Abstract
Cell therapies have yielded durable clinical benefits for patients with cancer, but the risks associated with the development of therapies from manipulated human cells are understudied. For example, we lack a comprehensive understanding of the mechanisms of toxicities observed in patients receiving T cell therapies, including recent reports of encephalitis caused by reactivation of human herpesvirus 6 (HHV-6)1. Here, through petabase-scale viral genomics mining, we examine the landscape of human latent viral reactivation and demonstrate that HHV-6B can become reactivated in cultures of human CD4+ T cells. Using single-cell sequencing, we identify a rare population of HHV-6 ‘super-expressors’ (about 1 in 300–10,000 cells) that possess high viral transcriptional activity, among research-grade allogeneic chimeric antigen receptor (CAR) T cells. By analysing single-cell sequencing data from patients receiving cell therapy products that are approved by the US Food and Drug Administration2 or are in clinical studies3–5, we identify the presence of HHV-6-super-expressor CAR T cells in patients in vivo. Together, the findings of our study demonstrate the utility of comprehensive genomics analyses in implicating cell therapy products as a potential source contributing to the lytic HHV-6 infection that has been reported in clinical trials1,6–8 and may influence the design and production of autologous and allogeneic cell therapies.
During the first years of life, humans become exposed to endemic pathogens that infect nearly the entire population9,10. Although primary infection typically clears with subclinical symptoms, viruses from the Herpesviridae, Polyomaviridae, Adenoviridae and Parvoviridae families, among others, can become quiescent, resulting in a latent phase of the virus that is stably maintained in healthy individuals10. Under acute stress conditions, such as immunosuppressive states following haematopoietic stem cell transplantation11 or trauma9, latent viruses may become reactivated, leading to a variety of complex clinical manifestations12 (Fig. 1a). Although there have been anecdotal reports of latent viral reactivation, we lack a complete understanding of the factors and mechanisms underlying latency and the settings in which latent viruses become reactivated. Notably, viral reactivation has more recently been described as a complication in patients receiving CAR T cell therapies1,13, an emerging therapeutic modality that redirects T cells towards a specific antigen using a transgene receptor typically containing an antibody fragment and T cell receptor signalling domains14. One recent study prospectively tested patients receiving CAR T cell therapy for viral reactivation, identifying that approximately one in ten patients had detectable HHV-6 viraemia following CAR T cell infusion with a median of 21 days to the first positive test13. However, the factors driving viral latency and reactivation in these emerging cell therapies that are most often given with lymphodepleting chemotherapy have not been fully characterized.
Fig. 1 |. Petabase-scale analysis of viral nucleic acids reveals that HHV-6 is reactivated in human T cells.

a, Schematic of life cycles of endogenous human viruses. Viruses can enter a latent phase after primary infection and become reactivated following environmental cues, including drugs, immunization and other infections. b, Enumeration of BioSamples with human viruses showing signs of reactivation based on expressed viral nucleic acids in human Sequence Read Archive samples. c, Proportion of BioSamples with viral transcriptional expression annotated as T cells. These eight viruses showed evidence of reactivation specifically in T cells whereas the remaining viruses from b showed no such evidence of reactivation. d, Reanalysis of RNA-seq data from ref. 22. CD4+ T cells from three separate donors were either infected with HIV or mock infected and cultured for about 2 weeks. Shown is the percentage of RNA molecules aligning to the HHV-6B reference transcriptome e, Reanalysis of the data from ref. 18. Naive and memory CD4+ T cells were separated and cultured for 2 weeks. Shown is the percentage of RNA molecules aligning to the HHV-6B reference transcriptome. f, Quantification of HHV-6B episomal DNA from a reanalysis of ChIP–seq data from ref. 18. Shown are the percentages of DNA reads uniquely mapping to the HHV-6B transcriptome, noting the percentage of total DNA reads mapping to the HHV-6B reference genome. g, Coverage of the HHV-6B genome across two ChIP–seq libraries from different donors from ref. 18. Repetitive regions on the ends of the chromosome show no uniquely mapping reads.
Here we use comprehensive genomics analyses to link specific human cell states to viral reactivation events. Through reanalysis of existing sequencing datasets, we identify that HHV-6, an endemic beta-herpesvirus, becomes reactivated in standard CD4+ T cell cultures. Using single-cell genomics, we characterize a rare CAR T cell state with high expression of HHV-6 in both cell cultures in vitro and patients in vivo. Together, our analyses demonstrate the ability of genomics technologies to uncover new associations in cell therapy settings that may affect future clinical and manufacturing guidelines.
Landscape of human virus reactivation
In an effort to study human viral reactivation systematically, we mined a petabase-scale resource, Serratus15, that quantified viral abundance from RNA-sequencing (RNA-seq) and DNA-sequencing data from the Sequence Read Archive16. We considered the occurrence of high-confidence viral RNA from 129 curated human viruses17, focusing on the 17 viruses across the Herpesviridae, Polyomaviridae, Adenoviridae and Parvoviridae families that are known to have latent replication cycles and can reactivate in vivo10 (Methods and Supplementary Table 1). For 15 of these 17 viruses, we observed annotation of viral RNA in at least one BioSample, totalling 8,617 libraries over 6,902 distinct BioSamples with plausible viral RNA expression, and in turn, potential lytic reactivation (Fig. 1b, Methods and Supplementary Table 2). After defining this landscape, we examined samples with the highest levels of RNA per virus, including Epstein–Barr virus (EBV) expression in B cells (Supplementary Table 3). We observed that the top positive samples for HHV-6 came from primary T cell cultures for which no record of the HHV-6 virus was noted in the original study18 and an overall enrichment of T cell BioSamples with HHV-6 compared with other viruses (Fig. 1c and Supplementary Table 3).
Although HHV-6 latency is generally established through a primary infection in the first 2 years of life, approximately 1% of the population has chromosomally integrated HHV-6 that is transmitted following Mendelian principles19. For >90% of the population, primary infection of HHV-6 occurs during infancy and may manifest as roseola infantum, characterized by high fever and exanthema. Although complications from these primary infections in infancy are rare, the reactivation of HHV-6 in adulthood can cause severe disease, including encephalitis. In immunocompromised individuals, including those receiving chemotherapy, HHV-6 reactivation can be lethal. Two strains of HHV-6 are endemic worldwide: HHV-6A infections are largely confined to sub-Saharan Africa20, whereas HHV-6B accounts for 97–100% of HHV-6 infections reported in the USA, Europe and Japan. Notably, the receptor for HHV-6B, OX40 (also known as CD134; encoded by the gene TNFRSF4)21, is specifically upregulated during CD4+ T cell activation (and, to a lesser extent, during CD8+ T cell activation), and the TNFRSF4 transcript is detectable in a variety of healthy tissues (Extended Data Figs. 1 and 2). Comparing the viral abundances from T cell libraries showing plausible HHV-6 expression, we observed substantially more annotated HHV-6B viral RNA than HHV-6A viral RNA for all libraries, consistent with the epidemiological prevalence of these viruses (Methods and Extended Data Fig. 3a). Thus, we used the HHV-6B reference genome (Genbank AF157706) for subsequent analyses.
HHV-6 is reactivated in T cell cultures
To better understand the kinetics of HHV-6 reactivation in T cell cultures, we reanalysed sequencing data from the two studies nominated by our computational screen that showed high levels of viral reactivation18,22. For both studies, T cells were cultured under standard conditions (activation with anti-CD3 and anti-CD28 beads plus interleukin-2 (IL-2)) and were profiled longitudinally by RNA- and DNA-based sequencing. In ref. 22, total CD4+ T cells from three separate donors were infected with HIV or mock infected and cultured for 14 days. Two donors (14 and 49) showed clear evidence of HHV-6B reactivation in both conditions over the 2 weeks in culture, including a sample in which nearly 3.5% of all RNA molecules were derived from HHV-6B and mapped across the entire HHV-6B transcriptome (Fig. 1d and Extended Data Fig. 3b). Analysis of the data from ref. 18, in which naive and memory CD4+ T cells were separated and cultured for 2 weeks before profiling with RNA-seq (Fig. 1e and Extended Data Fig. 3c) and chromatin immunoprecipitation with sequencing (ChIP–seq) (Fig. 1f,g), similarly indicated HHV-6B reactivation. Cells in culture from donor 5,131 expressed HHV-6B transcripts at high levels (about 4.27% of sequenced RNA molecules) after 2 weeks of memory CD4+ T cell culture in vitro. As this study used the same cell cultures to carry out ChIP–seq targeting histone modifications, we remapped ChIP–seq reads to estimate the abundance of HHV-6-derived DNA from these samples (Methods). We observed that all three donors showed evidence of HHV-6B DNA copy number increase throughout the cultures, including a sample from donor 5,131 that had the highest viral copy number for both DNA and RNA (Fig. 1e,f and Methods). We confirmed that these mapped DNA molecules were derived from the HHV-6B virus, again noting the uniform coverage across the HHV-6B reference genome outside repeat regions and across all transcripts (Fig. 1g, Extended Data Fig. 3b and Methods). Here we note that detection of HHV-6 was robust to viral polymorphisms, as Serratus detected these BioSamples despite hundreds of single nucleotide variants differing from the reference genome (Extended Data Fig. 3d and Methods). Finally, public data mining provided additional support for HHV-6 reactivation in other contexts, including BioSamples with high HHV-6B derived from patients with bronchiolitis23, graft-versus-host disease24, purified T cell populations from patients with cutaneous T cell lymphoma25 and therapeutic regulatory T cell cultures26 (Extended Data Fig. 3e–g and Supplementary Table 4). In total, analysis of these public datasets links the reactivation and transcriptional activity of HHV-6B in T cells in vitro and in vivo.
CAR T cells reactivate HHV-6 in vitro
In light of eight cases of HHV-6 encephalitis from patients receiving CAR T cell therapy, including in three recent clinical trials1,6–8, we reasoned that cell therapy products could present a potential source of HHV-6 reactivation responsible for the subsequent clinical presentation. To assess this, we monitored research-grade allogeneic CAR T cells over a 19-day manufacturing window, a standard duration for allogeneic cultures, and screened cultures with quantitative PCR (qPCR) to detect the U31 DNA from the HHV-6B virus (Methods). We identified four exemplar peripheral blood mononuclear cell (PBMC) lots derived from healthy donors that by day 19 expressed a range of HHV-6B from 0.0015 to 0.90 copies of U31 per cell (Fig. 2a). Two of these donors, 34 and 38, were monitored longitudinally, which demonstrated low, if any, early HHV-6 DNA before a rapid increase after approximately 2 weeks in culture (Fig. 2b). These data provide direct support corroborating our observations from the Serratus analysis and demonstrate that HHV-6 nucleic acid can become more abundant in T cell cultures over time, consistent with viral reactivation.
Fig. 2 |. scRNA-seq identifies rare HHV-6-expressing cells in CAR T cell culture.

