Abstract
2,5,6-Trichloro-1-β-d-ribofuranosyl benzimidazole (TCRB) is a potent and selective inhibitor of human cytomegalovirus (HCMV) replication. TCRB acts via a novel mechanism involving inhibition of viral DNA processing and packaging. Resistance to the 2-bromo analog (BDCRB) has been mapped to the UL89 open reading frame (ORF), and this gene product was proposed as the viral target of the benzimidazole nucleosides. In this study, we report the independent isolation of virus that is 20- to 30-fold resistant to TCRB (isolate C4) and the characterization of the virus. The six ORFs known to be essential for viral DNA cleavage and packaging (UL51, UL52, UL56, UL77, UL89, and UL104) were sequenced from wild-type HCMV, strain Towne, and from isolate C4. Mutations were identified in UL89 (D344E) and in UL56 (Q204R). The mutation in UL89 was identical to that previously reported for virus resistant to BDCRB, but the mutation in UL56 is novel. Marker transfer analysis demonstrated that each of these mutations individually caused ∼10-fold resistance to the benzimidazoles and that the combination of both mutations caused ∼30-fold resistance. The rate and extent of replication of the mutants was the same as for wild-type virus, but the viruses were less sensitive to inhibition of DNA cleavage by TCRB. Mapping of resistance to UL56 supports and extends recent work showing that UL56 codes for a packaging motif binding protein which also has specific nuclease activity (E. Bogner et al., J. Virol. 72:2259–2264, 1998). Resistance which maps to two different genes suggests that their putative proteins interact and/or that either or both have a benzimidazole ribonucleoside binding site. The results also suggest that the gene products of UL89 and UL56 may be antiviral drug targets.
Human cytomegalovirus (HCMV) can cause significant morbidity and mortality in immunocompromised populations (3). It is a common opportunistic disease in patients with AIDS and is often a factor in their death (38). HCMV infection has been implicated in increased risk of organ rejection following heart (28) and kidney transplants (8) and in restenosis of diseased arteries following angioplasty (41, 63). It is also a leading cause of birth defects (16).
Current therapies for HCMV infection include ganciclovir (GCV) (22), cidofovir (30), and foscarnet (20). Each of these drugs has several limitations to its use: none are orally bioavailable, all have dose-limiting toxicity, and resistance has developed to each (26). Because all three of these drugs inhibit viral replication through an interaction with the virally encoded DNA polymerase (25, 31, 37), the possibility of cross-resistance exists. Thus, additional drugs with unique mechanisms of action are needed for the treatment of HCMV infections.
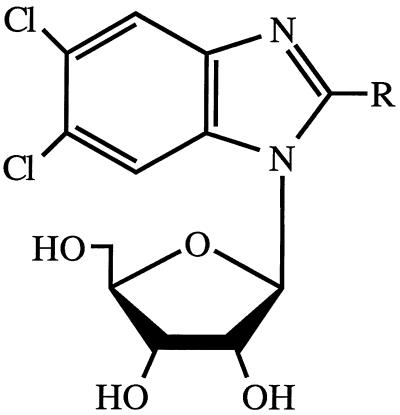
In 1995, we reported that 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole (BDCRB; Fig. 1) and the 2-chloro analog [2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole TCRB] are potent and selective inhibitors of HCMV replication (55). These compounds have a novel mechanism of action, which unlike the current therapies for HCMV infection, does not involve inhibition of DNA synthesis. The benzimidazole ribonucleosides prevent the cleavage of high-molecular-weight viral DNA concatemers to monomeric genomic lengths (57). Resistance to BDCRB has been mapped to the HCMV UL89 open reading frame (ORF), which, by analogy to gene gp17 from bacteriophage T4, may be a terminase (23, 57). Consequently, we have proposed that the benzimidazole ribonucleosides inhibit the product of this gene and that the UL89 gene product is involved in the viral DNA concatemer cleavage process (57).
FIG. 1.
Structure of benzimidazole ribonucleosides. TCRB, R = Cl; BDCRB, R = Br.
HCMV replication proceeds in a manner which is conserved among herpesviruses. The virally encoded DNA polymerase produces large, complex head-to-tail concatemers (10, 29, 33) which must be cleaved into genomic-length pieces before insertion into preformed capsids (59). With herpes simplex virus type 1 (HSV-1), temperature-sensitive mutants which are unable to cleave and package the concatemeric DNA have been derived (1, 2, 4, 45, 49, 50, 61). By this process, six HSV-1 genes have been found to be involved in concatemer cleavage and packaging. They are UL6, UL15, UL25, UL28, UL32, and UL33. In addition, recent studies in Homa’s laboratory have established that the product of UL25 is required for viral DNA encapsidation but not cleavage (39). Homologs of these genes exist in HCMV and are UL104, UL89, UL77, UL56, UL52, and UL51, respectively (18).
In our continuing investigation of the mode of action of benzimidazole nucleosides, we report herein the independent isolation of HCMV strains resistant to TCRB, characterization of these strains, and identification of the mutations responsible for the development of resistance. The results demonstrate that the mechanism of action of the benzimidazole ribonucleosides is more complex than previously proposed and that a second gene product implicated in DNA cleavage and packaging is involved.
MATERIALS AND METHODS
Chemicals.
BDCRB and TCRB (Fig. 1) were synthesized in the laboratory of L. B. Townsend as previously described (55). GCV was provided through the courtesy of Syntex Laboratories, Inc., Palo Alto, Calif.
Cell and viral culture.
Monolayer cultures of human foreskin fibroblasts (HFF cells) and MRC-5 cells were grown in minimal essential medium with Earle’s salts [MEM(E)] plus 10% fetal bovine serum at 37°C in a humidified atmosphere of 3% CO2–97% air. They were regularly passaged at 1:2 dilutions, using conventional techniques, with 0.05% trypsin plus 0.02% EDTA in HEPES-buffered saline (51). HCMV strain Towne was kindly provided by Mark Stinski, University of Iowa. Stocks of HCMV were prepared by infecting HFF cells at a multiplicity of infection (MOI) of <0.01 PFU per cell, and stock viral titers were determined in monolayer cultures of HFF cells as described earlier (46, 56). Viral assays, but not the preparation of viral stocks, were done in the presence of penicillin-streptomycin (100 U of penicillin G per ml, 100 μg of streptomycin per ml).
HCMV antiviral assays.
