Abstract
Hypoxia is a critical feature of solid tumors and exerts crucial roles in cancers, including breast cancer. However, the detailed relationship between lncRNA-miRNA-mRNA triple network and hypoxia in breast cancer is still indistinct. In this study, a series of in silico analyses and online databases or tools were employed to establish a hypoxia-related lncRNA-miRNA-mRNA network in breast cancer based on competing endogenous RNA mechanism at single-cell resolution. RAMP2-AS1 was, eventually, identified as the most potential lncRNA, which was significantly negatively associated with hypoxia in breast cancer. Compared with normal controls, RAMP2-AS1 was markedly downregulated in breast cancer. Moreover, survival analysis revealed favorable prognostic values of RAMP2-AS1 in total or in specific clinicopathological breast cancer patients. Next, miR-660-5p, miR-2277-5p and miR-1301-3p, upregulated and possessed poor prognostic values in breast cancer, were identified as three potential downstream miRNAs of RAMP2-AS1. Then, the most potential downstream hypoxia-related genes (ATM and MYH11) of RAMP2-AS1/miRNA axis in breast cancer were screened out. Intriguingly, in vitro experiments confirmed that RAMP2-AS1 was a hypoxia-suppressed lncRNA and miR-660-5p/ATM was a potential downstream axis of RAMP2-AS1 in breast cancer. Collectively, our current data elucidated a key hypoxia-suppressed lncRNA RAMP2-AS1 and its possible miRNA-mRNA regulatory mechanism in breast cancer.
Keywords: Hypoxia, Single-cell resolution, RAMP2-AS1, Breast cancer, Prognosis
1. Introduction
Breast cancer is the most frequent cancer type and also ranks one of the most leading causes of cancer-associated deaths among women all over the world [1]. Despite tremendous advancements in the aspects of diagnosis and therapy during the past decades, the prognosis or outcome of patients with breast cancer is still dismal [2]. Therefore, more efforts are required to further study the molecular action mechanism of breast cancer’s initiation and progression.
It has been widely acknowledged that hypoxia is an important cancer characteristic in multiple solid tumors, including breast cancer [3]. Several lines of evidence have confirmed close correlation of hypoxia with invasion [4], metastasis [5], inflammation [6] and poor prognosis [7] in breast cancer.
More than 90% of human genome is transcribed into noncoding RNAs (ncRNAs) which can be generally classified into two groups according to transcript size, consisting of short ncRNAs (such as microRNA, miRNA) and long ncRNAs (such as long noncoding RNA, lncRNA) [8]. With the usage of competing endogenous RNA (ceRNA) mechanism, molecular interaction among different ncRNAs has been widely studied in human cancers during the past decades [9,10]. However, the specific correlation and mechanism between ncRNA and hypoxia in breast cancer are still unclear.
In this study, we firstly screened out potential hypoxia-related lncRNAs in breast cancer by CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp) and validated expression of these identified hypoxia-related lncRNAs in breast cancer using starBase (http://starbase.sysu.edu.cn/) and GEPIA (http://gepia.cancer-pku.cn/). Next, the prognostic value of RAMP2-AS1 in breast cancer was assessed through Kaplan-Meier plotter (http://kmplot.com/analysis/). Finally, the downstream miRNAs of RAMP2-AS1 and the target hypoxia-related genes of potential miRNAs were further predicted and analyzed by usage of a series of bioinformatic analyses and in vitro experiments. Consequently, a potential lncRNA-miRNA-mRNA regulatory network related to hypoxia in breast cancer were successfully established.
2. Results
2.1. Identification of hypoxia-related lncRNAs in breast cancer
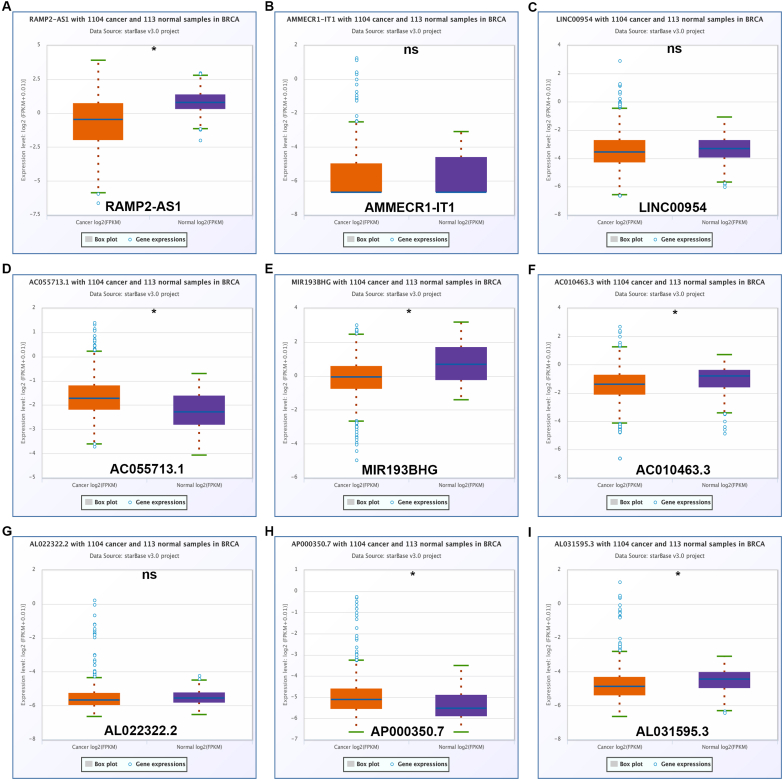
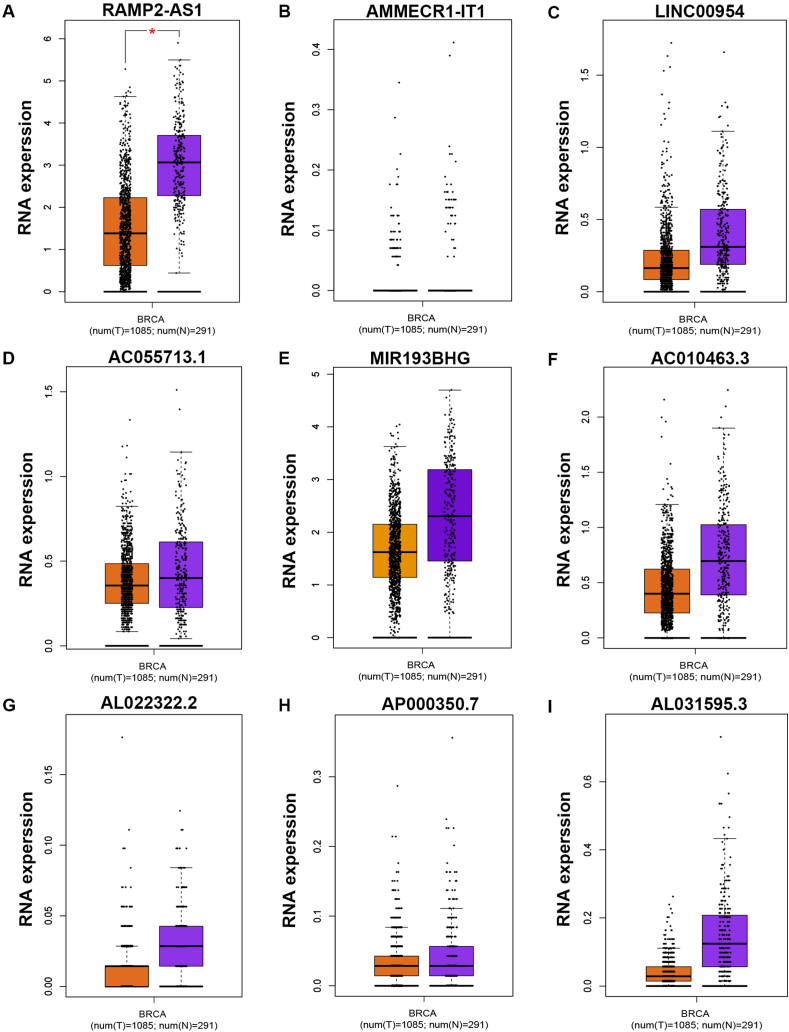
In order to screen out those lncRNAs that might be associated with hypoxia in breast cancer, a single cell database, namely CancerSEA, was employed. Consequently, a total of 2455 hypoxia-related lncRNAs were identified (data not shown). Among these lncRNAs, only 9 lncRNAs were presented in two datasets (four datasets in total), consisting of RAMP2-AS1, AMMECR1-IT1, LINC00954, AC055713.1, MIR193BHG, AC010463.3, AL022322.2, AP000350.7 and AL031595.3. All the 9 lncRNAs were significantly negatively correlated with hypoxia in breast cancer (Table 1). Next, expression levels of the 9 hypoxia-related lncRNAs in breast cancer were determined by usage of TCGA breast cancer and normal data. As presented in Fig. 1, RAMP2-AS1 (Fig. 1A), MIR193BHG (Fig. 1E), AC010463.3 (Fig. 1F) and AL031595.3 (Fig. 1I) were markedly downregulated but AC055713.1 (Fig. 1D) and AP000350.7 (Fig. 1H) were obviously upregulated in breast cancer when compared with normal controls. To further improve the analytic accuracy, another database GEPIA was also used to analyze their expression in breast cancer (Fig. 2). Among the 9 hypoxia-related lncRNAs, only RAMP2-AS1's expression was statistically decreased in TCGA breast cancer tissues compared with TCGA and GTEx breast normal tissues (Fig. 2A). Taken together, RAMP2-AS1 was a downregulated lncRNA which was also inversely correlated with hypoxia in breast cancer.
