Abstract
Bovine viral diarrhea virus (BVDV), strain NADL, was originally isolated from an animal with fatal mucosal disease. This isolate is cytopathic in cell culture and produces two forms of NS3-containing proteins: uncleaved NS2-3 and mature NS3. For BVDV NADL, the production of NS3, a characteristic of cytopathic BVDV strains, is believed to be a consequence of an in-frame insertion of a 270-nucleotide cellular mRNA sequence (called cIns) in the NS2 coding region. In this study, we constructed a stable full-length cDNA copy of BVDV NADL in a low-copy-number plasmid vector. As assayed by transfection of MDBK cells, uncapped RNAs transcribed from this template were highly infectious (>105 PFU/μg). The recovered virus was similar in plaque morphology, growth properties, polyprotein processing, and cytopathogenicity to the BVDV NADL parent. Deletion of cIns abolished processing at the NS2/NS3 site and produced a virus that was no longer cytopathic for MDBK cells. This deletion did not affect the efficiency of infectious virus production or viral protein production, but it reduced the level of virus-specific RNA synthesis and accumulation. Thus, cIns not only modulates NS3 production but also upregulates RNA replication relative to an isogenic noncytopathic derivative lacking the insert. These results raise the possibility of a linkage between enhanced BVDV NADL RNA replication and virus-induced cytopathogenicity.
Bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and border disease virus are members of the pestivirus genus, a group of important animal pathogens in the family Flaviviridae (24, 32). The spread and maintenance of BVDV in cattle involves two kinds of infections (1, 32). Most infections are acute and self-limiting, with effective clearance of the virus. In contrast, infection of pregnant animals early in gestation can lead to efficient transplacental transmission of the virus to the fetus and birth of a persistently infected, BVDV-immunotolerant calf. Such animals are the main reservoir for BVDV, shedding virus for the life of the animal. Sporadically, these animals develop a uniformly fatal pathology called mucosal disease (MD). Two types of BVDV, distinguishable by their ability to cause cytopathic effect (CPE) in cell culture, can be isolated from animals with MD. Strains responsible for establishing persistent infections are typically noncytopathic (non-CP), whereas both non-CP and CP strains can be isolated from animals exhibiting MD. Considerable data suggest that CP strains are derived from non-CP strains by rare RNA recombination events (17).
The typical non-CP pestivirus genome is approximately 12.5 kb in length and consists of a 5′ nontranslated region (NTR), a single open reading frame encoding all viral polypeptides, and a nonpolyadenylated 3′ NTR (17). Uncapped pestivirus mRNA is translated via internal initiation (23, 26) to produce a polyprotein that is cleaved into 11 to 12 polypeptides by host and viral proteases (17). The first protein, Npro, possesses an autoproteolytic activity responsible for cleavage at its own C terminus. Downstream cleavages producing the structural components of the virion, C, Erns, E1, and E2, are mediated mainly by cellular signal peptidase (although the enzyme responsible for cleavage at the Erns/E1 junction has not been defined). The nonstructural (NS) portion of the polyprotein is processed at four sites (3/4A, 4A/4B, 4B/5A, and 5A/5B) by a BVDV-encoded serine protease activity (29, 35, 36). The catalytic domain of this enzyme resides in the NS3 region and requires the NS4A protein as a cofactor for cleavage of at least two sites (4B/5A and 5A/5B) (36).
Surprisingly, processing of the NS2-3 region differs between non-CP and CP BVDV isolates (see reference 17 for a review). Cleavage at the NS2/NS3 junction is not observed for non-CP BVDV. In contrast, a discrete NS3 protein is observed for all CP BVDV strains studied to date (10, 11). Depending on the CP isolate, processing at the NS2/NS3 junction is accomplished by several different strategies, but most appear to involve RNA recombinational events. These observations have led to the hypothesis that MD pathogenesis is linked to the generation of CP BVDV and, in particular, to the recombination events which lead to NS3 production. RNA recombination events linked to NS3 production include duplication and rearrangement of pestivirus sequences, insertion of cellular sequences, and large in-frame deletions resulting in subgenomic defective interfering (DI) RNAs (see reference 17 for a review). For some CP isolates, the mechanism by which NS3 is produced is clear. In several strains, in-frame insertion of cellular ubiquitin (Ub) sequences adjacent to the NS3 N terminus provides a processing site for cellular Ub carboxyl-terminal hydrolase. In other cases, a duplicated Npro autoprotease sequence fused to NS3 mediates the cleavage producing the NS3 N terminus. Recently, subgenomic (∼7.5-kb) CP BVDV DI RNAs with large in-frame deletions were identified (14, 31). These CP DI RNAs require a non-CP helper virus for spread and/or replication. For CP9, the sequences encompassing the coding region of C through NS2 have been deleted such that Npro is fused directly to NS3 (31). In CP13, two deletions have resulted in the fusion of 13 Npro residues and 10 E1 residues to NS3, with the NS3 N terminus truncated by five residues relative to the Ub- and Npro-NS3 fusion junctions (14). A CP DI RNA for CSFV in which all sequences between the methionine initiating the open reading frame and NS3 have been deleted has also been identified (18–20).
