Abstract
ABT-378, a new human immunodeficiency virus type 1 (HIV-1) protease inhibitor which is significantly more active than ritonavir in cell culture, is currently under investigation for the treatment of AIDS. Development of viral resistance to ABT-378 in vitro was studied by serial passage of HIV-1 (pNL4-3) in MT-4 cells. Selection of viral variants with increasing concentrations of ABT-378 revealed a sequential appearance of mutations in the protease gene: I84V-L10F-M46I-T91S-V32I-I47V. Further selection at a 3.0 μM inhibitor concentration resulted in an additional change at residue 47 (V47A), as well as reversion at residue 32 back to the wild-type sequence. The 50% effective concentration of ABT-378 against passaged virus containing these additional changes was 338-fold higher than that against wild-type virus. In addition to changes in the protease gene, sequence analysis of passaged virus revealed mutations in the p1/p6 (P1′ residue Leu to Phe) and p7/p1 (P2 residue Ala to Val) gag proteolytic processing sites. The p1/p6 mutation appeared in several clones derived from early passages and was present in all clones obtained from passage P11 (0.42 μM ABT-378) onward. The p7/p1 mutation appeared very late during the selection process and was strongly associated with the emergence of the additional change at residue 47 (V47A) and the reversion at residue 32 back to the wild-type sequence. Furthermore, this p7/p1 mutation was present in all clones obtained from passage P17 (3.0 μM ABT-378) onward and always occurred in conjunction with the p1/p6 mutation. Full-length molecular clones containing protease mutations observed very late during the selection process were constructed and found to be viable only in the presence of both the p7/p1 and p1/p6 cleavage-site mutations. This suggests that mutation of these gag proteolytic cleavage sites is required for the growth of highly resistant HIV-1 selected by ABT-378 and supports recent work demonstrating that mutations in the p7/p1/p6 region play an important role in conferring resistance to protease inhibitors (L. Doyon et al., J. Virol. 70:3763–3769, 1996; Y. M. Zhang et al., J. Virol. 71:6662–6670, 1997).
The human immunodeficiency virus (HIV) contains an aspartyl protease whose function is required for the proper processing of Gag and Gag-Pol polypeptide precursors into the structural proteins of the virus (MA [p17], CA [p24], NC [p7], and p6) as well as the enzymes necessary for viral propagation (reverse transcriptase [RT], integrase [IN], and protease) (29). Because inhibition of the HIV protease is known to result in the formation of noninfectious viral particles (18, 21), the HIV protease has long been considered a good therapeutic target for the treatment of patients with AIDS (10). Much progress has been made in recent years in the development of compounds that specifically inhibit this enzyme, and there are currently four protease inhibitors licensed for the treatment of patients infected with HIV. These compounds have greatly enhanced the repertoire of drugs available to HIV patients and have helped foster the hope that infection with HIV may someday be a successfully treated condition.
In spite of this remarkable progress, one of the most serious hurdles facing the successful clinical use of these compounds is the suppression of drug-resistant variants of HIV. Resistance to protease inhibitors has been observed in vitro and is due to specific mutations in the protease that lead to decreased drug sensitivity (11, 13, 16, 17, 20, 22, 23, 25, 28, 36). Not unexpectedly, similar mutations have also been observed in vivo, leading to viral resistance in patients receiving therapy with these compounds (6, 7, 15, 24, 40). Because of these limitations, it is important to investigate novel protease inhibitors that are not only more potent, but whose resistance profiles differ from those of the currently available compounds.
ABT-378 is a novel protease inhibitor, structurally related to ritonavir (ABT-538) (19, 22, 24), that is currently in clinical development (Fig. 1). This compound is significantly more active than ritonavir in cell culture, even in the presence of human serum proteins (35). Although ABT-378 produces a plasma drug profile that is similar to that of most other protease inhibitors when dosed alone, when codosed with small amounts of ritonavir, this compound achieves and maintains plasma drug levels that are highly suppressive of HIV replication in vitro (35). Additionally, this compound retains high antiviral activity against ritonavir-resistant strains of HIV (35). In this study, we describe the in vitro selection and characterization of HIV-1 variants having increased resistance to ABT-378. Specific mutations in the protease as well as in two of the gag proteolytic cleavage sites were characterized and were shown to be important in conferring resistance to this compound. The results observed during in vitro selection with ABT-378 may be predictive of possible resistance patterns observed in vivo and may aid in the clinical development and therapeutic utility of this compound.
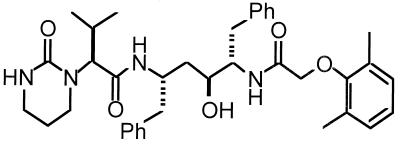
FIG. 1.
Structure of ABT-378. Ph, phenyl.
MATERIALS AND METHODS
Cells and viruses.
MT-4 cells and CEM cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. Uninfected peripheral blood mononuclear cells (PBMCs) were purified from the whole blood of human donor volunteers by Ficoll gradient centrifugation. After 3 days of stimulation with phytohemagglutinin (PHA [5 μg/ml]), PBMCs were maintained in PHA-free RPMI 1640 medium supplemented with 10% FBS, antibiotics, and interleukin-2 (50 U/ml). COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and antibiotics. The HIV-1 pNL4-3 proviral DNA clone (1) was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, and was contributed by Malcolm Martin.
Generation of ABT-378-resistant HIV-1 by in vitro passage.
MT-4 cells (2 × 106) were infected with NL4-3 at a multiplicity of infection (MOI) of 0.003 for 2 h, washed, and then cultured in the presence of ABT-378 at an initial concentration of 0.02 μM. Viral replication was monitored by determination of p24 antigen levels in the culture supernatants by a commercial assay (Abbott Laboratories), as well as by observation for any cytopathic effect (CPE) present in the cultures. When p24 antigen levels exceeded 100 ng/ml, the viral supernatants were filtered and frozen at −80°C for subsequent analysis. Infected cells were washed, lysed, and then stored at −20°C for subsequent analysis of proviral DNA sequences. Virus was serially passaged by using one aliquot of viral supernatant from the preceding passage to infect fresh MT-4 cells in the presence of increasing concentrations of ABT-378, leading to the generation of viral stocks having increased resistance to ABT-378. The drug concentrations used in the selection protocol varied, depending on the level of viral replication present in the preceding passage. Typically, virus from the preceding passage was used to infect fresh MT-4 cells in the subsequent passage at three different drug concentrations. The cultures were monitored for viral replication, and supernatants from successfully infected cultures grown in the presence of the highest drug concentration were used to infect fresh MT-4 cells in the following passage. The selection was carried out for a total of 29 passages, with ABT-378 drug concentrations ranging from 0.02 μM (passage 1 [P1]) to 5.0 μM (P29) (Table 1). The titers of all viral stocks were determined on MT-4 cells, and the profile of cross-resistance of selected viral stocks to several protease inhibitors was determined with the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (27).