Table 1.
The correlation of lncRNAs with hypoxia in breast cancer determined by CancerSEA database.
| lncRNA name | ExpID and R-value | |
|---|---|---|
| RAMP2-AS1 | EXP0053–0.31 | EXP0054–0.51 |
| AMMECR1-IT1 | EXP0053–0.33 | EXP0054–0.64 |
| LINC00954 | EXP0053–0.30 | EXP0054–0.73 |
| AC055713.1 | EXP0053–0.31 | EXP0054–0.55 |
| MIR193BHG | EXP0053–0.33 | EXP0054–0.45 |
| AC010463.3 | EXP0053–0.31 | EXP0054–0.65 |
| AL022322.2 | EXP0053–0.32 | EXP0054–0.66 |
| AP000350.7 | EXP0053–0.30 | EXP0054–0.52 |
| AL031595.3 | EXP0053–0.30 | EXP0054–0.43 |
Fig. 1.
Expression of hypoxia-related lncRNAs in breast cancer determined by starBase database. (A) RAMP2-AS1's expression in breast cancer compared with normal controls. (B) AMMECR1-IT1's expression in breast cancer compared with normal controls. (C) LINC00954's expression in breast cancer compared with normal controls. (D) AC055713.1's expression in breast cancer compared with normal controls. (E) MIR193BHG's expression in breast cancer compared with normal controls. (F) AC010463.3's expression in breast cancer compared with normal controls. (G) AL022322.2's expression in breast cancer compared with normal controls. (H) AP000350.7's expression in breast cancer compared with normal controls. (I) AL031595.3′ expression in breast cancer compared with normal controls. *P < 0.05. nsP>0.05.
Fig. 2.
Expression of hypoxia-related lncRNAs in breast cancer determined by GEPIA database. (A) RAMP2-AS1's expression in breast cancer compared with normal controls. (B) AMMECR1-IT1's expression in breast cancer compared with normal controls. (C) LINC00954's expression in breast cancer compared with normal controls. (D) AC055713.1's expression in breast cancer compared with normal controls. (E) MIR193BHG's expression in breast cancer compared with normal controls. (F) AC010463.3's expression in breast cancer compared with normal controls. (G) AL022322.2's expression in breast cancer compared with normal controls. (H) AP000350.7's expression in breast cancer compared with normal controls. (I) AL031595.3′ expression in breast cancer compared with normal controls. *P < 0.05. nsP>0.05.
2.2. RMAP2-AS1 was a prognostic biomarker in breast cancer
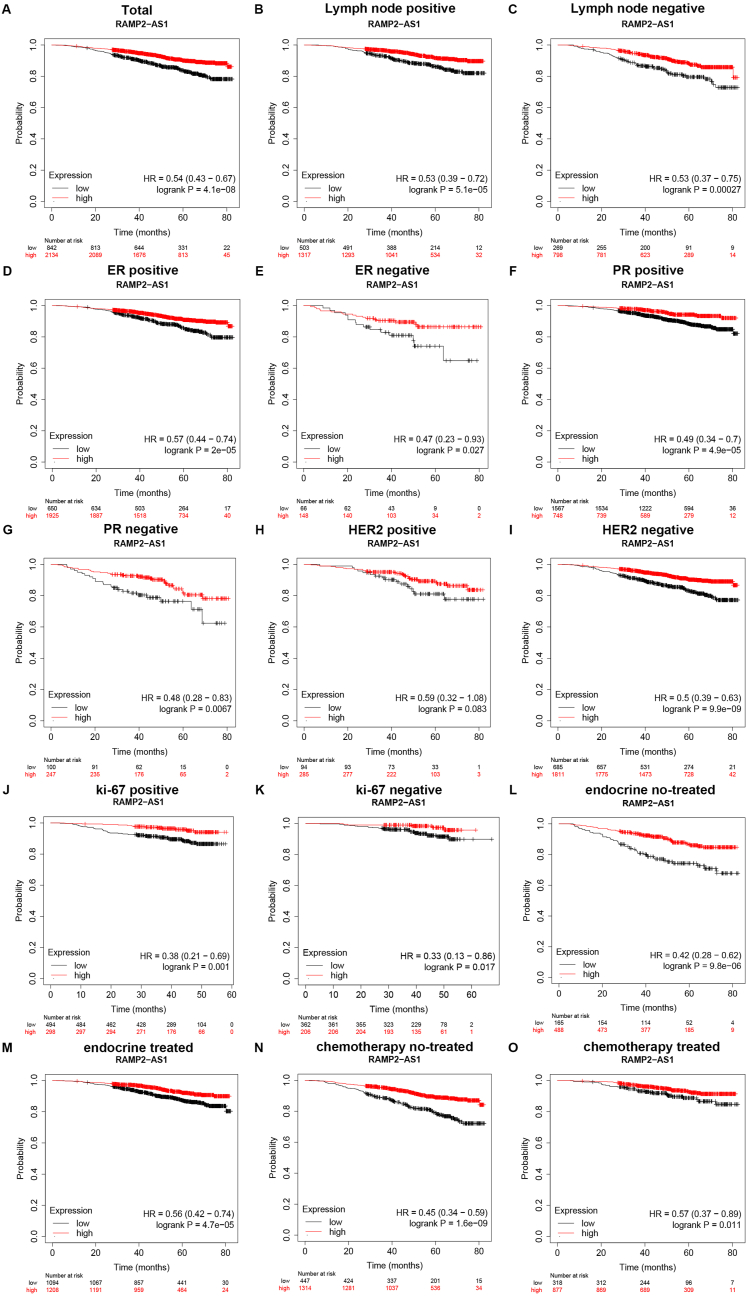
Subsequently, survival analysis for RAMP2-AS1 in breast cancer was performed by using Kaplan-Meier plotter. Firstly, the prognostic value of RAMP2-AS1 in total breast cancer patients was evaluated. As suggested in Fig. 3A, breast cancer patients with higher expression of RAMP2-AS1 presented better prognosis. Next, we conducted survival analysis for RAMP2-AS1 in breast cancer based on different clinicopathological features, including lymph node status (Fig. 3B–C), ER status (Fig. 3D–E), PR status (Fig. 3F–G), HER2 status (Fig. 3H–I), ki-67 status (Fig. 3J–K), with or without endocrine treatment (Fig. 3L-M) and with or without chemotherapy (Fig. 3N-O). Intriguingly, RAMP2-AS1 had significant favorable prognostic roles in almost all these subgroups except no statistical prognostic predictive value was observed in HER2 positive breast cancer subgroup. All these findings suggest that RAMP2-AS1 was a potential prognostic biomarker in breast cancer.