a, Quantification of HHV-6B by qPCR at day 19 for four donors normalized to the cell count. Bars are shown in order of increasing abundance. b, Longitudinal qPCR surveillance of HHV-6B U31 gene copies in CAR T cell culture from two donors. c, Schematic of the single-cell sequencing workflow to detect HHV-6+ cells from the CAR T culture. Two models are presented that would explain HHV-6 reactivation: model 1 (top), in which all cells express HHV-6 transcripts; and model 2 (bottom), in which only a subset of cells express HHV-6B. Both the host and HHV-6B viral RNA can be directly quantified using the 10x Genomics scRNA-seq workflow. d, Summary of HHV-6B expression from an individual donor (98). The top 0.2% of cells contain 99% of the HHV-6B transcript UMIs from this experiment. e, Tabulated summary of scRNA-seq profiling for four CAR T donors, including number of cells profiled, percentage expressing HHV-6, U31 qPCR value and number of shared TCR clones between the HHV-6B+ cells. N/A, not applicable. f, Extended longitudinal sampling of HHV-6B through U31 qPCR. Number at right end of each plot line indicates the fold (×) increase from the first qPCR measurement (day 21) to the final measurement (day 27; black outline) for each donor. g, Schematic and summary of HHV-6B expression in donor 34 after 19 and 25 days, showing evidence of HHV-6B spreading in the culture as depicted in the schematic. h, Correlation analyses of host factor gene expression with HHV-6B RNA abundance in individual cells for donor 34 on day 25. The per-gene correlation statistics are shown in black against a permutation of the HHV-6B expression in grey. Select genes are indicated. i, Pathway enrichment analysis of Molecular Signatures Database Hallmark gene sets for gene set enrichment analysis. A positive normalized enrichment score corresponds to genes that are overexpressed in cells with large amounts of HHV-6B transcript.
To better understand the dynamics and cell states of reactivated HHV-6B during cell culture, we considered whether HHV-6 would be uniformly reactivated across a cell population (model 1) or if only a subset of cells may spread the virus throughout the culture (model 2; Fig. 2c). In line with recent efforts that quantify both viral and host gene expression at single-cell resolution27, we established an efficient pseudoalignment pipeline after single-cell RNA-seq (scRNA-seq) to quantify HHV-6B transcripts with no detectable false positive quantification that would ordinarily occur owing to homology of viral transcripts with the human KDM2A gene (Methods). Our single-cell characterization of CAR T cell cultures revealed the presence of rare HHV-6+ cells that we term ‘super-expressors’ (that is, cells that have ≥10 viral unique molecular identifiers (UMIs)), supporting our model 2, in which only a subset of cells initially express HHV-6B (Fig. 2c,d and Extended Data Fig. 4a). For these cells, we detected expression of 90 of 103 HHV-6B transcripts, including immediate-early viral genes that were enriched among a subset of super-expressors, suggesting that these cells had recently reactivated or been infected by HHV-6B (Extended Data Fig. 4b). In total, these rare super-expressors accounted for only 0.2% of total cells in culture for donor 98 despite expressing most of the total HHV-6 viral RNA detected (99.2%; Fig. 2d). We confirmed the existence of such rare cell populations, finding that between 0.01% and 0.3% of cells (or 1 in 360–10,000) reactivated or expressed HHV-6 transcripts at high levels across three donors (34, 97 and 98). Using the host (T cell) gene expression signatures, we carried out standard dimensionality reduction and annotation, revealing that super-expressors were confined to the CD4+ T cell compartment, consistent with relative overexpression of the HHV-6B entry receptor OX40 in CD4+ T cell subsets (Methods and Extended Data Figs. 1a and 4c). For a fourth donor (38), no HHV-6B+ cells were detected, corresponding to a negative U31 qPCR value (Fig. 2e). Sequencing of T cell receptor αβ chain (TCRαβ) transcripts revealed that none of the HHV-6B+ cells from these donors shared a clonotype receptor sequence, indicating that reactivation of HHV-6 is of a polyclonal origin and is not restricted to a single expanding clonotype (Fig. 2e).
For one donor (98), we sought to further characterize viral RNA and DNA abundance in the same cell using single-cell multi-omics. Here we used a concomitant assay for transposase-accessible chromatin (DNA) and RNA-seq from the same cells as active HHV-6 DNA exists as an episome in the nucleus (Extended Data Fig. 5a). Profiling 13,952 cells from this donor, we detected HHV-6B DNA or RNA in 22.5% cells (n = 3,139). At the single-cell level, our profiling revealed a strong correlation between viral DNA and RNA detection in individual CAR T cells (Pearson r = 0.57), directly linking our quantification of viral RNA from scRNA-seq to viral DNA abundance (Extended Data Fig. 5b). After stratifying the HHV-6B transcripts as immediate early, early and late using a previous annotation28, we computed a per-cell metric of viral gene expression to identify a population of cells with high expression of immediate-early transcripts, in addition to populations that spanned variable levels of early and late gene expression (Methods and Extended Data Fig. 5c). Cells with high early viral RNA expression had the highest viral DNA abundance, consistent with the known role of early genes in facilitating viral replication29 (Extended Data Fig. 5d,e). Thus, these single-cell analyses directly link our observed viral transcription to viral replication, which was further corroborated by correlating measurements at the donor level (Extended Data Fig. 5f).
For products from two donors, we extended the time of T cell expansion and collected scRNA-seq libraries after a total of 25 and/or 27 days from the initiation of the culture. Analysis of data from these later time points revealed an increase in the abundance of HHV-6B transcripts by about 1,200–2,000 fold, compared to day 20 (Fig. 2f). Notably, 49% and 62% of all T cells were HHV-6 super-expressors, including both CD4+ and CD8+ cells, indicating lytic reactivation and viral spreading, which we confirmed using both scRNA-seq and immunofluorescence (Fig. 2g and Extended Data Fig. 6a,b). The wide prevalence of the virus in vitro verifies that reactivated HHV-6 spread across the culture, probably facilitated by the fact that its entry receptor OX40 is a co-stimulatory molecule expressed at high levels on activated T cells.
Next we sought to link host gene expression programs with HHV-6B activation (Methods). Using the day-19 transcriptomic profiles, HHV-6 super-expressors compared to other HHV-6B negative cells showed a consistent upregulation of lymphotoxin-α (LTα) and downregulation of LTβ (Methods and Extended Data Fig. 6c), indicative of host cell gene expression programs responding to the virus. However, we were unable to nominate host factors, such as surface markers, that would segregate cells capable of HHV-6 reactivation from other cells. For the three samples profiled with scRNA-seq after extended culture, we further used a correlation statistic coupled with permutation testing to identify hundreds of genes associated with HHV-6B infection following reactivation in culture (Fig. 2h, Methods and Supplementary Table 5). Of note, this analysis identified proviral genes such as HSPD1 and SLC3A2, which have been characterized as positive regulators of other infections30,31 (Fig. 2h). Conversely, we identified upregulation of genes such as LY6E (ref. 32), STAT1 (ref. 33) and IL32 (ref. 34) in HHV-6Blow cells, each of which has previously been implicated in antiviral control in other infection contexts (Fig. 2h). After stratifying the HHV-6B transcripts into immediate early, early and late, the only gene signature correlating with OX40 expression in single cells was the immediate-early signature, aligning with the expectation that recently infected cells would express higher levels of the viral receptor (Methods and Extended Data Fig. 6d). Integrated analyses of altered gene expression signatures associated with HHV-6B expression levels corroborated the role of various proviral and antiviral pathways, including type I interferon response35 and oxidative phosphorylation36 (Fig. 2i and Methods). Our host–viral association analyses indicate that a subset of CAR T cells in culture that resists infection show transcriptional signatures driving an antiviral response, spanning metabolic transporters, transcription factors, cytokines and surface molecules.
To determine whether the process of manufacturing CAR T cells is sufficient to drive HHV-6 reactivation, we obtained cells from a donor with chromosomally integrated HHV-6 and assessed HHV-6 DNA copy number using the standard or modified versions of the manufacturing protocol (Extended Data Fig. 7a). Over the 19-day manufacturing window, we did not observe a meaningful increase in HHV-6 DNA copy number; nor was there detectable viral RNA at either the beginning or end of the manufacturing window (Extended Data Fig. 7b,c). These results indicate that no individual step in the engineering of the CAR T product is sufficient to drive HHV-6 reactivation, even if HHV-6 is present in every cell and OX40 is broadly expressed on activated CAR T cells. Instead, our data suggest a model in which HHV-6B reactivation is a stochastic event that may be affected by various cues in the cell culture and intrinsic differences in host cell state.
CAR T cells reactivate HHV-6 in vivo
Having characterized this phenomenon in vitro, we reasoned that these cells could be a potential source of HHV-6 reactivation in patients. To assess this, we reanalysed three cohorts of scRNA-seq datasets collected from patients with B cell lymphoma or leukaemia receiving one of three autologous CAR T cell products (all targeting the CD19 antigen), including US Food and Drug Administration (FDA)-approved axicabtagene ciloleucel and tisagenlecleucel, as well as from one early-phase clinical study (SJCAR19; NCT03573700). Across these three cohorts, we screened single-cell expression profiles of 619,392 CAR T cells from pre-infusion products and 829,535 cells from peripheral blood draws weeks after the infusion (Fig. 3a and Table 1). Of the 72 patient products, we did not detect any HHV-6 viral transcripts through reanalysis of the pre-infusion products (Fig. 3a and Table 1). However, we detected a total of 28 cells in post-infusion samples that expressed one or more HHV-6B transcripts, a significant increase in HHV-6+ cells over the pre-infusion profiles (P = 1.98 × 10−8; two-sided Fisher’s exact test). Out of 35 donors with high-quality post-infusion scRNA-seq profiles, we detected HHV-6+ cells in four donors, including a total of thirteen ‘super-expressor’-like cells (Fig. 3b and Methods). Using CAR protein (from flow-enriched cells), CAR transcript, TCRαβ transcript and CD3 expression, we verified the cellular identity of 28 total HHV-6B+ cells as engineered CAR T cells that expressed a diversity of HHV-6B transcripts, indicating viral reactivation in vivo (Fig. 3b,c, Methods and Supplementary Table 6). To verify the specificity of our results, we applied our single-cell HHV-6-quantification algorithm to about 1,000 PBMC samples, each stemming from a different donor, and more than 60 tissues from healthy donors (Methods). Although a variety of cells express OX40 throughout the body, we did not detect a single HHV-6B transcript in these datasets, despite the high seroprevalence of previous HHV-6 infection and the presence of detectable viral DNA37. These results indicate that HHV-6 is not transcriptionally active in PBMCs at steady state.
Fig. 3 |. HHV-6 reactivation in FDA-approved CAR T cells and clinically used CAR T cells in vitro and in vivo.

a, Schematic and summary of patient cohorts for scRNA-seq profiling, noting different cell therapy products (axicabtagene ciloleucel (axi-cel) and tisagenlecleucel (tisa-cel)) and two different time points for sampling (pre-infusion product and post-infusion blood draws). The numbers of total cells analysed and HHV-6+ cells detected are noted underneath. b, Summary heatmap of gene expression in vivo for 22 HHV-6+ cells from cohort 2 follow up and 6 HHV-6+ cells from cohort 3 post-infusion samples. The number of HHV-6 transcripts is in the blue rectangles; green rectangles indicate nonzero host expression (Methods). c, Heatmap of 86 detected HHV-6B transcripts (columns) across the described 28 individual cells (rows, as in b) grouped by viral gene class (immediate-early, early and late expressing genes as previously described28). d, Summary of the clinical timecourse of patient axi-R-15, noting four time points of scRNA-seq data collection (red dots) and the clinical period corresponding to altered mental status. SEs, super-expressors. ***P = 0.00047, day 7 versus day 0; P = 0.00032, day 7 versus day 14 (two-sided binomial tests). e, Summary of the clinical in vivo timecourse of patient SJCAR19–09, including PCR of HHV-6 and T cell scRNA-seq abundance at the indicated time points. Treatment regimen with foscarnet is noted with the initial dose administered on day +24. CPM, counts per million. f, Schematic (left) of an in vitro reculture experiment in which CAR T cells were recovered from the infusion product from SJCAR19–09 and then recultured with TransAct, IL-7 and IL-15, and HHV-6B RNA counts per million over five time points, measured by CAR T cell scRNA-seq (right). HHV-6 was not detected (n.d.) at day 0 but was observed as soon as 3 days following reculture and persisted for the full 14 days of additional culture.
