also known as RbAp48 or NURF55, is integral to multiple chromatin-modifying and remodeling complexes. It was originally discovered as a binding partner of the tumor suppressor retinoblastoma protein (RB) in yeast (16), and subsequently revealed to cofractionate with histone deacetylase (HDAC)1 (17). Located on chromosome 1p35.1, the RBBP4 gene encodes a 425-amino acid protein ubiquitously present across human tissues (18). The molecular structure of the RBBP4 protein is shown in Fig. 1A.
Figure 1.
Crystal structure of histone-binding protein RBBP4/7. (A) Crystal Structure of RBBP4 (PDB ID: 3GFC). (B) crystal structure of the Apo form of human RBBP7 (PDB ID: 7M3X). RBBP, RB binding protein; PDB, Protein Data Bank.
RBBP7, alternatively known as RbAp46, is a nuclear protein ubiquitously expressed across various cell types. Located on chromosome 3p25.1, the RBBP7 gene encodes a 425-amino acid protein that is universally expressed in human tissues (18). RBBP4 and RBBP7 are 92% identical (9); however, they differ in certain amino acid sequences, which may lead to subtle structural variations, especially in domains that interact with other proteins. The molecular structure of RBBP7 is displayed in Fig. 1B. Structurally, both RBBP4 and RBBP7 possess a seven-bladed WD40 repeat domain, indicating that RBBP4 and RBBP7 can serve multiple roles in chromatin remodeling, histone modification and transcriptional regulation (19). Notably, both RBBP4 and RBBP7 are integral subunits of the NuRD complex (7), the switch-independent 3A complex (20) and PRC2 (8). Additionally, RBBP4 has been identified as a subunit of the chromatin assembly factor 1 (CAF-1) complex (21) and a core component of the MuvB complex (22), while RBBP7 is known to be an essential component of the histone acetyltransferase 1 (HAT1) complex (23). As part of these multisubunit protein complexes, RBBP4 and RBBP7 are believed to function as chromatin adapters, mediating direct interactions with histone H3/H4 (24).
3. The biological functions of RBBP4/7
The functions of RBBP4/7 in the cell cycle
RBBP4/7 have a pivotal role in cell cycle regulation, with their absence leading to dysregulation of cell cycle genes and cycle arrest (25). Specifically, RBBP4 deficiency results in S phase defects and inhibits mitotic exit (M to G1 transition) (26), whereas RBBP7 deficiency causes G2/M phase arrest in 293 cells (27). In addition, RBBP4/7 (LIN-53) are crucial for centromere protein A (CENP-A) localization to centromeres (28). It has also been suggested that the Cullin-4 (CUL4) RING ligase (CRL4) complex containing RBBP7 might regulate mitosis by promoting ubiquitin-dependent loading of newly synthesized CENP-A during the G1 phase (29).
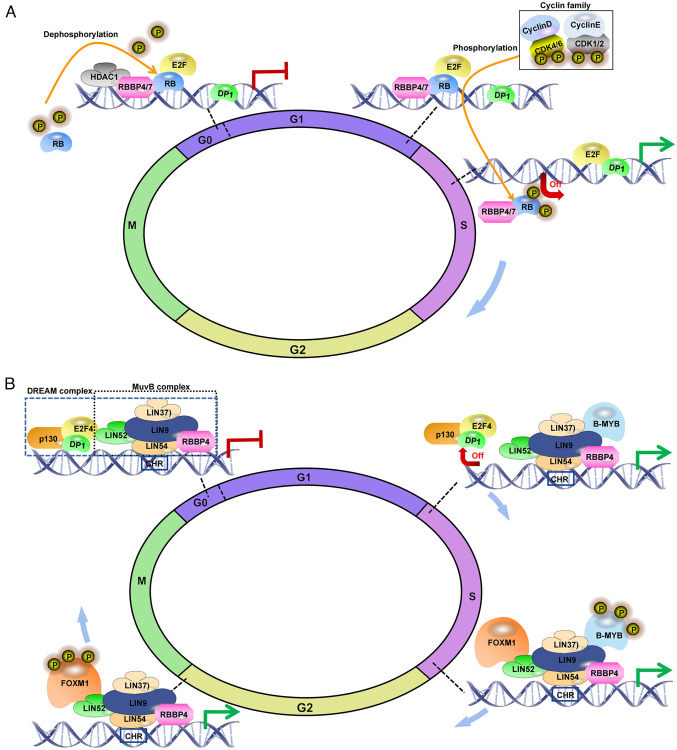
RBBP4/7 are members of the RB family (30). Studies conducted in yeast and cultured cells have shown that RBBP4 appears to function as a tumor suppressor along with RB, leading to inhibition of cell cycle progression and cell growth (16,18). Nevertheless, Schultz-Rogers et al (31) indicated that RBBP4 is essential for the cell cycle progression of neural progenitor cells and the initiation of G0, irrespective of the involvement of RB. The E2F family plays a key role in cell cycle regulation, and all RB family members interact with typical E2F proteins to form transcriptional inhibition complexes (32). RB primarily inhibits E2F transcription factor 1, whereas RBBP4 can be directed to RB via HDAC1 (33). In the G1 phase, RBBP4/7 and RB inhibit E2F target gene activation, preventing entry into S phase. Cyclin D-cyclin-dependent kinase (CDK)4/6 can monophosphorylate RB, whereas cyclin E-CDK1/2 are involved in poly-phosphorylation or hyper-phosphorylation of RB (34). Once hyperphosphorylated, RBBP4/7 and RB dissociate from E2F, activating target genes and recruiting chromatin remodelers (35) (Fig. 2A).
Figure 2.
Molecular mechanism underlying how RBBP4/7 affects the cell cycle. (A) Involvement of RBBP4/7 in cell cycle regulation via RB interaction: Guided by HDAC1, RBBP4/7 is oriented to interact with RB. During the G0/G1 phase of the cell cycle, RB restrains the expression of genes regulated by transcription factors of the E2F family via binding to them, thus the dephosphorylated RB prevents cell entry into S phase. As the cell prepares to transition from the G1 phase to the S phase, RB is phosphorylated by CDKs, and cyclins D/E. This phosphorylation leads to the disassociation of RB together with RBBP4 from E2F and gene promotors. Through dephosphorylation, RB regains its activity, binds to E2F once again, and inhibits excessive progression of the cell cycle. (B) During the G0/G1 phase, the MuvB complex, composed of LIN9, LIN37, LIN52, LIN54 and RBBP4, forms the DREAM complex through association with the RB-like protein p130, E2F4 and DP1 under quiescent conditions. This complex suppresses the expression of cell cycle regulatory genes during the G0 phase. RBBP4, through its interactions with complex members, modulates the localization of the MuvB complex and suppresses the transcription of specific genes. When transitioning to the S phase, the p130/E2F4/DP1 module dissociates from the DREAM complex, resulting in the loss of suppression. Subsequently, the MuvB complex, with RBBP4 as the core complex, cooperates with B-MYB to form the MMB complex, activating the expression of genes required for the S/G2 phase. FOXM1 is then recruited to MMB, forming the MMB-FOXM1 complex during the transition of S/G2. B-MYB undergoes phosphorylation during the late S phase, leading to its dissociation from the MMB-FOXM1 complex as the cell cycle advances, while MuvB and FOXM1 persist in the DNA until mitosis. RBBP, RB binding protein.
In addition, RBBP4 is a core component of the MuvB complex, collaboratively working with LIN9, LIN37, LIN52 and LIN54 to regulate the cell cycle (22). During the G0/G1 phase, the RBBP4-containing MuvB complex associates with p130/E2F4/DP1 to form the dimerization partner, RB-like, E2F and MuvB (DREAM) complex (36), which inhibits expression of cell cycle regulatory genes, maintaining the cell in a quiescent state. Within the DREAM complex, RBBP4 directly binds to LIN9 and LIN37 within the complex, playing a pivotal role in its assembly process (22). Additionally, RBBP4 collaborates with p130, E2F4 and DP1 to inhibit the transcriptional activity of E2F target genes (36). As the cell cycle progresses, phosphorylation by CDK leads to the disassembly of the DREAM complex, releasing p130/E2F4/DP1 from MuvB. Subsequently, the MuvB complex, inclusive of RBBP4, interacts with activated transcription factors B-MYB (also known as MYB proto-oncogene like 2) and forkhead box M1 (FOXM1), culminating in the formation of the activated MYB-MuvB-FOXM1 complex. This complex further regulates the expression of cell cycle genes, especially during the G2 phase and mitosis (37) (Fig. 2B). Thus, RBBP4, through its positioning and recruitment roles in the MuvB complex, serves a critical role in the regulation of various stages of cell cycle gene expression. Notably, the DREAM complex collaborates with RB during quiescence to suppress cell cycle gene expression (38,39). Furthermore, in response to the p53 tumor suppressor and genotoxic stress, the involvement of RBBP4 within the DREAM complex and its collaboration with RB becomes evident. Specifically, the DREAM complex and RB utilize the p53-p21 pathway to induce p21 and consequently arrest the cell cycle (39,40). The dual association of RBBP4 with the RB family and the DREAM complex indicates that it has a crucial role in maintaining cell cycle arrest.
Finally, RBBP4/7 serve as components of chromatin-remodeling complexes, such as NuRD and PRC2, further modulating the cell cycle via nucleosome acetylation and methylation regulation (8,41).
In summary, RBBP4/7, as components of the RB family or MuvB complex, have a pivotal role in cell cycle regulation. Their association with chromatin remodeling further influences cell cycle progression. While the role of RBBP4/7 in the cell cycle is evident, the specifics of their mechanism require further exploration.
The multifaceted role of RBBP4/7 in chromatin remodeling
Chromatin remodeling is a critical process that governs the accessibility of DNA to various cellular machineries, influencing gene transcription, replication and repair (42). RBBP4/7 can affect histone conformation by directly binding to histone H4 and H3 (43).
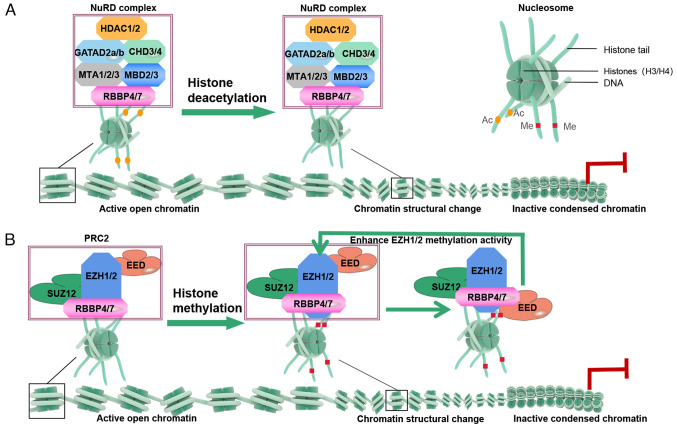
The NuRD complex, comprising several subunits, including the HDAC complex, chromodomain helicase DNA binding protein 3/4 ATPase, methyl-CpG binding domain protein 2/3, RBBP4/7, metastasis associated 1/2/3 (MTA1/2/3) and GATA zinc finger domain containing 2A/B, is a multifunctional entity with nucleosome remodeling and deacetylase activities (44). These activities enable the NuRD complex to alter chromatin structure, thereby regulating gene expression. Mu et al (45) suggested that RBBP4 may contribute to controlling the acetylation (ac) of lysine 27 on histone H3 (H3K27ac) levels at enhancer elements by promoting the deacetylase activity of the HDAC complex, effectively removing acetyl groups from H3K27 (Fig. 3A). RBBP4/7 also have regulatory chromatin remodeling effects independent of NuRD complexes. RBBP4 promotes H3K27ac by maintaining p300 levels (46), and together with RBBP7, mediates H4K5ac and H4K12ac to enable CENP-A deposition into centromeres (47). A network including SIN3 transcription regulator family member A-HDSAC3-RBBP4-H4 recognizes and deacetylates histones during chromatin assembly (48). RBBP4 also controls histone deacetylation at H3K4, H4K8, H4K12 and H4K16 during meiosis I (49). Additionally, RBBP4/7 collaborate with HAT1 in the site-specific de novo acetylation of histone H4 (50), facilitating its nuclear delivery and folding (51), which may be crucial in chromatin assembly and gene expression regulation.
Figure 3.
Mechanism of RBBP4/7 in chromatin remodeling. (A) Deacetylation: RBBP4/7 interacts with the tails of histones H3 and H4, promoting the deacetylase activity of the HDAC complex, resulting in the removal of acetyl groups from H3/H4. This leads to chromatin compaction, preventing gene promoter transcription and inhibiting gene expression. (B) Methylation: In PRC2, RBBP4 recruits SUZ12 to PRC2 target sites. The EZH2 subunit serves as the methyltransferase active center, performing primary, secondary and tertiary methylation on the 27th lysine of histone H3, resulting in H3K27me3 formation. EED binds to the H3K27me3 mark and further enhances the methyltransferase activity of EZH2. PRC2-mediated methylation induces chromatin compaction, limiting the binding of transcription factors and RNA polymerases, thus enabling gene silencing. RBBP, RB binding protein.
PRC2, consisting of core subunits SUZ12 polycomb repressive complex 2 subunit (SUZ12), embryonic ectoderm development (EED), RBBP4/7 and enhancer of zeste homolog (EZH)2 or EZH1, is the sole confirmed methyltransferase responsible for the mono-, di- and trimethylation of H3K27, generating the H3K27me3 mark (52). RBBP4/7 interact with various molecules to facilitate PRC2 recruitment and activity modulation. Studies have shown that RBBP4 can recruit SUZ12 to PRC2 target sites, and methylate H3K27 or H1K26 with the histone lysine N-methyltransferase EZH2 (45,53). Simultaneously, EED can interact with the H3K27me3 mark, thereby activating the methyltransferase activity of EZH2 and influencing the overall activity of PRC2. Consequently, this process promotes the 'spreading' of H3K27me3 (54) (Fig. 3B). Notably, the absence of the SUZ12-RBBP4 complex influences H3K27me3 (55). Therefore, as pivotal constituents of PRC2, RBBP4/7 emerge as determinants of site-specific H3K27me3 and other histone methylations across the genome.
RBBP4/7 also exhibit methyltransferase activity independent of PRC2. Kitange et al (56) showed that RBBP4 knockdown can enhance the levels of H3K9 trimethylation. RBBP4/7 interact with SUV39H1 to specifically methylate lysine 9 on histone H3, leading to heterochromatin silencing or RB transcriptional repression (57). Furthermore, RBBP7 inhibits DNA methyltransferase 1, affecting DNA methylation and limiting transcription factor access to promoters (58).
In summary, RBBP4/7 serve an important role in regulating histone deacetylation and methylation, which are key processes in chromatin remodeling and the regulation of gene expression. The precise mechanisms underlying these regulatory activities of RBBP4/7 both inside and outside the aforementioned complexes remain an area of ongoing research and exploration.
The role of RBBP4/7 in the DNA damage response
RBBP4/7 have been identified as crucial components of several protein complexes involved in DNA repair, making them essential players in maintaining genome integrity.
