Abstract
Human immunodeficiency virus type 1 (HIV-1) entry into target cells involves sequential binding of the gp120 exterior envelope glycoprotein to CD4 and to specific chemokine receptors. Soluble CD4 (sCD4) is thought to mimic membrane-anchored CD4, and its binding alters the conformation of the HIV-1 envelope glycoproteins. Two cross-competing monoclonal antibodies, 17b and CG10, that recognize CD4-inducible gp120 epitopes and that block gp120-chemokine receptor binding were used to investigate the nature and functional significance of gp120 conformational changes initiated by CD4 binding. Envelope glycoproteins derived from both T-cell line-adapted and primary HIV-1 isolates exhibited increased binding of the 17b antibody in the presence of sCD4. CD4-induced exposure of the 17b epitope on the oligomeric envelope glycoprotein complex occurred over a wide range of temperatures and involved movement of the gp120 V1/V2 variable loops. Amino acid changes that reduced the efficiency of 17b epitope exposure following CD4 binding invariably compromised the ability of the HIV-1 envelope glycoproteins to form syncytia or to support virus entry. Comparison of the CD4 dependence and neutralization efficiencies of the 17b and CG10 antibodies suggested that the epitopes for these antibodies are minimally accessible following attachment of gp120 to cell surface CD4. These results underscore the functional importance of these CD4-induced changes in gp120 conformation and illustrate viral strategies for sequestering chemokine receptor-binding regions from the humoral immune response.
Human immunodeficiency virus type 1 (HIV-1), the etiologic agent of AIDS (6, 26, 49), infects cells that express CD4 and particular chemokine receptor molecules, which serve as coreceptors for the virus (1, 12, 14, 16, 18, 19, 28, 31, 59). The initial attachment of HIV-1 to target cells occurs via specific binding of the HIV-1 surface glycoprotein gp120 to CD4 (36, 38, 39, 42), creating a high-affinity binding site for the CCR5 chemokine receptor (73). Receptor binding facilitates fusion of the virus and cell membranes by an unknown mechanism. The fusion event probably involves insertion of the hydrophobic amino-terminal fusion peptide of the HIV-1 transmembrane protein, gp41, into the target cell membrane (7, 24, 25, 33). The core structure of gp41 has been solved; it exhibits a striking similarity to the low-pH-induced (fusion-active) conformation of influenza virus hemagglutinin HA2, which also possesses an amino-terminal fusion peptide thought to interact with target cell membranes (11, 70). In the native HIV-1 envelope glycoprotein complex, the gp41 fusion peptide, like most of the gp41 ectodomain, is not accessible to antibodies (5, 17, 25, 55). It is therefore likely that, as has been documented for the influenza virus HA2 protein, conformational changes in the HIV-1 envelope glycoproteins are required to allow exposure of the fusion peptide (25). While viral endocytosis and a decreased pH trigger these conformational changes in the influenza virus hemagglutinin (9, 61; reviewed in reference 71), the ability of the HIV-1 envelope glycoproteins to mediate virus entry at the plasma membrane and to cause cell-cell fusion (syncytium formation) suggests that HIV-1-induced membrane fusion does not require a drop in pH (36–38).
It is likely that conformational changes in the HIV-1 envelope glycoproteins are induced by binding to both CD4 and the chemokine receptors. While there is no information on the effects of chemokine receptor binding on the HIV-1 envelope glycoproteins, soluble CD4 (sCD4) binding has been shown to initiate changes in envelope glycoprotein conformation (2–4, 15, 45, 52, 54, 55). The binding of sCD4 to the envelope glycoprotein complexes of particular HIV-1 strains results in dissociation of gp120 from the gp41 glycoprotein (23, 29, 42, 44, 45, 66, 72). Some of the variable loops (V1/V2 and V3) on the HIV-1 gp120 glycoprotein change conformation or become more exposed upon sCD4 binding (8, 52, 54, 72, 74). Movement of the V1/V2 loops results in the exposure of conserved, discontinuous structures on the HIV-1 gp120 glycoprotein recognized by the 17b and 48d monoclonal antibodies (67, 74). Another monoclonal antibody, CG10, recognizes gp120-sCD4 complexes, but neither gp120 nor sCD4 alone, suggesting the creation or improved exposure of the antibody epitope upon formation of the ligand-receptor complex (27).
The functional relevance to the membrane fusion process of the sCD4-induced changes in HIV-1 envelope glycoprotein structure is uncertain. That at least some of the sCD4-mediated conformational changes are functionally important is suggested by the observation that some primary patient HIV-1 isolates as well as HIV-2 and simian immunodeficiency virus isolates exhibit increases in either virus entry or syncytium formation in the presence of sCD4 (2, 3, 13, 57, 63). Conformational changes relevant to envelope glycoprotein-mediated membrane fusion would be expected to exhibit the following features: (i) all HIV-1 strains demonstrate the changes, (ii) amino acid substitutions in the envelope glycoproteins or CD4 that compromise the conformational change result in decreases in virus entry or syncytium formation, and (iii) inhibitors of the conformational change likewise interfere with the function of the envelope glycoproteins.
In this study, we investigated the potential functional relevance of the CD4-induced exposure of the gp120 epitope for the 17b monoclonal antibody. The probable importance of this conformational change is implied by the conserved nature of the 17b epitope among HIV-1 strains, by the ability of the 17b antibody to interfere with the chemokine receptor interaction in in vitro binding assays, and by the neutralizing activity of the antibody against T-cell line-adapted (TCLA) HIV-1 strains (53, 67, 73). Here we examine the induction of the 17b epitope on different HIV-1 strains by sCD4 and investigate the structural basis and temperature dependence of this induction in the context of the intact oligomeric envelope glycoprotein complex. Amino acid changes in the gp120 glycoprotein that partially attenuate the induction of the 17b epitope without affecting CD4 binding are identified, and the effects of these changes on HIV-1 envelope glycoprotein function are examined. Finally, we compare the virus-neutralizing abilities of the 17b antibody and another antibody, CG10, that competes with 17b for binding to gp120-sCD4 complexes but does not recognize gp120 in the absence of CD4. These studies provide insights into antibody accessibility to the chemokine receptor-interactive moieties on the functional HIV-1 envelope glycoprotein complex.
MATERIALS AND METHODS
Antibodies and sCD4.
Human monoclonal antibody (HMAb) 17b was derived by Epstein-Barr virus (EBV) transformation of peripheral blood B cells obtained from an asymptomatic HIV-1-infected patient by using a previously described protocol (51). This was the same patient (N70) from whom several other HMAbs had been derived previously (51). These included HMAbs 19b to the V3 loop (58) and 15e to the CD4 binding region (34). Freshly prepared peripheral blood mononuclear cells were exposed to EBV and plated at low cell density (approximately 104 cells per well) in microtiter plates containing irradiated cord blood lymphocytes as feeder cells (51). Screening was performed for antibodies reactive with concanavalin A-captured gp120 from strain J62. Because the EBV-transformed cell line producing the 17b antibody grew poorly, we constructed a hybridoma by fusing the cells with a mouse-human fusion partner (HMMA), which was developed and kindly provided by Marshall Posner (50). sCD4 was a gift from Raymond Sweet, SmithKline Beecham. MIP-1β was obtained from R & D Systems, Minneapolis, Minn.
Cells and cell lines.
COS-1 cells were grown in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS). The T-cell lines Molt 4 clone 8, Jurkat, and SupT1 were maintained in RPMI 1640 medium containing 10% FBS. Peripheral blood mononuclear cells were purified over a Ficoll gradient and stimulated with phytohemagglutinin at 1 μg/ml. Forty-eight hours later, the cells were activated and maintained in RPMI 1640 medium containing 10% FBS and 20 U of interleukin-2 per ml.
Plasmids for envelope glycoprotein and sCD4 expression.
