Abstract
Vaccinia virus NPH-II is the prototypal RNA helicase of the DExH box protein family, which is defined by six shared sequence motifs. The contributions of conserved amino acids in motifs I (TGVGKTSQ), Ia (PRI), II (DExHE), and III (TAT) to enzyme activity were assessed by alanine scanning. NPH-II-Ala proteins were expressed in baculovirus-infected Sf9 cells, purified, and characterized with respect to their RNA helicase, nucleic acid-dependent ATPase, and RNA binding functions. Alanine substitutions at Lys-191 and Thr-192 (motif I), Arg-229 (motif Ia), and Glu-300 (motif II) caused severe defects in RNA unwinding that correlated with reduced rates of ATP hydrolysis. In contrast, alanine mutations at His-299 (motif II) and at Thr-326 and Thr-328 (motif III) elicited defects in RNA unwinding but spared the ATPase. None of the mutations analyzed affected the binding of NPH-II to RNA. These findings, together with previous mutational studies, indicate that NPH-II motifs I, Ia, II, and VI (QRxGRxGRxxxG) are essential for nucleoside triphosphate (NTP) hydrolysis, whereas motif III and the His moiety of the DExH-box serve to couple the NTPase and helicase activities. Wild-type and mutant NPH-II-Ala genes were tested for the ability to rescue temperature-sensitive nph2-ts viruses. NPH-II mutations that inactivated the phosphohydrolase in vitro were lethal in vivo, as judged by the failure to recover rescued viruses containing the Ala substitution. The NTPase activity was necessary, but not sufficient, to sustain virus replication, insofar as mutants for which NTPase was uncoupled from unwinding (H299A, T326A, and T328A) were also lethal. We conclude that the phosphohydrolase and helicase activities of NPH-II are essential for virus replication.
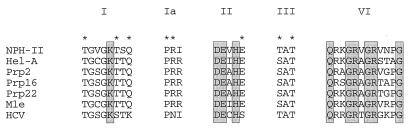
Vaccinia virus NPH-II (nucleoside triphosphate [NTP] phosphohydrolase II) is an NTP-dependent helicase that catalyzes unidirectional unwinding of 3′-tailed duplex RNAs (27). NPH-II is a member of the DExH family of nucleic acid-dependent NTPases. The DExH proteins are defined by six conserved motifs arrayed in a collinear fashion (7, 9, 16); five of these motifs are shown in Fig. 1. The size of the DExH family is expanding rapidly as the signature elements are encountered in newly cloned genes and during genome sequencing efforts. The products of these genes are typically designated putative helicases, yet relatively few DExH proteins have actually been shown to unwind duplex DNA or RNA. Vaccinia virus NPH-II was the first RNA-dependent NTPase to be purified (21, 22), and its substrate specificity and nucleic unwinding properties have been thoroughly characterized (10, 22, 27, 28). NPH-II exemplifies a subgroup of structurally related DExH proteins that includes human RNA helicase A, the yeast pre-mRNA splicing factors Prp2, Prp16, and Prp22, Drosophila maleless, and hepatitis C virus (HCV) RNA helicase NS3 (Fig. 1) (1, 12–14, 18, 19, 26).
FIG. 1.
Conserved sequence elements define the DExH protein family. The amino acid sequence of motifs I, Ia, II, III, and VI of vaccinia virus NPH-II (GenBank accession no. M35027) is aligned with the sequences (with GenBank accession numbers in parentheses) of the corresponding motifs of six other DExH-box proteins: human helicase A (Hel-A) (L13848); Saccharomyces cerevisiae splicing factors Prp2 (X55936), Prp16 (M31524), and Prp22 (X58681); Drosophila MLE protein (M7412); and HCV RNA helicase NS3 (M62385). Amino acids that have been shown by mutagenesis to be important for the ATPase or helicase activities of NPH-II are shown in shaded boxes. New residues mutated in this study are marked by asterisks.
Insights into the function of the conserved motifs have emerged from structure-function analyses of NPH-II (8, 9). We demonstrated previously that alanine substitutions for Lys-191 in the GKT element (motif I) or Asp-296 and Glu-297 in the DExH box (motif II) severely impaired ATP hydrolysis and RNA unwinding without affecting the binding of the mutated proteins to RNA. Changing the DExH-box His residue to Ala resulted in a protein that was constitutively active for ATPase in the absence of a nucleic acid effector; the H299A enzyme did not unwind duplex RNA, even though RNA binding was unaffected (8). We have also performed an alanine scan of 10 residues within motif VI of NPH-II (491-QRKGRVGRVNPG-502). Alanine substitutions at invariant residues Gln-491, Arg-492, Gly-494, Arg-495, Gly-497, Arg-498, and Gly-502 caused severe defects in RNA unwinding that correlated with reduced rates of ATP hydrolysis (9). None of these mutations in motif VI significantly affected the binding of NPH-II to RNA. The residues shown to be important for helicase activity of NPH-II are highlighted in shaded boxes in Fig. 1.
In this study, we extended the mutational analysis to motifs Ia and III and to additional conserved residues in motifs I and II. The eight new positions targeted for alanine substitution are marked by asterisks in Fig. 1. We found that mutations (underlined) of Thr-192 in motif I (GKT), Arg-229 in motif Ia (PRI), and Glu-300 in motif II (DExHE) coordinately inactivated RNA unwinding and ATP hydrolysis without affecting RNA binding. In contrast, mutations of the two threonines in motif III (TAT) abolished helicase activity with only modest effects on ATP hydrolysis. The phosphohydrolase activity of the T326A and T328A proteins remained nucleic acid dependent. These results suggest that motif III acts to couple NTP hydrolysis to duplex unwinding.
