Abstract
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) has been shown to redirect the site of virus assembly in polarized epithelial cells. To test whether localization of the glycoprotein exclusively to the endoplasmic reticulum (ER) could redirect virus assembly to that organelle in nonpolarized cells, an ER -retrieval signal was engineered into an epitope-tagged variant of Env. The epitope tag, attached to the C terminus of Env, did not affect the normal maturation and transport of the glycoprotein or the incorporation of Env into virions. The epitope-tagged Env was also capable of mediating syncytium formation and virus entry with a similar efficiency to that of wild-type Env. When the epitope was modified to contain a consensus K(X)KXX ER retrieval signal, however, the glycoprotein was no longer proteolytically processed into its surface and transmembrane subunits and Env could not be detected at the cell surface by biotinylation. Endoglycosidase H analysis revealed that the modified Env was not transported to the Golgi apparatus. Immunofluorescent staining patterns were also consistent with the exclusion of Env from the Golgi. As expected, cells expressing the modified Env failed to form syncytia with CD4+ permissive cells. Despite this tight localization of Env to the ER, when the modified Env was expressed in the context of virus, virions continued to be produced efficiently from the plasma membrane of transfected cells. However, these virions contained no detectable glycoprotein and were noninfectious. Electron microscopy revealed virus budding from the plasma membrane of these cells, but no virus was seen assembling at the ER membrane and no assembled virions were found within the cell. These results suggest that the accumulation of Env in an intracellular compartment is not sufficient to redirect the assembly of HIV Gag in nonpolarized cells.
Human immunodeficiency virus type 1 (HIV-1), like a type C retrovirus, assembles its capsid at the plasma membrane just prior to, or concomitant with, budding from the cell (21). HIV-1 acquires its envelope from a portion of the plasma membrane enriched with viral glycoproteins as it buds. Most retroviral envelope glycoproteins accumulate at the plasma membrane and are incorporated into virions in this manner. However, one exception appears to be the envelope glycoproteins of foamy viruses (spumaviruses). These retroviruses, which have been known to mature primarily at intracellular membranes, were recently found to encode a potential K(X)KXX consensus endoplasmic reticulum (ER) retrieval signal at the C terminus of the Env protein (18). This cytoplasmic signal sequence has been identified in several ER-resident membrane proteins (23, 30) and has been shown to bind coatomer, a cytosolic protein component of the coat complex involved in retrograde vesicular cycling of proteins back to the ER (4). It was recently shown that the spumaviruses do accumulate their Env proteins within the ER and that this signal is responsible for Env retrieval (17). The way in which accumulation of Env in the ER affects the assembly of these viruses is poorly understood. Nevertheless, foamy virus mutants lacking a functional ER retrieval signal have been found to bud predominantly from the plasma membrane whereas wild-type foamy virus assemble largely intracellularly (18a).
The method used by retroviruses to enrich their envelopes with Env protein is poorly understood. One possible mechanism that appears to be used during assembly of HIV-1 involves active incorporation of Env into the assembling virion via an interaction with Gag protein (21). Mutations within the matrix protein (MA) of HIV-1 which block the incorporation of Env into virus (8, 14, 15, 39) have been described, suggesting a necessary interaction between the MA and Env proteins.
The Env sequence requirements for incorporation appear to differ among different retroviruses, even among retroviruses using the same assembly pathway and within the same retrovirus family (21). The Mason-Pfizer monkey virus requires a full-length Env cytoplasmic tail for normal levels of incorporation into virions (1), while Rous sarcoma virus appears to have no requirement for a specific Env cytoplasmic tail, incorporating tailless native Env and foreign glycoproteins containing long cytoplasmic tails (6, 7, 34, 38). Simian immunodeficiency virus and HIV-2 incorporate their Env proteins more efficiently if the cytoplasmic tail is truncated (24, 29). Conversely, their close relative, HIV-1, requires a full cytoplasmic tail for efficient incorporation of its Env protein in most cell types (11, 14, 16, 40).
The earliest evidence for a specific interaction between HIV-1 Env and Gag during virus assembly was the ability of Env to redirect the site of virus budding in polarized epithelial cells (32). HIV-1 Env is preferentially transported to the basolateral membrane of polarized epithelial cells (31). In the absence of Env, HIV-1 Gag assembled and budded from both the apical and basolateral membranes of these cells. However, when coexpressed with Env, Gag assembled and budded preferentially from the site of Env accumulation, the basolateral membrane. When truncated forms of Env were analyzed in similar experiments (28), it was found that the ability of Env to redirect the site of virus budding was lost following truncation of its cytoplasmic tail. This indicated that the C terminus of the Env cytoplasmic tail either interacts with Gag or is important to maintain a specific conformation of the cytoplasmic domain necessary for Env-Gag interaction. Additional evidence for an interaction between Env and Gag is discussed below.
In this report, we address the potential for HIV-1 Env containing a K(X)KXX cytoplasmic ER retrieval signal to redirect the site of virus assembly and budding from the plasma membrane to the ER in nonpolarized cells. An initial Env mutant containing a C-terminal epitope tag was generated to test whether the cytoplasmic tail could accommodate exogenous sequences at its C terminus and remain functional. The epitope tag was then modified to contain the ER retrieval signal. The characterization of both of these mutant glycoproteins and their effects on virus assembly are described.
MATERIALS AND METHODS
Cell culture.
COS-1, CV-1, and 293T cells were obtained from the American Type Culture Collection. HeLa-CD4/LTR–β-gal cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, and were originally contributed by Michael Emerman. All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, penicillin, and streptomycin. HeLa-CD4/LTR–β-gal cells were additionally maintained in G418 and hygromycin as recommended by the contributor.
Mutagenesis and construction of plasmids.
