Abstract
Background and Aim:
Among Streptococcus suis serotypes, S. suis serotype 2 is the most significant serotype that causes serious diseases in pigs and humans worldwide. The present study aimed to estimate the global prevalence of S. suis serotype 2 isolated from pigs, determine its trend, and explore the factors associated with this serotype.
Materials and Methods:
We retrieved relevant published studies from PubMed, Scopus, and the Web of Science. The retrieved citations were screened for possible inclusion. Relevant data were then extracted from the included studies. The random-effects model was used for all meta-analyses. A subgroup meta-analysis was used to assess the heterogeneity of the prevalence for four characteristics (continents, sampling organs, reporting unit, and pig’s health status). A cumulative meta-analysis was performed to determine the cumulative prevalence over time. Meta-regression analysis was used to determine the trend of pooled prevalence of S. suis serotype 2 over time.
Results:
Of 600 articles retrieved, 36 studies comprising a total sample size of 6939 isolates or samples from 16 countries of four continents were included for meta-analysis. The pooled prevalence of S. suis serotype 2 isolated from pigs was 13.6% (95% confidence interval [CI], 10.7%–17.1%), with high heterogeneity among the included studies (Cochran’s Q, 431.6; p < 0.001; I2 = 91.9%; Table-1). No statistical significance was observed among subgroups of the four characteristics examined. However, the pooled prevalence of S. suis serotype 2 was as high as 16.0% (95% CI, 12.5%–20.3%; n = 16) in diseased pigs compared with 9.9% (95% CI, 5.6%–17.0%; n = 15) in healthy pigs. The pooled prevalence of S. suis serotype 2 isolated from pigs did not significantly decrease over time [regression coefficient = −0.020 (95% CI, 0.046–0.006, p = 0.139)]. The pooled prevalence of S. suis serotype 2 isolated from pigs fluctuated slightly between 13.2% and 17.8% from 2007 to 2023, although the pooled prevalence gradually decreased from 30.6% in 1987 to over 20% in 2003.
Conclusion:
The global prevalence of S. suis serotype 2 isolated from pigs was estimated to be 13.6% (approximately 10% in healthy pigs and around 16% in diseased pigs). S. suis serotype 2 isolated from pigs did not change significantly over time. These results indicate that S. suis serotype 2 remains a problem for the pig industry and poses a threat to human health.
Keywords: meta-analysis, pigs, prevalence, serotype 2, Streptococcus suis
Introduction
Streptococcus suis, which originated from pigs, is an emerging zoonotic pathogen that causes severe illnesses such as meningitis, deafness, septicemia, and even death in humans [1–4]. Although S. suis is considered as a commensal bacterium residing mainly in the palatine tonsil and nasal cavity of pigs [5], some strains of S. suis can also cause severe diseases, especially in susceptible weaning pigs, resulting in a huge economic loss in the pig industry worldwide [6, 7]. In humans, the burden of S. suis infection is highly pronounced in Southeast Asian countries such as Thailand, Vietnam, and Indonesia [8, 9]. Almost all human cases in this region are associated with raw or undercooked pork or pork products [1, 3, 4]. Several sociocultural factors, such as traditional culture, shared beliefs, socioeconomic level, and personal attitudes, play a role in human S. suis infection in Southeast Asian countries [10]. In North America and Europe, people typically get infected through close contact with pigs or pork products; therefore, pig farmers and butchers are the occupations at risk [11].
S. suis is currently classified into 29 serotypes (1–19, 21, 23–25, 27–31, and 1/2) on the basis of capsular polysaccharide antigens [11–14]. Of these 29 serotypes, S. suis serotype 2 has the greatest significance because it can cause severe diseases in both humans and pigs worldwide [1, 11, 13]. S. suis serotype 2 is responsible for more than 70% and 25% of clinical cases in humans and pigs, respectively [2, 11, 13]. The prevalence of S. suis serotype 2 in healthy and diseased pigs from several regions of the world, including Asia, Europe, and North America [15–25] has been reported. Individual studies are limited in sample size, study location, and study period. Combining individual data (using meta-analytic techniques) from relevant studies would help us to comprehend the global picture of S. suis serotype 2 isolated from pigs.
Therefore, the study aimed to use a meta-analysis to estimate the global prevalence and trend of S. suis serotype 2 isolated from pigs and to explore some characteristics associated with the heterogeneity of the prevalence.
Materials and Methods
Ethical approval
This study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (http://www.prisma-statement.org/). Ethical approval was not required as no animals or animal-derived products were used in this study.
Study period and location
The literature search, data collection, and data analysis were conducted at the Faculty of Veterinary Medicine, Khon Kaen University, from June 2022 to July 2023. The included studies were published between 1987 and 2023. The included studies were conducted in 16 countries from Asia, Australia, Europe, and North America.
Search strategy
Two authors independently performed data search from PubMed, Scopus, and Web of Science to identify the relevant citations reporting the prevalence of S. suis serotype 2 isolated from pigs. The search was originally conducted in June 2022 and was updated in May 2023. The PECO structure [26] was used to formulate the research question (P, population = pigs or swine; E, exposure = positive case of S. suis infection; C, comparator = negative cases of S. suis infection; and O, outcome = the prevalence of S. suis isolated from pigs). The most important search keywords were “swine, Streptococcus suis, prevalence.” For each database, the search was not limited to date but limited to English. The medical subject heading (MeSH) was checked for appropriate and complete keywords. An example search algorithm from the databases was follows: (“swine”[MeSH Terms] OR “swine”[All Fields] OR “swines”[All Fields] OR (“swine”[MeSH Terms] OR “swine”[All Fields] OR “pigs”[All Fields]) OR (“swine”[MeSH Terms] OR “swine”[All Fields] OR “suidae”[All Fields]) OR (“swine”[MeSH Terms] OR “swine”[All Fields] OR (“wart”[All Fields] AND “hogs”[All Fields]) OR “wart hogs”[All Fields]) OR (“swine”[MeSH Terms] OR “swine”[All Fields] OR “warthogs”[All Fields] OR “warthog”[All Fields])) AND “Streptococcus suis”[All Fields] AND (“epidemiology”[MeSH Subheading] OR “epidemiology”[All Fields] OR “prevalence”[All Fields] OR “prevalence”[MeSH Terms] OR “prevalance”[All Fields] OR “prevalences”[All Fields] OR “prevalences”[All Fields] OR “prevalent”[All Fields] OR “prevalently”[All Fields] OR “prevalents”[All Fields]).
Inclusion and exclusion criteria
Two independent reviewers carefully screened the titles and abstracts for initial inclusion after the duplicated citations were removed. Full-text articles that passed the first screening step were carefully examined under specific inclusion and exclusion criteria for final inclusion. This study was included if it reported the prevalence of S. suis serotype 2 isolated from pigs. The studies were excluded if (1) they were not related to the prevalence of S. suis serotype 2 isolated from pigs, (2) they were reviewed articles, (3) they were case reports, (4) they were experimental studies, and (5) they contained unclear information concerning the prevalence of S. suis serotype 2 isolated from pigs. Because S. suis serotype 2 is genetically very similar to S. suis serotype 1/2, conventional or multiplex polymerase chain reaction (PCR) cannot distinguish between these two serotypes. Therefore, agglutination and coagglutination tests using serotype-specific antisera or additional PCR techniques such as PCR-restriction fragment length polymorphism and whole-genome sequencing data are required for confirmation of S. suis serotype 2 [27–29]. Therefore, studies reporting S. suis serotype 2 using PCR techniques without additional techniques for serotype confirmation, as mentioned above, were excluded from the study. To evaluate the effect of this exclusion, a sensitivity analysis was performed as described in the statistical analysis section. Disagreements regarding inclusion or exclusion were resolved by discussion.