In addition, we exploited our collection of biochemically defined NPH-II alanine substitution mutants and temperature sensitive (ts) nph2-ts viruses isolated by Fathi and Condit (4, 5) to examine if the NTPase and helicase functions of NPH-II are essential for vaccinia virus replication in vivo. This was done by DNA-mediated marker rescue of the nph2-ts viruses with wild-type and mutant NPH-II genes. Rescued viruses were genotyped to determine if the targeted alanine mutation had been incorporated during recombination to restore the wild-type codon at the ts lesion. We found that alanine mutations that inactivated the NTPase of NPH-II were invariably lethal, as judged by the failure to recover rescued viruses containing the Ala substitution. The NTPase activity of NPH-II was necessary, but not sufficient, to sustain virus replication, insofar as mutants for which ATPase was uncoupled from unwinding were also lethal. We conclude that the helicase activity of NPH-II is essential for vaccinia virus replication. A possible role of NPH-II-mediated strand displacement during vaccinia virus transcription is discussed.
MATERIALS AND METHODS
Alanine mutagenesis of His-NPH-II.
Phagemid pTM-His10-NPH-II containing a His-tagged NPH-II gene under the control of a T7 promoter and a picornavirus translational enhancer was described previously (8). Uracil-substituted single-stranded pTM-His10-NPH-II DNA was used as a template for oligonucleotide-directed mutagenesis. Mutagenic DNA primers were designed to create alanine substitutions at residues T187, T192, Q194, P228, R229, E300, T326, and T328. The presence of the desired mutations was screened initially by the gain or loss of a restriction site and confirmed by DNA sequencing. The mutated NPH-II genes were exchanged for the corresponding wild-type gene in the recombinant His10-NPH-II baculovirus transfer vector pBacPAK-9 (10). The entire NPH-II gene was sequenced in each case to ensure that no unwanted mutations were present in the expression vectors. The recombinant baculoviruses expressing the His10-NPH-II protein were constructed and selected according to protocols supplied by Clontech.
Expression and purification of His10-NPH-II.
Wild-type and alanine-substituted His10-NPH-II proteins were each expressed by infection of 30 150-cm2 dishes of Sf9 cell monolayers with recombinant baculovirus at a multiplicity of 10. Infected cells were incubated at 27°C for 72 h. Soluble cell lysates were prepared as described previously (8, 10). The His10-NPH-II protein was adsorbed to Ni-nitrilotriacetic acid agarose (10). After extensive washing of the resin with buffer containing 50 mM imidazole, His10-NPH-II was recovered by elution with 500 mM imidazole. Aliquots (0.3 ml) of each Ni-agarose fraction were layered onto a 4.7-ml 15 to 30% glycerol gradient containing 0.3 M NaCl in buffer A (50 mM Tris HCl, pH 8.0, 2 mM dithiothreitol (DTT), 1 mM EDTA, 10% glycerol, 0.1% Triton X-100). The gradients were centrifuged for 20 h at 50,000 rpm in a Beckman SW50 rotor. Fractions (0.17 ml) were collected from the bottoms of the tubes. The concentrations of NPH-II in the peak glycerol gradient fractions were determined by quantitative Western blotting as described previously (9).
Helicase assay.
Reaction mixtures (20 μl) contained 40 mM Tris HCl (pH 8.0), 2 mM DTT, 2 mM MgCl2, 2 mM ATP, and 25 fmol of [α32P]GMP-labeled tailed double-stranded RNA (dsRNA) substrate (prepared as described in reference 8). After incubation for 15 min at 37°C, the reactions were halted by addition of 5 μl of 0.1 M Tris HCl (pH 7.4)–5 mM EDTA–0.5% sodium dodecyl sulfate (SDS)–50% glycerol–0.1% xylene cyanol–0.1% bromophenol blue. Aliquots (20 μl) were electrophoresed at 15-mA constant current through an 8% polyacrylamide gel containing 45 mM Tris-borate and 1.2 mM EDTA. Labeled RNAs were visualized by autoradiography. The extent of unwinding (displaced RNA/total RNA) was quantitated by scanning the gel with a Fuji BAS1000 phosphorimager.
RNA binding assay.
Reaction mixtures (20 μl) contained 40 mM Tris HCl (pH 8.0), 2 mM DTT, 1 mM MgCl2, and 25 fmol of [α32P]GMP-labeled 98-mer single-stranded RNA (ssRNA) (corresponding to the 98-mer strand of the helicase substrate and labeled to high specific activity). After incubation for 15 min at 37°C, samples were adjusted to 8% glycerol, and 20-μl aliquots were electrophoresed at 15 mA through a native 8% polyacrylamide gel containing 22 mM Tris-borate and 0.6 mM EDTA. The extent of RNA-protein complex formation (bound RNA/total RNA) was quantitated with a phosphorimager.
Marker rescue.
Confluent BSC40 cell monolayers (35-mm-diameter dishes) grown at 31°C in Dulbecco modified Eagle (DME) medium with 5% fetal bovine serum (FBS) were infected with the ts vaccinia viruses at a multiplicity of 5. The inoculum was removed after 30 min, and the cells were washed twice and overlaid with DME–5% FBS. After 30 min of incubation at 31°C, the monolayers were treated with 1 ml of 0.05% trypsin–0.5 mM EDTA. The cell suspensions were mixed with 4 ml of DME and then harvested by centrifugation. The cell pellets were washed with 3 ml of HEPES-buffered saline (20 mM HEPES [pH 7.0], 150 mM NaCl, 0.7 mM Na2HPO4, 5 mM KCl, 6 mM dextrose) and recentrifuged. The pellets were resuspended in 0.8 ml of cold HEPES-buffered saline by gentle pipetting and transferred to a chilled tube containing 10 μg of pTM-His10-NPH-II plasmid DNA. The suspension were mixed by gentle agitation and chilled on ice for 10 min. The contents were transferred to chilled Bio-Rad 0.4-cm electrogap cuvettes. The cell suspensions were electroporated at 200 V (capacitance, 960 μF) in a Bio-Rad Gene Pulser equipped with a Bio-Rad capacitance extender and then placed on ice for 10 min. The cells were diluted into 7 ml of medium at room temperature. An aliquot (3.5 ml) was applied to a confluent cell monolayer of BSC40 cells (35-mm-diameter well) maintained at 40°C. After 48 h of incubation at 40°C, cells were harvested by scraping and centrifugation. The cell pellets were resuspended in 1 ml of DME. Lysis was achieved by three rounds of freezing and thawing, followed by brief sonication. The extent of marker rescue was determined by applying serial 10-fold dilutions of the lysates onto BSC40 monolayers at 40°C. After 48 h at 40°C, the cells were stained with 0.1% crystal violet in order to visualize plaques.