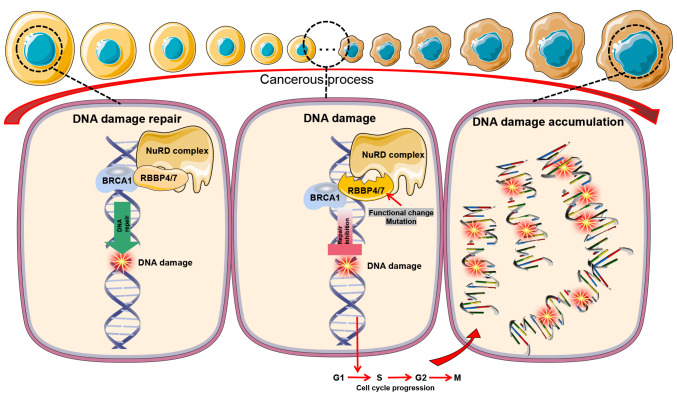
The NuRD complex governs gene expression and DNA damage repair by modulating nucleosome RNA polymerase accessibility at transcription factor binding sites, enhancers and promoters (59). RBBP4/7, as subunits of the NuRD complex, mediate the interaction of NuRD with histone tails and transcription factors (59). Yang et al (60) showed that breast cancer (BC) anti-estrogen resistance 1 and RBBP4 can form a complex, be recruited to chromatin, and jointly occupy the promoter regions of some DNA repair genes, and promote DNA damage repair. Similarly, RBBP4/7 specifically interact with the C-terminal domain of BC type 1 susceptibility protein (BRCA1) and inhibit its transactivation activity (61). The association between BRCA1 and RBBP7 is disrupted in cells treated with DNA-damaging agents (62). Therefore, the interaction between RBBP4/7 and BRCA1 might be the key to regulating DNA damage repair.
Li et al (63) demonstrated that RBBP4 disruption results in heightened DNA damage and apoptosis in glioblastoma (GBM) cells post-temozolomide (TMZ) and radiotherapy. Additionally, in MCF10AT3B cells, which are neoplastigenic breast epithelial cells derived from a model of human proliferative breast disease, high levels of RBBP7 might induce the growth arrest- and DNA damage-induced (GADD) gene, GADD45 (64). Consequently, aberrant expression of RBBP4/7 has implications for the DNA damage repair response.
In summary, RBBP4/7 serve a pivotal role in DNA damage repair and gene regulation, particularly in collaboration with BRCA1. Their interaction with BRCA1 facilitates DNA repair, and aberrant expression may affect this response, leading to the accumulation of DNA damage, as cell cycle progression and increased carcinogenic risk (Fig. 4). The complexity of these interactions warrants further investigation.
Figure 4.
RBBP4/7 participate in the DNA damage response. As a subunit of the NuRD complex, RBBP4/7 facilitate DNA repair, especially in collaboration with BRCA1 at chromatin promoter regions. Disruption of RBBP4/7 can lead to accumulated DNA damage during the cell cycle, potentially giving rise to cancer cells. RBBP, RB binding protein.
The role of RBBP4/7 in cell development, differentiation, maturation and senescence
In mouse oocytes, RBBP4 is crucial for bipolar spindle formation, and its deficiency can lead to mitotic abnormalities (49), suggesting its importance in cell division, a fundamental process of cell growth. RBBP7 is strongly expressed in the kidneys and brain from embryonic day 9.5 (65), regulating Kisspeptin-1 expression to participate in reproductive development (66). Giri et al (67) revealed that RBBP7 can be suppressed in relation to Ras-associated cell proliferation through stabilization by small ubiquitin like modifier 1.
RBBP4 is indispensable in heterochromatin assembly and serves as a crucial barrier in inducing cell fate transition from pluripotency to totipotency. Ping et al (68) demonstrated that the deletion of RBBP4 enhances the transition of mouse embryonic stem cells to trophectoderm cells. In the NuRD complex, RBBP4/7 and MTA interact with friend of GATA protein 2 (FOG-2) and are involved in FOG-2-mediated inhibition of GATA binding protein 4 activity, preventing the aberrant cell differentiation that leads to cardiac malformations (69). Moreover, the RBBP4 homolog DjRbAp48 in planarians (Dugesia japonica) regulates stem cell differentiation (70). In addition, notable discoveries have been made in the study of RBBP7. RBBP7 is involved in regulating histone acetylation and the expression of cyclin D3 in post-implantation trophoblast matrix cells (71), and it interacts with the pregnancy-induced non-coding RNA, inhibiting the differentiation of alveolar cells during pregnancy (72). Xin et al (73) showed that RBBP4/7 may be indirectly involved in the differentiation process of bone marrow cells by affecting the expression of the long noncoding RNA HOTAIRM1. Finally, in kidney development, RBBP7, as a target gene of the transcription factor Wilms tumor 1, exhibits decreased expression, reflecting podocyte dedifferentiation (74). These data suggested that RBBP4/7 may have a key role in cell differentiation processes.
During cell growth, RBBP4 influences cell morphology and cytoskeleton organization by enhancing K-Ras activity and mitogen-activated protein kinase signaling (75). Gasca et al (76) proposed that RBBP7 is involved in the maturation of oocytes. Likewise, RBBP7 also contributes to histone deacetylation during oocyte maturation (77). By contrast, Guan et al (78) showed that RBBP7 has strong growth inhibitory activity in the developing kidney and gonads. In summary, RBBP4/7 play diverse roles in cell maturation, influencing cell morphology, oocyte maturation and organ growth.
In aging human fibroblasts, a decrease in RBBP4 expression leads to chromatin defects (79). Concurrently, Hunt et al (80) demonstrated that ubiquitin protein ligase E3 component N-recognin 4 deficiency prevents skeletal muscle cell aging and atrophy by reducing the ubiquitination and degradation of the HAT1/RBBP4/RBBP7 histone-binding complex. Additionally, decreased RBBP4 expression in the aging hippocampus is associated with memory loss (81). Tsujii et al (82) further illustrated that RBBP4 knockdown might inhibit nuclear transport and induce cellular aging. Furthermore, RBBP7 has been reported to be consistently upregulated in the lobules of degenerated mammary glands and to be associated with hormone induction (83). Collectively, RBBP4/7 are closely associated with cellular senescence, affecting chromatin defects, skeletal muscle cell aging and memory loss.
In summary, RBBP4/7 exhibit diverse functions throughout cell growth and development, impacting essential cellular processes and providing valuable insights into cellular differentiation, aging and chromatin regulation.
4. Expression and function of RBBP4/7 in diseases
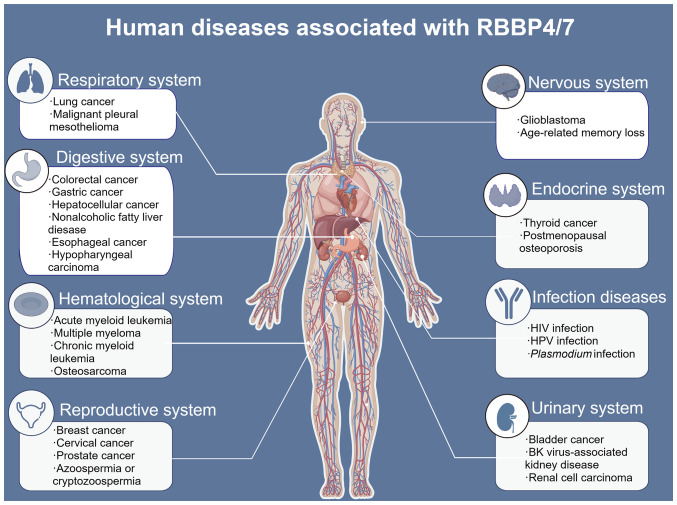
Aberrant expression of RBBP4/7 has been observed in several diseases. The present review summarizes the various diseases regulated by RBBP4/7 (Fig. 5), the clinical characteristics of RBBP4 and RBBP7 in human diseases (Tables I and II), and their roles and mechanisms in disease development (Tables III and IV).
Figure 5.
Human diseases associated with RBBP4/7. The diseases associated with RBBP4/7 are distributed across multiple systems, including the respiratory system, nervous system, digestive system, endocrine system, hematological system, reproductive system and urinary system. RBBP4/7 are also associated with infectious diseases. RBBP, RB binding protein.
Table I.
Expression of RB binding protein 4 in human diseases and relative clinical significance.
| First author, year | Disease type | Expression | Samples | Clinical characteristics | (Refs.) |
|---|---|---|---|---|---|
| Gao M, 2023 | NSCLC | Upregulated | LUAD: 54 adjacent normal, 497 tumor tissues; LUSC: 49 adjacent normal, 502 tumor tissues | Poor prognosis | (84) |
| Cao X, 2021 | NSCLC | Upregulated | / | / | (13) |
| Hao D, 2023 | NSCLC | Upregulated | 54 NSCLC tissues and 54 adjacent normal tissues | / | (85) |
| Wang N, 2021 | NSCLC | / | mRNA expression profiles of 43 patients with NSCLC | Poor prognosis, tumor recurrence | (86) |
| Vavougios GD, 2015 | MPM | Upregulated | 40 MPM tissues and 9 control tissues (5 pleura tissues and 4 lung tissues) | / | (92) |
| Shou J, 2021 | GBM | Upregulated | 33 GBM tissues and adjacent normal tissues | / | (94) |
| Li J, 2023 | GBM | Upregulated | / | Poor prognosis | (63) |
| Li D, 2018 | NB | Upregulated | Tissues from 42 primary cases of NB | Poor prognosis, tumor stage, survival rate | (97) |
| Pavlopoulos E, 2013 | Age-related memory loss | Downregulated | Entorhinal cortex and DG of 10 healthy human brains, mouse DG tissues | Memory loss | (81) |
| Kosmidis S, 2018 | Discriminative | Downregulated | Mouse DG tissues memory deficits | Discriminative memory, spatial memory | (100) |
| Khateb A, 2021 | AML | Upregulated | / | Overall survival, tumor development | (106) |
| Casas S, 2003 | AML | Upregulated | Bone marrow aspirate of 15 patients with AML and 5 healthy individuals | / | (107) |
| Sakhinia E, 2005 | AML | Upregulated | Bone marrow aspirate of 26 patients with AML, 12 patients with AML in remission and 9 individuals with morphologically normal bone marrow | AML remission | (108) |
| Sakhinia E, 2005 | ALL | Upregulated | Bone marrow aspirate of 5 patients with ALL and 9 individuals with morphologically normal bone marrow | / | (108) |
| Li YD, 2019 | CRC | Upregulated | Colon cancer tissues, para-colon cancer tissues and haptic metastatic cancer tissues from 80 patients with CRC | Haptic metastases, poor prognosis | (117) |
| Ding L, 2019 | GC | Upregulated | 142 GC tissues and adjacent normal tissues | / | (120) |
| Song H, 2004 | HCC | Upregulated | Tissue from a patient with primary HCC | / | (124) |
| Zhi S, 2022 | NAFLD | Downregulated | Liver tissues of patients with NAFLD | / | (130) |
| Chen L, 2022 | ESCC | Upregulated | ESCC tissues and corresponding normal tissues from 111 patients | / | (131) |
| Pacifico F, 2007 | TC | Upregulated | / | / | (135) |
| Guo Q, 2020 | BC | Upregulated | 240 BC tumor tissues | Overall survival, lymph node metastasis, tumor development | (141) |
| Gong X, 2020 | BC | Upregulated | / | / | (142) |
| Zheng Z, 2022 | TNBC | Upregulated | / | / | (144) |
| Barreiro-Alonso A, 2021 | PCa | Upregulated | 494 prostate adenocarcinoma tissues | Progression-free survival | (160) |
| Lohavanichbutr P, 2009 | OSCC | Upregulated | 124 OSCC patient tissues and 45 normal tissues | Radiosensitivity, chemosensitivity | (174) |
| Wurlitzer M, 2020 | HPV-positive OPSCC | Upregulated | 8 HPV-positive and 9 HPV-negative oropharyngeal tumor tissues | / | (175) |
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MPM, malignant pleural mesothelioma; GBM, glioblastoma; NB, neuroblastoma; DG, dentate gyrus; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; NAFLD, non-alcoholic fatty liver disease; ESCC, esophageal squamous cell carcinoma; TC, thyroid cancer; BC, breast cancer; TNBC, triple-negative BC; PCa, prostate cancer; OSCC, oral squamous cell carcinoma; HPV, human papillomavirus; OPSCC, oropharyngeal squamous cell carcinoma; / indicates data not specified or applicable in the sources.
Table II.
Expression of RB binding protein 7 in human diseases and relative clinical significance.
| First author, year | Disease type | Expression | Samples | Clinical characteristics | (Refs.) |
|---|---|---|---|---|---|
| Wang CL, 2009 | NSCLC | Upregulated | 154 lung cancer tissues and adjacent normal tissues | Distant metastasis | (90) |
| Wang H, 2022 | LUAD | Upregulated | 334 LUAD samples and 59 adjacent normal lung samples, and 23 cancer tissues and adjacent normal tissues from patients with LUAD | Relapse-free survival, poor prognosis, TNM stage | (89) |
| Zhu H, 2022 | LUAD | / | / | Poor prognosis | (88) |
| Vavougios GD, 2015 | MPM | Upregulated | 40 MPM tissues and 9 control tissues (5 pleura tissues and 4 lung tissues) | / | (92) |
| Crea F, 2010 | Anaplastic astrocytoma | Upregulated | / | / | (98) |
| Crea F, 2010 | Anaplastic oligodendroglioma | Upregulated | / | / | (98) |
| Dave N, 2021 | AD | Downregulated | 89 AD brain tissues and 98 normal brain tissues | CERAD (neuritic plaque density), Braak stage, brain weight | (105) |
| Hu SY, 2005 | AL | Upregulated | Bone marrow cells from 98 patients with AL, 5 patients with relapsing AL, 8 patients with CR-AL and 32 healthy individuals | / | (113) |
| Hu SY, 2005 | CML-BC | Upregulated | Bone marrow cells from 13 patients with CML-CP, patients with CML-BC and 32 healthy individuals | Tumor progression | (113) |
| Yu N, 2018 | ESCC | Upregulated | 126 ESCC tissues, 72 of which had adjacent non-neoplastic tissues | Poor differentiation, lymph node invasion and progression, pathological TNM stage, poor prognosis, overall survival | (14) |
| Wang R, 2022 | EC | Upregulated | 182 EC tissues and 286 normal tissues | Overall survival, relapse-free survival, tumor stage | (133) |
| Thakur A, 2007 | BC | Upregulated | 20 breast cancer and adjacent benign or normal breast tissue | / | (150) |
| Ebata A, 2012 | pDCIS | Upregulated | 53 pDCIS and 27 IDC tissues | / | (151) |
| Barreiro-Alonso A, 2021 | PCa | Upregulated | 494 prostate adenocarcinoma tissues | Progression-free survival | (160) |
| Riera-Escamilla A, 2022 | Azoospermia | / | X-linked protein-coding genes in 2,354 men with idiopathic NOA/cryptozoospermia | Spermatogenesis | (163) |
| Yeh HH, 2015 | Bladder cancer | Upregulated | Tissues from 4 patients with clinical bladder cancer | / | (166) |
| Wang Y, 2022 | BKVN | Upregulated | / | Immune cell infiltration, graft rejection, diagnosis | (167) |
| Wurlitzer M, 2020 | HPV-positive OPSCC | Upregulated | 8 HPV-positive and 9 HPV-negative oropharyngeal tumor tissues | / | (175) |
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; TNM, tumor-node-metastasis; MPM, malignant pleural mesothelioma; AD, Alzheimer's disease; CERAD, Consortium to Establish a Registry for AD; AL, acute leukemia; BM, bone marrow; CR-AL, complete remission acute leukemia; CML-CP, chronic myeloid leukemia in chronic phase; CML-BC, chronic myeloid leukemia in blast crisis; ESCC, esophageal squamous cell carcinoma; EC, esophageal cancer; BC, breast cancer; pDCIS, pure ductal carcinoma in situ; IDC, invasive ductal carcinoma; PCa, prostate cancer; NOA, non-obstructive azoospermia; BKVN, BK virus-associated nephropathy; HPV, human papillomavirus; OPSCC, oropharyngeal squamous cell carcinoma; / indicates data not specified or applicable in the sources.