Both plaque and yield reduction assays were used to compare the sensitivities of isolates to BDCRB and TCRB. For plaque reduction assays, HFF cells in 24-well cluster dishes were infected with approximately 100 PFU of HCMV per well as described earlier (56). Following virus adsorption, compounds dissolved in growth medium were added to duplicate wells at four to eight selected concentrations. After incubation at 37°C for 10 days, cell monolayers were stained with crystal violet, and plaques were enumerated at 40-fold magnification. Drug effects were calculated as a percentage of reduction in number of plaques observed in the presence of each drug concentration compared to the number observed in the absence of drug. For yield reduction assays, the procedure devised by us (46) was used. Briefly, HFF cells were planted in 96-well cluster dishes, incubated overnight, and infected with HCMV at an MOI of 0.5 to 1. After virus adsorption, inoculum was replaced with 0.2 ml of fresh medium containing test compounds in quadruplicate on a single plate in a manner which provided drug concentrations from 100 to 0.14 μM, using one-half log10 dilutions. Plates were incubated at 37°C for 4 days and subjected to one cycle of freezing and thawing. Aliquots from each of the wells were transferred to a fresh 96-well monolayer culture of HFF cells and serial diluted across the plate. Cultures were incubated, cells were stained, plaques were enumerated at 40-fold magnification, and titers were calculated as described.
For both plaque and yield assays, dose-response relationships were constructed by plotting the percent inhibition of plaque number or the log10 of the percent inhibition of virus titer against log10 drug concentrations. The 50 or 95% inhibitory concentration (IC50 or IC95) and corresponding 95% confidence intervals were calculated by using the variable-slope dose-response algorithm of GraphPad Prism (GraphPad, San Diego, Calif.). Samples containing GCV as a positive control were used in all assays.
Selection of TCRB-resistant virus.
HFF cells were infected with HCMV strain Towne, and the virus was grown in 10 μM TCRB for 45 days. The progeny were collected and grown for an additional 7 days in 10 μM TCRB. The resulting virus was passaged in 30 μM TCRB for 13 days. The duration of each passage was a function of the viral input and the rate of viral growth. The resulting virus stock was plaque purified three times by limiting dilution, and purified isolates were designated D10 and B11. Isolate D10 was further passaged three times in 50 μM TCRB, and the highly resistant viral stock was plaque purified three times by limiting dilution; this isolate was designated C4. Virus isolates D10 and C4 were tested for drug resistance by plaque reduction assay and yield reduction assay and were further characterized by pulsed-field electrophoresis as described previously (57).
Growth characteristics.
Viral growth rates were measured both over a single cycle of viral replication and over multiple cycles of replication. For observation of a single cycle, HFF cells were plated at 10,000 cells per well in 96-well cell culture plates and infected at an MOI of 2 PFU/cell. At time points spaced 8 to 12 h apart, the cultures were frozen at −80°C. Samples were collected for 4 days, and after all were collected, they were titered across microplates as described previously (46). For assays involving multiple cycles of replication, growth was determined by infecting 96-well cell culture plates with 10,000 HFF cells per well at an MOI of 0.01 PFU/cell and freezing samples every 12 h over a course of 10 days.
Analysis of DNA processing.
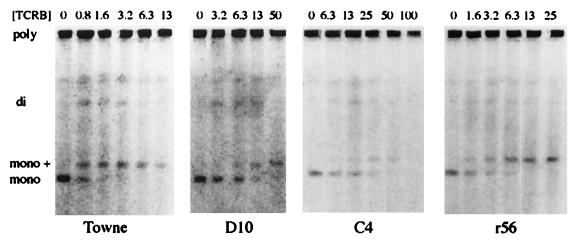
Samples were prepared for contour-clamped homogeneous electric field electrophoresis with a ChefMapper (Bio-Rad, Hercules, Calif.) as described previously (57). Briefly, HFF cells were infected at an MOI of 3 PFU/cell and treated with selected drug concentrations for 3 days. Cells were removed from monolayers by treatment with a solution of 0.5 mM EDTA and 0.05% trypsin, suspended in 0.8% agarose, and digested with 100 mg of proteinase K (PK) per ml in 1% sarkosine for 48 h. Aliquots were loaded into the wells of a 1% agarose gel, and electrophoresis was performed as detailed previously (52). Upon completion of the electrophoresis procedure, gels were stained with ethidium bromide and photographed. Figure 4 was obtained by scanning the photographs with a Microtek Scanmaker III using Adobe Photoshop. The colors were inverted and selected portions of the gels were transferred to Abobe Pagemaker, aligned, and labeled.
FIG. 4.
Pulsed-field electrophoresis study of monomer formation by the mutant viruses in the presence of TCRB. HFF cells were infected at an MOI of 3 PFU/cell and treated with selected concentrations (micromolar) of TCRB for 3 days. Cells were embedded in agarose and digested with PK. DNA was separated with a ChefMapper, stained with ethidium bromide, and photographed. Bands: poly, polygenomic, concatemeric HCMV DNA; di, digenomic HCMV DNA; mono +, 270-kb HCMV DNA; mono, 230-kb genomic-length HCMV DNA.
DNA sequencing.
ORFs UL51, UL52, UL56, UL77, UL89, and UL104 were sequenced from wild-type HCMV, strain Towne, and isolate C4. Additionally, the UL56 and UL89 ORFs were sequenced from isolate D10 and, later, from recombinant isolates r56 and rC4. Viral stocks (∼106 PFU/ml) were lysed in an equal volume of 1× PCR buffer (Perkin-Elmer, Foster City, Calif.)–0.05% Nonidet P-40–0.05% Tween 20. These were digested with PK (100 μg/ml) at 55°C for 1 h, and the enzyme was inactivated at 95°C for 15 min. Specific viral genes were amplified from this lysate by PCR. Primers for PCR and sequencing were designed by using the PRIME program in the Genetics Computer Group package and are based on the published sequence of the AD169 strain of HCMV (18, 27). A primer list is available upon request. The PCR mixtures included 20% viral lysate, 10% dimethyl sulfoxide, deoxynucleoside triphosphates at 250 nM (Gibco BRL, Grand Island, N.Y.), primers at ∼1.5 μM (Midland Certified Reagents, Midland, Tex.), and 2.5 U of AmpliTaq (Perkin-Elmer). The reagents were prepared in an ice bath, and the tubes were placed in a Perkin-Elmer GeneAmp 2400 PCR system preheated to 95°C. The reactions were cycled 35 times as follows: 95°C for 45 s, 58°C for 1 min, and 72°C for 3 min. PCR products were purified by using a QiaQuick PCR purification kit (Qiagen, Chatsworth, Calif.) or a Centricon 100 column (Amicon, Beverly, Mass.). DNA was quantified by comparison with DNA mass markers (Gibco BRL) and sequenced using a Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer). The sequencing reactions were purified with a CentriSep spin column (Princeton Separations, Aldephia, N.J.) and were separated and detected with an Applied Biosystems Inc. 377 DNA sequencer. The data were aligned and edited by using Sequencher software (GeneCodes, Ann Arbor, Mich.).
Marker transfer studies.
