Introduction
Pharmacogenetic studies of anti-diabetic drugs have focused largely on metformin, which is now first-line therapy for type 2 diabetes. Prior to 2011, metformin pharmacogenetics studies were largely based on candidate genes, focused on genes encoding transporters that are involved in metformin pharmacokinetics. Since 2011, several genome wide association studies (GWAS) have been published, in large multi-ethnic cohorts. These studies have revealed some genes at genome-wide level significance, which are associated with glycemic response to metformin in patients with Type 2 diabetes. For other anti-diabetic drugs, there have been few pharmacogenetic studies, and all have been focused on candidate genes. In general, the studies have been small and largely centered on associations between drug metabolizing enzymes and transporters with the pharmacokinetics or pharmacodynamics of the drugs. In this chapter, we begin by reviewing the pharmacogenetic studies of metformin disposition and response, mostly focused on pharmacogenetic associations in healthy volunteers and diabetic patients on metformin. Afterwards we include a brief review of pharmacogenetics studies of other classes of anti-diabetic drugs including sulfonylureas, thiazolidinediones, meglinitides, acarbose and SGLT2 inhibitors (Figure 1). A brief conclusion is included.
FIGURE 1.
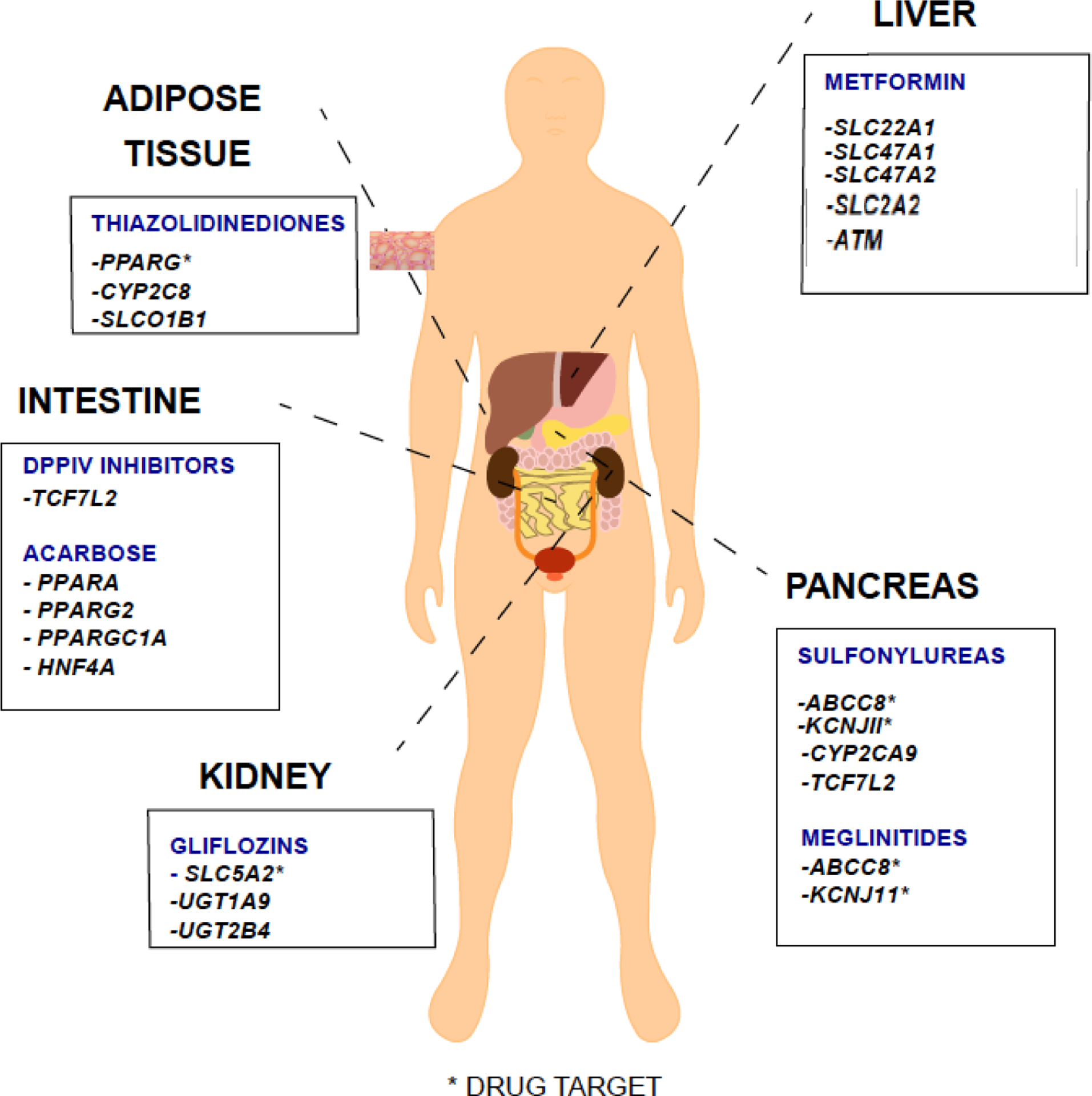
MAJOR SITES OF ACTION OF ANTI-DIABETIC DRUGS AND GENES ASSOCIATED WITH PHARMACOKINETICS OR CLINICAL RESPONSE
Note: Listed drugs may have other secondary sites of action
DISCOVERY OF GENETIC LOCI RELATED TO METFORMIN RESPONSE THROUGH GENOMEWIDE APPROACHES
Genomewide association studies (GWAS) have revealed many loci, genes and individual SNPs, which are critical for therapeutic and adverse drug response and drug disposition. A recent commentary reported that only 216 pharmacogenomics GWAS are available in the GWAS Catalog (MacArthur et al., 2017), and that the majority of the pharmacogenomic GWAS have focused on drugs used in the treatment of cancer, neuropsychiatric disorders and cardiovascular diseases (Giacomini et al., 2017). One of the major challenges to pharmacogenomic GWAS has been sample size. In particular, pharmacogenomic phenotypes are more difficult to obtain than phenotypes for other human traits, since multiple kinds of information are needed to assess drug response. For example, drug dosage, adherence to medications, and baseline and on-drug measurements of the phenotype of interest, need to be collected in each individual for a robust study. This information may be difficult to procure in large numbers of patients. Thus, to enhance sample size of primary and replication cohorts, several pharmacogenomics consortia have been established. These include the International Consortium on Lithium Genetics (Schulze et al., 2010), the International SSRI Pharmacogenomics Consortium (ISPC) (Biernacka et al., 2015), the Pharmacogenetics of Drug-Induced Liver Injury group (DILIGEN) (Urban et al., 2017), and the Pharmacogenomics in Childhood Asthma (PiCA) Consortium (Farzan et al., 2017). For anti-diabetic drugs, two consortia largely centered on metformin genetics, have been established: the metformin genetics consortium, MetGen (http://www.pgrn.org/metgen.html) and the DIRECT (DIabetes REsearCh on patient straTification) (http://www.direct-diabetes.org/). The effort of MetGen consortium is to engage researchers from around the world who have datasets for metformin and also other anti-diabetic drugs to enhance the discovery of genetic biomarkers of clinical response to antidiabetic drugs. Importantly, this consortium encourages the inclusion of other ethnic populations in metformin and other anti-diabetic drug pharmacogenomics studies. MetGen has already published GWAS focused on metformin, which are described below.
In terms of identification of genetic factors that associate with response to anti-diabetic drugs, only two GWAS have been published. Both studies focused on metformin and one involved the MetGen consortium (K. Zhou et al., 2011), (K. Zhou et al., 2016). Given that over 100 risk alleles for Type 2 diabetes have been discovered, with the inclusion of clinical data collected from diverse ethnic populations (Florez, 2017)(Flannick & Florez, 2016), the fact that genomewide association methods have only been applied to metformin response underscores the immense need for pharmacogenomics GWAS in patients on anti-diabetic drugs.
In both of the metformin GWAS focused on response, retrospective observational approaches, using information in the electronic health record (EHR) were employed. For response measurements, glycated hemoglobin (HbA1c), a reliable biomarker for assessing average plasma glucose levels, was obtained from the EHR. The first metformin GWAS, published in 2011 (K. Zhou et al., 2011), identified genetic determinants of metformin response, where response was defined as achieving the treatment target goal of < 7% HbA1c level within 18 months after starting metformin. The study identified a SNP, rs11212617, in a locus, which included ATM (Ataxia-Telangiectasia). The C-allele of rs11212617 was associated with treatment success (n = 3,920 European ancestry, P = 2.9 × 10−9, odds ratio = 1.35, 95% CI 1.22–1.49). Further replication of the SNP revealed a significant association in a meta-analysis of five cohorts of European ancestry (van Leeuwen et al., 2012), and conflicting results in other ethnic groups (Shokri et al., 2016),(Y. Zhou et al., 2014). However, in prediabetes patients, the SNP did not appear to be associated with response to metformin (Florez et al., 2012).