TABLE 1.
In vitro selection and phenotypic susceptibility of passaged HIV-1 to ABT-378
| Virus | ABT-378 concn used in selection (μM) | Total no. of days in culture | ABT-378 EC50 (μM)a | Fold resis- tanceb |
|---|---|---|---|---|
| NL4-3 | NAc | NA | 0.028 | 1 |
| P1 | 0.02 | 8 | NDd | ND |
| P2 | 0.02 | 16 | 0.025 | 1 |
| P3 | 0.04 | 26 | ND | ND |
| P4 | 0.04 | 37 | 0.032 | 1 |
| P5 | 0.06 | 44 | 0.140 | 5 |
| P6 | 0.12 | 58 | 0.149 | 5 |
| P7 | 0.15 | 65 | 0.201 | 7 |
| P8 | 0.21 | 75 | 0.295 | 11 |
| P9 | 0.30 | 82 | 0.333 | 12 |
| P10 | 0.30 | 93 | 0.464 | 17 |
| P11 | 0.42 | 100 | 0.346 | 12 |
| P12 | 0.62 | 107 | 0.291 | 10 |
| P13 | 0.80 | 114 | 1.001 | 36 |
| P14 | 1.10 | 121 | 1.115 | 40 |
| P15 | 1.50 | 128 | 1.337 | 48 |
| P16 | 2.00 | 135 | 1.426 | 51 |
| P17 | 3.00 | 142 | 9.475e | 338e |
| P25 | 3.00 | 268 | ND | ND |
| P29 | 5.00 | 332 | ND | ND |
The resistance of each viral passage to ABT-378 was determined with MT-4 cells by the MTT colorimetric assay (27).
Fold resistance relative to the NL4-3 EC50.
NA, not applicable.
ND, not determined.
The EC50 for P17 was determined in separate experiments (see Table 3) and is included in this table for comparison.
Titration and drug sensitivity of ABT-378-passaged viral stocks.
Titers of passaged viral stocks were determined by infecting 2 × 105 MT-4 cells with six serial half-log dilutions of virus for 3 h at 37°C. Following infection, the cells were washed, and 104 cells were plated into 96-well plates in 10-fold replicates for each dilution of virus. Five days later, the level of virus-induced CPE was measured in each culture by using the MTT colorimetric assay (27), and the 50% tissue culture infective dose (TCID50) of each passaged viral stock was determined by the Spearman-Karber method. For drug sensitivity assays, 106 MT-4 cells were infected with titered viral stocks at an MOI of 0.003 for 3 h at 37°C. The cells were then washed, and 104 cells were plated into 96-well plates in the presence of eight serial half-log dilutions of drug (threefold replicates for each drug dilution). On day 5, the virus-induced CPE in each culture was measured by the MTT colorimetric assay (27), and the concentration of each compound which protected 50% of the cells from viral killing (50% effective concentration [EC50]) was determined by linear regression analysis.
Sequence analysis of the protease coding region and proteolytic cleavage sites from selected passages.
Infected cells from each passage were washed once with phosphate-buffered saline and then resuspended in 400 μl of lysis buffer (100 mM KCL, 10 mM Tris-HCL [pH 8.0], 25 mM MgCl2, 1% Tween 20, 1% Nonidet P-40), followed by addition of proteinase K to a final concentration of 300 μg/ml. Samples were then heated at 65°C for 1 h (or 37°C overnight) and then boiled at 100°C for 20 min. Proviral DNA sequences were amplified from infected-cell lysates representing 13 of the 29 total viral passages (P4 to P7, P11 to P17, P25, and P29) by a nested-PCR protocol. In the initial PCR, an 870-bp fragment containing the entire protease coding region and flanking regions in p7, p1, p6, and RT was amplified with primers 1 and 2 (5′-GCAAGAGTTTTGGCTGAAGC-3′ and 5′-GGCAAATACTGGAGTATTGTATGGA-3′, respectively). An aliquot from this reaction was then used in the nested PCR to amplify a 620-bp fragment with internal primers 3 and 4 (5′-AAAATTGCAGGGCCCCTAGGAAAAAGGGCTG-3′ and 5′-GTTTAACGTCTCGGCCATCCATTCCTGGC-3′ containing ApaI and BsmBI sites, respectively). Two additional sets of primers were used to determine the sequence of the p17/p24/p2/p7 and RT/RNase/IN cleavage sites. Primers A and B (5′-AGTCCTCTATTGTGTGCATC-3′ and 5′-GCCTGTCTCTCAGTACAATC-3′, respectively) were used to amplify a 1,043-bp fragment spanning the p17/p24 and p2/p7 junctions. Primers C and D (5′-TAGCCACAGAAAGCATAGTA-3′ and 5′-TGTGTACAATCTAGCTGCCA-3′, respectively) were used to amplify a 756-bp fragment spanning the RT/RNase and RNase/IN junctions. All PCRs were performed for 30 cycles under the following conditions: melting at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The amplified products were purified and then blunt-end ligated directly into the pCR-Script cloning vector (Stratagene). The ligation mixture was then used to transform Epicurian Coli supercompetent cells (Stratagene). Individual bacterial colonies were picked, and mini-prep DNA was purified and then sequenced with the T7 sequencing kit (Pharmacia Biotech, Inc.). For each of the 13 different virus passages examined, protease sequences from 5 to 11 individual clones were obtained and analyzed (Fig. 2).
FIG. 2.
Sequence analysis of the protease coding region from HIV-1 passaged with ABT-378. The amino acid sequence of the protease coding region from clones derived from 13 different passages is indicated. The fraction of clones containing each unique protease sequence is indicated on the right. The top line shows the protease sequence of the wild-type pNL4-3 clone. Identity with this sequence at individual amino acid positions is indicated by dashes.
Construction of full-length mutant HIV-1 DNA clones.
Viral DNA amplified with primers 3 and 4 contains protease coding regions that are flanked by an upstream ApaI site (primer 3) and by a downstream BsmBI site (primer 4). This allows for direct isolation of ApaI-BsmBI fragments containing protease genes with specific mutations. ApaI-BsmBI fragments of interest (620 bp) were cloned into the 5′-NE shuttle vector (38), which contains the 6,027-bp StuI-EcoRI fragment from pNL4-3, as well as an engineered BsmBI site at position 2593 of the HIV-1 sequence. ApaI-EcoRI fragments (3,737 bp) were then isolated and cloned into the pNL4-3 vector to generate full-length infectious DNA clones. For generation of clones containing cleavage-site mutations at the p1/p6 and p7/p1 junctions, mini-prep DNAs containing the desired protease mutations were first amplified with primer 5 (5′-GAGCTTCAGGTTTGGGG-3′) and primer 4 (described above) to generate 440-bp fragments containing the protease gene. Small aliquots of these PCR products were then used, along with upstream primer 3 (described above), in a second PCR to prime DNA synthesis off template DNAs containing mutations at the p1/p6 or p1/p6 plus p7/p1 junctions. ApaI-BsmBI fragments were then cloned and used to generate full-length infectious DNA clones as described above.