For construction of the FLAG mutant, a nucleotide sequence encoding the 8-amino-acid FLAG epitope (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys [Kodak-IBI]), preceded by a 3-amino-acid spacer peptide (Asn-Ser-Gly), was introduced into the env gene of the HIV-1 HXB2 immediately preceding the stop codon by using the pALTER mutagenesis system (Promega). The regions complementary to and flanking the mutagenic oligonucleotide were sequenced (Sequenase; U.S. Biochemicals). A BamHI-XhoI HXB2 env fragment was then subcloned into pSRHS3. This plasmid expresses the HXB2 env, rev, and tat genes under control of the simian virus 40 (SV40) late promoter. The vector also expresses the SV40 large T antigen, contains the SV40 origin of replication, and uses the Mason-Pfizer monkey virus 3′ long terminal repeat (LTR) for transcriptional termination and polyadenylation (10).
For conversion of the FLAG mutant to the ER retention signal (ERRS) mutant, a PCR product was generated by using a mutagenic primer and pSRHS3 containing the FLAG mutation as the template. The resulting product was then digested with BspEI and XhoI and subcloned into a derivative of pSRHS3 in which a unique BspEI site in the M-PMV LTR had been deleted by digestion with BspEI followed by filling in by the Klenow fragment of Escherichia coli DNA polymerase I and religation. The entire coding region originating from the PCR product was sequenced, and two separate clones were selected for phenotypic characterization in parallel. For construction of proviral clones, BamHI-XhoI fragments containing each of the two mutations were subcloned into pNL4-3, a proviral clone of the HIV-1 NL4-3. HXB2 and NL4-3 are highly homologous clade B virus strains.
Glycoprotein expression and radioimmunoprecipitation.
COS-1 cells at approximately 80% confluency on 60-mm plates were transfected with 3 μg of pSRHS3 wild-type or mutant plasmid by using DEAE-dextran. At 36 to 48 h posttransfection, the cells were starved for 20 min in cysteine- and methionine-deficient DMEM and then metabolically radiolabelled for 30 min with 125 μCi of [35S]cysteine-[35S]methionine in 350 μl of cysteine- and methionine-deficient DMEM per plate. The cells were then washed twice in DMEM and chased in 1.2 ml of complete growth medium for 3 h. After the chase, the growth medium was collected and the cells were lysed in lysis buffer for 10 min on ice. Both the cell lysate and the culture medium were microcentrifuged for 5 min to remove cellular debris, and the supernatants were transferred to fresh tubes for immunoprecipitation by incubation with serum from an AIDS patient for 1 h at 4°C, followed by incubation with fixed Staphylococcus aureus (Staph A) for 30 min at room temperature. The immunoprecipitate was pelleted and washed three times in lysis buffer containing 0.1% sodium dodecyl sulfate (SDS) and once in 20 mM Tris-HCl (pH 6.8) before being subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell-cell fusion assay.
COS-1 cells in 35-mm plates were transfected with 1 μg of pSRHS3 wild-type or mutant plasmid by using DEAE-dextran. At 24 h posttransfection, the cells were trypsinized, mixed with HeLa-CD4/LTR–β-gal cells at a ratio of approximately 1:10, and replated. Since the pSRHS3 vector expresses both Env and Tat, any cells transfected with this vector that fuse with a MAGI cell will provide Tat for transactivation of β-galactosidase gene transcription. At 24 h later, the cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyronoside (X-Gal) as described previously (25). The total number of blue (β-galactosidase-positive) syncytia per plate was determined microscopically by using a counting grid.
Cell surface biotinylation.
COS-1 cells transfected and metabolically radiolabeled as described above were washed three times in ice-cold phosphate-buffered saline (PBS) containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS-C/M) at 36 to 72 h posttransfection. The cells were then incubated in 0.5 mg of sulfo-NHS-biotin (Pierce) per ml of PBS-C/M for 30 min on ice. To quench the reaction, the cells were washed once with DMEM for 10 min on ice and then twice with ice-cold PBS-C/M containing 20 mM glycine. The cells were lysed in lysis buffer containing 20 mM glycine and were immunoprecipitated as described above. The immunoprecipitate was boiled for 5 min in PBS–2% SDS to denature the antibody, and the Staph A was removed by pelleting. The supernatant was diluted with PBS to a final SDS concentration of 0.05% and incubated for 1 h with streptavidin-agarose beads (Pierce). The beads were then washed as described above for an immunoprecipitate, and the resulting pellet was resuspended by boiling in protein-loading buffer–12% SDS for 5 min before being subjected to SDS-PAGE.
Endo H treatment.
COS-1 cells transfected and metabolically radiolabeled as described above were lysed immediately after the pulse or were chased for 1, 2, or 3 h. Lysates were immunoprecipitated as described above, and the resulting washed pellets were boiled for 5 min in 0.04% SDS–2% 2-mercaptoethanol–50 mM sodium citrate (pH 5.3). The Staph A was removed by pelleting at room temperature, and the supernatant was divided into two tubes. An equal volume of 2× endoglycosidase H (endo H) buffer (50 mM sodium citrate [pH 5.3], 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 200 μg of soybean trypsin inhibitor per ml, 2 μg of leupeptin per ml, 2 μg of pepstatin A per ml) was added to each tube, and 1 mU of endo H (Boehringer Mannheim) was added to one of the tubes. The samples were incubated for 12 h at 37°C. An equal volume of 2× protein-loading buffer was then added, and the samples were reboiled for 2 min before being loaded onto SDS-PAGE gels.
Immunofluorescent staining.
CV-1 cells transfected as described above were plated 24 h posttransfection onto glass coverslips. The cells were fixed and stained for total-cell immunofluorescence 48 h later with an anti-gp120 monoclonal antibody as described previously (36). They were photographed at a magnification of ×400 by using a fluorescence microscope (Zeiss).
Virus production and entry.
For analysis of virus production and virus entry, 293T cells in 100-mm plates were transfected with 10 μg of pNL4-3 wild-type or mutant plasmid by a modified calcium phosphate-mediated method (2). At 48 h posttransfection, the culture medium was filtered through a 0.45-μm-pore-size filter. To quantitate the relative amount of virus production, 25-μl aliquots were removed for reverse transcriptase (RT) assays in triplicate as described previously (10).