Study quality assessment
A checklist for quality assessment adapted from the previous study that had five questions [30]. These were as follows: (1) Was the research objective clearly described and stated? (2) Were the period and location of the study clearly stated? (3) Was the sample categorized into different subgroups? (4) Was the sampling method described in detail? (5) Were the diagnostic techniques clearly pointed out? Scoring of each question was based on a simple scale system (“2” for yes, “0” for no, or “1” for unsure); therefore, a total score for each included study was 10.
Data extraction
Two authors extracted and verified the data from the included studies. Disagreement regarding data extraction was resolved by discussion. Details of the extracted data were as follows: (1) study identification (name of the first author and publication year), (2) study year, (3) study location, (4) pig’s health status, (5) sampling organs, (6) number of total S. suis isolates or samples (sample size), (7) number of S. suis serotype 2 positive isolates or samples, and (8) detection methods.
Statistical analysis
Comprehensive meta-analysis program version 3 (Biostat, Englewood, NJ, USA) was used for all meta-analyses, including overall meta-analysis, subgroup meta-analysis, meta-regression analysis, cumulative meta-analysis, and assessment of publication bias. p < 0.05 was considered statistically significant for all analyses unless otherwise stated. To stabilize the variance of the raw prevalence before pooling, logit transformation was used as follows: logit(p) = ln(p/[1-p]), where “p” is the proportion and “ln” is the natural log [30].
Overall meta-analysis
A random-effects model was used to estimate the overall pooled prevalence and its 95% confidence interval (CI). Cochran’s Q and I2 statistics were used to assess the heterogeneity. A Cochran’s Q test p < 0.05 implied significant heterogeneity. I2 values of 25%, 50%, and 75% were considered low, moderate, and high heterogeneity, respectively [31]. Individual studies were used as a unit of analysis for the overall pooled prevalence.
Subgroup meta-analysis
A subgroup meta-analysis was used to assess the heterogeneity of the pooled prevalence of S. suis serotype 2 isolated from pre-defined pig characteristics. Subgroup meta-analysis was performed for the following four characteristics: (1) location of the study (among continents), (2) sampling organs (tonsils versus other organs), (3) unit of the report (isolates versus samples), and (4) pig’s health status (healthy versus diseased). For all subgroup meta-analyses, the subgroup within the study was used as a unit of analysis.
Meta-regression and cumulative meta-analysis
Meta-regression analysis was used to determine the pooled prevalence trend of S. suis serotype 2 isolated from pigs. Meta-regression of the pooled prevalence was performed against the study year. A median of multiple years was used for analysis in a study reporting a multiple-year study. The expected study year was used in a study that did not report the study year. We calculated the expected study year by subtracting 2 years from the publication year (median difference between the publication year and study year of the included studies). A cumulative meta-analysis was also used to determine the cumulative evidence of the pooled prevalence of S. suis serotype 2 isolated from pigs over time (the publication year).
Sensitivity analysis
Sensitivity analysis was conducted to assess the robustness of the pooled prevalence estimation of S. suis serotype 2 infections in pigs. First, we compared the results of the fixed-effects model with those of the random-effects model. Second, we compared the results of using a study as a unit of analysis with those of using a subgroup as a unit of analysis. Third, we compared the results of the included studies with those of the excluded studies reporting S. suis serotype 2 but did not describe additional serotype confirmation techniques. Finally, we conducted a leave-one-out meta-analysis to evaluate whether each individually included study influenced the results.
Assessment of publication bias
Publication bias was assessed using Begg’s test and Egger’s test with p < 0.1, indicating the presence of publication bias [32, 33]. Publication bias was also visually assessed using a funnel plot. Duval and Tweedie’s trim-and-fill method [34] was used to estimate the adjusted prevalence of S. suis serotype 2 isolated from pigs in the case of asymmetrical funnel plots.
Results
Identification and selection of studies
A total of 600 articles were identified from three electronic databases (PubMed, 410 articles; Scopus, 133 articles; and Web of Science, 57 articles). Of these, 145 were duplicated. After screening the titles and abstracts, there were 67 full-text articles remaining, of which 388 studies were excluded from the study. This exclusion also included 10 studies reporting S. suis serotype 2 but did not describe additional serotype confirmation techniques [35–44]. Ultimately, 36 studies that met the inclusion criteria were included in the meta-analysis [15–25, 45–69]. The study selection process is shown in the flow chart (Figure-1).
Figure-1.
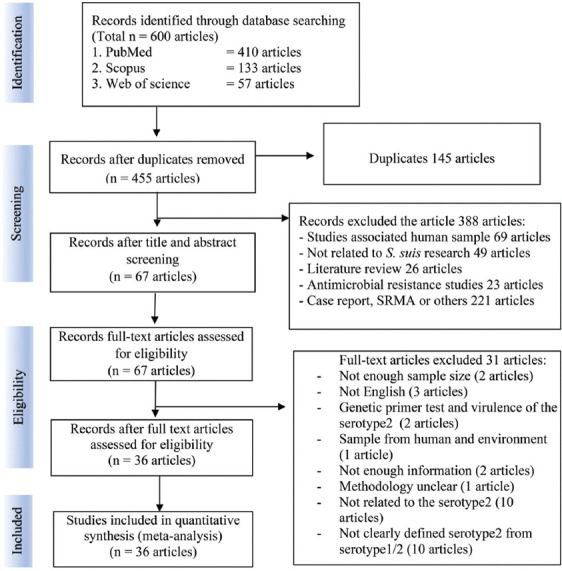
PRISMA flow diagram for the study selection.
Characteristics of the included studies
Table-1 [15–25, 45–69] summarizes the characteristics of the 36 studies included in the study. Of the 36 included studies, 6939 isolates or samples were examined, resulting in 962 isolates or samples positive for S. suis serotype 2. The median sample size was 118 isolates (range, 15–680 isolates). Data were reported from 16 countries from four continents (12 studies from Asia, one study from Australia, 14 studies from Europe, and nine studies from North America) (five studies each from Canada, Spain, and Thailand, four studies from China and the United States, and 13 studies from other countries). Of the 6939 isolates or samples examined, 2032 were from Asia, 2617 from Europe, and 1749 from North America. Of the 6939 isolates or samples examined, 2388 were from healthy pigs, 2973 were from diseased pigs, and 1578 were from pigs with mixed or unspecified health status.
Table-1.
Characteristics of the included studies.