Plasmid pSVIIIenv, expressing the HIV-1 envelope glycoproteins from the HXBc2 isolate, has been described previously (32). The YU2 molecular clone was a gift from the AIDS Research and Reference Reagent Program (source, Beatrice Hahn). The YU2 envelope glycoprotein expression plasmid was made by introducing a BamHI site into the envelope gene by PCR and substituting the KpnI (6347)-BamHI (8475) fragment of plasmid pSVIIIenv with the amplified fragment as described previously (63). Amino acid changes were introduced into the HXBc2 envelope glycoprotein by site-directed mutagenesis, following the method of Kunkel, as previously described (7).
Envelope glycoprotein expression.
COS-1 cells were transfected by the DEAE-dextran method with pSVIIIenv DNA expressing envelope glycoproteins as described previously (7). To measure envelope protein expression at the cell surface, cells were radiolabeled with [35S]cysteine overnight. The cells were washed once with phosphate-buffered saline (PBS) containing 2% FBS and incubated for 90 min with an excess of a mixture of sera derived from HIV-1-infected individuals. Unbound patient serum was removed, the cells were washed four times with PBS containing 2% FBS and lysed in 0.75 ml of Nonidet P-40 (NP-40) buffer (0.5% NP-40, 0.5 M NaCl, 10 mM Tris HCl [pH 7.5]), and antiserum-gp120 complexes were precipitated on protein A-Sepharose. The amount of radiolabeled HIV-1 envelope glycoproteins bound to serum antibodies was measured by densitometric analysis of autoradiograms from sodium dodecyl sulfate (SDS)-polyacrylamide gels after SDS-polyacrylamide gel electrophoresis (PAGE).
sCD4 binding assay.
To measure the CD4 binding ability of envelope glycoproteins, envelope-transfected COS-1 cells were metabolically labeled with [35S]cysteine overnight. The radioactive supernatant was removed; the cells were washed with PBS containing 2% FBS and incubated with sCD4, at various concentrations, in 1 ml of PBS for 90 min at room temperature. The cells were washed four times with ice-cold PBS containing 2% FBS and lysed in 0.75 ml of NP-40 buffer, and the sCD4-gp120 complexes were immunoprecipitated with the OKT4 anti-CD4 antibody (Ortho Diagnostics). The amount of radiolabeled HIV-1 envelope glycoproteins bound to sCD4 was measured by densitometric analysis of autoradiograms from SDS-polyacrylamide gels.
Binding of 17b antibody to transfected COS-1 cells.
17b binding to cell surface-expressed HIV-1 envelope glycoproteins was measured in two ways. In the first method, COS-1 cells expressing HIV-1 envelope glycoproteins were radiolabeled with [35S]cysteine. Radioactive medium was removed, and the cells were washed with PBS containing 2% FBS (PBS-FBS). The cells were incubated with PBS containing 17b (5 μg/ml) or 17b plus sCD4 (1 μg/ml) for 90 min at room temperature. The cells were washed four times with PBS-FBS, and the cells were lysed in 0.75 ml of NP-40 buffer. The 17b-envelope glycoprotein complexes were precipitated by using protein A-Sepharose and quantitated by densitometry of SDS-polyacrylamide gels.
In the second method, hybridoma cells producing the 17b antibody were metabolically labeled with [35S]methionine and [35S]cysteine. Radiolabeled supernatants were incubated with COS-1 cells expressing the HIV-1 envelope glycoproteins as described above. The cells were washed and lysed as described above. The bound radiolabeled 17b antibody was detected by incubation of the cell lysates with protein A-Sepharose and analyzed as described above.
Envelope complementation and virus neutralization assay.
Complementation of a single round of replication of the env-deficient chloramphenicol acetyltransferase (CAT)-expressing provirus by the various envelope glycoproteins was performed as described previously (7, 32). To inhibit viral replication, monoclonal antibody was incubated with recombinant virus for 90 min at 37°C before addition of the virus to target lymphocytes (Molt 4 clone 8, Jurkat, or SupT1). Three days after infection, the target cells were lysed and CAT activity was measured as described previously. The standard deviation in this assay was experimentally determined and was less than 10% of the mean (data not shown).
Enzyme-linked immunosorbent assay (ELISA) determination of the effects of gp120 amino acid changes on CG10 antibody recognition.
A previously described panel of HIV-1 envelope glycoprotein mutants (69) was used to assess the effects of gp120 amino acid changes on recognition of gp120-CD4 complexes by the CG10 antibody.
COS-1 cells were transfected with 10 μg of pSVIIIenv DNA expressing wild-type or mutant HXBc2 envelope glycoproteins and a Tat-expressing plasmid, pSVTat. Seventy-two hours after transfection, cell supernatants were collected and frozen. For analysis of antibody recognition, various amounts of the supernatants (1 to 100 μl), supplemented with Tris-buffered saline–10% fetal calf serum to a total volume of 100 μl, were incubated in wells of Immulon II ELISA plates (Dynatech, Ltd.) coated with sheep antibody D7324 (Aalto BioReagents, Dublin, Ireland) to the carboxyl-terminal 15 amino acids of gp120.
The CD4 binding ability of the captured mutant glycoproteins was determined by incubating CD4-immunoglobulin G (IgG) (Genentech), diluted in Tris-buffered saline containing 2% nonfat milk powder and 20% sheep serum (TMSS buffer) at a concentration of 0.5 μg/ml, with the captured gp120 glycoproteins, followed by detection with alkaline phosphate-conjugated goat anti-human immunoglobulin (Accurate Chemicals, Westbury, N.Y.) and the AMPAK system (Dako Diagnostics).
To study the effects of gp120 amino acid changes on the ability of the CG10 antibody to recognize the gp120-CD4 complex, CD4-IgG at a final concentration of 0.5 μg/ml and CG10 at a final concentration of 2 μg/ml were incubated in TMSS buffer with the captured gp120 glycoproteins, followed by detection with alkaline phosphatase-conjugated goat anti-human IgG (Accurate Chemicals) and the AMPAK amplification system (Dako Diagnostics).
To determine the specific binding of the CG10 antibody to each mutant gp120 glycoprotein, the ratio of the binding of the CG10 antibody in the presence of CD4-IgG to that of CD4-IgG alone was calculated for each mutant. The average ratio for the entire panel of mutants was calculated, and any individual ratio deviating from the mean by less than 0.5 times was considered an indication of decreased gp120 recognition by the CG10 antibody, independent of effects of the amino acid change on CD4 binding. Likewise, those individual ratios deviating from the mean by more than 1.5 times were considered indications of gp120 residue changes that specifically increased CG10 recognition. Wells containing each mutant envelope glycoprotein were tested in triplicate, and mutants that exhibited binding ratios 0.5 times below or 1.5 times above the mean ratio in duplicate experiments are reported.
Analysis of HMAb competition for CG10 antibody binding.
Antigen capture ELISA was used for these studies. Briefly, BH10 gp120 was captured onto plastic (Immulon 2 Microplates; Dynatech), using the sheep antibody D7324 to the gp120 C terminus (Aalto BioReagents). Competitor HMAbs were added at 10 μg/ml for 15 min in a volume of 50 μl of TMSS buffer. Then sCD4 in 10 μl of TMSS buffer was added to a final concentration of 1 μg/ml. After incubation for 1 h, the bound CG10 antibody was detected by an alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulin antibody and the AMPAK system (Dako Diagnostics). Wells without gp120 and without sCD4 were used as controls. The mean and standard deviation of the optical density from triplicate ELISA wells were calculated, and the data reported are typical of results obtained in three similar experiments.
Induction of CG10 epitope by human sCD4 or rat-human sCD4 chimeras.
H9 cells were infected with the TCLA Hx10 clone of HIV-1 as previously described (53). Approximately 5 × 105 infected H9 cells were then incubated for 2 h at 4°C with 10 μg of human sCD4 or chimeric rat-human sCD4 per ml. The human sCD4 and human-rat sCD4 chimeras were obtained from J. Simon (The Sir William Dunn School of Pathology, Oxford, England) as protein purified from transfected CHO supernatants, as previously described (56). Under these saturating conditions, similar levels of binding were achieved for all forms of sCD4 as demonstrated by indirect immunofluorescent staining with the rat CD4/D4-specific monoclonal antibody OX71 (data not shown). The cells were washed twice in PBS–1% fetal calf serum–0.02% sodium azide and then incubated for 1 h at 4°C with serial dilutions of CG10. After washing, the cells were labeled with anti-mouse phycoerythrin (Immunotech, Marseille, France) for 30 min at 4°C, then washed a final time, and analyzed for fluorescence by using a FACScan with Lysis II software (Becton Dickinson, Mountain View, Calif.). Each point represents the mean of duplicate samples of 10,000 accumulated events gated on forward- and side-angle light scatter. Background staining represented by the signal obtained with the phycoerythrin-conjugated antibody alone was subtracted from the signal obtained in the presence of the primary antibody.