Genotyping of rescued viruses.
Viral revertants were selected by plating ∼10 PFU from each marker rescue onto BSC40 monolayers at 40°C under an agar overlay. Individual plaque isolates were amplified once by infection of BSC40 cell monolayers at 40°C. Infected cells were harvested by scraping and centrifugation. The infected cell pellets were resuspended in 20 mM Tris HCl (pH 8.0)–10 mM NaCl–10 mM EDTA and lysed by addition of 0.75% SDS. The samples were digested with proteinase K and then extracted twice with phenol-chloroform (1:1) and once with chloroform. DNA was recovered from the aqueous phase by ethanol precipitation and resuspended in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA. These DNAs were used as templates for PCR amplification of the NPH-II gene. PCR was carried out with Taq polymerase (Boehringer Mannheim) and oligonucleotide primers flanking the NPH-II coding sequence. The sequence of the sense-strand primer was 5′-CGTGATAGTTTCTCATTGGCCG-3′, and that of the antisense-strand flanking primer was 5′-GTTAATTCTCCCGTCCTCTC-3′. The PCR products were screened for the presence of diagnostic restriction endonuclease cleavage sites linked to the targeted alanine mutations in the transfected NPH-II-Ala genes.
RESULTS
Expression and purification of wild-type and mutated NPH-II proteins.
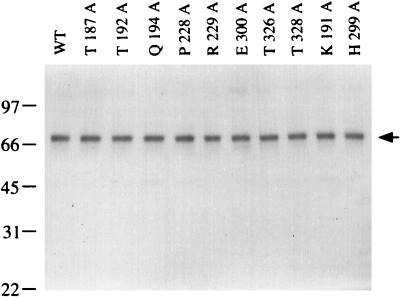
Alanine substitutions were introduced at conserved residues Thr-187, Thr-192, and Gln-194 in motif I, Pro-228 and Arg-229 in motif Ia, Glu-300 in motif II, and Thr-326 and Thr-328 in motif III (Fig. 1). Wild-type NPH-II, the eight new mutant proteins, and two other mutants, K191A (motif I) and H299A (motif II), were expressed in cultured insect cells infected with recombinant baculoviruses carrying an inserted copy of the NPH-II gene under the control of a polyhedrin promoter (10). Earlier studies of the K191A and H299A mutants had been performed with protein expressed in vaccinia virus-infected mammalian cells (8). Each protein was expressed as a N-terminal His-tagged derivative. We showed previously that recombinant wild-type His-tagged NPH-II is functionally identical to the native enzyme purified from vaccinia virions (8). The recombinant wild-type and mutant NPH-II proteins were purified from soluble lysates of baculovirus-infected cells by adsorption to Ni-agarose and elution with 0.5 M imidazole. The proteins were then purified further by glycerol gradient sedimentation. The peak gradient fractions from each of the enzyme preparations were used for biochemical studies of NPH-II activity. The concentration of His-NPH-II protein was gauged by quantitative Western blotting (10). To assess overall purity, equivalent amounts of each protein preparation were analyzed by SDS-polyacrylamide gel electrophoresis, and the gel was stained with Coomassie brilliant blue. A single 70-kDa polypeptide corresponding to His-NPH-II was detected in each case (Fig. 2).
FIG. 2.
NPH-II purification. Aliquots (0.23 μg) of the glycerol gradient preparations of wild-type (WT) NPH-II and the indicated NPH-II-Ala mutants were electrophoresed through an 8% polyacrylamide gel containing 0.1% SDS. Polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated at the left. The polypeptide corresponding to NPH-II is indicated by the arrow on the right.
Helicase activities of mutated His-NPH-II proteins.
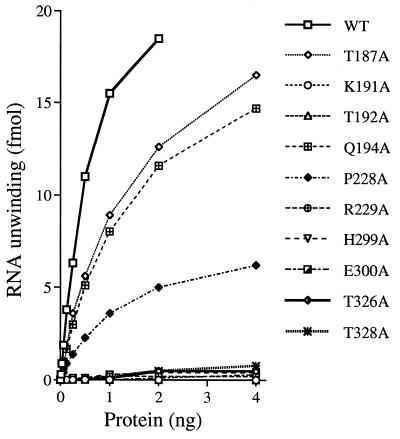
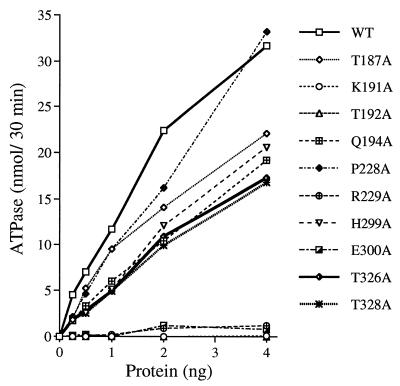
Helicase activity was tested by using a dsRNA substrate formed by annealing a 98-nucleotide RNA strand to a 38-nucleotide radiolabeled RNA strand to produce a 29-bp duplex with 5′ and 3′ tails (27). The extent of RNA unwinding by wild-type NPH-II was proportional to the level of input protein (Fig. 3). Five of the eight new mutants displayed severe reductions in helicase activity: T192A (motif I), R229A (motif Ia), E300A (motif II), and T326A and T328A (motif III). Their specific activities were <1% of the wild-type value (Fig. 3). The K191A and H299A proteins were also defective in RNA unwinding, as noted previously (Fig. 1). In contrast, mutants T187A and Q194A displayed ∼50% of the wild-type specific activity, and P228A was 20% as active as wild-type NPH-II (Fig. 3). RNA unwinding by wild-type NPH-II and each of the catalytically active mutants was completely dependent on added ATP (data not shown).
FIG. 3.