Genomic DNA was prepared using two different methods. For wild-type (wt) virus, strain Towne, a modification of the package insert from a RecoverEase DNA isolation kit (Stratagene, La Jolla, Calif.) was used. Supernatant virus from HCMV-infected cells was pelleted at ∼10,000 × g for 2.5 h. Pellets were suspended in 1 volume of digestion buffer plus 1 volume of PK solution and were incubated at 55°C for 2 h with gentle rocking every 15 min. The resulting suspension was dialyzed against TE (10 mM Tris-HCl [pH 7.5], and 1 mM EDTA) overnight. For wt virus, strain AD169, and isolate D10, a second method was used (52). Briefly, virus pellets were suspended in TNE (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) with 0.05% sodium dodecyl sulfate and digested overnight with PK (100 μg/ml) at 37°C. The resulting DNA-containing solutions were gently extracted twice with 1 volume of phenol and once with 1 volume of 24:1 chloroform-isoamyl alcohol. The DNA was precipitated overnight at −20°C with 2.5 volumes of ice-cold 95% ethanol. Precipitates were collected by centrifugation at 12,000 × g for 30 min and pellets were dissolved in TE by standing undisturbed at room temperature for several days.
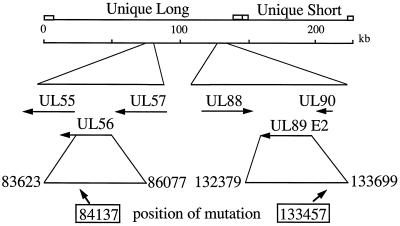
Calcium phosphate transfections were performed as described by Sambrook et al. (48). Infectious DNA (0.3 to 2 μg) from either wt Towne or D10 virus isolate was coprecipitated with 12- to 25-fold molar excess of a PCR product encoding the portion of interest of UL56 from either wt Towne or C4 isolate (Fig. 2). Low (10 to 15)-passage HFF cells were exposed to the precipitate for 4 to 5 h, and then the precipitate was removed. The cells were treated with 15% glycerol in buffered saline for 90 s, washed briefly with MEM(E) containing 5% fetal bovine serum and penicillin-streptomycin, and incubated overnight in this medium. The medium was replaced with fresh medium the following morning. A PCR product from exon II of UL89 (Fig. 2) from B11 was similarly transfected with infectious wt AD169 DNA into MRC-5 cells. Transfected cultures were incubated in the absence of drug until maximum cytopathic effect was observed (usually around 21 days), and the supernatant progeny virus was clarified by centrifugation (2,000 × g for 5 min) and stored in liquid nitrogen. Titers of viral stocks were determined as described previously (46).
FIG. 2.
Schematic of the genome of HCMV showing the locations and orientations of the UL56 and UL89 ORFs. A PCR product encoding exonII of UL89 was used in the marker transfer experiments, and a 3′-truncated portion of UL56 was similarly used. The numbers shown are from the published sequence of HCMV strain AD169 and are only relative for the Towne-derived viruses reported in this paper.
Viral plating efficiencies were determined by infecting five flasks (75 cm2) each with 20,000 to 70,000 infectious progeny virions under a methylcellulose overlay containing BDCRB. The concentrations of BDCRB used to screen the various recombinants are given in Table 3. A parallel experiment was performed in the absence of BDCRB to determine the actual number of infectious virions plated in a given experiment. Five additional flasks each were infected with 1/10 the virus used in the first experiment. Recombination efficiencies were calculated by subtracting the number of plaques produced by wt progeny virus in the presence of BDCRB from the number of plaques produced by recombinant progeny virus populations in the presence of BDCRB. The resulting number of plaques observed under drug was divided by 10 times the number of plaques observed in the absence of drug, and the result was converted to a percentage.
TABLE 3.
Efficiencies of marker transfer experiments
| Virus strain for:
|
Gene for PCR product | BDCRB concn (μM) | Plating efficiencya (%) | Recombinant strain | |
|---|---|---|---|---|---|
| Parental DNA | PCR product | ||||
| AD169 | wt (Towne) | UL89 | 2 | 0.070 | |
| AD169 | B11 (Towne) | UL89 | 2 | 0.50 | rT89E2-4 |
| D10 | wt (Towne) | UL56 | 20 | <0.003 | |
| D10 | C4 (Towne) | UL56 | 20 | 0.29 | rC4 |
| Towne | wt (Towne) | UL56 | 5 | <0.005 | |
| Towne | C4 (Towne) | UL56 | 5 | 5.4 | r56 |
HFF cells were cotransfected with the indicated sources of parental and mutated DNA (PCR product). Following transfections, stocks of progeny virus were grown and titers were determined. Five flasks (20,000 to 70,000 PFU per flask) were infected with this virus and incubated in the presence of the indicated concentrations of BDCRB. Five flasks also were infected in the absence of drug, using 1/10 the amount of virus. Plating efficiency is the percentage of plated recombination progeny which formed plaques under the indicated concentration of BDCRB as described in the text.
The recombinant, mutant viruses were isolated and plaque purified by the method of Klein (34). The first round was performed in the presence of 2, 5, or 15 μM BDCRB, depending on the resistance of the virus, and two subsequent rounds were performed in the absence of drug.
Nucleotide sequence accession numbers.
Nucleotide and amino acid sequences were obtained from GenEMBL. Accession numbers are as follows: HCMV strain AD169, X17403; HSV-1, X14112; human herpesvirus 6, X83413; human herpesvirus 7, U43400; and varicella-zoster virus, X04370. The sequences from HCMV strain Towne of UL51, UL52, UL56, UL77, UL89 exon 1, UL89 exon 2, and UL104 as reported herein are AF039234, AF047521, AF047523, AF047522, AF047525, AF047526, and AF047524, respectively.
RESULTS
Initial isolation and characterization of resistant virus.
Prior studies in our laboratories have shown that BDCRB inhibits viral DNA packaging through an interaction with the gene product of UL89 (57). The resistant isolates (1038rA, -B, and -C) were obtained by growth of HCMV strain AD169 in the presence of BDCRB. Independently, another clone of resistant virus was isolated by passaging HCMV strain Towne in the presence of increasing concentrations of TCRB up to 30 μM. Isolates resulting from plaque purification of the heterogeneous mixture have been designated D10 and B11. Isolates D10 and B11 were approximately 10-fold resistant to the benzimidazole ribonucleosides in plaque reduction assays (Table 1). This was nearly identical to resistance observed with strains 1038rA and 1038rC (57). Moreover, all four isolates were sensitive to GCV and grew in the absence of drugs at rates nearly identical to those for wt HCMV (data not presented), further illustrating that a unique mechanism of action was involved.
TABLE 1.
