Abstract
Background
Allergic conjunctivitis (AC) afflicts a significant portion of the global populace. Yet, its metabolic foundations remain largely unexplored.
Methods
We applied Mendelian Randomization (MR) and Linkage Disequilibrium Score Regression (LDSC) to scrutinize a cohort comprising 20 958 AC cases and 356 319 controls. Data were amalgamated from the metabolomics GWAS server and the FinnGen project, under strict quality control protocols.
Results
Using two-sample MR analysis, 486 blood metabolites were investigated in relation to AC. The IVW approach highlighted 18 metabolites as closely tied to AC risk; of these, 16 retained significance post sensitivity assessments for heterogeneity and horizontal pleiotropy. LDSC analysis, adopted to bolster our findings and negate confounders from shared genetic markers, revealed 8 metabolites with marked heritability, including: palmitate (OR = 0.614), 3-methoxytyrosine (OR = 0.657), carnitine (OR = 1.368), threonate (OR = 0.828), N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide (OR = 1.257), metoprolol acid metabolite (OR = 0.982), oleoylcarnitine (OR = 0.635), and 2-palmitoylglycerophosphocholine (OR = 1.351).
Conclusion
AC is precipitated by ocular responses to environmental allergens. Our study unveils a causal link between 8 blood metabolites and AC. This insight accentuates the role of metabolites in AC onset, suggesting novel avenues for its early prediction, targeted prevention, and tailored therapeutic interventions.
Keywords: Allergic conjunctivitis, Metabolites, Mendelian randomization, GWAS, Causal associations
Introduction
Allergic conjunctivitis (AC), an immune-driven hypersensitivity disorder, afflicts approximately 40% of the global population.1 Manifestations of this condition predominantly include ocular pruritus, conjunctival erythema, and edema.2 Such disturbances invariably deteriorate the individual's quality of life. Persistent exposure to allergens and continual ocular irritation often necessitate repetitive medical consultations, leading to significant economic implications. AC remains a predominant concern for both allergists and ophthalmologists, marking its significant clinical prevalence.3 The etiology of AC encompasses genetic predisposition, regional air quality, pet exposure, and individual susceptibility.4 However, the metabolic alterations associated with AC have been relatively underexplored.
Metabolomics, an emergent dimension within systems biology, heralds a novel avenue in comprehending disease etiopathogenesis. Modern analytical techniques facilitate exhaustive profiling of metabolites in various biological matrices, fortifying the essence of metabolomics. Metabolites, wielding roles as signaling agents, immunomodulators, intrinsic toxins, and environmental sensors, profoundly influence disease pathophysiology.5 For instance, research by Liu et al elucidated the relationship between fatty acid unsaturation levels and skin aging.5 Yun et al discerned correlations between certain circulating metabolites and colorectal cancer,6 while Guo et al unveiled the significance of blood metabolites in non-alcoholic fatty liver disease.7 Metabolomic markers have proven indispensable in disease diagnosis, therapeutic monitoring, and prognostication. Contemporary medical research ardently pursues human serum and plasma metabolomics for biomarker discovery and disease detection. Recent insights suggest that neuropeptide CGRP, activated by interleukin-33, interacts with somatosensory neurons, precipitating conjunctival irritation in AC patients.8 Literature attests that AC symptoms primarily emanate from Ig E and type 2 immune responses, characterized by inflammatory cell influx, notably eosinophils and mast cells.9 Several studies underscore the anti-inflammatory prowess of omega-3 fatty acids and their metabolites in countering allergic afflictions. Notably, lipid mediators are pivotal in AC pathogenesis.10 Prostaglandin (PG) D2, an influential cyclooxygenase metabolite derived from activated mast cells, is implicated in allergic disorders' etiology.10 Research by Arimura et al. advocates the therapeutic potential of DP receptor antagonists in allergy syndromes precipitated by mast cell hyperactivity.11 The prospect of leveraging blood metabolite inhibitors could revolutionize therapeutic strategies, especially in chemotherapy resistance contexts. Recognizing the blood metabolites integral to AC predisposition could significantly advance its early detection, prophylaxis, and treatment modalities. Yet, the dearth of prospective studies delineating the interplay between blood metabolites and AC leaves a pressing lacuna in our understanding.
Mendelian randomization (MR) offers a cutting-edge, analytical paradigm to decipher the causal nexus between exposure variables and outcomes using single nucleotide polymorphism (SNP) conglomerates.12 MR harnesses genetic variance to formulate instrumental variables for exposure, thereby elucidating exposure-disease causality.13 Although randomized controlled trials (RCTs) are the gold standard, their feasibility constraints render MR a potent alternative.14 Moreover, MR can identify disease biomarkers by investigating the genetic predisposition's ripple effects on other biological parameters. A case in point is the study by Zheng et al., which established a causal link between cardiovascular metabolic markers and chronic renal disease via MR.15 This study pioneers the exploration of the nexus between blood metabolites and AC. Leveraging genome-wide association study (GWAS) derived summary data, we employ MR to assiduously discern the causal influence of 11 pivotal blood metabolites on AC. The crux of our investigation orbits around unraveling the causal interplay between human blood metabolites and AC predisposition.