Table 1 |.
Summary of HHV-6B expression in large-scale single-cell datasets
| Description | No. of cells | No. of reads | No. of HHV-6B UMIs | No. of HHV-6+ cells (No. of HHV-6 super-expressors) | Ref. |
|---|---|---|---|---|---|
| 24-patient CAR T cell pre-infusion product | 137,326 | 8,972,716,245 | 0 | 0 (0) | 42 |
| 32-patient (CAR) T cell, pre and post infusion | 602,577 | 15,403,016,654 | 919 | 7 (4) | 2 |
| 16-patient (CAR) T cell, pre and post infusion | 798,618 | 66,076,393,465 | 374 | 6 (3) | 3 |
| 1-patient (axi-R-15) pre- and post-infusion follow-up | 31,953 | 3,086,842,082 | 789 | 15 (6) | This study |
| Human cell landscape, 60 tissue types | 599,926 | 15,680,467,351 | 0 | 0 (0) | 43 |
| About 1,000 donor PBMCs | 1,449,385 | 53,872,337,003 | 6 | 0 (0) | 44 |
we more closely examined the timecourse-6+ CAR T cells in vivo, we more closely examined the timecourse of two patients with super-expressors in the post-infusion blood draws. First, we examined additional samples from the patient, axi-R-15, whose cells showed the highest number of captured HHV-6B transcripts. We complemented the existing scRNA-seq profiles by sequencing another 31,953 cells for this patient spanning four time points, including weekly blood draws to day +21 (Fig. 3d and Methods). Along this timecourse, we observed HHV-6+ cells only at day 7, when the HHV-6+ super-expressors were present at an overall frequency of about 1 in 1,000 T cells (a total of 10 super-expressors were detected among 9,757 T cell scRNA-seq profiles). Notably, this patient was clinically diagnosed with immune effector-associated neurotoxicity syndrome (ICANS) following axicabtagene ciloleucel treatment and subsequently developed altered mental status with tremulousness and word-finding difficulties beginning on day +9, which peaked on day +12 (grade 3a by Common Terminology Criteria for Adverse Events criteria; grade 2 by American Society for Transplantation and Cellular Therapy criteria) before returning to baseline by day +14 (Fig. 3d). Similarly, we observed a spike in HHV-6+ cells at day +7 that was resolved by day +14, noting that the dynamics of viral reactivation and clearance could be observed within 1 week of each other (Fig. 3d). Although the symptoms correlated with the presence of HHV-6+ CAR T cells in the blood, and are consistent with clinical symptoms of HHV-6 encephalitis following CAR treatment1, we note that we cannot conclude that the detection of HHV-6 in peripheral blood is causal for the patient’s neurotoxicity. As delirium and neurocognitive decline are common symptoms in patients with HHV-6 viraemia receiving haematopoietic stem cell transplantation, whereas ICANS is not38, we emphasize that HHV-6 may be a pathogenic agent for ICANS in some—but probably not most—patients receiving CAR T cell therapy.
In a second participant, we detected three HHV-6-super-expressor cells from patient SJCAR19–09 in cohort 3. Of two patients tested for HHV-6 by qPCR from this cohort, only SJCAR19–09 tested positive, aligning with our single-cell-resolved detection of HHV-6+ CAR T cells in this individual. SJCAR19–09 continued to test positive by qPCR at day +21 post-CAR T cell infusion and was started on foscarnet at day +24 (Fig. 3e). After another positive qPCR test on day +27, the patient no longer had detectable levels of HHV-6 at day +34 (Fig. 3e). In line with these findings, we detected HHV-6+ super-expressors at day +14 that persisted in vivo until day +21 before these cells were undetectable at day +27. As the CAR T cells used in cohorts 2 and 3 were cultured only for 7–10 days before infusion, we reasoned that HHV-6 may reactivate in vivo only after the therapy is administered. We reasoned that an extension of the culture, as is carried out in many allogeneic or gene-modification manufacturing contexts to achieve higher cell numbers or efficacy, could lead to viral reactivation in vitro. To test this, we extended the culture of the SJCAR19–09 infusion for an additional 2 weeks and examined the culture using scRNA-seq at five time points (Methods). Indeed, between 5 and 14 days of additional culture, we consistently detected expression of HHV-6B spanning 58 of the 103 distinct viral genes (Fig. 3f). These data support a model in which HHV-6 can be transcriptionally detected in vitro or in vivo between 2 and 3 weeks after the initial manufacture of the CAR T cell product. For SJCAR19–09 and axi-R-15, the CAR T cell infusion occurs before this window in which detectable reactivation typically occurs. As a consequence, HHV-6+ CAR T cells could be detected in peripheral blood draws from these donors only after an additional 1–2 weeks of propagation in vivo.
Mitigation of HHV-6 spreading in culture
The analysis of SJCAR19–09 suggests that foscarnet may be a promising agent to mitigate the effects of viral infection during therapy. We reasoned that the addition of foscarnet to our allogeneic CAR T cell cultures could mitigate viral reactivation and spreading. On the basis of previous reports that included foscarnet in cell culture39, we thawed day-19 vials from our allogeneic CAR T cell donors and compared cultures extended another 5 days with and without foscarnet at concentrations previously described (1, 2.5 and 5 mM; Methods). Quantifying viral abundance through qPCR with reverse transcription, our results confirmed that the addition of foscarnet during culture led to a lower viral RNA abundance compared to that of the untreated control (P = 0.00026; ordinary least squares linear model; Methods and Extended Data Fig. 8a,b). Using scRNA-seq to study another donor treated with 1 mM foscarnet (Supplementary Table 7), we replicated the reduction of HHV-6+ in the culture, observing that 63.6% of cells were HHV-6+ in the control compared to 21.8% in the foscarnet-treated cells. Notably, we observed minimal changes in cell states after the treatment (Methods and Extended Data Fig. 8c–f). Although further investigation is needed to determine optimal dosing, duration and intervention for antiretroviral drugs in cell therapy product engineering, our results suggest a potential path to mitigating viral reactivation in cell manufacturing.
Allogeneic CAR T cells reactivate HHV-6
Although autologous CAR T cells are the most widely used, emerging engineered cell therapy products, particularly hypoimmunogenic allogeneic products, are being increasingly evaluated in clinical trials14. In these settings, patients may receive advanced lymphodepletion regiments that could increase immunosuppression and the risk of complications from HHV-6 reactivation. Thus, we sought to examine the HHV-6 reactivation in the context of patients receiving allogeneic CAR T cells in vivo. We examined single-cell genomics data from a phase I clinical trial (NCT04538599) targeting CD7, an antigen expressed in T acute lymphoblastic leukaemia5. To assess the potential for HHV-6 reactivation in this CD7 CAR T product, we reanalysed the scRNA-seq data generated for one CAR T product and CAR T-enriched PBMCs from one patient (drawn at days 14 and 19), representing all genomics data available in the study5 (Fig. 4a and Methods). Here we observed HHV-6 transcripts in the infusion product and in PBMCs at both time points and a marked increase in viral transcripts at day 19 versus 14 (Fig. 4b). We attribute our detection of HHV-6 transcripts in allogeneic products to their longer culture durations, compared with that of the autologous products5,40.
Fig. 4 |. Allogeneic CAR T cells discriminate donor and host reactivation of HHV-6B in vivo.

a, Schematic of data generation for CD7 allogeneic CAR T cells, including the infusion product and PBMCs derived at day 14 and 19 following patient infusion. b, Summary of HHV-6 pseudobulk expression across the three samples from RD13–01-treated patient 5 assayed in the study. c, Uniform manifold approximation and projection (UMAP) of day-19 scRNA-seq data, showing the annotation from genetic demultiplexing (left) and log10[HHV-6 UMIs] across all cells in the experiment (right). d, Summary of HHV-6+ cells from day-19 infusion product. Host and CAR T cells were separated using SouporCell as shown in c. More HHV-6+ cells were detected among the CAR T cells, but the host cell shad more super-expressors (red) and a higher overall UMI count.
Next we sought to characterize the underlying cell states associated with high HHV-6 levels and carried out genetic demultiplexing analyses to annotate each cell as a host or allogeneic CAR T cell (Methods). Of the 6,468 cells with a high-confidence donor assignment at the day-19 blood draw, we annotated 5,017 as donor-derived cells, which were largely a homogeneous population of CD8+ CAR T cells, and 1,401 as host-derived cells across various haematopoietic lineages (Extended Data Fig. 9). Between both host endogenous and allogeneic CAR T cells, we identified a total of 369 cells with detectable HHV-6 transcripts, including 216 cells from the CAR T product (Fig. 4c,d). We observed a rare population of host CD4+ T cells that expressed hundreds of viral UMIs, which collectively encompassed more super-expressors (n = 22; max: 1,492 HHV-6 UMI counts per 10,000) than the allogeneic CAR T cells (n = 2; max: 1,136) but with similar magnitudes of HHV-6 expression. As there were no genetic differences between the host-derived and allogeneic-derived HHV-6, consistent with the genetic stability of HHV-6 (ref. 41), we could not discriminate between whether the host-derived HHV-6 was attributable to an independent reactivation event in vivo (rather than infection from the CAR T product that would be genetically identical; Methods). Ultimately, these data suggest a model in which both host cells and cell therapy products can amplify the overall lytic viral pool, with the relative timing, proportions and duration of the reactivation likely to vary from patient to patient.
Viral reactivation in other cell therapies
Beyond the association between T cell cultures and HHV-6 reactivation, latent viral reactivation may extend to other viruses or forms of cell therapy. From our inferences from mining the Serratus database, we observed reactivation of herpes simplex virus 1 from reprogrammed induced pluripotent stem cells (Supplementary Table 8) and EBV expression in T cells (Methods and Supplementary Table 4). We did not detect reactivation of any viruses in natural killer cells or their products (Methods). Ultimately, further mechanistic studies are required to explicitly link the reactivation of other latent viruses to any other cell therapy product. Although our existing screen identified thousands of high-confidence associations, future work that systematically accounts for genetic variation in viruses will enhance the sensitivity to detect viral reactivation using sequencing approaches.