Table III.
The functions and mechanisms of RB binding protein 4 in diseases.
| First author, year | Disease type | Cell lines | Role | Functions | Upstream regulators | Target/interacting genes | (Refs.) |
|---|---|---|---|---|---|---|---|
| Cao X, 2021 | NSCLC | A549, HC827 | Oncogene | Promotes cell proliferation, invasion and migration | hsa_circ_0102231, miR-145 | / | (13) |
| Hao D, 2023 | NSCLC | A549, H1299 | Oncogene | Promotes cell proliferation, migration, invasion, glycolysis and drug resistance | circ_0110498, miR-1287-5p | / | (85) |
| Jin X, 2022 | LUAD | A549, H1299 | Oncogene | / | / | CBX3, ARHGAP24 | (87) |
| Shou J, 2021 | GBM | U87, U251 | Oncogene | Promotes cell proliferation, viability and adhesion, inhibits cell apoptosis | HOXA-AS2, miR-885-5p | / | (94) |
| Mladek AC, 2022 | GBM | U251 | Oncogene | TMZ resistance | / | p300, c-MYC, RAD51 | (95) |
| Kitange GJ, 2016 | GBM | GBM12, GBM22 | Oncogene | Promotes tumor growth in orthotopic xenografts and TMZ resistance | / | p300, CBP, RAD51, MGMT | (56) |
| Li J, 2023 | GBM | U87MG, T98G, U118, U251 | Oncogene | Reduces sensitivity to RT and TMZ, promotes DNA damage repair | / | MRE11, RAD50, NBS1, ELF-1 | (63) |
| Li D, 2018 | NB | BE (2)-C, IMR32, SH-SY5Y | Oncogene | Promotes cell growth and invasion | / | ARMC12 | (97) |
| Kosmidis S, 2018 | Age-related memory loss | / | / | Maintains normal brain function | / | GPR158, BDNF | (100) |
| Khateb A, 2021 | AML | MOLM-13, U937 | Oncogene | Promote cell growth | RNF5 | ANXA1, NCF1, CDKN1A | (106) |
| Hu SY, 2006 | CML | K562 | Tumor suppressor | Inhibits cell growth, arrests cell cycle | / | IGFBP-rP1 | (111) |
| Zhu X, 2023 | CRC | HT29, HCT116, LOVO, SW620, RKO | Oncogene | Promotes cell proliferation and invasion | circAGO2, miR-1-3p | HSPB8 | (118) |
| Li YD, 2020 | CRC | SW620, HT29, LoVo, SW480, HCT-116 | Oncogene | Promotes cell proliferation, migration and invasion, inhibits cell apoptosis | / | Wnt/β-catenin | (12) |
| Zhuo FF, 2022 | CRC | HCT116 | Oncogene | Promotes cell proliferation and migration | / | / | (119) |
| Ding L, 2019 | GC | BGC-823, AGS | Oncogene | Promotes cell growth and reduces apoptosis | circ-DONSON | SOX4 | (120) |
| Jin X, 2018 | GC | AGS | Tumor suppressor | Promotes apoptosis, inhibits cell proliferation, enhances radiosensitivity and cell cycle arrest | / | PI3K/Akt | (121) |
| Song H, 2004 | HCC | L02, Bel-7404, HepG2, Bel-7402 and HuH7 | / | / | / | / | (124) |
| Li L, 2015 | HCC | EPCAM+ and EPCAM− HCCLM3 | Oncogene | Promotes cell proliferation, self-renewal, chemotherapy resistance and tumorigenesis | miR-429 | E2F1, OCT4 | (126) |
| Zhi S, 2022 | NAFLD | Mouse AML12 hepatocytes | Reduces hepatic steatosis | Regulates lipid metabolism and reduces lipid accumulation in liver cells | / | Cpt1α, Acox1 | (130) |
| Chen L, 2022 | ESCC | Kyse150, Kyse170, Eca109, TE1 | Oncogene | Promotes EMT transition | KTN1-AS1 | HDAC1 | (131) |
| Bai X, 2015 | Hypopharyngeal carcinoma | FaDu | Tumor suppressor | Inhibits cell proliferation, colony formation and tumor formation. Promotes apoptosis and regulates tumor suppressors | / | p53, RB, Bax, caspase-3, caspase-8, caspase-9 | (132) |
| Pacifico F, 2007 | TC | FRO | Oncogene | Promotes cell proliferation and cell cycle arrest | NF-κB | / | (135) |
| Yang C, 2019 | PMOP | / | / | Regulates mitochondrial function | / | ESR1 | (138) |
| Ishimaru N, 2006; Ishimaru N, 2008 | Autoimmune exocrine disease | / | / | Promotes exocrine cell apoptosis | / | p53 | (139, 140) |
| Gong X, 2020 | BC | MCF-7, MDA-MB-231 | Oncogene | Promotes cell proliferation, migration and invasion, inhibits cell apoptosis | / | LCPAT1, MFAP2 | (142) |
| Creekmore AL, 2008 | BC | MCF-7 | / | Regulates the expression of estrogen-responsive genes and estrogen signaling | / | ERα | (143) |
| Zheng Z, 2022 | TNBC | / | Oncogene | Promotes cell proliferation, invasion and migration, and regulates EMT activity | / | / | (144) |
| Moody RR, 2018 | TNBC | / | Oncogene | Promotes tumorigenesis | / | BCL11A | (145) |
| Kong L, 2007 | Cervical cancer | Caski, H8 | Tumor suppressor | Inhibits cell proliferation and colony formation, promotes cell senescence and prevents tumor formation | / | RB, p53, caspase-3, caspase-8, E6, E7, CCND1, c-MYC | (153) |
| Wu S, 2017 | Cervical cancer | SiHa, HeLa | Tumor suppressor | Promotes cell apoptosis and inhibits cell proliferation | / | HPV E6/E7, RB, p53, caspase-3 | (155) |
| Zhong J, 2015 | Cervical cancer | MS751 | Oncogene | Promotes cell migration and invasion, inhibits EMT | / | SNAIL, TWIST | (156) |
| Zheng L, 2013 | Cervical cancer | SiHa, Caski, HeLa | Tumor suppressor | Promotes apoptosis, inhibits cell proliferation and enhances radiosensitivity | / | / | (157) |
| Cai L, 2014 | PCa | LNCaP | Tumor suppressor | Promotes apoptosis and inhibits cell proliferation | / | EAF2 | (159) |
| Wang J, 2016 | HIV infection | 293T, TZM-bl, CEM-ss | Antiviral | Inhibits the expression of HIV-1 | / | / | (169) |
| Wang J, 2019 | HIV infection | HIV-1 infected T cells, J-lat | Antiviral | Suppresses transcription and remodels chromatin | / | NR2F1, HDAC1/2 | (170) |
| Biswas S, 2018 | HIV infection | HIV-2-infected MDMs | / | / | / | / | (171) |
| Xu W, 2020 | HIV infection | 293T | antiviral | Inhibits the activity of LTR, affect the nuclear translocation of p65 | / | P65, NF-κB pathway | (172) |
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; GBM, glioblastoma; TNM, tumor-node-metastasis; TMZ, temozolomide; RT, radiotherapy; NB, neuroblastoma; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; NAFLD, non-alcoholic fatty liver disease; ESCC, esophageal squamous cell carcinoma; TC, thyroid cancer; BC, breast cancer; PMOP, postmenopausal osteoporosis; TNBC, triple-negative BC; EMT, epithelial-mesenchymal transition; HPV, human papillomavirus; PCa, prostate cancer; LTR, long terminal repeat; / indicates data not specified or applicable in the sources.
Table IV.
The functions and mechanisms of RB binding protein 7 in diseases.
| First author, year | Disease type | Cell lines | Roles | Functions | Upstream regulators | Target/interacting genes | (Refs.) |
|---|---|---|---|---|---|---|---|
| Wang CL, 2009 | NSCLC | CL1-0, CL1-5 | Oncogene | Promotes cell migration | / | / | (90) |
| Dave N, 2021 | AD | HT-22 | / | Inhibits cell death | / | tau, p300 | (105) |
| Duan WM, 2004 | / | U937 | Tumor suppressor | Inhibits leukemic tumor growth | / | / | (114) |
| Zhang TF, 2003 | Osteosarcoma | Saos-2 | Tumor suppressor | Promotes apoptosis and inhibits the growth of tumor grafts | / | JNK | (116) |
| Guo L, 2023 | CRC | / | Tumor suppressor | Promotes apoptosis | / | p53, FAS, FIT | (15) |
| Yu N, 2018 | ESCC | TE1, KYSE30, KYSE150, Eca109 | Oncogene | Promotes cell invasion and migration | / | / | (14) |
| Wang R, 2022 | EC | Eca109, KYSE450 | Oncogene | Promotes cell viability and proliferation | HIF1α | CDK4 | (133) |
| Xie ZF, 2020 | EC | ECA-109, KYSE-510c | Oncogene | Promotes cell proliferation, migration, invasion and glycolysis | circ_0006168, miR-384 | S6K/S6, | (134) |
| Creekmore AL, 2008 | BC | MCF-7 | / | Regulates the expression of estrogen-responsive genes and estrogen signaling | / | ERα | (143) |
| Moody RR, 2018 | TNBC | / | Oncogene | Promotes tumorigenesis | / | BCL11A | (145) |
| Zhang TF, 2003 | BC | MCF-7, MDA-MB-231, MDA-MB-43 | Tumor suppressor | Inhibits tumor formation | / | / | (146) |
| Zhang TF, 2007 | BC | MCF10AT3B | Tumor suppressor | Inhibits tumor formation and cell proliferation | / | β-catenin | (147) |
| Li Gc, 2003 | BC | MCF10AT3B | Tumor suppressor | Promotes apoptosis and inhibits cell growth | / | JNK | (64) |
| Li GC, 2006 | BC | MCF10AT3B | Oncogene | Promotes cell migration, invasion and EMT | / | N-cadherin, E-cadherin, α/β/γ-catenin | (149) |
| Fu J, 2011 | BC | MDA-MB-435 | Oncogene | Promotes cell migration, invasion and EMT | / | E-cadherin, TWIST | (148) |
| Li HJ, 2016 | Cervical cancer | HeLa, SiHa | Tumor suppressor | Inhibits cell invasion and EMT | NKX6.1 | Vimentin, N-cadherin | (158) |
| Cai L, 2014 | PCa | LNCaP | Tumor suppressor | Promotes apoptosis and inhibits cell proliferation | / | EAF2 | (159) |
| Wang J, 2020 | PCa | DU145 | Tumor suppressor | Inhibits cell migration and EMT | / | HNF1B | (161) |
| Yeh HH, 2015 | Bladder cancer | T24, NIH3T3 | Oncogene | Promotes cell invasion and tumor metastasis | Ras | RECK, MMP-9, HDAC1, Sp1 | (166) |
NSCLC, non-small cell lung cancer; AD, Alzheimer's disease; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; EC, esophageal cancer; BC, breast cancer; TNBC, triple-negative BC; EMT, epithelial-mesenchymal transition; PCa, prostate cancer; / indicates data not specified or applicable in the sources.
Respiratory system Lung cancer (LC)
RBBP4 is associated with genetic susceptibility to LC (84). Elevated RBBP4 expression in non-small cell LC (NSCLC) tissues amplifies cell proliferation and invasion, and is concomitantly linked to an adverse clinical outcome (13,84). RBBP4 is also increased in cisplatin-resistant NSCLC, affecting drug resistance (85), and is associated with enhanced DNA damage sensitivity and repair pathway activity (86). Additionally, in lung adenocarcinoma (LUAD) cells, RBBP4 interacts with chromobox homolog 3, which is found to be upregulated in current smokers with LUAD, thereby promoting LUAD progression (87). Thus, RBBP4 could be a potential biomarker or therapeutic target for LUAD recurrence and prognosis post-platinum treatment.
Research has revealed that the expression levels of RBBP7 in NSCLC are higher than those in normal lung tissues, and this elevated expression is associated with distant metastasis, poor prognosis and tumor immune response in NSCLC, serving as a predictor for the recurrence of early-stage NSCLC (88-90).
Despite these significant findings, the understanding of the functions of RBBP4/7 in LC is limited and more in-depth research is needed to develop new treatment strategies.
Malignant pleural mesothelioma (MPM)
MPM has an average survival of 1 year post-diagnosis, urgently necessitating improved treatment methods (91). Vavougios et al (92) showed that RBBP4/7 interact with Parkinson disease protein 7 and are upregulated in an array of 18 different sarcoma types. However, the mechanism by which RBBP4/7 functions in MPM remains unknown.
Nervous system
GBM and other brain tumors
RBBP4 plays an indispensable role in the disease development of GBM (93), with its mRNA expression universally upregulated in GBM tissues where it acts as an oncogene to promote GBM malignancy, countering the tumor-suppressive effects of microRNA (miR)-885-5p (94). In GBM cells, the RBBP4/p300 complex governs pro-survival genes and influences the responsiveness to TMZ (95). This suggests that disrupting RBBP4 or p300 might enhance sensitivity to TMZ. In addition, RBBP4 increases TMZ resistance by regulating the expression of the MRN complex (MRE11 homolog, RAD50 double-strand break repair protein, and nibrin), thereby reducing sensitivity to radiotherapy and TMZ (63). Therefore, RBBP4 potentially enhances cancer progression or drug resistance through DNA repair and might be considered a new therapeutic target for future GBM treatment.
RBBP4 is required during brain development, and RBBP4 is upregulated in RB1-mutated embryonic brain tumors, serving as a potential target for inducing apoptosis in RB1-mutated brain cancer cells (96). Additionally, in neuroblastoma, RBBP4 is upregulated and is associated with poor patient prognosis (97).