Resistance of HCMV strains to benzimidazole ribonucleosides and GCV
| HCMV strain | mean IC50 (μM)a ± SD
|
||
|---|---|---|---|
| TCRB | BDCRB | GCV | |
| Towne | 3.4 ± 1.4 | 0.92 ± 0.61 | 4.9 ± 2.8 |
| D10 | 29 ± 9.9 | 7.4 ± 5.7 | 5.8 ± 2.8 |
| B11 | 28 ± 15 | 14.3 ± 7.4 | 3.5 ± 0.4 |
| AD169 | 1.8 ± 0.1 | 0.3 ± 0.1 | 4.6 ± 3.3 |
| rT89E2-4 | NDb | 6.3 (4.9–8.1)c | ND |
Results from plaque reduction assays performed in duplicate, using the indicated strains of HCMV as described in the text. IC50s were calculated from 2 to 20 experiments using at least four drug concentrations each.
ND, not determined.
Results presented with 95% confidence interval (in parentheses) because data are from a single experiment performed in duplicate which used eight drug concentrations.
Sequence analysis of B11 and D10.
Based on the similarity of phenotypic characteristics among B11, D10, and 1038rA and 1038rC plus the known mutation of aspartate to glutamate at position 344 (D344E) in UL89, which caused the resistance of the AD169 strains (57), UL89 of B11 and D10 was sequenced. A change of C to G in nucleotide 1032 of UL89 resulting in an inferred D344E amino acid mutation was found in B11 and D10 (Table 2). This change was identical to the C-to-G nucleotide change found in AD169 strains 1038rA and 1038rC but different from the C-to-A change found in strain 1038rB, which conferred the identical D344E change (57). In addition to the C-to-G change, a difference between AD169 and Towne strains was found in the next nucleotide (1033, T to G), resulting in a coding change of serine to alanine in position 345.
TABLE 2.
Sequence of the UL89 ORF from drug-resistant isolates
| HCMV strain | Position no.a | Sequenceb |
|---|---|---|
| AD169c | 1021 | ACT ACC AGT GAC TCC ACG TGT |
| 341 | Thr Thr Ser Asp Ser Thr Cys | |
| Towne | 1021 | ACT ACC AGT GAC GCC ACG TGT |
| 341 | Thr Thr Ser Asp Ala Thr Cys | |
| D10 | 1021 | ACT ACC AGT GAG GCC ACG TGT |
| 341 | Thr Thr Ser Glu Ala Thr Cys | |
| B11 | 1021 | ACT ACC AGT GAG GCC ACG TGT |
| 341 | Thr Thr Ser Glu Ala Thr Cys | |
| rT89E2-4 | 1021 | ACT ACC AGT GAG TCC ACG TGT |
| 341 | Thr Thr Ser Glu Ser Thr Cys | |
| C4 | 1021 | ACT ACC AGT GAG GCC ACG TGT |
| 341 | Thr Thr Ser Glu Ala Thr Cys | |
| rC4 | 1021 | ACT ACC AGT GAG GCC ACG TGT |
| 341 | Thr Thr Ser Glu Ala Thr Cys | |
| r56 | 1021 | ACT ACC AGT GAC GCC ACG TGT |
| 341 | Thr Thr Ser Asp Ala Thr Cys |
For the first nucleotide or amino acid listed.
The nucleotide sequence of the entire UL89 ORF was determined as described in the text. Only the portion of exon 2 which contained the mutation is presented. Nucleotide and inferred amino acid changes are shown in boldface.
Nucleotide sequence from GenBank.
To determine if the D344E change was necessary and sufficient for the Towne strain to be resistant to the benzimidazole ribonucleosides, drug resistance was transferred by transfecting exon 2 of the UL89 ORF from isolate B11 into MRC-5 cells with infectious, genomic-length AD169 DNA. The plating efficiency difference of 0.43% between populations transfected with wt and B11 UL89 DNA (Table 3) represents the percentage of drug-resistant virions in the heterogeneous progeny and demonstrated that the mutation in UL89 generated resistance to the benzimidazoles. This recombinant (termed rT89E2-4) was isolated, and its resistance to the benzimidazole ribonucleosides was similar to that of isolates B11 and D10 (Table 1), demonstrating that the mutation in UL89 was sufficient to cause the level of resistance observed. The recombinant was sequenced and had the expected C-to-G change in nucleotide 1032 in exon 2 of UL89 resulting in the D344E mutation (Table 2). Surprisingly, T was found in position 1033, inferring serine in position 345 which is characteristic of the parental AD169 DNA used in the transfection, not an alanine which would be expected based on Towne strain as the source of the drug-resistant DNA. We speculate that because there were two adjacent mismatches due to homologous recombination between genomic DNA from wt AD169 and PCR product of mutant UL89 from Towne, the second mismatched nucleotide (position 1033) was repaired in the recombinant DNA.
Isolation and characterization of highly resistant virus.
In our prior study, isolate 1038rB was more resistant to the benzimidazole ribonucleosides than 1038rA and 1038rC due to an additional mutation from Ala to Thr at position 355 (57). To explore the possibility that the region of UL89 around amino acids 344 to 355 was critically involved in the binding of benzimidazole ribonucleosides, we attempted to isolate additional drug-resistant mutants by passaging strain D10 (with the single D344E mutation) in a higher concentration of TCRB. Upon further passage of isolate D10 in the presence of 50 μM TCRB, progeny virus eventually grew; another mutant was selected and plaque purified, and the resulting isolate was designated C4. It was ∼30-fold resistant to the benzimidazole ribonucleosides (Table 4). Based on the prior results and the increased resistance to the benzimidazoles, virus isolate C4 was expected to encode additional mutations in UL89, but none were detected (Table 2). C4 was further characterized genotypically by sequencing the other five genes known to be involved in viral DNA processing and packaging (ORFs UL51, UL52, UL56, UL77, and UL104). The only ORF of these six that had any mutation was UL56. A mutation from A to G in position 611 resulted in a coding change from glutamine to arginine at position 204 (Q204R) (Table 5).
TABLE 4.
Susceptibilities of HCMV strains and recombinants to benzimidazole ribonucleosides and GCVa
| HCMV strain | Plaque reduction assay (IC50 [μM])
|
Yield reduction assay (IC95 [μM])
|
||||
|---|---|---|---|---|---|---|
| TCRB | BDCRB | GCV | TCRB | BDCRB | GCV | |
| Towne | 2.4 (2.2–2.7) | 2.3 (1.5–3.5) | 4.9 (3.7–6.4) | 1.1 (0.9–1.2) | 0.18 (0.15–0.22) | 3.1 (2.1–4.7) |
| D10 | 18 (14–22) | 13 (11–16) | 5.4 (3.9–7.5) | 7.9 (5.5–11) | 2.0 (1.4–2.9) | 3.4 (2.6–4.4) |
| C4 | 57 (43–76) | 33 (29–38) | 6.1 (4.9–7.7) | 25 (18–34) | 6.8 (4.5–10) | 1.8 (0.85–3.7) |
| rC4 | 68 (28–163) | 32 (24–42) | 2.5 (2.1–2.9) | 39 (32–48) | 4.8 (2.8–8.3) | 3.7 (2.7–5.0) |
| r56 | 12 (10–15) | 7.6 (6.0–9.6) | 7.0 (5.0–9.8) | 5.2 (4.0–7.0) | 3.0 (2.4–3.8) | 2.9 (1.9–4.4) |
Plaque and yield reduction assays were performed with the indicated strains of HCMV as described in the text; 95% confidence intervals (in parentheses) were calculated by using the variable-slope dose-response algorithm of GraphPad Prism. Plaque assay data are averages of three experiments, each done in duplicate except for r56, in which case two experiments were done. Yield reduction assays were done once, in quadruplicate.