RESULTS
Binding of the 17b antibody to envelope glycoproteins of TCLA and primary HIV-1 isolates.
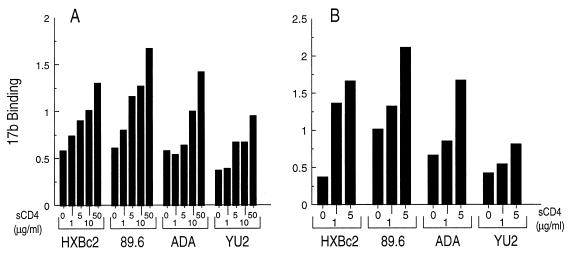
The induction of 17b binding to monomeric gp120 glycoprotein from a TCLA HIV-1 isolate (HXBc2) by sCD4 has been previously studied (67). The neutralizing activity of sCD4 and monoclonal antibodies is best predicted by studies that examine binding of these ligands to gp120 oligomers present on virions or cell surfaces (22, 40, 53, 63). Therefore, we examined the ability of sCD4 to induce the exposure of the 17b epitope on envelope glycoproteins expressed on the surface of COS-1 cells. Two methods were used to estimate sCD4-induced 17b binding to envelope glycoproteins expressed on cell surfaces. In Fig. 1A, purified 17b monoclonal antibody was bound to metabolically labeled COS-1 cells expressing the HIV-1 envelope glycoproteins in the absence or presence of increasing concentrations of sCD4. In a separate experiment (Fig. 1B), metabolically labeled 17b antibody from hybridoma supernatants was bound to unlabeled COS-1 cells expressing HIV-1 envelope glycoproteins. As was observed for gp120 monomers, the 17b antibody bound to the HXBc2 envelope glycoproteins in the absence of sCD4 but exhibited preferential binding when sCD4 was present. There was a dose-dependent increase in 17b binding in response to sCD4.
FIG. 1.
Soluble CD4 induction of the 17b epitope on the envelope glycoproteins of primary HIV-1 isolates. (A) Binding of 17b to radiolabeled envelope glycoproteins. COS-1 cells were transfected with plasmids expressing the different HIV-1 envelope glycoproteins and metabolically labeled. 17b antibody (5 μg/ml) was bound in the absence or presence of sCD4 for 90 min at room temperature, the cells were washed, and the amount bound was determined by precipitation of antibody-gp120 complexes with protein A-Sepharose. (B) Binding of radiolabeled 17b to unlabeled envelope glycoproteins. COS-1 cells were transfected as for panel A. The 17b antibody was metabolically labeled and binding was carried out as for panel A. Bound 17b was determined by precipitation with protein A-Sepharose and quantitated by SDS-PAGE. Data were normalized for the cell surface expression of each envelope glycoprotein, as determined by immunoprecipitation with pooled HIV-1-infected patient serum. The experiment shown is representative of at least six independent experiments.
Phenotypic differences, including sCD4 or antibody binding and neutralization, exist between TCLA and primary macrophage-tropic HIV-1 (20, 21, 64, 65). Therefore, we tested the ability of sCD4 to induce the 17b conformational change on the envelope glycoproteins of one primary dualtropic (89.6) and two macrophagetropic (ADA and YU2) HIV-1 isolates. Figure 1 shows that sCD4 induced a dose-dependent increase in 17b binding to the envelope glycoproteins of all the isolates examined.
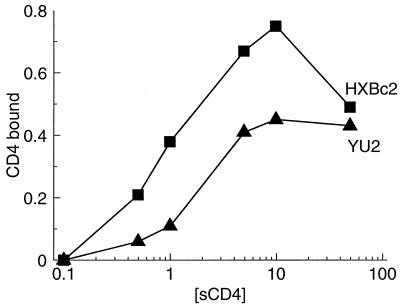
The YU2 envelope glycoproteins consistently exhibited lower induction of 17b binding than the other isolates tested. The binding of sCD4 to the oligomeric envelope glycoproteins of primary HIV-1 isolates is typically lower than that to TCLA isolates (42, 43, 48, 63). Since induction of 17b binding is a function of sCD4 binding, we tested the hypothesis that a reduced affinity of the YU2 envelope glycoproteins for sCD4 might explain the lower induction of 17b binding for this isolate. Different concentrations of sCD4 were incubated with the labeled HXBc2 and YU2 envelope glycoproteins expressed on the surface of COS-1 cells, and sCD4-gp120 complexes were precipitated from cell lysates with the OKT4 anti-CD4 antibody. As shown in Fig. 2, the YU2 envelope glycoproteins bound sCD4 less efficiently than the HXBc2 envelope glycoproteins, consistent with previous results (63). The apparent decrease in sCD4 binding to the HXBc2 envelope glycoproteins at the highest sCD4 concentration is due to sCD4-induced dissociation of the gp120 glycoprotein from the envelope glycoprotein complex (data not shown).
FIG. 2.
Binding of sCD4 to HIV-1 envelope glycoproteins. COS-1 cells were transfected with plasmids expressing either the HXBc2 or YU2 envelope glycoproteins and metabolically labeled. Soluble CD4 was bound at room temperature for 90 min, and the amount of sCD4 bound was determined by immunoprecipitation of the sCD4-envelope glycoprotein complexes with the OKT4 anti-CD4 antibody. Data were normalized for the cell surface expression of each envelope glycoprotein as described in the legend to Fig. 1.
The experiments described above demonstrate that the sCD4-induced conformational change resulting in exposure of the 17b epitope occurs on envelope glycoproteins expressed on the surface of COS-1 cells. The results also show that the envelope glycoproteins of primary HIV-1 isolates, including those that have not undergone propagation in tissue culture, undergo a similar conformational change. Similar results have also been obtained by 17b staining of HIV-1-infected cells in the presence or absence of sCD4 (results not shown).
Temperature dependence of 17b binding and induction by sCD4.
Membrane fusion mediated by the HIV-1 envelope glycoproteins is a temperature-dependent process (10). The rate of formation of syncytia between HIV-1 envelope glycoprotein-expressing cells and CD4-positive cells increases with temperature, up to 45°C. It has been proposed (30) that the envelope glycoproteins undergo a transition to activation intermediates after preincubation of envelope glycoprotein-expressing cells at 16°C.
We analyzed the temperature dependence of 17b binding to HIV-1 envelope glycoproteins expressed on the surface of COS-1 cells, in the absence and presence of sCD4. The levels of binding of the 17b antibody to the HXBc2 envelope glycoproteins in the absence of sCD4 were approximately equivalent at 4, 16, 22, and 37°C (data not shown). The levels of sCD4-induced increases in 17b binding were also similar at the four temperatures tested, demonstrating that the CD4-induced structural changes in the HIV-1 envelope glycoproteins associated with exposure of the 17b epitope are permitted over a wide temperature range.
Contribution of the V1/V2 variable loops to the exposure of the 17b epitope.
Previous work has implicated the gp120 V1/V2 variable loops in determining the exposure of the 17b epitope on the monomeric HIV-1 gp120 glycoprotein (74). The V1/V2 deletion (Δ128-194) includes residues 128 to 194, as previously described (74). A deletion of the V1/V2 loops led to a level of 17b epitope exposure equivalent to that observed for the wild-type gp120 glycoprotein in the presence of sCD4. Exposure of the 17b epitope on the V1/V2-deleted gp120 monomer was not increased further by sCD4 binding. These results suggested that, at least on the gp120 monomer, most of the effect of sCD4 binding on 17b epitope exposure could be explained by a CD4-induced movement of the V1/V2 loops.