For quantitation of virus entry, after determination of the mean levels of RT activity for each culture, aliquots of filtered medium were normalized for relative RT activity by dilution in complete growth medium. DEAE-dextran was added to a final concentration of 5 μg per ml, and 0.3 ml was added to 8 × 105 HeLa-CD4/LTR–β-gal cells in triplicate wells of a 12-well plate. The virus was allowed to adsorb for 2 h and then an additional 2 ml of complete growth medium was added. At 48 h postinfection, the cells were stained with X-Gal as described previously (25). The number of blue syncytia per 16-mm2 field was determined for three random, nonoverlapping low-power (×100) microscopic fields in each of the triplicate wells by using a grid. The total number of blue syncytia per well was then estimated by multiplying the mean number of blue syncytia per 16 mm2 by the total number of fields per well.
Virus pelleting and EM analysis.
For analysis of virus assembly, 293T cells were transfected as described above, except that after transfection, the cells were washed twice in DMEM and split between two 100-mm plates.
For analysis of glycoprotein expression and incorporation into virions, one of the plates was metabolically radiolabelled 24 h posttransfection (48 h after the addition of DNA) with 625 μCi of [35S]cysteine-[35S]methionine (DuPont-NEN) in 2.5 ml of cysteine- and methionine-deficient DMEM plus 10% complete growth medium for 16 h. The culture medium was then filtered through a 0.45-μm filter, and 0.8 ml was transferred to each of three microcentrifuge tubes for centrifugation at 13,000 × g for 1 h. The resulting virus pellets were resuspended in lysis buffer and combined into one tube. The cells were lysed in lysis buffer, and cellular debris was removed by centrifugation for 5 min. Viral proteins were immunoprecipitated from each sample and analyzed by SDS-PAGE as described above.
For analysis of the site of virus assembly and budding, cells from the other plate were suspended in PBS, pelleted, and fixed in 1% glutaraldehyde in preparation for analysis by electron microscopy (EM).
RESULTS
Modification of HIV-1 Env to contain an ER retrieval signal.
Linear peptide signals have been identified which permit errant ER-resident proteins to be retrieved from the Golgi and cycled back to the ER (30). The best characterized of these signals is the KDEL retrieval signal commonly found on resident soluble proteins in the lumen of the ER. More recently, a K(X)KXX consensus sequence found at the terminus of the cytoplasmic tail of ER-resident type I membrane proteins has been identified to function as an ER retrieval signal (23).
To address whether the assembly of HIV-1 can be redirected by the localization of Env to an intracellular compartment, we modified the C terminus of HIV-1 Env to contain a consensus K(X)KXX ER retrieval signal (Fig. 1). The C terminus of HIV-1 Env is hydrophobic and has been proposed to interact with membranes (19). For this reason, we first engineered an epitope tag onto the C-terminus of the cytoplasmic tail of Env to facilitate the recognition of the KKXX signal. We chose the highly charged 8-amino-acid FLAG marker peptide as an epitope because of its high degree of surface accessibility. We also inserted a 3-amino-acid spacer sequence between the Env C terminus and the FLAG marker peptide to avoid disrupting any membrane interaction or structural conformation crucial to the function of the Env cytoplasmic tail. The sequence of the epitope was then modified to contain the consensus dilysine motif which constitutes the ER retrieval signal, as well as an additional lysine residue. The additional lysine was added because a previous study suggested that it facilitates the ER localization of the human foamy virus Env protein (17).
FIG. 1.
Conversion of a C-terminal epitope tag to an ER retrieval signal. The five C-terminal amino acid residues of the HIV-1 envelope glycoprotein cytoplasmic tail are shown at the top. The 8-amino-acid FLAG epitope (DYKDDDDK) plus an additional 3-amino-acid spacer peptide (NSG) were added to the C terminus of Env by site-directed mutagenesis. The FLAG epitope sequence was mutated to introduce the consensus ER retrieval signal plus an additional lysine residue at position −4 to −5, which is not a necessary part of the consensus signal motif (bottom).
HIV-1 Env containing an ER retrieval signal is not proteolytically processed to yield gp120.
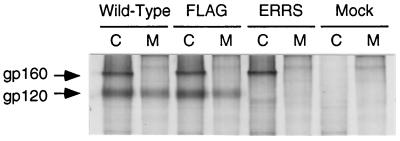
The epitope-tagged (FLAG) and ER retrieval signal-containing (ERRS) Env precursor glycoproteins (gp160) were expressed and maintained at levels similar to wild-type Env after a 3-h chase period (Fig. 2), indicating that neither Env mutant was degraded significantly. Although the wild-type and FLAG Env proteins were proteolytically processed to yield the surface glycoprotein subunit (gp120) (Fig. 2, lanes Wild-Type C and FLAG C), no normal gp120 band was detected for the ERRS Env (lane ERRS C). A faint band at approximately 100 kDa, migrating faster than gp120, was produced with the ERRS mutant, which was not present in the mock-transfected cell lysate (compare lane ERRS C with other C lanes). The identity of this band is unknown, but it is suspected to be an Env cleavage product, perhaps the gp120 region of Env which has had a subset of its mannose-rich oligosaccharide side chains trimmed by mannosidases located in the cis compartment of the Golgi apparatus before being cycled back to the ER. Evidence for the identity of this band is discussed in later sections. Similarly, gp120 was released into the media from cells expressing either wild-type or FLAG Env but not from cells expressing ERRS Env (Fig. 2, lanes M). The 100-kDa band seen in the lysates of cells expressing ERRS Env was also absent from the culture media (lane ERRS M). Finally, no gp160 could be detected in the media of cells expressing any of the three Env constructs, confirming that the C-terminal modifications in Env did not in some way disrupt the stable association of the glycoprotein with the membrane. The lack of production of a normal gp120 product following a 3-h chase suggests that the ER retrieval signal is functional in ERRS Env, preventing its transport through the Golgi.
FIG. 2.
Expression and processing of envelope glycoproteins. COS-1 cells expressing glycoproteins were metabolically radiolabeled and chased for 3 h. The cell lysate (C) and culture medium (M) were collected, and HIV proteins were immunoprecipitated for analysis by SDS-PAGE (8% polyacrylamide).
ERRS Env does not mediate syncytium formation.