Fig. 3.
Survival analysis (overall survival) for RAMP2-AS1 in breast cancer according to different clinicopathological characteristics by Kaplan-Meier plotter database. The prognostic value of RAMP2-AS1 in total (A), in lymph node positive (B), in lymph node negative (C), in ER positive (D), in ER negative (E), in PR positive (F), in PR negative (G), in HER2 positive (H), in HER2 negative (I), in ki-67 positive (J), in ki-67 negative (K), in endocrine no-treated (L), in endocrine treated (M), in chemotherapy no-treated (N), or in chemotherapy treated (O) breast cancer assessed by Kaplan-Meier plotter database.
2.3. MiR-660-5p, miR-2277-5p and miR-1301-3p were identified as potential downstream miRNAs of RAMP2-AS1 in breast cancer
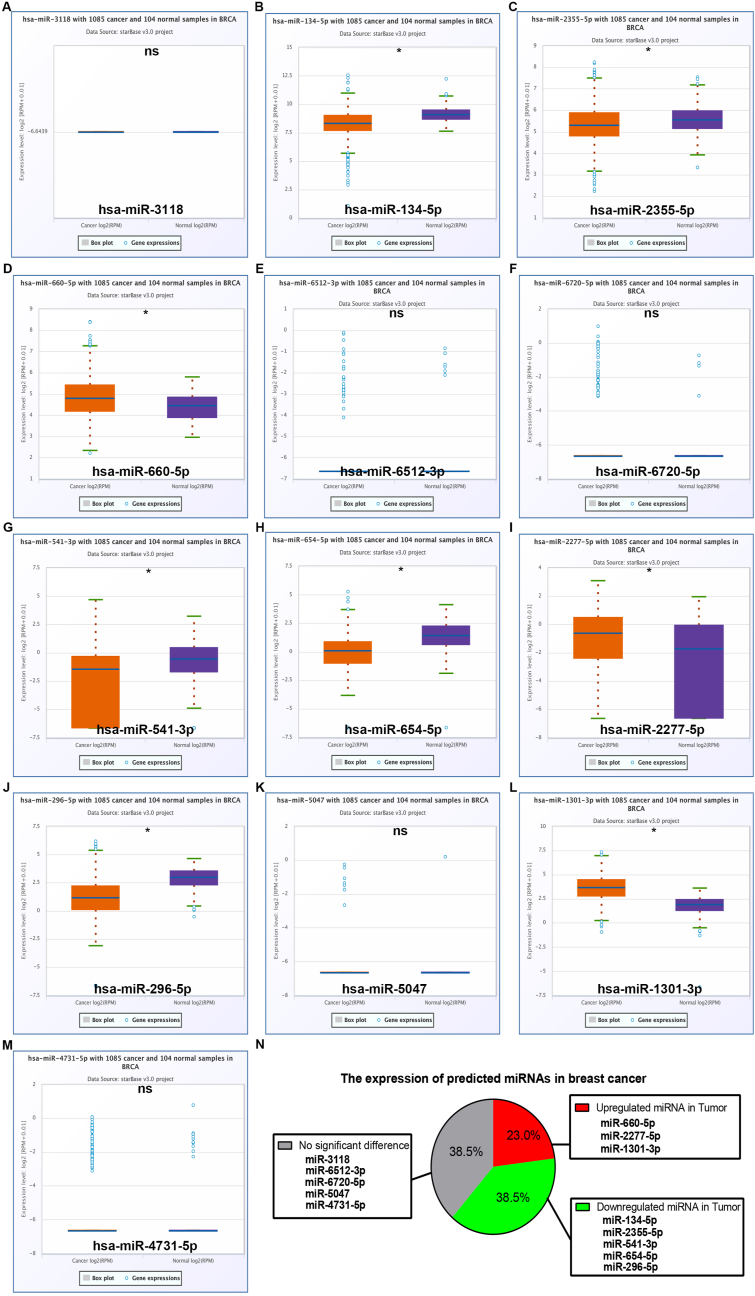
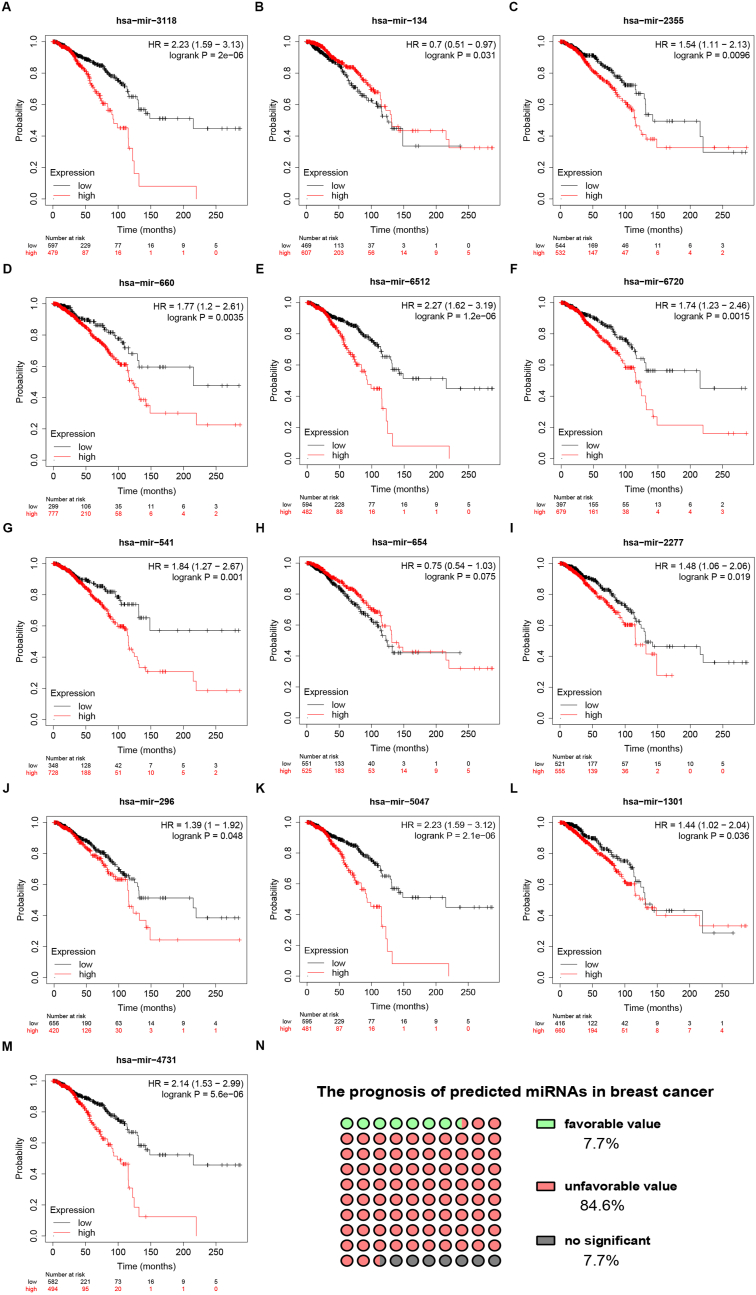
Lots of lines of evidence have indicated that lncRNAs might exert their roles by sponging or/and decreasing miRNAs. To ascertain if RAMP2-AS1 could potentially bind to miRNAs, bioinformatic prediction was performed by usage of starBase database. As a consequent, a total of 13 miRNAs were acquired, involving miR-3118, miR-134-5p, miR-2355-5p, miR-660-5p, miR-6512-3p, miR-6720-5p, miR-541-3p, miR-654-5p, miR-2277-5p, miR-296-5p, miR-5047, miR-1301-3p and miR-4731-5p. Expression correlation of RAMP2-AS1 with the 13 predicted miRNAs in breast cancer were listed in Table 2. Expression levels of the 13 miRNAs in breast cancer were determined (Fig. 4). Among these miRNAs, miR-134-5p (Fig. 4B), miR-2355-5p (Fig. 4C), miR-541-3p (Fig. 4G), miR-654-5p (Fig. 4H) and miR-296-5p (Fig. 4J) were downregulated but miR-660-5p (Fig. 4D), miR-2277-5p (Fig. 4I) and miR-1301-3p (Fig. 4L) were upregulated in breast cancer tissues. As shown in Figs. 4I and 23.0% and 38.5% of miRNAs were significantly upregulated and downregulated in breast cancer. Next, survival analysis for the 13 miRNAs was performed. As presented in Fig. 5, 11 of 13 miRNAs (84.6%) possessed unfavorable prognostic values in breast cancer. However, only high expression of miR-134-5p (7.7%) indicated favorable prognosis in breast cancer (Fig. 5B). Altogether, three miRNAs, consisting of miR-660-5p, miR-2277-5p and miR-1301-3p, were significantly upregulated and had poor prognostic values in breast cancer. The three miRNAs were regarded as the most potential downstream miRNAs of RAMP2-AS1 in breast cancer.