Conclusion
Our work has implications for current preclinical and future applications of cell therapies. We observed HHV-6 reactivation in both native and engineered T cell states as well as in FDA-approved CAR T cells following their clinical delivery. These data indicate that HHV-6 reactivation in vitro occurs in a CAR-independent capacity and is linked to T cell activation, proliferation and culture duration, perhaps giving the virus the ability to spread under more favourable conditions. Further, we observed the increase of lytic HHV-6 over time both in vitro and in vivo, indicating that screening cell therapy products for the virus at a single time point (for example, pre-infusion product) may not be fully adequate to identify virus-positive cells from the final therapeutic product or its fate in vivo. Although we use single-cell genomics to study HHV-6 reactivation in this work, we emphasize that any clinical procedure could use PCR-based methods to test products and patients. To our knowledge, no explicit recommendations are in place that require cell therapy products to be screened for potential viruses before infusion. Ultimately, our results do not warrant changes to patient monitoring or standards of care at present, but we suggest that the possibility of latent viral reactivation in cell therapies could be broadly considered in current and future products. These include infusion products with high viral DNA levels that could prompt post-infusion monitoring, including in individuals with chromosomally integrated HHV-6. More broadly, our efforts demonstrate the utility of comprehensive genomics sequencing and analyses in disentangling complex clinical phenotypes and ultimately nominate an unappreciated route of viral infection through cell therapy.
Methods
Serratus viral screen analyses
To examine human viruses that may become transcriptionally activated in a systematic approach, we queried the Serratus15 database (v210225 nucleotide release containing 789,929 libraries; accessed 1 August 2022). For 129 human viruses curated from ViralZone (accessed 1 August 2022)17, we used the NC genome identification provided on the ViralZone webpage as input in the Serratus API. The values for the hyperparameters in the call (scoreMin, scoreMax, identityMin and identityMax) were selected for inclusiveness and were later filtered programmatically. An example of the API call for the HHV-6B genome (NC_000898.1) is as follows: wget –O NC_000898.1. txt “https://api.serratus.io/matches/nucleotide?scoreMin=0&scoreMax=100&identityMin=75&identityMax=100&sequence=NC_000898.1”.
To focus on viral reactivation, we selected 17 DNA viruses from the Herpesviridae, Polyomaviridae, Adenoviridae and Parvoviridae families. We categorized a sample as a potential hit (Fig. 1b) if the Serratus output returned a minimum of 100 reads with an average mapped read identity of at least 50% for the individual BioSample–virus pair. We note that if viral genomes or segments were not reported in the ViralZone annotation, the potential viral tropism association would not be detected. As the ViralZone genome annotation for HHV-6 corresponded to HHV-6A (NC_001664), the most common strain in sub-Saharan Africa20, we further queried the Serratus database for entries for HHV-6B (NC_000898) for all subsequent analyses. This decision was corroborated by a head-to-head comparison of all 16 T cell-derived libraries that had high-confidence expression in either HHV-6A or HHV-6B (Extended Data Fig. 3a). Additionally we note that the identification of viruses with many polymorphisms may not be as sensitively detected in our current framework. Future work that enhances the bioinformatics screen, including the automatic inclusion of different strains and polymorphisms, will facilitate more robust detection of viral reactivation in heterogeneous human tissues. Ultimately, for HHV-6, we note that our inferences seem robust to viral nucleotide diversity despite the widely used reference genome for HHV-6B being a relatively uncommon sequence for the virus41 (Extended Data Fig. 3d).
Next, to annotate BioSamples as belonging to specific cell types, we used the NCI Thesaurus for T cells and induced pluripotent stem cells (iPSCs) to define a comprehensive set of search terms for each cell type (for example, ‘T Lymphocyte’ for T cells). Using the union of applicable terms per cell type, we queried Sequence Read Archive and National Center for Biotechnology Information BioSamples for matches to any of the terms in the ontogeny, yielding a list of BioSamples and/or sequencing libraries annotated with either of these cell types of interest. These queries (date: 13 March 2023) yielded a total of 46,374 T cell libraries and 31,433 iPSC libraries present in Serratus, which are specified as part of our online resources (in the ‘Code Availability’ section). These were used as a high-confidence annotation of T cell and iPSC samples from our screen to subset the Serratus library annotations. The full code needed to reproduce the screen for additional viruses is available as part of our online resources in the ‘Code Availability’ section.
Notably, we did observe expression of EBV from annotated T cell populations (Fig. 1c), including intestinal intraepithelial cytotoxic CD8+ T cells45. As EBV infects cells through CD21, a canonical B cell marker, we examined the expression of CD21 across all resting and stimulated immune cell subsets, revealing that γδ T cells can express CD21 at higher levels than any B cell subset (Extended Data Fig. 1b). This result led us to speculate that EBV reactivation may occur in γδ T cells, but future work is needed to corroborate these inferences. Of note, our screen did not nominate a reactivation candidate virus in natural killer cells. Regardless, we suggest that natural killer cell therapies may warrant further investigation for potential viral latency, as longitudinal cultures were more commonly profiled for T cells and iPSCs than for natural killer cells. Overall, we also suggest that the systematic approach pairing viral expression to specific BioSamples through Serratus could enable the identification of cell types to become infected that have not been previously characterized.
We note that the two primary studies that showed HHV-6 reactivation in T cell culture18,22 took place on different continents, making it highly unlikely that other factors enabled the reactivation of HHV-6 in their cultures (for example, same donor) aside from T cell culture-induced HHV-6 reactivation. For our deeper analyses of these datasets (Fig. 1d,e), we paired the Serratus read abundance with metadata from the Gene Expression Omnibus (GEO), including the total number of reads per library. After the experiments from these two manuscripts were identified using Serratus, we downloaded the raw.fastq files to directly quantify the proportion of molecules that could be derived from the virus as opposed to host transcripts. In the dataset of ref. 22, cells were treated with an HIV infection. We reanalysed the expression from these data at the per-gene level, verifying that a diversity of transcripts were expressed and that regions of high homology between HHV-6B and HIV were not the only transcripts detected (also noting that not all HIV-infected cells showed HHV-6B expression). Thus, the inference of HHV-6B reactivation in this setting is not attributable to off-target viral read mapping.
To study the impact of viral polymorphisms and to benchmark their detection for deconvolving viral heterogeneity in allogeneic settings, we used the two BioSamples with the most HHV-6 expression in the studies of ref. 22 (donor 49, day 14) and ref. 18 (donor 5,131, day 14, memory CD4+) and mapped RNA-seq libraries to the HHV-6B reference genome using bwa46. To determine the per-position nucleic acid count, we used bam-readcount (v1.0.1) on each.bam file from this mapping. We empirically called variants for which the reference position differed from the most abundant nucleotide, and this alternative nucleotide was present in at least 95% of molecules. Positions annotated as ‘well-covered’ in the 5′ scRNA-seq dataset (Extended Data Fig. 3d, right diagram) were determined with a coverage of at least ×100 over the positions in the day-19 CD7 allogeneic dataset (Fig. 4).
For assay for transposase-accessible chromatin with sequencing and ChIP–seq libraries, DNA was not processed through the Serratus workflow, requiring realignment to the HHV-6B reference genome (for example, Fig. 1f,g). Thus, we downloaded raw.fastq files from GEO and aligned reads to the HHV-6B reference genome (Genbank AF157706) using bwa and retaining only uniquely mapped reads (MAPQ > 30) for downstream analysis and quantification. To screen for reads outside the repetitive regions (which have high homology with regions of the human genome), we required the read to map within the (8788, 153321) coordinates.
Although we cannot exclude the possibility of contamination or de novo infections yielding the HHV-6B RNA detected in our reanalysis, the recurrence of our observation indicates that reactivation is the most plausible source of the viral transcripts. Further, many of the samples analysed by the bulk RNA-seq and DNA-seq in this workflow may contain low-frequency cells that would explain the viral reactivation. We thus emphasize the importance of well-controlled experiments to corroborate any computational inferences as we have done to link HHV-6B reactivation to T cell cultures.
Primary human T cell cultures
For the research-grade allogeneic CAR T cultures, PBMCs were isolated from leukopaks obtained from healthy donors and stored in liquid nitrogen before use. Donors were deidentified with Institutional Review Board-approved consent forms and protocols, and as donors could not be re-identified, they do not constitute human subjects research. PBMC donor lots were screened at Allogene Therapeutics for HHV-6 DNA following CAR T cell culture conditions. PBMCs were thawed, resuspended in X-vivo medium (Lonza) supplemented with 5% human serum (Access Cell Culture) and IL-2 (Miltenyi), and stimulated with T Cell TransAct (Miltenyi). Cells were transduced with a lentivirus containing proprietary CAR sequences early in culture followed by further genetic manipulation for immune evasion and prevention of graft rejection. Additional details regarding the engineering of these cells through Allogene’s allogeneic CAR T manufacturing are available as previously described40,47. In line with the manufacturing details previously reported in preclinical reports, cells were activated at day +2, transduced with the CAR at day +4, and further electroporated with TALEN mRNA at day +6 before continued expansion for the duration of the culture to day 19 (ref. 40). These T cell cultures were allowed to expand for up to 19 days and then cryopreserved using a controlled rate freezer before storage in liquid nitrogen. For selected donor PBMCs (Fig. 2f), the 19-day cryovial was thawed, recultured and expanded up to 7 additional days. A subset of the lots screened were included in the manuscript (Fig. 2) to demonstrate a range of HHV-6B abundance. To assess reactivation of HHV-6 in the SJ19–09 culture, 2 × 106 cells from the infusion product from donor SJ19–09 were thawed and seeded into 30-ml XVIVO15–5% human serum containing 10 ng ml−1 IL-7 and IL-15 and activated with TransAct (Miltenyi). Additional cytokines were added at day +10, and vials of cells were frozen at days +5, +7, +10 and +14 before processing the cells with scRNA-seq.
To assess the impact of culture with foscarnet, day-19 cryovials of the allogeneic CAR T cell products for donors 34 and 97 were cultured in AIM-V medium for 5 additional days with 10% FBS, 1% Pen-Strep and 10 ng ml−1 IL-2. Cells from donor 97 were divided into four subgroups, for treatment with foscarnet, at a final concentration of 1 mM, 2.5 mM or 5 mM, or control. These concentrations were motivated by previous reports of adding foscarnet to other cell culture contexts39. RNA was isolated with Trizol reagent (Ambion). RNA was reverse-transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s protocol. Transcript quantification was carried out by qPCR with reverse transcription using Taqman Master Mix (Thermo Scientific). For a separate donor, we repeated the 1 mM and control conditions and quantified viral RNA abundance using scRNA-seq.
U31 qPCR protocol
Primary human T cells were centrifuged at 540g for 5 min before removal of culture supernatant, leaving the cell pellet for qPCR. Cells were processed for DNA using the manufacturer’s protocol from the Qiagen DNeasy Blood & Tissue Kit. The concentration and 260/280-nm ratio of the final product were measured using a NanoDrop spectrophotometer (ThermoFisher).