Proteolytic cleavage of gp160 is required for the function of Env in mediating membrane fusion following binding to the CD4 receptor. Since no gp120 was detected in cells expressing ERRS Env in the preceding experiment, these cells would not be expected to form syncytia with cells expressing CD4. To test this, COS-1 cells expressing either wild-type or mutant Env were mixed at a ratio of approximately 1:10 with HeLa-CD4/LTR–β-gal cells and replated. HeLa-CD4/LTR–β-gal cells express high levels of CD4 and contain a lacZ reporter gene under control of the HIV-1 LTR (25). If the COS-1 cells express an Env molecule which is capable of mediating fusion with the HeLa-CD4/LTR–β-gal cells, the Tat coproduced from the pSRHS Env expression plasmids within the COS-1 cells will activate expression of the lacZ reporter gene. Upon fixation and staining with X-Gal, these cultures should contain clearly visible blue syncytia. The blue staining is most intense within the nuclei, since the β-galactosidase encoded by these cells has been modified to contain a nuclear localization signal (25). This nuclear staining facilitates the quantitation of syncytia as well as the number of nuclei they contain.
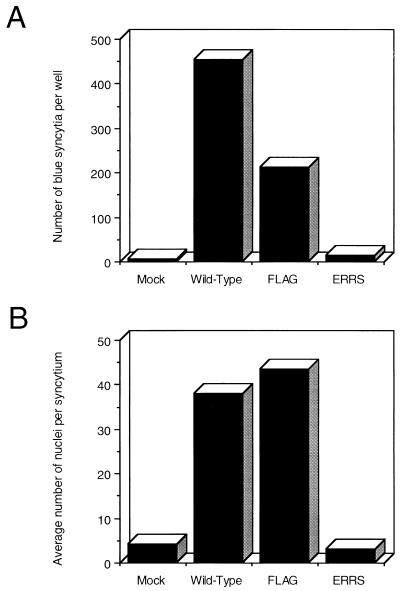
Figure 3 shows the results of a representative experiment. Both wild-type and FLAG Env led to the production of large numbers of blue syncytia (Fig. 3A). Although the FLAG mutant resulted in about half as many syncytia as did the wild-type Env in this experiment, the syncytia produced by both Env proteins were of similar size (Fig. 3B). Since the absolute number of syncytia can be affected by both relative transfection efficiencies and variations in cell mixing, the average sizes of syncytia produced in different cultures provide a more accurate measure of the relative fusogenicities of different Env proteins. Based on this criteria, the FLAG mutant Env appears to be as fusogenic as wild-type Env.
FIG. 3.
Envelope glycoprotein-mediated syncytium formation. COS-1 cells expressing glycoprotein were mixed 1:10 with HeLa-CD4/LTR–β-gal indicator cells and replated. The cells were stained in situ with X-Gal, and blue syncytia were quantitated microscopically. (A) The average number of syncytia per 35-mm well was determined from two wells, counting the entire well with the aid of a grid. (B) The average number of nuclei per syncytium was determined by quantitating 25 syncytia in each of two wells and averaging the total.
In contrast to the FLAG mutant, however, the ERRS mutant produced only three times as many blue syncytia as were found in the mock culture as background and only 7% of the number of syncytia produced by the FLAG mutant (Fig. 3A). Perhaps more importantly, the average size of these syncytia (based on the number of nuclei per syncytium) was smaller than that of the background syncytia in the mock culture (Fig. 3B), indicating that these are unlikely to be the result of true Env-mediated cell-cell fusion events. Indeed, no syncytia were apparent in unstained cultures when the ERRS construct was transfected directly into HeLa-T4 cells (data not shown), suggesting that the blue syncytia observed here with the ERRS mutant represent the background level of the β-galactosidase assay. Therefore, the ERRS mutant appears to be unable to mediate syncytium formation, presumably due to its inability to be transported to the cell surface and to the lack of normal processing of gp160 to gp120.
ERRS Env is not transported to the cell surface.
Although gp120 was not produced from cells expressing the ERRS Env mutant, it was possible that the mutant was transported through the Golgi but not processed. In this case, the preceding cell-cell fusion assay would still have failed to indicate the presence of ERRS Env on the cell surface, since the Env would not have been cleaved and thus would have been nonfusogenic. Therefore, to determine whether the mutant glycoprotein was transported to the cell surface, we biotinylated proteins exposed on the cell surface with the membrane-impermeable reagent Sulfo-NHS-biotin (27). This reagent reacts with primary amines, forming a covalent linkage. After quenching of the biotinylation reaction, radiolabeled Env was purified from cell lysates by immunoprecipitation. The immunoprecipitate was then boiled in 2% SDS to denature antibodies, and the Staph A was removed by pelleting. Biotinylated Env proteins were then separated from nonbiotinylated Env proteins by binding to streptavidin-agarose beads.
As can be seen in Fig. 4, equivalent amounts of gp120 are present at the cell surface for both wild-type Env and FLAG Env. In addition, a small amount of uncleaved gp160 precursor could be detected on the cell surface. This is consistent with our previous findings which demonstrate that in this expression system, some gp160 can escape proteolytic processing and be transported to the cell surface (36). In contrast, however, no glycoprotein, cleaved or uncleaved, could be detected on the surface of cells expressing the ERRS mutant. Therefore, it appears that the ER retrieval signal prevents the processing of Env to gp120 by preventing its transport through the Golgi and on to the cell surface.
FIG. 4.
Biotinylation of envelope glycoproteins expressed on the cell surface. Cells expressing radiolabeled glycoprotein were incubated with biotinylation reagent on ice. Following lysis and immunoprecipitation with patient serum to isolate viral proteins, pellets were boiled in SDS to denature antibody and biotinylated proteins were isolated by binding to streptavidin-agarose. The biotinylated viral proteins were then analyzed by SDS-PAGE (8% polyacrylamide).
ERRS Env is localized to the ER.