Table 2.
Expression correlation of RAMP2-AS1 with miRNAs in breast cancer.
| lncRNA name | miRNA name | R-value | P-value |
|---|---|---|---|
| RAMP2-AS1 | miR-3118 | 0.00 | 1.00E+00 |
| RAMP2-AS1 | miR-134-5p | −0.07 | 1.52E-02 |
| RAMP2-AS1 | miR-2355-5p | −0.26 | 7.94E-18 |
| RAMP2-AS1 | miR-660-5p | −0.13 | 1.88E-05 |
| RAMP2-AS1 | miR-6512-3p | −0.01 | 7.95E-01 |
| RAMP2-AS1 | miR-6720-5p | −0.12 | 1.09E-04 |
| RAMP2-AS1 | miR-541-3p | 0.06 | 6.19E-02 |
| RAMP2-AS1 | miR-654-5p | 0.01 | 8.33E-01 |
| RAMP2-AS1 | miR-2277-5p | −0.17 | 4.48E-08 |
| RAMP2-AS1 | miR-296-5p | −0.03 | 3.43E-01 |
| RAMP2-AS1 | miR-5047 | 0.02 | 4.63E-01 |
| RAMP2-AS1 | miR-1301-3p | −0.27 | 1.10E-19 |
| RAMP2-AS1 | miR-4731-5p | 0.01 | 7.82E-01 |
Fig. 4.
Expression analysis for predicted miRNAs of RAMP2-AS1 in breast cancer determined by starBase database. Expression levels of miR-3118 (A), miR-134-5p (B), miR-2355-5p (C), miR-660-5p (D), miR-6512-3p (E), miR-6720-5p (F), miR-541-3p (G), miR-654-5p (H), miR-2277-5p (I), miR-296-5p (J), miR-5047 (K), miR-1301-3p (L) and miR-4731-5p (M) in breast cancer when compared with normal breast tissues. (N) The dysregulated expression distribution of these miRNAs in breast cancer. *P < 0.05. nsP>0.05.
Fig. 5.
Survival analysis (overall survival) for predicted miRNAs of RAMP2-AS1 in breast cancer by Kaplan-Meier plotter database. The prognostic values of miR-3118 (A), miR-134-5p (B), miR-2355-5p (C), miR-660-5p (D), miR-6512-3p (E), miR-6720-5p (F), miR-541-3p (G), miR-654-5p (H), miR-2277-5p (I), miR-296-5p (J), miR-5047 (K), miR-1301-3p (L) and miR-4731-5p (M) in breast cancer. (N) The prognostic distribution of these miRNAs in breast cancer.
2.4. Identification of hub hypoxia-related genes in breast cancer
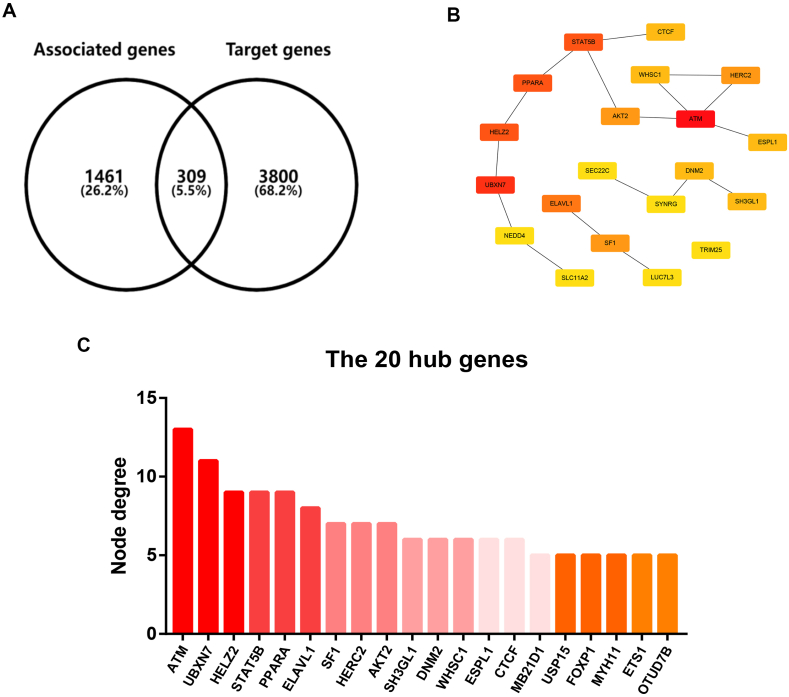
MiRNAs play key roles in biological and pathological processes by binding to target gene expression and inhibiting target gene function. To find downstream target genes of miR-660-5p, miR-2277-5p and miR-1301-3p, seven target gene prediction tools, including PITA, RNA22, miRmap, microT, miRanda, PicTar and TargetScan, were utilized by starBase database. Finally, 1661, 218 and 2959 target genes were predicted to potentially bind to miR-660-5p, miR-2277-5p and miR-1301-3p, respectively (Data not shown). Furthermore, 1770 genes that were negatively correlated with hypoxia in breast cancer were also screened out using CancerSEA database. By intersection of the two gene sets, a total of 309 genes were obtained (Fig. 6A). In order to better understand the interaction among these genes, protein-protein interaction (PPI) network analysis was performed by STRING database as shown in Table S1. After calculated by CytoHubba according to node degree, the top 20 hypoxia-related hub genes were obtained. For better visualization, a sub-PPI network were constructed (Fig. 6B). As shown in Fig. 6C, ATM and UBXN7 were the top 2 hub genes related to hypoxia among all the 309 genes.
Fig. 6.
Identification of hub genes of candidate hypoxia-related genes in breast cancer. (A) The intersection analysis for hypoxia-related genes and target genes by VENNY 2.1. (B) The protein-protein interaction network of the top 20 hub genes constructed by Cytoscape software. (C) The node degrees of the top 20 hub genes calculated by CytoHubba.