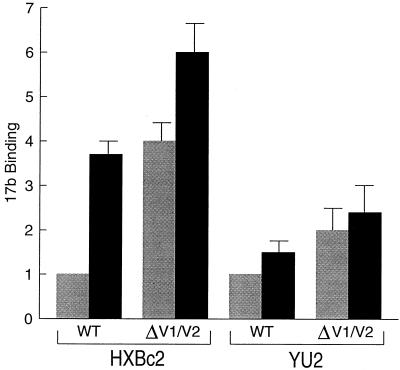
We wished to determine the effect of deletion of the V1/V2 loops on the exposure and sCD4 inducibility of the 17b epitope in the context of the native envelope glycoprotein oligomer. Therefore, wild-type and V1/V2-deleted (Δ128-194) envelope glycoproteins of the HXBc2 and YU2 HIV-1 isolates were expressed on the surface of COS-1 cells, and the binding of the 17b antibody in the absence or presence of sCD4 was measured. In the absence of sCD4, both HXBc2 and YU2 envelope glycoproteins containing the Δ128-194 deletion exhibited increased recognition by the 17b antibody, compared with the wild-type glycoproteins (Fig. 3). The binding of the 17b antibody to the Δ128-194 envelope glycoproteins exceeded the level of 17b binding to wild-type glycoproteins incubated with sCD4. The V1/V2-deleted envelope glycoproteins exhibited only a modest induction of 17b binding by sCD4. The induction ratio between wild-type and V1/V2-deleted glycoproteins was significantly different for the HXBc2 (P < 0.001) but not for the YU2 proteins. These experiments demonstrate that either sCD4 binding or deletion of the V1/V2 loops exposes the 17b epitope on the oligomeric envelope glycoproteins. These results are consistent with a model in which CD4 binding results in a movement of the V1/V2 loops from a position in which the 17b epitope is partially masked. These results also show that the gp120 structures involved in the CD4-induced conformational change relevant to 17b epitope exposure are similar for both a T-cell line and primary HIV-1 isolate.
FIG. 3.
Binding of the 17b antibody to HIV-1 envelope glycoproteins containing deletions in the V1/V2 loops. COS-1 cells were transfected with plasmids expressing either wild-type (WT) or V1/V2-deleted HXBc2 envelope glycoproteins and incubated at room temperature for 90 min with metabolically labeled 17b antibody in the absence (shaded bars) or presence (black bars) of sCD4 (1 μg/ml). Unbound antibody was removed, and the amount of 17b antibody bound was determined by precipitation with protein A-Sepharose and quantitation by SDS-PAGE. Cell surface expression levels of the wild-type and V1/V2-deleted envelope glycoproteins were equivalent. Data represent the average of three experiments and for each isolate are normalized to the level of 17b binding to the wild-type envelope glycoproteins in the absence of sCD4.
Effect of single gp120 amino acid changes on exposure of the 17b epitope.
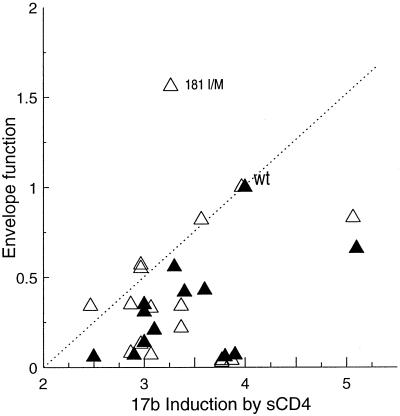
The hypothesis that sCD4-induced exposure of the 17b epitope is relevant to receptor-activated membrane fusion predicts that amino acid changes in the viral envelope glycoproteins that diminish the conformational change detected by the 17b antibody will also decrease envelope glycoprotein function. To test this, we expressed a panel of HXBc2 envelope glycoproteins containing single or double amino acid modifications (67) on the surface of COS-1 cells and assessed 17b antibody binding to the envelope glycoproteins in the absence or presence of sCD4. The function of each of these envelope glycoproteins in mediating the formation of syncytia and in supporting HIV-1 entry was examined. The efficiency with which sCD4-induced exposure of the 17b epitope, syncytium-forming ability, and function in virus entry for each mutant envelope glycoprotein is shown in Table 1 and Fig. 4.
TABLE 1.
17b induction and envelope fusion function for envelope glycoproteins containing changes in the variable loopsa
| gp120 region | Envelope glycoprotein | Virus entry | Syncytium formation | Induction of 17b binding |
|---|---|---|---|---|
| V1/V2 | Wild-type | 1 | 1 | 4 |
| 152/153 GE/SM | 0.33 | ND | 3.1 | |
| 168 K/L | 0.22 | ND | 3.4 | |
| 177 Y/F | 0.34 | 0.06 | 2.5 | |
| 178 K/E | 0.83 | 0.66 | 5.1 | |
| 179 L/V | 0.55 | 0.35 | 3 | |
| 179/180 LD/DL | 0.35 | ND | 2.9 | |
| 181 I/M | 1.56 | 0.56 | 3.3 | |
| 191/192/193 YSL/GSS | 0.06 | ND | 3.1 | |
| V3 | 298 R/G | 0.82 | 0.43 | 3.6 |
| 306/308 RR/SH | 0.07 | 0.21 | 3.1 | |
| 308 R/H | 0.13 | 0.14 | 3 | |
| 313 P/S | 0.34 | 0.42 | 3.4 | |
| 313/315 PR/LG | 0.03 | 0.06 | 3.8 | |
| 314 G/Q | 0.57 | 0.31 | 3 | |
| 314 G/W | 0.08 | 0.07 | 2.9 | |
| 315 R/G | 0.04 | 0.07 | 3.9 | |
| MT33+1 | 0.04 | 0.06 | 3.8 | |
| C4 | 420 I/R | 0.15 | ND | 1.8 |
| 475 M/S | 0.73 | ND | 2.5 |
17b induction is the ratio of 17b bound in the presence of sCD4 (1 μg/ml) to 17b binding in the absence of sCD4. Syncytium-forming ability was determined by cocultivation of envelope glycoprotein-expressing COS-1 cells with CD4+ SupT1 cells and is expressed relative to the wild-type level of 1. Virus entry was measured by using an envelope complementation assay described in Materials and Methods. Values represent virus entry relative to the level of entry by viruses with the wild-type envelope glycoprotein. The values presented for 17b binding, syncytium formation, and virus entry are representative of at least three experiments except for a subset of the virus entry values, which are the means of two experiments. ND, not determined.
FIG. 4.
Relationship between sCD4 induction of the 17b epitope and envelope glycoprotein function. Binding of the 17b antibody was measured as described in the legend to Fig. 4. Data were normalized for cell surface expression of each of the envelope glycoproteins as described in the legend to Fig. 1. Envelope glycoprotein function represents the ability of each envelope glycoprotein to mediate syncytium formation (closed symbols) or virus entry (open symbols) as described in Materials and Methods. Each data point represents the average of at least three experiments. The dashed line is for illustrative purposes and is not a linear regression of the data. WT, wild type.
Several mutant envelope glycoproteins with changes in the V1/V2 or V3 gp120 loops exhibited decreases in the efficiency with which the 17b epitope was exposed following incubation with sCD4. sCD4 binding to these mutant glycoproteins expressed on the surface of COS-1 cells was at least 80% of wild-type binding (data not shown). Invariably, these mutants exhibited decreases in the ability to form syncytia or to support virus entry compared with the wild-type HIV-1 envelope glycoproteins. One mutant (181 I/M) exhibited a modest decrease in the efficiency of 17b induction following sCD4 incubation but supported virus entry at least as well as the wild-type envelope glycoprotein. The syncytium-forming ability of the 181 I/M mutant, however, was reduced compared with that of the wild-type envelope glycoproteins. It is possible that the structure of this mutant envelope glycoprotein differs on the cell surface from that on the virion surface or that the consequences of this change differ in these two contexts. Some mutant envelope glycoproteins that exhibited reduced function in syncytium formation or virus entry assays exposed the 17b epitope in response to sCD4 equivalently to, or more efficiently than, the wild-type envelope glycoproteins. These mutant envelope glycoproteins may have defects that influence their ability to mediate fusion unrelated to exposure of the 17b epitope. Overall, these results support the hypothesis that HIV-1 envelope glycoproteins that inefficiently undergo the CD4-induced structural modification associated with 17b epitope exposure are less fusion competent.