Methods
Metabolite data source overview
The Metabolomics GWAS Server (http://metabolomics.helmholtz-muenchen.de/gwas/) provided summary-type GWAS data for human serum metabolites. Shin et al. conducted an extensive metabolic analysis coupled with genome-wide association scanning, unveiling metabolites intrinsically linked to human genetic variance.16 We used data from the UK Twin Study and KORA F4 cohorts, adhering to their fasting and blood processing protocols, ensuring the validity of our metabolite/SNP associations. This pivotal study encompassed a cohort of 7824 individuals of European descent, with 6056 drawn from the UK Twin Study and an additional 1768 from the German-centric KORA F4 cohorts. Among the 491 metabolites scrutinized, 200 remained enigmatic, labeled as unidentified due to their yet-to-be deciphered chemical properties. Conversely, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database has elucidated 309 metabolites, chemically authenticating them and subsequently classifying them into 8 distinct metabolic pathways: amino acid, carbohydrate, cofactors and vitamins, energy, lipid, nucleotide, peptide, and xenobiotic metabolism.17 To ensure the robustness of our data, we selected the Metabolomics GWAS Server due to its comprehensive coverage and quality of metabolic data, and its data is widely acknowledged and utilized in the field for high-quality genetic association studies.
Outcome sources of AC
The summary data for the GWAS on AC was sourced from the FinnGen research project.18 This expansive project, which can be accessed at https://r9.finngen.fi/, provided the foundational statistics for our analysis. The FinnGen project was chosen for its extensive sample size, diversity in data, and rigorous data quality control standards, making it an ideal resource for our GWAS summary data. The AC study comprised a substantial dataset, including 20 958 cases juxtaposed with 356 319 controls. The analysis underwent meticulous adjustments to mitigate potential confounders, taking into consideration factors such as age, sex, genetic relatedness, genotyping batch, and the first 10 principal components.
The process of selecting and validating instrumental variables
Genome-wide association significance was set traditionally at p < 1 × 10−5 to identify SNPs linked with metabolite biomarkers. This stringent significance threshold ensures that only SNPs with a strong and reliable association with the metabolite levels are considered as instrumental variables, thereby increasing the validity of our MR analysis. Within the analytical framework, SNPs exhibiting pronounced linkage disequilibrium (either R2 > 0.01 or situated within a 500 kb proximity) were pinpointed and subsequently excluded via the linkage disequilibrium clumping methodology. This approach minimizes bias from potential SNP interactions and ensures that each instrumental variable independently represents a specific genetic variation. For this procedure, the European-ancestry Reference Panel from the 1000 Genomes project served as the foundation. Furthermore, to evaluate the robustness of the instruments in use, mean F-statistics were computed, as alluded to in previous discussions.19
MR analysis
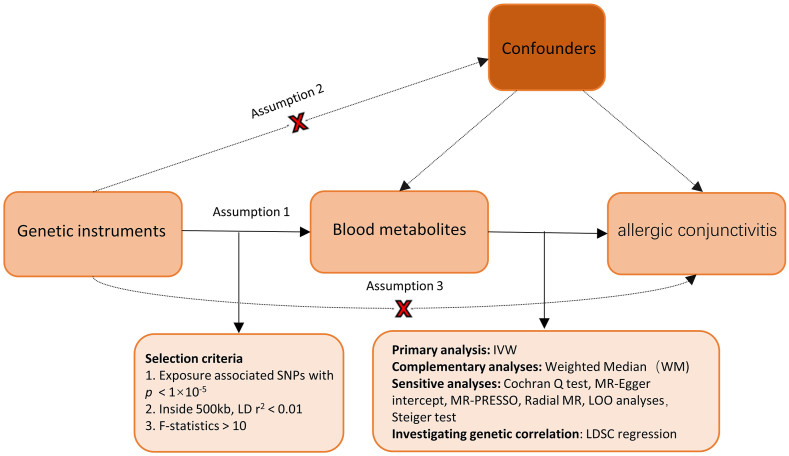
A minimum of 3 distinct instrumental variables (IVs) were employed to delineate these metabolites, facilitating rigorous statistical analyses to account for potential pleiotropic effects. We applied the conventional inverse variance technique, with the principal analysis centered around inverse variance weighted (IVW) estimates. The aggregated Wald ratio for each SNP was linked to the outcome, subsequently producing a consolidated causal estimate.20 Our emphasis was predominantly on metabolites that displayed statistically significant estimates via the inverse-variance weighted approach (p < 0.05). Within the scope of this investigation, which meticulously assessed the 486 serum metabolites concerning AC risk, 3 cardinal prerequisites for a genuine MR study were adhered to: 1) the genetic instruments must have a strong association with the pertinent exposures; 2) the genetic variations should be free from influence by any outcome-related confounding variables; and 3) the genetic instruments should impact the outcome solely through the exposure.21 We conducted comprehensive MR analyses using tools such as Two Sample MR, MRPRESSO, and Radial MR in the R environment (version 4.2.1). Fig. 1 provides an illustrative overview of the research methodology.
Fig. 1.