To confirm that the ER retrieval signal is preventing the transport of Env through the Golgi, we subjected the ERRS glycoprotein mutant to digestion with endoglycosidase H (endo H). This enzyme removes the high-mannose-type carbohydrate side chains which are added to glycoproteins in the ER but cannot remove carbohydrate side chains which have been modified in the Golgi apparatus (20). For this experiment, cells expressing FLAG or ERRS glycoprotein mutants were pulse-labeled and then chased for 1, 2, or 3 h before being subjected to lysis and immunoprecipitation. The immunoprecipitates were then divided into two tubes for either digestion with endo H or mock treatment.
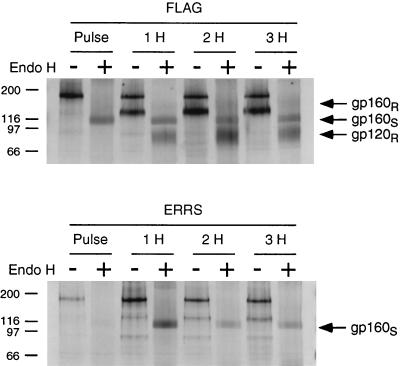
As seen in Fig. 5, epitope-tagged Env was completely sensitive to endo H digestion following pulse-labeling, resulting in a product of approximately 95 kDa, as would be expected for the fully deglycosylated 867-amino-acid epitope-tagged Env protein (Fig. 5, FLAG, lane Pulse +). After a chase period of 1 h or more, a second band which migrated at approximately 80 kDa appeared after endo H treatment, consistent with partially endo H-resistant gp120 resulting from the modification of 13 of the 24 carbohydrate side chains of gp120 in the Golgi (26). In addition, a faint and diffuse band could be seen migrating at approximately 130 to 150 kDa following the 3-h chase, which is consistent with a partially endo H-resistant gp160 species (FLAG, lane 3H +). In contrast to the epitope-tagged Env, the ERRS-containing Env remained completely sensitive to endo H digestion even after a 3-h chase period, with only a single band, corresponding to a fully deglycosylated gp160, resulting from endo H digestion (ERRS, lane 3H +). Thus, the ERRS signal prevented the acquisition of endo H-resistant carbohydrate side chain modifications, presumably because transport of the Env protein through the Golgi was prevented.
FIG. 5.
Endo H treatment of envelope glycoproteins. Cells were radiolabeled and chased for 1 h (lanes 1H), 2 h (lanes 2H), or 3 h (lanes 3H). Glycoproteins were immunoprecipitated from the resulting cell lysates and split in half for either endo H digestion (lanes +) or mock treatment (lanes −). Results for the FLAG and ERRS mutants are shown in the top and bottom panels, respectively. gp160S indicates glycoprotein precursor which was sensitive to endo H digestion. gp120R and gp160R indicate processed glycoprotein and glycoprotein precursor, respectively, which was resistant to endo H digestion.
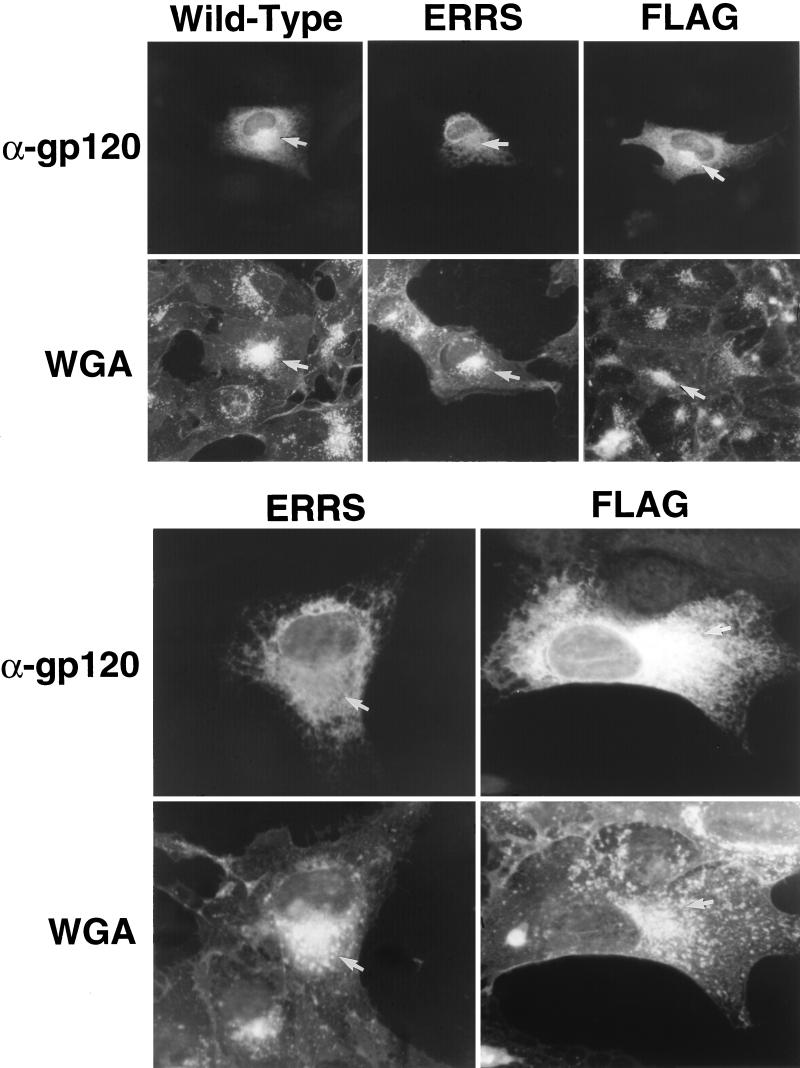
If the ERRS-containing Env is indeed not transported to the Golgi or at least is not allowed to accumulate there, it should be possible to observe this lack of Env in the Golgi through immunofluorescent staining of Env in cells expressing the mutant. To examine this, CV-1 cells expressing each of the Env constructs were fixed and stained by indirect immunofluorescence with a monoclonal antibody to the V3 loop of gp120/gp160 and a Texas Red-conjugated secondary antibody. The location of the Golgi complex was determined by costaining with fluorescein-conjugated wheat germ agglutinin (WGA).