Since the discovery of the association of ATM with metformin response, several follow-up studies have been performed to evaluate (a) the effect of metformin in cultured cells on the activation or phosphorylation of ATM and its downstream targets (Storozhuk et al., 2013; Vazquez-Martin, Oliveras-Ferraros, Cufi, Martin-Castillo, & Menendez, 2011,{Feng, 2015 #224), (Corominas-Faja et al., 2012), (Woods, Leiper, & Carling, 2012); (b) the effect of patients with ATM mutations on glucose-insulin homeostasis and indices of insulin secretion and sensitivity (Connelly et al., 2016); (c) the association of ATM variants with metformin response in pre-diabetic patients (Florez et al., 2012) and in patients with polycystic ovary syndrome (Pedersen, Stage, Glintborg, Andersen, & Christensen, 2018). These studies highlight the importance of follow-up studies to elucidate the mechanisms by which ATM modulates response to metformin in diabetes and other conditions. However, two independent studies showed that the ATM inhibitor, KU-55933 used to elucidate the mechanism by which ATM modulates response to metformin, reduces metformin uptake (Yee, Chen, & Giacomini, 2012), (Woods et al., 2012), and in particularly, inhibits OCT1, the primary transporter for metformin in liver cells (Yee et al., 2012). Since metformin uptake via OCT1 was shown to be independent of ATM (Yee et al., 2012), the studies question the effect of ATM on modulating response to metformin via activation of AMPK, and suggest that other mechanisms may be involved (Yee et al., 2012),(Woods et al., 2012).
Five years after the first GWAS of clinical response to metformin in people with type 2 diabetes, a second GWAS involving MetGen was published. The study identified a minor C-allele in an intron SNP (rs8192675) of a glucose transporter, GLUT2, SLC2A2, which was associated with greater change in HbA1c from baseline (Table 1) as a quantitative trait in discovery and replication cohorts (K. Zhou et al., 2016). A meta-analysis of the SNP was performed in MetGen (http://www.pgrn.org/metgen.html), which included over 10,000 participants in 10 cohorts from Europe and the United States. Strikingly, the results reached genomewide level significance in both models of change in HbA1c (on metformin) adjusted (beta 0.075, p=2.0×10−8) and non-adjusted (beta 0.17, p=6.6×10−14) with baseline HbA1c (n=10,557). Since SLC2A2 is known to play a role in the development of type 2 diabetes and to have an effect on fasting glucose, genetic variation in the transporter may impact the glucose-lowering effect of anti-diabetic therapy. The result showed that the SNP that associated with higher baseline HbA1c was associated with greater HbA1c reduction after initiation of metformin. This phenomenon has been observed in other pharmacogenetic studies, where a SNP, which has an effect on a baseline trait, also impacts drug response. For example, SNPs in PCSK9 and APOE, which are associated with baseline LDL-C levels, are also significantly associated with LDL-C reduction and fractional LDL-C reduction in response to rosuvastatin (Chasman et al., 2012). These studies suggest that pharmacogenomic studies of drug response should consider SNPs known to be associated with disease as those SNPs may also play a role in drug response. Further, the studies suggest that performing the association analyses with and without adjusting for baseline is important particularly for genes known to play a role in baseline trait measured. Other results worth mentioning from the more recent GWAS of metformin response (K. Zhou et al., 2016) include:
The rs8192675 minor C-allele of SLC2A2 was associated with lower expression levels of SLC2A2 in the human liver.
There was a stronger HbA1c reduction induced by metformin in individuals who were obese (BMI ≥ 30 kg/m2) and who harbored the C-allele of rs8192675 compared to individuals with the C-allele but who have BMI < 30 kg/m2.
The rs8192675 minor C-allele of SLC2A2 was associated with metformin response in non-European populations. The combined p-values in the non-European populations were p=0.006 and p=0.005 in baseline adjusted and non-adjusted models, respectively. Both models showed that the minor C-allele was associated with greater HbA1c reduction upon metformin treatment, similar to the results obtained in the European cohorts.
TABLE 1:
GENES ASSOCIATED WITH METFORMIN PHARMACOKINETICS OR RESPONSE
| Gene | Study Population | Metformin dose | Variant | Phenotype |
|---|---|---|---|---|
| SLC22A1 (OCT1) | 20 healthy volunteers | 2 doses 1000mg and 850 mg | R61C (rs12208357), G401S (rs34130495), 420del (rs72552763 and G4665R/rs34059508) | Impaired response to oral glucose tolerance test (Shu et al., 2007) |
| 20 healthy volunteers | 2 doses 1000mg and 850 mg | R61C (rs12208357), G401S (rs34130495), 420del (rs72552763 and G4665R/rs34059508) | Higher AUC, higher Cmax, lower oral volume of distribution (Shu et al., 2008) | |
| 103 healthy male Caucasian volunteers | 1 dose of 500 mg | G465R | Increased renal clearance and decreased hepatic uptake (Tzvetkov et al., 2009) | |
| 1531 GoDARTS subjects with type 2 diabetes | Average dose 1260 mg/day | R61C and M420del | No effect on HbA1c reduction(K. Zhou et al., 2009) | |
| 159 Danish South Danish Diabetes Study participants with type 2 diabetes | 1000 mg twice a day | R61C (rs12208357), G401S (rs34130495), 420del (rs72552763 and G4665R/rs34059508) | Decrease in trough-steady state metformin concentration and less change in HbA1c over 6 months (Christensen et al., 2011) | |
| 1915 metformin tolerant and 251 metformin intolerant GoDARTS participants | Average dose 1000 mg/day | R61C (rs12208357), G401S (rs34130495), 420del (rs72552763 and G4665R/rs34059508) | 2 reduced function alleles associated with metformin intolerance. Higher odds of intolerance in the presence of OCT-1 interacting drugs (Dujic et al., 2015) | |
| 102 subjects with type 2 diabetes from Rotterdam Study | Average dose 677 mg/day | rs622342 | Less reduction in HbA1c level(Becker et al., 2009b) | |
| 148 drug naïve Caucasian patients with type 2 diabetes | Average dose ~ 1400mg/day | rs622342 | No effect on HbA1c reduction (Tkac et al., 2013) | |
| 990 multi-ethnic DPP participants with pre-diabetes | 850 mg twice a day | rs622342 | No evidence of interaction with metformin (Jablonski et al., 2010) | |
| SLC22A2(OCT2) | 15 healthy Chinese participants | A single dose of 500 mg | 808G>T | Reduced metformin renal tubular clearance (Wang et al., 2008) |
| 26 healthy Korean subjects | A single dose of 500 mg of metformin | 596C>T, 602C>T, and 808G>T | Higher Cmax, AUC and lower renal clearance (Song et al., 2008) | |
| 23 healthy Caucasian and African American volunteers | A single dose of 850 mg of metformin | 808G>T | Decreased renal clearance and renal clearance by secretion of metformin (Chen et al., 2009) | |
| 371 Danish participants with type 2 diabetes from South Danish Diabetes Study | 1000 mg twice a day | rs316019 | No effect on metformin concentration or HbA1c response (Christensen et al., 2011) | |
| 990 multi-ethnic DPP participants with pre-diabetes | 850 mg twice a day | rs316019 | No evidence of interaction with metformin (Jablonski et al., 2010) | |
| SLC47A1(MATE1) and SLC47A2(MATE2) | 57 healthy volunteers | 2 doses 1000mg and 850 mg | rs2252281 (MATE1) rs12943590 (MATE2) |
Renal and secretory clearance of metformin higher in MATE2 carriers who were MATE1 reference. Enhanced OGTT response with MATE1, reduced response with MATE2 (Stocker et al., 2013) |
| 145 patients with type 2 diabetes | Average dose 938 mg/day | rs2252281 (MATE1) | Greater change in HbA1c level with MATE1(Stocker et al., 2013) | |
| 253 Caucasian and African American subjects with type 2 diabetes | Average dose 938 mg/day | rs12943590 (MATE2-K) | Less reduction in HbA1c (Choi et al., 2011) | |
| 116 Caucasian subjects with type 2 diabetes | Average dose 741 mg | rs2289669 (MATE1) | Decrease in HbA1c (Becker et al., 2009a) | |
| 148 drug naïve Caucasian patients with type 2 diabetes | Average dose ~ 1400mg/day | rs2289669 (MATE1) | Greater reduction in HbA1c(Tkac et al., 2013) | |
| 990 multi-ethnic DPP participants with pre-diabetes | 850 mg twice a day | rs8065082 (In LD r2 0.8 with rs2289669) (MATE1) | Poorer response to metformin(Jablonski et al., 2010) | |
| 371 Danish participants with type 2 diabetes from South Danish Diabetes Study | 1000 mg twice a day | rs2252281, rs2289669 (MATE1) rs34399035 (MATE2) |
No effect on metformin concentration or change in HbA1c (Christensen et al., 2011) | |
| ATM | 1,024 Scottish subjects with type 2 diabetes (GWAS Discovery cohort) 2,896 Caucasian subjects with type 2 diabetes (2 Replication cohorts) |
The average daily dose during the 3 months prior to the minimum HbA1c was achieved | rs11212617 (intron of C11orf65), in LD with r2>0.8 with SNPs in several genes including ATM, NPAT, KDELC2. | Minor C-allele of rs11212617 is associated with treatment success (K. Zhou et al., 2011). Treatment success was defined as achieving HbA1c <7% in the first 18 months of metformin initiation (n = 3,920, P = 2.9×10−9, odds ratio = 1.35). |
| SLC2A2 (GLUT2) | 3,103 Scottish subjects with type 2 diabetes (GWAS Discovery cohort) 7,454 Caucasian subjects with type 2 diabetes (9 Replication cohorts) 2,526 Non-European subjects with type 2 diabetes (3 different ethnic groups) |
The average daily dose during the 3 months prior to the minimum HbA1c was achieved | rs8192675 (intron of SLC2A2) | Minor C-allele of rs8192675 is associated with greater response to metformin (K. Zhou et al., 2016). Response to metformin was defined as baseline HbA1c minus the minimum treatment HbA1c in the first 18 months of metformin initiation (n = 10,557 Caucasians, P = 2.0×10−8, beta = 0.075 (baseline-adjusted model); P=6.6×10−14, beta=0.17 (baseline non-adjusted model)). Also significant in the combined non-European subjects (n = 2,566 Non-Europeans, P = 0.006, beta = 0.077 (baseline-adjusted model); P = 0.005, beta = 0.15 (baseline non-adjusted model)). Minor C-allele of rs8192675 is associated with lower SLC2A2 transcript levels in human liver tissue samples (n=1,226, P<5×10−8)) and other tissues (fibroblasts, islet, intestine) (K. Zhou et al., 2016) |
The fact that GLUT2 is the major transporter for hepatic glucose output (Seyer et al., 2013) and metformin has a primary pharmacologic effect of reducing hepatic glucose output (Madiraju et al., 2014), (Foretz et al., 2010)suggests a strong mechanism by which the variant may modulate response to metformin. Further studies are warranted to replicate these findings and to elucidate whether genetic information can be used in precision medicine to guide therapy with metformin.