| Reference | Pig’s health status | Country | Study year | Events | Sample size | Prevalence (%) |
|---|---|---|---|---|---|---|
| Aradanas et al. [15] | Healthy | Canada | 2013–2018 | 8 | 90 | 8.9 |
| Diseased | Canada | 2013–2018 | 15 | 183 | 8.2 | |
| Zouharová et al. [16] | Diseased | Czech Republic | 2018–2022 | 49 | 528 | 9.3 |
| Matiašovic et al. [17] | Diseased | Czech Republic | 2020–2021 | 5 | 39 | 12.8 |
| Nicholson and Bayles [18] | Unspecified | United States | 2015–2017 | 28 | 106 | 26.4 |
| Scherrer et al. [19] | Diseased | Switzerland | 2006–2019 | 4 | 88 | 4.5 |
| Kerdsin et al. [20] | Unspecified | Thailand | 2010–2011 | 39 | 204 | 19.1 |
| Bojarska et al. [21] | Diseased | Poland, Belarus | 2003–2012 | 21 | 96 | 21.9 |
| Niazy et al. [22] | Diseased | Canada | NA | 8 | 64 | 12.5 |
| Lacouture et al. [23] | Diseased | Canada | 2015–2020 | 85 | 680 | 12.5 |
| Cucco et al. [24] | Diseased | Italy | 2017–2019 | 7 | 78 | 9.0 |
| Petrocchi-Rilo et al. [25] | Diseased | Spain | 2019–2020 | 45 | 207 | 21.7 |
| Amass et al. [45] | Healthy | United States | 1997 | 0 | 21 | 0.0 |
| Baele et al. [46] | Unspecified | Belgium | NA | 0 | 60 | 0.0 |
| Boetner et al. [47] | Unspecified | Denmark | 1984–1985 | 33 | 108 | 30.6 |
| Brisebois et al. [48] | Healthy | Canada | NA | 15 | 164 | 9.1 |
| Han et al. [49] | Healthy | Korea | 1999 | 2 | 55 | 3.6 |
| Maneerat et al. [50] | Diseased | Thailand | 2007 | 2 | 24 | 8.3 |
| Healthy | Thailand | 2007 | 7 | 194 | 3.6 | |
| Marois et al. [51] | Healthy | France | NA | 12 | 406 | 3.0 |
| Martinez et al. [52] | Mixed | Canada | NA | 40 | 133 | 30.1 |
| Meekhanon et al. [53] | Healthy | Thailand | 2014–2015 | 3 | 135 | 2.2 |
| Mogollon et al. [54] | Mixed | United States | NA | 26 | 66 | 39.4 |
| Ngo et al. [55] | Healthy | Vietnam | 2006–2007 | 45 | 317 | 14.2 |
| Oh et al. [56] | Diseased | Korea | 2009–2010 | 36 | 240 | 15.0 |
| Paterson et al. [57] | Unspecified | Papua New Guinea | NA | 126 | 541 | 23.3 |
| Sánchez Del Rey et al. [58] | Unspecified | Spain | 2007–2010 | 1 | 320 | 0.3 |
| Sánchez Del Rey et al. [59] | Diseased | Spain | 2010–2011 | 13 | 15 | 86.7 |
| Healthy | Spain | 2010–2011 | 11 | 128 | 8.6 | |
| Tarradas et al. [60] | Healthy | Spain | NA | 41 | 81 | 50.6 |
| Thongkamkoon et al. [61] | Healthy | Thailand | 2010 | 11 | 196 | 5.6 |
| Torremorell et al. [62] | Diseased | United States | NA | 45 | 242 | 18.6 |
| van Leengoed et al. [63] | Diseased | Netherlands | 1983–1985 | 44 | 161 | 27.3 |
| Vela et al. [64] | Diseased | Spain | 1999–2002 | 44 | 302 | 14.6 |
| Wang et al. [65] | Healthy | China | 2008–2011 | 7 | 61 | 11.5 |
| Wang et al. [66] | Diseased | China | 2007–2010 | 9 | 26 | 34.6 |
| Healthy | China | 2007–2010 | 31 | 36 | 86.1 | |
| Wongsawan et al. [67] | Unspecified | Thailand | 2001–2002 | 5 | 40 | 12.5 |
| Zhang et al. [68] | Healthy | China | 2005–2007 | 31 | 421 | 7.4 |
| Zheng et al. [69] | Healthy | China | 2011–2012 | 8 | 83 | 9.6 |
NA=Not available
Study quality assessment
This study’s quality assessment tool was based on the full 10-point rating scale. The mean ± standard deviation of the overall quality scores of all included studies was 9.03 ± 1.38. Regarding the range of scores from 5 to 10, the median score was 9. All study quality assessment results are presented in Table-2.
Table-2.
Study quality assessment showing the number of the included studies in each category of the simple rating scale based on a checklist of five items [30].
| Items | No. of the included studies | ||
|---|---|---|---|
|
| |||
| Yes | No | Unsure | |
| Was the research objective clearly described and stated? | 33 | 0 | 3 |
| Was the period and location of the study clearly stated? | 25 | 0 | 11 |
| Was the sample categorized into different subgroups? | 27 | 4 | 5 |
| Was the sampling method described in detail? | 30 | 1 | 5 |
| Was the diagnostic technique clearly pointed out? | 35 | 0 | 1 |
Overall meta-analysis
The overall pooled prevalence of S. suis serotype 2 isolated from pigs was 13.6% (95% CI, 10.7%–17.1%), (n = 36 studies). High heterogeneity was observed among the included studies (Cochran’s Q = 431.6; p < 0.001; I2 = 91.9%).
Subgroup meta-analysis
The subgroup analysis results are presented in Table-3. No significant difference was observed among subgroups among the four characteristics analyzed. However, the pooled prevalence of S. suis serotype 2 was as high as 16.0% (95% CI, 12.5%–20.3%) in diseased and 9.9% (95% CI, 5.6%–17.0%) in healthy pigs. The pooled prevalence of S. suis serotype 2 ranged from 12.2% (95% CI, 7.8%–18.6%) in Asia to 16.1% (95% CI, 11.1%–22.8%) in North America. Figure-2 shows the prevalence of S. suis serotype 2 isolated from pigs from the most reported countries.
Table-3.
The overall pooled prevalence of S. suis serotype 2 isolated from pigs and subgroup meta-analysis.
| Categories | No. of studies or subgroups | Prevalence (%) | Heterogeneity | p-value for subgroup difference | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Estimate | 95% CI | Q | p-value | I2 (%) | |||
| Overall | 36 | 13.6 | 10.7–17.1 | 43.6 | <0.001 | 91.9 | |
| Continenta | 0.631 | ||||||
| Asia | 14 | 12.2 | 7.8–18.6 | 125.7 | <0.001 | 89.7 | |
| Europe | 15 | 14.1 | 8.8–21.7 | 208.8 | <0.001 | 93.3 | |
| North America | 10 | 16.1 | 11.1–22.8 | 76.6 | <0.001 | 88.2 | |
| Sampling organsb | 0.701 | ||||||
| Tonsil | 20 | 13.2 | 8.9–19.0 | 277.5 | <0.001 | 93.2 | |
| Others | 19 | 14.4 | 10.9–18.9 | 143.9 | <0.001 | 87.5 | |
| Unit of report | 0.070 | ||||||
| Isolates | 33 | 13.6 | 10.3–17.7 | 371.2 | <0.001 | 91.4 | |
| Samples | 7 | 19.7 | 14.6–26.1 | 31.9 | <0.001 | 81.2 | |
| Pig’s health statusc | 0.114 | ||||||
| Healthy | 15 | 9.9 | 5.6–17.0 | 218.6 | <0.001 | 93.6 | |
| Diseased | 16 | 16.0 | 12.5–20.3 | 89.8 | <0.001 | 83.3 | |
Two subgroups were removed from analysis because low number of sample (Australia n = 1, Mixed [Asia-Europe] n = 1).
One subgroup was removed from analysis due to an unspecified sampling organ.
Nine subgroups were removed from analysis due to mixed or unspecified health status, CI=Confidence interval
Figure-2.
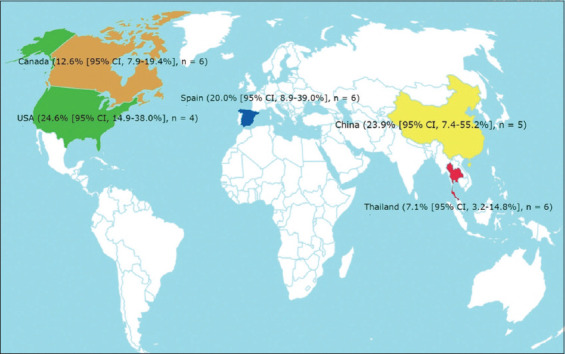
The prevalence estimates of Streptococcus suis serotype 2 isolated from pigs from the most reported countries.
Meta-regression and cumulative meta-analysis
Meta-regression analysis showed that the global prevalence of S. suis serotype 2 isolated from pigs did not significantly decrease over time [regression coefficient = −0.020 (95% CI, −0.046–0.006, p = 0.139)]. The meta-regression equation is expressed as the logit event rate = 37.49−0.020 (year), where year is the study year (Figure-3).
Figure-3.