RNA unwinding by wild-type and mutant NPH-II proteins. Helicase assays were performed as described in Materials and Methods. The extent of RNA unwinding by wild-type (WT) and mutated NPH-II proteins is plotted as a function of the amount of input enzyme. Each data point represents the average of two independent determinations. The source of the protein preparation is indicated in the key to symbols.
RNA binding by His-NPH-II mutants.
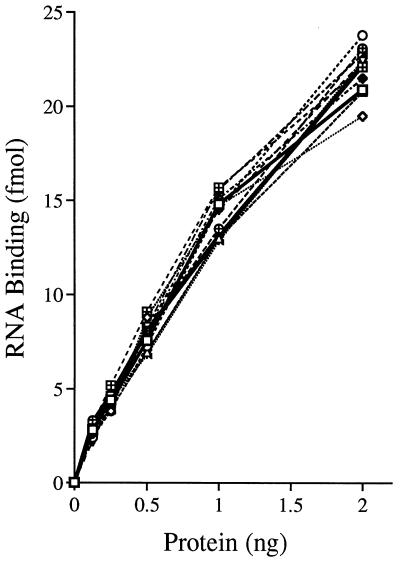
NPH-II binds to single-stranded nucleic acid in the absence of NTPs. The complex of NPH-II bound to ssRNA is stable and can be easily resolved from free RNA by native gel electrophoresis (8, 10, 27, 28). The radiolabeled RNA ligand used in standard binding assays corresponds to the 98-mer strand of the helicase substrate. The amount of the shifted protein-RNA complex formed by wild-type NPH-II and by each of the Ala mutants varied linearly with input protein; every one of the Ala-substituted proteins retained the ability to bind RNA (Fig. 4). The electrophoretic mobility of the RNA-protein complex formed by each of the mutant His-NPH-II proteins was similar to that of the wild-type enzyme analyzed in parallel (not shown). All of the NPH-II-Ala proteins displayed an affinity for ssRNA that was essentially identical to that of the wild-type enzyme (Fig. 4).
FIG. 4.
Binding of wild-type and mutated His-NPH-II proteins to ssRNA. Binding of NPH-II to a radiolabeled 98-mer ssRNA was measured in a gel shift assay as described in Materials and Methods. The extent of protein-RNA complex formation is plotted as a function of input enzyme. The symbols denoting identities of the proteins used in the titration experiments are as shown in Fig. 3.
To relate the RNA binding properties of the mutant proteins more directly to the observed effects on helicase activity, we performed native gel shift assays of the binding of the NPH-II mutants to the helicase substrate itself (10, 28). All 10 mutated proteins bound to the helicase substrate with affinity similar to that of wild-type NPH-II (not shown). Thus, the deleterious effects of the K191A, T192A, R229A, H299A, E300A, T326A, and T328A mutations on RNA unwinding could not be attributed to lack of binding to the helicase substrate.
Mutational effects on nucleic acid-dependent ATPase.
The extent of ATP hydrolysis by NPH-II in the presence of a ssDNA cofactor was proportional to input enzyme (Fig. 5). The specific activities of the mutants, expressed in parentheses as the percentage of the wild-type value, were as follows: T187A (68%), K191A (<0.2%), T192A (<0.2%) Q194A (55%), P228A (91%), R229A (2%), H299A (58%), E300A (1%), T326A (51%), and T328A (52%). The wild-type and mutant proteins were also assayed for ATP hydrolysis in the presence of a ssRNA cofactor (not shown). The specific activities of the mutants, expressed in parentheses as the percentage of the wild-type value, were as follows: T187A (56%), K191A (<0.2%), T192A (<0.2%), Q194A (67%), P228A (61%), R229A (4%), E300A (2%), T326A (61%), and T328A (61%). In light of our previous findings that the H299A mutation rendered the ATPase activity of NPH-II nucleic acid independent (8), we assayed each of the mutants for ATPase in the presence and absence of polynucleotide cofactors. ATP hydrolysis by T187A, Q1941, P228A, R229A, E300A, T326A, and T328A was stimulated 5- to 30-fold by nucleic acid (not shown). The K191A and T192A proteins displayed no detectable activity up to 60 ng of input enzyme in the presence or absence of a nucleic acid cofactor.
FIG. 5.
ATP hydrolysis by wild-type (WT) and mutated His-NPH-II proteins. ATP hydrolysis by NPH-II in the presence of single-stranded M13mp18 DNA was assayed as described elsewhere (8). ATPase activity is expressed as nanomoles of 32Pi released from [γ32P]ATP during a 30-min incubation at 37°C and is plotted as a function of input enzyme. Each data point represents the average of two independent determinations. The protein preparations used for each titration experiment are indicated in the key to symbols.
The mutational effects on the DNA-dependent ATPase, RNA-dependent ATPase, and helicase activities of NPH-II are summarized in Tables 1 and 2. We operationally define as nonessential those residues at which alanine substitution elicited less than an order of magnitude effect on helicase or ATP specific activities. By this criterion, conserved residues Thr-187 in motif I and Pro-228 in motif Ia are nonessential. The eight other residues analyzed above are defined as important for NPH-II function, because alanine substitutions reduced ATPase or helicase activities to less than 10% of the wild-type values. Two functional classes of important residues are illuminated by our results: (i) those at which side chain removal coordinately inactivates the ATPase and helicase functions, e.g., Lys-191 and Thr-192 in motif I, Arg-229 in motif Ia, and Glu-300 in motif II; and (ii) those at which side chain removal preserves phosphohydrolase activity but eliminates the capacity for duplex unwinding, e.g., His-299 in motif II and Thr-326 and Thr-328 in motif III.
TABLE 1.
Rescue of ts10 by NPH-II alleles containing alanine mutations in motifs II and III
| NPH-II gene | Virus titer at 40°C (PFU/ml) | Sp activity of protein (% of wild-type activity)
|
|
|---|---|---|---|
| ATPase | Helicase | ||
| Wild type | 1 × 107 | 100 | 100 |
| D296A | 2 × 104 | 5a | <5a |
| E297A | 3 × 104 | 5a | <5a |
| H299A | 3 × 104 | 58 | <1 |
| E300A | 1 × 104 | 1 | <1 |
| T326A | 1 × 104 | 51 | <1 |
| T328A | 1 × 104 | 52 | 1 |
| Plasmid | <10 | ||
From reference 8.