GENOMEWIDE SCREENING TO ELUCIDATE METFORMIN ACTION THROUGH IN VITRO ASSAYS
The mechanism of action of metformin is complex. In brief, the drug appears to reduce mitochondrial energy production via inhibition of complex 1. The lower levels of ATP (and higher AMP to ATP ratio) lead to phosphorylation or activation of the energy sensor, AMP-Kinase, AMPK (G. Zhou et al., 2001). The molecular components LKB1/STK11 and ATM have been shown to play a role in the phosphorylation of AMPK in the presence of metformin, though these are not direct targets of metformin (Foretz & Viollet, 2011). Activation of AMPK results in reduced gluconeogenesis, increased insulin sensitivity, and enhanced peripheral glucose uptake. There is also evidence to suggest that metformin inhibits hepatic gluconeogenesis independent of the LKB1/AMPK pathway(Foretz et al., 2010). Another way by which gluconeogenesis is reduced is by increased AMP levels which functions as a key signaling mediator that inhibits CAMP-PKA signaling through suppression of adenylate cyclase , by allosteric inhibition of fructose 1,6 bisphosphatase and by activation of AMPK (Rena, Pearson, & Sakamoto, 2013). Metformin also likely has beneficial effects beyond the treatment of type 2 diabetes. An increasing number of observational studies suggest that treatment with metformin (relative to other glucose-lowering therapies) is associated with reduced risk of cancer(Giovannucci et al., 2010).
Different genomewide approaches using in vitro cell based assays have been used to identify genes associated with metformin action (Luizon et al., 2016),(Niu et al., 2016). For example, RNA-seq and ChIP-seq (chromatin immunoprecipitation with massively parallel DNA sequencing) have been used to identify genes and DNA regions regulated by metformin after exposing primary human hepatocytes to metformin. These methods demonstrated that the transcript levels of ATF3, a transcription factor that plays an important role in modulating glucose homeostasis, are significantly increased by metformin (Luizon et al., 2016),(Jadhav & Zhang, 2017). Further the studies also add mechanistic support to the finding that rs11212617 in the ATM gene is associated with response to metformin. That is, the SNP lies in an important enhancer region which is regulated by treatment with metformin (K. Zhou et al., 2011), (Luizon et al., 2016). Application of GWAS to 266 human lymphoblastoid cell lines treated with metformin resulted in the identification of genetic biomarkers associated with metformin cytotoxicity (Niu et al., 2016). Some of the genes identified in the cytotoxicity assays may also influence metformin’s anti-diabetic actions through common pathways, such as the LKB1/AMPK pathway and PI3K/mTOR signaling pathway which are important for cellular growth as well as energy homeostasis (Niu et al., 2016), (Snima, Pillai, Cherian, Nair, & Lakshmanan, 2014). For example, STUB1, a E3 ubiquitin ligase, is involved in activation of AMPK through the LKB pathway (Niu et al., 2016).
STUDIES RELATED TO METFORMIN CANDIDATE GENES
Prior to the onset of GWAS that offered the ability to interrogate the entire genome, research related to metformin pharmacogenetics was limited to pre-existing biological knowledge of metformin through a candidate gene approach. Studies were mainly centered on the cellular transporters that regulated metformin disposition, and the majority of work focused on organic cation transporters. In order to understand the role of candidate genes in metformin response, it is helpful to have an understanding of the pharmacokinetics and pharmacodynamics of metformin.
Metformin Pharmacokinetics
Metformin is a hydrophilic molecule that diffuses poorly across biological membranes. As such, for its disposition, metformin requires membrane transporters, and in particular, organic cation transporters in the intestine, liver and kidney. Metformin is excreted unchanged in the urine and has an elimination half-life of approximately 5 hours in patients with normal renal function. Metformin is primarily eliminated through active tubular secretion in the kidney. The oral absorption, hepatic uptake and renal excretion of metformin are largely mediated by organic cation transporters (OCTs) (Graham et al., 2011). Though the exact transporters that contribute to the absorption, distribution and elimination of metformin in patients are not known, in vitro studies and studies in animal models provide support for several transporters. For example, the intestinal absorption of metformin may involve the plasma membrane monoamine transporter (PMAT encoded by gene SLC29A4), the organic cation transporter 3 (OCT3 encoded by gene SLC22A3), the thiamine transporter (THTR2 encoded by SLC19A3) (Liang et al., 2015) and the serotonin transporter (SERT encoded by SLC6A4)(Han et al., 2015; M. Zhou, Xia, & Wang, 2007). These transporters are expressed on the luminal membrane of enterocytes. To mediate flux from the enterocyte into the portal circulation, OCT1 (gene SLC22A1), which is expressed on the basolateral surface, appears to be involved. For hepatic distribution of metformin to targets in the liver, evidence in OCT1 knockout mice and humans using imaging of 11C-metformin suggests that OCT1 (SLC22A1) is primarily involved (Jensen et al., 2016; Sundelin et al., 2017). Additionally, in OCT1 deficient mice the glucose lowering effects of metformin are completely abolished (Shu et al., 2007). Metformin excretion into bile appears to involve multidrug and toxin extrusion 1 (MATE1 encoded by SLC47A1) (Otsuka et al., 2005), though in humans the bulk of metformin is excreted unchanged into the urine (Graham et al., 2011). The uptake of metformin from the circulation into renal epithelial cells is primarily facilitated by OCT2 (SLC22A2) located on the basolateral membrane of renal proximal tubule cells (Takane, Shikata, Otsubo, Higuchi, & Ieiri, 2008). MATE1 (SLC47A1) and MATE2-K (SLC47A2), expressed on the apical membrane appear to be responsible for excretion of metformin into urine (Masuda et al., 2006; Otsuka et al., 2005; Tanihara et al., 2007). OCT1 and PMAT are also expressed on the apical membrane of renal epithelial cells and these transporters may also play a role in metformin renal disposition (Gong, Goswami, Giacomini, Altman, & Klein, 2012).
Metformin pharmacodynamics
Metformin lowers both fasting and post-prandial glucose levels primarily by reducing excessive hepatic glucose production through suppression of gluconeogenesis. Metformin may also increase peripheral glucose utilization and has downstream effects on decreasing fatty acid and triglyceride production (Hundal et al., 2000; Rena et al., 2013). As metformin does not stimulate endogenous insulin production, it does not cause hypoglycemia, which is an adverse effect that is associated with several anti-diabetic medications.
The molecular mechanisms of metformin action are described above. Below we describe candidate gene studies that have provided information on genes, proteins and pathways involved in the absorption, disposition and response to metformin in healthy volunteers and in patients with type 2 diabetes (Table 1).