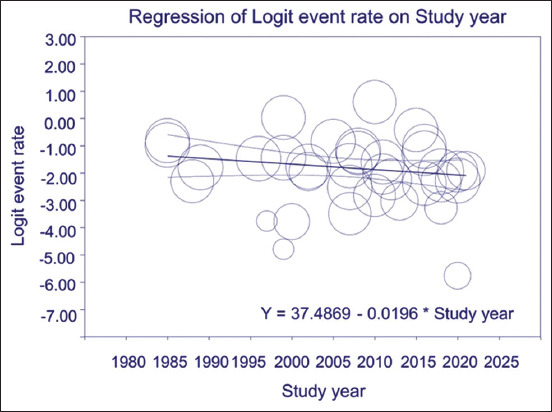
Scatter plot showing results of meta-regression analysis for the correlation between logit event rate (Y-axis) and study year (X-axis). A circle represents each included study (n = 36).
The cumulative prevalence of S. suis serotype 2 isolated from pigs gradually decreased from 30.6% in 1987 to >20% in 2003. From 2007 to 2023, the cumulative prevalence fluctuated between 13.2% and 17.8% (Figure-4).
Figure-4.
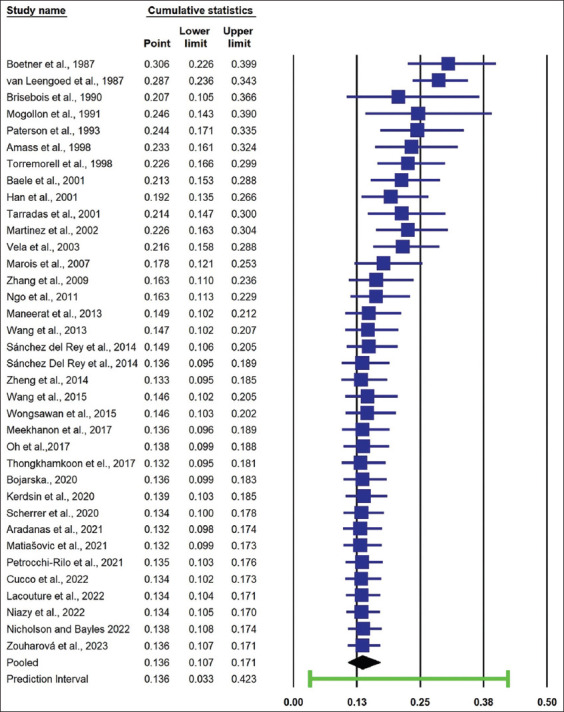
Forest plot of cumulative prevalence and 95% CI of S. suis serotype 2 isolated from pigs over time. From 2007 to 2023, the pooled prevalence of S. suis serotype 2 isolated from pigs fluctuated between 13.2% and 17.8%.
Sensitivity analysis
The sensitivity analysis results are shown in Table-4. The overall pooled prevalence of S. suis serotype 2 isolated from pigs in the fixed-effects model was higher than that in the random-effects model (16.9% [95% CI; 15.9%–17.9%] vs. 13.6% [95% CI, 10.7%–17.1%]). Leave-one-out analysis showed that the pooled prevalence of S. suis serotype 2 isolated from pigs slightly changed from 12.9% [95% CI, 10.3%–16.0%] to 14.3% [95% CI, 11.4%–17.8%].
Table-4.
Sensitivity analysis to assess the robustness of the result estimates.
| Categories | No. of studies or subgroups | Prevalence (%) | |
|---|---|---|---|
|
| |||
| Estimate | 95%CI | ||
| Model | |||
| Fixed effects | 36 | 16.9 | 15.9–17.9 |
| Random effects | 36 | 13.6 | 10.7–17.1 |
| Unit of analysis | |||
| Studies | 36 | 13.6 | 10.7–17.1 |
| Subgroups | 40 | 14.2 | 11.3–17.1 |
| Serotyping confirmationa | |||
| Confirmed | 36 | 13.6 | 10.7–17.1 |
| Unconfirmed | 10 | 14.1 | 7.8–24.1 |
| Leave-one-out analysis | |||
| The lowest prevalenceb | 35 | 12.9 | 10.3–16.0 |
| The highest prevalencec | 35 | 14.3 | 11.4–17.8 |
Specific methods or techniques that used to differentiate S. suis serotype 2 from serotype 1/2 were clearly described (confirmed) or (unconfirmed). The unconfirmed 10 studies were excluded from the data synthesis.
Removed the study of Wang et al., [66],
Removed the study of Marois et al., [51], CI=Confidence interval
Assessment of publication bias
Egger’s test (p = 0.027) and Begg’s test (p = 0.072) indicated publication bias. Funnel plot showed an asymmetrical distribution among the included studies (Figure-5). The trim-and-fill method revealed nine missing (hypothetical) studies. The adjusted point estimate of the overall pooled prevalence of S. suis serotype 2 isolated from pigs somewhat increased from 13.6% (95% CI, 10.7%–17.1%) to 18.3% (95% CI, 14.5%–22.8%) after these nine studies were imputed in the model.
Figure-5.
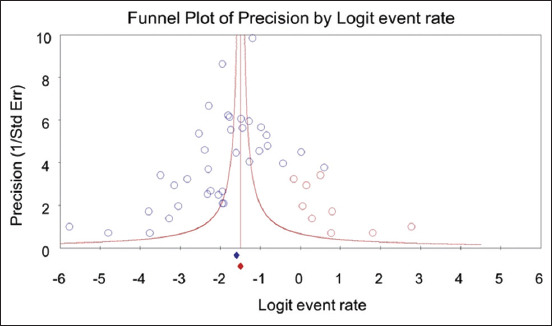
Funnel plot for visual assessment of publication bias. X-axis is the effect size (the logit of the event rate). Y-axis is the precision (standard error). A blue circle represents each included study. A red circle represents an imputed study (or missing study) to make the funnel plot symmetry.
Discussion
Our study synthesized the pooled prevalence of S. suis serotype 2 isolated from pigs from 36 included studies worldwide with a total sample size of 6939 isolates or samples. The overall pooled prevalence of S. suis serotype 2 isolated from pigs in our study was estimated to be 13.6% (95% CI, 10.7%–17.1%) with high heterogeneity. The point estimate of the pooled prevalence in our study is quite high, indicating that pigs remain a potential source of S. suis serotype 2 contamination. The high heterogeneity of the result was within our expectations. Therefore, subgroup meta-analysis and meta-regression were used to assess the heterogeneity or variability of the prevalence among the included studies. Below is our detailed discussion.
Our subgroup meta-analysis did not find a significant difference among the three continents (Asia, Europe, and North America; Australia was excluded from the analysis because it had only one study). The pooled prevalence of S. suis serotype 2 isolated from pigs did not significantly differ among continents, ranging from 12.2% (95% CI, 7.8%–18.6%) in Asia, 14.1% (95% CI, 8.8%–21.7%) in Europe, and to 16.1% (95% CI, 11.1%–22.8%) in North America. For all continents, the 95% CI of the point estimate was very wide. This indicated that there was a high variability in the prevalence data from the included studies within each continent. In Asia, the prevalence of S. suis serotype 2 in pigs was as low as 2.2% in Thailand [53] and as high as 64.5% in China [66]. In Europe, the prevalence of S. suis serotype 2 was as low as 0% [46] in Belgium and as high as 50.6% [60] in Spain. The prevalence of S. suis serotype 2 in the United States was as low as 0% in the study of Amass et al. [45] and as high as 39.4% in the study of Mogollon et al. [54] in North America. This phenomenon has also been observed in other countries, such as Thailand [20, 53], Spain [58, 60], and Canada [15, 52]. The great variability in the prevalence of S. suis serotype 2 isolated from pigs among the included studies, even in the same region, reflected the difference in study settings of the included studies and depended on several factors such as coinfection with porcine reproductive and respiratory syndrome virus at weaning, introduction of pigs into the nursery, sow parity, relative humidity, and temperature [7, 70, 71].