TABLE 2.
Rescue of ts18 by NPH-II alleles containing alanine mutations in motifs I and Ia
| NPH-II gene | Virus titer at 40°C (PFU/ml) | Sp activity of protein (% of wild-type activity)
|
|
|---|---|---|---|
| ATPase | Helicase | ||
| Wild type | 2 × 107 | 100 | 100 |
| T187A | 3 × 106 | 68 | 52 |
| T192A | 4 × 104 | <0.2 | <1 |
| Q194A | 2 × 106 | 55 | 46 |
| P228A | 8 × 105 | 91 | 21 |
| R229A | 2 × 104 | 2 | <1 |
| Plasmid | <10 | ||
NPH-II RNA unwinding activity is essential for vaccinia virus replication.
Three ts vaccinia virus mutants isolated by Fathi and Condit (ts10, ts18, and ts39) are unable to produce infectious virus at 40°C because they encode mutated versions of NPH-II (4, 5). The nph2-ts viruses assemble normal-appearing progeny virions at the restrictive temperature. These particles contain a wild-type complement of virion polypeptides (including vaccinia virus RNA polymerase and other enzymatic components of the virus core), but they lack NPH-II (11). We previously correlated the loss of infectivity of the mutant virions with a defect in the production of early mRNAs (11). In the present study, we exploited the nph2-ts viruses to test the biological activity of NPH-II-Ala mutants whose biochemical defects have been determined. This was done by DNA-mediated marker rescue of the nph2-ts viruses with wild-type and mutant NPH-II genes.
ts39 encodes a G469S mutation (4); the lesion is located 22 amino acids upstream of motif VI (QRKGRVGRVNPG). The biochemical consequences of alanine substitutions at eight amino acids in this motif are summarized in Table 3. Marker rescue was performed as follows: NPH-II-Ala DNA was transfected by electroporation into ts39-infected cells, and the cells were then plated onto virgin monolayers at the restrictive temperature. Formation of infectious centers is contingent on reversion of ts39 to the wild-type Gly at codon 469; this occurs via recombination between the transfected NPH-II-Ala plasmids and the newly replicated viral genomes in the transfected cells. The revertants can then spread to the uninfected cells of the monolayer at 40°C. The monolayers were harvested 2 days after transfection and tested for marker rescue by determining the titer of virus capable of plaque formation at 40°C. It is expected that mutants encoding functional proteins will rescue like wild-type NPH-II but that those encoding proteins that are nonfunctional (or ts) in vivo will display rescue frequencies several orders of magnitude lower than wild-type NPH-II. For NPH-II alleles containing alanine substitutions in motif VI, there was a strong correlation between the extents of marker rescue and the specific ATPase and helicase activities of the encoded gene products. N500A and P501A, which are as active as wild-type NPH-II in vitro, rescue ts39 as effectively as the wild-type gene, yielding 5 × 106 to 8 × 106 revertants per transfection. Rescue was absolutely dependent on introduction of a functional NPH-II gene, insofar as transfection of the plasmid vector yielded <10 viruses capable of plaque formation at 40°C (Table 3). This finding attests to the low level of spontaneous reversion by ts39. Mutant alleles Q491A, G494A, R495A, G497A, R498A, and G502A, which encode catalytically impaired ATPases/helicases (9), yielded only 0.6 × 104 to 1 × 104 revertants per transfection (Table 3).
TABLE 3.
Rescue of ts39 by NPH-II alleles containing alanine mutations in motifs VI
| NPH-II gene | Virus titer at 40°C (PFU/ml) | Sp activity of proteina (% of wild-type activity)
|
|
|---|---|---|---|
| ATPase | Helicase | ||
| Wild type | 8 × 106 | 100 | 100 |
| Q491A | 1 × 104 | 9 | 2 |
| G494A | 1 × 104 | 10 | 6 |
| R495A | 1 × 104 | 3 | 7 |
| G497A | 1 × 104 | 18 | 10 |
| R498A | 1 × 104 | 3 | 7 |
| N500A | 8 × 106 | 100 | 100 |
| P501A | 5 × 106 | 100 | 100 |
| G502A | 6 × 103 | 18 | 6 |
| Plasmid | <10 | ||
From reference 9.
A defective allele can give rise to rescued virus by recombination between the engineered mutation and the ts lesion. Given the close physical linkage between the ts39 lesion and the Ala substitutions in motif VI, we expect such events to be relatively rare compared to events in which the Ala mutations are introduced and the ts lesions are reverted. Because the NPH-II Ala genes were engineered to gain or lose a diagnostic restriction site at the mutated codon, we were able to determine if the Ala mutation was incorporated into the revertant viruses. To do this, we plaque purified rescued viruses and then screened for the presence of the diagnostic restriction site in a 1,718-bp NPH-II gene fragment prepared by PCR amplification of DNA from a monolayer infected with the plaque-purified virus. If the NTPase or helicase activity of NPH-II is essential for replication, then we will not be able to recover any rescued viruses that have taken up the ATPase-inactivating alanine mutations; instead, all rescued viruses derived from such transfections will be wild type with respect to the restriction site polymorphism that overlaps the alanine mutation. On the other hand, if the engineered mutant supports replication, we will detect the restriction site in the majority of the rescued virus isolates.