By contrast, limited research has been conducted on the involvement of RBBP7 in GBM. Notably, Crea et al (98) explored the Oncomine database and observed an upregulation of RBBP7 in anaplastic astrocytoma and anaplastic oligodendroglioma.
Age-related memory loss
Loss of RBBP4 is key to age-related memory decline. Its expression in human and mouse brains declines with age, affecting memory formation (81). Another study demonstrated that RBBP4 regulates the expression of brain-derived neurotrophic factor and G protein-coupled receptor 158, key components of the mouse hippocampal osteocalcin (OCN) signaling pathway (99). Inhibition of RBBP4 disrupts the cognitive benefits of OCN and leads to discriminative memory deficits (100). Furthermore, certain genetic variants in a cAMP element binding protein-dependent histone acetylation pathway, associated with RBBP4, influence memory performance in cognitively healthy elderly individuals (101). Therefore, RBBP4 could serve as a potential therapeutic target for age-related memory loss.
Current research has explored RBBP4/7 as therapeutic targets to address age-related memory loss. The functional role of RBBP4 in Alzheimer's disease (AD) might be influenced by the instability of the RBBP4-FOG1 complex (102). Huang et al (103) identified three traditional Chinese medicine compounds (bittersweet alkaloid ii, eicosanedioic acid and perivine), which could enhance the stability of the RBBP4-FOG1 complex, offering potential therapeutic benefits for AD. However, another study showed that RBBP4/7 did not contribute to the neuroprotective effects of green tea polyphenols (104). Thus, the mechanism of RBBP4/7 as a target for age-related memory loss requires further investigation.
Dave et al (105) discovered that RBBP7 mRNA expression is diminished in AD cases, with significant negative correlations with the Consortium to Establish a Registry for AD and Braak stage. Moreover, this previous study revealed that high RBBP7 expression mitigates tau acetylation and phosphorylation, thereby preventing tau pathologies (105).
In summary, RBBP4/7 play crucial roles in age-related memory deficits, and present promising therapeutic targets for future interventions in cognitive aging and associated diseases.
Hematological (blood and bone marrow) system diseases Acute myeloid leukemia (AML)
In AML, elevated RBBP4 expression is linked to poorer survival and disease progression (106-108). Moreover, AML primary blasts with lower levels of ring finger protein 5/RBBP4 have demonstrated increased sensitivity to the HDAC inhibitor FK228. These findings suggest that the abundance of RBBP4 may serve as valuable marker to stratify patients with AML who might benefit from treatment with HDAC inhibitors (106). However, to the best of our knowledge, there is no currently research indicating an association between RBBP7 and AML.
Multiple myeloma (MM)
Gao et al (109) showed that RBBP4 is a core gene in MM, and its dysregulation is evident in the epigenetic modifications of MM. Thus, RBBP4 emerges as a potential focal point in MM research, providing promising avenues to understand the pathogenesis of MM and develop more effective therapeutic strategies. However, to the best of our knowledge, no studies have revealed a role for RBBP7 in MM.
Chronic myeloid leukemia (CML)
CML is a malignant hematological disorder. It has been shown that BMI1 (encoding B lymphoma Mo-MLV insertion region 1 homolog) transcript levels are significantly increased in CML cells, and the expression of RBBP4 is decreased after BMI1 silencing (110). In addition, RBBP7 in K562 leukemic cells inhibits growth by inducing IGFBP7 expression (111). These findings revealed that RBBP4/7 might serve an important role in the biological process of CML. However, the expression levels of RBBP7 were significantly elevated in patients with acute leukemia and CML in blast crisis compared with healthy donors and those with CML in chronic phase, indicating that it might be involved in the occurrence of leukemia (112,113). In addition, RBBP7 overexpression slows growth in the U937 leukemia cell line (114).
Osteosarcoma
Osteosarcoma originates from primitive mesenchymal cells in the bone, rarely in soft tissue, and if untreated, can lead to local and often metastatic progression (115). Zhang et al (116) showed that the inducible expression of RBBP7 can activate the c-Jun N-terminal kinase signaling pathway, and trigger apoptosis in Saos-2 osteosarcoma cells, while also strongly suppressing the formation of tumor grafts in nude mice and significantly reducing the growth of established osteosarcoma xenografts.
Digestive system
CRC
RBBP4 has been identified as a key factor in CRC, with studies showing its upregulation in colon cancer tissues, and linking increased RBBP4 expression to poor prognosis and liver metastasis (117). Reducing RBBP4 levels can hinder the growth and migration, and increase the apoptosis of HCT116 and SW620 colon cancer cells, and also suppress the Wnt/β-catenin pathway (12). Concurrently, knocking down RBBP4 also inhibits H3K27ac acetylation and HSPB8 gene transcription (118). Another study demonstrated that RBBP4 is a key target of protopanaxadiol (PPD), a major ginseng metabolite (119), suggesting its potential as a new diagnostic and therapeutic target for CRC.
RBBP7 has been reported to form a trimeric complex with long non-coding RNA FIT and p53, enhancing p53-mediated FAS gene transcription, which promotes CRC cell apoptosis (15). This suggests the apoptosis-inducing role of RBBP7 in CRC, which differs from the role of RBBP4. However, the precise expression and mechanism of RBBP7 in CRC warrant further investigation.
Gastric cancer (GC)
Ding et al (120) revealed that the expression of RBBP4 is increased in GC tissues, and knocking down RBBP4 can significantly inhibit GC cell proliferation, migration and invasion, and promote cell apoptosis. Radiation can also increase RBBP4 expression in AGS GC cells, leading to G2 phase arrest. Moreover, RBBP4 enhances the radiosensitivity of these cells by inhibiting the PI3K/Akt pathway (121). These results underscore the potential of RBBP4 as a promising target for future gene therapy interventions in the treatment of GC.
Currently, research on the role of RBBP7 in GC is limited. Src-suppressed C-kinase substrate (SSeCKS), a crucial substrate for protein kinase C, is significantly downregulated in GC (122). Liu et al (123) demonstrated that the re-expression of SSeCKS induces RBBP7, suggesting a potential connection. However, the specific function and significance of RBBP7 in GC remains to be further determined.
Hepatocellular carcinoma (HCC)
It has been shown that RBBP4 is highly expressed in liver tumor tissues, and is associated with clinical severity and disease prognosis (124). In HCC cells, Liu et al (125) identified that RBBP4 interacts with the N-terminal peptide of Sal-like protein 4 (SALL4), contributing to the silencing of tumor suppressor genes, such as PTEN. Furthermore, a potent SALL4 peptide antagonist (FFW) targeting RBBP4 significantly inhibits HCC cell growth. These studies have shown that RBBP4 plays a role in promoting HCC, which is expected to be a potential target for future HCC treatment.
By contrast, Li et al (126) observed a decrease in RBBP4 expression in HCC tissues; it was revealed that RBBP4 knockdown may enhance the self-renewal and tumorigenic potential of epithelial cell adhesion molecule-positive liver tumor-initiating cells, suggesting that RBBP4 serves as a downstream target of miR-429. Furthermore, epigallocatechin gallate (EGCG) can reverse multidrug resistance (MDR) in HCC, and RBBP4 has been reported to be significantly upregulated after EGCG treatment (127), suggesting that RBBP4 could be a potential anti-MDR target in HCC.
Nonalcoholic fatty liver disease
Hepatic steatosis, crucial in nonalcoholic steatohepatitis development, elevates the risk of cirrhosis and HCC (128). Within the NuRF complex, the subunit RBBP4 assumes a role in the regulation of lipid droplet size by transcriptionally suppressing target genes (129). Zhi et al (130) suggested that RBBP4 exerts a favorable influence on liver cell steatosis by promoting the expression of genes associated with fatty acid β-oxidation. These findings illustrate a protective role of RBBP4 in the context of hepatic steatosis.
Esophageal cancer (EC) and hypopharyngeal carcinoma
Research has shown that RBBP4 is upregulated in ESCC and promotes the epithelial-mesenchymal transition (EMT) process (131). By contrast, Bai et al (132) demonstrated that in hypopharyngeal carcinoma, a distinct squamous cell carcinoma impacting the upper aerodigestive tract, RBBP4 overexpression curtails proliferation, colony formation and tumorigenesis in the FaDu hypopharyngeal carcinoma cell line. The role of RBBP4 in hypopharyngeal carcinoma growth appears linked to its modulation of tumor suppressors. This contrasting role of RBBP4, from facilitating tumor progression in ESCC to inhibiting growth in hypopharyngeal carcinoma, demonstrates its functional variability across different types of cancer, highlighting its potential complexity as a therapeutic target.
Yu et al (14) observed that higher RBBP7 expression in ESCC tissues is correlated with poor differentiation, advanced lymph node involvement, higher tumor-node-metastasis stage, reduced survival, and increased cell invasion and migration. Mechanistically, hypoxia can induce high expression of RBBP7, which in turn upregulates CDK4 expression and promotes tumor progression (133). Furthermore, RBBP7 is upregulated in EC. As a target of miR-384, RBBP7 mRNA levels are elevated by circ_0006168 through its interaction with miR-384. This, in turn, promotes cell proliferation, migration, invasion and glycolysis in EC (134).
Collectively, RBBP4/7 are crucial in EC progression and prognosis, with their complex molecular interactions making them promising for future therapeutic and diagnostic developments in esophageal oncology.
Endocrine system
Thyroid cancer (TC)
Pacifico et al (135) detected increased RBBP4 expression in primary human TC via immunohistochemical analysis, particularly in undifferentiated TC samples and cell lines. In addition, RBBP4 knockdown was shown to reduce FRO anaplastic thyroid carcinoma cell colony formation, suggesting RBBP4 as a target of nuclear factor (NF)-κB and a potential therapeutic target for NF-κB-dependent TC. Therefore, RBBP4 might be a potential target for TC therapy, particularly for NF-κB-dependent cases.
Postmenopausal osteoporosis (PMOP) and other estrogen-influenced conditions
PMOP represents a significant global public health concern. RBBP4 expression is elevated in estrogen-deficient rats and patients with PMOP, decreasing after treatment with Liuwei Dihuang pills (136,137). Moreover, RBBP4 has been reported to be upregulated in the blood of patients with PMOP, suggesting that it could serve as a potential diagnostic biomarker of PMOP (138). The interaction between RBBP4 and estrogen receptor 1 (ESR1) is believed to serve a crucial role in the pathogenesis of PMOP (138). In summary, RBBP4 serves as a potential biomarker for PMOP diagnosis and its interaction with ESR1 suggests a fundamental mechanism in PMOP pathogenesis.
Additionally, estrogen deficiency-induced overexpression of RBBP4 triggers p53-mediated apoptosis in exocrine cells, implying a link to autoimmune exocrine disorders in postmenopausal women (139,140). Therefore, RBBP4 represents a novel immunotherapeutic target for preventing the development of sex-based autoimmune exocrine disorders.
Reproductive system
BC
RBBP4 is highly expressed in BC and is associated with poorer overall survival and a greater likelihood of lymph node metastasis (141). Knockdown of RBBP4 inhibits the proliferation, migration and invasion of BC cells, while affecting the transcription of tumor-related genes, such as microfibril-associated protein 2, which is activated via the interaction between RBBP4 and the long noncoding RNA LCPAT1 (142). Another study showed that RBBP4/7 interact with DNA-bound estrogen receptor α to alter the expression of estrogen-responsive genes in MCF-7 cells (143). Thus, RBBP4 is implicated in BC progression, in which it influences survival, metastasis and estrogen-responsive gene expression.
RBBP4 expression has also been shown to be significantly elevated in triple-negative BC (TNBC) cells and tissues; and its knockdown markedly inhibits TNBC cell proliferation, invasion and migration, and concurrently downregulates EMT regulatory activities (144). In addition, Moody et al (145) demonstrated that BCL11 transcription factor A (BCL11A), which possesses the ability to promote BC progression, can interact with RBBP4/7; therefore, targeting RBBP4-BCL11A binding may have therapeutic potential.
The role of RBBP7 in BC appears complex and contradictory. Zhang et al (146) observed decreased expression levels of RBBP7 in BC cell lines, and its dysregulation was shown to contribute to BC tumorigenesis. Further research has revealed that RBBP7 is related to estrogen regulation and may affect the early development of BC (64,147). As a component of the Mi2/NuRD complex, RBBP7 regulates TWIST-mediated repression of E-cadherin expression and inhibits BC cell metastasis (148). By contrast, Li and Wang (149) found that recombinant RBBP7 induces EMT and enhances mammary epithelial cell migration. Thus, the mechanism of RBBP7 in BC progression and metastasis requires further investigation.
Notably, in contrast to the observation of Zhang et al (146) of decreased RBBP7 expression in BC, Thakur et al (150) showed that RBBP7 expression was upregulated in 79% of BC cases and its expression was positively correlated with malignancy. RBBP7 may also be involved in the pathogenesis of estrogen receptor-positive pure ductal carcinoma in situ (151). In addition, Mieczkowska et al (152) found that RBBP7 was downregulated in parental G-2 cells from the WAP-T transgenic breast cancer line after surviving traditional cytotoxic combination therapy, suggesting that RBBP7 might be considered a potential therapeutic target for BC in the future.
In summary, both RBBP4 and RBBP7 have demonstrated significant roles in the progression, metastasis and therapeutic potentialities of BC; however, their precise mechanisms and interactions in various BC subtypes warrant deeper exploration.
Cervical cancer
Kong et al (153) observed that RBBP4 overexpression inhibits cervical cancer growth and affects human papillomavirus (HPV)16 transformation by regulating tumor suppressors and oncogenes. Notably, 5-aminole-vulinic acid photodynamic therapy (ALA-PDT), an effective treatment for HPV-related conditions, has been reported to elevate RBBP4 expression in HPV16 immortalized cervical epithelial H8 cells (154). A subsequent decrease in RBBP4 can mitigate the inhibitory effects of ALA-PDT-induced cell proliferation and apoptosis in cervical cancer cells (155). These studies demonstrated that RBBP4 may function as a tumor suppressor in cervical cancer and could serve as a promising therapeutic target for future cervical cancer intervention.
However, studies have also indicated that RBBP4 promotes cervical cancer, influencing EMT and radiotherapy outcomes (156,157). This implicates RBBP4 as a prospective target to boost radiotherapeutic outcomes in patients with cervical cancer. In addition, RBBP7 can be recruited by NK6 homeobox 1, thereby inhibiting the invasive ability of cervical cancer cells (158).
In conclusion, RBBP4/7 have complex roles in cervical cancer and may be potential therapeutic targets. However, the role of RBBP4 in cervical cancer remains controversial, and its mechanism requires further study.
Prostate cancer
Cai et al (159) showed that there is a physical interaction between the tumor suppressor gene EAF2 (encoding ELL associated factor 2) and RBBP4/7, where their overexpression induces cell death in LNCaP prostate cancer cells. High RBBP4/7 expression in prostate adenocarcinoma is also linked to shorter progression-free survival, with RBBP7 interacting with high mobility group box 1 to regulate RNA processing (160). Furthermore, overexpression of RBBP7 suppresses SLUG1/EMT in DU145 cells and exerts tumor suppressive functions in presence of hepatocyte nuclear factor 1β (161). Thus, RBBP4/7 are pivotal in prostate cancer progression and potential therapeutic targets.