An exhaustive depiction of the research methodology
Statistical analysis and sensitivity analysis
All MR analyses were undertaken using the TwoSampleMR package (version 0.4.22). The odds ratio (OR) coupled with its associated 95% confidence interval (CI) were chosen as the primary metrics for effect assessment. Recognizing the potential limitations of the IVW approach, which assumes the validity of all genetic instruments, we have incorporated a nuanced discussion on its selection and the potential biases it may introduce. To fortify the reliability of our estimates, we also integrated the MR-Egger and Weighted Median (WM) techniques, each offering distinct advantages under specific conditions. The WM method, in particular, discerns causality by determining the median of the estimates.22 Notably, even when devoid of suitable genetic variants as instrumental variables, the MR-Egger regression can still deliver a consistent causal inference.23 For sensitivity exploration, we deployed an array of methodologies: the Cochran-Q test, MR-Egger intercept, leave-one-out analysis (LOO), and MR-PRESSO. A Cochran-Q p-value of 0.05 was deemed indicative of prevailing heterogeneity.24 Specifically, the Cochran-Q test evaluated heterogeneity among genetic instruments, and MR-Egger regression addressed horizontal pleiotropy, providing an intercept term as an indicator of its direction and magnitude. Moreover, by systematically excluding each SNP through the LOO analysis, we sought to reinforce the robustness of our determinations. In essence, our comprehensive investigation was centered on identifying blood metabolites potentially instrumental in the onset of AC, based on several stringent benchmarks: 1) A salient p-value in the main analysis (specifically, p < 0.05 as dictated by IVW); 2) The MR outcomes exhibiting no traces of heterogeneity or horizontal pleiotropy; and 3) The stability of MR estimates even in the face of individual SNP exclusions.
The search for causation and genetic correlation
To ensure the rigor of our MR estimates, we initiated further studies to address potential biases stemming from the genetic correlation between exposure and outcome variables. Despite meticulous efforts to exclude SNPs associated with AC during instrumental variable selection, there remains a latent risk that certain unrelated SNPs inadvertently influence the genetic underpinnings of AC. The Linkage disequilibrium score (LDSC) regression, a sophisticated statistical approach, discerns the magnitude of coinheritance between 2 attributes by leveraging Chi-squared statistics pertinent to SNPs.25,26 Accordingly, we deployed LDSC to ascertain any genetic affiliation between the assiduously screened metabolites and AC. This endeavor was geared towards ensuring that any overlap between exposure and outcome did not obfuscate genuine causal associations.
It is paramount to delineate whether metabolites directly influence AC onset, or conversely, if the manifestation of AC modulates metabolite concentrations. To substantiate this paradigm, we engaged in a Steiger test.27 Through this meticulous evaluation, we sought to eschew the pitfalls of reverse causality bias, thereby enriching our insights into the nuances of causative direction and magnitude.
Results
Evaluation of the extent and validity of instrumental variables
In the progression of this investigation, a comprehensive two-sample MR methodology was utilized to scrutinize 486 metabolites. Six metabolites (namely, fructose, xanthine, glutamate, X-11843, X-12776, and ergothioneine) were excluded due to their possessing fewer than 4 IVs. Hence, IVs were formulated for the residual 480 metabolites, with the count of SNPs ranging between 4 and 244 for each metabolite (Supplementary Tables 1 and 2). Notably, specific metabolites such as phosphate, aspartate, theobromine, phenylalanylphenylalanine, linolenate, isovalerate, and pyroglutamylglycine were characterized by the minimal IV variability, each registering a mere 4 SNPs. In stark contrast, 2-methoxyacetaminophen sulfate recorded the paramount SNP count, culminating at 478. Furthermore, empirical evidence suggests that all the IVs demonstrated commendable efficacy when employed for multiple regression evaluations across the 480 metabolites. This assertion is bolstered by the minimum F-statistic value of 15.243, which considerably exceeds the oft-cited benchmark of 10, traditionally regarded as a testament to a robust correlation.