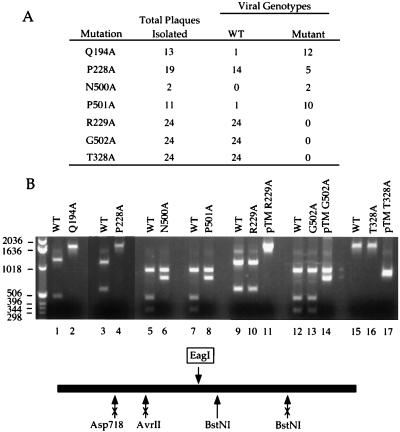
The DNA of plaque-purified revertants from the N500A and P501A transfections displayed a diagnostic loss of a BstNI site (Fig. 6B), indicating that the N500A and P501A mutations were present in viruses that grew at 40°C. Ten of eleven P501A viruses and two of two N500A viruses that were genotyped in this way had taken up the Ala mutations (Fig. 6A). We surmise that Pro-501 and Asn-500 are not important for vaccinia virus replication. In contrast, none of 24 plaque-purified revertants from cells transfected with the catalytically defective G502A allele had incorporated the restriction polymorphism linked to the G502A mutation. All 24 virus isolates retained the wild-type genotype (Fig. 6). Thus, the low frequency of marker rescue by G502A compared to N500A and P501A reflected the requirement for a crossover between the Ala-502 and Gly-496 codons.
FIG. 6.
Genotyping of rescued viruses. (A) Summary of viral genotypes determined by restriction analysis of PCR-amplified NPH-II genes from plaque-purified virus revertants. WT, wild type. (B) Genotyping by restriction digestion. Representative restriction endonuclease digests were analyzed by agarose gel electrophoresis. The NPH-II gene was PCR amplified from DNA isolated from cells infected with wild-type vaccinia virus or plaque-purified revertants recovered after transfection with the indicated NPH-II-Ala genes. The 1,718-bp PCR products were digested with restriction endonuclease Asp718 (lanes 1 and 2), AvrII (lanes 3, 4, 9, 10, and 11), BstNI (lanes 5, 6, 12, 13, and 14), or EagI (lanes 15 to 17). For those transfections where the mutant allele was not incorporated into any rescued viruses (R229A, G502A, and T328A), a control restriction digest was performed with the 1,718-bp PCR product amplified from the pTM-based plasmid containing the indicated alanine substitution (lanes 11, 14, and 17). DNA was visualized by staining the agarose gel with ethidium bromide. The positions and sizes (in base pairs) of linear DNA markers are indicated on the left. The 1.7-kbp PCR fragment is depicted as a horizontal bar. The locations of the restriction sites within the wild-type NPH-II gene are denoted below the bar. The Q194A mutation eliminates the Asp718 site. The P228A and R229A mutations destroy the AvrII site. The N500A, P501A, and G502A mutations eliminate the downstream BstNI site. The T328A mutation creates a unique EagI site; digestion at this site (indicated by the arrow above the bar) generates a doublet of 877- and 841-bp fragments.
To analyze the in vivo effects of mutations in motifs II and III, we performed marker rescue experiments with vaccinia strain ts10. The ts10 lesion (A310E) is situated 10 amino acids downstream of motif II (DEVHE) and 15 amino acids upstream of motif III (TAT). The seven mutant alleles (D296A, E297A, H299A, E300A, T326A, and T328A) encoding defective helicases rescued ts10 with 300- to 1,000-fold-lower efficiency than wild-type NPH-II (Table 1). The results are instructive insofar as all three mutations (H299A, T326A, and T328A) that abrogated the helicase function but spared ATPase activity were all lethal in vivo. Genotyping of 24 plaque-purified T328A revertants showed that every isolate was wild type with respect to the EagI restriction site at the T328 locus (Fig. 6). This finding argues that although the NTPase activity of NPH-II is essential for vaccinia virus replication, it is not sufficient if uncoupled from the capacity for duplex unwinding.
Similar rescue analyses were performed with ts18, which encodes a G208R mutation (4) located 16 amino acids downstream of motif I and 20 amino acids upstream of motif Ia. Mutants T192A in motif I and R229A in motif Ia displayed the lowest extent of ts18 rescue compared to the wild-type NPH-II gene (Table 2); each of these alleles encodes a catalytically defective ATPase/helicase. Genotyping of 24 plaque-purified R229A revertants showed that every isolate was wild type with respect to the R229 locus (Fig. 6). We surmise that the R229A mutation is lethal in vivo. Alleles T187A, Q194A, and P228A, which encode active NTPases/helicases, displayed the highest extents of ts18 rescue (Table 2). Genotyping of the Q194A revertant showed that 12 of 13 isolates had taken up the Q194A mutation (Fig. 6). In the case of P228A, 5 of 14 isolates had the mutant genotype. Thus, Gln-194 and Pro-228 are not critical for virus replication.
DISCUSSION
In this study, we identified four new amino acid side chains that are essential for the helicase activity of vaccinia virus NPH-II. This effectively completes our assessment by alanine scanning of the functions of conserved helicase motifs I, Ia, II, III, and VI. Of the 22 residues targeted here and in prior studies, 15 are essential for helicase activity. These are Lys-191 and Thr-292 in motif I, Arg-229 in motif Ia, Asp-296, Glu-297, His-299, and Glu-300 in motif II, Thr-326A and Thr-328A in motif III, and Gln-491, Arg-492, Gly-494, Arg-495, Gly-497, Arg-498, and Gly-502 in motif VI. Individual residues in motifs I, Ia, II, and VI participate in NTP hydrolysis, whereas motif III and the His moiety of the DExH box function to couple the energy to NTP hydrolysis to dsRNA unwinding. None of the mutations that inactivate the NTPase or helicase functions have a significant effect on nucleic acid binding.
Functions of the helicase motifs in NTP hydrolysis and duplex unwinding.
Comparison of our latest mutagenesis results for NPH-II with those reported for other DEAD- or DExH-box proteins underscores the theme that structure-function relationships at conserved residues in the helicase motifs are context dependent. This is especially so with respect to the role of motif III in coupling the phosphohydrolase and unwinding steps. For example, changing the SAT motif of translation initiation factor 4A to AAA preserves ATP hydrolysis but abrogates RNA helicase activity (23, 24). This result agrees with our findings for NPH-II. In contrast, a TAT-to-AAT change in motif III of HCV RNA helicase, which had no effect on RNA binding, coordinately reduced the phosphohydrolase and RNA unwinding activities (to 21 and 53% of the wild-type values, respectively); i.e., the NTPase and helicase activities remained coupled (13). The structure of the HCV RNA helicase NS3 has been solved by X-ray crystallography (33). Motif III is located on a β-strand that connects a protein domain containing the GKT and DExH elements to a second domain containing motif VI. Although Yao et al. (33) propose that the TAT element transmits an NTP-dependent conformational switch, the mutational findings for HCV RNA helicase do not yet reveal such a role. It is conceivable that the second threonine of the TAT motif is involved in energy coupling; the results of mutating that side chain have not, to our knowledge, been reported.