Characterization of the epitope for the CG10 monoclonal antibody.
The CG10 monoclonal antibody was raised by immunization with the HIV-1 gp120 glycoprotein from the T-cell line-tropic IIIB strain complexed with sCD4. The CG10 antibody recognizes neither gp120 nor sCD4 alone, but only the gp120-sCD4 complex (27, 35). This finding suggests that the CG10 antibody recognizes either a neoepitope created by the binding of gp120 and CD4 or an epitope on gp120 or CD4 that is exposed or induced only in the presence of the other ligand. To determine whether the CG10 antibody recognized a conserved epitope, the gp120 glycoproteins from seven clade B primary HIV-1 isolates (source, Steve Wolinsky) were incubated with sCD4 and tested for recognition by the CG10 antibody. All seven gp120-sCD4 complexes were recognized with identical affinity by the CG10 antibody (data not shown), indicating that the antibody epitope represents a well-conserved structure. Similar results were obtained for cells infected with HIV-1 isolates from clades A, B, and D (data not shown), demonstrating that this CG10 epitope is also recognized in the oligomeric form of the envelope glycoproteins.
We examined whether the binding of the CG10 antibody to gp120-sCD4 complexes captured on an ELISA plate would be competed by other antibodies directed against the gp120 glycoproteins. As shown in Table 2, the efficient binding of the CG10 antibody to gp120 captured in this manner was dependent on the presence of sCD4, as expected. An antibody, 15e, directed against the gp120 CD4 binding site, decreased CG10 binding, probably by disrupting gp120-sCD4 complexes. Both the 17b antibody and the 48d antibody, which recognizes an epitope proximal to that of 17b, efficiently competed for CG10 binding to the gp120-sCD4 complex. Interestingly, the A32 antibody, the binding of which has been shown to increase the binding of the 17b and 48d antibodies to the gp120 glycoprotein (47), also enhanced the binding of the CG10 antibody to gp120-sCD4 complexes. The A32 antibody did not enhance CG10 antibody binding to the gp120 glycoprotein in the absence of sCD4 (data not shown). Other antibodies directed against gp120 (C11, 2G12, and 212A) did not affect the binding of the CG10 antibody. These results indicate that the CG10 and 17b antibodies bind to overlapping structures on the gp120-CD4 complex and that both epitopes are similarly induced by binding of the A32 antibody.
TABLE 2.
Competition of HMAbs for CG10 binding to the gp120 glycoproteina
| BH10 gp120 | HMAb (10 μg/ml) | sCD4 (1 μg/ml) | CG10 bound |
|---|---|---|---|
| − | None | − | 0.057 ± 0.017 |
| + | None | − | 0.083 ± 0.014 |
| + | None | + | 0.324 ± 0.053 |
| + | C11 | + | 0.324 ± 0.019 |
| + | 2G12 | + | 0.354 ± 0.023 |
| + | 212A | + | 0.303 ± 0.045 |
| + | 15e | + | 0.117 ± 0.013 |
| + | 48d | + | 0.063 ± 0.012 |
| + | 17b | + | 0.053 ± 0.007 |
| + | A32 | + | 0.583 ± 0.016 |
Binding of the CG10 antibody to the BH10 gp120 glycoprotein was measured in an ELISA format. The gp120 glycoprotein was captured on ELISA plates with an antibody (D7324) that binds to the C terminus of gp120. Values expressed are means ± standard deviations for replicate wells.
A previously published panel of HIV-1 HXBc2 gp120 mutants (69) was tested for ability to bind CD4-IgG and to be recognized by the CG10 antibody. None of the gp120 mutants were recognized by the CG10 antibody in the absence of CD4-IgG (data not shown). Several of the gp120 mutants previously shown to affect CD4 binding (177 Y/F, 256 S/Y, 257 T/R, 262 N/T, 368 D/R, 370 E/Q, 370 E/R, 427 W/V, 427 W/S, 427 W/R, 457 D/R, and 457 D/A) were not recognized by the CG10 antibody. We confirmed that these mutant envelope glycoproteins bound CD4-IgG inefficiently (data not shown), suggesting that the effect of these gp120 amino acid changes on CG10 binding resulted from poor CD4 binding. The CG10 antibody bound to gp120 glycoproteins containing deletions of the V1/V2 loops and of the V3 loop (Δ128-194 and Δ298-327, respectively), indicating that the major gp120 variable loops are not necessary for the formation of the CG10 epitope. While the CG10 antibody recognized a complex of CD4-IgG and a gp120 glycoprotein lacking the V1/V2 loops (Δ128-194), the antibody did not bind to a complex of CD4-IgG and a gp120 mutant lacking the entire V1/V2 stem-loop structure (Δ119-205) (Table 3). The latter mutant efficiently binds CD4 (74) but is recognized inefficiently by the 17b and 48d antibodies. Other amino acid changes, 314 G/W (affecting the V3 loop) and 432 K/A (affecting the fourth conserved [C4] region of gp120), resulted in decreased recognition by the CG10 antibody, although CD4 binding was similar to that of the wild-type gp120 glycoproteins. Finally, a few mutants (Δ298-327, 384 Y/G, 298 R/A and 435 Y/S) displayed increases in CG10 binding, relative to that seen for the wild-type glycoprotein.
TABLE 3.
CG10 recognition of mutant HXBc2 envelope glycoproteins in the presence of sCD4
| Glycoprotein | Binding ratioa |
|---|---|
| Decreased recognition | |
| Δ119-205 | 0.07 |
| 314 G/W | 0.33 |
| 432 K/A | 0.26 |
| 183/184 PI/SG | 0.29 |
| Increased recognition | |
| Δ298-327 | 2.5 |
| 384 Y/E | 2.2 |
| 298 R/G | 1.7 |
| 435 Y/S | 1.7 |
Binding of CG10 to the mutant glycoprotein divided by the average CG10 binding for the entire panel of mutants.
Contribution of CD4 and gp120 to the CG10 epitope.
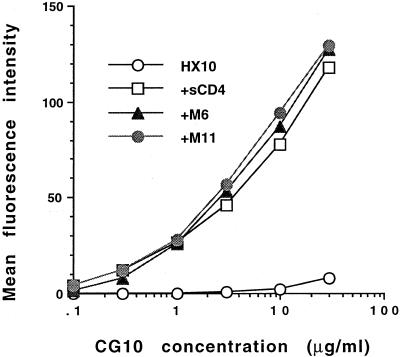
To examine the potential contribution of CD4 structures to the CG10 epitope, Hx10-infected H9 cells were pretreated with saturating concentrations of human sCD4 or rat-human sCD4 chimeras before the addition of the CG10 antibody. The chimera M6 is full-length rat CD4 with a substitution of human CD4 amino acid residues 33 to 62. This substitution allowed the chimeric CD4 protein to bind gp120 (60). The M11 chimera contains an additional substitution of human amino acids 80 to 95, which specify the CDR-3-like loop of CD4, and binds gp120 with an affinity similar to that of M6. The CG10 antibody bound equivalently to envelope glycoprotein-expressing cells incubated with all three sCD4 molecules (Fig. 5). Thus, the only human CD4 region required for induction of the CG10 epitope is the CDR-2-like loop. Therefore, any CD4 component of the CG10 epitope must either reside within the CDR2-like loop or be conserved between human and rat CD4.
FIG. 5.
Induction of the CG10 epitope by human sCD4 or rat-human sCD4 chimeras. Hx10-infected H9 cells were pretreated with sCD4 of human origin or rat sCD4 in which residues 33 to 62 (M6) or 33 to 62 and 80 to 95 (M11) were substituted for the human sequence. Both of these chimeric molecules bind gp120 with an affinity close to that observed for wild-type sCD4 (55). CG10 binding was subsequently measured by indirect immunofluorescence and flow cytometric analysis. The results are expressed as mean fluorescence units, and each data point represents the mean of duplicate samples of 10,000 gated events each.