OCT1
The most studied transporter in relation to metformin genetics and disposition is OCT1, which is essential for the hepatic uptake of metformin. OCT1 is encoded by the gene SLC22A1 which is highly polymorphic and a number of coding missense SNPs affect its activity (Kerb et al., 2002; Leabman et al., 2002; Sakata et al., 2004; Shu et al., 2003). In a study of 20 healthy volunteers, it was demonstrated that the presence of at least one of four reduced function variants (R61C/rs12208357, G401S/rs34130495, 420del/rs72552763 and/or G465R/rs34059508) attenuates the effects of a short course of metformin on glucose tolerance tests (Shu et al., 2007). A follow up study demonstrated that individuals carrying any of the reduced function OCT1 alleles showed a higher area under the concentration-time curve (AUC), higher peak serum concentration (Cmax), and a lower volume of distribution compared to individuals carrying wild-type alleles (Shu et al., 2008). A subsequent study in 103 healthy volunteers found that G465R was associated with increased renal clearance and decreased hepatic uptake. However, the reduced function allele did not lead to differences in metformin AUC (Tzvetkov et al., 2009). In a large retrospective observation study of 1531 subjects with type 2 diabetes assembled by GoDARTS, the two most common reduced function polymorphisms (R61C and M420del) were not associated with four different measures of clinical response whether defined as initial HbA1c reduction, odds of achieving a target HbA1c of less than 7%, average HbA1c or hazard of monotherapy failure. It is important to note that since the study was observational, the metformin dose, prescription of other anti-diabetic agents and timing of HbA1c measurements were at the discretion of the prescribing clinician (K. Zhou et al., 2009). In the South Danish Diabetes Study, which is a prospective interventional study of 159 participants, both trough and steady state metformin concentrations and 6 month change in HbA1c were lower with increasing number of reduced-function alleles (i.e., R61C, G401S, M420del and G465R) (Christensen et al., 2011). In terms of response, the retrospective Rotterdam study reported the association of SLC22A1 intronic SNP rs622342 with metformin response in subjects with type 2 diabetes (Becker et al., 2009b), but this association was not observed in several later studies (Christensen et al., 2011; Jablonski et al., 2010; Tkac et al., 2013). Despite consistent data from various studies (Shu et al., 2008) (Tzvetkov et al., 2009) 22,(Sundelin et al., 2017) that OCT1 reduced function variants are associated with metformin pharmacokinetics, their role in metformin response has not been observed in some studies (K. Zhou et al., 2009) including the recent large observational study in diabetic patients sponsored by MetGen (Dujic et al., 2017). Reasons for this discrepancy may be related to differences in analysis of the data or the fact that genes involved in the pharmacokinetics of metformin do not contribute much to variation in the pharmacodynamics of the drug.
OCT2
The renal transporter OCT2 appears to have less functional variation than OCT1 and the effect of variation in OCT2 with respect to metformin has been studied to a lesser extent than OCT1. A study of 15 healthy Chinese participants showed that the reduced-function 808G>T polymorphism (Ala270Ser) was associated with reduced metformin renal tubular clearance and that in the presence of cimetidine, metformin clearance was decreased in all participants, but the decrease was significantly lower in homozygous TT participants compared to GG participants (18.7 vs. 48.2%, P=0.029)(Wang, Yin, Tomlinson, & Chow, 2008). Similarly, a study in Korean healthy participants showed that reduced-function genetic variants of OCT2 (596C>T, 602C>T, and 808G>T) were associated with higher peak plasma concentration and area under the curve measurements and lower renal clearance (Song et al., 2008). In contrast, in a study of 23 healthy volunteers of Caucasian and African-American ancestries, though an association between the renal clearance of metformin and 808G>T was observed, the direction of effect was the opposite and subjects who were homozygous for the reference allele 808G>G (P<0.005) had reduced renal clearance of metformin (Chen et al., 2009). The reasons for these differences are likely related to small sample sizes and differences in the ethnicities of the participants. Studies of association between genetic variation in OCT2 and metformin response have not shown positive results (Christensen et al., 2011; Jablonski et al., 2010).
MATE1 and MATE2
Studies have examined the effects of promoter variants in both MATE1 and MATE2 on variation in metformin disposition and response. In a study of 57 healthy volunteers who received metformin, the renal and secretory clearances of metformin were higher in carriers of MATE2 variants who also carried the reference alleles for MATE1. Additionally, 145 patients with type 2 diabetes carrying the MATE1 variant showed enhanced response to metformin (Stocker et al., 2013). In the Rotterdam study, the intronic SNP rs2289669 in MATE1 was associated with change in HbA1c after metformin initiation, with the minor allele enhancing the reduction in HbA1c (Becker et al., 2009a). Similar findings were observed in Tkac et al (Tkac et al., 2013) and in the Diabetes prevention program where rs8065082 in MATE1 which is in high linkage disequilibrium with rs2289669 was associated with lower diabetes incidence in the metformin arm and not in the placebo arm in subjects who were at risk for the development of diabetes (Jablonski et al., 2010). However, this finding was not observed in the South Danish Diabetes Study (Christensen et al., 2011; Dujic et al., 2017) or in a large meta-analysis conducted by the Metformin Genetics (MetGen) Consortium.
Overall, the results of genetic associations between transporters and pharmacokinetics of metformin have led to conflicting results. Differences in data analyses, study design and in the characteristics of the participants such as their ethnicities or disease status, may have led to some of the discrepancies among studies. Most of the studies have been small and underpowered, so clearly larger well-powered studies in multiple ethnic groups are needed to understand which variants and genes may associate with the pharmacokinetics of metformin.
GENETIC STUDIES OF OTHER ANTI-DIABETIC DRUGS
Sulfonylureas
Sulfonylureas are among the most widely used anti-diabetic drugs and act by directly stimulating insulin secretion from the pancreatic beta cells. Serious side effects of these drugs are hypoglycemia and weight gain. Sulfonylureas were previously considered as a first line treatment for type 2 diabetes, but now are mainly used as a second line treatment in combination with metformin (Marathe, Gao, & Close, 2017; Nathan et al., 2009). Sulfonylureas stimulate insulin secretion by binding to the ATP-sensitive potassium channel (KATP) which is composed of four subunits of the sulfonylurea receptor (SUR1, ABCC8) and four subunits of the potassium inward rectifier channel (Kir) 6.2. Sulfonylurea binding leads to closure of the KATP channel which alters the resting potential of the cell, leading to calcium influx and stimulation of insulin secretion (Thule & Umpierrez, 2014). Sulfonylureas are metabolized in the liver primarily by the polymorphic cytochrome P450 isoenzyme 2C9 encoded by CYP2C9. Substitution of arginine with cysteine at amino acid position 144 (Arg144Cys) and isoleucine with leucine at position 359 (Ile359Leu) gives rise to mutant alleles CYP2C9*2 and CYP2C9*3 respectively which have reduced catalytic activity when compared to the wild-type CYP2C9*1.
Several studies have investigated the effect of genetic variation in CYP2C9 (the rate limiting step of metabolism) on the pharmacokinetics and pharmacodynamics of sulfonylureas. Studies in healthy subjects show that reduced function alleles of CYP2C9 are associated with increased plasma concentrations and decreased clearance of sulfonylureas after oral administration. While small-scale studies have shown differences in sulfonylurea metabolism, studies on actual measured drug response are few. In a large retrospective study of 1073 subjects from GoDARTs, carriers of loss of function CYP2C9*2 or CYP2C9*3 alleles had a 3.4 fold increased likelihood of achieving therapeutic target effects on blood glucose compared to carriers of the wild type alleles. This translated to a 0.5% greater HbA1c reduction in individuals with the variant CYP2C9 alleles (K. Zhou et al., 2010). In addition to CYP2C9 polymorphisms, CYP2C19 polymorphisms have been reported to be influential in gliclazide pharmacokinetics (Y. Zhang et al., 2007), (Shao et al., 2010).
The identification of SU-binding sites SUR1 and Kir6.2 (encoded by ABCC8 and KCNJ11, respectively) has led to various pharmacogenetic explorations of variations in these genes. Non-synonymous variants E23K in KCNJ11 and S1369A in ABCC8 form a haplotype and pharmacogenetic studies of these variants show conflicting results. Association of S1369A with glycemic control has been reported in 115 Chinese patients who were on gliclazide for 8 weeks with carriers of the minor allele demonstrating a greater reduction in HbA1c than the wild type carriers (Zhang, Liu, Kuang, Yi, & Xing, 2007). In a study that investigated the effect of the E23K variant KCNJ11 gene on gliclazide modified release treatment in 108 newly diagnosed patients with type 2 diabetes mellitus, at baseline, patients with the KK genotype had higher blood glucose and lower serum insulin levels after oral glucose administration than patients with the EE and EK genotypes (P < 0.05 for all). During treatment, individuals with the KK genotype had lower fasting glucose levels and were more likely to attain the target fasting glucose level than E allele carriers suggesting that the KCNJ11 E23K variant is associated with a greater effect of sulphonylurea treatment in newly diagnosed Chinese patients with type 2 diabetes (Li et al., 2014). However, results from UKPDS are conflicting. In this study they evaluated 364 subjects with type 2 diabetes randomized to sulfonylurea treatment and determined that the presence of the K allele in E23K did not predict failure to treatment with sulfonylureas at 1 year (Gloyn et al., 2001).
The KCNQ1 gene encodes the pore-forming subunit of a voltage-gated K+ channel (KvLQT1) and studies have indicated that KCNQ1 polymorphisms are related to impaired insulin secretion (Hu et al., 2009) and type 2 diabetes in several GWAS (see GWAS Catalog (https://www.ebi.ac.uk/gwas/home). A study evaluated the effect KCNQ1 genotype on sulphonylurea therapy in addition to metformin therapy on glycemic control in 87 patients with type 2 diabetes. The results showed that carriers of the T-allele achieved significantly lower fasting glucose levels in compared with patients with the GG genotype (6.95±0.13 vs. 7.50±0.21 mmol/l, P =0.033)(Schroner et al., 2011).