Elucidating causal associations between metabolites and allergic conjunctivitis
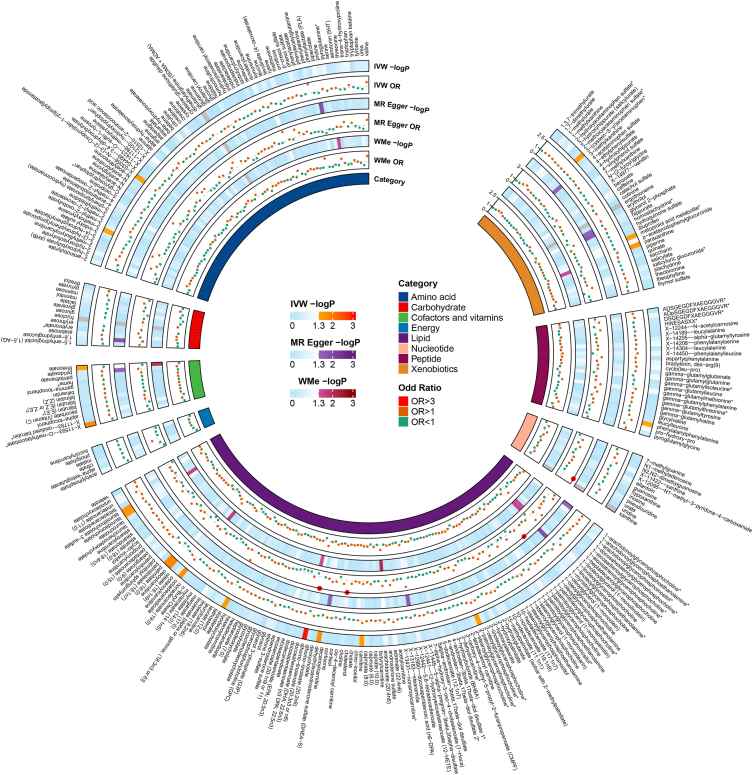
The IVW method was harnessed to discern the causal associations between 480 metabolites and AC, drawing on summary data derived from GWAS. Remarkably, 22 notable causal association patterns emerged through the IVW methodology (p < 0.05); this ensemble comprised 18 recognized metabolites (Fig. 2) and 4 metabolites of enigmatic provenance (Supplementary Table 2). The current investigation narrows its focus to the cohort of 18 metabolites previously identified and authenticated. Although the IVW strategy excels at elucidating correlations between exposure and disease manifestation, it remains susceptible to the nuances of weak instrument bias. Consequently, we undertook sensitivity and pleiotropy assessments to fortify the robustness of our causality determinations. Pronounced heterogeneity was observed in metabolites like myo-inositol and O-methylascorbate, as evidenced by Cochran's Q test (p < 0.05). Moreover, the MR-Egger regression facilitated the exploration of horizontal pleiotropy among the metabolites, unveiling no consequential intercepts (p > 0.05). Subsequent to this, the MR-PRESSO analysis was deployed for a more nuanced investigation of heterogeneity, findings that resonated with prior observations from both Cochran's Q test and MR-Egger regression (Supplementary Table 3). In addition, Steiger tests corroborated the absence of reverse causality, underscoring the integrity of the genetic inferences linking metabolites and AC (p < 0.05). It warrants mention that both the metoprolol acid metabolite and 3-(cystein-S-yl)acetaminophen were annotated as Not Available (NA).
Fig. 2.
A nuanced depiction of the SNP's influence on the causal relationship using 3 MR methodologies, highlighting prospective causal metabolites related to allergic conjunctivitis phenotypes within a heatmap. Shades of gray indicate the exclusion of 4 metabolites: fructose, xanthine, glutamate, and ergothioneine, due to their retention of fewer than 4 instrumental variables. IVW, inverse variance weighted; WMe, Weighted median; OR, odds ratio; P, p-value.
Analyzing the specific metabolite links with allergic conjunctivitis
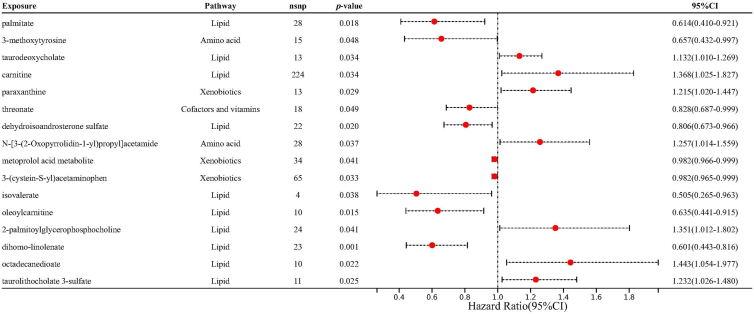
Upon controlling for the confounding effects of horizontal pleiotropy and heterogeneity, we successfully uncovered unique correlations between 16 particular metabolites and the incidence of AC by employing the IVW technique (Fig. 3). Among the Lipid metabolites, palmitate (OR = 0.614, 95% CI = 0.410–0.921, p = 1.84 × 10−2) and dehydroisoandrosterone sulfate (DHEA-S) (OR = 0.806, 95% CI = 0.673–0.966, p = 1.97 × 10−2) demonstrated a protective association, while taurodeoxycholate (OR = 1.132, 95% CI = 1.010–1.269, p = 3.35 × 10−2), carnitine (OR = 1.368, 95% CI = 1.025–1.827, p = 3.37 × 10−2), isovalerate (OR = 0.505, 95% CI = 0.265–0.963, p = 3.82 × 10−2), oleoylcarnitine (OR = 0.635, 95% CI = 0.441–0.915, p = 1.47 × 10−2), 2-palmitoylglycerophosphocholine (OR = 1.351, 95% CI = 1.012–1.802, p = 4.12 × 10−2), dihomo-linolenate (OR = 0.601, 95% CI = 0.443–0.816, p = 1.12 × 10−3), octadecanedioate (OR = 1.443, 95% CI = 1.054–1.977, p = 2.21 × 10−2), and taurolithocholate 3-sulfate (OR = 1.232, 95% CI = 1.026–1.480, p = 2.52 × 10−2) displayed risk associations. Among Amino acids, 3-methoxytyrosine (OR = 0.657, 95% CI = 0.432–0.997, p = 4.84 × 10−2) presented a protective effect, while N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide (OR = 1.257, 95% CI = 1.014–1.559, p = 3.72 × 10−2) acted as a risk factor. In the Xenobiotics category, paraxanthine (OR = 1.215, 95% CI = 1.020–1.447, p = 2.93 × 10−2) showed a positive correlation, whereas metoprolol acid metabolite (OR = 0.982, 95% CI = 0.966–0.999, p = 4.12 × 10−2) and 3-(cystein-S-yl)acetaminophen (OR = 0.982, 95% CI = 0.965–0.999, p = 3.31 × 10−2) indicated negative correlations. Lastly, threonate, under Cofactors and vitamins, displayed a protective correlation (OR = 0.828, 95% CI = 0.687–0.999, p = 4.92 × 10−2). These findings reveal the complex interaction between AAC and various classes of metabolites.