For two CP strains, CP7 and NADL, the mechanism(s) by which NS3 is produced remains obscure. Both isolates contain insertions in the NS2 region, apparently upstream of the NS2/NS3 (2/3) cleavage site. For CP7, the insertion is a duplicated viral sequence of 27 nucleotides which somehow promotes processing at the 2/3 site, NS3 production, and cytopathogenicity in cell culture (16, 30). In the case of the American prototype CP BVDV strain, NADL, the insert is a 270-base portion of a bovine mRNA of unknown function (called cIns [cellular insertion]) that results in an in-frame insertion of 90 amino acid residues.
To investigate the mechanism of NS3 production and cytopathogenicity by BVDV NADL, we constructed a stable, functional cDNA clone for this virus. Using this clone, we have gone on to engineer an isogenic derivative in which cIns has been deleted. Virus production, NS2-3 protein processing, accumulation of virus-specific proteins and RNA, and cytopathogenicity were then assessed. Our results indicate that cIns is necessary for NS3 production and the CP phenotype.
MATERIALS AND METHODS
Cells and viruses.
MDBK cells were propagated in Dulbecco’s modified minimal essential medium (DMEM) supplemented with sodium pyruvate and heat-inactivated 10% horse serum (HS). Cells were maintained at 37°C with 5% CO2.
The NADL strain of BVDV was obtained from the American Type Culture Collection, plaque purified, and amplified by growth in MDBK cells. For infection of MDBK cells, virus dilutions made in DMEM-HS were adsorbed for 1 h at 37°C; then the inoculum was removed and replaced with fresh DMEM-HS. Cultures were incubated at 37°C for 48 h, or until CPE was observed. Virus stocks were prepared by three freeze-thaw cycles of cells in their culture medium and clarified by centrifugation at 1,000 × g for 5 min.
BVDV plaque and focus-forming assays.
MDBK cells (70 to 80% confluent) were infected with 10-fold dilutions of virus as described above. Following 1 h of adsorption at 37°C, cells were washed once with DMEM, overlaid with 1.5% low-melting-point (LMP) agarose (Gibco-BRL) in MEM containing 5% HS, and incubated at 37°C. To assay for plaque-forming virus, after 3 days monolayers were fixed with 3.7% formaldehyde for 2 h at room temperature, the agarose plugs were removed, and the monolayers were stained with crystal violet (25). Foci produced by non-CP BVDV were visualized by immunostaining. After fixation with formaldehyde, agarose plugs were removed, and cells were permeabilized with Triton X-100 (0.25% in phosphate-buffered saline [PBS]) for 10 min, washed once with PBS, and then incubated with a bovine polyclonal anti-BVDV serum (α49; 1/1,000 dilution in PBS) (5) for 1.5 h. Monolayers were washed two times with PBS, incubated with peroxidase-conjugated rabbit anti-bovine immunoglobulin (1/1,000 dilution in PBS; catalog no. A-5295; Sigma Chemical Co.). After 1.5 h, excess second antibody was removed by washing the monolayer two times with PBS, and foci of BVDV-specific antigens were visualized by using the peroxidase substrate 3-amino-9-ethylcarbazole (catalog no. A-5754; Sigma).
Construction of a full-length BVDV NADL cDNA clone in a low-copy-number plasmid.