The TCF7L2 gene is strongly associated with the development of type 2 diabetes with one of the highest effect sizes for common variants (Grant et al., 2006; Helgason et al., 2007). Several studies have focused on the rs7903146 variant in TCF7L2 as a determinant of drug response. In a large GoDARTs retrospective study of subjects with type 2 diabetes, of 901 users of sulfonylureas, homozygotes for the type 2 diabetes T risk allele at TCF7L2 rs7903146 were less likely to respond to sulfonylureas, with an odds ratio of failure of 1.73 (95% CI 1.11–2.70, P=0.015)(Pearson et al., 2007). This was in contrast to results from the Study to Understand the Genetics of the Acute Response of metformin and glipizide in humans (SUGAR-MGH) study where 608 participants who were healthy or at risk for type 2 diabetes were given a single dose of 5 mg of glipizide with biochemical data measured after the dose. Results from SUGAR-MGH showed that the T allele of rs7903146 at TCF7L2 was associated with a significantly shorter time and a steeper slope to trough glucose levels after glipizide administration(Srinivasan et al., 2018). These contrasting results suggest that it is possible that the genotype has a differential effect in subjects in whom diabetes has not yet developed compared with subjects with established type 2 diabetes, in whom some degree of beta cell failure may have already occurred.
Thiazolidinediones
The thiazolidinediones (TZDs) are a group of drugs that increase insulin sensitivity by acting on adipose tissue, muscle, and liver to increase glucose utilization and decrease glucose production. The mechanism by which the thiazolidinediones exert their effect is not fully understood but is thought to relate to activation of one or more peroxisome proliferator-activated receptors (PPARs), which regulate gene expression in response to ligand binding. Hepatic uptake of TZDs is mediated by OATP1B1 and TZDs are metabolized by CYP2C8. Homozygous carriers of the gain of function allele for CYP2C8, CYP2C8*3 coding for the Arg139Lys and Lys399Arg amino acid substitutions have lower rosiglitazone plasma concentrations and higher weight adjusted clearance when compared to wild type carriers (Aquilante et al., 2008; Kirchheiner et al., 2006). In addition to the role of OATP1B1 encoded by SLCO1B1 and CYP2C8 in thiazolidinediones pharmacokinetics, a recent study in 833 Scottish patients showed that CYP2C8*3 and OATP1B1 non-synonymous variant (Val174Ala, rs4149056) alters drug efficacy (Dawed et al., 2016). In particular, gain of function CYP2C8*3 was associated with reduced glycemic response to rosiglitazone (P=0.01) and less weight gain (P=0.02) (27271184), which is in agreement with the effect of this variant to cause increase drug clearance and lower plasma levels (Aquilante et al., 2008; Kirchheiner et al., 2006). PPARG is a target of TZDs and has been explored in pharmacogenetic investigations. A study in 250 Chinese patients showed that carriers of the minor allele of variant rs1801282 in PPARG had a higher odds of being responders to pioglitazone when compared to carriers of wild type alleles (Hsieh et al., 2010). Studies have also looked at other adipokinins including adiponectin, leptin, resistin and tumor-necrosis-factor-alpha based on the role they play in insulin resistance with variable results obtained. A study of 42 Chinese patients with type 2 diabetes treated with 12 weeks of rosiglitazone monotherapy demonstrated an attenuated effect of rosiglitazone on fasting glucose, 2 hour glucose and HOMA-IR values in subjects with adiponectin variant C-11377G(rs266729) CG and GG genotypes compared to CC genotype, however, the effects were enhanced in patients with the diplotype C-11377( rs266729)/ T45G(rs2241766) CGTT in adiponectin (Sun et al., 2008). The investigators also evaluated the impact of genetic polymorphisms of leptin and TNF-alpha on rosiglitazone response in the same group of subjects. They found an enhanced rosiglitazone effect in patients with G-2548A (rs7799039)AA genotype of leptin on on fasting and 2 hour insulin values compared with GG and GA genotypes. They also showed an attenuated rosiglitazone effect in patients with GA andAA genotypes of TNF-alpha G-308A (rs1800629_ on fasting insulin values compared with GG genotype (Liu et al., 2008).
Meglinitides
The meglitinides, repaglinide and nateglinide, are short-acting glucose-lowering drugs that are structurally distinct from sulfonylureas and exert their effects via different pancreatic beta cell receptors, but they act similarly by regulating adenosine triphosphate (ATP)-sensitive potassium channels (K-ATP channels) in pancreatic beta cells, thereby increasing insulin secretion. Meglitinides have a rapid onset and short duration of action. They are transported into the liver by OATP1B1 encoded by SLCO1B1 after which metabolism occurs via CYP family isoenzymes. There have been a few pharmacogenetic studies of meglinitides. Genetic polymorphisms in CYP2C8 has been found to be associated with reduced plasma concentrations of repaglinide (Niemi et al., 2003). Repaglinide pharmacokinetics has also been shown to be modulated by genetic variants in SLCO1B1 (Kalliokoski, Backman, Neuvonen, & Niemi, 2008). Several candidate genes studies have evaluated the effect of genetic variantion in genes implicated in type 2 diabetes risk on response to meglinitides (Semiz, Dujic, & Causevic, 2013). These include SNPs in KCNQ1 ((Dai et al., 2012; W. Yu et al., 2011)), SLC30A8 (Huang et al., 2010), TCF7L2 and KCNJ11 ((M. Yu et al., 2010)). However, these studies have small sample size and were performed only in Chinese patients with type 2 diabetes and therefore the results cannot be extrapolated to other populations.
Acarbose
The alpha-glucosidase inhibitor, acarbose, when taken orally, inhibits the upper gastrointestinal enzymes (alpha-glucosidases) that convert complex polysaccharide carbohydrates into monosaccharides in a dose-dependent fashion thereby slowing the absorption of glucose. Interactions between acarbose and PPAR-alpha (PPARA) with PPAR-gamma2 (PPARG2), PPAR-gamma coactivator 1alpha (PPARGC1A), and hepatic nuclear factor 4alpha (HNF4A), were evaluated in the STOP-NIDDM trial with modest associations revealed (Andrulionyte, Kuulasmaa, Chiasson, & Laakso, 2007). The clinical significance of these associations remain to be tested.
SGLT2 inhibitors (Gliflozins)
SGLT2 inhibitors are a new class of drugs that promote the renal excretion of glucose and thereby modestly lower elevated blood glucose levels in patients with type 2 diabetes. They are mainly eliminated through O-glucouronidation by uridine diphosphate glucuronosyl transferases. A study of 134 subjects including both healthy and subjects with type 2 diabetes demonstrated that carriers of reduced function variant, UGT1A9*3 and UGT2B4*2 had an increased plasma concentration of canagliflozin, an SGLT2 inhibitor when compared to carriers of the wild type alleles (Francke et al., 2015). Recently, a cross-sectional study in a population at risk for type 2 diabetes in which 603 subjects received empagliflozin, an SGLT2 inhibitor and 305 received placebo showed no association of common SLC5A2 variants that encode SGLT2 with empagliflozin response (Zimdahl et al., 2017).
Glucagon-like peptide-1 (GLP-1) based therapies
GLP-1-based therapies (eg, GLP-1 receptor agonists, dipeptidyl peptidase-4 [DPP-4] inhibitors) are a very promising group of drugs that affect glucose control through several mechanisms, including enhancement of glucose-dependent insulin secretion, slowed gastric emptying, and reduction of postprandial glucagon and of food intake. There are very few pharmacogenetic studies of GLP-1 based therapies. In a study of the association of TCF7L2 on the response to treatment with the DPP-4 inhibitor linagliptin in 61 patients with type 2 diabetes, no significant treatment differences were seen between non risk variant CC carriers and heterozygous CT subjects, although HbA1c response was reduced in homozygous TT subjects when compared with CC subjects (Zimdahl et al., 2014)
Pharmacogenetics of adverse reactions and off target effects of anti-diabetic drugs
In addition to studies of pharmacokinetics and response, there have been candidate gene studies studies of pharmacogenetics of adverse reactions to anti-diabetic drugs. The most common side effect of metformin therapy is gastrointestinal distress with symptoms of taste alteration, nausea, abdominal discomfort , diarrhea or mild anorexia. Metformin therapy can cause gastrointestinal distress in upto 40 % of patients on treatment and around 5% of patients have to discontinue metformin because of side effects (C. J. Bailey, 2015), A study of adverse effects by GoDARTS investigators was undertaken in 1915 participants with type 2 diabetes who had evidence of metformin intolerance including gastrointestinal side effects and the need to discontinue the drug and 251 subjects who were metformin tolerant. Their results showed that the presence of two or more reduced function alleles of OCT1 (at R61C, C88R/rs55918055, G401S, M420del or G465R) increased the odds of metformin intolerance by more than two-fold. Further, concomitant use of OCT1 inhibiting medications such as verapamil, citalopram or proton pump inhibitors increased the odds by four-fold, presumably by increasing metformin concentrations in the intestine (Dujic et al., 2015). Hypoglycemia is the most common side effect of sulfonylureas. Loss of function CYP2C9 variants have been associated with increased drugs levels of gliclazide with significantly increased risk of hypoglycemia (Gokalp et al., 2011). The DREAM (Diabetes REduction Assessment with Ramipril and rosiglitazone Medication) trial evaluated adverse effects of rosiglitazone on edema in 4197 partcipants. A single SNP rs6123045 in NFATC2 was significantly associated with edema and the interaction between the SNP and rosiglitazone for edema was significant (S. D. Bailey et al., 2010). Studies are required to evaluate the pharmacogenetics of adverse effects of other newer drugs such as SGLT2 inhibitors and incretin based therapies.