Fig. 3.
Forest plots delineating the effect magnitudes of discerned candidate metabolite associations with allergic conjunctivitis. Solid lines demarcate the MR odds ratio accompanied by its 95% confidence interval
Assessing the genetic impact on AC and its metabolites
In our rigorous examination utilizing LDSC in the context of AC-related metabolites, we identified marked heritability for several compounds, accentuating the genetic imprint on their concentrations. Specifically, the heritability coefficients (hˆ2) were as follows: palmitate at 0.343 (p = 8.36 × 10−08), 3-methoxytyrosine at 0.211 (p = 4.90 × 10−3), carnitine at 1.176 (p = 3.11 × 10−38), threonate at 0.161 (p = 7.88 × 10−3), N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide at 0.363 (p = 2.88 × 10−09), metoprolol acid metabolite at 3.651 (p = 1.41 × 10−2), oleoylcarnitine at 0.168 (p = 3.23 × 10−2), and 2-palmitoylglycerophosphocholine at 0.227 (p = 4.50 × 10−4). These results potently affirm a significant genetic influence on these metabolites. However, for several metabolites, notably paraxanthine, 3-(cystein-S-yl)acetaminophen, and dihomo-linolenate, the elucidation of SNP-based heritability remained elusive, as evidenced by the NA results (Supplementary table 4). The meticulous nature of these findings reinforces the need for delving deeper into the genetic correlations among these metabolites and AC, emphasizing the nuanced interplay that shapes the disease's genesis.
In the subsequent stage of our research, we delved into the genetic correlations between AC and 8 metabolites that exhibited significant heritability. These included palmitate, 3-methoxytyrosine, carnitine, threonate, N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide, metoprolol acid metabolite, oleoylcarnitine, and 2-palmitoylglycerophosphocholine (Supplementary table 5). Intriguingly, none of these metabolites showcased a discernible genetic association with AC (p > 0.05). This suggests that the likelihood of a confounding influence due to overlapping genetic architectures affecting our MR assessments is minimal. These insights lend robust credence to the validity of our MR findings, underscoring that the delineated associations between the aforementioned metabolites and AC are unlikely to be significantly influenced by common genetic antecedents.
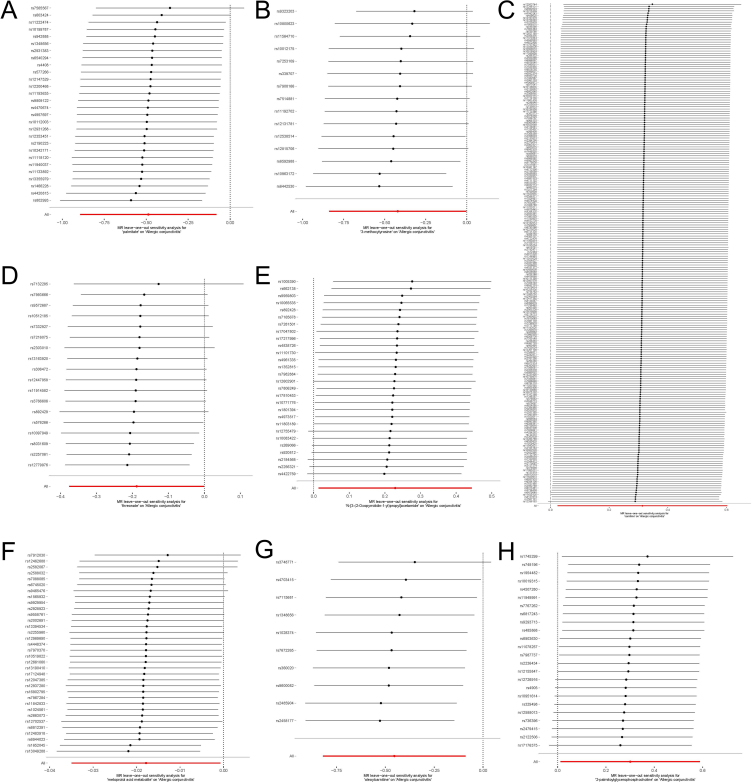
In order to provide more evidence about the causal relationship between 8 metabolites and AC, a leave-one-out analysis was performed (Fig. 4). The findings unambiguously demonstrated that the observed relationships were not attributable to a solitary SNP, but rather arose from the combined impact of several SNPs. Furthermore, the results of each sensitivity analysis were visually represented using scatter plots (Fig. 5), offering a clear and easily understandable depiction of the findings.
Fig. 4.