Titration and drug sensitivity of mutant HIV-1 molecular clones.
COS-7 cells at 60% confluence on 100-mm-diameter tissue culture plates were transfected by DEAE-dextran (Sigma Chemical Co.) with 15 μg of HIV-1 proviral DNAs containing the desired protease mutations. Five hours after the addition of DNA, the cells were shocked for 2 min with 10% dimethyl sulfoxide and then washed twice with phosphate-buffered saline (PBS) before undergoing refeeding with 10 ml of fresh medium. Forty-eight hours after transfection, the COS-7 cells were cocultivated with MT-4 cells for 3 days. Following the cocultivation, supernatants containing infectious virus were removed, centrifuged to remove cells, filtered, and then used to infect 2 × 106 fresh MT-4 cells. The viral supernatants were propagated in short-term cultures, and p24 antigen levels were monitored to determine peak viral activity, at which point viral supernatants were collected and aliquoted. Titers of molecularly cloned viral stocks were determined by infection of 2 × 105 MT-4 cells with six serial half-log dilutions of virus for 3 h at 37°C. Following infection, the cells were washed, and 104 cells were plated into 96-well plates in fourfold replicates for each dilution of virus. Six days later, p24 antigen levels in the culture supernatants were measured, and the TCID50 of each viral stock was determined by the Spearman-Karber method. For drug sensitivity assays, MT-4 cells were infected with titered stocks at an MOI of 0.003 for 3 h at 37°C and then plated in the presence of six serial half-log dilutions of drug (threefold replicates for each drug dilution). Six days postinfection, the p24 antigen levels in the culture supernatants were measured, and the EC50 for each compound was determined.
RESULTS
Selection for ABT-378-resistant HIV-1 by in vitro passage.
In order to select HIV-1 resistant to ABT-378 in vitro, MT-4 cells were infected with NL4-3, and the virus was serially passaged in the presence of increasing concentrations of ABT-378 (Table 1). Virus was initially grown in the presence of 0.02 μM ABT-378 (P1), and during the course of the selection procedure, the drug concentration was increased to 5.0 μM (P29). Following the selection, viral stocks from each passage were titered, and their susceptibility to ABT-378 was determined by the MTT colorimetric assay (27). The initially passaged viruses (P1 to P4) displayed a sensitivity to ABT-378 similar to that of the parental NL4-3. Viruses selected after 5 to 16 passages (P5 to P16) were between 5- and 51-fold more resistant to ABT-378 than was the wild-type NL4-3 strain. Starting with P17, a highly resistant population emerged that differed from NL4-3 in its susceptibility to ABT-378 by 338-fold. The emergence of this highly resistant virus correlated with the appearance of specific mutations in the protease, as well as mutations in two of the gag proteolytic cleavage sites, as described in detail below. This mutation pattern was maintained and remained almost identical even after prolonged culture in the presence of a 5.0 μM concentration of ABT-378 (P29, Table 1 and Fig. 2).
Sequence analysis of the protease coding region from selected passages.
Proviral DNA sequences from infected cells from P4 to P7, P11 to P17, P25, and P29 were cloned and sequenced as described in Materials and Methods. A complete listing of individual protease sequences and their frequency at each passage is shown in Fig. 2. The wild-type protease gene sequence was observed in a large majority of the clones obtained from the early P4 and P5 passages (11 of 13 clones) but was not observed in any other clone obtained after P5.
By P6, a predominant I84V mutation had emerged which was present in 7 of 11 clones. This mutation was present in all but one clone sequenced after P6, suggesting that it is a critical mutation selected at an early stage necessary to confer resistance to ABT-378. P6 was also marked by the appearance of two additional mutations (L10F and M46I), which were present in all clones sequenced after P7. Although only present in 4 of 11 clones from P6, the L10F mutation appeared in all 11 clones from P7, as well as in all subsequent clones. Similarly, the M46I mutation was present in 2 of 11 clones from P6, 4 of 11 clones from P7, and in all subsequent clones. The persistence of these additional mutations suggests that they too contribute to the ABT-378-resistant phenotype. Similar mutations at positions 10 and 46 have commonly been observed following in vitro or in vivo selection with other protease inhibitors (6, 7, 11, 13, 17, 20, 22, 23, 25, 28, 32, 36).
By P11, the emergence of a fourth highly conserved mutation, T91S, was observed in five of seven clones. Although clones from subsequent passages were obtained that did not contain this substitution, the T91S mutation was observed at a frequency of at least 60% in all passages after P11. Passaged viruses that were 36- to 51-fold more resistant to ABT-378 than was the parental NL4-3 virus (P13 to P16) were marked by the appearance of two additional mutations, V32I and I47V. Although only present in one of five clones from P13, both mutations were present at a frequency of 80% in each of the next three passages, and they always appeared together in the same clone, with the exception of a single clone obtained from P16.
Between P16 and P17, a significant change in genotype was observed that correlated with a >6-fold reduction in sensitivity. Comparison of the clones obtained at P17 with those obtained from P16 revealed two active-site changes: a reversion of residue 32 back to the wild-type sequence and an additional amino acid change at residue 47 from Val to Ala. These active-site changes occurred in tandem in all but 2 of 22 clones examined from P17 onward, suggesting an interplay of mutational changes at these two positions in the development of highly resistant virus.
In addition to the two active-site changes observed at P17, two additional non-active-site mutations appeared at P17 that persisted even after prolonged culture at a 5.0 μM concentration of ABT-378 (P29). The G16E mutation was present in all but one clone examined from P17 onward, while the H69Y mutation was present in two-thirds of the clones examined from P17 onward. Since neither residue lies in close proximity to the active site of the enzyme, it is not clear what role, if any, mutations at these positions may play in conferring resistance to the virus. The eight common residues that undergo mutation during ABT-378 selection are shown in Fig. 3, and the frequency of mutation observed for each viral passage is shown in Table 2. Based on this analysis, it appears that the increased resistance to ABT-378 obtained during in vitro selection may be attributed to the sequential accumulation of specific mutations in the protease.
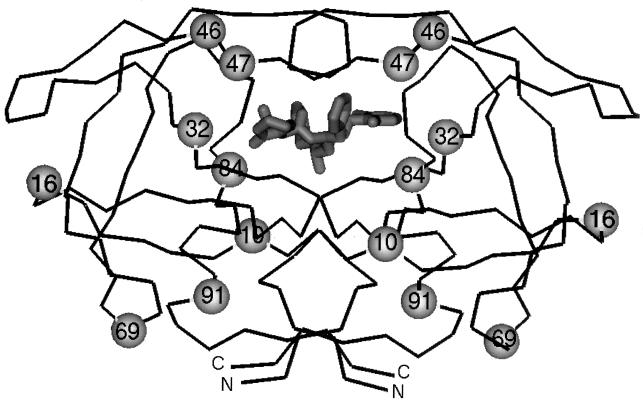
FIG. 3.