The histidine moiety of the DExH box of NPH-II functions in coupling NTP hydrolysis to nucleic acid binding. Changing the histidine to alanine elicits a gain of function, whereby the enzyme hydrolyzes ATP in the absence of a nucleic acid cofactor (8). We proposed that the histidine side chain exerts a negative effect on ATPase activity of the enzyme in the ground state and that this is relieved upon nucleic acid binding. The DExA mutation of NPH-II also uncouples the phosphohydrolase and helicase functions. In the HCV helicase structure (33), the histidine side chain participates in a network of hydrogen bond interactions that involves the threonine (boldface) of the TAT motif (which is critical for NTPase/helicase coupling in NPH-II) and the glutamate of the DExH box (which is essential for ATP hydrolysis in NPH-II). A recent study suggests that the DExH-box histidine couples the ATPase and helicase activities of HCV NS3. Heilek and Peterson (12) found that the His→Ala change abolished helicase activity but had no deleterious effect on nucleic acid-dependent ATPase. In fact, the specific activity of the DExA mutant was slightly higher than that of wild-type NS3 (12). However, Kim et al. (13) reported that the same His→Ala mutation of NS3 was active as an RNA helicase (with 60% of the wild-type specific activity). The reasons for the discordant findings of these two groups regarding the helicase activity of the DExA protein are unclear. Kim et al. (13) noted that the DExA mutant of NS3 was constitutively activated for ATP hydrolysis in the absence of nucleic acid. This feature is reminiscent of our findings for NPH-II.
Motifs I and II, which are essential for ATP hydrolysis, are in close proximity in the NS3 crystal structure; the Lys side chain in the GKT element makes a salt bridge to the Asp of the DExH box (33). The NS3 structure was solved in the absence of nucleotide. Structures are available for other helicases (Rep and PcrA) and NTPases (RecA and p21) with nucleotide bound (17, 20, 30, 31). In p21 bound to GMPPNP, the ɛ-amino group of lysine in the GKS element contacts the β and γ phosphates. The adjacent serine hydroxyl (equivalent to the essential Thr-192 of NPH-II) interacts with magnesium coordinated to the β and γ phosphates (20). Similar contacts are proposed for the GKT motif in the RecA-ADP cocrystal (30). The Lys and Thr side chains of the Rep and PcrA helicases complexed with ADP are also poised near the β phosphate (17, 31). The aspartic acid residue in the B motif of the p21 structure (equivalent to the Asp in the DExH box) interacts via a water molecule with the magnesium ion (20). The aspartate and glutamate residues of the PcrA DExx box are located near the bound ADP (17, 31). The Glu side chain of the DExx box is in the same position in space as Glu-96 of RecA (17, 31), which has been proposed to serve as general base to activate water during attack on the γ phosphate (30). The aspartate and glutamate side chains are both essential for ATP hydrolysis by NPH-II (8). A second glutamate (boldface) immediately adjacent to motif II (DEVHE) is also critical for ATP hydrolysis by NPH-II. This residue is conserved in a subset of DExH proteins that includes the helicase A and splicing factors Prp2, Prp16, and Prp22, but it is not conserved in HCV helicase (Fig. 1).
A glutamine side chain (Gln-194) in RecA is proposed to interact with the γ phosphate of ATP and thereby elicit a conformational change in the protein that enhances DNA binding affinity (30). A glutamine residue is conserved at the corresponding position of the PcrA structure (31). The analogous residue the DExH family appears to be the glutamine in motif VI (QRxGRxGRxxxG). Mutation of this glutamine to alanine in NPH-II reduced helicase specific activity to 2% of wild-type activity, and ATPase specific activity to 9% of wild-type activity, but had no effect on RNA binding (9). Note that NPH-II differs from RecA in that NPH-II binds to RNA with high affinity in the absence of NTPs. A Gln→His mutation in motif VI of NS3 abolished both ATPase and helicase functions, also without affecting RNA binding (13). There is complete agreement between our findings (9) and those of Kim et al. (13) that mutations of the conserved amino acids in motif VI have no significant effect on RNA binding affinity. In the NS3 crystal structure, the three conserved arginine side chains of motif VI are solvent exposed (33). We found that alanine substitution at each of the three conserved Arg residues of motif VI reduced phosphohydrolase activity by an order of magnitude (9). Kim et al. report that mutations of the second and third Arg residues abolished ATPase activity, but alanine replacement at the first Arg had no effect on NS3 function (13). This again points up the context dependence of mutational effects at conserved residues.
The ATPase and helicase activities of NPH-II are essential for vaccinia virus replication.
Rescue of ts vaccinia viruses by mutant alleles encoding biochemically characterized proteins provides a general method to assess whether the catalytic activities of a given gene product are critical for virus growth. Ellison et al. (3) have used this approach to demonstrate that the uracil-DNA glycosylase activity of the vaccinia virus D4 protein is essential for virus replication, i.e., by showing that mutations at the active site are incompatible with virus viability. The method is particularly powerful when the viral gene product has more than one biochemical activity. This is the case for NPH-II, which catalyzes both NTP hydrolysis and duplex unwinding. The two activities are interdependent, insofar as unwinding requires NTP hydrolysis, but not obligately coupled, as shown by our analyses of mutants that hydrolyze ATP but do not unwind dsRNA. Plasmid-mediated rescue experiments described above show that the phosphohydrolase and RNA unwinding activities of the NPH-II protein are both essential for vaccinia virus replication. NPH-II-Ala alleles encoding catalytically competent enzymes (e.g., N500A and P501A) rescued an nph2-ts virus at high frequency, and progeny revertants containing the Ala mutations were readily isolated. In contrast, NPH-II alleles encoding ATPase-defective proteins rescued at low frequency, and none of the isolated revertants contained the Ala mutation. Mutations that blocked RNA unwinding without significantly reducing ATPase specific activity were also lethal. The implication is that duplex unwinding by NPH-II is critical for vaccinia virus replication. The P228A allele, the product of which is one-fifth as active as wild type NPH-II in RNA unwinding, is capable of rescuing nph2-ts, whereas mutant alleles encoding proteins whose helicase activity is ≤10% of wild-type activity are lethal. This finding suggests that vaccinia replication is contingent on a threshold level of duplex unwinding by NPH-II.