2.5. Expression determination and survival analysis for the top 20 hypoxia-related hub genes in breast cancer
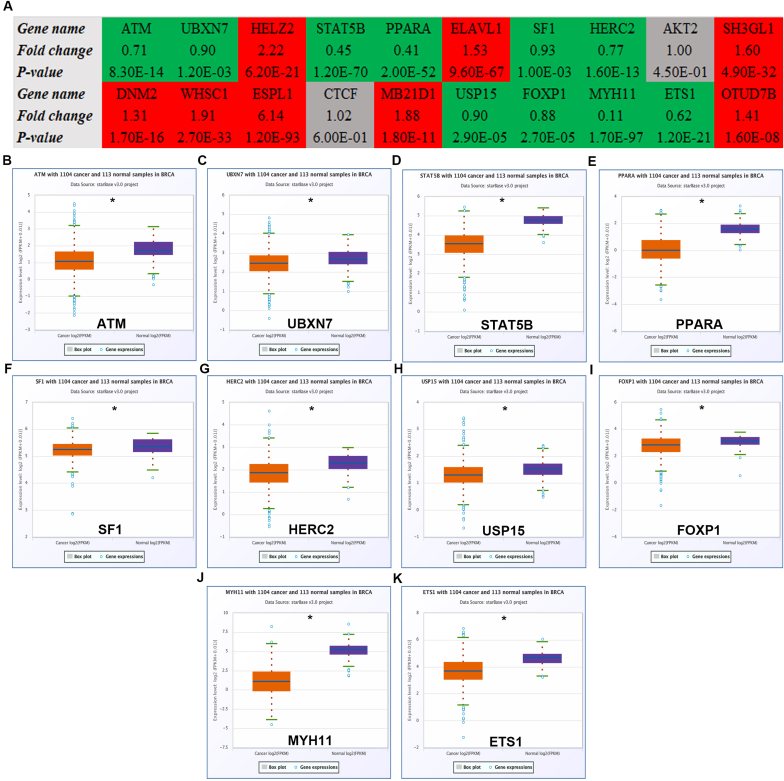
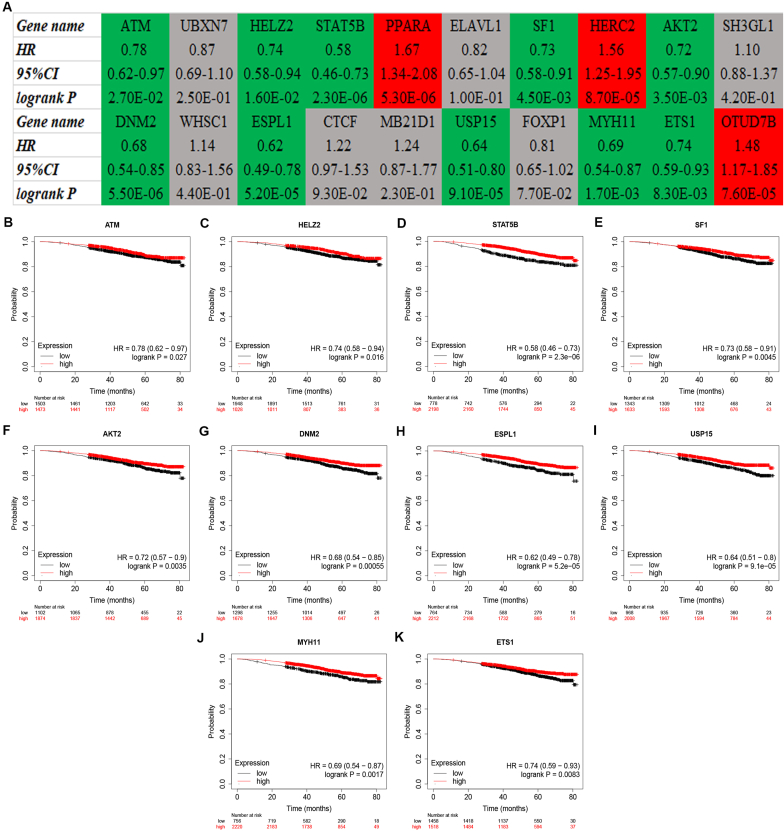
StarBase database was employed to conduct expression analysis for the top 20 hypoxia-related hub genes. The expression landscape of them were vividly shown in Fig. 7A, including 10 downregulated genes (ATM, UBXN7, STAT5B, PPARA, SF1, HERC2, USP15, FOXP1, MYH11 and ETS1) and 8 upregulated genes (HELZ2, ELAVL1, SH3GL1, DNM2, WHSC1, ESPL1, MB21D1 and OTUD7B). For better visualization, expression levels of the 10 downregulated genes in breast cancer tissues and normal controls were presented in Fig. 7B–K. Next, the prognostic values of the top 20 hypoxia-related hub genes in breast cancer were assessed using Kaplan-Meier plotter database. The prognosis landscape of the 20 genes were shown in Fig. 8A, involving 10 favorable genes (ATM, HELZ2, STAT5B, SF1, AKT2, DNM2, ESPL1, USP15, MYH11 and ETS1) and 3 unfavorable genes (PPARA, HERC2 and OTUD7B). And the overall survival plots of the 10 favorable genes in breast cancer were also shown in Fig. 8B–K. By combination of expression determination and survival analysis, only 6 genes, consisting of ATM, STAT5B, SF1, USP15, MYH11 and ETS1, were significantly downregulated and possessed favorable prognostic values in breast cancer.
Fig. 7.
Expression analysis for the top 20 hub genes in breast cancer determined by starBase database. (A) The expression landscape of top 20 hub genes in breast cancer. “Red” represents “High expression”; “Green” represents “Low expression”; “grey” represents “No statistical difference”. Expression levels of ATM (B), UBXN7 (C), STAT5B (D), PPARA (E), SF1 (F), HERC2 (G), USP15 (H), FOXP1 (I), MYH11 (J) and ETS1 (K) in breast cancer compared with normal controls determined by starBase database. *P < 0.05.
Fig. 8.
Survival analysis (overall survival) for the top 20 hub genes in breast cancer by Kaplan-Meier plotter database. (A) The prognosis landscape of top 20 hub genes in breast cancer. “Red” represents “Poor prognosis”; “Green” represents “Good prognosis”; “grey” represents “No statistical significance”. The prognostic roles of ATM (B), HELZ2 (C), STAT5B (D), SF1 (E), AKT2 (F), DNM2 (G), ESPL1 (H), USP15 (I), MYH11 (J) and ETS1 (K) in breast cancer compared with normal controls determined by Kaplan-Meier plotter database.