Backbone diagram of the dimeric HIV-1 protease. The backbone trace of the dimeric HIV-1 protease is denoted by the thin line. A model of ABT-378 bound in the active site is shown in thick lines at the center of the figure. The carboxy and amino termini of the protein are denoted by C and N, respectively. The eight residues which are commonly mutated during in vitro selection with ABT-378 are indicated by the spheres on both symmetry-related chains of the protein. Three of these residues lie within the active site of the protease (residues 32, 47, and 84), while the other five residues lie outside of the active site (residues 10, 16, 46, 69, and 91).
TABLE 2.
Frequency and sequential appearance of mutations during in vitro selection with ABT-378
| Viral passage (μM) | No. of clones | No. of clones containing mutationa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I84V — L10F — M46I —➛ T91S —➛ V32I — I47V —➛ V47A — G16E — H69Y | ||||||||||
| P4 (0.04) | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P5 (0.06) | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P6 (0.12) | 11 | 7 | 4 | 2 | 0 | 4 | 0 | 0 | 0 | 0 |
| P7 (0.15) | 11 | 10 | 11 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| P11 (0.42) | 7 | 7 | 7 | 7 | 5 | 0 | 0 | 0 | 0 | 0 |
| P12 (0.62) | 7 | 7 | 7 | 7 | 6 | 0 | 0 | 0 | 0 | 0 |
| P13 (0.80) | 5 | 5 | 5 | 5 | 3 | 1 | 1 | 0 | 0 | 1 |
| P14 (1.10) | 5 | 5 | 5 | 5 | 4 | 4 | 4 | 0 | 0 | 0 |
| P15 (1.50) | 5 | 5 | 5 | 5 | 5 | 4 | 4 | 0 | 0 | 0 |
| P16 (2.00) | 10 | 10 | 10 | 10 | 8 | 9 | 8 | 0 | 0 | 0 |
| P17 (3.00) | 5 | 5 | 5 | 5 | 4 | 0 | 0 | 5 | 5 | 3 |
| P25 (3.00) | 7 | 7 | 7 | 7 | 5 | 2 | 0 | 7 | 6 | 5 |
| P29 (5.00) | 10 | 10 | 10 | 10 | 8 | 0 | 0 | 10 | 10 | 6 |
Mutations at eight residues were consistently observed during in vitro selection with ABT-378. These mutations are listed at the top of the table and are shown as they appeared during the selection process. Dashes between mutations indicate changes that were initially observed during the same passage, making it difficult to determine an exact order of appearance. Arrows between mutations indicate a clear sequential appearance of a particular change. For each viral passage indicated on the left-hand side of the table, the concentration of drug used during selection and the total number of clones analyzed are indicated. Underneath each mutation, the number of clones in each passage containing that mutation is indicated, with numbers representing mutation frequencies of at least 50% indicated in boldface. Although the V32I mutation was initially seen in P6, it was then not seen again until P13, and is therefore listed after the T91S mutation. Residue 47 underwent a mutation from Ile to Val, which was then followed by an additional change from Val to Ala.
Sensitivity of HIV-1 selected in vitro by ABT-378 to ritonavir and saquinavir.
Four viral passages were examined to determine the phenotypic susceptibility to ABT-378 and the level of cross-resistance to ritonavir (19, 22, 24) and saquinavir (14–16, 32) (Table 3). The four passages (P7, P11, P14, and P17) were chosen not only to reflect a broad range of resistance to ABT-378 (Table 1) but also because sequence analysis of clones obtained from these passages revealed relatively homogeneous populations of virus that contained few sporadic mutations (Fig. 2 and Table 2). Furthermore, examination of the common amino acid substitutions observed during in vitro selection with ABT-378 (Table 2) revealed that these four passages represent distinct steps in the sequential accumulation of mutations in the protease. As expected, the resistance to ABT-378 was higher in later passages, reflecting the increasing concentration of drug used during viral selection. While P7 and P11 viruses displayed moderate resistance to ABT-378 (4- and 12-fold higher than NL4-3, respectively), the P14 virus was 46-fold more resistant to ABT-378 than was the NL4-3 strain. Very-high-level resistance (338-fold higher than NL4-3) was observed starting with the P17 virus. This sudden and dramatic increase in resistance was unexpected, but most likely reflects the important mutational changes which occur in the protease beginning with this passage as described above.
TABLE 3.
Sensitivity of passaged HIV-1 to ABT-378, ritonavir, and saquinavir
| Virus | EC50 [μM]a (fold resistance)b
|
||
|---|---|---|---|
| ABT-378 | Ritonavir | Saquinavir | |
| NL4-3 | 0.028 (1) | 0.098 (1) | 0.020 (1) |
| P7 | 0.116 (4) | 0.221 (2) | 0.033 (2) |
| P11 | 0.349 (12) | 0.478 (5) | 0.058 (3) |
| P14 | 1.278 (46) | 1.738 (18) | 0.048 (2) |
| P17 | 9.475 (338) | 2.094 (21) | 0.082 (4) |
The resistance of each viral passage to three protease inhibitors was determined with MT-4 cells by the MTT colorimetric assay (27). The values shown are representative of three separate experiments.
Fold resistance relative to the NL4-3 EC50.
The P17 virus was also more resistant to ritonavir (21-fold higher than NL4-3). However, this passaged virus remained very sensitive to saquinavir, showing only a modest fourfold increase in resistance compared to NL4-3. The lack of cross-resistance to saquinavir observed with ABT-378-resistant viruses reflects the difference in the pattern of protease mutations selected by this compound in comparison to other protease inhibitors (6, 16, 22, 24, 32).
In vitro selection with ABT-378 results in mutations in two proteolytic cleavage sites.
Recently, Doyon et al. (11) identified changes in the HIV-1 gag p7/p1 and p1/p6 proteolytic cleavage sites during in vitro selection with the protease inhibitor BILA 2185 BS. To see if similar changes occurred in response to selection with ABT-378, we sequenced a portion of the gag gene spanning the p7/p1 and p1/p6 junctions from clones obtained from 11 different passages (Fig. 4). A mutation identical to that reported in the previous study (11) was observed at the p1/p6 junction, in which the P1′ residue was altered from Leu to Phe. This mutation was seen in 30% of the P7 clones and in all clones examined after P7. Interestingly, this mutation was also found in one of eight clones obtained from P4 that contained none of the common protease gene mutations discussed above. In later passages, a second cleavage-site mutation was observed at the p7/p1 junction in which the P2 residue was altered from Ala to Val. This mutation differed from the mutation reported by Doyon et al. (11), in which both the P2 and P3 residues were altered at the p7/p1 junction, but was identical to a mutation observed in vivo by Zhang et al. (40) in six patients who developed drug resistance during therapy with indinavir.