Few studies have evaluated the pharmacogenetics factors involved in off-target effects of anti-diabetic drugs. In addition to its beneficial effect on glycemic control, metformin has been shown to have cardiovascular benefit in patients with type 2 diabetes (Lamanna, Monami, Marchionni, & Mannucci, 2011). A small Russian study of 52 subjects evaluated the type 2 diabetes associated PPRA-gamma2 pro12Ala polymorphism in relation to metformin therapy in patients with coronary heart disease and metabolic syndrome or type 2 diabetes. They found that patients with coronary artery disease who carried the Pro allele showed a significant metformin-induced reduction in weight, waist circumference, body mass index, and plasma levels of cardiovascular markers incuding total cholesterol, C-peptide, and cytokines, such IL-1beta, IL-6, IL-8, and TNF-alpha. Further studies are clearly needed to evaluate the pharmacogenetics of off-target effects of metformin and other anti-diabetic drugs(Lavrenko, Shlykova, Kutsenko, Mamontova, & Kaidashev, 2012).
Conclusions
In summary, there are few pharmacogenomics studies of anti-diabetic drugs other than metformin. For metformin, two GWAS have been conducted and have revealed that genetic variants in ATM and SLC2A2 are associated with response to the drug in large populations of patients with Type 2 diabetes treated with metformin. Though these results need replication in additional cohorts, they pave the way for precision medicine in guiding selection and dosing of metformin in diabetic patient populations. Candidate gene studies focused on metformin pharmacokinetics have been small, and there is a clear need for larger studies to identify the genes that play a role in variation in pharmacokinetics of the drug. Pharmacogenomic studies are clearly needed for other anti-diabetic drugs beyond metformin, as to date, most pharmacogenomic studies of other anti-diabetic drugs are small and focused on candidate genes. The goal of precision medicine is to tailor medical therapy according to patient characteristics to achieve optimal therapeutic response while minimizing adverse effects. For precision medicine to be used in guiding selection of anti-diabetic drugs for individual patients based on genetic (and other) factors, there is a clear need for larger studies focused on efficacy of both metformin and other anti-diabetic drugs. In addition, because many anti-diabetic drugs are associated with dose-limiting toxicities, further studies are needed to identify the genetic factors that underlie adverse drug reactions in order to enhance the optimal selection of drug and dose in the treatment of type 2 diabetes.
Footnotes
Conflict of interest: SS does not have any conflicts of interest.
References
- Andrulionyte L, Kuulasmaa T, Chiasson JL, & Laakso M (2007). Single nucleotide polymorphisms of the peroxisome proliferator-activated receptor-alpha gene (PPARA) influence the conversion from impaired glucose tolerance to type 2 diabetes: the STOP-NIDDM trial. Diabetes, 56(4), 1181–1186. doi: 10.2337/db06-1110 [DOI] [PubMed] [Google Scholar]
- Aquilante CL, Bushman LR, Knutsen SD, Burt LE, Rome LC, & Kosmiski LA (2008). Influence of SLCO1B1 and CYP2C8 gene polymorphisms on rosiglitazone pharmacokinetics in healthy volunteers. Hum Genomics, 3(1), 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CJ (2015). Safety of antidiabetes medications: An update. Clin Pharmacol Ther, 98(2), 185–195. doi: 10.1002/cpt.125 [DOI] [PubMed] [Google Scholar]
- Bailey SD, Xie C, Do R, Montpetit A, Diaz R, Mohan V, … Anand S (2010). Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study. Diabetes Care, 33(10), 2250–2253. doi: 10.2337/dc10-0452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, & Stricker BH (2009a). Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes, 58(3), 745–749. doi: 10.2337/db08-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, & Stricker BH (2009b). Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J, 9(4), 242–247. doi: 10.1038/tpj.2009.15 [DOI] [PubMed] [Google Scholar]
- Biernacka JM, Sangkuhl K, Jenkins G, Whaley RM, Barman P, Batzler A, … Weinshilboum R (2015). The International SSRI Pharmacogenomics Consortium (ISPC): a genome-wide association study of antidepressant treatment response. Transl Psychiatry, 5, e553. doi: 10.1038/tp.2015.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, & Ridker PM (2012). Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: the Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet, 5(2), 257–264. doi: 10.1161/circgenetics.111.961144 [DOI] [PubMed] [Google Scholar]
- Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, … Giacomini KM (2009). Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics, 19(7), 497–504. doi: 10.1097/FPC.0b013e32832cc7e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Yee SW, Ramirez AH, Morrissey KM, Jang GH, Joski PJ, … Giacomini KM (2011). A common 5’-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther, 90(5), 674–684. doi: 10.1038/clpt.2011.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen MM, Brasch-Andersen C, Green H, Nielsen F, Damkier P, Beck-Nielsen H, & Brosen K (2011). The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet Genomics, 21(12), 837–850. doi: 10.1097/FPC.0b013e32834c0010 [DOI] [PubMed] [Google Scholar]
- Connelly PJ, Smith N, Chadwick R, Exley AR, Shneerson JM, & Pearson ER (2016). Recessive mutations in the cancer gene Ataxia Telangiectasia Mutated (ATM), at a locus previously associated with metformin response, cause dysglycaemia and insulin resistance. Diabet Med, 33(3), 371–375. doi: 10.1111/dme.13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Faja B, Quirantes-Pine R, Oliveras-Ferraros C, Vazquez-Martin A, Cufi S, Martin-Castillo B, … Menendez JA (2012). Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany NY), 4(7), 480–498. doi: 10.18632/aging.100472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XP, Huang Q, Yin JY, Guo Y, Gong ZC, Lei MX, … Liu ZQ (2012). KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol, 39(5), 462–468. doi: 10.1111/j.1440-1681.2012.05701.x [DOI] [PubMed] [Google Scholar]
- Dawed AY, Donnelly L, Tavendale R, Carr F, Leese G, Palmer CN, … Zhou K (2016). CYP2C8 and SLCO1B1 Variants and Therapeutic Response to Thiazolidinediones in Patients With Type 2 Diabetes. Diabetes Care, 39(11), 1902–1908. doi: 10.2337/dc15-2464 [DOI] [PubMed] [Google Scholar]
- Dujic T, Zhou K, Donnelly LA, Tavendale R, Palmer CN, & Pearson ER (2015). Association of Organic Cation Transporter 1 With Intolerance to Metformin in Type 2 Diabetes: A GoDARTS Study. Diabetes, 64(5), 1786–1793. doi: 10.2337/db14-1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujic T, Zhou K, Yee SW, van Leeuwen N, de Keyser CE, Javorsky M, … Pearson ER (2017). Variants in Pharmacokinetic Transporters and Glycemic Response to Metformin: A Metgen Meta-Analysis. Clin Pharmacol Ther, 101(6), 763–772. doi: 10.1002/cpt.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan N, Vijverberg SJ, Andiappan AK, Arianto L, Berce V, Blanca-Lopez N, … Maitland-van der Zee AH (2017). Rationale and design of the multiethnic Pharmacogenomics in Childhood Asthma consortium. Pharmacogenomics, 18(10), 931–943. doi: 10.2217/pgs-2017-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, & Florez JC (2016). Type 2 diabetes: genetic data sharing to advance complex disease research. Nat Rev Genet, 17(9), 535–549. doi: 10.1038/nrg.2016.56 [DOI] [PubMed] [Google Scholar]
- Florez JC (2017). Mining the Genome for Therapeutic Targets. Diabetes, 66(7), 1770–1778. doi: 10.2337/dbi16-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez JC, Jablonski KA, Taylor A, Mather K, Horton E, White NH, … Pollin TI (2012). The C allele of ATM rs11212617 does not associate with metformin response in the Diabetes Prevention Program. Diabetes Care, 35(9), 1864–1867. doi: 10.2337/dc11-2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, … Viollet B (2010). Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest, 120(7), 2355–2369. doi: 10.1172/jci40671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, & Viollet B (2011). Regulation of hepatic metabolism by AMPK. J Hepatol, 54(4), 827–829. doi: 10.1016/j.jhep.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Francke S, Mamidi RN, Solanki B, Scheers E, Jadwin A, Favis R, & Devineni D (2015). In vitro metabolism of canagliflozin in human liver, kidney, intestine microsomes, and recombinant uridine diphosphate glucuronosyltransferases (UGT) and the effect of genetic variability of UGT enzymes on the pharmacokinetics of canagliflozin in humans. J Clin Pharmacol, 55(9), 1061–1072. doi: 10.1002/jcph.506 [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, & Kubo M (2017). Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov, 16(1), 1. doi: 10.1038/nrd.2016.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, … Yee D (2010). Diabetes and cancer: a consensus report. Diabetes Care, 33(7), 1674–1685. doi: 10.2337/dc10-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, & Turner RC (2001). Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53). Diabet Med, 18(3), 206–212. [DOI] [PubMed] [Google Scholar]