TABLE 5.
Sequences of UL56 from drug-resistant virus isolates
| HCMV strain | Position no.a | Sequenceb |
|---|---|---|
| AD169c | 601 | ATC CCG AAT CAG GGC CGC TCG |
| 201 | Ile Pro Asn Gln Gly Arg Ser | |
| Towne | 601 | ATC CCG AAT CAG GGC CGC TCG |
| 201 | Ile Pro Asn Gln Gly Arg Ser | |
| D10 | 601 | ATC CCG AAT CAG GGC CGC TCG |
| 201 | Ile Pro Asn Gln Gly Arg Ser | |
| C4 | 601 | ATC CCG AAT CGG GGC CGC TCG |
| 201 | Ile Pro Asn Arg Gly Arg Ser | |
| rC4 | 601 | ATC CCG AAT CGG GGC CGC TCG |
| 201 | Ile Pro Asn Arg Gly Arg Ser | |
| r56 | 601 | ATC CCG AAT CGG GGC CGC TCG |
| 201 | Ile Pro Asn Arg Gly Arg Ser |
For the first nucleotide or amino acid listed.
The nucleotide sequence of the entire UL56 ORF was determined as described in the text. Only the portion which contains the mutation is presented. Nucleotide and inferred amino acid changes are shown in boldface.
Nucleotide sequence from GenBank.
Marker transfer analysis of highly resistant virus.
Marker transfer techniques were used to explore the significance of the mutation identified in the UL56 ORF. This mutation was introduced into D10 virus through homologous recombination. The plating efficiency of the progeny (Table 3) strongly suggested that the Q204R mutation was responsible for the increase in drug resistance between isolates D10 and C4. Since none of the progeny from the recombination between D10 DNA and wt UL56 grew under the selection conditions (20 μM BDCRB), the increased drug resistance did not appear to develop spontaneously during the course of the experiment and the resistant virus observed was a result of the mutation in the UL56 ORF.
The drug-resistant recombinant was plaque purified and designated rC4. Its susceptibility to the benzimidazole ribonucleosides and to GCV was similar to that of C4 (Table 4). The similarity in the resistance patterns of virus isolates C4 and rC4 confirmed that the mutation in UL56 was responsible for the increase in drug resistance between isolates D10 and C4 and substantiated that it was the only additional mutation involved in the phenotype of a high level of drug resistance (20- to 30-fold) in isolate C4. UL89 and UL56 from virus isolate rC4 were sequenced, and each encoded only the expected mutations (Tables 2 and 5).
Homologous recombination also was used to selectively introduce the mutation in UL56 from isolate C4 into wt Towne virus. The progeny from this experiment had a plating efficiency of 5.4% in 5 μM BDCRB, which demonstrated that the single nucleotide change in the UL56 ORF could cause drug resistance in the absence of any changes in the UL89 ORF (Table 3). This recombinant virus was plaque purified and designated r56. It was 5- to 10-fold resistant to the benzimidazole nucleosides (Table 4), demonstrating that this mutation also was necessary and sufficient for drug resistance. Virus r56 was sequenced through ORFs UL89 and UL56 to verify that it had only one mutation in the UL56 ORF (Q204R) and did not have any mutations in UL89 (Tables 2 and 5).
Growth studies.
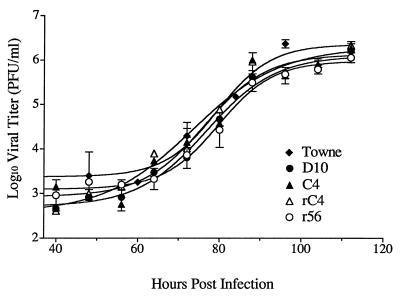
The replication characteristics of wt Towne, D10, C4, rC4, and r56 were measured to determine if any of the isolates were growth defective. The rate of replication of each virus isolate initially was measured following infection at a high MOI (2 PFU/cell). Figure 3 shows that there were no differences in the rates of replication of the various isolates over the course of a single cycle of replication. Statistical comparisons showed that the slopes of all curves were similar (slopes ± 95% confidence intervals = 7.1 ± 2.5, 5.7 ± 1.5, 6.0 ± 1.4, 7.1 ± 1.7, and 5.4 ± 1.4 h−1, respectively for Towne, D10, C4, rC4, and r56). Additionally, there were no statistically significant differences in the total amounts of infectious virus produced during the course of the infection (yield ± 95% confidence intervals = 6.4 ± 0.3, 6.0 ± 0.2, 6.1 ± 0.2, 6.1 ± 0.2, and 5.9 ± 0.1 log10 PFU/ml at 112 h postinfection, respectively, for Towne, D10, C4, rC4, and r56) (Fig. 3). Thus, all of the mutants were fully capable of replicating at normal levels. Each isolate was also observed over multiple cycles of replication by infecting cells at a low MOI (0.01 PFU/cell). Samples were frozen twice daily for 10 days, and the viral titer of each sample was determined. No statistically significant differences in the slopes of the growth curves or in the total yield of HCMV were detected among the mutants (slopes ± 95% confidence intervals = 25 ± 2.6, 35 ± 8.4, and 31 ± 7.3 h−1 and yield at plateaus = 7.4 ± 0.2, 8.2 ± 0.7, 7.0 ± 0.3, and 8.4 ± 0.9 log10 PFU/ml at 204 h postinfection, respectively, for D10, C4, rC4, and r56).
FIG. 3.
Single-cycle growth curves comparing mutant viruses. HFF cells were infected at an MOI of 2 PFU/cell and were harvested at time points spaced 8 to 12 h apart over the course of 4 days. After all samples were collected, they were titered across 96-well plates. Curves were fitted and data were analyzed by using the exponential growth algorithm of Prism.
Analysis of DNA processing by the recombinant viruses.