2.6. Construction of a potential hypoxia-related lncRNA-miRNA-mRNA triple ceRNA regulatory network in breast cancer
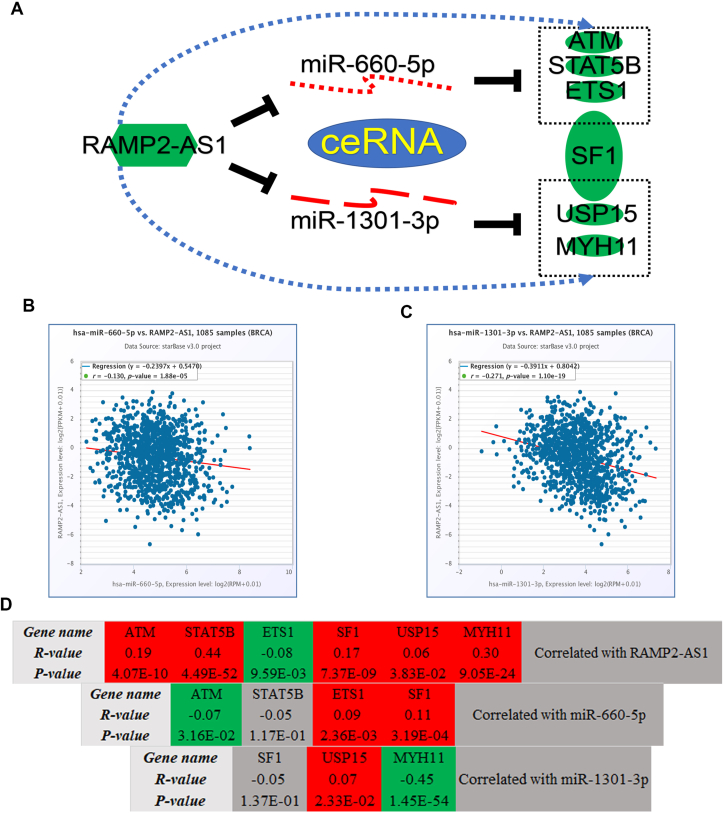
By pairing miRNAs (miR-660-5p, miR-2277-5p and miR-1301-3p) and mRNAs (ATM, STAT5B, SF1, USP15, MYH11 and ETS1), a hypoxia-related ceRNA regulatory network was established in breast cancer, consisting of 1 lncRNA (RAMP2-AS1), 2 miRNAs (miR-660-5p and miR-1301-3p) and 6 mRNAs (ATM, STAT5B, SF1, USP15, MYH11 and ETS1) (Fig. 9A). According to ceRNA hypothesis, miRNA should be negatively correlated with lncRNA and mRNA, and lncRNA should be positively associated with mRNA. As shown in Fig. 9B and C, RAMP2-AS1 was significantly negatively correlated with miR-660-5p and miR-1301-3p. For RAMP2-AS1-mRNA pairs, RAMP2-AS1 was markedly positively correlated with ATM, STAT5B, SF1, USP15 and MYH11 in breast cancer (Fig. 9D). After performing expression correlation analysis for miRNA-mRNA pairs (Fig. 9D), we found that there were only two miRNA-mRNA pairs with negative expression correlation in breast cancer, consisting of miR-660-5p-ATM and miR-1301-3p-MYH11.
Fig. 9.
Construction and analysis of a hypoxia-associated ceRNA network in breast cancer. (A) The potential hypoxia-associated ceRNA network in breast cancer. miR-660-5p (B) and miR-1301-3p (C) were negatively correlated with RAMP2-AS1 in breast cancer determined by starBase database. (D) The correlation analysis for RAMP2-AS1-gene pairs, miR-660-5p-gene pairs and miR-1301-3p-gene pairs evaluated by starBase database.
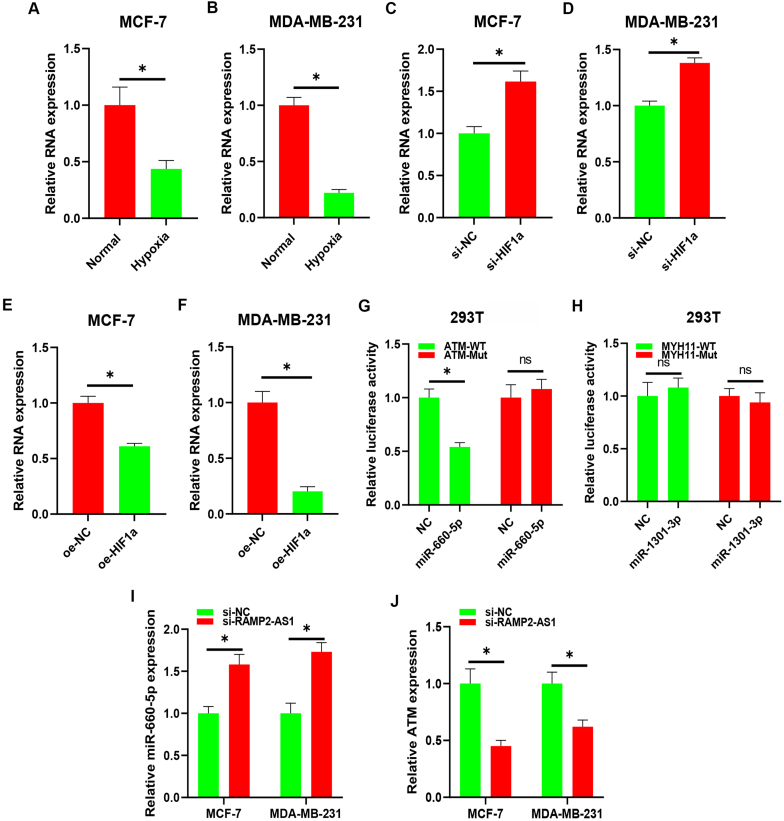
To validation the association between hypoxia and RAMP2-AS1, two experimental assays were performed. As shown in Fig. 10A–B, after exposing to hypoxia condition, RAMP2-AS1 expression was significantly decreased in breast cancer cells. Moreover, the specific siRNA targeting HIF-1α was employed. Compared with negative control, RAMP2-AS1 expression was markedly increased after administration of siRNA targeting HIF-1α in breast cancer cells (Fig. 10C–D) and was significantly decreased after overexpression of HIF-1α (Fig. 10E–F). Next, luciferase reporter assay showed that miR-660-5p could directly bind to ATM but miR-1301-3p could not directly bind to MYH11 (Fig. 10G–H). Finally, expression regulation assay was conducted. The result demonstrated that knockdown of RAMP2-AS1 markedly increased miR-660-5p expression and decreased ATM expression in both MCF-7 and MDA-MB-231 cells. Taken all these findings into consideration, RAMP2-AS1-miR-660-5p-ATM might be the most potential hypoxia-suppressed lncRNA-miRNA-mRNA triple ceRNA regulatory network in breast cancer.
Fig. 10.
Validation of RAMP2-AS1/miR-660-5p/ATM as a potential hypoxia-regulated axis in breast cancer. RAMP2-AS1 expression was significantly downregulated in breast cancer cells after exposing to hypoxia condition (A–B). (C–D) RAMP2-AS1 expression was significantly increased in breast cancer cells after knockdown of HIF-1α. (E–F) RAMP2-AS1 expression was significantly decreased in breast cancer cells after overexpression of HIF-1α. (G–H) Luciferase reporter assay explored the direct bind of miR-660-5p/ATM and miR-1301-3p/MYH11. (I) Knockdown of RAMP2-AS1 markedly upregulation of miR-660-5p in breast cancer cells. (J) Knockdown of RAMP2-AS1 markedly downregulation of ATM in breast cancer cells. nsP>0.05; *P < 0.05.
3. Discussion
Growing evidence has confirmed existence of hypoxia and its crucial roles in breast cancer [11,12]. During the past years, several lncRNAs induced by hypoxia in breast cancer have been reported, such as RBM5-AS1 [13], MTORT1 [14], MALAT1 [15] and IHAT [16]. However, a comprehensive study at single-cell resolution regarding the detailed correlation of ncRNA network with hypoxia in breast cancer is still absent and need to be explored.
In the first place, 9 potential lncRNAs negatively related to hypoxia in breast cancer were identified using single-cell research database CancerSEA. By combination of expression analytic results from starBase and GEPIA databases, downregulated RAMP2-AS1 was selected as the most potential hypoxia-related lncRNA in breast cancer. Moreover, survival analysis revealed that breast cancer patients with high expression of RAMP2-AS1 had favorable prognosis. RAMP2-AS1 has been found to act as key regulators in malignancies. For example, Cheng et al. suggested that exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells facilitated angiogenesis by miR-2355-5p/VEGFR2 axis [17]; Liu et al. indicated that lncRNA RAMP2-AS1 suppressed tumorigenesis of glioblastoma by indirect inhibition of NOTCH3 [18]. Additionally, a recent study reported the tumor suppressive role of lncRNA RAMP2-AS1 in breast cancer by DNMT1 and DNMT3B mediated downregulation of CXCL11 [19]. Nevertheless, correlation of RAMP2-AS1 with hypoxia and RAMP2-AS1-mediated ceRNA regulatory network in breast cancer are still unknown.