- Gokalp O, Gunes A, Cam H, Cure E, Aydin O, Tamer MN, … Dahl ML (2011). Mild hypoglycaemic attacks induced by sulphonylureas related to CYP2C9, CYP2C19 and CYP2C8 polymorphisms in routine clinical setting. Eur J Clin Pharmacol, 67(12), 1223–1229. doi: 10.1007/s00228-011-1078-4 [DOI] [PubMed] [Google Scholar]
- Gong L, Goswami S, Giacomini KM, Altman RB, & Klein TE (2012). Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics, 22(11), 820–827. doi: 10.1097/FPC.0b013e3283559b22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, … Williams KM (2011). Clinical pharmacokinetics of metformin. Clin Pharmacokinet, 50(2), 81–98. doi: 10.2165/11534750-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, … Stefansson K (2006). Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet, 38(3), 320–323. doi: 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- Han TK, Proctor WR, Costales CL, Cai H, Everett RS, & Thakker DR (2015). Four cation-selective transporters contribute to apical uptake and accumulation of metformin in Caco-2 cell monolayers. J Pharmacol Exp Ther, 352(3), 519–528. doi: 10.1124/jpet.114.220350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, … Stefansson K (2007). Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet, 39(2), 218–225. doi: 10.1038/ng1960 [DOI] [PubMed] [Google Scholar]
- Hsieh MC, Lin KD, Tien KJ, Tu ST, Hsiao JY, Chang SJ, … Chen HC (2010). Common polymorphisms of the peroxisome proliferator-activated receptor-gamma (Pro12Ala) and peroxisome proliferator-activated receptor-gamma coactivator-1 (Gly482Ser) and the response to pioglitazone in Chinese patients with type 2 diabetes mellitus. Metabolism, 59(8), 1139–1144. doi: 10.1016/j.metabol.2009.10.030 [DOI] [PubMed] [Google Scholar]
- Hu C, Wang C, Zhang R, Ma X, Wang J, Lu J, … Jia W (2009). Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia, 52(7), 1322–1325. doi: 10.1007/s00125-009-1335-6 [DOI] [PubMed] [Google Scholar]
- Huang Q, Yin JY, Dai XP, Wu J, Chen X, Deng CS, … Liu ZQ (2010). Association analysis of SLC30A8 rs13266634 and rs16889462 polymorphisms with type 2 diabetes mellitus and repaglinide response in Chinese patients. Eur J Clin Pharmacol, 66(12), 1207–1215. doi: 10.1007/s00228-010-0882-6 [DOI] [PubMed] [Google Scholar]
- Hundal RS, Krssak M, Dufour S, Laurent D, Lebon V, Chandramouli V, … Shulman GI (2000). Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes, 49(12), 2063–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, … Florez JC (2010). Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes, 59(10), 2672–2681. doi: 10.2337/db10-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav K, & Zhang Y (2017). Activating transcription factor 3 in immune response and metabolic regulation. Liver Res, 1(2), 96–102. doi: 10.1016/j.livres.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JB, Sundelin EI, Jakobsen S, Gormsen LC, Munk OL, Frokiaer J, & Jessen N (2016). [11C]-Labeled Metformin Distribution in the Liver and Small Intestine Using Dynamic Positron Emission Tomography in Mice Demonstrates Tissue-Specific Transporter Dependency. Diabetes, 65(6), 1724–1730. doi: 10.2337/db16-0032 [DOI] [PubMed] [Google Scholar]
- Kalliokoski A, Backman JT, Neuvonen PJ, & Niemi M (2008). Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet Genomics, 18(11), 937–942. doi: 10.1097/FPC.0b013e32830d733e [DOI] [PubMed] [Google Scholar]
- Kerb R, Brinkmann U, Chatskaia N, Gorbunov D, Gorboulev V, Mornhinweg E, … Koepsell H (2002). Identification of genetic variations of the human organic cation transporter hOCT1 and their functional consequences. Pharmacogenetics, 12(8), 591–595. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Thomas S, Bauer S, Tomalik-Scharte D, Hering U, Doroshyenko O, … Fuhr U (2006). Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Clin Pharmacol Ther, 80(6), 657–667. doi: 10.1016/j.clpt.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Lamanna C, Monami M, Marchionni N, & Mannucci E (2011). Effect of metformin on cardiovascular events and mortality: a meta-analysis of randomized clinical trials. Diabetes Obes Metab, 13(3), 221–228. doi: 10.1111/j.1463-1326.2010.01349.x [DOI] [PubMed] [Google Scholar]
- Lavrenko AV, Shlykova OA, Kutsenko LA, Mamontova TV, & Kaidashev IP (2012). [Pharmacogenetic features of the effect of metformin in patients with coronary heart disease in the presence of metabolic syndrome and type 2 diabetes mellitus in terms of PPAR-gamma2 gene polymorphism]. Ter Arkh, 84(9), 35–40. [PubMed] [Google Scholar]
- Leabman MK, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, … Giacomini KM (2002). Polymorphisms in a human kidney xenobiotic transporter, OCT2, exhibit altered function. Pharmacogenetics, 12(5), 395–405. [DOI] [PubMed] [Google Scholar]
- Li Q, Chen M, Zhang R, Jiang F, Wang J, Zhou J, … Jia W (2014). KCNJ11 E23K variant is associated with the therapeutic effect of sulphonylureas in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol, 41(10), 748–754. doi: 10.1111/1440-1681.12280 [DOI] [PubMed] [Google Scholar]
- Liang X, Chien HC, Yee SW, Giacomini MM, Chen EC, Piao M, … Giacomini KM (2015). Metformin Is a Substrate and Inhibitor of the Human Thiamine Transporter, THTR-2 (SLC19A3). Mol Pharm, 12(12), 4301–4310. doi: 10.1021/acs.molpharmaceut.5b00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Lin YG, Wu J, Sun H, Gong ZC, Hu PC, … Liu ZQ (2008). Impact of genetic polymorphisms of leptin and TNF-alpha on rosiglitazone response in Chinese patients with type 2 diabetes. Eur J Clin Pharmacol, 64(7), 663–671. doi: 10.1007/s00228-008-0483-9 [DOI] [PubMed] [Google Scholar]
- Luizon MR, Eckalbar WL, Wang Y, Jones SL, Smith RP, Laurance M, … Ahituv N (2016). Genomic Characterization of Metformin Hepatic Response. PLoS Genet, 12(11), e1006449. doi: 10.1371/journal.pgen.1006449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, … Parkinson H (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res, 45(D1), D896–d901. doi: 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, … Shulman GI (2014). Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature, 510(7506), 542–546. doi: 10.1038/nature13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe PH, Gao HX, & Close KL (2017). American Diabetes Association Standards of Medical Care in Diabetes 2017. J Diabetes, 9(4), 320–324. doi: 10.1111/1753-0407.12524 [DOI] [PubMed] [Google Scholar]
- Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, … Inui K (2006). Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol, 17(8), 2127–2135. doi: 10.1681/asn.2006030205 [DOI] [PubMed] [Google Scholar]
- Nathan DM, Buse JB, Davidson MB, Ferrannini E, Holman RR, Sherwin R, & Zinman B (2009). Medical Management of Hyperglycemia in Type 2 Diabetes: A Consensus Algorithm for the Initiation and Adjustment of Therapy: A consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care, 32(1), 193–203. doi: 10.2337/dc08-9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M, Leathart JB, Neuvonen M, Backman JT, Daly AK, & Neuvonen PJ (2003). Polymorphism in CYP2C8 is associated with reduced plasma concentrations of repaglinide. Clin Pharmacol Ther, 74(4), 380–387. doi: 10.1016/s0009-9236(03)00228-5 [DOI] [PubMed] [Google Scholar]
- Niu N, Liu T, Cairns J, Ly RC, Tan X, Deng M, … Wang L (2016). Metformin pharmacogenomics: a genome-wide association study to identify genetic and epigenetic biomarkers involved in metformin anticancer response using human lymphoblastoid cell lines. Hum Mol Genet, 25(21), 4819–4834. doi: 10.1093/hmg/ddw301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, & Moriyama Y (2005). A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci U S A, 102(50), 17923–17928. doi: 10.1073/pnas.0506483102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, … Palmer CN (2007). Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes, 56(8), 2178–2182. doi: 10.2337/db07-0440 [DOI] [PubMed] [Google Scholar]
- Pedersen AJT, Stage TB, Glintborg D, Andersen M, & Christensen MMH (2018). The Pharmacogenetics of Metformin in Women with Polycystic Ovary Syndrome: A Randomized Trial. Basic Clin Pharmacol Toxicol, 122(2), 239–244. doi: 10.1111/bcpt.12874 [DOI] [PubMed] [Google Scholar]
- Rena G, Pearson ER, & Sakamoto K (2013). Molecular mechanism of action of metformin: old or new insights? Diabetologia, 56(9), 1898–1906. doi: 10.1007/s00125-013-2991-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T, Anzai N, Shin HJ, Noshiro R, Hirata T, Yokoyama H, … Endou H (2004). Novel single nucleotide polymorphisms of organic cation transporter 1 (SLC22A1) affecting transport functions. Biochem Biophys Res Commun, 313(3), 789–793. [DOI] [PubMed] [Google Scholar]