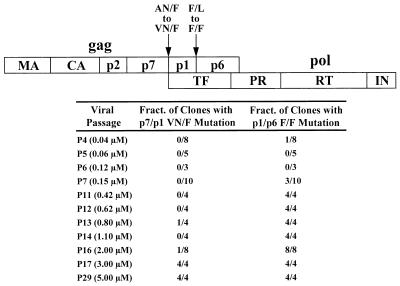
FIG. 4.
Appearance and frequency of p7/p1 and p1/p6 cleavage-site mutations during in vitro selection with ABT-378. A schematic diagram of the gag and pol open reading frames is shown at the top of the figure. The p7/p1 and p1/p6 cleavage sites are indicated by the arrows. During in vitro selection with ABT-378, the p7/p1 cleavage site was altered from AN/F to VN/F (P2 residue Ala to Val). The p1/p6 cleavage site was altered from F/L to F/F (P1′ residue Leu to Phe). For the viral passages indicated on the left-hand side of the table, the fraction of clones containing each cleavage-site mutation is shown. No mutations were observed at any of the other cleavage sites (3). MA, matrix; CA, capsid; TF, transframe protein; PR, protease.
The emergence of the p7/p1 mutation correlated with the active-site changes at residues 32 and 47 observed in P17 and described above. With the exception of one clone from P13 and one from P16, all clones containing the p7/p1 mutation also contained the wild-type reversion at residue 32 and the additional change at residue 47 from Val to Ala. Furthermore, all clones examined after the virus had acquired significant resistance to ABT-378 (P17 onward) contained these active-site alterations as well as mutations at both the p1/p6 and p7/p1 cleavage sites. Cleavage-site mutations may be limited to the p1/p6 and p7/p1 junctions, however, since sequence analysis of all remaining gag and pol cleavage sites from clones obtained from each of three different late viral passages (P16, P17, and P29) revealed no additional mutations (3). Similarly, mutations at the p1/p6 and p7/p1 junctions were the only two cleavage-site alterations observed with the protease inhibitor BILA 2185 BS (11).
Mutation of both p1/p6 and p7/p1 proteolytic cleavage sites is required for the growth of highly resistant HIV-1 selected by ABT-378.
Sequence analysis of multiple clones derived from viral passages can give a good approximation of the predominant genotypes present in a viral population. However, given the relatively small sample size of clones analyzed from each passage, we could not exclude the possibility that the resistance phenotype observed for the passaged virus (Table 3) was caused by viral species representing relatively minor components of the overall viral population that were not identified by clonal sequencing. To address this issue more directly, we constructed a series of molecular clones representing distinct steps in the sequential accumulation of protease mutations observed during selection with ABT-378 (Fig. 5). Constructs containing three, four, and six mutations in the protease were prepared (constructs c7, c11, and c14), paralleling the protease mutations observed at passages P7, P11, and P14, respectively. Since we were also interested in determining how mutations in the proteolytic cleavage sites might affect viral resistance, the p1/p6 mutation was introduced in the context of the same protease mutations (constructs c7.m1, c11.m1, and c14.m1). Constructs containing the consensus protease sequence observed in passages P17 to P29 were also prepared, either with (c17[69Y]) or without (c17) the H69Y mutation seen in two-thirds of clones starting with P17. Both of the latter constructs, as well as the wild-type protease sequence from pNL4-3, were prepared in the context of a wild-type cleavage site, a mutated p1/p6 cleavage site, and mutated p1/p6 plus p7/p1 cleavage sites (Fig. 5).
FIG. 5.
Panel of full-length HIV-1 DNA clones. Amino acid sequences for the wild-type (wt) pNL4-3 p7/p1 and p1/p6 cleavage sites are indicated above the schematic diagram of the p7/p1/p6/protease gene region, while the sequences of the mutated p7/p1 and p1/p6 cleavage sites are shown underneath. In addition, the wild-type sequences for the eight common amino acids which undergo mutation during in vitro selection with ABT-378 are indicated. Beneath the diagram, the protease (prot) sequences for the panel of HIV-1 DNA clones are shown. Clones were named to reflect the initial viral passage in which that particular protease sequence was observed. The presence of the p1/p6 mutation (mut) is designated by m1 in the name of the clone, while the presence of the p7/p1 mutation is designated by m2 in the name of the clone. Clones indicated by the asterisks failed to generate infectious virus in either MT-4 cells, CEM cells, or PBMCs and are therefore not included in Table 4.
Infectious virus was generated from the molecular clones by short-term passage in MT-4 cells, as described in detail in Materials and Methods. In the absence of both p1/p6 and p7/p1 cleavage-site mutations, we were unable to generate any virus from constructs containing the protease consensus sequence from the highly resistant passages P17 to P29. Multiple attempts performed with different cell types (MT-4 cells, CEM cells, and PBMCs) failed to generate infectious virus if the highly mutated proteases were placed in constructs containing either wild-type cleavage sites (c17 and c17[69Y]) or only a mutated p1/p6 cleavage site (c17.m1 and c17[69Y].m1), even after prolonged passage in culture (3). This is consistent with the observation that all clones sequenced from P17 onward contained both p1/p6 and p7/p1 mutations (Fig. 4) and underscores the important role proteolytic cleavage-site mutations may play in compensating for the decreased ability of virus containing multiple protease mutations to replicate.
Antiviral activity of three protease inhibitors against HIV-1 molecular clones.
The susceptibility of HIV-1 molecular clones to the three protease inhibitors ABT-378, ritonavir, and saquinavir was examined (Table 4). The EC50 values in these experiments were determined by measurement of p24 levels in culture supernatants, as described in Materials and Methods. This is a more sensitive method than determination by the MTT colorimetric assay, which utilizes cell cytopathicity as an end point (27), and as a result, the EC50 values in these experiments are about threefold lower than corresponding values obtained with the MTT colorimetric assay (compare NL4-3 EC50 values from Tables 3 and 4).
TABLE 4.