In cells transfected with either the wild-type or epitope-tagged Env expression vectors, reticular staining, characteristic of the ER, and bright perinuclear fluorescent staining, consistent with localization of Env in the Golgi apparatus, could be observed (Fig. 6, Wild-Type and FLAG, α-gp120). The latter was coincident with the bright staining of the Golgi complex seen with fluorescein-conjugated WGA (Fig. 6, Wild-Type and FLAG, WGA). In contrast, cells expressing ERRS Env lacked any discrete region of intense staining, displaying instead only a reticular staining of the cytoplasm (Fig. 6, ERRS, α-gp120). Indeed, the area of the cell most highly stained by fluorescein-conjugated WGA showed a reduced intensity of α-gp120 staining (Fig. 6, ERRS, α-gp120). This staining pattern is consistent with the localization of the ERRS mutant to the ER and its exclusion from the Golgi apparatus. These results confirm that the ER retrieval signal in the ERRS mutant is functional.
FIG. 6.
Internal immunofluorescent staining of envelope glycoproteins expressed in CV-1 cells. CV-1 cells expressing glycoprotein were fixed and stained with a monoclonal antibody to gp120 (α-gp120), followed by detection with a Texas Red-conjugated secondary antibody. They were then costained with fluorescein-conjugated WGA to identify the location of the Golgi complex. Cells in the upper panels are shown at a magnification of ×364, and those in the lower panels are shown at a magnification of ×910. Cells expressing the ERRS mutant glycoprotein lack the bright staining of the Golgi complex seen in control cells.
ERRS Env does not affect extracellular virus production.
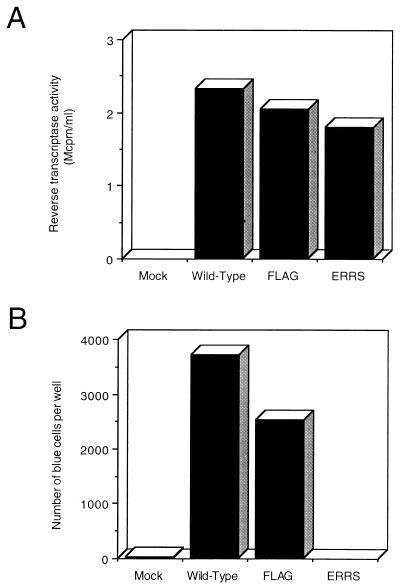
The preceding sections demonstrate that the ER retrieval signal incorporated into the C-terminal marker peptide is functional in retrieving Env before its transport through the Golgi and that, as a result, Env accumulates in the ER. To test how this intracellular localization of Env affects virus assembly and budding, both the FLAG and ERRS modifications were transferred to the env gene of an NL4-3 proviral clone. If the site of virus assembly and budding is dependent on the localization of Env, virus containing ERRS Env might be expected to assemble at the ER membrane and bud into the ER. Two days after the transfection of 293T cells with the proviral construct, the culture medium was collected and filtered to remove cellular debris. Aliquots of the filtered medium were assayed for RT activity as a measure of the relative level of virus production. As can be seen in Fig. 7A, each of the proviral constructs produced high levels of RT activity. The FLAG and ERRS proviruses produced only slightly reduced levels of RT activity compared to the wild-type provirus, indicating that these modifications do not significantly affect the production of virus from cells. Therefore, the localization of ERRS Env to the ER does not prevent the efficient release of virus from the cell.
FIG. 7.
Virus expression and entry. pNL4-3 proviral constructs were expressed in 293T cells. (A) Culture supernatants were collected and filtered, and the relative levels of virus were quantitated by measuring RT activity. (B) The volumes of the supernatants were normalized based on their relative levels of RT activity and used to infect HeLa-CD4/LTR–β-gal cells. The number of blue cells per well was estimated by counting the number of blue cells in each of three 16-mm2 fields at low power for three duplicate wells. The average number of blue cells per field was then multiplied by the total number of fields per well.
Virus produced from cells expressing ERRS Env is noninfectious.
To determine whether the virus produced from the FLAG and ERRS proviral constructs was infectious and thus contained functional Env, aliquots of the RT-assayed media were used to infect HeLa-CD4/LTR–β-gal cells. Aliquots were normalized according to their relative RT values and incubated with HeLa-CD4/LTR–β-gal cells for 2 h. Fresh medium was then added, and the cells were fixed and stained with X-Gal 2 days later.
Figure 7B shows that virus produced from both wild-type and FLAG proviral constructs resulted in equivalent numbers of blue cells, indicating that these viruses were infectious. In contrast to the FLAG provirus, however, the ERRS construct yielded only background numbers of blue cells. Therefore, the virus produced from cells expressing ER-localized Env is completely noninfectious.
ERRS Env is not incorporated into virions.
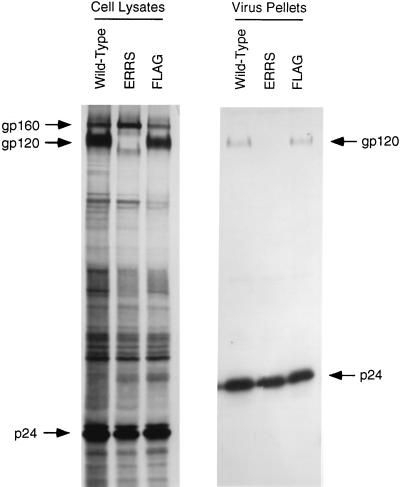
Since it was possible that virions were being assembled in the ER and released after transport through the secretory pathway, we determined, by pelleting virus for protein analysis, whether the virions produced from the ERRS provirus contained any glycoprotein. At 24 h after transfection with either wild-type or mutant provirus, 293T cells were metabolically radiolabeled overnight. The culture medium was then filtered through a 0.45-μm filter to remove cellular debris and centrifuged to pellet virions. The virus pellet was resuspended in lysis buffer for immunoprecipitation and analysis of proteins by SDS-PAGE.