Although our study showed that the pooled prevalence of S. suis serotype 2 was not significantly different between diseased pigs and healthy pigs, the pooled prevalence was as high as 16.0% in diseased pigs compared with 9.9% in healthy pigs (i.e., 1.6 times higher in diseased pigs). The non-significant result may arise from the small sample size within the subgroup resulting in a low statistical power. Our results indicated that S. suis serotype 2 was associated with diseased pigs because the prevalence of S. suis serotype 2 in diseased pigs was 1.6 times higher than that in healthy pigs. S. suis serotype 2 is known as a virulent strain that causes diseases in humans and pigs worldwide [11, 13]. Clinical manifestations in pigs include septicemia, meningitis, endocarditis, pneumonia, and even death [5, 11]. This impacted the health, welfare, and economic loss of pigs [6]. Most S. suis outbreaks in humans result from S. suis serotype 2 [1, 11]. Important risk factors for human infections include raw pork consumption, exposure to pigs or pork, pig-related occupation, and male sex [8]. The high pooled prevalence of S. suis serotype 2 (9.9%, 95% CI, 5.6%–17.0%; n = 15 subgroups) in healthy pigs was surprising. A high prevalence of S. suis serotype 2 in healthy pigs may be a potential source of infection for other pigs and humans [5, 72]. Horizontal transmission primarily occurs through the oro-nasal route or nose-to-nose contact in pigs [5]. In addition, many of the included studies used samples from healthy pigs or pork products from slaughterhouses or wet markets [20, 49, 53, 55, 65, 67]. This production step is close to- and represents a risk for consumers.
For ease of comparison, we divided the sampling organs into two main categories (tonsils vs. other organs). The pooled prevalence of S. suis serotype 2 in pigs was not significantly lower in tonsils than in other organs (13.2% and 14.4%, respectively). The palatine tonsil of a pig is a primary organ for the early colonization of S. suis [5]. S. suis can be found in other organs, such as the lung, brain, spleen, blood, joint fluid, saliva, oral swab, nasal swab, vaginal swab, tongue swab, and pleural effusion [25, 53]. Samples from other organs mainly come from studies of diseased pigs [15, 20, 21, 23, 24, 27, 52, 62]. Because organ collection sampling was confounded by the pig’s health status, further analysis was performed to reveal this confounding effect. Diseased pigs and healthy pigs were separately analyzed on the basis of the sampling organs. The pooled prevalence of S. suis serotype 2 was 28.6% (95% CI, 14.8%–48.0%, n = 4) in the tonsil samples and 13.9% (95% CI, 11.0%–17.4%, n = 12) in the other organ samples in diseased pigs. In healthy pigs, the pooled prevalence of S. suis serotype 2 was 8.2% (95% CI, 4.1%–15.6%, n = 10) in the tonsil samples and 14.4% (95% CI, 4.0%–40.4%, n = 5) in the other organ samples. These results reveal a confounding relationship between the sampling organs and the pig’s health status.
Most of the included studies reported isolates as a unit of report for the prevalence (n = 33 subgroups) compared with samples as a unit of report for the prevalence (n = 7 subgroups). The pooled prevalence of S. suis serotype 2 was not significantly different between isolates and samples (13.6% and 19.7%, respectively). Most of the included studies used isolates as a unit of report because they usually involved identifying all S. suis isolate serotype. Therefore, it is more convenient to report each serotype as a proportion of the total isolate. Some included studies [20, 45, 47] reported the prevalence of S. suis serotype 2 directly from a positive sample out of a total number of positive samples. A non-significant difference in the pooled prevalence of S. suis serotype 2 in the reporting unit indicates that both reporting units are satisfactory.
No significant difference among subgroups in our subgroup analysis does not mean that there is no heterogeneity among subgroups. The number (n) of subgroups in our analysis was small due to the small number of studies included. This may result in a low statistical power and result in a false negative error. If more data are available, we will be able to prove the existence of significant subgroup differences. For example, a significant difference in the pooled prevalence of S. suis serotype 2 between diseased pigs and healthy pigs (16.6% [95% CI, 13.2%–20.8%] vs. 9.6% [95% CI, 5.8%–15.4%], respectively) was found if we added ten more studies (that we excluded) reporting S. suis serotype 2 but without describing additional techniques for serotype confirmation in our analysis.
Our meta-regression analysis showed that the pooled prevalence of S. suis serotype 2 in pigs did not change significantly over time. Our cumulative evidence showed that the pooled prevalence of S. suis serotype 2 in pigs fluctuated slightly from 13.2% to 17.8% between 2007 and 2023. Both results indicate that S. suis serotype 2 remains a source of problems for pigs and poses a threat to human health. In addition, S. suis rapid antimicrobial resistance may exacerbate the problems [73–75].
Sensitivity analysis was used to check the robustness of the decision-making results during the systematic review and meta-analysis. Our sensitivity analysis indicates that the point estimate of the pooled prevalence of S. suis serotype 2 in pigs is robust for all categories except the model of choice used in the analysis. The point estimate of the random-effects model (the model of choice of our study) was 13.6%. It was somewhat lower when compared with the fixed-effects model (16.9%). However, the random-effects model was more appropriate in our study because the study settings, sampling designs, and other conditions of the included studies varied greatly [76]. The results of the leave-one-out analysis indicate that individual studies do not distort the point estimate of the pooled prevalence of S. suis serotype 2 isolated from pigs. When we removed the study of Wang et al. [66] and that of Marois et al. [51], the point estimate was the lowest (12.9%) and the highest (14.1%), respectively, from the analysis. These values were very close to the point estimate from all the included studies (13.5%).
Regarding publication bias, Egger’s test (p = 0.027) and Begg’s test (p = 0.072) indicated publication bias in our study. A funnel plot also showed an asymmetrical distribution of the included studies. In addition, the trim-and-fill method revealed nine missing (hypothetical) studies. After these nine hypothetical studies were imputed in the model, the point estimate of the overall pooled prevalence of S. suis serotype 2 isolated from pigs increased from 13.6% (95% CI, 10.7%–17.1%) to 18.3% (95% CI, 14.5%–22.8%). This indicates that the pooled prevalence of S. suis serotype 2 isolated from pigs would be higher than we estimated without publication bias.
Our study has some limitations. First, our study focused only on the pooled prevalence of S. suis serotype 2 because this serotype is very important in human disease. Therefore, the pooled prevalence of all S. suis serotypes would be much higher because S. suis has 29 specific serotypes [13]. A recent study estimated that the pooled prevalence of S. suis in pigs in China during 2000–2021 was as high as 40% [77]. This means that S. suis poses a potential threat to the health of pigs and humans. Second, the study locations were mainly from Asia, Europe, and North America. There was only one study from Australia and no study from Africa. Therefore, subgroup analysis for Australia and Africa is not feasible. Interpretation of the global prevalence and subgroup analysis of S. suis serotype 2 with regard to the study locations should be carefully performed. Additional information from other regions (if available) may affect the point estimate of the pooled prevalence. Third, we included only studies published in English and retrieved from three databases (PubMed, Scopus, and Web of Science) due to limitations in our research team. Therefore, we may have missed data from other languages, which may have changed the point estimate of the pooled prevalence.
Conclusion
A random-effects meta-analysis was used to estimate the pooled prevalence of S. suis serotype 2 isolated from pigs from 36 included studies containing a total of 6939 isolates or samples from 16 countries of four continents. The overall pooled prevalence of S. suis serotype 2 isolated from pigs was 13.6% (95% CI, 10.7%–17.1%) with high heterogeneity among the included studies. The point estimate may be lower than the real-world situation due to the presence of publication bias. The pooled prevalence of S. suis serotype 2 was as high as 16.0% in diseased pigs and 9.9% in healthy pigs. There was no significant change in the pooled prevalence of S. suis serotype 2 isolated from pigs over time. The results of this study indicate that S. suis serotype 2 remains a threat to pigs and humans worldwide.
Authors’ Contributions
KK, NHN, CS, PaS, PiS, SK, and PeS: Conception. KK and PeS: Extracted, verified, and analyzed the data and drafted and revised the manuscript. All authors have read, reviewed, criticized, and approved the final manuscript.