Antiviral activity of ABT-378, ritonavir, and saquinavir against HIV-1 molecular clones
| Virus | EC50 [μM]a (fold resistance)b
|
||
|---|---|---|---|
| ABT-378 | Ritonavir | Saquinavir | |
| NL4-3 | 0.009 (1) | 0.033 (1) | 0.006 (1) |
| NL4-3.m1 | 0.012 (1) | 0.014 (1) | 0.006 (1) |
| NL4-3.m1.m2 | 0.022 (2) | 0.070 (2) | 0.008 (1) |
| c7 | 0.054 (6) | 0.210 (6) | 0.006 (1) |
| c7.m1 | 0.074 (8) | 0.166 (5) | 0.007 (1) |
| c11 | 0.084 (9) | 0.106 (3) | 0.002 (1) |
| c11.m1 | 0.123 (14) | 0.164 (5) | 0.011 (2) |
| c14 | 0.229 (25) | 0.553 (17) | 0.003 (1) |
| c14.m1 | 0.903 (100) | 0.504 (15) | 0.005 (1) |
| c17.m1.m2 | 2.133 (237) | 0.182 (6) | 0.003 (1) |
| c17[69Y].m1.m2 | 2.189 (243) | 0.225 (7) | 0.003 (1) |
The resistance of HIV-1 molecular clones to three protease inhibitors was determined with MT-4 cells by measurement of p24 antigen levels in the culture supernatants. The values shown are representative of two separate experiments.
Fold resistance relative to the NL4-3 EC50.
As expected, the degree of resistance to ABT-378 increased in clones containing sequential protease mutations, paralleling a similar increase in resistance observed with the passaged virus (Table 3). Thus, the EC50 of ABT-378 against the c7, c11, and c14 clones differed by 6-, 9-, and 25-fold, respectively, from the EC50 against the wild-type clone. The effects of the p1/p6 cleavage-site mutation differed, depending on the nature of mutations present in the protease. In constructs containing the wild-type protease, addition of either the p1/p6 or p1/p6 plus p7/p1 cleavage-site mutations (NL4-3.m1 and NL4-3.m1.m2, respectively) did not significantly decrease the susceptibility to any of the three inhibitors. A similar observation was seen in constructs with or without the p1/p6 cleavage-site mutation and either three (c7 versus c7.m1) or four (c11 versus c11.m1) protease mutations. Addition of the p1/p6 mutation in the context of a protease bearing six mutations resulted in a modest (fourfold) increase in the resistance to ABT-378 (c14 versus c14.m1), although resistance to ritonavir and saquinavir remained unaffected. Constructs containing protease mutations observed very late during in vitro selection, which were viable only in the context of both the p1/p6 and p7/p1 cleavage-site mutations, displayed dramatically increased resistance to ABT-378 (approximately 240-fold), although the presence or absence of the H69Y mutation (c17[69Y].m1.m2 versus c17.m1.m2, respectively) did not affect the phenotypic susceptibility. The substantial increase in EC50 observed with these molecular clones parallels the results observed with the P17 virus (Table 3) and suggests that in the context of a highly mutated protease, mutation of both the p1/p6 and p7/p1 proteolytic cleavage sites plays an important role in the development of high-level resistance to ABT-378.
With the exception of the c17.m1.m2 and c17[69Y].m1.m2 clones, the susceptibility of the viral clones to ritonavir also mirrored that of the corresponding passaged virus (Tables 3 and 4). The reason for this discrepancy is not entirely clear, but may be attributed to highly ritonavir-resistant viral species present in relatively minor amounts in the P17 viral population that differ from either the c17.m1.m2 or the c17[69Y].m1.m2 clone. All clones, including those highly resistant to ABT-378, showed a similarly high sensitivity to saquinavir, as observed with the passaged virus. However, because of the limitations in correlating drug selection data obtained in vitro with possible resistance patterns observed in vivo, the effectiveness of saquinavir in suppressing the emergence of ABT-378-resistant variants in vivo remains unknown.
DISCUSSION
In this study, we have shown that in vitro selection of HIV-1 with increasing concentrations of the protease inhibitor ABT-378 leads to the sequential accumulation of specific mutations in the protease. Of the eight residues which were found to be commonly mutated during in vitro selection, three lie within the active site of the protease (residues 32, 47, and 84), while the other five lie outside of the active site (residues 10, 16, 46, 69, and 91). The mutations in the protease were accompanied by mutations in two of the gag proteolytic cleavage sites (p1/p6 and p7/p1). Highly ABT-378-resistant viruses emerging late during in vitro selection contained multiple protease mutations, along with mutations at both the p1/p6 and p7/p1 cleavage sites. Mutations at both of these cleavage sites were required for the growth of highly resistant infectious molecular clones constructed to parallel the protease mutations observed during in vitro selection with ABT-378. Both the highly ABT-378-resistant viruses and molecular clones retained high sensitivity to saquinavir and partial sensitivity to ritonavir.
Many studies have shown that strains of HIV-1 containing protease mutations that developed in response to the selective pressure of protease inhibitors also display impaired growth kinetics (8, 11, 22, 40). Although mutations in the active site of the protease can lead to the development of drug resistance by increasing the Ki of the inhibitor (13, 25, 26), these mutations can also result in impaired protease function and polyprotein processing, thus leading to slower viral growth. A recent study by Schock et al. (34) indicates that non-active-site mutations in the protease may partially circumvent the problem of impaired protease function by enhancing the catalytic efficiency of the enzyme. In that study, the presence of two non-active-site mutations (M46I/L63P) enhanced the catalytic efficiency of both the wild-type protease and a protease containing two active-site mutations (V82A/I84V). Similarly, deficient polyprotein processing by mutant proteases appears to be partially alleviated by mutations in gag proteolytic cleavage sites, resulting in the generation of better substrates for proteolytic cleavage. Doyon et al. (11) have shown that peptides containing mutant p1/p6 and p7/p1 cleavage sites are more efficiently cleaved in vitro by both mutant and wild-type proteases than are the corresponding peptides containing wild-type cleavage sites. This observation correlates with earlier studies showing that peptides containing wild-type p1/p6 and p7/p1 proteolytic cleavage sites are relatively inefficient substrates for proteolytic cleavage (9, 37, 39). Nevertheless, the natural variation of HIV-1 strains observed in vivo at the p1/p6 and p7/p1 cleavage sites appears to be extremely low (2) and suggests there is a strong evolutionary advantage in maintaining these inefficiently processed sequences intact, perhaps mandated by the strict amino acid sequence requirements near the scissile bonds, which generally tend to be hydrophobic in nature (30, 31).
The in vitro resistance to ABT-378 is associated with the appearance of both active-site mutations (residues 32, 47, and 84) and non-active-site mutations (residues 10, 16, 46, 69, and 91) in the protease (Fig. 3), as well as mutations in two of the gag proteolytic cleavage sites (p1/p6 and p7/p1). Residues 10, 16, 69, and 91 are found on the surface of the protein distant from the active site and are not in direct contact with the inhibitor. The mutations at these positions may have subtle effects on the catalytic efficiency and/or inhibitor binding affinity of the enzyme. Residue 46 is also not in direct contact with the inhibitor, but is located in the flexible flap loop of the enzyme (4, 12). Mutations at this position have been shown to affect the dynamics of flap movement (5), which may influence enzyme kinetics by altering the on/off rate of the substrate or by affecting inhibitor access to the active site.