Azoospermia or cryptozoospermia
Male infertility, affecting ~7% of men in the general population, is often due to factors such as azoospermia or cryptozoospermia (162). The X chromosome is vital for male reproductive health. Riera-Escamilla et al (163) linked RBBP7 mutations to early spermatogenic failure in an analysis of 2,354 men with azoospermia/cryptozoospermia, revealing that these mutations were more prevalent in this infertile group compared with in control individuals with normozoospermia. RBBP7 forms the CRL4B-RBBP7 complex with CUL4B (encoded by another mutated gene found in infertile men), which contributes to the degradation of HUWE1 and is associated with non-obstructive azoospermia (164). In conclusion, RBBP7 has a central role in male infertility, highlighting the importance of genetic factors in reproductive health.
Urinary system
Bladder cancer
Bladder cancer, affecting >440,000 individuals annually worldwide (165), shows high RBBP7 expression in specimens. Mechanistically, RBBP7 can bind to HDAC1 and specificity protein 1 (SP1), and then bind to the RECK (encoding reversion inducing cysteine rich protein with kazal motifs) promoter at the SP1 site, thereby inhibiting the expression of RECK, which in turn leads to matrix metalloproteinase-9 activation and metastasis, thereby participating in Ras-induced experimental lung metastasis (166). Therefore, RBBP7 could be used as a therapeutic target for Ras-related cancer; however, to the best of our knowledge, there are currently no studies on RBBP4 in bladder cancer.
BK virus-associated kidney disease
Wang et al (167) demonstrated that RBBP7 is highly enriched in BK virus-associated nephropathy (BKVN) tissues and is associated with alterations in various immune cells, such as CD8 naïve cells, induced regulatory T cells, neutrophils and CD8+ T cells. Furthermore, RBBP7 serves as a molecular biomarker for the precise diagnosis of BKVN, effectively distinguishing transplant rejection responses. Thus, targeting RBBP7 as a diagnostic tool may offer novel therapeutic and prognostic opportunities for BKVN in transplant recipients.
Renal cell carcinoma (RCC)
Kim et al (168) showed that RBBP7 is highly expressed in the chromaffin subtype of RCC, but not in traditional RCC. Therefore, RBBP7 could be used as a candidate biomarker in RCC, and its existence and expression patterns might be related to the pathological characteristics of RCC subtypes, providing a novel direction for the diagnosis and treatment of RCC.
Infectious diseases
HIV infection
Wang et al (169) reported increased RBBP4 expression following HIV-1 infection in cell culture models, with RBBP4 knockdown enhancing HIV infection and viral production. RBBP4 suppresses HIV-1 transcriptionally by binding to its long terminal repeats, recruiting nuclear receptor subfamily 2 F group member 1 and HDAC1/2, leading to H3 deacetylation and replication control (170). Similarly, Biswas et al (171) observed elevated RBBP4 levels in HIV-2-infected monocyte-derived macrophages, and Xu et al (172) reported that thieno[3,4-d] pyrimidine treatment in infected cells increases RBBP4 levels and activates the NF-κB pathway, suppressing HIV-1. Collectively, these findings demonstrate a critical role for RBBP4 in the regulation of HIV infection and suggest its potential as a therapeutic target for HIV management.
HPV infection
Oral squamous cell carcinoma (OSCC) and oropharyngeal squamous cell carcinoma (OPSCC) constitute a major global public health burden, and there is an association between infection with high-risk types of HPV and OSCC risk (173). Lohavanichbutr et al (174) identified differential expression of RBBP4 in HPV-positive vs. HPV-negative oropharyngeal cancer. Wurlitzer et al (175) performed a mass spectrometric comparison of eight HPV-positive and nine HPV-negative OPSCC cases, and found that RBBP4/7 was expressed at higher levels in HPV-positive OPSCC.
In cervical cancer, a major HPV-related cancer, RBBP4 mediates the transforming activity of HPV16 (153) and is upregulated by ALA-PDT in HPV16 immortalized cervical cells (154). These findings indicated that RBBP4 plays a key role in HPV infection; however, the specific mechanism still needs further exploration. Moreover, current research on the role of RBBP7 in HPV infection is insufficient.
Plasmodium infection
Kaushik et al (176) discovered that the homologs of RBBP4/7 in Plasmodium falciparum (PfRBBP4/7, PF3D7_0110700) retain the β-helical conformation and binding interfaces, exhibit significant interspecies differences, and show stage-specific expression in the asexual blood stages of the parasite, increasing from the ring stage to the schizont stage, and localizing in the nucleus. Furthermore, PfRBBP4/7 have been shown to interact with histone H4, suggesting their role in chromatin assembly and remodeling pathways in P. falciparum. As CAF-1 family members, they show structural and functional consistency. PfRBBP4, central in malaria biology with 108 PPIs, emerges as a potential antimalarial drug target (177). Thus, the function of PfRBBP4/7 in P. falciparum illustrates their potential as targets to develop novel antimalarial interventions.
5. RBBP4/7 as potential targets for human disease treatment
In the realm of targeted therapy research focused on RBBP4/7, these proteins have demonstrated significant potential in the treatment of various diseases, particularly in modulating therapeutic outcomes. For example, increased expression of RBBP4 has been linked to mitigating lead-induced neuronal apoptosis, suggesting a potential role in alleviating lead poisoning and related neurological disorders (178). Additionally, the interaction of RBBP4 with the efficacy of multiple drugs has been extensively studied, including its role in enhancing the sensitivity of GBM cells to TMZ (56,95), suppression of LC cell malignancy via ropivacaine by downregulating RBBP4 (179), and the identification of the circ-0110498/miR-1287-5p/RBBP4 axis as a novel target for overcoming cisplatin resistance in NSCLC (85). RBBP4 is also considered a potential target for treating CRC with PPD (119). In therapeutic contexts, RBBP4 expression is significantly increased in cervical cancer cell lines treated with ALA-PDT (155), and upregulation of RBBP4 has been found to induce radiosensitivity in BC, melanoma and TNBC (180). Conversely, reduced levels of RBBP7 may be associated with survival rates and chemoresistance phenotypes in basal-like BC (152).
PPIs play a pivotal role in cellular functions, and modulating PPIs offers a novel therapeutic avenue. It has been reported that blocking the interaction between BCL11A and RBBP4 reduces the cancer stem cell population in TNBC (145). Furthermore, compounds, such as bittersweet alkaloid II, may aid in AD treatment by stabilizing the RBBP4-FOG1 complex (103). Additionally, peptides designed by Hart et al targeting the RBBP4/MTA1 interaction interface show potential as future therapeutic strategies for disrupting epigenetic regulation mechanisms in various types of cancer (181). Despite the potential of small molecules or peptides targeting RBBP4/7, challenges such as low oral bioavailability and poor in vivo stability remain, necessitating further research to overcome these obstacles.
Emerging research has consistently linked elevated RBBP4/7 expression to poorer prognosis across various cancer types (Table V), underscoring their pivotal role in disease therapy. Despite the evident potential of RBBP4/7 in treating various diseases, the success of targeted strategies remains elusive, possibly due to the complex biological roles of RBBP4/7 and their involvement in multiple protein complexes. Future research should investigate the mechanisms of RBBP4/7 to develop targeted and effective treatment approaches. This includes a deeper understanding of the specific roles of RBBP4/7 in different cell types and disease states, identifying the molecular networks interacting with RBBP4/7, studying their expression and functional variations across diseases, and validating therapeutic interventions targeting RBBP4/7 in preclinical and clinical studies. Through these efforts, the scientific groundwork may be laid for novel treatment methods based on RBBP4/7, offering more personalized and effective therapeutic options for patients.
Table V.
Prognostic significance of RBBP4/7 in cancer.
| A, RBBP4
| ||||||
|---|---|---|---|---|---|---|
| First author, year | Disease type | Omics type | Samples or sample sources | Analysis methods | Prognostic relevance | (Refs.) |
| Gao M, 2023 | NSCLC | Transcriptomics | LUAD: 54 adjacent normal, 497 tumor samples; LUSC: 49 adjacent normal, 502 tumor samples | KM-plotter | Poor prognosis: Reduced OS | (84) |
| Wang N, 2021 | LUAD | Transcriptomics | KM website | KM-plotter | Poor prognosis: Reduced OS | (86) |
| Jia W, 2023 | LUAD | Transcriptomics | Patients with LUAD from the KM-plotter database | KM-plotter | Poor prognosis: Reduced OS, FP and PPS | (179) |
| Li J, 2023 | GBM | Transcriptomics | Samples from patients withMGMT-negative GBM in TCGA database | KM-plotter | Poor prognosis: Reduced OS and PFS | (63) |
| Li D, 2018 | NB | Transcriptomics, proteomics | Tissues from 42 primary cases of NB, GSE14340 and GSE16476 datasets | Log-rank test | Poor prognosis: Poor differentiation, reduced survival probability | (97) |
| Khateb A, 2021 | AML | Transcriptomics | GEPIA and TCGA | Log-rank test | Poor prognosis: Reduced OS | (106) |
| Li YD, 2019 | CRC | Proteomics | Tumor tissues of 80 patients with CRC | KM-plotter, log-rank test | Poor prognosis: Reduced OS | (117) |
| Guo Q, 2020 | BC | Proteomics | 240 BC tumor tissues | KM-plotter, log-rank test | Poor prognosis: Reduced OS | (141) |
| Barreiro-Alonso A, 2021 | PCa | Transcriptomics | 494 prostate adenocarcinoma tissues | Log-rank test | Poor prognosis: Reduced PFS | (160) |
| B, RBBP7
| ||||||
|---|---|---|---|---|---|---|
| First author, year | Disease type | Omics type | Samples or sample sources | Analysis methods | Prognostic relevance | (Refs.) |
| Wang H, 2022 | LUAD | Transcriptomics | Samples of patients with early-stage LUAD from different cohorts (TCGA, GSE30219, GSE31210, GSE37745, GSE50081) | KM-plotter, log-rank test, meta-analysis | Poor prognosis: Reduced RFS | (89) |
| Zhu H, 2022 | LUAD | Transcriptomics | / | LASSO regression, forest plots | Poor prognosis | (88) |
| Yu N, 2018 | EC | Transcriptomics, proteomics | 126 ESCC tissues, 182 patients with EC from TCGA database | KM-plotter, log-rank test | Poor prognosis: Reduced OS and DFS | (14) |
| Wang R, 2022 | EC | Transcriptomics | Patients with EC in GEPIA database and TCGA database | KM-plotter | Poor prognosis: Reduced OS and DFS | (133) |
| Barreiro-Alonso A, 2021 | PCa | Transcriptomics | 494 prostate adenocarcinoma tissues | Log-rank test | Poor prognosis: Reduced PFS | (160) |
RBBP, RB binding protein; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; KM, Kaplan-Meier; OS, overall survival; FP, first progression; PPS, post-progression survival; GBM, glioblastoma; MGMT, O-6-methylguanine-DNA methyltransferase; TCGA: The Cancer Genome Atlas; PFS, progression-free survival; NB, neuroblastoma; AML, acute myeloid leukemia; GEPIA: Gene Expression Profiling Interactive Analysis; CRC, colorectal cancer; BC, breast cancer; PCa, prostate cancer; EC, esophageal cancer; ESCC, esophageal squamous cell carcinoma; DFS, disease-free survival; LASSO, least absolute shrinkage and selection operator; / indicates data not specified or applicable in the sources.
6. Conclusion and future perspectives
RBBP4/7 are conserved proteins ubiquitously present in various organisms, which function in chromatin modification and gene regulation across species. However, their specific structure, expression patterns and molecular mechanisms may differ depending on the organism. For example, in P. falciparum, Drosophila, zebrafish and Saccharomyces cerevisiae, the structure of RBBP4 might resemble that in humans; however, there could be unique structural domains or adjustments (18,80,96,176). Moreover, in these organisms, RBBP4/7 primarily function during developmental and reproductive stages. By contrast, in humans, RBBP4/7 are expressed across diverse cells and tissues, and are associated with cell cycle regulation and gene transcription.
As histone chaperones, RBBP4/7 regulate various cellular processes and are implicated in a variety of human diseases, thus the future of RBBP4/7 research is promising. However, the expression patterns of RBBP4/7 exhibit significant variability in certain tumor types. This variability is attributed to the diverse roles of RBBP4/7 within multiple functional complexes, whose impact on tumorigenesis is intricately linked to the specific actions of these complexes, which vary with the cellular context and tumor type. In addition, research into gene mutations and DNA methylation abnormalities of RBBP4/7 in diseases remains limited, with mutations in RBBP7 identified only in cases of early spermatogenic failure (163). This underscores an important area for further investigation. Further studies of the complex molecular functions of RBBP4/7 may improve the understanding of cellular processes and disease pathways, leading to the development of innovative therapies for a variety of human diseases and cancers. Furthermore, exploring RBBP4/7 as potential biomarkers could improve diagnostic accuracy, enabling early detection and personalized medicine approaches.
Acknowledgments
Not applicable.
Abbreviations
- RBBP
RB binding protein
- NuRD
nucleosome remodeling and deacetylase
- PRC2
polycomb repressive complex 2
- RB
retinoblastoma protein
- CAF-1
chromatin assembly factor 1
- HAT1
histone acetyltransferase 1
- CENP-A
centromere protein A
- HDAC
histone deacetylase
- CDK
cyclin-dependent kinase
- FOXM1
forkhead box M1
- MTA1
metastasis associated 1
- SUZ12
SUZ12 polycomb repressive complex 2 subunit
- EED
embryonic ectoderm development
- EZH
enhancer of zeste homolog
- BRCA1
breast cancer type 1 susceptibility protein
- GBM
glioblastoma
- TMZ
temozolomide
- FOG-2
friend of GATA protein 2
- LC
lung cancer
- NSCLC
non-small cell LC
- CBX3
chromobox homolog 3
- LUAD
lung adenocarcinoma
- MPM
malignant pleural mesothelioma
- NB
neuroblastoma
- OCN
osteocalcin
- AD
Alzheimer's disease
- AML
acute myeloid leukemia
- MM
multiple myeloma
- CML
chronic myeloid leukemia
- CRC
colorectal cancer
- PPD
protopanaxadiol
- GC
gastric cancer
- SSeCKS
Src-suppressed C-kinase substrate
- HCC
hepatocellular carcinoma
- SALL4
Sal-like protein 4
- EGCG
epigallocatechin gallate
- MDR
multidrug resistance
- ESCC
esophageal squamous cell carcinoma
- EMT
epithelial-mesenchymal transition
- EC
esophageal cancer
- TC
thyroid cancer
- NF-κB
nuclear factor κB
- PMOP
postmenopausal osteoporosis
- ESR1
estrogen receptor 1
- BC
breast cancer
- TNBC
triple-negative BC
- BCL11A
BCL11 transcription factor A
- HPV
human papillomavirus
- ALA-PDT
5-aminole-vulinic acid photodynamic therapy
- SP1
specificity protein 1
- BKVN
BK virus-associated nephropathy
- RCC
renal cell carcinoma
- OSCC
oral squamous cell carcinoma
- OPSCC
oropharyngeal squamous cell carcinoma
- PPI
protein-protein interaction
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant nos. 82004007 and 81774026).