Acknowledgments
This study was supported by a research grant from the Faculty of Veterinary Medicine, Khon Kaen University (Grant No. VM 011/2564).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
References
- 1.Brizuela J, Kajeekul R, Roodsant T.J, Riwload A, Boueroy P, Pattanapongpaibool A, Thaipadungpanit J, Jenjaroenpun P, Wongsurawat T, Batty E.M, van der Putten B.C.L, Schultsz C, Kerdsin A. Streptococcus suis outbreak caused by an emerging zoonotic strain with acquired multi-drug resistance in Thailand. Microb. Genomics. 2023;9(2):mgen000952. doi: 10.1099/mgen.0.000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji L, Chen Z, Li F, Hu Q, Xu L, Duan X, Wu H, Xu S, Chen Q, Wu S, Qiu S, Lu H, Jiang M, Cai R, Qiu Y, Li Y, Shi X. Epidemiological and genomic analyses of human isolates of Streptococcus suis between 2005 and 2021 in Shenzhen, China. Front. Microbiol. 2023;14:1118056. doi: 10.3389/fmicb.2023.1118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerdsin A. Human Streptococcus suis infections in Thailand:Epidemiology, clinical features, genotypes, and susceptibility. Trop. Med. Infect. Dis. 2022;7(11):359. doi: 10.3390/tropicalmed7110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarini N.M.A, Susilawathi N.M, Sudewi A.A.R, Soejitno A, Fatmawati N.N.D, Mayura I.P.B, Lestari A.A.W, Suputra G, Subrata I.K, Astiti C.I.S.D, Besung I.N.K, Mahardika G.N. A large cluster of human infections of Streptococcus suis in Bali, Indonesia. One Health (Amsterdam, Netherlands) 2022;14:100394. doi: 10.1016/j.onehlt.2022.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vötsch D, Willenborg M, Weldearegay Y.B, Valentin-Weigand P. Streptococcus suis - The “two faces”of a pathobiont in the porcine respiratory tract. Front. Microbiol. 2018;9:480. doi: 10.3389/fmicb.2018.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neila-Ibáñez C, Casal J, Hennig-Pauka I, Stockhofe-Zurwieden N, Gottschalk M, Migura-García L, Pailler-García L, Napp S. Stochastic assessment of the economic impact of Streptococcus suis-associated disease in German, Dutch and Spanish swine farms. Front. Vet. Sci. 2021;8:676002. doi: 10.3389/fvets.2021.676002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neila-Ibáñez C, Napp S, Pailler-García L, Franco-Martínez L, Cerón J.J, Aragon V, Casal J. Risk factors associated with Streptococcus suis cases on pig farms in Spain. Vet. Rec. 2023;193(5):e3056. doi: 10.1002/vetr.3056. [DOI] [PubMed] [Google Scholar]
- 8.Rayanakorn A, Goh B.H, Lee L.H, Khan T.M, Saokaew S. Risk factors for Streptococcus suis infection:A systematic review and meta-analysis. Sci. Rep. 2018;8(1):13358. doi: 10.1038/s41598-018-31598-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayanakorn A, Ademi Z, Liew D, Lee L.H. Burden of disease and productivity impact of Streptococcus suis infection in Thailand. PLoS Negl. Trop. Dis. 2021;15(1):e0008985. doi: 10.1371/journal.pntd.0008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerdsin A, Segura M, Fittipaldi N, Gottschalk M. Sociocultural factors influencing human Streptococcus suis disease in Southeast Asia. Foods (Basel, Switzerland) 2022;11(9):1190. doi: 10.3390/foods11091190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyette-Desjardins G, Auger J.P, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014;3(6):e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, Takamatsu D. Current taxonomical situation of Streptococcus suis. Pathogens (Basel, Switzerland) 2016;5(3):45. doi: 10.3390/pathogens5030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segura M, Aragon V, Brockmeier S.L, Gebhart xC, Greeff A, Kerdsin A, O'Dea M.A, Okura M, Saléry M, Schultsz C, Valentin-Weigand P, Weinert L.A, Wells J.M, Gottschalk M. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction:4th International Workshop on S. suis. Pathogens (Basel, Switzerland) 2020;9(5):374. doi: 10.3390/pathogens9050374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinert L.A, Chaudhuri R.R, Wang J, Peters S.E, Corander J, Jombart T, Baig A, Howell K.J, Vehkala M, Välimäki N, Harris D, Chieu T.T.B, Van Vinh Chau N, Campbell J, Schultsz C, Parkhill J, Bentley S.D, Langford P.R, Rycroft A.N, Wren B.W, Farrar J, Baker S, Hoa N.T, Holden M.T.G, Tucker A.W, Maskell D.J. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015;6:6740. doi: 10.1038/ncomms7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aradanas M, Poljak Z, Fittipaldi N, Ricker N, Farzan A. Serotypes, virulence-associated factors, and antimicrobial resistance of Streptococcus suis isolates recovered from sick and healthy pigs determined by whole-genome sequencing. Front. Vet. Sci. 2021;8:742345. doi: 10.3389/fvets.2021.742345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zouharová M, Šimek B, Gebauer J, Králová N, Kucharovičová I, Plodková H, Pecka T, Brychta M, Švejdová M, Nedbalcová K, Matiašková K, Matiašovic J. Characterisation of Streptococcus suis isolates in the Czech republic collected from diseased pigs in the years 2018–2022. Pathogens (Basel, Switzerland) 2023;12(1):5. doi: 10.3390/pathogens12010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matiašovic J, Nedbalcová K, Žižlavský M, Fleischer P, Pokludová L, Kellnerová D, Nechvátalová K, Šimek B, Czanderlová L, Zouharová M, Bernardy J, Králová N, Šlosárková S. Streptococcus suis isolates-serotypes and susceptibility to antimicrobials in terms of their use on selected repopulated Czech pig farms. Pathogens (Basel, Switzerland) 2021;10(10):1314. doi: 10.3390/pathogens10101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson T.L, Bayles D.O. Comparative virulence and antimicrobial resistance distribution of Streptococcus suis isolates obtained from the United States. Front. Microbiol. 2022;13:1043529. doi: 10.3389/fmicb.2022.1043529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherrer S, Rosato G, Spoerry Serrano N, Stevens M.J.A, Rademacher F, Schrenzel J, Gottschalk M, Stephan R, Peterhans S. Population structure, genetic diversity and pathotypes of Streptococcus suis isolated during the last 13 years from diseased pigs in Switzerland. Vet. Res. 2020;51(1):85. doi: 10.1186/s13567-020-00813-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerdsin A, Takeuchi D, Nuangmek A, Akeda Y, Gottschalk M, Oishi K. Genotypic comparison between Streptococcus suis isolated from pigs and humans in Thailand. Pathogens (Basel, Switzerland) 2020;9(1):50. doi: 10.3390/pathogens9010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bojarska A, Janas K, Pejsak Z, Otulak-Kozieł K, Garbaczewska G, Hryniewicz W, Sadowy E. Diversity of serotypes and new cps loci variants among Streptococcus suis isolates from pigs in Poland and Belarus. Vet. Microbiol. 2020;240:108534. doi: 10.1016/j.vetmic.2019.108534. [DOI] [PubMed] [Google Scholar]
- 22.Niazy M, Hill S, Nadeem K, Ricker N, Farzan A. Compositional analysis of the tonsil microbiota in relationship to Streptococcus suis disease in nursery pigs in Ontario. Anim. Microbiome. 2022;4(1):10. doi: 10.1186/s42523-022-00162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacouture S, Olivera Y.R, Mariela S, Gottschalk M. Distribution and characterization of Streptococcus suis serotypes isolated from January 2015 to June 2020 from diseased pigs in Québec, Canada. Can. J. Vet. Res. 2022;86(1):78–82. [PMC free article] [PubMed] [Google Scholar]