Benzimidazole ribonucleosides inhibit the cleavage of concatemeric, viral DNA to monomeric, genomic-length units at concentrations at and above the IC50 for both wt virus and HCMV isolates resistant to BDCRB due to the mutation in UL89 (57). To explore if this also were true for resistant isolates with mutations in UL56, HFF cells were infected with the mutant HCMV isolates and grown for a single cycle of viral replication. Intracellular DNA was separated by pulsed-field electrophoresis and stained with ethidium bromide. Figure 4 shows gels obtained from wt Towne and three mutant viruses. The concentration of TCRB required to inhibit monomer formation by each of the mutants was proportional to the IC50 of the isolate. The phenotype did not differ between D10, with a single mutation in UL89, and r56, with a single mutation in UL56, nor was there any difference with C4, which contains both mutations. These data further confirm that the benzimidazole ribonucleosides target viral DNA processing.
We also observed a band migrating slightly slower than the monomer band (labeled “mono +” in Fig. 4), similar to previous studies with BDCRB-resistant viruses (57). The amount of this apparently higher-molecular-weight DNA species first appeared at drug concentrations around the IC50 and increased with increasing drug concentrations. At particularly high concentrations of TCRB, this band disappeared with wt virus and isolate C4. With isolates D10 and r56, it persisted at the highest drug level tested, but we have no data to explain why this persistence was observed only with the single-mutation viruses. The identity and structure of this DNA species is not known but is under investigation (57).
DISCUSSION
There is substantial evidence from our laboratories indicating that the benzimidazole ribonucleosides inhibit viral replication during the DNA processing and packaging stage (57). Most significant is that polygenomic concatemeric viral DNA is not cleaved to monomeric lengths and is not encapsidated (57). This is a phenotype consistent with that of temperature-sensitive and null mutant HSV-1 isolates with aberrations in one of six ORFs: UL6, UL15, UL25, UL28, UL31, or UL32 (1, 2, 4, 5, 45, 49, 50, 54, 61, 62). Each of these genes is essential for viral replication and is presumed to be involved in viral DNA maturation. The similarity in phenotype between these mutants and wt HCMV treated with the benzimidazole ribonucleosides is consistent with the hypothesis that benzimidazole ribonucleosides inhibit viral DNA processing.
HSV-1 ORFs UL15 and UL28 are homologs of HCMV ORFs UL89 and UL56, respectively. Previous studies have shown that null mutants of HSV-1 UL15 and UL28 have the same phenotype of inability to process viral DNA (5, 54, 62). Our isolates, D10 and r56, respectively, have similar resistance to the benzimidazole ribonucleosides in antiviral assays. Thus, our mutants with a single mutation in either HCMV UL89 or UL56 also have identical phenotypes. These studies suggest that the proteins encoded by UL56 and UL89 either have very similar functions or participate in an intertwined, concerted series of activities.
Antiviral drug resistance that maps to two different genes is not unprecedented. With GCV, resistance maps both to the kinase responsible for activation of the drug (53) and to the DNA polymerase which GCV triphosphate inhibits (52). However, this is very different from our current results in that these two steps in GCV action are not directly related in viral replication. Furthermore, BDCRB does not require phosphorylation for activity, and there is no evidence for any other form of activation (36). Consequently, this does not explain the pattern of resistance to the benzimidazoles.
Mutations in proteins known to interact with the target protein of a drug have been shown to cause altered drug sensitivity (21). For example, selected mutations in the gene for viral DNA polymerase that result in resistance to phosphonoacetic acid concurrently result in hypersensitivity to aphidicolin (9, 21). In addition, temperature sensitive mutants with lesions in the single-stranded DNA binding protein (UL29) have increased sensitivity to phosphonoacetic acid and aphidicolin (19). That mutations in one protein (UL29) could affect the activity of another (DNA polymerase) led to the proposal of an interaction between the viral DNA polymerase and the single-stranded DNA binding protein (reviewed in reference 42).
Likewise, the fact that drug resistance maps to both UL89 and UL56 strongly suggests an interaction between their two gene products. This is consistent with the proposed role of each in viral DNA maturation and with observations of the pseudorabies virus (PRV) homolog of UL56 (UL28). This protein is found primarily in the nuclei of infected cells (44), but it is limited to the cytoplasm in the absence of other viral proteins. When it is coexpressed with HSV-1 UL15, it regains nuclear localization (35). These findings support an interaction between the two proteins which is required for proper intracellular localization of the PRV UL28 gene product. It is possible that in HCMV such an interaction between UL56 and UL89 proteins results in a complex which functions during viral DNA maturation.
There also are similarities to bacteriophage. Bacteriophage replication has long been proposed as a model for that of the herpesviruses because of the similarity in genome size and the large number of gene products required for DNA replication and packaging (60). Both synthesize high-molecular-weight DNA concatemers, cleave these concatemers into genomic-length units, and package the pieces into preformed capsids. A two-subunit terminase complex is common among bacteriophages (12). In bacteriophage T4, gp17 is part of a two-subunit terminase complex with gp16 (12). The gp17 subunit appears to have constitutive endonucleolytic activity which seems to be regulated by an association with gp16 (47). gp17 also interacts directly with another protein—p20, the bacteriophage capsid vertex protein (32). A mutant with a lesion in p20 displays the same phenotype as the HSV-1 packaging mutants; viral DNA is synthesized but not cleaved or encapsidated.
HCMV UL89 and homologs are highly conserved among herpesviruses and are invariably expressed as the spliced product of two or more exons. The HSV-1 homolog, UL15, has been studied extensively (5–7, 24, 45, 57, 62) but the function of this protein has not been observed directly. In 1992, Davison reported some homology between UL89 and the endonucleolytic portion of the terminase complex of bacteriophage T4, protein gp17 (23), thereby suggesting that UL89 could be the HCMV terminase. Both proteins encode a potential nucleotide binding motif, HCMV amino acids GTK219 to DE310. In a linear sense, this motif is distal to the mutations in UL89 that confer resistance. It is possible that protein folding juxtaposes the two areas and allows the mutations to alter the nucleotide binding site and thereby affect endonuclease activity (57). However, we know of no direct evidence for nuclease activity of the UL89 protein.
In contrast, the recent studies of Bogner et al. (13) provide direct evidence for specific nuclease activity of the UL56 gene product. Until now, little has been known about the function of this protein. It is a minor structural protein and can be detected with human convalescent serum (14, 15). There were early suggestions that the HSV-1 homolog could be involved in both DNA processing (2) and glycoprotein expression (43). However, later studies negated its involvement in glycoprotein trafficking, and a null mutant of the HSV-1 UL28 ORF was fully capable of glycoprotein expression (17, 54). This finding suggests that UL28 may have multiple, independently functioning domains. Bogner et al. also provide direct evidence for such a suggestion by establishing that the gene product of UL56 has not only nuclease activity but also specific DNA binding affinity (13).