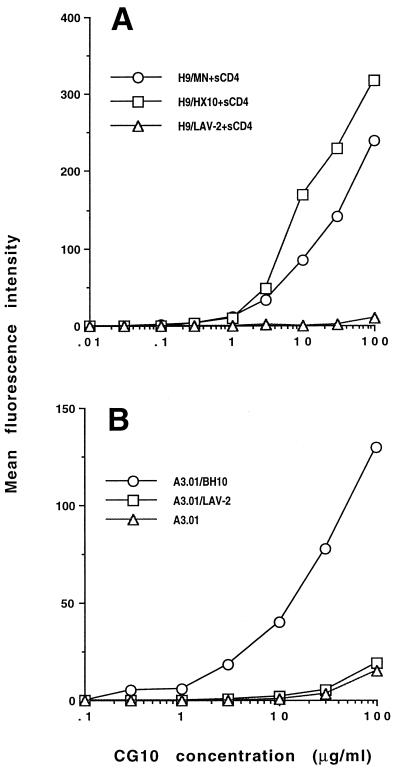
To study the CG10 epitope further, we compared the exposure of the CG10 epitope by human sCD4 on gp120 from different viral origins: the Hx10 and MN strains of HIV-1 and the HIV-2 isolate LAV-2. The CG10 antibody efficiently recognized cells expressing the HIV-1 envelope glycoproteins but did not appreciably bind cells expressing the HIV-2 glycoproteins (Fig. 6A). Similar levels of sCD4 binding were observed for each type of Env expressed on the infected cells (data not shown); thus, differences in CD4 binding did not account for the different CG10 binding observed.
FIG. 6.
CG10 binding to cells expressing HIV-1 or HIV-2 envelope glycoproteins. (A) HIV-infected H9 cells that express the envelope glycoproteins of HIV-1 MN and Hx10 strains or HIV-2 LAV-2 at the cell surface were incubated with sCD4 for 2 h at 4°C, washed, and incubated with increasing concentrations of CG10 as described in Materials and Methods. CG10 binding was quantitated as described in the legend to Fig. 5. (B) CG10 binding to CD4+ A3.01 cells untreated or pretreated with soluble gp120 from HIV-1 BH10 or gp105 from HIV-2 LAV-2.
To confirm this result with cell-associated CD4, we bound soluble gp120 and soluble gp105 from BH10 and LAV-2, respectively, to the CD4+ T-cell line A3.01 and then examined the binding of the CG10 antibody to these cells. Despite the equivalent levels of Env bound to the cells, no binding of CG10 to LAV-2-carrying cells was detected, whereas strong CG10 binding was observed for BH10-coated cells (Fig. 6B). The most likely reason for the lack of CG10 binding to the LAV-2 gp105-CD4 complex is that an important component of the CG10 epitope is not conserved on HIV-2 gp120. This result supports those obtained by examination of CG10 binding to a panel of HIV-1 gp120 mutants and suggest that a major element of the CG10 epitope resides on gp120. This gp120 element must be reasonably conserved, since the envelope glycoproteins of viruses from HIV-1 clades A, B, and D are able to expose the CG10 epitope when complexed with sCD4 (data not shown).
HIV-1-neutralizing activity of the 17b and CG10 antibodies.
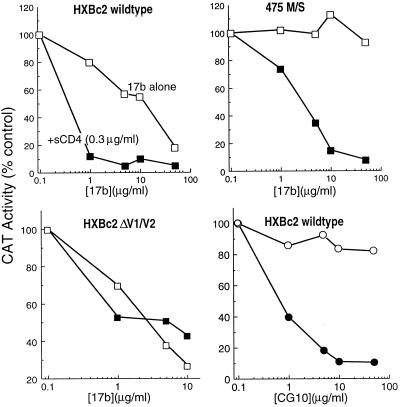
We wished to examine the relationship between the binding of antibodies to the CD4-induced epitopes on gp120 and virus-neutralizing activity. The 17b antibody has been shown to neutralize HIV-1, although the potency of this antibody is relatively low, requiring more than 30 μg/ml to achieve 90% inhibition of TCLA HIV-1 entry (67). One possible explanation for this weak neutralizing activity is that the 17b epitope is poorly exposed on the envelope glycoproteins in their native configuration. Since sCD4 induces the exposure of the 17b epitope, we hypothesized that a subinhibitory concentration of sCD4 might enhance the binding of the 17b antibody to HIV-1 virions, thereby increasing the efficiency of neutralization. To test this, we used an env complementation assay to examine neutralization by the 17b antibody in the absence or presence of 0.3 μg of sCD4 per ml, which alone exhibited no neutralizing activity (Fig. 7). In the absence of sCD4, the 17b antibody neutralized virions containing the wild-type HXBc2 envelope glycoproteins with a 50% inhibitory concentration (IC50) slightly greater than 10 μg/ml. When the experiment was performed in the presence of 0.3 μg of sCD4 per ml, the 17b antibody neutralized the virus with an IC50 of 0.04 μg/ml. These results indicate that sCD4, even at concentrations that are not neutralizing, can significantly increase the neutralizing capacity of the 17b antibody.
FIG. 7.
Neutralization by the 17b and CG10 antibodies of viruses containing wild-type or mutant HXBc2 envelope glycoproteins. Envelope glycoprotein-mediated virus entry was determined in a single-round replication assay as described in Materials and Methods. Viruses bearing wild-type or mutant HXBc2 envelope glycoproteins were incubated for 90 min at room temperature with the 17b or CG10 antibody (open symbols) at the concentrations indicated. In parallel samples (closed symbols), viruses were incubated with both 17b or CG10 antibody and a subinhibitory concentration of sCD4 at 0.3 μg/ml. In the absence of added antibody, CAT conversion was the same in the absence or presence of sCD4 (0.3 μg/ml).
We wished to examine neutralization by the 17b antibody of viruses with envelope glycoproteins that exhibited changes in antibody recognition. We examined 17b antibody neutralization of viruses with the Δ128-194 envelope glycoprotein, which lacks the V1/V2 loops and is recognized almost equivalently by the 17b antibody in the absence or presence of sCD4 (Fig. 7). The virus with the Δ128-194 envelope glycoprotein was neutralized more efficiently than the wild-type virus in the absence of sCD4 and did not become more sensitive to neutralization by the 17b antibody when sCD4 was present. We also examined inhibition of viruses containing mutant envelope glycoproteins that are recognized less efficiently by the 17b antibody. It has been reported that amino acid changes at gp120 residues 420 and 475 resulted in a marked reduction in recognition of monomeric envelope glycoproteins by the 17b antibody and allowed escape from neutralization by the 17b antibody (67). Recognition of these envelope glycoproteins by the 17b antibody was partially restored by sCD4 binding (reference 67 and Table 1). In the absence of sCD4, 17b antibody concentrations of up to 50 μg/ml failed to neutralize viruses bearing envelope glycoproteins with the 420 I/R or 475 M/S change (Fig. 7 and data not shown). However, addition of sCD4 at 0.3 μg/ml allowed the 17b antibody to neutralize both viruses, with IC50 values of less than 10 μg of the 17b antibody per ml (Fig. 7 and data not shown).
For the wild-type and mutant glycoproteins examined, the efficiency of 17b antibody binding to the cell surface envelope glycoprotein complex was predictive of 17b neutralization, in both the absence and presence of sCD4 (not shown). These results provide evidence that the neutralization potential of the 17b antibody can be enhanced by CD4 binding, suggesting that accessibility of this epitope on the functional envelope glycoprotein complex can be increased by sCD4 in a manner not achieved upon virion binding to cell surface CD4.
To examine this issue further, we studied the neutralizing ability of the CG10 antibody in the absence and presence of subinhibitory sCD4 concentrations. As expected from the inability of the CG10 antibody to recognize the HIV-1 envelope glycoproteins in the absence of CD4, the CG10 antibody exhibited no neutralizing ability at concentrations of up to 50 μg/ml. However, in the presence of sCD4 at 0.3 μg/ml, the CG10 antibody neutralized the virus with the HXBc2 envelope glycoproteins with an IC50 of less than 1 μg of antibody per ml. Thus, the CG10 antibody can bind and neutralize the functional HIV-1 envelope glycoprotein complex in the presence of sCD4 even though it does not do so in the context of virion binding to cell surface CD4.