It is worth emphasizing that essentiality of the catalytic activities of DExH proteins cannot be taken for granted. For example, the yeast DExH-box helicase Rad3 is essential for cell growth (i.e., it cannot be deleted), but the ATPase activity of Rad3 is not critical for cell growth, insofar as a rad3 mutant with no ATPase activity is viable (32). The ATPase and helicase activities of the Escherichia coli DExH-box helicase PriA are not necessary for PriA to assemble the DNA replication primosome in vitro or for PriA to function in homologous recombination and double-strand break repair in vivo (15, 25, 34). Thus, it was not a foregone conclusion that NPH-II helicase activity would be essential for vaccinia virus replication.
We showed previously that NPH-II plays an important role during the transcription of early mRNAs by the vaccinia virus particle (11). In contrast, NPH-II is not required for transcription of early genes in a reconstituted in vitro transcription system. We suggested that NPH-II facilitates transcription in the virion by preventing R-loop formation behind the elongating RNA polymerase (11). R-loop formation is favored by the negative superhelicity generated during transcription of divergently oriented genes (2), as is the case with early transcription by vaccinia virions. The encapsidated vaccinia virus DNA topoisomerase (29) might be expected to relieve some of this superhelical tension and thereby limit R-loop formation. However, the findings of Fernandez-Beros and Tse-Dinh (6) that vaccinia virus topoisomerase preferentially removes positive supercoils, and that its action on actively transcribed circular plasmids results in the accumulation of negative supercoils, raises the prospect that the topoisomerase might actually promote RNA loop formation during mRNA synthesis by virions. We posit that the RNA-DNA helicase activity of NPH-II (10) serves to disrupt these R loops. This hypothesis is consistent with the present findings that the duplex unwinding activity of NPH-II is essential in vivo. Our findings do not exclude other functions for the NPH-II helicase activity during vaccinia virus gene expression.
REFERENCES
- 1.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 2.Drolet M, Phoenix P, Menzel R, Masse E, Liu L F, Crouch R J. Overexpression of RNase H partially complements the growth defect of an Escherichia coli topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellison K S, Peng W, McFadden G. Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J Virol. 1996;70:7965–7973. doi: 10.1128/jvi.70.11.7965-7973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fathi Z, Condit R C. Genetic and molecular biological characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991;181:258–272. doi: 10.1016/0042-6822(91)90491-s. [DOI] [PubMed] [Google Scholar]
- 5.Fathi Z, Condit R C. Phenotypic characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991;181:273–276. doi: 10.1016/0042-6822(91)90492-t. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Beros M, Tse-Dinh Y C. Vaccinia virus DNA topoisomerase I preferentially removes positive supercoils from DNA. FEBS Lett. 1996;384:265–268. doi: 10.1016/0014-5793(96)00317-1. [DOI] [PubMed] [Google Scholar]
- 7.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 8.Gross C H, Shuman S. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J Virol. 1995;69:4727–4736. doi: 10.1128/jvi.69.8.4727-4736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross C H, Shuman S. The QRxGRxGRxxxG motif of the vaccinia virus DExH box RNA helicase NPH-II is required for ATP hydrolysis and RNA unwinding but not for RNA binding. J Virol. 1996;70:1706–1713. doi: 10.1128/jvi.70.3.1706-1713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross C H, Shuman S. Vaccinia virus RNA helicase: nucleic acid specificity in duplex unwinding. J Virol. 1996;70:2615–2619. doi: 10.1128/jvi.70.4.2615-2619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross C H, Shuman S. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J Virol. 1996;70:8549–8557. doi: 10.1128/jvi.70.12.8549-8557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilek G M, Peterson M G. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D W, Kim J, Gwack Y, Han J H, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S-H, Smith J, Claude A, Lin R-J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogoma T, Cadwell G W, Barnard K G, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin E V, Senkevich T G. Vaccinia virus encodes four putative DNA and/or RNA helicases distantly related to each other. J Gen Virol. 1992;73:989–993. doi: 10.1099/0022-1317-73-4-989. [DOI] [PubMed] [Google Scholar]
- 17.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by crystal structure of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 18.Lee C G, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 19.Lee C G, Chang K A, Kuroda M I, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;15:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paoletti E, Moss B. Two nucleic acid-dependent nucleoside triphosphate phosphohydrolases from vaccinia virus. Nucleoside substrate and polynucleotide cofactor specificities. J Biol Chem. 1974;249:3281–3286. [PubMed] [Google Scholar]
- 22.Paoletti E, Rosemond-Hornbeak H, Moss B. Two nucleic acid-dependent nucleoside triphosphate phosphohydrolases from vaccinia virus: purification and characterization. J Biol Chem. 1974;249:3273–3280. [PubMed] [Google Scholar]
- 23.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandler S J, Samra H S, Clark A J. Differential suppression of priA::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 27.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuman S. Vaccinia virus RNA helicase: directionality and substrate specificity. J Biol Chem. 1993;268:11798–11802. [PubMed] [Google Scholar]
- 29.Shuman S, Moss B. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc Natl Acad Sci USA. 1987;84:7478–7482. doi: 10.1073/pnas.84.21.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Story R M, Steitz T A. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 31.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 32.Sung P, Higgins D, Prakash L, Pakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7:3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 34.Zavitz K H, Marians K J. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]