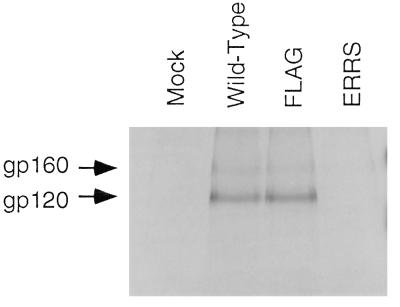
Figure 8 demonstrates that glycoprotein was efficiently expressed by all three proviral constructs in the cell lysates. However, once again, no gp120 was produced by the ERRS construct. The faint 100-kDa band, as well as a distinct band migrating just ahead of gp160, was also once again detected for this mutant. In the viral pellets, similar amounts of gp120 were detected for both wild-type and FLAG constructs, but no form of glycoprotein was detected in the pellet from the ERRS construct. p24 levels were similar in each of the viral pellets, indicating that the lack of glycoprotein in the ERRS lane was not due to a decreased amount of pelleted virus. These results demonstrate that the localization of Env to the ER results in the efficient production of virions which lack envelope glycoprotein and are thus noninfectious.
FIG. 8.
Incorporation of glycoprotein into virions. Virus was pelleted from the filtered culture supernatant of radiolabeled 293T cells. The viral pellet was resuspended in lysis buffer, and viral proteins were immunoprecipitated from both the viral and cell lysates in parallel by using patient serum prior to analysis by SDS-PAGE.
Virus produced from cells expressing ERRS Env buds from the plasma membrane.
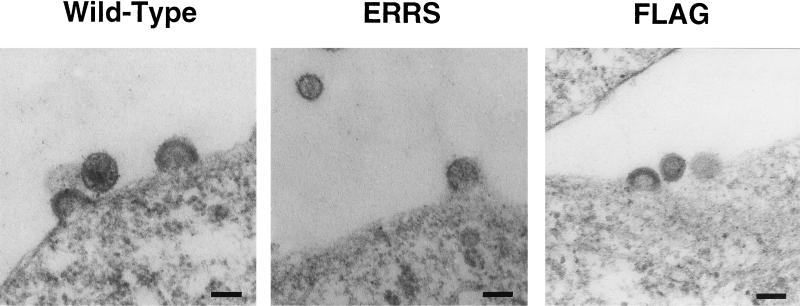
The lack of incorporation of ERRS Env into virions suggested that virus was budding exclusively from the plasma membrane, which was shown in Fig. 4 to be devoid of glycoprotein. Alternatively, virus could be budding into the ER but failing to incorporate Env for some other reason. Yet another possibility was that only a subset of virus was assembling at the ER membrane but was unable to completely bud for some reason. To distinguish between these different possibilities, sections of 293T cells expressing high levels of virus were examined by EM for budding structures. Representative photomicrographs are shown in Fig. 9. In each cell culture, regardless of the construct being expressed, virions were found budding from the plasma membrane. No intracellular budding structures could be found in the cells expressing the ERRS construct, which were distinguishable from mock-transfected cells, and no budded virus could be found in an intracellular compartment. These results confirm that the lack of incorporation of ERRS Env into virions was due to the exclusive budding of virus from the plasma membrane.
FIG. 9.
Budding of virus from the plasma membrane. 293T cells expressing pNL4-3 proviral constructs were fixed and sectioned for EM analysis. Bar, 100 nm.
DISCUSSION
We have shown that accumulation of the HIV envelope glycoprotein within the ER is insufficient to redirect the assembly of virions at the ER membrane. The introduction of an 8-amino-acid highly charged epitope tag and a 3-amino-acid spacer peptide to the C terminus of Env did not affect the ability of the protein to mediate both cell-cell fusion and virus entry. However, when this tag was modified to contain an ER retrieval signal, the glycoprotein was no longer transported through the Golgi and thus was not processed normally or expressed on the cell surface. As a result, expression of this Env mutant (ERRS) did not result in cell-cell fusion. Importantly, expression of the ERRS mutant did not affect the normal budding of virions from the plasma membrane, and the virus which was released did not contain glycoprotein and thus was noninfectious. EM analysis of these cells failed to reveal any structures of assembling or budding virus at the ER membrane, and no virions were found within an intracellular compartment. These results, which confirm and extend similar studies by Pfeiffer et al. (35), do not provide any evidence for a functional interaction between Env and Gag proteins at this early step in glycoprotein transport.
While no mature gp120 was produced in COS-1 cells or 293T cells expressing the ERRS Env construct, a faint band migrating at approximately 100 kDa was apparent. In addition, a similarly faint band was seen migrating in a similar position ahead of gp160 following proviral expression in 293T cells (Fig. 8). This is consistent with a subset of the glycoprotein reaching the cis Golgi, where the high-mannose side chains are trimmed, before being retrieved back to the ER, resulting in a higher mobility of the glycoproteins. Since the bands of partially processed gp160 and the 100-kDa underglycosylated surface subunit are of similar intensity in 293T cells, it seems possible that the cellular enzyme which cleaves the glycoprotein precursor is at least partially active in the early compartments of the Golgi.
Evidence for the association of HIV-1 Env with Gag has increased in recent years. Deletions within the HIV-1 MA protein prevent the incorporation of glycoprotein into virions (8, 39). More recently, individual amino acid substitutions within MA were found to be sufficient for this effect, and the block to glycoprotein incorporation by these mutants was further shown to be overcome by truncating the cytoplasmic tail of Env (14, 15). These results suggested that regions near the center of the cytoplasmic tail were critical for an interaction with Gag. The existence of such an interaction was also strongly supported by a recent study demonstrating that an endocytosis signal within the cytoplasmic tail of Env was down-regulated by coexpression of the Gag precursor protein, Pr55 (12). In addition, an interaction between immobilized Env cytoplasmic domain and soluble MA has been demonstrated in vitro (3).