Availability of data and materials
Not applicable.
Authors' contributions
YZ was primarily responsible for writing, reviewing and revising this review. AY, XS, NT, ZZ and YC participated in the literature review and provided feedback for this review. JW and WW provided guidance throughout the preparation of this manuscript and made significant revisions to the text. Data authentication is not applicable. All authors read and approved the final version of the manuscript
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 3.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 4.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C, Shao Y, Wang X, Wang J, Wang P, Huang C, Wang W, Wang J. The effect of the histone chaperones HSPA8 and DEK on tumor immunity in hepatocellular carcinoma. Int J Mol Sci. 2023;24:2653. doi: 10.3390/ijms24032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millard CJ, Varma N, Saleh A, Morris K, Watson PJ, Bottrill AR, Fairall L, Smith CJ, Schwabe JW. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Elife. 2016;5:e13941. doi: 10.7554/eLife.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YD, Lv Z, Zhu WF. RBBP4 promotes colon cancer malignant progression via regulating Wnt/β-catenin pathway. World J Gastroenterol. 2020;26:5328–5342. doi: 10.3748/wjg.v26.i35.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Li F, Shao J, Lv J, Chang A, Dong W, Zhu F. Circular RNA hsa_circ_0102231 sponges miR-145 to promote non-small cell lung cancer cell proliferation by up-regulating the expression of RBBP4. J Biochem. 2021;169:65–73. doi: 10.1093/jb/mvaa093. [DOI] [PubMed] [Google Scholar]
- 14.Yu N, Zhang P, Wang L, He X, Yang S, Lu H. RBBP7 is a prognostic biomarker in patients with esophageal squamous cell carcinoma. Oncol Lett. 2018;16:7204–7211. doi: 10.3892/ol.2018.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L, Xia Y, Li H, Wang Z, Xu H, Dai X, Zhang Y, Zhang H, Fan W, Wei F, et al. FIT links c-Myc and P53 acetylation by recruiting RBBP7 during colorectal carcinogenesis. Cancer Gene Ther. 2023;30:1124–1133. doi: 10.1038/s41417-023-00624-z. [DOI] [PubMed] [Google Scholar]
- 16.Qian YW, Wang YC, Hollingsworth RE, Jr, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 17.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 18.Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 19.Brasen C, Dorosz J, Wiuf A, Boesen T, Mirza O, Gajhede M. Expression, purification and characterization of the human MTA2-RBBP7 complex. Biochim Biophys Acta Proteins Proteom. 2017;1865:531–538. doi: 10.1016/j.bbapap.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/S0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 21.Sauer PV, Gu Y, Liu WH, Mattiroli F, Panne D, Luger K, Churchill ME. Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 2018;46:9907–9917. doi: 10.1093/nar/gky823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller GA, Asthana A, Rubin SM. Structure and function of MuvB complexes. Oncogene. 2022;41:2909–2919. doi: 10.1038/s41388-022-02321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/S0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 24.Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008;22:1265–1268. doi: 10.1101/gad.1676208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao L, Dang Y, Hu B, Luo L, Zhao P, Wang S, Zhang K. Overlapping functions of RBBP4 and RBBP7 in regulating cell proliferation and histone H3.3 deposition during mouse preimplantation development. Epigenetics. 2022;17:1205–1218. doi: 10.1080/15592294.2021.1999006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satrimafitrah P, Barman HK, Ahmad A, Nishitoh H, Nakayama T, Fukagawa T, Takami Y. RbAp48 is essential for viability of vertebrate cells and plays a role in chromosome stability. Chromosome Res. 2016;24:161–173. doi: 10.1007/s10577-015-9510-8. [DOI] [PubMed] [Google Scholar]
- 27.Guan LS, Li GC, Chen CC, Liu LQ, Wang ZY. Rb-associated protein 46 (RbAp46) suppresses the tumorigenicity of adenovirus-transformed human embryonic kidney 293 cells. Int J Cancer. 2001;93:333–338. doi: 10.1002/ijc.1338. [DOI] [PubMed] [Google Scholar]
- 28.Lee BC, Lin Z, Yuen KW. RbAp46/48(LIN-53) is required for Holocentromere assembly in Caenorhabditis elegans. Cell Rep. 2016;14:1819–1828. doi: 10.1016/j.celrep.2016.01.065. [DOI] [PubMed] [Google Scholar]
- 29.Mouysset J, Gilberto S, Meier MG, Lampert F, Belwal M, Meraldi P, Peter M. CRL4(RBBP7) is required for efficient CENP-A deposition at centromeres. J Cell Sci. 2015;128:1732–1745. doi: 10.1242/jcs.162305. [DOI] [PubMed] [Google Scholar]
- 30.Huang S, Lee WH, Lee EY. A cellular protein that competes with SV40 T antigen for binding to the retinoblastoma gene product. Nature. 1991;350:160–162. doi: 10.1038/350160a0. [DOI] [PubMed] [Google Scholar]
- 31.Schultz-Rogers LE, Thayer ML, Kambakam S, Wierson WA, Helmer JA, Wishman MD, Wall KA, Greig JL, Forsman JL, Puchhalapalli K, et al. Rbbp4 loss disrupts neural progenitor cell cycle regulation independent of Rb and leads to Tp53 acetylation and apoptosis. Dev Dyn. 2022;251:1267–1290. doi: 10.1002/dvdy.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 34.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3 doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gubern A, Joaquin M, Marquès M, Maseres P, Garcia-Garcia J, Amat R, González-Nuñez D, Oliva B, Real FX, de Nadal E, Posas F. The N-Terminal phosphorylation of RB by p38 bypasses its inactivation by CDKs and prevents proliferation in cancer cells. Mol Cell. 2016;64:25–36. doi: 10.1016/j.molcel.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Guiley KZ, Liban TJ, Felthousen JG, Ramanan P, Litovchick L, Rubin SM. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015;29:961–974. doi: 10.1101/gad.257568.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller GA, Quaas M, Schümann M, Krause E, Padi M, Fischer M, Litovchick L, DeCaprio JA, Engeland K. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40:1561–1578. doi: 10.1093/nar/gkr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mages CF, Wintsche A, Bernhart SH, Müller GA. The DREAM complex through its subunit Lin37 cooperates with Rb to initiate quiescence. Elife. 2017;6:e26876. doi: 10.7554/eLife.26876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uxa S, Bernhart SH, Mages CFS, Fischer M, Kohler R, Hoffmann S, Stadler PF, Engeland K, Müller GA. DREAM and RB cooperate to induce gene repression and cell-cycle arrest in response to p53 activation. Nucleic Acids Res. 2019;47:9087–9103. doi: 10.1093/nar/gkz635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schade AE, Fischer M, DeCaprio JA. RB, p130 and p107 differentially repress G1/S and G2/M genes after p53 activation. Nucleic Acids Res. 2019;47:11197–11208. doi: 10.1093/nar/gkz961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, et al. Structural plasticity of histones H3-H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat Struct Mol Biol. 2013;20:29–35. doi: 10.1038/nsmb.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015;72:2491–2507. doi: 10.1007/s00018-015-1880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mu W, Murcia NS, Smith KN, Menon DU, Yee D, Magnuson T. RBBP4 dysfunction reshapes the genomic landscape of H3K27 methylation and acetylation and disrupts gene expression. G3 (Bethesda) 2022;12:jkac082. doi: 10.1093/g3journal/jkac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei S, Liu Y, Bao Y, Zhang Y, Min S, Liu Y, Huang Y, Yuan X, Feng Y, Shi J, Yang R. Dendritic cell-associated miRNAs are modulated via chromatin remodeling in response to different environments. PLoS One. 2014;9:e90231. doi: 10.1371/journal.pone.0090231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, et al. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun. 2016;7:13465. doi: 10.1038/ncomms13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847–5860. doi: 10.1128/MCB.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balboula AZ, Stein P, Schultz RM, Schindler K. RBBP4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod. 2015;92:105. doi: 10.1095/biolreprod.115.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue Y, Yang WS, Zhang L, Liu CP, Xu RM. Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex. Genes Dev. 2022;36:408–413. doi: 10.1101/gad.349099.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pardal AJ, Bowman AJ. A specific role for importin-5 and NASP in the import and nuclear hand-off of monomeric H3. Elife. 2022;11:e81755. doi: 10.7554/eLife.81755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42–48. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/S1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 54.Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. Capturing the onset of PRC2-Mediated repressive domain formation. Mol Cell. 2018;70:1149–1162.e5. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S, Jiao L, Liu X, Yang X, Liu X. A Dimeric structural scaffold for PRC2-PCL targeting to CpG island chromatin. Mol Cell. 2020;77:1265–1278.e7. doi: 10.1016/j.molcel.2019.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitange GJ, Mladek AC, Schroeder MA, Pokorny JC, Carlson BL, Zhang Y, Nair AA, Lee JH, Yan H, Decker PA, et al. Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 2016;14:2587–2598. doi: 10.1016/j.celrep.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marin TL, Gongol B, Zhang F, Martin M, Johnson DA, Xiao H, Wang Y, Subramaniam S, Chien S, Shyy JY. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci Signal. 2017;10:eaaf7478. doi: 10.1126/scisignal.aaf7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li DQ, Kumar R. Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle. 2010;9:2071–2079. doi: 10.4161/cc.9.11.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang B, Zhang W, Sun L, Lu B, Yin C, Zhang Y, Jiang H. Creatine kinase brain-type regulates BCAR1 phosphorylation to facilitate DNA damage repair. iScience. 2023;26:106684. doi: 10.1016/j.isci.2023.106684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarden RI, Brody LC. BRCA1 interacts with components of the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:4983–4988. doi: 10.1073/pnas.96.9.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen GC, Guan LS, Yu JH, Li GC, Choi Kim HR, Wang ZY. Rb-associated protein 46 (RbAp46) inhibits transcriptional transactivation mediated by BRCA1. Biochem Biophys Res Commun. 2001;284:507–514. doi: 10.1006/bbrc.2001.5003. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Song C, Gu J, Li C, Zang W, Shi L, Chen L, Zhu L, Zhou M, Wang T, et al. RBBP4 regulates the expression of the Mre11-Rad50-NBS1 (MRN) complex and promotes DNA double-strand break repair to mediate glioblastoma chemoradiotherapy resistance. Cancer Lett. 2023;557:216078. doi: 10.1016/j.canlet.2023.216078. [DOI] [PubMed] [Google Scholar]
- 64.Li GC, Guan LS, Wang ZY. Overexpression of RbAp46 facilitates stress-induced apoptosis and suppresses tumorigenicity of neoplastigenic breast epithelial cells. Int J Cancer. 2003;105:762–768. doi: 10.1002/ijc.11148. [DOI] [PubMed] [Google Scholar]
- 65.Yang J, Kiefer S, Rauchman M. Characterization of the gene encoding mouse retinoblastoma binding protein-7, a component of chromatin-remodeling complexes. Genomics. 2002;80:407–415. doi: 10.1006/geno.2002.6844. [DOI] [PubMed] [Google Scholar]
- 66.Horihata K, Inoue N, Uenoyama Y, Maeda KI, Tsukamura H. Retinoblastoma binding protein 7 is involved in Kiss1 mRNA upregulation in rodents. J Reprod Dev. 2020;66:125–133. doi: 10.1262/jrd.2019-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giri R, Yeh HH, Wu CH, Liu HS. SUMO-1 overexpression increases RbAp46 protein stability and suppresses cell growth. Anticancer Res. 2008;28:3749–3756. [PubMed] [Google Scholar]
- 68.Ping W, Sheng Y, Hu G, Zhong H, Li Y, Liu Y, Luo W, Yan C, Wen Y, Wang X, et al. RBBP4 is an epigenetic barrier for the induced transition of pluripotent stem cells into totipotent 2C-like cells. Nucleic Acids Res. 2023;51:5414–5431. doi: 10.1093/nar/gkad219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garnatz AS, Gao Z, Broman M, Martens S, Earley JU, Svensson EC. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev Biol. 2014;395:50–61. doi: 10.1016/j.ydbio.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonuccelli L, Rossi L, Lena A, Scarcelli V, Rainaldi G, Evangelista M, Iacopetti P, Gremigni V, Salvetti A. An RbAp48-like gene regulates adult stem cells in planarians. J Cell Sci. 2010;123:690–698. doi: 10.1242/jcs.053900. [DOI] [PubMed] [Google Scholar]
- 71.He H, Kong S, Liu F, Zhang S, Jiang Y, Liao Y, Jiang Y, Li Q, Wang B, Zhou Z, et al. Rbbp7 is required for uterine stromal decidualization in mice. Biol Reprod. 2015;93:13. doi: 10.1095/biolreprod.115.129015. [DOI] [PubMed] [Google Scholar]
- 72.Shore AN, Kabotyanski EB, Roarty K, Smith MA, Zhang Y, Creighton CJ, Dinger ME, Rosen JM. Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation. PLoS Genet. 2012;8:e1002840. doi: 10.1371/journal.pgen.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xin J, Li J, Feng Y, Wang L, Zhang Y, Yang R. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. Onco Targets Ther. 2017;10:1307–1315. doi: 10.2147/OTT.S124201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rico M, Mukherjee A, Konieczkowski M, Bruggeman LA, Miller RT, Khan S, Schelling JR, Sedor JR. WT1-interacting protein and ZO-1 translocate into podocyte nuclei after puromycin aminonucleoside treatment. Am J Physiol Renal Physiol. 2005;289:F431–F441. doi: 10.1152/ajprenal.00389.2004. [DOI] [PubMed] [Google Scholar]
- 75.Scuto A, Zhang H, Zhao H, Rivera M, Yeatman TJ, Jove R, Torres-Roca JF. RbAp48 regulates cytoskeletal organization and morphology by increasing K-Ras activity and signaling through mitogen-activated protein kinase. Cancer Res. 2007;67:10317–10324. doi: 10.1158/0008-5472.CAN-06-3313. [DOI] [PubMed] [Google Scholar]