Of more direct importance to inhibition by ABT-378 are the active-site mutations at positions 32, 47, and 84. Residues 32 and 47 comprise the symmetry-related S2 and S2′ pockets, while residue 84 lies at the interface of the S1′ and S2 pockets (and symmetry-related S1 and S2′ pockets) and forms part of each pocket. Binding of ABT-378 places hydrophobic substituents, specifically an isopropyl group and a 2,6-dimethylphenyl group (Fig. 1), near these side chains. A single change at residue 84 (I84V) impacts the binding of ABT-378 into all four hydrophobic pockets of the enzyme active site. This alteration opens up an unfilled volume approximately the size of a methyl group within the active-site cavity when bound with ABT-378 (compare Fig. 6A and B with C and D). Because of the resulting decreased Van der Waal contacts and decreased hydrophobic interactions, the I84V mutant enzyme would be expected to be less sensitive to ABT-378 than the wild-type enzyme. The appearance of viruses containing the I84V mutation very early during in vitro selection with ABT-378 (P6) is in accord with the key location and orientation of this residue relative to ABT-378.
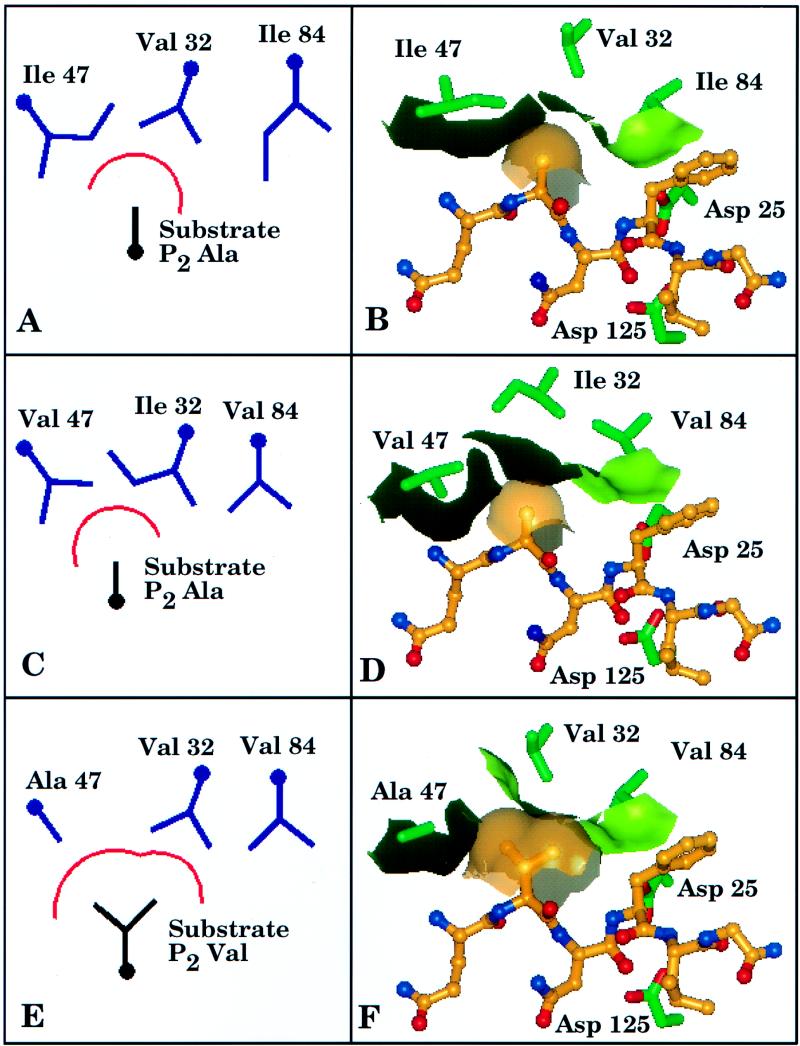
FIG. 6.
Model of the HIV-1 protease active site and p7/p1 substrate. The left-hand side of the figure (A, C, and E) depicts schematic diagrams of the active-site residues 32, 47, and 84 present in the wild-type protease (A), P16 virus (C), and P17 virus (E). The approximate outline of the S2 pocket is indicated by a red arc, underneath which a schematic of the corresponing substrate P2 residue present at each stage is shown. The right-hand side of the figure (B, D, and F) depicts atomic representations derived from three-dimensional molecular modeling of the interactions shown in the matching left-hand side of the figure and present in the wild-type protease (B), P16 virus (D), and P17 virus (F). Active-site residues 32, 47, and 84 of the crystal structure of the HIV-1 protease complexed with the inhibitor MVT-101 (Protein Data Bank entry 4HVP) are shown in green, and the boundary of the S2 pocket is delineated by the light-green–black surface. A hexapeptide model of the p7/p1 substrate (Gln-Ala-Asn-Phe-Leu-Gly), based on the MVT-101 inhibitor structure, is shown with brown carbons, blue nitrogens, and red oxygens. A brown transparent surface over the side chain of the substrate P2 residue is shown. The side chains of the active-site aspartate residues (Asp 25 and Asp 125) are also shown near the Asn-Phe scissile bond of the substrate.
Dual changes at positions 32 and 47 were observed beginning at P13. The interplay between the side chains of these two residues has been discussed previously (33) and is further supported by crystallographic evidence indicating that the side chains of these two residues are in direct contact with one another in the S2 pocket of the HIV protease (12). The mutations occurring at P13 (V32I and I47V) are the equivalent of a transfer of a methyl group from one side chain to another, which results in no net change in the active-site volume (compare Fig. 6A with C). While the total volume of the S2 pocket does not change, modeling studies suggest that there is a subtle reorganization of that volume such that a slight steric clash with ABT-378 is induced, resulting in decreased affinity of ABT-378 for the V32I/I47V mutant enzyme (compare Fig. 6B with D).
Beginning with passage P17, residue 32 reverts to wild type, while residue 47 undergoes an additional alteration from Val to Ala. This results in the loss of one methylene group at residue 32 and the loss of two methylene groups at residue 47, for a net loss of three methylene groups (Fig. 6E). These alterations create a large change in the volume and shape of the active site, which would be expected to dramatically diminish the interaction between the protease and ABT-378, producing a significant decline in inhibitor potency (Fig. 6F). This hypothesis is supported by the dramatic (greater than sixfold) difference in ABT-378 sensitivity observed between the P16 and P17 viruses (Table 1). Coincident with the changes in the enzyme active site, a substrate change in the P2 residue at the p7/p1 junction from Ala to Val was observed. Therefore, in addition to an increase in the enzyme active-site volume, there has been a corresponding increase in the volume required by the substrate (Fig. 6E and F). This change is consistent with the significant drop in ABT-378 potency and presumably allows enzyme activity that is sufficient for polyprotein processing. The nonviability of clones c17, c17.m1, c17[69Y], and c17[69Y].m1 (Fig. 5), which contain the active-site alterations discussed above but lack the substrate modification at the p7/p1 junction, supports this conclusion.