- Schroner Z, Dobrikova M, Klimcakova L, Javorsky M, Zidzik J, Kozarova M, … Tkac I (2011). Variation in KCNQ1 is associated with therapeutic response to sulphonylureas. Med Sci Monit, 17(7), Cr392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, Alda M, Adli M, Akula N, Ardau R, Bui ET, … McMahon FJ (2010). The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology, 62(1), 72–78. doi: 10.1159/000314708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiz S, Dujic T, & Causevic A (2013). Pharmacogenetics and personalized treatment of type 2 diabetes. Biochem Med (Zagreb), 23(2), 154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyer P, Vallois D, Poitry-Yamate C, Schutz F, Metref S, Tarussio D, … Thorens B (2013). Hepatic glucose sensing is required to preserve beta cell glucose competence. J Clin Invest, 123(4), 1662–1676. doi: 10.1172/jci65538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Ren XM, Liu NF, Chen GM, Li WL, Zhai ZH, & Wang DW (2010). Influence of CYP2C9 and CYP2C19 genetic polymorphisms on pharmacokinetics and pharmacodynamics of gliclazide in healthy Chinese Han volunteers. J Clin Pharm Ther, 35(3), 351–360. doi: 10.1111/j.1365-2710.2009.01134.x [DOI] [PubMed] [Google Scholar]
- Shokri F, Ghaedi H, Ghafouri Fard S, Movafagh A, Abediankenari S, Mahrooz A, … Omrani MD (2016). Impact of ATM and SLC22A1 Polymorphisms on Therapeutic Response to Metformin in Iranian Diabetic Patients. Int J Mol Cell Med, 5(1), 1–7. [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, … Giacomini KM (2008). Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther, 83(2), 273–280. doi: 10.1038/sj.clpt.6100275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, … Giacomini KM (2003). Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci U S A, 100(10), 5902–5907. doi: 10.1073/pnas.0730858100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, … Giacomini KM (2007). Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest, 117(5), 1422–1431. doi: 10.1172/jci30558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snima KS, Pillai P, Cherian AM, Nair SV, & Lakshmanan VK (2014). Anti-diabetic drug metformin: challenges and perspectives for cancer therapy. Curr Cancer Drug Targets, 14(8), 727–736. [DOI] [PubMed] [Google Scholar]
- Song IS, Shin HJ, Shim EJ, Jung IS, Kim WY, Shon JH, & Shin JG (2008). Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin Pharmacol Ther, 84(5), 559–562. doi: 10.1038/clpt.2008.61 [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Kaur V, Chamarthi B, Littleton KR, Chen L, Manning AK, … Florez JC (2018). TCF7L2 Genetic Variation Augments Incretin Resistance and Influences Response to a Sulfonylurea and Metformin: The Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH). Diabetes Care. doi: 10.2337/dc17-1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker SL, Morrissey KM, Yee SW, Castro RA, Xu L, Dahlin A, … Giacomini KM (2013). The effect of novel promoter variants in MATE1 and MATE2 on the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacol Ther, 93(2), 186–194. doi: 10.1038/clpt.2012.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhuk Y, Hopmans SN, Sanli T, Barron C, Tsiani E, Cutz JC, … Tsakiridis T (2013). Metformin inhibits growth and enhances radiation response of non-small cell lung cancer (NSCLC) through ATM and AMPK. Br J Cancer, 108(10), 2021–2032. doi: 10.1038/bjc.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Gong ZC, Yin JY, Liu HL, Liu YZ, Guo ZW, … Liu ZQ (2008). The association of adiponectin allele 45T/G and −11377C/G polymorphisms with Type 2 diabetes and rosiglitazone response in Chinese patients. Br J Clin Pharmacol, 65(6), 917–926. doi: 10.1111/j.1365-2125.2008.03145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelin E, Gormsen LC, Jensen JB, Vendelbo MH, Jakobsen S, Munk OL, … Jessen N (2017). Genetic Polymorphisms in Organic Cation Transporter 1 Attenuates Hepatic Metformin Exposure in Humans. Clin Pharmacol Ther, 102(5), 841–848. doi: 10.1002/cpt.701 [DOI] [PubMed] [Google Scholar]
- Takane H, Shikata E, Otsubo K, Higuchi S, & Ieiri I (2008). Polymorphism in human organic cation transporters and metformin action. Pharmacogenomics, 9(4), 415–422. doi: 10.2217/14622416.9.4.415 [DOI] [PubMed] [Google Scholar]
- Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, & Inui K (2007). Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem Pharmacol, 74(2), 359–371. doi: 10.1016/j.bcp.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Thule PM, & Umpierrez G (2014). Sulfonylureas: a new look at old therapy. Curr Diab Rep, 14(4), 473. doi: 10.1007/s11892-014-0473-5 [DOI] [PubMed] [Google Scholar]
- Tkac I, Klimcakova L, Javorsky M, Fabianova M, Schroner Z, Hermanova H, … Tkacova R (2013). Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab, 15(2), 189–191. doi: 10.1111/j.1463-1326.2012.01691.x [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Vormfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, … Brockmoller J (2009). The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther, 86(3), 299–306. doi: 10.1038/clpt.2009.92 [DOI] [PubMed] [Google Scholar]
- Urban TJ, Nicoletti P, Chalasani N, Serrano J, Stolz A, Daly AK, … Fontana RJ (2017). Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B *35:02 as a risk factor. J Hepatol, 67(1), 137–144. doi: 10.1016/j.jhep.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen N, Nijpels G, Becker ML, Deshmukh H, Zhou K, Stricker BH, … Pearson ER (2012). A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia, 55(7), 1971–1977. doi: 10.1007/s00125-012-2537-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Martin-Castillo B, & Menendez JA (2011). Metformin activates an ataxia telangiectasia mutated (ATM)/Chk2-regulated DNA damage-like response. Cell Cycle, 10(9), 1499–1501. doi: 10.4161/cc.10.9.15423 [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Yin OQ, Tomlinson B, & Chow MS (2008). OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics, 18(7), 637–645. doi: 10.1097/FPC.0b013e328302cd41 [DOI] [PubMed] [Google Scholar]
- Woods A, Leiper JM, & Carling D (2012). The role of ATM in response to metformin treatment and activation of AMPK. Nat Genet, 44(4), 360–361. doi: 10.1038/ng.2235 [DOI] [PubMed] [Google Scholar]
- Yee SW, Chen L, & Giacomini KM (2012). The role of ATM in response to metformin treatment and activation of AMPK. Nat Genet, 44(4), 359–360. doi: 10.1038/ng.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Xu XJ, Yin JY, Wu J, Chen X, Gong ZC, … Liu ZQ (2010). KCNJ11 Lys23Glu and TCF7L2 rs290487(C/T) polymorphisms affect therapeutic efficacy of repaglinide in Chinese patients with type 2 diabetes. Clin Pharmacol Ther, 87(3), 330–335. doi: 10.1038/clpt.2009.242 [DOI] [PubMed] [Google Scholar]
- Yu W, Hu C, Zhang R, Wang C, Qin W, Lu J, … Jia W (2011). Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clin Pharmacol Ther, 89(3), 437–442. doi: 10.1038/clpt.2010.351 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu X, Kuang H, Yi R, & Xing H (2007). Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res Clin Pract, 77(1), 58–61. doi: 10.1016/j.diabres.2006.10.021 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Si D, Chen X, Lin N, Guo Y, Zhou H, & Zhong D (2007). Influence of CYP2C9 and CYP2C19 genetic polymorphisms on pharmacokinetics of gliclazide MR in Chinese subjects. Br J Clin Pharmacol, 64(1), 67–74. doi: 10.1111/j.1365-2125.2007.02846.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, … Moller DE (2001). Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest, 108(8), 1167–1174. doi: 10.1172/jci13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, … Pearson ER (2011). Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet, 43(2), 117–120. doi: 10.1038/ng.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Donnelly L, Burch L, Tavendale R, Doney AS, Leese G, … Pearson ER (2010). Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin Pharmacol Ther, 87(1), 52–56. doi: 10.1038/clpt.2009.176 [DOI] [PubMed] [Google Scholar]
- Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, … Pearson ER (2009). Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes, 58(6), 1434–1439. doi: 10.2337/db08-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Yee SW, Seiser EL, van Leeuwen N, Tavendale R, Bennett AJ, … Pearson ER (2016). Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet, 48(9), 1055–1059. doi: 10.1038/ng.3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xia L, & Wang J (2007). Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos, 35(10), 1956–1962. doi: 10.1124/dmd.107.015495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Guo Y, Ye W, Wang Y, Li X, Tian Y, … Yan J (2014). RS11212617 is associated with metformin treatment response in type 2 diabetes in Shanghai local Chinese population. Int J Clin Pract, 68(12), 1462–1466. doi: 10.1111/ijcp.12534 [DOI] [PubMed] [Google Scholar]
- Zimdahl H, Haupt A, Brendel M, Bour L, Machicao F, Salsali A, … Staiger H (2017). Influence of common polymorphisms in the SLC5A2 gene on metabolic traits in subjects at increased risk of diabetes and on response to empagliflozin treatment in patients with diabetes. Pharmacogenet Genomics, 27(4), 135–142. doi: 10.1097/fpc.0000000000000268 [DOI] [PubMed] [Google Scholar]
- Zimdahl H, Ittrich C, Graefe-Mody U, Boehm BO, Mark M, Woerle HJ, & Dugi KA (2014). Influence of TCF7L2 gene variants on the therapeutic response to the dipeptidylpeptidase-4 inhibitor linagliptin. Diabetologia, 57(9), 1869–1875. doi: 10.1007/s00125-014-3276-y [DOI] [PMC free article] [PubMed] [Google Scholar]