For qPCR, the master mix used was TaqMan Fast Virus 1-Step Master Mix (ThermoFisher). The manufacturer’s protocol was followed to add the appropriate volumes of master mix, water, DNA and primers. All samples were prepared in triplicate and run on a QuantStudio 6/7 Flex Real-Time PCR System (ThermoFisher). Primer sequences for the designated HHV-6 marker (U31) were designed on the basis of previous reports48 and were modified as follows: forward sequence: 5′-CGACTCTCACCCTACTGAACGA-3′; reverse sequence: 5′-GAGGCTGGCGTCGTAGTAGAA-3′; probe sequence: 5′-/5YakYel/AGCCACAGC/ZEN/AGCCATCTACATCTGTCAA/3IABkFQ/−3′. Measurements were taken in triplicate and the mean value is shown for qPCR plots. The same probes were used for both qPCR and qPCR with reverse transcription experiments in this manuscript. Internal controls were not included in these experiments as we normalize to total genomic DNA input, micrograms per reaction, for each sample, and we report the absolute quantification of HHV-6 based on previously published methods48.
scRNA-seq
Libraries for scRNA-seq were generated using the 10x Chromium Controller and the Chromium Single Cell 5′ Library Construction Kit and human B cell and T cell V(D)J enrichment kit according to the manufacturer’s instructions. Briefly, the suspended cells were loaded on a Chromium controller Single-Cell Instrument to generate single-cell gel bead-in-emulsions (GEMs) followed by reverse transcription and sample indexing using a C1000 Touch thermal cycler with a 96-Deep Well Reaction Module (BioRad). After breaking the GEMs, the barcoded cDNA was purified and amplified, followed by fragmenting, A-tailing and ligation with adaptors. Finally, PCR amplification was carried out to enable sample indexing and enrichment of scRNA-seq libraries. For T cell receptor sequencing, target enrichment from cDNA was conducted according to the manufacturer’s instructions. The final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent). 10x scRNA-seq libraries were sequenced as recommended by the manufacturer (about 20,000 reads per cell) using a Nova-seq 6000 with an S4 flow cell. Raw sequencing data were demultiplexed using CellRanger mkfastq and aligned to the host reference genome using CellRanger v6.0 and TCR sequences were processed using the CellRanger vdj pipeline with default settings. Key summary metrics associated with libraries generated in this work are included in Extended Data Fig. 7.
10x Genomics Multiome
Single-cell Multiome libraries of healthy paediatric control PBMCs were generated using the 10x Genomics Chromium Next GEM Single Cell Multiome ATAC + Gene Expression reagent bundle (no. 100285) and the Chromium controller according to the manufacturer’s instructions (CG000338-Rev E). Briefly, following the sorting of live cells, 1.5 ml DNA LoBind tubes (Eppendorf) were used to wash cells in PBS and for downstream processing steps. After washing, cells were lysed for 3 min in lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% NP40, 0.01% digitonin, 1% BSA, 1 mM DTT, 1 U μl−1 RNase inhibitor). Following lysis, cells were washed three times with 1 ml wash buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20, 1 mM DTT, 1 U μl−1 RNase inhibitor) before centrifugation at 500g, for 5 min at 4 °C. The supernatant was discarded and cells were diluted in 1× diluted nuclei buffer (10x Genomics) before counting using trypan blue and a Countess II FL automated cell counter. If large cell clumps were observed, a 40-μm Flowmi cell strainer was used before processing cells according to the Chromium Next GEM Single Cell Multiome ATAC plus Gene Expression user guide with no further modifications. Briefly, after transposition and chip loading, the cells were loaded into the Chromium Controller instrument to generate single-cell GEMs followed by incubation as described in the protocol using a C1000 Touch thermal cycler with a 96-deep well reaction module (BioRad). After breaking the GEMs, the barcoded DNA was purified and further amplified before separate ATAC and cDNA library construction. For the ATAC part, the purified DNA was further amplified to enable sample indexing and enrichment of the DNA. The cDNA was further amplified, purified and quantified using a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent). The cDNA was subsequently fragmented, PCR-amplified and purified as depicted in the Chromium Next GEM Single Cell Multiome ATAC plus Gene Expression user guide with no further modifications. All final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
Single-cell HHV-6B quantification workflow
To quantify HHV-6B reads, we sought to develop a workflow that: would use pseudoalignment rather than standard transcriptomic alignment; could be mapped to only the HHV-6B reference transcriptome (rather than a mixed transcriptome); and had no detectable off-target expression (that is, 0 reported HHV-6B expression for scRNA-seq samples from human transcripts). Our rationale for these constraints was to optimize computational efficiency to reprocess massive-scale scRNA-seq libraries, noting billions of paired-end reads were processed in the studies listed in Table 1.
To satisfy these computational constraints, we developed a kallisto bustools49,50 workflow to rapidly quantify reads from either single-cell transcriptomes or bulk transcriptomes. We downloaded the GenBank AF157706 reference transcriptome and created a kallisto index using the default -k 31 (kmer) parameter. For single-cell libraries, raw sequencing reads were processed using the kallisto bus command with appropriate hyperparameters for each version of the single-cell chemistry (either 14 or 16-base-pair sequence barcode and 10 or 12 bases of UMI sequence). After barcode and UMI correction, a plain text sparse matrix was emitted, corresponding to unique HHV-6B reads mapping to individual cells in the single-cell sequencing library. For bulk libraries, the same index could be utilized with the standard kallisto quant execution.
To assess our specificity (that is, no human transcript expression being reported as an HHV-6B transcript), we quantified four healthy PBMC libraries that had no expected HHV-6B expression. We recurrently observed nonzero expression at only the AF157706.1_cds_AAD49614.1_1 and AF157706.1_cds_AAD49682.1_98 coding features, both genic transcripts that encode for the DR1 gene, which corresponded to AF157706 equivalence classes 6 and 120 in the kallisto index. By examining reads that mapped to both HHV-6B transcriptome and the human genome, we determined that a region of the DR1 gene is highly homologous to the human gene KDM2A, which is broadly expressed across tissues, including lymphocytes, leading to about 1–3 reads per million errantly aligning to the HHV-6B transcriptome. By excluding these two equivalence classes, we consistently observed exactly 0 HHV-6B UMIs across libraries derived from PBMCs44 and across 60 healthy tissue types from various donors43. The number of UMIs per cell shown in Figs. 2 and 3 represents the sum of all other equivalence classes that match to the viral transcriptome. Finally, verification of the in vitro expression of the CAR transgene was confirmed through single-cell quantification of the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) in all sequencing libraries and single cells.
To assign a cell as a super-expressor, we used a semi-arbitrary cutoff of 10 HHV-6B UMIs per cell based on empirical expression (for example, Fig. 2d) and the ease of use and interpretation of the number 10. We note that one cell in the in vivo dataset expressed 8 UMIs (Fig. 3b), so it is probably a super-expressor but was under-sequenced on the basis of the number of overall UMIs detected for this cell (Supplementary Table 4). For visualization across the day-19 samples (Extended Data Fig. 4a), we show both the rank-ordered expression per cell based on total HHV-6B UMIs and a null model in which HHV-6B reads are allocated proportionally to the number of total UMIs detected for that particular cell barcode. For HHV-6B transcript heatmaps (Fig. 3c and Extended Data Fig. 4b), we use the per-gene annotation from a previous report28. For simplicity in viewing expression signatures, we collapsed the ‘intermediate early early’ and ‘immediate early early’ into a single gene set for visualization in these two heatmaps.
To quantify HHV-6B DNA copy number within single cells, we used Chromap (v0.2.3) for fast single-cell processing51. Using the same GenBank ID for HHV-6B (AF157706), we established a Chromap index for rapid alignment of the ATAC arm of the Multiome datasets generated here. Briefly, the R2.fastq file was used to indicate the cell barcode whereas R1 and R3 were used for inference of viral-derived DNA as is standard for the ATAC arm of the Multiome kit. To quantify per-cell signatures of viral gene expression, we estimated a simple proportion of UMIs of all viral genes based on four classes of viral gene annotation previously described28. Cells were included for analysis of relative viral signature (for example, Extended Data Fig. 5c,d) with at least 10 viral RNA UMIs detected per cell.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde and permeabilized using 0.5% saponin before incubating with antibodies to HHV-6 p41 (clone 3E3, HHV-6 Foundation) or gB (Clone H-AR-1, HHV-6 Foundation) at 1:1,000. Cells were then stained with goat anti-mouse IgG conjugated with Alexa Fluor 488 (ThermoFisher Scientific; catalogue no. A-11001) at 1:1,000 and DAPI before imaging on an epifluorescence microscope at ×20 magnification.
Integrated viral–host gene expression analyses
To determine differentially expressed genes between HHV-6+ cells and HHV-6− cells from the day-19 time points, we considered donors 34 and 38, who had the highest number of HHV-6+ cells. We achieved this using the ‘FindMarkers’ function from Seurat after segregating HHV-6+ cells as those that have a minimum of 2 UMIs (to increase power) and HHV-6− as those with identically 0 HHV-6 UMIs. From the FindMarkers output, we computed a statistic, −log10[P value] × log2[FC], that preserves the direction of overexpression or underexpression while partially scaling the gene expression difference by the magnitude of the statistical association, resulting in the plot show in Extended Data Fig. 6c. From this association, the lymphotoxin genes LTA and LTB were distinguished as the consistent associations. As the homotrimer LTα3 is secreted but the heterotrimer (LTα1β2) is membrane-bound52, this suggests that changes in the LT expression following HHV-6 reactivation lead to a secreted signal through LTα3, but owing to the pleiotropic effects of this cytokine trimer52, the impact of its possible increased secretion is not evident.
To comprehensively examine host gene association with HHV-6B transcript abundance, we also computed Pearson correlation statistics per gene between the log-transformed total HHV-6B UMIs with the Seurat-normalized host expression. We performed pathway analyses using the correlation statistic to rank order genes linked to HHV-6B activity. These analyses were carried out for all three recultured samples, which were selected as these were the only samples that had more than 100 super-expressors. Using the permuted distribution of correlations to establish a background distribution, we identified 6,722 (donor 34 day 27), 4,259 (donor 34 day 27) and 1,006 (donor 61 day 27) genes correlated with total HHV-6 RNA levels at a 1% false discovery rate. After broadly verifying that the top genes were concordant between these different donors and time points, we presented the data for donor 34 at day 25 total of culture, as it was the earliest time point. From the pathway analyses, we identified an enrichment of genes downstream of type I interferon activity as being more expressed in cells with low levels of HHV-6B, consistent with the antiviral properties of this pathway35. By contrast, targets of the E2F transcription factor were upregulated as the HHV-6B infection spread throughout the culture, consistent with previous reports of E2F1-induced HHV-6A expression in T cells53. Furthermore, we observed an increase in genes associated with oxidative phosphorylation, a pathway previously reported to be co-opted during active viral infection36. Overall, our results largely recapitulate known proviral and antiviral responses, indicating that the host transcriptome at the single-cell level permits or prevents HHV-6B infection from spreading within the T cell culture. For the foscarnet analyses, we used all barcodes passing the knee filter with at least 1,000 UMIs in our analysis to assess HHV-6 abundance. Dimensionality reduction, including principal component analysis and uniform manifold approximation and projection, was carried out using default parameters. Genes with differential expression between treatment and control were assessed for host genes using the FindMarkers function with a more lenient log[FC] threshold (where ‘FC’ represents ‘fold change’) of 0.05 to include more genes for analyses (n = 2,164 tested). No genes that were differentially expressed required an abs(log2[FC]) > 1, suggesting that the foscarnet treatment did not meaningfully alter the transcriptomes of these cells.
Reanalysis of public single-cell data
The data presented in Table 1 represent a summation of four large datasets reanalysed for HHV-6B expression. In brief, the number of cells and raw sequencing reads analysed are reported, alongside the number of high-confidence HHV-6B UMIs from the quantification pipeline. The number of HHV-6+ cells are barcodes annotated as cells before quality-control filtering in the original publication with at least one HHV-6 UMI. We note that many super-expressors contain high levels of mitochondrial and ribosomal transcripts, potentially owing to these cells nearing apoptosis caused by viral infection or reactivation.