Two previous studies have now shown that resistance to protease inhibitors correlates with the appearance of mutations not only in the protease, but also in at least one of the proteolytic cleavage sites (11, 40). The present study extends these observations to a third protease inhibitor and suggests that in response to the selective pressure exhibited on the virus by this class of compounds, mutation of proteolytic cleavage sites may be a somewhat general strategy employed by HIV-1 to facilitate the growth of virus containing impaired protease function. While limitations exist in the extent to which in vitro observations can be extrapolated to predict results seen in vivo, it is interesting to note that the p7/p1 mutation associated with the emergence of HIV-1 that is highly resistant to ABT-378 is identical to the mutation observed in six of six patients who developed drug resistance during therapy with indinavir (40). Although the results from the in vitro studies shown here and by Doyon et al. (11) indicate that mutations at proteolytic cleavage sites occur after mutations in the protease have developed, results from the in vivo study indicate that cleavage-site mutations can appear at approximately the same time as mutations in the protease (40). The results of the present study, along with those of earlier studies, suggest that the p7/p1/p6 region may be an important determinant of viral resistance that should be examined in patient populations receiving therapy with other protease inhibitors.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within gag/pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 3.Carrillo, A., and A. Molla. Unpublished data.
- 4.Chen Z, Li Y, Schock H B, Hall D, Chen E, Kuo L C. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J Biol Chem. 1995;270:21433–21436. doi: 10.1074/jbc.270.37.21433. [DOI] [PubMed] [Google Scholar]
- 5.Collins J R, Burt S K, Erickson J W. Activated dynamics of flap opening in HIV-1 protease. Adv Exp Med Biol. 1995;362:455–460. doi: 10.1007/978-1-4615-1871-6_59. [DOI] [PubMed] [Google Scholar]
- 6.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 8.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darke P L, Nutt R F, Brady S F, Garsky V M, Ciccarone T M, Leu C T, Lumma P K, Freidinger R M, Veber D F, Sigal I S. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem Biophys Res Commun. 1988;156:297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- 10.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retroviruses. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 11.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson J, Neidhart D J, VanDrie J, Kempf D J, Wang X C, Norbeck D W, Plattner J J, Rittenhouse J W, Turon M, Wideburg N, et al. Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science. 1990;249:527–533. doi: 10.1126/science.2200122. [DOI] [PubMed] [Google Scholar]
- 13.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C-M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobsen H, Haenggi M, Ott M, Duncan I B, Andreoni M, Vella S, Mous J. Reduced sensitivity to saquinavir: an update on genotyping from phase I/II trials. Antiviral Res. 1996;29:95–97. doi: 10.1016/0166-3542(95)00927-2. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen H, Hanggi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 16.Jacobsen H, Yasargil K, Winslow D L, Craig J C, Krohn A, Duncan I B, Mous J. Characterization of human immunodeficiency virus type 1 mutants with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology. 1995;206:527–534. doi: 10.1016/s0042-6822(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan A H, Michael S F, Wehbie R S, Knigge M F, Paul D A, Everitt L, Kempf D J, Norbeck D W, Erickson J W, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan A H, Zack J A, Knigge M, Paul D A, Kempf D J, Norbeck D W, Swanstrom R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67:4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kempf D J, Marsh K C, Denissen J F, McDonald E, Vasavanonda S, Flentge C A, Green B E, Fino L, Park C H, Kong X P, et al. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King R W, Garber S, Winslow D L, Reid C, Bacheler L T, Anton E, Otto M J. Multiple mutations in the human immunodeficiency virus protease gene are responsible for decreased susceptibility to protease inhibitors. Antiviral Chem Chemother. 1995;6:80–88. [Google Scholar]
- 21.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo H, Markowitz M, Majer P, Burt S K, Gulnik S V, Suvorov L I, Erickson J W, Ho D D. Design, synthesis, and resistance patterns of MP-134 and MP-167, two novel inhibitors of HIV type 1 protease. AIDS Res Hum Retroviruses. 1996;12:55–61. doi: 10.1089/aid.1996.12.55. [DOI] [PubMed] [Google Scholar]
- 24.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 25.Partaledis J A, Yamaguchi K, Tisdale M, Blair E E, Falcione C, Maschera B, Myers R E, Pazhanisamy S, Futer O, Cullinan A B, Stuver C M, Byrn R A, Livingston D J. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patick A K, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P-F. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 28.Pazhanisamy S, Stuver C M, Cullinan A B, Margolin N, Rao B G, Livingston D J. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 29.Peng C, Ho B K, Chang T W, Chang N T. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit S C, Michael S F, Swanstrom R. The specificity of the HIV-1 protease. Perspect Drug Discov Des. 1993;1:69–83. [Google Scholar]
- 31.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison C A D, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 32.Roberts N A. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS. 1995;9:S27–S32. [PubMed] [Google Scholar]
- 33.Sardana V V, Schlabach A J, Graham P, Bush B L, Condra J H, Culberson J C, Gotlib L, Graham D J, Kohl N E, LaFemina R L, Schneider C L, Wolanski B S, Wolfgang J A, Emini E A. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino acid substitutions in the enzyme’s substrate binding site. Biochemistry. 1994;33:2004–2010. doi: 10.1021/bi00174a005. [DOI] [PubMed] [Google Scholar]
- 34.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 35.Sham, H. L., D. J. Kempf, A. Molla, K. C. Marsh, G. N. Kumar, C.-M. Chen, W. Kati, K. Stewart, R. Lal, A. Hsu, D. Betebenner, M. Korneyeva, S. Vasavanonda, E. McDonald, A. Saldivar, N. Wideburg, X. Chen, P. Niu, C. Park, V. Jayanti, B. Grabowski, G. R. Granneman, E. Sun, A. J. Japour, J. M. Leonard, J. J. Plattner, and D. W. Norbeck. ABT-378, a potent second generation inhibitor of the human immunodeficiency virus protease. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 36.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tozser J, Blaha I, Copeland T D, Wondrak E M, Oroszlan S. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 1991;281:77–80. doi: 10.1016/0014-5793(91)80362-7. [DOI] [PubMed] [Google Scholar]
- 38.Winslow D L, Anton E D, Horlick R A, Zagursky R J, Tritch R J, Scarnati H, Ackerman K, Bacheler L T. Construction of infectious molecular clones of HIV-1 containing defined mutations in the protease gene. Biochem Biophys Res Commun. 1994;205:1651–1657. doi: 10.1006/bbrc.1994.2857. [DOI] [PubMed] [Google Scholar]
- 39.Wondrak E M, Louis J M, de Rocquigny H, Chermann J C, Roques B P. The gag precursor contains a specific HIV-1 protease cleavage site between the NC (P7) and P1 proteins. FEBS Lett. 1993;333:21–24. doi: 10.1016/0014-5793(93)80367-4. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]