Leave-one-out assessments related to 8 metabolites associated with allergic conjunctivitis: (A) palmitate, (B) 3-methoxytyrosine, (C) carnitine, (D) threonate, (E) N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide, (F) metoprolol acid metabolite, (G) oleoylcarnitine, and (H) 2-palmitoylglycerophosphocholine. The panel illustrates the leave-one-out analysis, underscoring the robustness of the causal estimate when each instrumental SNP is singularly omitted
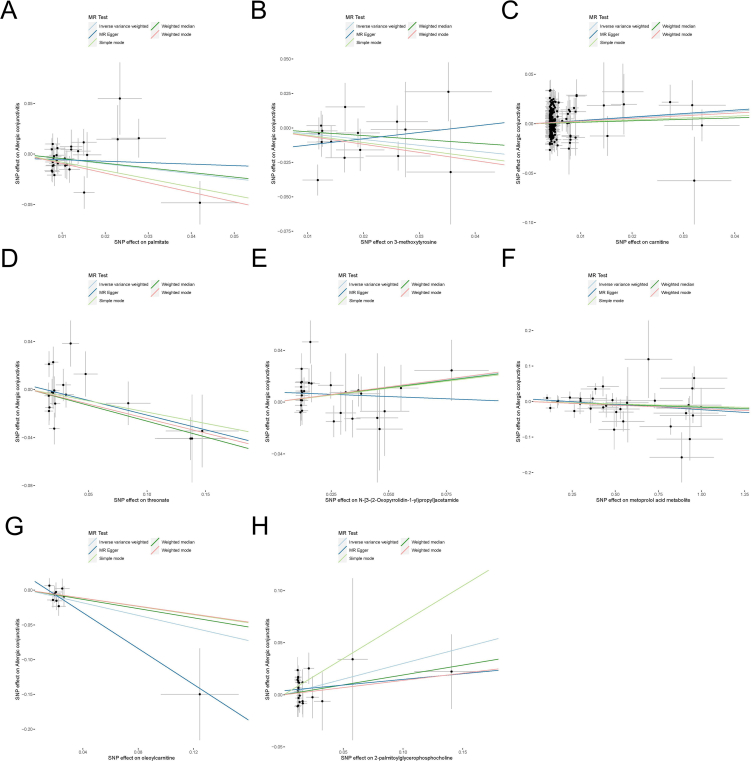
Fig. 5.
Scatter plots depicting associations of 8 metabolites with allergic conjunctivitis: (A) palmitate, (B) 3-methoxytyrosine, (C) carnitine, (D) threonate, (E) N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide, (F) metoprolol acid metabolite, (G) oleoylcarnitine, and (H) 2-palmitoylglycerophosphocholine. The panel presents scatter plots contrasting SNP-exposure effects with SNP-outcome effects, where each distinct point signifies an instrumental SNP
Discussion
In this rigorous study, we adeptly merged expansive individual-level data with GWAS summary statistics, aiming to methodically decipher the genetic architecture underlying the association between blood metabolites and AC. This investigation culminated in robust evidence supporting a causal nexus between select blood metabolites and AC susceptibility.
The pathophysiology of AC is thought to be a complex interplay of genetic, environmental, and metabolic factors. Our recent examination of serum metabolites and their association with AC has brought forth interesting revelations that could unravel key metabolic pathways influencing the disease. Among the lipids, palmitate and oleoylcarnitine presented protective odds ratios, hinting at their potential roles in stabilizing cell membranes or modulating inflammatory responses in the eye. Palmitate, a saturated fatty acid, is known to exert varied biological effects.28 In the context of our study, its association with a lowered risk suggests it might modulate certain inflammatory pathways crucial to AC.29 In contrast, carnitine's elevated risk could suggest its involvement in fostering a pro-inflammatory environment or dysregulated lipid oxidation processes linked with AC's pathogenesis.30 The lipid metabolite 2-palmitoylglycerophosphocholine's association might underline lipid membrane alterations or its role in inflammatory signaling.31 Amino acids, often tied with immunomodulatory functions, showcased their importance via 3-methoxytyrosine and N-[3-(2-Oxopyrrolidin-1-yl)propyl]acetamide. While 3-methoxytyrosine might regulate immune cell responses by modulating enzyme activities, the latter's role remains elusive but could be tied to neurotransmitter pathways and their influence on allergic reactions.32,33 In the realm of cofactors and vitamins, threonate emerged with its association, potentially reflecting its antioxidant capabilities or its integral role in collagen synthesis in ocular tissues,34 both of which can be pivotal in maintaining ocular health during allergenic assaults. Interestingly, the xenobiotic, metoprolol acid metabolite, underscores the possible interactions between medication metabolites and AC, suggesting that certain drug breakdown products could modulate the allergic response,35 either by influencing immune responses or by altering ocular tissue's receptivity to allergens. However, we recognize that these findings, while statistically significant, represent an initial foray into understanding the complex metabolic contributions to AC. In sum, these serum metabolite associations with AC accentuate the need for a holistic understanding, integrating genetics, environment, and metabolism. As we disentangle these intricate threads, we remain cautious in our conclusions, recognizing that the identified associations lay the groundwork for future research rather than serving as definitive evidence of causality.
Our study's findings, particularly the identification of specific metabolites associated with AC, hold significant potential for enhancing current clinical approaches. By deepening the understanding of AC's pathophysiology, our research could aid in refining diagnostic markers and lead to more precise and earlier diagnosis. Furthermore, identifying new therapeutic targets among these metabolites may offer innovative avenues for treatment, advocating for a personalized approach based on individual metabolic profiles. We propose clinical trials to assess the therapeutic potential of targeting the metabolites identified in our study and suggest extensive research into the interactions between genetic predispositions, metabolite levels, and environmental factors.