- 76.Gasca S, Pellestor F, Assou S, Loup V, Anahory T, Dechaud H, De Vos J, Hamamah S. Identifying new human oocyte marker genes: A microarray approach. Reprod Biomed Online. 2007;14:175–183. doi: 10.1016/S1472-6483(10)60785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balboula AZ, Stein P, Schultz RM, Schindler K. Knockdown of RBBP7 unveils a requirement of histone deacetylation for CPC function in mouse oocytes. Cell Cycle. 2014;13:600–611. doi: 10.4161/cc.27410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guan LS, Rauchman M, Wang ZY. Induction of Rb-associated protein (RbAp46) by Wilms' tumor suppressor WT1 mediates growth inhibition. J Biol Chem. 1998;273:27047–27050. doi: 10.1074/jbc.273.42.27047. [DOI] [PubMed] [Google Scholar]
- 79.Pegoraro G, Kubben N, Wickert U, Göhler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunt LC, Stover J, Haugen B, Shaw TI, Li Y, Pagala VR, Finkelstein D, Barton ER, Fan Y, Labelle M, et al. A key role for the ubiquitin ligase UBR4 in myofiber hypertrophy in drosophila and mice. Cell Rep. 2019;28:1268–1281.e1266. doi: 10.1016/j.celrep.2019.06.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pavlopoulos E, Jones S, Kosmidis S, Close M, Kim C, Kovalerchik O, Small SA, Kandel ER. Molecular mechanism for age-related memory loss: The histone-binding protein RbAp48. Sci Transl Med. 2013;5:200ra115. doi: 10.1126/scitranslmed.3006373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsujii A, Miyamoto Y, Moriyama T, Tsuchiya Y, Obuse C, Mizuguchi K, Oka M, Yoneda Y. Retinoblastoma-binding protein 4-regulated classical nuclear transport is involved in cellular senescence. J Biol Chem. 2015;290:29375–29388. doi: 10.1074/jbc.M115.681908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ginger MR, Gonzalez-Rimbau MF, Gay JP, Rosen JM. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol. 2001;15:1993–2009. doi: 10.1210/mend.15.11.0724. [DOI] [PubMed] [Google Scholar]
- 84.Gao M, Li Y, Huang H, Fan Y, Shi R, Su L, Chen C, Li X, Zhu G, Wu D. Exploring the association between PRC2 genes variants and lung cancer risk in chinese han population. Onco Targets Ther. 2023;16:499–513. doi: 10.2147/OTT.S417190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hao D, Li Y, Shi J, Jiang J. Circ_0110498 facilitates the cisplatin resistance of non-small cell lung cancer by mediating the miR-1287-5p/RBBP4 axis. Thorac Cancer. 2023;14:662–672. doi: 10.1111/1759-7714.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang N, Wang W, Mao W, Kuerbantayi N, Jia N, Chen Y, Zhou F, Yin L, Wang Y. RBBP4 enhances platinum chemo resistance in lung adenocarcinoma. Biomed Res Int. 2021;2021:6905985. doi: 10.1155/2021/6905985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin X, Zhang B, Zhang H, Yu H. Smoking-associated upregulation of CBX3 suppresses ARHGAP24 expression to activate Rac1 signaling and promote tumor progression in lung adenocarcinoma. Oncogene. 2022;41:538–549. doi: 10.1038/s41388-021-02114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu H, Zheng C, Liu H, Kong F, Kong S, Chen F, Tian Y. Significance of macrophage infiltration in the prognosis of lung adenocarcinoma patients evaluated by scRNA and bulkRNA analysis. Front Immunol. 2022;13:1028440. doi: 10.3389/fimmu.2022.1028440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Lu X, Chen J. Construction and experimental validation of an acetylation-related gene signature to evaluate the recurrence and immunotherapeutic response in early-stage lung adenocarcinoma. BMC Med Genomics. 2022;15:254. doi: 10.1186/s12920-022-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang CL, Wang CI, Liao PC, Chen CD, Liang Y, Chuang WY, Tsai YH, Chen HC, Chang YS, Yu JS, et al. Discovery of retinoblastoma-associated binding protein 46 as a novel prognostic marker for distant metastasis in nonsmall cell lung cancer by combined analysis of cancer cell secretome and pleural effusion proteome. J Proteome Res. 2009;8:4428–4440. doi: 10.1021/pr900160h. [DOI] [PubMed] [Google Scholar]
- 91.Tsao AS, Pass HI, Rimner A, Mansfield AS. New era for malignant pleural mesothelioma: Updates on therapeutic options. J Clin Oncol. 2022;40:681–692. doi: 10.1200/JCO.21.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vavougios GD, Solenov EI, Hatzoglou C, Baturina GS, Katkova LE, Molyvdas PA, Gourgoulianis KI, Zarogiannis SG. Computational genomic analysis of PARK7 interactome reveals high BBS1 gene expression as a prognostic factor favoring survival in malignant pleural mesothelioma. Am J Physiol Lung Cell Mol Physiol. 2015;309:L677–L686. doi: 10.1152/ajplung.00051.2015. [DOI] [PubMed] [Google Scholar]
- 93.Barzegar Behrooz A, Latifi-Navid H, da Silva Rosa SC, Swiat M, Wiechec E, Vitorino C, Vitorino R, Jamalpoor Z, Ghavami S. Integrating Multi-Omics analysis for enhanced diagnosis and treatment of glioblastoma: A comprehensive Data-Driven approach. Cancers (Basel) 2023;15:3158. doi: 10.3390/cancers15123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shou J, Gao H, Cheng S, Wang B, Guan H. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int. 2021;21:39. doi: 10.1186/s12935-020-01690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mladek AC, Yan H, Tian S, Decker PA, Burgenske DM, Bakken K, Hu Z, He L, Connors MA, Carlson BL, et al. RBBP4-p300 axis modulates expression of genes essential for cell survival and is a potential target for therapy in glioblastoma. Neuro Oncol. 2022;24:1261–1272. doi: 10.1093/neuonc/noac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schultz LE, Haltom JA, Almeida MP, Wierson WA, Solin SL, Weiss TJ, Helmer JA, Sandquist EJ, Shive HR, McGrail M. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Dis Model Mech. 2018;11:dmm034124. doi: 10.1242/dmm.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li D, Song H, Mei H, Fang E, Wang X, Yang F, Li H, Chen Y, Huang K, Zheng L, Tong Q. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat Commun. 2018;9:2829. doi: 10.1038/s41467-018-05286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crea F, Hurt EM, Farrar WL. Clinical significance of Polycomb gene expression in brain tumors. Mol Cancer. 2010;9:265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khrimian L, Obri A, Ramos-Brossier M, Rousseaud A, Moriceau S, Nicot AS, Mera P, Kosmidis S, Karnavas T, Saudou F, et al. Gpr158 mediates osteocalcin's regulation of cognition. J Exp Med. 2017;214:2859–2873. doi: 10.1084/jem.20171320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kosmidis S, Polyzos A, Harvey L, Youssef M, Denny CA, Dranovsky A, Kandel ER. RbAp48 protein is a critical component of GPR158/OCN signaling and ameliorates Age-Related memory loss. Cell Rep. 2018;25:959–973.e6. doi: 10.1016/j.celrep.2018.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barral S, Reitz C, Small SA, Mayeux R. Genetic variants in a 'cAMP element binding protein' (CREB)-dependent histone acetylation pathway influence memory performance in cognitively healthy elderly individuals. Neurobiol Aging. 2014;35:2881.e7–2881.e10. doi: 10.1016/j.neurobiolaging.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, Laue ED, Mackay JP. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48•FOG-1 complex. J Biol Chem. 2011;286:1196–1203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang HJ, Lee CC, Chen CY. Lead discovery for Alzheimer's disease related target protein RbAp48 from traditional Chinese medicine. Biomed Res Int. 2014;2014:764946. doi: 10.1155/2014/764946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ramis MR, Sarubbo F, Tejada S, Jiménez M, Esteban S, Miralles A, Moranta D. chronic Polyphenon-60 or catechin treatments increase brain monoamines syntheses and hippocampal SIRT1 levels improving cognition in aged rats. Nutrients. 2020;12:326. doi: 10.3390/nu12020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dave N, Vural AS, Piras IS, Winslow W, Surendra L, Winstone JK, Beach TG, Huentelman MJ, Velazquez R. Identification of retinoblastoma binding protein 7 (Rbbp7) as a mediator against tau acetylation and subsequent neuronal loss in Alzheimer's disease and related tauopathies. Acta Neuropathol. 2021;142:279–294. doi: 10.1007/s00401-021-02323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khateb A, Deshpande A, Feng Y, Finlay D, Lee JS, Lazar I, Fabre B, Li Y, Fujita Y, Zhang T. The ubiquitin ligase RNF5 determines acute myeloid leukemia growth and susceptibility to histone deacetylase inhibitors. Nat Commun. 2021;12:5397. doi: 10.1038/s41467-021-25664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Casas S, Ollila J, Aventín A, Vihinen M, Sierra J, Knuutila S. Changes in apoptosis-related pathways in acute myelocytic leukemia. Cancer Genet Cytogenet. 2003;146:89–101. doi: 10.1016/S0165-4608(03)00102-X. [DOI] [PubMed] [Google Scholar]
- 108.Sakhinia E, Faranghpour M, Liu Yin JA, Brady G, Hoyland JA, Byers RJ. Routine expression profiling of microarray gene signatures in acute leukaemia by real-time PCR of human bone marrow. Br J Haematol. 2005;130:233–248. doi: 10.1111/j.1365-2141.2005.05594.x. [DOI] [PubMed] [Google Scholar]
- 109.Gao H, Wang H, Yang W. Identification of key genes and construction of microRNA-mRNA regulatory networks in multiple myeloma by integrated multiple GEO datasets using bioinformatics analysis. Int J Hematol. 2017;106:99–107. doi: 10.1007/s12185-017-2216-2. [DOI] [PubMed] [Google Scholar]
- 110.Merkerova M, Bruchova H, Kracmarova A, Klamova H, Brdicka R. Bmi-1 over-expression plays a secondary role in chronic myeloid leukemia transformation. Leuk Lymphoma. 2007;48:793–801. doi: 10.1080/10428190601186002. [DOI] [PubMed] [Google Scholar]
- 111.Hu SY, Chen ZX, Cen JN, Gu M, Zhao Y, Shen HL, Wang W. RbAp46 gene activates the expression of IGFBP-rP1 gene in K562 leukemic cells. Zhonghua Xue Ye Xue Za Zhi. 2006;27:107–110. In Chinese. [PubMed] [Google Scholar]
- 112.Hu SY, Chen ZX, Gu WY, Cen JN, Zhao Y, Gu M. Detection of RbAp46 expression in bone marrow cells of leukemia patients by real-time quantitative RT-PCR. Zhonghua Xue Ye Xue Za Zhi. 2005;26:417–420. In Chinese. [PubMed] [Google Scholar]
- 113.Hu SY, Chen ZX, Gu WY, Cen JN, Zhao Y. High expression of RbAp46 gene in patients with acute leukemia or chronic myelogenous leukemia in blast crisis. Chin Med J (Engl) 2005;118:1295–1298. [PubMed] [Google Scholar]
- 114.Duan WM, Chen ZX, Wang W, Fu JX, Bai X, Zhao XJ, Yao L. Establishment and characterization of leukemic cell line U937 stably expressing exogenous RbAp46. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2004;12:416–419. In Chinese. [PubMed] [Google Scholar]
- 115.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 116.Zhang TF, Yu SQ, Loggie BW, Wang ZY. Inducible expression of RbAp46 activates c-Jun NH2-terminal kinase-dependent apoptosis and suppresses progressive growth of tumor xenografts in nude mice. Anticancer Res. 2003;23:4621–4627. [PubMed] [Google Scholar]
- 117.Li YD, Lv Z, Xie HY, Zheng SS. Retinoblastoma binding protein 4 up-regulation is correlated with hepatic metastasis and poor prognosis in colon cancer patients. Hepatobiliary Pancreat Dis Int. 2019;18:446–451. doi: 10.1016/j.hbpd.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 118.Zhu X, Luo X, Long X, Jiang S, Xie X, Zhang Q, Wang H. CircAGO2 promotes colorectal cancer progression by inhibiting heat shock protein family B (small) member 8 via miR-1-3p/retinoblastoma binding protein 4 axis. Funct Integr Genomics. 2023;23:78. doi: 10.1007/s10142-023-00990-9. [DOI] [PubMed] [Google Scholar]
- 119.Zhuo FF, Guo Q, Zheng YZ, Liu TT, Yang Z, Xu QH, Jiang Y, Liu D, Zeng KW, Tu PF. Photoaffinity Labeling-Based chemoproteomic strategy reveals RBBP4 as a cellular target of protopanaxadiol against colorectal cancer cells. Chembiochem. 2022;23:e202200038. doi: 10.1002/cbic.202200038. [DOI] [PubMed] [Google Scholar]
- 120.Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, Li Z, Wei J, Liu M, Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jin X, Jiang R, Xiang Y, Fan Z, Wu Z, Yang B, Yang L, Wei S, Yang Y. Overexpression of retinoblastoma-binding protein 4 contributes to the radiosensitivity of AGS gastric cancer cells via phosphoinositide3-kinase/protein kinase B pathway suppression. Mol Med Rep. 2018;18:1571–1581. doi: 10.3892/mmr.2018.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim TY, Kim NK, Bang YJ. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095–7103. doi: 10.1038/sj.onc.1207932. [DOI] [PubMed] [Google Scholar]
- 123.Liu Y, Gao L, Gelman IH. SSeCKS/Gravin/AKAP12 attenuates expression of proliferative and angiogenic genes during suppression of v-Src-induced oncogenesis. BMC Cancer. 2006;6:105. doi: 10.1186/1471-2407-6-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol. 2004;10:509–513. doi: 10.3748/wjg.v10.i4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu BH, Jobichen C, Chia CSB, Chan THM, Tang JP, Chung TXY, Li J, Poulsen A, Hung AW, Koh-Stenta X, et al. Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide. Proc Natl Acad Sci USA. 2018;115:E7119–E7128. doi: 10.1073/pnas.1801253115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R, Tan Y, Fan J, Chang Y, Fu J, et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut. 2015;64:156–167. doi: 10.1136/gutjnl-2013-305715. [DOI] [PubMed] [Google Scholar]
- 127.Tang HH, Zhou M, Liang G. Impact of epigallocatechin gallate on gene expression profiles of human hepatocellular carcinoma cell lines BEL7404/ADM and BEL7402/5-FU. Ai Zheng. 2008;27:1056–1064. In Chinese. [PubMed] [Google Scholar]