- 24.Cucco L, Paniccià M, Massacci F.R, Morelli A, Ancora M, Mangone I, Di Pasquale A, Luppi A, Vio D, Cammà C, Magistrali C.F. New sequence types and antimicrobial drug-resistant strains of Streptococcus suis in diseased pigs, Italy, 2017–2019. Emerg. Infect. Dis. 2022;28(1):139–147. doi: 10.3201/eid2801.210816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrocchi-Rilo M, Martínez-Martínez S, Aguarón-Turrientes Á, Roca-Martínez E, García-Iglesias M.J, Pérez-Fernández E, González-Fernández A, Herencia-Lagunar E, Gutiérrez-Martín C.B. Anatomical site, typing, virulence gene profiling, antimicrobial susceptibility and resistance genes of Streptococcus suis isolates recovered from pigs in Spain. Antibiotics. 2021;10(6):707. doi: 10.3390/antibiotics10060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mintzker Y, Blum D, Adler L. Replacing PICO in non-interventional studies. BMJ Evid. Based Med. 2023;28(4):284. doi: 10.1136/bmjebm-2021-111889. [DOI] [PubMed] [Google Scholar]
- 27.Athey T.B.T, Teatero S, Lacouture S, Takamatsu D, Gottschalk M, Fittipaldi N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016;16(1):162. doi: 10.1186/s12866-016-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matiasovic J, Zouharova M, Nedbalcova K, Kralova N, Matiaskova K, Simek B, Kucharovicova I, Gottschalk M. Resolution of Streptococcus suis serotypes 1/2 versus 2 and 1 versus 14 by PCR-Restriction Fragment Length Polymorphism Method. J. Clin. Microbiol. 2020;58(7):e00480–20. doi: 10.1128/JCM.00480-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottschalk M, Higgins R, Boudreau M. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J. Clin. Microbiol. 1993;31(8):2192–2194. doi: 10.1128/jcm.31.8.2192-2194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukon P, Nam N.H, Kittipreeya P, Sara-In A, Wawilai P, Inchuai R, Weerakhun S. Global prevalence of chlamydial infections in birds:A systematic review and meta-analysis. Prev. Vet. Med. 2021;192:105370. doi: 10.1016/j.prevetmed.2021.105370. [DOI] [PubMed] [Google Scholar]
- 31.Higgins J.P.T, Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 32.Begg C.B, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 33.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duval S, Tweedie R. Trim and fill:A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 35.Arndt E.R, Farzan A, Slavic D, MacInnes J.I, Friendship R.M. An epidemiological study of Streptococcus suis serotypes of pigs in Ontario determined by a multiplex polymerase chain reaction. Can. Vet. J. 2018;59(9):997–1000. [PMC free article] [PubMed] [Google Scholar]
- 36.Baums C.G, Verkühlen G.J, Rehm T, Silva L.M.G, Beyerbach M, Pohlmeyer K, Valentin-Weigand P. Prevalence of Streptococcus suis genotypes in wild boars of Northwestern Germany. Appl. Environ. Microbiol. 2007;73(3):711–717. doi: 10.1128/AEM.01800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denich L.C, Farzan A, Friendship R, Arndt E, Gottschalk M, Poljak Z. A case-control study to investigate the serotypes of S. suis isolates by multiplex PCR in nursery pigs in Ontario, Canada. Pathogens (Basel, Switzerland) 2020;9(1):44. doi: 10.3390/pathogens9010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vishva K.V, Gangwar P, Thakor J.C, Dinesh M, Sahoo M, Singh R, Mahajan S, Qureshi S, Laddika L, Sahoo N.R, De U.K. Carrier status of Streptococcus suis in the palatine tonsils of apparently healthy slaughtered pigs of India. J. Immunoassay Immunochem. 2022;43(5):557–578. doi: 10.1080/15321819.2022.2048011. [DOI] [PubMed] [Google Scholar]
- 39.Lunha K, Chumpol W, Samngamnim S, Jiemsup S, Assavacheep P, Yongkiettrakul S. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018–2020. Antibiotics (Basel, Switzerland) 2022;11(3):410. doi: 10.3390/antibiotics11030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padungtod P, Tharavichitkul P, Junya S, Chaisowong W, Kadohira M, Makino S, Sthitmatee N. Incidence and presence of virulence factors of Streptococcus suis infection in slaughtered pigs from Chiang Mai, Thailand. Southeast Asian J. Trop. Med. Public Health. 2010;41(6):1454–1461. [PubMed] [Google Scholar]
- 41.Peng L, Lin M, Huang Z, Guo S, Sun H, Yang X. Genetic analysis and pathogenicity of different sequence types of Streptococcus suis isolated from pigs in southern China. FEMS Microbiol. Lett. 2020;367(6):fnaa049. doi: 10.1093/femsle/fnaa049. [DOI] [PubMed] [Google Scholar]
- 42.Silva L.M.G, Baums C.G, Rehm T, Wisselink H.J, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 2006;115(1–3):117–127. doi: 10.1016/j.vetmic.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, Ku X, Yu X, Sun Q, Wu H, Chen F, Zhang X, Guo L, Tang X, He Q. Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci. Rep. 2019;9(1):9908. doi: 10.1038/s41598-019-45482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou G, Zhou J, Xiao R, Zhang L, Cheng Y, Jin H, Li L, Zhang L, Wu B, Qian P, Li S, Ren L, Wang J, Oshota O, Hernandez-Garcia J, Wileman T.M, Bentley S, Weinert L, Maskell D.J, Tucker A.W.D, Zhou R. Effects of environmental and management-associated factors on prevalence and diversity of Streptococcus suis in clinically healthy pig herds in China and the United Kingdom. Appl. Environ. Microbiol. 2018;84(8):e02590–17. doi: 10.1128/AEM.02590-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amass S.F, Kreisle R.A, Clark L.K, Wu C.C. A pilot study of the prevalence of Streptococcus suis in pigs and personnel at five Indiana swine operations. J. Agromedicine. 1998;5:17–24. [Google Scholar]
- 46.Baele M, Chiers K, Devriese L.A, Smith H.E, Wisselink H.J, Vaneechoutte M, Haesebrouck F. The gram-positive tonsillar and nasal flora of piglets before and after weaning. J. Appl. Microbiol. 2001;91(60):997–1003. doi: 10.1046/j.1365-2672.2001.01463.x. [DOI] [PubMed] [Google Scholar]
- 47.Boetner A.G, Binder M, Bille-Hansen V. Streptococcus suis infections in Danish pigs and experimental infection with Streptococcus suis serotype 7. Acta Pathol. Microbiol. Immunol. Scand. B. 1987;95(4):233–239. doi: 10.1111/j.1699-0463.1987.tb03118.x. [DOI] [PubMed] [Google Scholar]
- 48.Brisebois L.M, Charlebois R, Higgins R, Nadeau M. Prevalence of Streptococcus suis in four to eight week old clinically healthy piglets. Can. J. Vet. Res. 1990;54(1):174–177. [PMC free article] [PubMed] [Google Scholar]
- 49.Han D.U, Choi C, Ham H.J, Jung J.H, Cho W.S, Kim J, Higgins R, Chae C. Prevalence, capsular type and antimicrobial susceptibility of Streptococcus suis isolated from slaughter pigs in Korea. Can. J. Vet. Res. 2001;65(3):151–155. [PMC free article] [PubMed] [Google Scholar]
- 50.Maneerat K, Yongkiettrakul S, Kramomtong I, Tongtawe P, Tapchaisri P, Luangsuk P, Chaicumpa W, Gottschalk M, Srimanote P. Virulence genes and genetic diversity of Streptococcus suis serotype 2 isolates from Thailand. Transbound. Emerg. Dis. 2013;60(Suppl 2):69–79. doi: 10.1111/tbed.12157. [DOI] [PubMed] [Google Scholar]