Next, downstream miRNAs of RAMP2-AS1 were predicted using starBase and a total of 13 miRNAs were obtained, after which correlation analysis, expression analysis and survival analysis for these miRNAs were conducted. Three miRNAs, consisting of miR-660-5p, miR-2277-5p and miR-1301-3p, were identified as the most potential downstream miRNAs of RAMP2-AS1 in breast cancer. Among the three miRNAs, miR-660-5p and miR-1301-3p have been validated to function as two oncogenic miRNAs in breast cancer [[20], [21], [22], [23]], which was in accordance with ceRNA hypothesis [24].
Our team and other groups have well documented that miRNAs exert their biological roles mainly by negative modulation of downstream target genes [[25], [26], [27]]. Subsequently, downstream target genes of miR-660-5p, miR-2277-5p or miR-1301-3p were forecasted by several online target gene prediction tools, and these predicted target genes were intersected with the genes positively correlated with hypoxia from CancerSEA database. At the end, a total of 309 genes were commonly appeared in both two gene sets. By performing PPI network analysis and hub gene identification, the top 20 hypoxia-related hub genes were screened out. By combination of expression analysis and survival analysis, six genes were considered as the potential hypoxia-related genes of RAMP2-AS1/miRNA axis, consisting of ATM, STAT5B, ETS1, SF1, USP15 and MYH11. Some of the six genes have been found to be suppressive genes in malignancies, including breast cancer. For example, the group of Kim GC showed that ETS1 inhibited tumorigenesis of breast cancer by transactivation of canonical tumor suppressive genes [28].
Subsequently, by conducting correlation analysis for RNA-RNA pairs, a lncRNA-miRNA-mRNA regulatory network related to hypoxia of breast cancer was constructed based on ceRNA mechanism, involving RAMP2-AS1, miR-660-5p, miR-1301-3p, ATM and MYH11. Finally, by performing some in vitro experiments, RAMP2-AS1/miR-660-5p/ATM was identified as the most potential hypoxia-regulated axis in breast cancer. Despite some interesting findings have been obtained, this study also possessed several limitations: (1) this study lacked of hypoxia-related miRNA data in breast cancer, causing this single cell-based analytic work not so good; (2) although multiple databases or tools were employed to explore and confirm, this study was mainly based on bioinformatic analysis; (3) in order to improve the analytic accuracy, some positive results might be lost throughout the entire analytic process. In the future, more basic experiments and large clinical trials need to be further validated the current findings.
4. Materials and methods
4.1. CancerSEA analysis
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp) is a database that aims to study the different functional states of cancer cells at single-cell resolution [29]. In this study, CancerSEA database, containing four breast cancer datasets, was employed to obtain hypoxia-related lncRNAs and genes in breast cancer. And only these lncRNAs and genes that were significantly positively or negatively correlated with hypoxia in breast cancer were included for subsequent analysis. P-value <0.05 was considered as statistically significant.
4.2. Expression analysis
Expression levels of lncRNAs, miRNAs and genes in breast cancer were determined by starBase database (http://starbase.sysu.edu.cn/), which is an online tool for exploring miRNA-ncRNA, miRNA-ceRNA and protein-RNA interaction networks from large-scale CLIP-seq data [30,31]. Furthermore, another database, namely GEPIA (http://gepia.cancer-pku.cn/), was also used to analyze lncRNAs’ expression in breast cancer [32,33]. P-value <0.05 was considered as statistically significant.
4.3. Survival analysis
Survival analysis was performed to assess prognostic values of RAMP2-AS1, miRNAs and genes in breast cancer by usage of Kaplan-Meier plotter (http://kmplot.com/analysis/), which is a widely-used database to access the effects of genomic features on survival in more than 20 cancer types (such as breast cancer) [34]. Logrank P-value <0.05 was considered as statistically significant.
4.4. MiRNA prediction
The downstream miRNAs of RAMP2-AS1 were predicted using starBase database (http://starbase.sysu.edu.cn/). Simply, the name of RAMP2-AS1 was first typed into starBase database, section “miRNA-lncRNA”. Then, a list of possible miRNAs was automatically generated and presented in the webpage. Finally, these miRNAs that could potentially bind to RAMP2-AS1 were directly downloaded from the webpage.
4.5. Target gene prediction
The downstream target genes of miR-660-5p, miR-2277-5p and miR-1301-3p were forecasted by a series of online target gene prediction tools using starBase database (http://starbase.sysu.edu.cn/), including PITA, RNA22, miRmap, microT, miRanda, PicTar and TargetScan. The target genes presented in any above tools were included and used for subsequent analyses.
4.6. Intersection analysis
To obtain the potential hypoxia-related genes involved in RAMP2-AS1-mediated ceRNA network, intersection analysis was conducted by using VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html). Only the genes that were common in hypoxia-associated gene set and target gene set were screened out for subsequent analyses.
4.7. Protein-protein interaction analysis
The relationship among hypoxia-related genes that were prioritized based on previous analysis was assessed using protein-protein interaction network analysis through STRING database (https://cn.string-db.org) [35]. Only the interactions with a combined score more than 0.4 were regarded as statistically significant.
4.8. Hub gene analysis
The significant protein-protein interactions obtained from STRING database were re-entered into Cytoscape software (version 3.6.0) and a protein-protein interaction sub-network was re-constructed. As previously described [36,37], the hub genes among this network were identified by calculating node degree using CytoHubba [38].
4.9. Correlation analysis
The correlation between expression of lncRNA-miRNA, lncRNA-mRNA or miRNA-mRNA pairs was assessed by starBase database (http://starbase.sysu.edu.cn/) as mentioned above. The correlated plots were downloaded from online webpage. P-value <0.05 was considered as statistically significant.
4.10. Cell culture, cell transfection, RNA extraction and RT-qPCR
MCF-7 and MDA-MB-231 cell lines were purchased from the Chinese Academy of Science's Cell Bank. MCF-7 cells were cultured in RPMI-1640 medium and MDA-MB-231 cell were cultured in DMEM medium supplemented with 10% FBS under a 5% CO2 atmosphere at 37 °C.
To stimulate the hypoxia condition, the breast cancer cells were cultured with a gas mixture containing 1% O2, 5% CO2 and 94% nitrogen for 2 min at 2 psi.
To explore the association of hypoxia with lncRNA, the specific siRNA targeting HIF-1α and overexpressed plasmid of HIF-1α were transfected into the breast cancer cells using Lipofectamine 3000 according to the manufacturer's instructions.
The total RNA of cells was extracted by usage of RNAiso plus Reagent, after which was reversely transcribed into complementary DNA using the PrimeScript RT Reagent Kit and qPCR was successively performed using the SYBR Premix Ex Taq. Then, gene or miRNA expression was detected by calculated using the method of 2−ddCt.
4.11. Statistical analysis
The bioinformatic statistical analyses used in this study were directly performed by usage of the online databases or tools as mentioned above. The experimental data were shown as mean ± standard deviation (SD). For example, student's t-test, kaplan-meier and regression analysis were used for expression analysis, survival analysis and correlation analysis, respectively. P-value<0.05 or logrank P-value<0.05 was considered as statistically significant.
Funding
This work was supported by the National Natural Science Foundation of China (82203239) and was funded by the Science and Technology Project of Wenzhou (Y2023489).
CRediT authorship contribution statement
Weiyang Lou: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Data curation. Shuyuan Xiao: Methodology, Investigation. Kuailu Lin: Writing – review & editing, Validation, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2024.02.007.
Contributor Information
Weiyang Lou, Email: 11718264@zju.edu.cn.