While our results offer strong indications of a causal relationship between certain metabolites and AC, the inherent limitations of MR studies, such as potential confounding and measurement inaccuracies, prompt a cautious interpretation. We've implemented rigorous measures to counteract these pitfalls, including LDSC regression and Steiger testing. However, our study's reliance on the FinnGen study database, primarily focused on individuals of European descent, may limit the generalizability of our findings. This dataset's lack of detailed subtype classification for AC (such as Seasonal AC, Perennial AC, Vernal Keratoconjunctivitis and Atopic Keratoconjunctivitis) presents another limitation, as these subtypes represent distinct clinical manifestations with unique diagnostic and treatment considerations. This ethnic homogeneity, while reducing population stratification, necessitates further research involving more diverse cohorts and more detailed AC subtype data to confirm these associations across different populations and AC subtypes. Additionally, the potential for residual confounding exists, as factors like lifestyle, diet, and environmental exposures, not fully captured in our dataset, could influence metabolite levels and AC susceptibility. Acknowledging these limitations is crucial, and future studies should aim to address these gaps by including more diverse cohorts, detailed subtype classifications of AC, and a broader range of factors to validate our findings further and enhance our understanding of AC.
In conclusion, our study paints a complex picture of the serum metabolite landscape in allergic conjunctivitis. The diverse roles of these molecules, from protective agents to potential risk factors, shed light on the multifactorial nature of the disease. Further studies are imperative to elucidate the precise molecular mechanisms and translate these insights into potential therapeutic interventions, advancing our quest for personalized medicine in the treatment of allergic conjunctivitis.
Abbreviations
AC, allergic conjunctivitis; MR, Mendelian Randomization; LDSC, Linkage Disequilibrium Score Regression; SNP, single nucleotide polymorphism; GWAS, genome-wide association study; KEGG, Kyoto Encyclopedia of Genes and Genomes; IVs, instrumental variables; IVW, inverse variance weighted; OR, odds ratio; CI, confidence interval; NA, Not Available.
Acknowledgments
The authors would like to acknowledge the participants and investigators of the FinnGen study, as well as the valuable data provided by the Metabolomics GWAS Server.
Funding
This study was supported by the Postdoctoral Fellowship Program of CPSF (GZC20233180).
Availability of data and materials
The datasets analyzed during the current study are available in the FinnGen study (https://www.finngen.fi/en) and the Metabolomics GWAS Server (http://metabolomics.helmholtz-muenchen.de/gwas/).
Authors’ contributions
Xuyan Zou: The main author of the manuscript, concept and design of the study, analysis and interpretation of data, writing and editing the manuscript, visualization, final approval of the submitted version.
Haiyan Huang: Concept and design of the study, data acquisition, provided guidance throughout the research process, critically reviewed and edited the manuscript, final approval of the submitted version.
Yao Tan: Concept and design of the study, data acquisition, supervision, critically reviewed and edited the manuscript, final approval of the submitted version.
Ethics statement
As the study was conducted using publicly available data, there was no requirement for informed consent or ethics committee approval.
Authors’ consent for publication
All authors agreed to the publication of this work in the World Allergy Organization Journal.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Full list of author information is available at the end of the article
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2024.100894.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Berger W.E., Granet D.B., Kabat A.G. Diagnosis and management of allergic conjunctivitis in pediatric patients. Allergy Asthma Proc. 2017;38:16–27. doi: 10.2500/aap.2017.38.4003. [DOI] [PubMed] [Google Scholar]
- 2.Kuruvilla M., Kalangara J., Lee F.E.E. Neuropathic pain and itch mechanisms underlying allergic conjunctivitis. J Investig Allergol Clin Immunol. 2019;29:349–356. doi: 10.18176/jiaci.0320. 20180917 . [DOI] [PubMed] [Google Scholar]
- 3.La Rosa M., Lionetti E., Reibaldi M., et al. Allergic conjunctivitis: a comprehensive review of the literature. Ital J Pediatr. 2013;39:18. doi: 10.1186/1824-7288-39-18. 20130314 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonardi S., Miraglia del Giudice M., La Rosa M., et al. Atopic disease, immune system, and the environment. Allergy Asthma Proc. 2007;28:410–417. doi: 10.2500/aap.2007.28.2954. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z., Mi J., Wu H. Relationships between circulating metabolites and facial skin aging: a Mendelian randomization study. Hum Genom. 2023;17:23. doi: 10.1186/s40246-023-00470-y. 20230317 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun Z., Guo Z., Li X., et al. Genetically predicted 486 blood metabolites in relation to risk of colorectal cancer: a Mendelian randomization study. Cancer Med. 2023;12:13784–13799. doi: 10.1002/cam4.6022. 20230503 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z., Zhang T., Yun Z., et al. Assessing the causal relationships between human blood metabolites and the risk of NAFLD: a comprehensive mendelian randomization study. Front Genet. 2023;14 doi: 10.3389/fgene.2023.1108086. 20230328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okano M., Hirahara K., Kiuchi M., et al. Interleukin-33-activated neuropeptide CGRP-producing memory Th2 cells cooperate with somatosensory neurons to induce conjunctival itch. Immunity. 2022;55:2352–2368.e2357. doi: 10.1016/j.immuni.2022.09.016. 20221021 . [DOI] [PubMed] [Google Scholar]