A potentially corroborating observation for specific DNA binding emerges from a comparison of the predicted amino acid sequences of some of the homologs of the UL56 ORF (Table 6). First, the amino acid change which resulted in drug resistance (Q204R) is in a position that is completely conserved but the change did not affect the viability of the virus. Second, three cysteines and one histidine also are completely conserved in all 12 herpesviruses that have been compared. These amino acids could represent a metal binding motif (11); in accord with observations of Bogner et al. (13), this suggests DNA sequence-specific binding capability for the UL56 protein.
TABLE 6.
Amino acid sequence comparison of homologs of UL56
| Virus |
Amino acid sequencea |
|---|---|
| HCMV | EVYVEGTT-CAQCYEELMAVPNQGRSLNKRLQGLLCNHIAVHRPSS |
| HSV-1 | ELS-DPSHPCAVCFEELCVTANQGATIARRLADRICNHVTQQAQVR |
| Varicella-zoster virus | VELFDPAHPCAICFEELCITANQGETLHRRLLGCICDHVTKQVRVN |
| Human herpesvirus 6 | ESYLETKT-CMKCYEELTLTPNQGKSLRRRLHGKFCNHLTEQKAFF |
| Human herpesvirus 7 | EVFSETTT-CLKCYEELSLVPNQGKSIRKRLAGKFCNHLTETHMVS |
Each amino acid in boldface represents the location of the mutation in virus isolate C4. Amino acids that are conserved among listed herpesviruses plus PRV, equine herpesvirus type 1, bovine herpesviruses 1 and 2, murine cytomegalovirus, rat cytomegalovirus, and herpesvirus saimiri are underlined. The cysteines and histidine suggest a potential metal binding site. From the Swiss Protein database.
Such specific binding may account for one of the major differences in DNA packaging between bacteriophage T4 and the herpesviruses. Bacteriophage T4 DNA is packaged in a head-full manner (12), whereas herpesvirus DNA is cleaved and packaged in a manner that is both sequence specific and measured (40, 58, 59). The protein encoded by UL56 apparently binds the pac sequences (cleavage recognition sites) and endonucleolytically cleaves viral DNA. The role of the gene product of UL89 is unknown but it may be an accessory protein to the UL56 protein. This may account for the putative interaction between these two proteins that is implied by the results reported herein.
For TCRB or BDCRB to inhibit viral DNA processing, it is possible that both UL89 and UL56 proteins interact directly with the benzimidazole ribonucleoside. Consequently resistance to the benzimidazoles would be a function of altered binding of the drug to each protein or to a common binding pocket formed by a complex of two (or more) proteins. On the other hand, the situation may be similar to that of the DNA polymerase/single-stranded DNA binding protein where a mutation in one protein affects the activity of the other protein (21). In this case, we hypothesize that the UL89 protein is the direct target of the benzimidazole ribonucleosides because drug resistance has mapped to this gene in most of the mutants found to date. When the UL89 protein binds to BDCRB, binding may alter the interaction between this protein and the UL56 protein, thereby decreasing nuclease activity and/or DNA binding. The mutation in UL56 may compensate for the interference by altering the stringency of UL56 protein binding to pac sites thereby facilitating a more productive interaction.
In summary, resistance to the benzimidazole ribonucleosides has been mapped to both HCMV ORFs UL56 and UL89. Mapping of resistance to UL56 complements and extends the recent work by Bogner et al. (13), who showed that UL56 encodes a pac motif binding protein which has specific nuclease activity. Our results also suggest that the proteins encoded by UL56 and UL89 interact and that both are potential antiviral drug targets.
ACKNOWLEDGMENTS
We thank Faris Albayya for expert technical assistance, Jonathon Winger for help with DNA sequencing, Jennie Jacobson for assistance with aligning sequences and isolating infectious DNA, and David Mindell for use of an ABI model 377 sequencer.
These studies were supported by research grant UO1-AI31718 from the National Institute of Allergy and Infectious Diseases and Research Agreement DRDA-942921 with Glaxo Wellcome Inc. Use of the GCG programs was supported by grant M01RR00042 to the University of Michigan General Clinical Research Center. P.M.K. was supported in part by NIH training grant GM07767.
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, O’Hara M, Preston V G. Characterization of a herpes simplex virus type 1 mutant which has a temperature sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Alford C A, Britt W J. Cytomegalovirus. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 227–255. [Google Scholar]
- 4.al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 5.Baines J D, Cunningham C, Nalwanga D, Davidson A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines J D, Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus type 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992;66:5621–5626. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balfour H H, Chace B A, Stapleton J T, Simmons R L, Fryd D S. A randomized, placebo-controlled trial of oral acyclovir for the prevention of cytomegalovirus disease in recipients of renal allographs. N Engl J Med. 1989;320:1381–1387. doi: 10.1056/NEJM198905253202105. [DOI] [PubMed] [Google Scholar]
- 9.Bastow K F, Derse D D, Cheng Y-C. Susceptibility of phosphonoformic acid-resistant herpes simplex virus variants to arabinosyl nucleosides and aphidicolin. Antimicrob Agents Chemother. 1983;23:914–917. doi: 10.1128/aac.23.6.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Porat T, Rixon F J. Replication of herpesvirus DNA. IV. Analysis of concatemers. Virology. 1979;94:61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- 11.Berg J M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 12.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;42:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 13.Bogner E, Radsak M, Stinski M. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72:2259–2264. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogner E, Reschke M, Reis B, Mockenhaupt T, Radsak K. Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology. 1993;196:290–293. doi: 10.1006/viro.1993.1477. [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw P A, Duran-Guarino M R, Perkins S, Rowe J I, Fernandez J, Fry K E, Reyes G R, Young L, Foung S K H. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology. 1994;205:321–328. doi: 10.1006/viro.1994.1648. [DOI] [PubMed] [Google Scholar]
- 16.Britt W J, Pass R F, Stagno S, Alford C A. Pediatric cytomegalovirus infection. Transplant Proc. 1991;23:115–117. [PubMed] [Google Scholar]
- 17.Cavalcoli J D, Baghian A, Homa F L, Kousoulas K G. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant (KOS) tsZ47: evidence for allelic complementation in the UL28 gene. Virology. 1993;197:23–34. doi: 10.1006/viro.1993.1563. [DOI] [PubMed] [Google Scholar]
- 18.Chee M S, Bankeir A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnel T, Hutchinson C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S P, Tomlinson P, Weston K M, Barrel B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 19.Chiou H C, Weller S K, Coen D M. Mutations in the herpes simplex virus DNA-binding protein gene leading to altered sensitivity to DNA polymerase inhibitors. Virology. 1985;145:213–226. doi: 10.1016/0042-6822(85)90155-2. [DOI] [PubMed] [Google Scholar]