- 128.Younossi ZM. Non-alcoholic fatty liver disease-A global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 129.Yao Y, Li X, Wang W, Liu Z, Chen J, Ding M, Huang X. MRT, Functioning with NURF complex, regulates lipid droplet size. Cell Rep. 2018;24:2972–2984. doi: 10.1016/j.celrep.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 130.Zhi S, Congcong Z, Zhiling G, Yihan Q, Yijing X, Guanjie L, Fang W, Xuehua S, Hongjie L, Xiaoni K, Yueqiu G. Quantitative proteomics of HFD-induced fatty liver uncovers novel transcription factors of lipid metabolism. Int J Biol Sci. 2022;18:3298–3312. doi: 10.7150/ijbs.71431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen L, Lu J, Xu T, Yan Z, Guo Y, Dong Z, Guo W. KTN1-AS1, a SOX2-mediated lncRNA, activates epithelial-mesenchymal transition process in esophageal squamous cell carcinoma. Sci Rep. 2022;12:20186. doi: 10.1038/s41598-022-24743-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bai X, Wang D, Ji H, Zheng L, Lu Y, Tang W, Zhang H, Xu W, Li J, Fei Z, Wang H. RbAp48 is critical for the proliferation of hypopharyngeal carcinoma. ORL J Otorhinolaryngol Relat Spec. 2015;77:310–319. doi: 10.1159/000438761. [DOI] [PubMed] [Google Scholar]
- 133.Wang R, Huang Z, Lin Z, Chen B, Pang X, Du C, Fan H. Hypoxia-induced RBBP7 promotes esophagus cancer progression by inducing CDK4 expression. Acta Biochim Biophys Sin (Shanghai) 2022;54:179–186. doi: 10.3724/abbs.2021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xie ZF, Li HT, Xie SH, Ma M. Circular RNA hsa_ circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci. 2020;24:151–163. doi: 10.26355/eurrev_202001_19906. [DOI] [PubMed] [Google Scholar]
- 135.Pacifico F, Paolillo M, Chiappetta G, Crescenzi E, Arena S, Scaloni A, Monaco M, Vascotto C, Tell G, Formisano S, Leonardi A. RbAp48 is a target of nuclear factor-kappaB activity in thyroid cancer. J Clin Endocrinol Metab. 2007;92:1458–1466. doi: 10.1210/jc.2006-2199. [DOI] [PubMed] [Google Scholar]
- 136.Saul D, Ninkovic M, Komrakova M, Wolff L, Simka P, Gasimov T, Menger B, Hoffmann DB, Rohde V, Sehmisch S. Effect of zileuton on osteoporotic bone and its healing, expression of bone, and brain genes in rats. J Appl Physiol (1985) 2018;124:118–130. doi: 10.1152/japplphysiol.01126.2016. [DOI] [PubMed] [Google Scholar]
- 137.Gong R, Ren S, Chen M, Wang Y, Zhang G, Shi L, Zhang C, Su R, Li Y. Bioinformatics analysis reveals the altered gene expression of patients with postmenopausal osteoporosis using liuweidihuang pills treatment. Biomed Res Int. 2019;2019:1907906. doi: 10.1155/2019/1907906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang C, Ren J, Li B, Jin C, Ma C, Cheng C, Sun Y, Shi X. Identification of gene biomarkers in patients with postmenopausal osteoporosis. Mol Med Rep. 2019;19:1065–1073. doi: 10.3892/mmr.2018.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ishimaru N, Arakaki R, Omotehara F, Yamada K, Mishima K, Saito I, Hayashi Y. Novel role for RbAp48 in tissue-specific, estrogen deficiency-dependent apoptosis in the exocrine glands. Mol Cell Biol. 2006;26:2924–2935. doi: 10.1128/MCB.26.8.2924-2935.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ishimaru N, Arakaki R, Yoshida S, Yamada A, Noji S, Hayashi Y. Expression of the retinoblastoma protein RbAp48 in exocrine glands leads to Sjögren's syndrome-like autoimmune exocrinopathy. J Exp Med. 2008;205:2915–2927. doi: 10.1084/jem.20080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guo Q, Cheng K, Wang X, Li X, Yu Y, Hua Y, Yang Z. Expression of HDAC1 and RBBP4 correlate with clinicopathologic characteristics and prognosis in breast cancer. Int J Clin Exp Pathol. 2020;13:563–572. [PMC free article] [PubMed] [Google Scholar]
- 142.Gong X, Dong T, Niu M, Liang X, Sun S, Zhang Y, Li Y, Li D. lncRNA LCPAT1 upregulation promotes breast cancer progression via enhancing MFAP2 transcription. Mol Ther Nucleic Acids. 2020;21:804–813. doi: 10.1016/j.omtn.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Creekmore AL, Walt KA, Schultz-Norton JR, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. The role of retinoblastoma-associated proteins 46 and 48 in estrogen receptor alpha mediated gene expression. Mol Cell Endocrinol. 2008;291:79–86. doi: 10.1016/j.mce.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zheng Z, Yao X, Liu Y. RBBP4 plays a vital role in the malignant progression of triple-negative breast cancer by regulating epithelial-mesenchymal transition. Genes Genomics. 2022;44:1301–1309. doi: 10.1007/s13258-022-01262-9. [DOI] [PubMed] [Google Scholar]
- 145.Moody RR, Lo MC, Meagher JL, Lin CC, Stevers NO, Tinsley SL, Jung I, Matvekas A, Stuckey JA, Sun D. Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes. J Biol Chem. 2018;293:2125–2136. doi: 10.1074/jbc.M117.811463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang TF, Yu SQ, Deuel TF, Wang ZY. Constitutive expression of Rb associated protein 46 (RbAp46) reverts transformed phenotypes of breast cancer cells. Anticancer Res. 2003;23:3735–3740. [PubMed] [Google Scholar]
- 147.Zhang TF, Yu SQ, Wang ZY. RbAp46 inhibits estrogen-stimulated progression of neoplastigenic breast epithelial cells. Anticancer Res. 2007;27:3205–3209. [PubMed] [Google Scholar]
- 148.Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li GC, Wang ZY. Constitutive expression of RbAp46 induces epithelial-mesenchymal transition in mammary epithelial cells. Anticancer Res. 2006;26:3555–3560. [PubMed] [Google Scholar]
- 150.Thakur A, Rahman KW, Wu J, Bollig A, Biliran H, Lin X, Nassar H, Grignon DJ, Sarkar FH, Liao JD. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res. 2007;5:171–181. doi: 10.1158/1541-7786.MCR-06-0071. [DOI] [PubMed] [Google Scholar]
- 151.Ebata A, Suzuki T, Takagi K, Miki Y, Onodera Y, Nakamura Y, Fujishima F, Ishida K, Watanabe M, Tamaki K, et al. Oestrogen-induced genes in ductal carcinoma in situ: Their comparison with invasive ductal carcinoma. Endocr Relat Cancer. 2012;19:485–496. doi: 10.1530/ERC-11-0345. [DOI] [PubMed] [Google Scholar]
- 152.Mieczkowska IK, Pantelaiou-Prokaki G, Prokakis E, Schmidt GE, Müller-Kirschbaum LC, Werner M, Sen M, Velychko T, Jannasch K, Dullin C, et al. Decreased PRC2 activity supports the survival of basal-like breast cancer cells to cytotoxic treatments. Cell Death Dis. 2021;12:1118. doi: 10.1038/s41419-021-04407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kong L, Yu XP, Bai XH, Zhang WF, Zhang Y, Zhao WM, Jia JH, Tang W, Zhou YB, Liu CJ. RbAp48 is a critical mediator controlling the transforming activity of human papillomavirus type 16 in cervical cancer. J Biol Chem. 2007;282:26381–26391. doi: 10.1074/jbc.M702195200. [DOI] [PubMed] [Google Scholar]
- 154.Wu SX, Ren XY, Pan YL, Guan DD, Liu S, Zou XY, Shi CG, Li YZ, Zhang YZ. Effects of ALA-PDT on HPV16-immortalized cervical epithelial cell. Neoplasma. 2017;64:175–181. doi: 10.4149/neo_2017_202. [DOI] [PubMed] [Google Scholar]
- 155.Wu S, Wang L, Ren X, Pan Y, Peng Y, Zou X, Shi C, Zhang Y. Involvement of retinoblastoma-associated protein 48 during photodynamic therapy of cervical cancer cells. Mol Med Rep. 2017;15:1393–1400. doi: 10.3892/mmr.2017.6151. [DOI] [PubMed] [Google Scholar]
- 156.Zhong J, Yang X, Mai M, Wang D, Lv L, Rao J. Effect of RbAp48 knockdown on migration and invasion of human cervical cancer cell line MS751 in vitro. Nan Fang Yi Ke Da Xue Xue Bao. 2015;35:1564–1569. In Chinese. [PubMed] [Google Scholar]
- 157.Zheng L, Tang W, Wei F, Wang H, Liu J, Lu Y, Cheng Y, Bai X, Yu X, Zhao W. Radiation-inducible protein RbAp48 contributes to radiosensitivity of cervical cancer cells. Gynecol Oncol. 2013;130:601–608. doi: 10.1016/j.ygyno.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 158.Li HJ, Yu PN, Huang KY, Su HY, Hsiao TH, Chang CP, Yu MH, Lin YW. NKX6.1 functions as a metastatic suppressor through epigenetic regulation of the epithelial-mesenchymal transition. Oncogene. 2016;35:2266–2278. doi: 10.1038/onc.2015.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cai L, Wang D, Fisher AL, Wang Z. Identification of a genetic interaction between the tumor suppressor EAF2 and the retinoblastoma protein (Rb) signaling pathway in C. elegans and prostate cancer cells. Biochem Biophys Res Commun. 2014;447:292–298. doi: 10.1016/j.bbrc.2014.03.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barreiro-Alonso A, Lamas-Maceiras M, Lorenzo-Catoira L, Pardo M, Yu L, Choudhary JS, Cerdán ME. HMGB1 Protein interactions in prostate and ovary cancer models reveal links to RNA processing and ribosome biogenesis through NuRD, THOC and septin complexes. Cancers (Basel) 2021;13:4686. doi: 10.3390/cancers13184686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang J, He C, Gao P, Wang S, Lv R, Zhou H, Zhou Q, Zhang K, Sun J, Fan C, et al. HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene. 2020;39:1335–1346. doi: 10.1038/s41388-019-1065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tournaye H, Krausz C, Oates RD. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544–553. doi: 10.1016/S2213-8587(16)30040-7. [DOI] [PubMed] [Google Scholar]
- 163.Riera-Escamilla A, Vockel M, Nagirnaja L, Xavier MJ, Carbonell A, Moreno-Mendoza D, Pybus M, Farnetani G, Rosta V, Cioppi F, et al. Large-scale analyses of the X chromosome in 2,354 infertile men discover recurrently affected genes associated with spermatogenic failure. Am J Hum Genet. 2022;109:1458–1471. doi: 10.1016/j.ajhg.2022.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu F, Cao L, Zhang T, Chang F, Xu Y, Li Q, Deng J, Li L, Shao G. CRL4BRBBP7 targets HUWE1 for ubiquitination and proteasomal degradation. Biochem Biophys Res Commun. 2018;501:440–447. doi: 10.1016/j.bbrc.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 165.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 166.Yeh HH, Tseng YF, Hsu YC, Lan SH, Wu SY, Raghavaraju G, Cheng DE, Lee YR, Chang TY, Chow NH, et al. Ras induces experimental lung metastasis through up-regulation of RbAp46 to suppress RECK promoter activity. BMC Cancer. 2015;15:172. doi: 10.1186/s12885-015-1155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y, Wang Y, Zhang D, Zhang H, Wang W, Hu X. Identifying RBBP7 as a promising diagnostic biomarker for BK virus-associated nephropathy. J Immunol Res. 2022;2022:6934744. doi: 10.1155/2022/6934744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim DS, Choi YP, Kang S, Gao MQ, Kim B, Park HR, Choi YD, Lim JB, Na HJ, Kim HK, et al. Panel of candidate biomarkers for renal cell carcinoma. J Proteome Res. 2010;9:3710–3719. doi: 10.1021/pr100236r. [DOI] [PubMed] [Google Scholar]
- 169.Wang J, Yang J, Yang Z, Lu X, Jin C, Cheng L, Wu N. RbAp48, a novel inhibitory factor that regulates the transcription of human immunodeficiency virus type 1. Int J Mol Med. 2016;38:267–274. doi: 10.3892/ijmm.2016.2598. [DOI] [PubMed] [Google Scholar]
- 170.Wang J, Yang Z, Cheng L, Lu L, Pan K, Yang J, Wu N. Retinoblastoma binding protein 4 represses HIV-1 long terminal repeat-mediated transcription by recruiting NR2F1 and histone deacetylase. Acta Biochim Biophys Sin (Shanghai) 2019;51:934–944. doi: 10.1093/abbs/gmz082. [DOI] [PubMed] [Google Scholar]
- 171.Biswas S, Haleyurgirisetty M, Ragupathy V, Wang X, Lee S, Hewlett I, Devadas K. Differentially expressed host long intergenic noncoding RNA and mRNA in HIV-1 and HIV-2 infection. Sci Rep. 2018;8:2546. doi: 10.1038/s41598-018-20791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu W, Wu Y, Zhao J, Chen J, Zhang W. Human immunodeficiency virus type 1 transcription is regulated by thieno[3,4-d] pyrimidine. Exp Ther Med. 2020;19:3090–3096. doi: 10.3892/etm.2020.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schwartz SM, Daling JR, Doody DR, Wipf GC, Carter JJ, Madeleine MM, Mao EJ, Fitzgibbons ED, Huang S, Beckmann AM, et al. Oral cancer risk in relation to sexual history and evidence of human papillomavirus infection. J Natl Cancer Inst. 1998;90:1626–1636. doi: 10.1093/jnci/90.21.1626. [DOI] [PubMed] [Google Scholar]
- 174.Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, Doody DR, Upton MP, Farwell DG, Schwartz SM, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: Potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–188. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wurlitzer M, Möckelmann N, Kriegs M, Vens M, Omidi M, Hoffer K, Bargen CV, Möller-Koop C, Witt M, Droste C, et al. Mass spectrometric comparison of HPV-Positive and HPV-Negative oropharyngeal cancer. Cancers (Basel) 2020;12:1531. doi: 10.3390/cancers12061531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kaushik M, Nehra A, Gakhar SK, Gill SS, Gill R. The multifaceted histone chaperone RbAp46/48 in Plasmodium falciparum: Structural insights, production, and characterization. Parasitol Res. 2020;119:1753–1765. doi: 10.1007/s00436-020-06669-5. [DOI] [PubMed] [Google Scholar]
- 177.Kaushik M, Nehra A, Gill SS, Gill R. Unraveling CAF-1 family in Plasmodium falciparum: Comparative genome-wide identification and phylogenetic analysis among eukaryotes, expression profiling and protein-protein interaction studies. 3 Biotech. 2020;10:143. doi: 10.1007/s13205-020-2096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen H, Zhang W, Luo S, Li Y, Zhu Q, Xia Y, Tan H, Bian Y, Li Y, Ma J, et al. Lead exposure induces neuronal apoptosis via NFκB p65/RBBP4/Survivin signaling pathway. Toxicology. 2023;499:153654. doi: 10.1016/j.tox.2023.153654. [DOI] [PubMed] [Google Scholar]
- 179.Jia W, Shen J, Wei S, Li C, Shi J, Zhao L, Jia H. Ropivacaine inhibits the malignant behavior of lung cancer cells by regulating retinoblastoma-binding protein 4. PeerJ. 2023;11:e16471. doi: 10.7717/peerj.16471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, Cantor AB, Scuto A, Li C, Zhang S, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 181.Hart P, Hommen P, Noisier A, Krzyzanowski A, Schüler D, Porfetye AT, Akbarzadeh M, Vetter IR, Adihou H, Waldmann H. Structure based design of bicyclic peptide inhibitors of RbAp48. Angew Chem Int Ed Engl. 2021;60:1813–1820. doi: 10.1002/anie.202009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.