For the dataset of about 1,000 PBMC donors44, we similarly downloaded fastq files from GEO and quantified, with the resulting libraries, reads that pseudoaligned to the HHV-6B transcriptome. For either dataset, we did not detect a single HHV-6B read annotated by a barcode that was a cell in the processed metadata for either study, although we did observe 6 total HHV-6B reads from the PBMC cohort (all mapping to different gel emulsion bead barcodes).
For the public CAR T cell datasets, all fastq files were either downloaded through the European Genome/Phenome Archive portal2 or reanalysed using a Terra workspace42. Each dataset was processed with the same HHV-6B kallisto bustools workflow. We observed no cells annotated in the complete metadata in the pre-infusion-only study2, resulting in an annotation of 0 HHV-6+ cells from this dataset. Requiring a donor to have at least 1,000 cells profiled at at least one time point, we could consider HHV-6 reactivation in a total of 35 donors, four of whom had detectable HHV-6 UMIs. For the 7-days post-infusion blood draw in cohort 2, we observed a total of 26 unique 10x barcodes with at least 1 HHV-6B UMI whereas only 7 of the 26 barcodes were annotated as cells in the original study42. Although the high UMI abundance for the donor axi-R-15 has no ambiguous interpretation, we note that cells for the other two donors (tisa-R-32 and axi-N-07) were pooled with cells from axi-R-15 and then were demultiplexed using genetic variants42. As a consequence, we cannot exclude the possibility that the residual 1–2 UMIs detected in these three cells are from barcode contamination from cells from the donor axi-R-15. Regardless, our characterization of the axi-R-15 cells confirms the presence of HHV-6hi cells in an FDA-approved CAR T cell product. For axi-R-15, we note that the participant was not treated with steroids for ICANS and received one dose of dexamethasone for cytokine release syndrome on day +7 before the onset of ICANS. Although we detected HHV-6 from the CAR T cells, we note that it is unlikely that the virus contributed to the development of ICANS in this patient. For the SJCAR19 analyses, the same kallisto bustools pipeline was run locally at St. Jude to infer HHV-6 expression in the SJCAR19 cohort (cohort 3). The full clinical timecourse of SJCAR19–09 has been previously reported3,4,54 as well as the relevant clinical timecourses for axi-R-15, tisa-R-32 and axi-N-07 (ref. 2).
To confirm that HHV-6+ cells from the in vivo fusion product were from CAR T cells, we assessed the presence of the CAR transgene through RNA (kallisto pseudoalignment to the axicabtagene ciloleucel transcript) as well as noted host gene expression values (Fig. 3 and Supplementary Table 6). The CAR ‘FACS’ cell was from the 10x channel that enriched for CAR T cells through flow cytometry, but owing to the mechanisms of the 10x Genomics library preparation protocol, the specific protein abundance for this individual cell is not discernible. The TCRαβ transcript number was the total number of UMIs aligning per barcode in the TCR vdj libraries.
For the CD7 allogeneic CAR T cell data, we downloaded fastq files from GEO under the accession provided in the manuscript5. Fastqs were processed with CellRanger v6.0 to derive gene expression counts matrices, and our kallisto|bustools pipeline was used to quantify gene expression across the HHV-6 transcripts. To determine host or allogeneic CAR T cell annotations, we used souporcell55 v2.0 for genetic demultiplexing and annotated each donor after examining the cell state embeddings using the gene expression features.
Expression of viral receptors across atlases
To evaluate other possible sources of HHV-6 reactivation and expression, we considered the landscape of cell types that express OX40, the canonical receptor of HHV-6B, and CR2 (CD21), the canonical receptor of EBV. We downloaded processed count data and/or summary plots from The Genotype-Tissue Expression portal56, a fetal development atlas57, profiles of sorted resting and stimulated immune cells58, and profiling of endothelial cell lines59.
Analyses of these atlases revealed that OX40 is expressed specifically in activated CD4+ and CD8+ T cells in the immune system but had detectable levels across all tissues (Extended Data Fig. 1). When examining single-cell and single-nucleus expression across The Genotype-Tissue Expression atlas, we unexpectedly observed consistent expression in endothelial cells across various tissues, including in the pancreas and brain. We further corroborated that 15 of the top 25 cell types or tissues were endothelial. Although the overall expression of OX40 seems to be 1–2 orders of magnitude higher in activated T cells, we suggest that the application of our atlas-level analyses of the HHV-6B viral receptor as a possibility indicates that endothelial cells may be targets of active HHV-6B infection, either from the primary infection or from cell therapy-mediated reactivation. Moreover, we speculate that the expression of OX40 by brain endothelial cells mediates a possible mechanism underlying the encephalitis observed in severe infections and reactivation.
Extended Data
Extended Data Fig. 1 |. Characterization of OX40 and CD21 gene expression in bulk sequencing experiments.

(a) Expression of TNFRSF4 (OX40), the canonical receptor of HHV-6B, in unstimulated and stimulated immune cell populations. OX40 is not expressed in unstimulated immune cells but highly expressed in CD4 and CD8 T cells after activation/stimulation via anti-CD3/anti-CD28 and IL-2. (b) Same as in (a) but for CD21, the canonical receptor for EBV. Gamma-delta T cells are highlighted for their high baseline expression of this receptor. (c) Broad expression of OX40 across healthy tissues from the GTEx bulk atlas. Data is summarized over 948 donors across the tissues shown from the GTEx v8 release. Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5× interquartile range.
Extended Data Fig. 2 |. Characterization of OX40 gene expression in resting and stimulated endothelial cells.
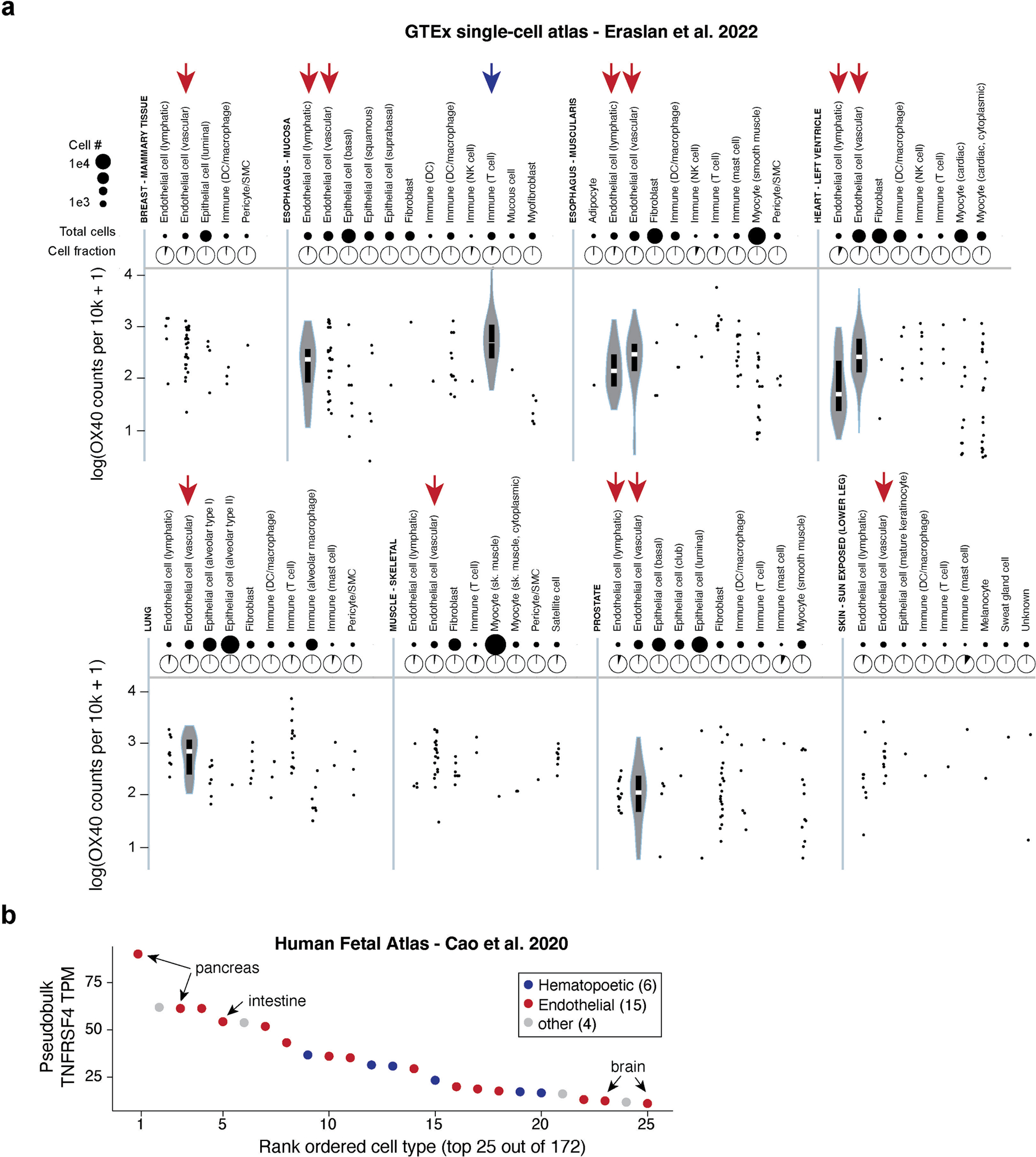
(a) Refinement of cell-type specific HHV-6B expression patterns using the single-cell GTEx atlas. Data is summarized over 25 donors across the tissues shown from the GTEx v8 release Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5× interquartile range. (b) Pseudobulk expression patterns of OX40 from the Human Fetal Atlas.
Extended Data Fig. 3 |. Supporting analyses for HHV-6 reactivation using Serratus.

(a) Comparison of read number matching to any of the annotated 16 T-cell libraries from Serratus for HHV-6A or HHV-6B. A distinct library is compared for HHV-6A and HHV-6B, connected by a line, quantification from Serratus. Libraries are sorted by HHV-6B expression, which was higher for all libraries than HHV-6A. (b) Heatmap of HHV-6B transcripts across the four samples with highest RNA expression in libraries from Serratus. Shown are the first 40 genes (based on genomic coordinate order) from the HHV-6B transcriptome and the number of reads that pseudoalign to each transcript. (c) Summary of % RNA molecules aligning to the HHV-6B reference in the naive CD4+ culture from the LaMere et al. dataset; compare to Fig. 1e. (d) Summary of HHV-6-SNV analysis and overlap across the two RNA-seq samples with highest HHV-6 reactivation (from Fig. 1d,e) (e) Summary of HHV-6B expression in a previously reported ATAC-seq atlas25, showing sorted T cells from Patient 59, an individual with CTCL, who had detectable levels of HHV-6B DNA within cells. (f) Smoothed coverage (rolling mean of 500 base pairs) over the four libraries from Patient 59, showing the coverage across the HHV-6B reference genome. (g) Schematic and results of previously described26 adoptive Treg therapy culture showing HHV-6 reactivation after reanalysis of primary data.
Extended Data Fig. 4 |. Supporting analyses for HHV-6 expression during in vitro CAR T cell culture.

(a) Summary of observed (red) and permuted (gray) HHV-6B expression for four donors at day 19 in culture. The dotted line is at 10 UMIs, the threshold for a super-expressor. (b) Heatmap of HHV-6B expression for selected cells across 3 donors with detectable super-expressors. Columns are grouped based on HHV-6B gene programs (immediate early; early; late). (c) UMAP for 3 samples, noting marker genes and HHV-6B UMI expression (log transformed). The WPRE feature indicates the presence of the CAR transgene.