DISCUSSION
Quantitatively, the increased binding of the 17b antibody represents one of the most dramatic of the conformational changes in the HIV-1 gp120 glycoprotein induced by interaction with the CD4 receptor (54, 55, 67). We show that this consequence of CD4 binding is a property of envelope glycoproteins from both TCLA and primary HIV-1 isolates and that the 17b antibody binds to native, oligomeric HIV-1 envelope glycoproteins more efficiently in the presence of sCD4. Deletion of the V1/V2 gp120 loops from the native envelope glycoprotein complex results in an increase of the 17b antibody bound, reaching a level equivalent to that seen for the wild-type glycoproteins in the presence of sCD4. Addition of sCD4 minimally affects the binding of the 17b antibody to the V1/V2-deleted mutant. These observations support the model previously proposed (74) based on studies of the monomeric gp120 glycoprotein; i.e., sCD4 interaction with the HIV-1 envelope glycoproteins results in a movement of the V1/V2 loops, demasking the 17b epitope. The 17b epitope exposure occurs over a wide temperature range, consistent with a model in which the energy derived from CD4 binding is sufficient to drive the V1/V2 loops into a new conformation. In the wild-type envelope glycoproteins, this new conformation allows increased exposure of the 17b epitope; however, in a number of gp120 mutants in which the V2 or V3 loop is altered, the level of induction in 17b antibody binding is lower. Since the V2 and V3 loops are not absolutely essential for the integrity of the 17b epitope, the simplest explanation is that the altered conformation of the V2 and V3 loops does not allow complete demasking of the 17b epitope. Previous studies suggesting structural and functional interactions between V1/V2 and V3 gp120 loops support the possibility that either of these structures resides proximal to the 17b epitope and could, with rather minimal conformational shifts, restrict antibody access to this epitope (46, 47, 62).
While it is not possible to prove conclusively that a given conformational change in the HIV-1 envelope glycoproteins has functional significance, all of the available evidence supports an important functional role for exposure of the 17b epitope. The 17b antibody is neutralizing and has been shown to block the interaction of gp120-sCD4 complexes with chemokine receptors (68, 73). The HIV-1 envelope glycoprotein mutants examined here that exhibit reductions in exposure of the 17b epitope invariably exhibit decreases in membrane fusion-related functions. A few of the envelope glycoprotein mutants exhibited decreases in function greater than might be expected based on the observed reductions in exposure of the 17b epitope, indicating that fusion-related processes other than 17b epitope exposure may be affected by these changes. It is possible, for example, that chemokine receptor interactive sites are altered by some of the amino acid changes.
There are a number of similarities between the CG10 antibody and the 17b antibody. Both antibodies block the interaction of gp120-CD4 complexes with chemokine receptors (73). The 17b and related 48d antibodies compete with the CG10 antibody for binding gp120-sCD4 complexes. Both the 17b and CG10 epitopes are induced by binding of the A32 antibody, although in the case of CG10, such induction is evident only if sCD4 is also bound to the gp120 glycoprotein. The binding of both 17b and CG10 antibodies is not dependent on the presence of the V1/V2 loops but is dramatically affected by deletion of the V1/V2 stem. Furthermore, some changes in the C4 gp120 region, which has previously been implicated in interactions with the V2 and V3 loops (46, 47, 72), alter 17b and CG10 binding independently of effects of the changes on CD4 binding. These results make it likely that both 17b and CG10 antibodies recognize closely related, conserved structures on the gp120 glycoprotein. The strict dependence of CG10 binding on CD4 suggests either that CG10 recognizes a gp120 element that is extremely well masked or not formed in the absence of bound CD4 or that one or more CD4 residues constitute a necessary component of the epitope.
In both the absence and presence of sCD4, a good correlation was observed between 17b antibody binding to mutant and wild-type HIV-1 envelope glycoprotein complexes expressed on the cell surface and the ability of the 17b antibody to neutralize virus. These results support a growing body of data that suggest that the HIV-1 envelope glycoproteins expressed on the cell surface reasonably represent those on virions (53). The ability of subneutralizing concentrations of sCD4 to enhance the neutralizing ability of the 17b antibody suggests that increased binding of the 17b antibody to native HIV-1 envelope glycoproteins occurs more efficiently in the context of sCD4 binding compared with cell surface CD4 binding, or, if binding is equivalent, the neutralizing potency of the antibody is less in the latter context. The neutralization studies with the CG10 antibody, which recognizes a related but absolutely CD4-dependent epitope, support the existence of significant differences in antibody accessibility or neutralization potency between a situation in which sCD4 is added to virions and a situation in which virus is binding to cell surface CD4. In both contexts, it is likely that one or more CD4 molecules interacting with the oligomeric envelope glycoproteins, as well as the envelope glycoprotein variable loops, contribute to the steric exclusion of antibodies from gp120 regions destined for chemokine receptor interactions. In addition, in the interaction of virions with cells, the target cell membrane in which CD4 is anchored could contribute to impeding antibody access to the viral envelope glycoprotein surface. Following virus binding to CD4, the 17b and CG10 epitopes are likely to face the target cell membrane, since antibodies to these structures efficiently block chemokine receptor interaction (68, 73). It is also noteworthy that in the neutralization experiments, subinhibiting concentrations of sCD4 allowed effective neutralization by the 17b and CG10 antibodies. This finding implies either that the binding of a subsaturating amount of sCD4 to the envelope glycoprotein oligomer induces multiple binding sites for the 17b and CG10 antibodies or that these antibodies are more effective at virus neutralization than is sCD4 for each molecule bound to the viral glycoproteins. Binding of a single antibody molecule to the region of the envelope glycoprotein spike facing the target cell membrane prior to interaction of the virus with the cell surface could explain this potency. Thus, while the chemokine receptor binding region of the HIV-1 envelope glycoproteins represent attractive targets, antibodies directed against this region may be most effective when accessing these sites prior to the time that virus attachment to the target cell occurs. There are considerable advantages for HIV-1 isolates that sequester the chemokine receptor interactive regions away from the humoral immune response until proximity to the target cell and the above-described steric factors allow exposure of these regions with impunity. Further studies may suggest a means to circumvent this viral strategy.
ACKNOWLEDGMENTS
We thank James Binley for helpful comments and Yvette McLaughlin for manuscript preparation.