Nonetheless, the lack of incorporation of glycoprotein into virions, the lack of inhibition of virus budding, and the lack of virus assembly and budding within the ER, reported here and by Pfeiffer et al. (35), all suggest that localization of Env to the ER is not sufficient to target Gag assembly to this site. How might one explain the discrepancy between our results and the ability of Env to redirect the site of virus budding in polarized epithelial cells? It is unlikely that the lack of retargeting is due to the C-terminal modification of the protein, since the FLAG modification, which is just as highly charged as the ERRS sequence, had no discernible effect on the functional association of Env and Gag. Therefore, it is likely that the inability of Env to productively interact with Gag is a consequence of its localization to the ER. It is important to note that the composition of the ER membrane is likely to be very different from that of the plasma membrane. It is possible that Gag interacts with ER-localized Env but that virions cannot assemble and bud at the ER membrane because of the absence of necessary cellular factors or because certain factors in and around the ER membrane disrupt assembly and budding. This seems unlikely, however, since an MA deletion mutant of HIV-1 does bud efficiently through the ER membrane (13).
An alternate explanation for our results is that the cellular machinery which is responsible for binding to the ER retrieval signal and rerouting the protein back to the ER inhibited the interaction of Gag with the Env cytoplasmic tail. The consensus K(X)KXX signal sequence has been shown to bind and drive the polymerization of microtubules in vitro (5). In addition, coatomer protein binds to ER-retrieval signal sequences (4), and it is possible that this binding sterically interferes with Gag association. This would also seem to be unlikely, however, since the primate spumaviruses have a similar ERRS at the C terminus of a much shorter cytoplasmic domain and yet efficiently bud into this membrane compartment of the cell (17, 18). Moreover, Pal et al. (33) have reported a similar Env-independent release of Gag from cells treated with brefeldin A, in which the transport of the Env precursor is blocked at the ER. Although brefeldin A-treated cells have a profoundly disturbed Golgi complex, these results are consistent with a lack of Gag-Env interaction at the early stages of the secretory pathway.
A more likely rationale for our inability to observe redirection of Gag assembly to the ER in these experiments is that the necessary protein-protein interactions cannot take place until the env gene product has traversed the Golgi complex. Protein sorting in polarized epithelial cells appears to occur in the trans Golgi complex (37), and this is also the site of retroviral glycoprotein precursor cleavage (22). It is possible that following cleavage, conformational changes in gp41 allow a specific interaction with the assembling Gag precursors to take place. Consistent with this hypothesis, we have previously observed that cleavage-deficient mutants of HIV-1 Env are inefficiently incorporated into virions (9) whereas glycophospholipid-linked forms of the same proteins, which lack the long cytoplasmic domain, are efficiently incorporated (36). Thus, it is possible that Gag-Env interactions require conformational changes in the cytoplasmic domain that occur only after transport to the trans Golgi and proteolytic cleavage of the mature glycosylated gp160.
ACKNOWLEDGMENTS
We thank Eric Freed (NIH) for supplying the pNL4.3 proviral clone, Susan Dubay for assistance with immunofluorescence experiments, and Eugene Arms (UAB) for help with the EM analysis.
This work was supported by research grants AI-33319 and AI-33784 from the National Institutes of Health. Virus culture was performed in the UAB Center for AIDS Research Central Virus Core Facility, supported by program grant AI-27767 from the National Institutes of Health. K. Salzwedel was supported in part by training grant CA09467 from the National Institutes of Health.
REFERENCES
- 1.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;17:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosson P. Direct interaction between the envelope and matrix protein of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson P, Letourner F. Coatomer interaction with dilysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 5.Dahllof B, Wallins M, Kvist S. The endoplasmic reticulum retention signal of the E3/19K protein of adenovirus-2 is microtubule binding. J Biol Chem. 1991;266:1804–1808. [PubMed] [Google Scholar]
- 6.Dong J, Hunter E. Analysis of retroviral assembly using a vaccinia/T7-polymerase complementation system. Virology. 1993;194:192–199. doi: 10.1006/viro.1993.1249. [DOI] [PubMed] [Google Scholar]
- 7.Dong J, Roth M G, Hunter E. A chimeric avian retrovirus containing the influenza virus hemagglutinin gene has an expanded host range. J Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubay J W, Dubay S R, Shin H J, Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69:4675–4682. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goepfert P A, Shaw K L, Ritter G, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Goepfert, P. A., and M. J. Mulligan. Unpublished data.
- 19.Haffar O K, Dowbenko D J, Berman P W. The cytoplasmic tail of HIV-1 gp160 contains regions that associate with cellular membranes. Virology. 1991;180:439–441. doi: 10.1016/0042-6822(91)90054-f. [DOI] [PubMed] [Google Scholar]
- 20.Hubbard S C, Ivatt R J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- 21.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 22.Hunter E, Swanstrom R. Retroviral envelope glycoproteins. In: Swanstrom R, Vogt P K, editors. Retroviruses—strategies for replication. Berlin, Germany: Springer-Verlag KG; 1990. pp. 187–253. [Google Scholar]
- 23.Jackson M R, Nilsson T, Peterson P A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard C K, Spellman M W, Riddle L, Harris R J, Thomas J N, Gregory T J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 27.Lisanti M P, Sargiacomo M, Graeve L, Saltiel A R, Rodriguez-Boulan E. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc Natl Acad Sci USA. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulligan M J, Yamshchikov G V, Ritter G, Jr, Gao F, Jin M J, Nail C D, Spies C P, Hahn B H, Compans R W. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson T, Warren G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Current Opinion in Cell Biology. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens R J, Compans R W. Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J Virol. 1989;63:978–982. doi: 10.1128/jvi.63.2.978-982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal R, Mumbauer S, Hoke G M, Takatsuki A, Sarngadharan M G. Brefeldin A inhibits the processing and secretion of envelope glycoproteins of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1991;7:707–712. doi: 10.1089/aid.1991.7.707. [DOI] [PubMed] [Google Scholar]
- 34.Perez L G, Davis G L, Hunter E. Mutants of the Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J Virol. 1987;61:2981–2988. doi: 10.1128/jvi.61.10.2981-2988.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 36.Salzwedel K, Johnston P B, Roberts S J, Dubay J W, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons K, Wandinger-Ness A. Polarized sorting in epithelia. Cell. 1990;62:207–210. doi: 10.1016/0092-8674(90)90357-k. [DOI] [PubMed] [Google Scholar]
- 38.Young J A, Bates P, Willert K, Varmus H E. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science. 1990;250:1421–1423. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- 39.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope glycoproteins into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]