Kuailu Lin, Email: Kuailu.Lin@outlook.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Xiao S., Chen J. Development of an inflammation-related lncRNA-miRNA-mRNA network based on competing endogenous RNA in breast cancer at single-cell resolution. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.839876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tutzauer J., Sjöström M. vol. 126. 2022. pp. 1145–1156. (Breast Cancer Hypoxia in Relation to Prognosis and Benefit From Radiotherapy After Breast-Conserving Surgery in a Large, Randomised Trial With Long-Term Follow-Up). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Liu S. LncRNA GHET1 promotes hypoxia-induced glycolysis, proliferation, and invasion in triple-negative breast cancer through the hippo/YAP signaling pathway. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.643515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimeault M., Batra S.K. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J. Cell Mol. Med. 2013;17:30–54. doi: 10.1111/jcmm.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lappano R., Talia M., Cirillo F., Rigiracciolo D.C., Scordamaglia D., Guzzi R., Miglietta A.M., De Francesco E.M., Belfiore A., Sims A.H., Maggiolini M. The IL1β-IL1R signaling is involved in the stimulatory effects triggered by hypoxia in breast cancer cells and cancer-associated fibroblasts (CAFs) J. Exp. Clin. Cancer Res. : CR. 2020;39:153. doi: 10.1186/s13046-020-01667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu P., Zhang L., Wang R., Ding W., Wang W., Liu Y., Wang W., Li Z., Yan B., Sun X. Development and validation of a novel hypoxia-related long noncoding RNA model with regard to prognosis and immune features in breast cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.796729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lertampaiporn S., Thammarongtham C., Nukoolkit C., Kaewkamnerdpong B., Ruengjitchatchawalya M. Identification of non-coding RNAs with a new composite feature in the Hybrid Random Forest Ensemble algorithm. Nucleic Acids Res. 2014;42:e93. doi: 10.1093/nar/gku325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Zhao Z., Wang X., Zhang Q., Lyu L., Tang B. The predictive competing endogenous RNA regulatory networks and potential prognostic and immunological roles of cyclin A2 in pan-cancer analysis. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.809509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng T., Heyward C.A., Chen X., Zheng M., Yang Y., Reseland J.E. vol. 14. 2022. (Comprehensive Analysis Identifies Ameloblastic-Related Competitive Endogenous RNA as a Prognostic Biomarker for Testicular Germ Cell Tumour). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giatromanolaki A., Gkegka A.G., Pouliliou S., Biziota E., Kakolyris S., Koukourakis M. vol. 194. 2022. pp. 13–23. (Hypoxia and Anaerobic Metabolism Relate with Immunologically Cold Breast Cancer and Poor Prognosis). [DOI] [PubMed] [Google Scholar]
- 12.Xu A.L., Xue Y.Y., Tao W.T., Wang S.Q., Xu H.Q. Oleanolic acid combined with olaparib enhances radiosensitization in triple negative breast cancer and hypoxia imaging with (18)F-FETNIM micro PET/CT. Biomed. Pharmacother. 2022;150 doi: 10.1016/j.biopha.2022.113007. [DOI] [PubMed] [Google Scholar]
- 13.Li X., Yang J., Ni R., Chen J., Zhou Y., Song H., Jin L. Vol. 13. 2022. p. 95. (Hypoxia-Induced lncRNA RBM5-AS1 Promotes Tumorigenesis Via Activating Wnt/β-Catenin Signaling in Breast Cancer). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y.C., Su L.Y., Chen L.H., Lu T.P., Chuang E.Y., Tsai M.H., Chuang L.L., Lai L.C. Regulatory mechanisms and functional roles of hypoxia-induced long non-coding RNA MTORT1 in breast cancer cells. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.663114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih C.H., Chuang L.L., Tsai M.H., Chen L.H., Chuang E.Y., Lu T.P., Lai L.C. Hypoxia-induced MALAT1 promotes the proliferation and migration of breast cancer cells by sponging MiR-3064-5p. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.658151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Bao L., Niu Y., Wang J.E. LncIHAT is induced by hypoxia-inducible factor 1 and promotes. Breast Cancer Prog. 2021;19:678–687. doi: 10.1158/1541-7786.MCR-20-0383. [DOI] [PubMed] [Google Scholar]
- 17.Cheng C., Zhang Z., Cheng F., Shao Z. Exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells promotes angiogenesis through miR-2355-5p/VEGFR2 Axis. OncoTargets Ther. 2020;13:3291–3301. doi: 10.2147/OTT.S244652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S., Mitra R., Zhao M.M., Fan W., Eischen C.M., Yin F., Zhao Z. The potential roles of long noncoding RNAs (lncRNA) in glioblastoma development. Mol. Cancer Therapeut. 2016;15:2977–2986. doi: 10.1158/1535-7163.MCT-16-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Gan Y.P., Peng H. RAMP2-AS1 inhibits CXCL11 expression to suppress malignant phenotype of breast cancer by recruiting DNMT1 and DNMT3B. Exp. Cell Res. 2022 doi: 10.1016/j.yexcr.2022.113139. [DOI] [PubMed] [Google Scholar]
- 20.Li C., Li R., Hu X., Zhou G., Jiang G. Vol. 192. 2022. pp. 353–368. (Tumor-Promoting Mechanisms of Macrophage-Derived Extracellular Vesicles-Enclosed MicroRNA-660 in Breast Cancer Progression). [DOI] [PubMed] [Google Scholar]
- 21.Peng B., Li C. Vol. 53. 2020. (miR-660-5p Promotes Breast Cancer Progression Through Down-Regulating TET2 and Activating PI3K/AKT/mTOR Signaling). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Ye Y.F., Ruan L.W., Bao L., Wu M.W., Zhou Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet. Mol. Res. : GMR. 2017;16 doi: 10.4238/gmr16019479. [DOI] [PubMed] [Google Scholar]
- 23.Lin W.H., Li J., Zhang B., Liu L.S., Zou Y., Tan J.F., Li H.P. MicroRNA-1301 induces cell proliferation by downregulating ICAT expression in breast cancer. Biomed. Pharmacother. 2016;83:177–185. doi: 10.1016/j.biopha.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou W., Chen J., Ding B., Fan W. XIAP, commonly targeted by tumor suppressive miR-3607-5p and miR-3607-3p, promotes proliferation and inhibits apoptosis in hepatocellular carcinoma. Genomics. 2021;113:933–945. doi: 10.1016/j.ygeno.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Gao S., Ding B., Lou W. microRNA-dependent modulation of genes contributes to ESR1's effect on ERα positive breast cancer. Front. Oncol. 2020;10:753. doi: 10.3389/fonc.2020.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao G., Guo X., Gu W., Lu Y., Chen Z. miRNA-142-3p functions as a potential tumor suppressor directly targeting FAM83D in the development of ovarian cancer. Aging. 2022;14:3387–3399. doi: 10.18632/aging.203998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim G.C., Lee C.G., Verma R., Rudra D., Kim T., Kang K., Nam J.H., Kim Y., Im S.H., Kwon H.K. ETS1 suppresses tumorigenesis of human breast cancer via trans-activation of canonical tumor suppressor genes. Front. Oncol. 2020;10:642. doi: 10.3389/fonc.2020.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z., Shi A., Zhao T., Xiao Y., Li X. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–d908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C., Tang Z., Zhang W., Ye Z., Liu F. Vol. 49. 2021. pp. W242–w246. (GEPIA2021: Integrating Multiple Deconvolution-Based Analysis into GEPIA). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021;19:4101–4109. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–d612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou W., Chen J., Ding B., Chen D., Zheng H., Jiang D., Xu L., Bao C., Cao G., Fan W. Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation. J. Transl. Med. 2018;16:266. doi: 10.1186/s12967-018-1639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lou W., Liu J., Ding B., Chen D., Xu L., Ding J., Jiang D., Zhou L., Zheng S., Fan W. Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. J. Transl. Med. 2019;17:7. doi: 10.1186/s12967-018-1761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.