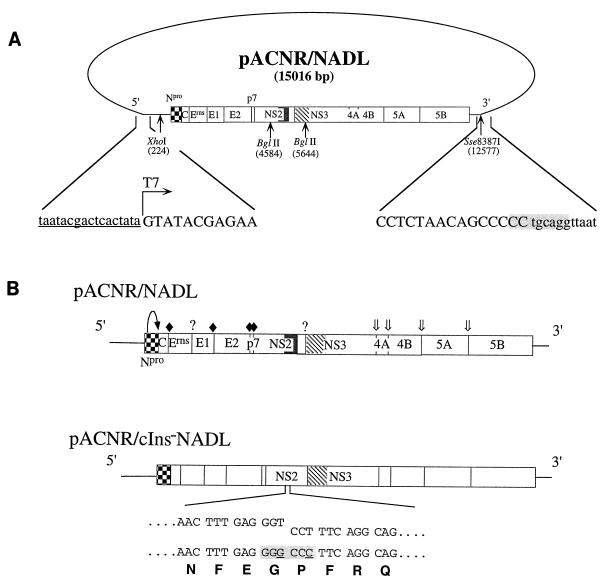
Initial attempts to assemble stable full-length BVDV NADL cDNA clones in medium-copy-number pBR322-derived vectors were unsuccessful. By using the low-copy-number plasmid vector pACNR1180 (27), standard recombinant DNA techniques and a series of intermediate plasmids were used to successfully assemble a full-length functional clone, called pACNR/NADL. Details of the assembly steps are available upon request. The salient features of the plasmid include a T7 promoter sequence fused to the BVDV 5′ terminus and the full-length BVDV cDNA sequence followed by an engineered Sse8387I site for production of runoff RNA transcripts corresponding to the precise BVDV genome RNA 3′ terminus. The T7-5′ and 3′-Sse8387I junction sequences are shown in Fig. 1. The clone was assembled by using previously constructed and sequenced NADL cDNA clones (5, 6, 35) or synthetic oligonucleotides and PCR (5′ and 3′ ends and an internal region to correct a single-base deletion at nucleotide 2702) (2). These regions and the clones from which they were derived include 1 to 223 (pBV-B55; by PCR), 224 to 1291 (pBV-18; XhoI-MscI fragment), 1292 to 2479 (pBV-116b; MscI-EcoRI), 2480 to 2826 (reverse transcription-PCR [RT-PCR] of NADL RNA; EcoRI-RsrII), 2827 to 3200 (pBV-116b; RsrII-MscI), 3201 to 4175 (pBV-D79; MscI-MunI), 4176 to 5173 (pBV-F2; MunI-EcoRV), 5174 to 12537 (pBV-SD2-3′; EcoRV-AatII), and 12538 to 12578 (pBV-C37; by PCR). Regions amplified by PCR were verified by sequence analysis. At nucleotide position 2653, a G residue was found in multiple independent clones instead of the previously reported A residue (6). This change is silent and was present in the BVDV NADL RNA preparation used for RT-PCR, as shown by direct sequencing of the PCR product. The full-length BVDV cDNA is positioned (sense orientation) in the pACNR-DraIII− backbone (created by filling in the unique DraIII site in pACNR1180 and religating) between the AatII and XhoI sites in the polylinker (which were treated with T4 DNA polymerase prior to cloning).
FIG. 1.
Diagram of plasmid pACNR/NADL, sequences surrounding transcription initiation and runoff sites, and engineered pACNR/NADL derivatives. (A) pACNR/NADL (15,016 bp) with the BVDV cDNA insert and the positions of BVDV-encoded polyprotein cleavage products are indicated. The Npro autoproteinase (checkered box), the cellular sequence insert (cIns; solid box), and serine proteinase domain (hatched box) are highlighted. Also shown are restriction sites used for subsequent constructions (positions are given in the NADL nucleotide sequence) and production of runoff RNA transcripts (Sse8387I). Sequences shown below include the T7 promoter (lowercase, underlined), the T7 transcription start site (arrow), the 5′- and 3′-terminal BVDV cDNA sequences (positive sense; uppercase), and the Sse8387I runoff site (shaded). (B) Structures of RNAs transcribed from pACNR/NADL (top) and pACNR/cIns−NADL (below). 5′ and 3′ NTRs are indicated by lines, and polyprotein cleavage products are represented by boxes. Processing sites for Npro (curved arrow), signal peptidase (solid diamonds), the serine proteinase (double arrows), and unidentified proteinases (question marks) are also shown. For the cIns− deletion mutant, the parental (upper staggered sequences) and mutant (below) nucleotide and amino acid sequences at the deletion breakpoints are shown. Silent nucleotide changes (underlined) were used to create a novel ApaI restriction site (shaded) to facilitate plasmid constructions and to serve as a convenient marker for distinguishing between each mutant and the parent.
We consistently found that bacterial colonies harboring the correct full-length plasmid were tiny and required 18 to 20 h to become visible. Of several bacterial hosts analyzed (see Results), Escherichia coli SURE cells (Stratagene) were most reliable for pACNR/NADL propagation. This host was therefore used for subsequent plasmid constructions. Large-scale DNA preparations were obtained from bacterial cultures grown in Terrific broth with carbenicillin (28), and plasmids were purified either by CsCl banding or by using Nucleobond AX columns (catalog no. 740-574; The Nest Group).
pACNR/cIns−NADL derivative.