J.M. and J.S. were supported by NIH award AI 39420. J.M. was supported by NIH award AI 36082. J.S. was supported by NIH awards AI 24755 and AI 31783 and by gifts from the late William McCarty-Cooper, the Friends 10, the Mathers Charitable Foundation, and Douglas and Judy Krupp. Q.S. was supported by the Centre Nationale de la Recherche Scientifique and the Agence Nationale de la Recherche sur le SIDA of France. The Dana-Farber Cancer Institute was a recipient of a Center for AIDS Research grant.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Allan J S. Receptor-mediated activation of immunodeficiency viruses in viral fusion. Science. 1991;252:1322–1323. doi: 10.1126/science.1925547. [DOI] [PubMed] [Google Scholar]
- 3.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 4.Allan J S, Whitehead E M, Strout K, Short M, Kanda P, Hart T K, Bugelski P J. Strong association of simian immunodeficiency virus (SIVagm) envelope glycoprotein heterodimers: possible role in receptor-mediated activation. AIDS Res Hum Retroviruses. 1992;8:2011–2020. doi: 10.1089/aid.1992.8.2011. [DOI] [PubMed] [Google Scholar]
- 5.Back N K, Smit L, Schutten M, Nara P L, Tersmette M, Goudsmit J. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J Virol. 1993;67:6897–6902. doi: 10.1128/jvi.67.11.6897-6902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 7.Bergeron L, Sullivan N, Sodroski J. Target cell-specific determinants of membrane fusion within the human immunodeficiency virus type 1 gp120 third variable region and gp41 amino terminus. J Virol. 1992;66:2389–2397. doi: 10.1128/jvi.66.4.2389-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bugelski P J, Ellens H, Hart T K, Kirsh R L. Soluble CD4 and dextran sulfate mediate release of gp120 from HIV-1: implications for clinical trials. J Acquired Immune Defic Syndr. 1991;4:923–924. [PubMed] [Google Scholar]
- 9.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 10.Busso M, Thornthwaite J, Resnik L. HIV-induced syncytium formation requires the formation of conjugates between virus-infected and uninfected T-cells in vitro. AIDS. 1991;5:1425–1432. doi: 10.1097/00002030-199112000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 15.Denisova G, Raviv D, Mondor I, Sattentau Q J, Gershoni J M. Conformational transitions in CD4 due to complexation with HIV envelope glycoprotein gp120. J Immunol. 1997;158:1157–1164. [PubMed] [Google Scholar]
- 16.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 17.Earl P L, Broder C C, Doms R W, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 20.Fenyo, E. M., J. Albert, and B. Asjo. 1989. Replicative capacity, cytopathic effect and cell tropism of HIV. AIDS 3(Suppl. 1):S5–S12. [DOI] [PubMed]
- 21.Fenyo E M, Morfeldt-Manson L, Chiodi F, Lind B, Von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y K, Hart T K, Jonak Z L, Bugelski P J. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:3818–3825. doi: 10.1128/jvi.67.7.3818-3825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallaher W R. Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- 25.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 26.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, Palker T J, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 27.Gershoni J M, Denisova G, Raviv D, Smorodinsky N I, Buyaner D. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J. 1993;7:1185–1187. doi: 10.1096/fasebj.7.12.7690724. [DOI] [PubMed] [Google Scholar]
- 28.Granelli-Piperno A, Moser B, Pope M, Chen D, Wei Y, Isdell F, O’Doherty U, Paxton W, Koup R, Mojsov S, Bhardwaj N, Clark-Lewis I, Baggiolini M, Steinman R M. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hart T K, Kirsh R, Ellens H, Sweet R W, Lambert D M, Petteway S R, Jr, Leary J, Bugelski P J. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart T K, Truneh A, Bugelski P J. Characterization of CD4-gp120 activation intermediates during human immunodeficiency virus type 1 syncytium formation. AIDS Res Hum Retroviruses. 1996;12:1305–1313. doi: 10.1089/aid.1996.12.1305. [DOI] [PubMed] [Google Scholar]
- 31.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 32.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho D D, McKeating J A, Li X L, Moudgil T, Daar E S, Sun N C, Robinson J E. A conformational epitope on gp120 important in CD4 binding and HIV-1 neutralization identified by a human monoclonal antibody. J Virol. 1991;65:489–493. doi: 10.1128/jvi.65.1.489-493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Peden K, Dimitrov D S, Broder C C, Manischewitz J, Denisova G, Gershoni J M, Golding H. Enhancement of human immunodeficiency virus type 1 envelope-mediated fusion by a CD4-gp120 complex-specific monoclonal antibody. J Virol. 1997;71:6037–6043. doi: 10.1128/jvi.71.8.6037-6043.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 37.Maddon P J, McDougal J S, Clapham P R, Dalgleish A G, Jamal S, Weiss R A, Axel R. HIV infection does not require endocytosis of its receptor, CD4. Cell. 1988;54:865–874. doi: 10.1016/s0092-8674(88)91241-x. [DOI] [PubMed] [Google Scholar]
- 38.McDougal J S, Maddon P J, Dalgleish A G, Clapham P R, Littman D R, Godfrey M, Maddon D E, Chess L, Weiss R A, Axel R. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harbor Symp Quant Biol. 1986;51(Pt. 2):703–711. doi: 10.1101/sqb.1986.051.01.083. [DOI] [PubMed] [Google Scholar]
- 39.McDougal J S, Maddon P J, Orloff G, Clapham P R, Dalgleish A G, Jamal S, Weiss R A, Axel R A. Role of CD4 in the penetration of cells by HIV. Adv Exp Med Biol. 1991;300:145–154. doi: 10.1007/978-1-4684-5976-0_9. [DOI] [PubMed] [Google Scholar]
- 40.McKeating J A, Shotton C, Cordell J, Graham S, Balfe P, Sullivan N, Charles M, Page M, Bolmstedt A, Olofsson S, Kayman S C, Wu Z, Pinter A, Dean C, Sodroski J, Weiss R A. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:4932–4944. doi: 10.1128/jvi.67.8.4932-4944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore J P, Klasse P J. Thermodynamic and kinetic analysis of sCD4 binding to HIV-1 virions and of gp120 dissociation. AIDS Res Hum Retroviruses. 1992;8:443–450. doi: 10.1089/aid.1992.8.443. [DOI] [PubMed] [Google Scholar]
- 43.Moore J P, McKeating J A, Huang Y X, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore J P, McKeating J A, Norton W A, Sattentau Q J. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J Virol. 1991;65:1133–1140. doi: 10.1128/jvi.65.3.1133-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 46.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S W, Fung M S, Traincard F, Pinkus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien W A, Mao S H, Cao Y, Moore J P. Macrophage-tropic and T-cell line-adapted chimeric strains of human immunodeficiency virus type 1 differ in their susceptibilities to neutralization by soluble CD4 at different temperatures. J Virol. 1994;68:5264–5269. doi: 10.1128/jvi.68.8.5264-5269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 50.Posner M, Elboim H, Santos D. The construction and use of a human-mouse myeloma analogue suitable for the routine production of hybridomas secreting human monoclonal antibodies. Hybridoma. 1987;6:611–625. doi: 10.1089/hyb.1987.6.611. [DOI] [PubMed] [Google Scholar]
- 51.Robinson J, Holton D, Pacheco-Morell S, Liu J, McMurdo H. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV transformed cell lines. AIDS Res Hum Retroviruses. 1990;6:567–579. doi: 10.1089/aid.1990.6.567. [DOI] [PubMed] [Google Scholar]
- 52.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 56.Schockmel G A, Somoza C, Davis S J, Williams A F, Healey D. Construction of a binding site for human immunodeficiency virus type 1 gp120 in rat CD4. J Exp Med. 1992;175:301–304. doi: 10.1084/jem.175.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schutten M, Andeweg A C, Bosch M L, Osterhaus A D. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol. 1995;41:18–22. doi: 10.1111/j.1365-3083.1995.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 58.Scott C F, Silver S, Profy A T, Putney S D, Langlois A, Weinhold K, Robinson J. Human monoclonal antibody which recognizes the V3 region of HIV-1 gp120 and neutralizes the HTLV-III MN strain. Proc Natl Acad Sci USA. 1990;87:8597–8601. doi: 10.1073/pnas.87.21.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon J H M, Stumbles P, Signoret N, Somoza C, Puklavec M, Sattentau Q J, Barclay A N, James W. Role of CD4 epitopes outside the gp120-binding site during entry of human immunodeficiency virus type 1. J Virol. 1997;71:1476–1484. doi: 10.1128/jvi.71.2.1476-1484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skehel J J, Bizebard T, Bullough P A, Hughson F M, Knossow M, Steinhauer D A, Wharton S A, Wiley D C. Membrane fusion by influenza hemagglutinin. Cold Spring Harbor Symp Quant Biol. 1995;60:573–580. doi: 10.1101/sqb.1995.060.01.061. [DOI] [PubMed] [Google Scholar]
- 62.Sodroski J, Wyatt R, Olshevsky U, Olshevsky V, Moore J. Conformation of the HIV-1 gp 120 envelope glycoprotein. Antibiot Chemother. 1996;48:184–187. [PubMed] [Google Scholar]
- 63.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tersmette M, de Goede R E, Al B J, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tersmette M, Gruters R A, de Wolf F, de Goede R E, Lange J M, Schellekens P T, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thali M, Furman C, Helseth E, Repke H, Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 69.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 71.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 72.Willey R L, Martin M A, Peden K W. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardins E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 74.Wyatt R, Moore J, Accola M, Desjardins E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]