- 9.Metz D.P., Hingorani M., Calder V.L., et al. T-cell cytokines in chronic allergic eye disease. J Allergy Clin Immunol. 1997;100:817–824. doi: 10.1016/s0091-6749(97)70279-3. [DOI] [PubMed] [Google Scholar]
- 10.Chen W., Luo J., Ye Y., et al. The roles of type 2 cytotoxic T cells in inflammation, tissue remodeling, and prostaglandin (PG) D(2) production are attenuated by PGD(2) receptor 2 antagonism. J Immunol. 2021;206:2714–2724. doi: 10.4049/jimmunol.2001245. 20210519 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimura A., Yasui K., Kishino J., et al. Prevention of allergic inflammation by a novel prostaglandin receptor antagonist, S-5751. J Pharmacol Exp Therapeut. 2001;298:411–419. [PubMed] [Google Scholar]
- 12.Burgess S., Foley C.N., Allara E., et al. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11:376. doi: 10.1038/s41467-019-14156-4. 20200117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P., Wang H., Guo L., et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 2022;20:443. doi: 10.1186/s12916-022-02657-x. 20221115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuccolo L., Holmes M.V. Commentary: mendelian randomization-inspired causal inference in the absence of genetic data. Int J Epidemiol. 2017;46:962–965. doi: 10.1093/ije/dyw327. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J., Zhang Y., Rasheed H., et al. Trans-ethnic Mendelian-randomization study reveals causal relationships between cardiometabolic factors and chronic kidney disease. Int J Epidemiol. 2022;50:1995–2010. doi: 10.1093/ije/dyab203. 20211020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin S.Y., Fauman E.B., Petersen A.K., et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. 20140511 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S., Furumichi M., et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. 20091030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez-Beeftink T., Guillen-Guio B., Lorenzo-Salazar J.M., et al. A genome-wide association study of survival in patients with sepsis. Crit Care. 2022;26:341. doi: 10.1186/s13054-022-04208-5. 20221105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K., Dunn D.W., Li W., et al. Linkage disequilibrium under polysomic inheritance. Heredity. 2022;128:11–20. doi: 10.1038/s41437-021-00482-1. 20220104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Hong X., Gao W., et al. Causal relationship between physical activity, leisure sedentary behaviors and COVID-19 risk: a Mendelian randomization study. J Transl Med. 2022;20:216. doi: 10.1186/s12967-022-03407-6. 20220513 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glymour M.M., Tchetgen Tchetgen E.J., Robins J.M. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am J Epidemiol. 2012;175:332–339. doi: 10.1093/aje/kwr323. 20120112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden J., Davey Smith G., Haycock P.C., et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. 20160407 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D., Xu H., Sun L., et al. Assessing causality between osteoarthritis with urate levels and gout: a bidirectional Mendelian randomization study. Osteoarthritis Cartilage. 2022;30:551–558. doi: 10.1016/j.joca.2021.12.001. 20211208 . [DOI] [PubMed] [Google Scholar]
- 24.Zhao H., Zhu J., Ju L., et al. Osteoarthritis & stroke: a bidirectional mendelian randomization study. Osteoarthritis Cartilage. 2022;30:1390–1397. doi: 10.1016/j.joca.2022.06.006. 20220705 . [DOI] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazal S., Finucane H.K., Furlotte N.A., et al. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu L., Wan B., Li L., et al. Hypothyroidism has a protective causal association with hepatocellular carcinoma: a two-sample Mendelian randomization study. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.987401. 20220930 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzeng H.-T., Chyuan I.-T., Chen W.-Y. Shaping of innate immune response by fatty acid metabolite palmitate. Cells. 2019;8:1633. doi: 10.3390/cells8121633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korbecki J., Bajdak-Rusinek K. The effect of palmitic acid on inflammatory response in macrophages: an overview of molecular mechanisms. Inflamm Res. 2019;68:915–932. doi: 10.1007/s00011-019-01273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahlin K. Boosting fat burning with carnitine: an old friend comes out from the shadow. J Physiol. 2011;589:1509. doi: 10.1113/jphysiol.2011.205815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Q., Xiao X., Fogle P., et al. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein D.S., Eisenhofer G., Kopin I.J. Sources and significance of plasma levels of catechols and their metabolites in humans. J Pharmacol Exp Therapeut. 2003;305:800–811. doi: 10.1124/jpet.103.049270. [DOI] [PubMed] [Google Scholar]
- 33.Qin C., Chen Z., Cao R., et al. Integrated analysis of the fecal metagenome and metabolome in bladder cancer in a Chinese population. Genes. 2022;13:1967. doi: 10.3390/genes13111967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anastassakis K. Androgenetic Alopecia from A to Z: Vol 2 Drugs, Herbs, Nutrition and Supplements. Springer; 2022. Vit C (L-Ascorbic acid) pp. 329–336. [Google Scholar]
- 35.Bodor N., Buchwald P. Ophthalmic drug design based on the metabolic activity of the eye: soft drugs and chemical delivery systems. AAPS J. 2005;7:E820–E833. doi: 10.1208/aapsj070479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the FinnGen study (https://www.finngen.fi/en) and the Metabolomics GWAS Server (http://metabolomics.helmholtz-muenchen.de/gwas/).