This deletion construct was produced by using specific oligonucleotide pairs (Table 1) (based on the BVDV/NADL nucleotide sequence [6]) to PCR amplify subregions of pACNR/NADL and produce a convenient restriction site at or near the deletion breakpoint. These junctions are detailed in Fig. 1B. Fragments were then subcloned into pACNR/NADL to produce the desired deletion mutants. All regions amplified by PCR were verified by sequence analysis.
TABLE 1.
Oligonucleotides used for construction of pACNR/NADL, pACNR/cIns−NADL, and RT-PCR analyses
| Oligonucleotide | NADL nucleotide position | Sense |
|---|---|---|
| 183a | 1–27 | |
| 343 | 4509–4531 | + |
| 344 | 5810–5835 | − |
| 345 | 4980-4993/5264-5287b | + |
| 346 | 4974-4993/5264-5276b | − |
| 353 | 4754–4774 | + |
Consists of a 5′ XbaI restriction enzyme recognition sequence and the T7 promoter, followed by BVDV NADL nucleotides 1 to 27.
Mutations relative to the BVDV NADL sequence are noted in Fig. 1B.
For pACNR/cIns−NADL, in which the cIns sequence between nucleotides 4994 and 5263 was deleted, oligonucleotides 343 and 346 were used to amplify the 4509–4993 region; oligonucleotides 345 and 344 were used to amplify the 5264–5835 region. Oligonucleotides 345 and 346 contained silent nucleotide changes (underlined in Fig. 1B) to create an ApaI site at the deletion breakpoint. PCR-amplified fragments were digested with ApaI, ligated with T4 DNA ligase, digested with BglII, purified by separation on an LMP agarose gel, and cloned into BglII-digested pACNR/NADL that had been treated with calf intestinal alkaline phosphatase. Potentially correct clones were first identified by supercoiled plasmid size (28) and then by digestion with appropriate restriction enzymes and finally were verified by sequence analysis.
Standard in vitro transcription reaction.
pACNR/NADL or pACNR/cIns−NADL were digested to completion with Sse8387I, extracted with phenol and then chloroform, and precipitated with ethanol. One microgram of linearized plasmid DNA was transcribed in 20 μl, using the T7-MEGAscript kit (Ambion) with 0.5 μCi of added [3H]UTP (Dupont). Reaction mixtures were incubated at 37°C for 2 h in the absence of the cap analog. After transcription, the template DNA was degraded by using DNase I (2 U per 20-μl reaction; 37°C for 20 min) followed by extraction and precipitation as described above. RNAs were quantified on the basis of [3H]UTP incorporation and resuspended at a concentration of 2 μg/μl. The fraction of full-length RNA transcripts was checked by agarose gel electrophoresis, and aliquots for transfection were stored at −80°C.
Transfection of MDBK cells.
MDBK cells (70 to 80% confluent) were trypsinized, washed three times with ice-cold RNase-free PBS, and resuspended at 2 × 107 cells/ml in PBS. Unless otherwise indicated, 1 to 5 μg of transcribed RNA was mixed with 0.4 ml of the cell suspension and immediately pulsed with a Bio-Rad Gene Pulser (1.5 kV, 25 μF, infinite resistance, 2 pulses) or a BTX ElectroSquarePorator (0.9 kV, 99-μs pulse length, 10 pulses). The electroporated mixture was diluted with 10 ml of DMEM-HS. Depending on the particular experiment, samples were diluted further and plated in multiple wells or tissue culture dishes. An infectious center assay (12), with slight modifications, was used to quantify RNA specific infectivity. Tenfold dilutions of electroporated MDBK cells (in DMEM-HS) were plated (2 ml per 35-mm-diameter well) on monolayers of MDBK cells grown to 50 to 60% confluence. To permit recovery and attachment of the electroporated cells, plates were incubated for 4 h at 37°C, after which the medium was replaced with a 1.5% LMP agarose overlay as described above. Plates were incubated for 3 days at 37°C, and infectious centers were visualized and counted by staining for plaques or foci as described above.
Radioimmunoprecipitation and SDS-PAGE.