- 20.Chrisp P, Clissold S P. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 21.Coen D M, Furman P A, Aschman D P, Schaffer P A. Mutations in the herpes simplex virus DNA polymerase gene conferring hypersensitivity to aphidicolin. Nucleic Acids Res. 1983;11:5287–5297. doi: 10.1093/nar/11.15.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crumpacker C S. Ganciclovir. Drug Ther. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 23.Davidson A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 24.Dolan A, Arbuckle M, McGeoch D J. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 1991;20:97–104. doi: 10.1016/0168-1702(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 25.Erriksson B, Öberg B, Wahren B. Pyrophosphate analogs as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim Biophys Acta. 1982;696:115–123. doi: 10.1016/0167-4781(82)90018-5. [DOI] [PubMed] [Google Scholar]
- 26.Field A K, Biron K K. “The end of innocence” revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev. 1991;7:1–13. doi: 10.1128/cmr.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genetics Computer Group. Program manual for the GCG package. 7th ed. Madison, Wis: Genetics Computer Group; 1991. [Google Scholar]
- 28.Grattan M T, Moreno-Cabral C E, Starnes V A, Stinson E B, Shumway N E. Cytomegalovirus infection is associated with cardiac allograph rejection and atherosclerosis. JAMA. 1989;261:3561–3566. [PubMed] [Google Scholar]
- 29.Hirsch I, Cabral G, Patterson M, Biswal N. Studies on intracellular replicating DNA of herpes simplex virus type 1. Virology. 1977;81:48–61. doi: 10.1016/0042-6822(77)90057-5. [DOI] [PubMed] [Google Scholar]
- 30.Hitchcock M J M, Jaffe H S, Martin J C, Stagg R J. Cidofovir, a new agent with potent antiherpesvirus activity. Antimicrob Agents Chemother. 1996;7:115–127. [Google Scholar]
- 31.Ho H-T, Woods K L, Bronson J J, DeBoeck H, Martin J C, Hitchcock M J M. Intracellular metabolism of the antiherpes agent (S)-1-[3-hydroxy-2-(phosphonylmethoxy)propyl]cytosine. Mol Pharmacol. 1992;41:197–202. [PubMed] [Google Scholar]
- 32.Hsiao C L, Black L W. DNA packaging and the pathway of bacteriophage T4 head assembly. Proc Natl Acad Sci USA. 1977;74:3652–3656. doi: 10.1073/pnas.74.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in the nuclei of infected cells and their role in the generation of four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein R J. Isolation of herpes simplex virus clones and drug resistant mutants in microculture. Arch Virol. 1975;49:73–80. doi: 10.1007/BF02175598. [DOI] [PubMed] [Google Scholar]
- 35.Koslowski K M, Shaver P R, Wang X-Y, Tenney D J, Pederson N E. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krosky, P. M., K. Z. Borysko, M. R. Nassiri, R. G. Ptak, S. S. Good, M. R. Underwood, K. K. Biron, L. B. Townsend, and J. C. Drach. 1997. Unpublished data.
- 37.Mar E, Chiou J, Cheng Y, Huang E. Inhibition of cellular DNA polymerase α and human cytomegalovirus-induced DNA polymerase by the triphosphates of 9-(2-hydroxyethoxymethyl)guanine and 9-(1,3-dihydroxy-2-propoxymethyl)guanine. J Virol. 1985;53:776–780. doi: 10.1128/jvi.53.3.776-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenzie R, Travis M W D, Dolan S A, Pittaluga S, Feuerstein I M, Shelhamer J, Yarchoan R, Masur H. The cause of death in patients with human immunodeficiency virus infection: a clinical and pathological study with emphasis on the role of pulmonary disease. Medicine. 1991;70:326–343. doi: 10.1097/00005792-199109000-00004. [DOI] [PubMed] [Google Scholar]
- 39.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization, and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 41.Nieto F J, Adam E, Sorlie P, Farzadegan H, Melnick J L, Comstock G W, Szklo M. Cohort study of cytomegalovirus infection as a risk factor for carotid intimal-medial thickening, a measure of subclinical atherosclerosis. Circulation. 1996;94:922–927. doi: 10.1161/01.cir.94.5.922. [DOI] [PubMed] [Google Scholar]
- 42.Olivo P D, Challberg M D. Functional analysis of the herpes simplex virus gene products involved in DNA replication. In: Wagner E, editor. Herpesvirus transcription and its replication. Boca Raton, Fla: CRC Press; 1990. pp. 137–150. [Google Scholar]
- 43.Pancake B A, Aschman D P, Schaffer P A. Genetic and phenotypic analysis of herpes simplex virus type 1 mutants conditionally resistant to immune cytolysis. J Virol. 1983;47:568–585. doi: 10.1128/jvi.47.3.568-585.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pederson N E, Enquist L W. Overexpression in bacteria and identification in infected cells of the pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5. J Virol. 1991;65:3746–3758. doi: 10.1128/jvi.65.7.3746-3758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon A W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard M N, Turk S R, Coleman L A, Englehardt S L, Shipman C, Jr, Drach J C. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus. J Virol Methods. 1990;28:101–106. doi: 10.1016/0166-0934(90)90091-s. [DOI] [PubMed] [Google Scholar]
- 47.Rao V B, Black L W. Cloning, overexpression, and purification of the terminase proteins gp16 and gp17 of bacteriophage T4: construction of a defined in vitro DNA packaging system using purified proteins. J Mol Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 50.Sherman G, Bachenheimer S L. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 51.Shipman C., Jr Evaluation of 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) as a tissue culture buffer. Proc Soc Exp Biol Med. 1969;130:305–310. doi: 10.3181/00379727-130-33543. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan V, Biron K K, Talarico C, Stanat S, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 54.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation requires the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Townsend L B, Devivar R V, Turk S R, Nassiri M R, Drach J C. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(β-d-ribofuranosyl)benzimidazoles. J Med Chem. 1995;38:4098–4105. doi: 10.1021/jm00020a025. [DOI] [PubMed] [Google Scholar]
- 56.Turk S R, Shipman C, Jr, Nassiri M R, Genzlinger G, Krawczyk S H, Townsend L B, Drach J C. Pyrrolo[2,3-d]pyrimidine nucleosides as inhibitors of human cytomegalovirus. Antimicrob Agents Chemother. 1987;31:544–550. doi: 10.1128/aac.31.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Underwood M R, Harvey R J, Stanat S C, Hemphill M L, Miller T, Drach J C, Townsend L B, Biron K K. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J Virol. 1998;72:717–725. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlazny D A, Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci USA. 1981;78:742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vlazny D A, Kwong A, Frenkel N. Site specific cleavage and packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin’s scientific legacy. Washington D.C: American Society for Microbiology; 1995. pp. 189–213. [Google Scholar]
- 61.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 62.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y F, Leon M B, Waclawiw M A, Popma J J, Yu Z X, Finkel T, Epstein S E. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med. 1994;335:624–630. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]