Extended Data Fig. 5 |. Association of viral DNA and RNA in CAR T cell cultures.
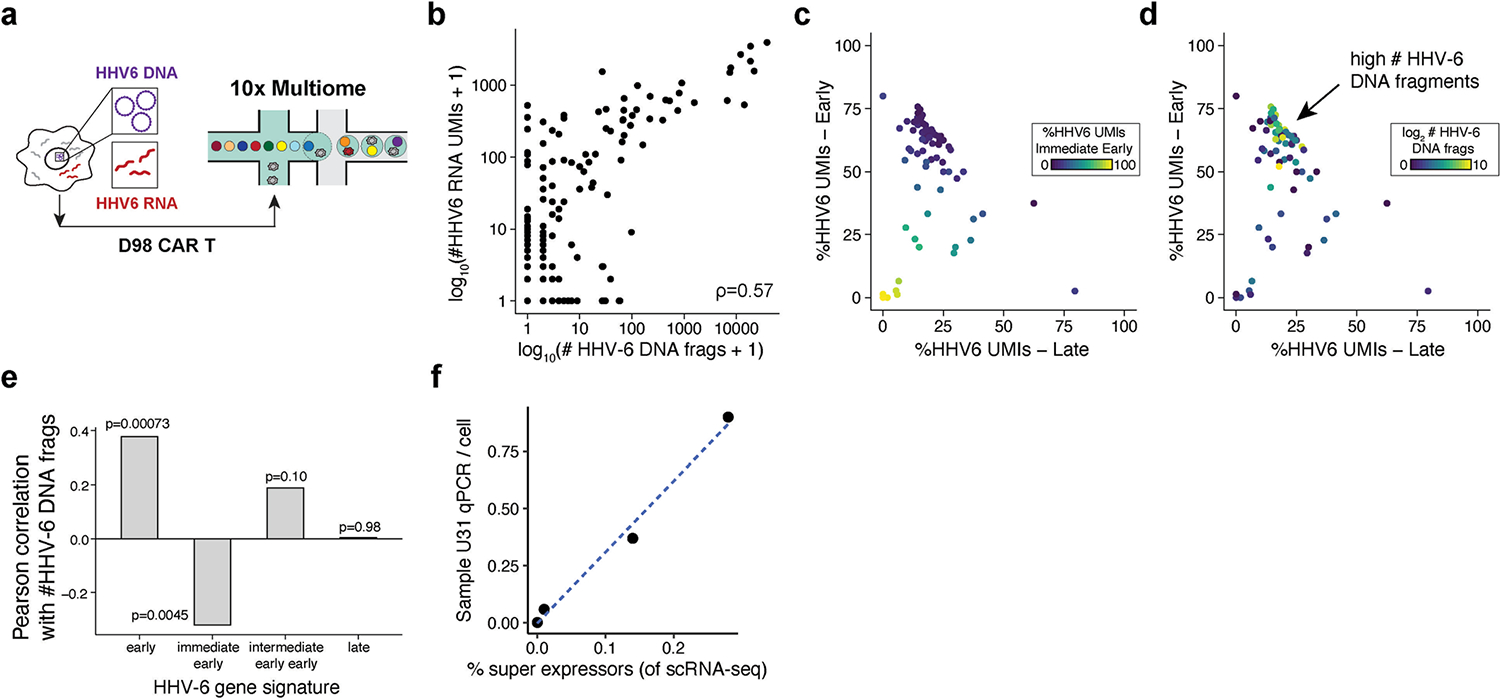
(a) Schematic of the experiment where CAR T cells from D98 were profiled using the 10x Genomics Multiome workflow to detect both viral DNA and RNA. (b) Scatter plot of the abundance of viral DNA and RNA at single-cell resolution. Pearson correlation between the log10 abundances is shown. (c) Per-cell viral gene expression signatures. The proportion of viral gene expression belonging to each class (late, early, immediate early) per cell is shown. (d) Same plot as in (c) but colored by the log2 number of viral DNA fragments. The population of cells highly expressing early HHV-6 transcripts show a corresponding high HHV-6 DNA copy number is highlighted by the arrow. (e) Pearson correlation of HHV-6 transcript signatures with their log abundance of DNA fragments per cell. The two-sided p-value for the Pearson correlation test is noted by each bar. (f) Bulk-level RNA and DNA correspondence in the four donors studied in the day 19 allogeneic CAR products.
Extended Data Fig. 6 |. Supporting analyses of HHV-6 expression dynamics during in vitro CAR T cell culture.
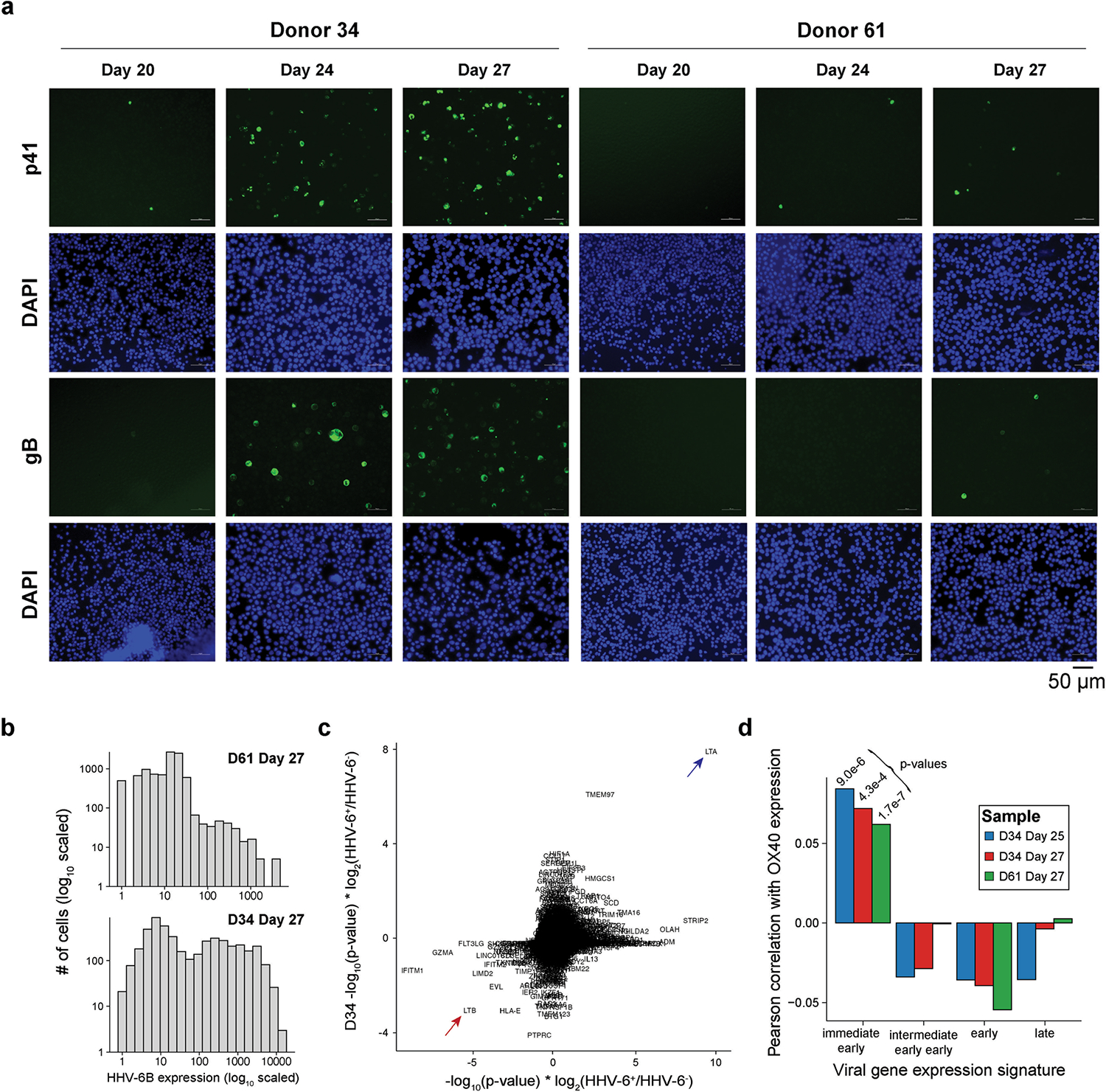
(a) Immunofluorescence staining of two viral proteins (p41 and gB) and nuclei (DAPI) over the course of the cell therapy product culture. The donor and day of culture are noted for each panel. Images are representative of independent experiments that confirmed results of qPCR assay. (b) Summary of HHV-6B transcript expression in re-cultured samples for donors D61 and D34. (c) Summary of HHV-6B +/− cells from differential expression testing for host factors. Arrows indicate lymphotoxin α (LTα/LTA) and downregulation of lymphotoxin β (LTβ/LTB). (d) Correlation statistics of HHV-6B transcript signatures with OX40 expression across 3 recultured samples, including p-values from Pearson’s product-moment correlation coefficient. The consistently positive, significant correlation statistic represents an association uniquely between OX40 expression and the immediate early HHV-6B gene signature.
Extended Data Fig. 7 |. Quantification of HVV6 expression during the manufacturing of allogeneic CAR T cells from a donor with ciHHV-6 under different conditions.

(a) Schematic of experimental design. PBMCs from a ciHHV-6 donor were activated and subjected to four different conditions to assess the impact of individual steps of the CAR T cell manufacturing process. (b) Quantification of conditions in (a) over the 19 day culture process. HHV-6 copy number is shown and stable over the culture. (c) Per-gene HHV-6 viral RNA abundance at Day 1 and Day 19 of the full CAR T manufacturing process for the ciHHV-6 donor (left) and a donor previously analyzed with scRNA-seq (D98, right).
Extended Data Fig. 8 |. Mitigation of HHV-6 reactivation and spreading via foscarnet treatment in vitro.

(a) Schematic of CAR T product reculture experiment. Donor D97, which at day 19 showed a low but detectable level of HHV-6, was selected for reculture for five days. (b) Summary of RT-qPCR at the control and two treatment levels of Foscarnet. Each dot represents a technical replicate over one biological replicate per condition (validated in panel d). HHV-6 was not detected (n.d.) at the 1 mM concentration. Error bars represent the standard error of the mean. Comparison of foscarnet treated to untreated resulted in significantly lower abundance of HHV-6 RNA (p = 0.00026; two-sided ordinary least squares linear model). (c) Schematic of D34 reculture +/− foscarnet at 1 mM. (d) Difference between untreated and treated in the abundance of HHV-6+ cells. Comparing the two 10x Genomics scRNA-seq data channels, foscarnet-treated cells had a lower incidence of HHV-6 positive cells (OR = 6.25; p = 8.3e-122; Fisher’s exact test, two-sided). (e) Reduced dimensionality analysis of treated and untreated D34 cells profiled with scRNA-seq. Host gene expression was used for the analysis, showing overlapping clustering of populations irrespective of treatment status. (f) Differential gene expression analysis comparing foscarnet treated and control CAR T cells. The three most significant differential genes are noted. 0 genes were differentially expressed with a minimum log2 fold-change exceeding 1 (noted by the red).
Extended Data Fig. 9 |. Gene expression annotation for the Day 19 PBMC allogeneic CD7 dataset.

Expression of select marker genes supporting the annotations contained in Fig. 4c.