- 51.Marois C, Le Devendec L, Gottschalk M, Kobisch M. Detection and molecular typing of Streptococcus suis in tonsils from live pigs in France. Can. J. Vet. Res. 2007;71(1):14–22. [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez G, Harel J, Lacouture S, Gottschalk M. Genetic diversity of Streptococcus suis serotypes 2 and 1/2 isolates recovered from carrier pigs in closed herds. Can. J. Vet. Res. 2002;66(4):240–248. [PMC free article] [PubMed] [Google Scholar]
- 53.Meekhanon N, Kaewmongkol S, Phimpraphai W, Okura M, Osaki M, Sekizaki T, Takamatsu D. Potentially hazardous Streptococcus suis strains latent in asymptomatic pigs in a major swine production area of Thailand. J. Med. Microbiol. 2017;66(5):662–669. doi: 10.1099/jmm.0.000483. [DOI] [PubMed] [Google Scholar]
- 54.Mogollon J.D, Pijoan C, Murtaugh M.P, Collins J.E, Cleary P.P. Identification of epidemic strains of Streptococcus suis by genomic fingerprinting. J. Clin. Microbiol. 1991;29(4):782–787. doi: 10.1128/jcm.29.4.782-787.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ngo T.H, Tran T.B.C, Tran T.T.N, Nguyen V.D, Campbell J, Pham H.A, Huynh H.T, Nguyen V.V.C, Bryant J.E, Tran T.H, Farrar J, Schultsz C. Slaughterhouse pigs are a major reservoir of Streptococcus suis serotype 2 capable of causing human infection in southern Vietnam. PLoS One. 2011;6(3):e17943. doi: 10.1371/journal.pone.0017943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh S.I, Jeon A.B, Jung B.Y, Byun J.W, Gottschalk M, Kim A, Kim J.W, Kim H.Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017;79(4):780–787. doi: 10.1292/jvms.16-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paterson R.A, Robertson I.D, Sanders R.C, Siba P.M, Clegg A, Hampson D.J. The carriage of Streptococcus suis type 2 by pigs in Papua New Guinea. Epidemiol. Infect. 1993;110(1):71–78. doi: 10.1017/s095026880005069x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sánchez del Rey V.S, Fernández-Garayzábal J.F, Bárcena C, Briones V, Domínguez L, Gottschalk M, Vela A.I. Molecular typing of Streptococcus suis isolates from Iberian pigs:A comparison with isolates from common intensively-reared commercial pig breeds. Vet. J. 2014;202(3):597–602. doi: 10.1016/j.tvjl.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Sánchez del Rey V, Fernández-Garayzábal J.F, Mentaberre G, Briones V, Lavín S, Domínguez L, Gottschalk M, Vela A.I. Characterisation of Streptococcus suis isolates from wild boars (Sus scrofa) Vet. J. 2014;200(3):464–467. doi: 10.1016/j.tvjl.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Tarradas C, Luque I, de Andrés D, Abdel-Aziz Shahein Y.E, Pons P, González F, Borge C, Perea A. Epidemiological relationship of human and swine Streptococcus suis isolates. J. Vet. Med. Ser. B. 2001;48(5):347–355. doi: 10.1046/j.1439-0450.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 61.Thongkamkoon P, Kiatyingangsulee T, Gottschalk M. Serotypes of Streptococcus suis isolated from healthy pigs in Phayao Province, Thailand. BMC Res. Notes. 2017;10(1):53. doi: 10.1186/s13104-016-2354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torremorell M, Calsamiglia M, Pijoan C. Colonization of suckling pigs by Streptococcus suis with particular reference to pathogenic serotype 2 strains. Can. J. Vet. Res. 1998;62(1):21–26. [PMC free article] [PubMed] [Google Scholar]
- 63.van Leengoed L.A, Vecht U, Verheyen E.R. Streptococcus suis type 2 infections in pigs in the Netherlands (part two) Vet. Q. 1987;9(2):111–117. doi: 10.1080/01652176.1987.9694087. [DOI] [PubMed] [Google Scholar]
- 64.Vela A.I, Goyache J, Tarradas C, Luque I, Mateos A, Moreno M.A, Borge C, Perea J.A, Domínguez L, Fernández-Garayzábal J.F. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 2003;41(6):2498–2502. doi: 10.1128/JCM.41.6.2498-2502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Zhang W, Li X, Lu C, Chen J, Fan W, Huang B. Characterization of Streptococcus suis isolates from slaughter swine. Curr. Microbiol. 2013;66(4):344–349. doi: 10.1007/s00284-012-0275-4. [DOI] [PubMed] [Google Scholar]
- 66.Wang S, Gao M, An T, Liu Y, Jin J, Wang G, Jiang C, Tu Y, Hu S, Li J, Wang J, Zhou D, Cai X. Genetic diversity and virulence of novel sequence types of Streptococcus suis from diseased and healthy pigs in China. Front. Microbiol. 2015;6:173. doi: 10.3389/fmicb.2015.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wongsawan K, Gottschalk M, Tharavichitkul P. Serotype- and virulence-associated gene profile of Streptococcus suis isolates from pig carcasses in Chiang Mai Province, Northern Thailand. J. Vet. Med. Sci. 2015;77(2):233–236. doi: 10.1292/jvms.14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C.P, Ning Y.B, Zhang Z.Q, Song L, Qiu H.S, Gao H.Y, Fan X.Z. Prevalence of Streptococcus suis isolated from clinically healthy sows in China. Agric. Sci. China. 2009;8(5):638–642. [Google Scholar]
- 69.Zheng H, Ji S, Lan R, Liu Z, Bai X, Zhang W, Gottschalk M, Xu J. Population analysis of Streptococcus suis isolates from slaughtered swine by use of minimum core genome sequence typing. J. Clin. Microbiol. 2014;52(10):3568–3572. doi: 10.1128/JCM.00536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obradovic M.R, Segura M, Segalés J, Gottschalk M. Review of the speculative role of co-infections in Streptococcus suis-associated diseases in pigs. Vet. Res. 2021;52(1):49. doi: 10.1186/s13567-021-00918-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giang E, Hetman B.M, Sargeant J.M, Poljak Z, Greer A.L. Examining the effect of host recruitment rates on the transmission of Streptococcus suis in nursery swine populations. Pathogens (Basel, Switzerland) 2020;9(3):174. doi: 10.3390/pathogens9030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clifton-Hadley F.A, Alexander T.J. The carrier site and carrier rate of Streptococcus suis type II in pigs. Vet. Rec. 1980;107(2):40–41. doi: 10.1136/vr.107.2.40. [DOI] [PubMed] [Google Scholar]
- 73.Dechêne-Tempier M, Marois-Créhan C, Libante V, Jouy E, Leblond-Bourget N, Payot S. Update on the mechanisms of antibiotic resistance and the mobile resistome in the emerging zoonotic pathogen Streptococcus suis. Microorganisms. 2021;9(8):1765. doi: 10.3390/microorganisms9081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uruén C, García C, Fraile L, Tommassen J, Arenas J. How Streptococcus suis escapes antibiotic treatments. Vet. Res. 2022;53(1):91. doi: 10.1186/s13567-022-01111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi L, Jin M, Li J, Grenier D, Wang Y. Antibiotic resistance related to biofilm formation in Streptococcus suis. Appl. Microbiol. Biotechnol. 2020;104(20):8649–8660. doi: 10.1007/s00253-020-10873-9. [DOI] [PubMed] [Google Scholar]
- 76.Dettori J.R, Norvell D.C, Chapman J.R. Fixed-effect vs random-effects models for meta-analysis:3 points to consider. Glob. Spine J. 2022;12(7):1624–1626. doi: 10.1177/21925682221110527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu P, Zhang Y, Tang H, Wang Y, Sun X. Prevalence of Streptococcus suis in pigs in China during 2000–2021:A systematic review and meta-analysis. One Health (Amsterdam, Netherlands) 2023;16:100513. doi: 10.1016/j.onehlt.2023.100513. [DOI] [PMC free article] [PubMed] [Google Scholar]