Supplementary Material
Acknowledgements
We thank members of the laboratory of A.T.S. for helpful discussions; M. Green and G. Syal for assistance with public sequencing data; and S. Schell for assistance with clinical samples at St. Jude. C.A.L. is supported by a Stanford Science Fellowship, a Parker Institute for Cancer Immunotherapy Scholarship, a seed award from the Center for Human Systems Immunology and NIH K99 HG012076. A.T.S. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, the Parker Institute for Cancer Immunotherapy, a Pew-Stewart Scholars for Cancer Research Award, a Cancer Research Institute Lloyd J. Old STAR Award and a Baxter Foundation Faculty Scholar Award. S.L. is supported by the NCI Research Specialist Award (R50CA251956). This work was supported by a sponsored research agreement with Allogene Therapeutics. Part of the analysis was carried out by the Center for Translational Immunology and Immunotherapy (CeTI2), which is supported by St. Jude Children’s Research Hospital.
Footnotes
Competing interests A.T.S. is a founder of Immunai and Cartography Biosciences and receives research funding from Allogene Therapeutics and Merck Research Laboratories. C.A.L. and L.S.L. are consultants to Cartography Biosciences. N.J.H. is a consultant for Constellation Pharmaceuticals. C.J.W. holds equity in BioNTech Inc and receives research funding from Pharmacyclics. G.G. receives research funds from IBM and Pharmacyclics, and is a founder of, consultant for and holder of private equity in Scorpion Therapeutics. M.V.M. is an inventor on patents related to CAR T cell therapies held by the University of Pennsylvania (some licensed to Novartis), holds equity in TCR2, Century Therapeutics, Genocea, Oncternal and Neximmune, has served as a consultant for multiple companies involved in cell therapies, receives research support from KitePharma and Moderna, and serves on the Board of Directors of 2Seventy Bio. M.J.K. has received research funding from Kite, a Gilead Company, as well as honoraria from Kite, a Gilead Company, Novartis, Takeda, Miltenyi Biotec, BMS/Celgene, Beigene and Adicet Bio. S.G. is a co-inventor on patent applications in the fields of cell or gene therapy for cancer, a consultant for TESSA Therapeutics and a member of the Data and Safety Monitoring Board of Immatics, and has received honoraria from Tidal, Catamaran Bio, Sanofi and Novartis within the past 2 years. J.C.C. and P.G.T. are named on patent applications in the fields of T cell or gene therapy for cancer. G.Y., J.P., R.S., W.L., A.S., A.M., Z.J.R., T.P. and H.D. are employees and shareholders of Allogene Therapeutics. A.K. is a scientific cofounder of Ravel Biotechnology Inc., is on the scientific advisory board of PatchBio Inc., SerImmune Inc., AINovo Inc., TensorBio Inc. and OpenTargets, is a consultant with Illumina Inc. and owns shares in DeepGenomics Inc., Immunai Inc. and Freenome Inc. R.G.M. is a cofounder of and holds equity in Link Cell Therapies and CARGO Therapeutics, and is a consultant for Lyell Immunopharma, NKarta, Arovella Pharmaceuticals, Innervate Radiopharmaceuticals, Aptorum Group, Gadeta, FATE Therapeutics (Data and Safety Monitoring Board) and Waypoint Bio. All other authors declare no competing interests.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-023-06704-2.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-023-06704-2.
Peer review information Nature thanks Artem Babaian, Joshua Hill and Richard O’Neil for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at http://www.nature.com/reprints.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Code to reproduce all custom analyses in this manuscript is available at https://github.com/caleblareau/hhv6-reactivation and archived at https://doi.org/10.5281/zenodo.8218129.
Data availability
Raw and processed sequencing data used in this study are available at GEO (accession GSE210063). The HHV-6B reference genome and transcriptome used in this work is available at Genbank (accession AF157706).
References
- 1.Spanjaart AM, van der Valk FM, van Rooijen G, Brouwer MC & Kersten MJ Confused about confusion. N. Engl. J. Med. 386, 80–87 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Haradhvala NJ et al. Distinct cellular dynamics associated with response to CAR-T therapy for refractory B-cell lymphoma. Nat. Med.28, 1848–1859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson TL et al. Common trajectories of highly effective CD19-specific CAR T cells identified by endogenous T cell receptor lineages. Cancer Discov. 10.1158/2159-8290.CD-21-1508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talleur A et al. Preferential expansion of CD8+ CD19-CAR T cells postinfusion and the role of disease burden on outcome in pediatric B-ALL. Blood Adv. 10.1182/bloodadvances.2021006293 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y et al. Genetically modified CD7-targeting allogeneic CAR-T cell therapy with enhanced efficacy for relapsed/refractory CD7-positive hematological malignancies: a phase I clinical study. Cell Res. 32, 995–1007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke FL et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol. 20, 31–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baird JH et al. Immune reconstitution and infectious complications following axicabtagene ciloleucel therapy for large B-cell lymphoma. Blood Adv. 5, 143–155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maude SL et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JI Herpesvirus latency. J. Clin. Invest. 130, 3361–3369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traylen CM et al. Virus reactivation: a panoramic view in human infections. Future Virol. 6, 451–463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Düver F et al. Viral reactivations following hematopoietic stem cell transplantation in pediatric patients - a single center 11-year analysis. PLoS One 15, e0228451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grinde B Herpesviruses: latency and reactivation – viral strategies and host response. J. Oral Microbiol. 5, 22766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampouri E et al. CMV and HHV-6 after chimeric antigen receptor (CAR)-T-cell immunotherapy for B-cell malignancies: a prospective study. Transplant. Cell. Ther. 29, S210–S211 (2023). [Google Scholar]
- 14.Labanieh L & Mackall CL CAR immune cells: design principles, resistance and the next generation. Nature 614, 635–648 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Edgar RC et al. Petabase-scale sequence alignment catalyses viral discovery. Nature 602, 142–147 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Leinonen R, Sugawara H & Shumway M, International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 39, D19–D21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulo C et al. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 39, D576–D582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaMere SA, Thompson RC, Komori HK, Mark A & Salomon DR Promoter H3K4 methylation dynamically reinforces activation-induced pathways in human CD4 T cells. Genes Immun. 17, 283–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellett PE et al. Chromosomally integrated human herpesvirus 6: questions and answers. Rev. Med. Virol. 22, 144–155 (2012).22052666 [Google Scholar]
- 20.Ablashi D et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch. Virol. 159, 863–870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H et al. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc. Natl Acad. Sci. USA 110, 9096–9099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shytaj IL et al. Alterations of redox and iron metabolism accompany the development of HIV latency. EMBO J. 39, e102209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones AC et al. Personalized transcriptomics reveals heterogeneous immunophenotypes in children with viral bronchiolitis. Am. J. Respir. Crit. Care Med. 199, 1537–1549 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Holtan SG et al. Stress responses, M2 macrophages, and a distinct microbial signature in fatal intestinal acute graft-versus-host disease. JCI Insight 5, e129762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu K. et al. Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell 32, 27–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reading JL et al. Augmented expansion of Treg cells from healthy and autoimmune subjects via adult progenitor cell co-culture. Front. Immunol. 12, 716606 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bost P et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 181, 1475–1488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gravel A et al. Mapping the human herpesvirus 6B transcriptome. J. Virol. 10.1128/JVI.01335-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White EA & Spector DH in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (eds Arvin A et al. ) (Cambridge Univ. Press, 2007). [PubMed] [Google Scholar]
- 30.Wyżewski Z, Gregorczyk KP, Szczepanowska J & Szulc-Dąbrowska L Functional role of Hsp60 as a positive regulator of human viral infection progression. Acta Virol. 62, 33–40 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Nguyen NNT et al. Hepatitis C virus modulates solute carrier family 3 member 2 for viral propagation. Sci. Rep. 8, 15486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaender S et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 5, 1330–1339 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriesel JD, Jones BB, Dahms KM & Spruance SL STAT1 binds to the herpes simplex virus type 1 latency-associated transcript promoter. J. Neurovirol. 10, 12–20 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Mesquita PMM et al. Role of interleukin 32 in human immunodeficiency virus reactivation and its link to human immunodeficiency virus–herpes simplex virus coinfection. J. Infect. Dis. 215, 614–622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukhele S, Boukhaled GM & Brooks DG Type I interferon signaling, regulation and gene stimulation in chronic virus infection. Semin. Immunol. 43, 101277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foo J, Bellot G, Pervaiz S & Alonso S Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol. 10.1016/j.tim.2021.12.011 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Zerr DM et al. A population-based study of primary human herpesvirus 6 infection. N. Engl. J. Med. 352, 768–776 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Zerr DM et al. HHV-6 reactivation and its effect on delirium and cognitive functioning in hematopoietic cell transplantation recipients. Blood 117, 5243–5249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenberg K, Skog S & Tribukait B Concentration-dependent effects of foscarnet on the cell cycle. Antimicrob. Agents Chemother. 28, 802–806 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panowski SH et al. Preclinical development and evaluation of allogeneic CAR T cells targeting CD70 for the treatment of renal cell carcinoma. Cancer Res. 82, 2610–2624 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Telford M, Navarro A & Santpere G Whole genome diversity of inherited chromosomally integrated HHV-6 derived from healthy individuals of diverse geographic origin. Sci. Rep. 8, 3472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Q et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han X et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Yazar S et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science 376, eabf3041 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Ciszewski C et al. Identification of a γc receptor antagonist that prevents reprogramming of human tissue-resident cytotoxic T cells by IL15 and IL21. Gastroenterology 158, 625–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer C et al. Allogeneic FLT3 CAR T cells with an off-switch exhibit potent activity against AML and can be depleted to expedite bone marrow recovery. Mol. Ther. 28, 2237–2251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sánchez-Ponce Y et al. Simultaneous detection of beta and gamma human herpesviruses by multiplex qPCR reveals simple infection and coinfection episodes increasing risk for graft rejection in solid organ transplantation. Viruses 10, 730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bray N, Pimentel H, Melsted P & Pachter L Near-optimal RNA-seq quantification with kallisto. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Melsted P, Ntranos V & Pachter L The barcode, UMI, set format and BUStools. Bioinformatics 35, 4472–4473 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Zhang H et al. Fast alignment and preprocessing of chromatin profiles with Chromap. Nat. Commun. 12, 6566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang K et al. T cell-derived lymphotoxin limits Th1 response during HSV-1 infection. Sci. Rep. 8, 17727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharon E, Volchek L & Frenkel N Human herpesvirus 6 (HHV-6) alters E2F1/Rb pathways and utilizes the E2F1 transcription factor to express viral genes. Proc. Natl Acad. Sci. USA 111, 451–456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hines MR et al. Hemophagocytic lymphohistiocytosis-like toxicity (carHLH) after CD19-specific CAR T-cell therapy. Br. J. Haematol. 194, 701–707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heaton H et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17, 615–620 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Eraslan G et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science 376, eabl4290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao J et al. A human cell atlas of fetal gene expression. Science 370, eaba7721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calderon D et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 51, 1494–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richards RM et al. NOT-gated CD93 CAR T cells effectively target AML with minimized endothelial cross-reactivity. Blood Cancer Discov. 2, 648–665 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data used in this study are available at GEO (accession GSE210063). The HHV-6B reference genome and transcriptome used in this work is available at Genbank (accession AF157706).