Rabbit polyclonal antiserum specific for BVDV NS3 (G40) or bovine anti-BVDV antiserum (α49) have been described elsewhere (5, 7). Depending on the antiserum, sodium dodecyl sulfate (SDS) (G40)- or Triton X-100 (α49)-solubilized cell lysates were used for immunoprecipitations. Following labeling of MDBK cells, the medium was removed, cells were washed twice with ice-cold PBS, and cell extracts were prepared by lysis (0.3 ml per 35-mm-diameter well) with either 0.5% SDS or 0.5% Triton X-100 in TNE (50 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.15 M NaCl, 20 μg of phenylmethylsulfonyl fluoride). SDS-solubilized lysates were sheared, heated to 75°C for 10 min, and clarified by centrifugation at 12,000 × g for 10 min. Triton X-100-solubilized lysates were also clarified. Clarified lysates were diluted 1:5 in TNE containing 0.5% Triton X-100, 2.5 μl (G40) or 5 μl (α49) of antiserum was added, and then the mixture was incubated overnight at 4°C with rocking. Protein A-agarose (Sigma), washed five times with TNE containing 0.1% Triton X-100, was added, and incubation was continued for 2 h at 4°C. Immunoprecipitates were washed three times with the same solution and then finally once with TNE lacking Triton X-100. Washed immunoprecipitates were resuspended in Laemmli sample buffer, heated to 85°C for 10 min, and centrifuged at 12,000 × g for 1 min. Immunoprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% polyacrylamide gel and visualized by fluorography (15).
Analysis of the cIns genetic marker.
Virus (culture media and freeze-thaw lysates) from ACNR/cIns-NADL- and control virus-infected MDBK cells was treated with 2 U of DNase I (RQI; Promega) and 1 μg of RNase A (catalog no. 1119915; Boehringer) for 30 min at 37°C and then used for infection of MDBK cells. Multiple sequential passages were conducted in duplicate, using these conditions. At each passage, RNA was obtained from the infected cells of one sample by using the RNAzol method as instructed by the manufacturer (Tel-Test, Inc.). RNA samples were used for RT-PCR with oligonucleotides 353 and 344 (Table 1). Amplified PCR products were extracted with phenol-chloroform and precipitated with ethanol before restriction enzyme digestion with ApaI or other enzymes. Passaged samples of wild-type (wt) BVDV/NADL and ACNR/NADL were used as controls for the absence of the ApaI site and presence of cIns.
Western blotting.
SDS-solubilized MDBK cell lysates were separated by SDS-PAGE (10% gel) and transferred to Immobilon P nitrocellulose membranes by using the semidry Multiphor II Nova blot system (LKB). The membranes were then stained for 90 s with 0.25% (wt/vol) fast green FCF in 10% acetic acid and then destained for 10 min in 10% acetic acid. Nonspecific binding sites were blocked overnight at 4°C with 5% milk in 20 mM Tris-Cl–137 mM NaCl–0.1% Tween 20, pH 7.6 (TBS-T). All following serum dilutions and washing steps were carried out in TBS-T. The membranes were incubated for 1 h at room temperature with primary rabbit polyclonal antisera specific for BVDV NS3 (G40) and E2 (D31) (5, 7) diluted 1/400 each, ensuring antibody saturation (data not shown), followed by a secondary horseradish peroxidase-conjugated goat anti-rabbit serum. Extensive wash steps were performed before primary and secondary antibodies and prior to detection with SuperSignal chemiluminescent substrate (Pierce) and exposure to X-ray film.
Northern blotting.
Total RNA was extracted from MDBK cells by using TRIZOL reagent (Gibco-BRL). Northern blotting and hybridization was performed essentially as described by Sambrook et al. (28). RNA from 106 cells was denatured with glyoxal for 1 h at 50°C, separated by sodium phosphate-buffered 1% agarose gel electrophoresis, and blotted overnight onto positively charged nylon membranes (Boehringer Mannheim), using the TurboBlotter system (Schleicher & Schuell) and alkaline transfer buffer (3 M NaCl, 8 mM NaOH). The membranes were then washed with 0.2 M sodium phosphate (pH 7.0), and the RNA was cross-linked by irradiation with a 254-nm light source (Stratalinker UV cross-linker; Stratagene). A 32P-labeled antisense RNA probe hybridizing to nucleotides 5413 to 5648 of the NADL genome was transcribed in vitro from the BamHI-linearized cDNA clone pGEM-3Zf(+)/NADLΔcIns-Bgl, which was constructed by inserting the 790-bp BglII fragment of pNADL/cIns−NADL into the BamHI site of pGEM-3Zf(+). One microgram of DNA was transcribed with SP6 polymerase in the presence of 0.5 mM each ATP, GTP, and CTP, 12.5 μM UTP, and 3.12 μM [α-32P]UTP (800 Ci/mmol; Amersham). After treatment with DNase I, the RNA was purified from unincorporated ribonucleoside triphosphates using a Quick Spin G-50 Sephadex column (Boehringer Mannheim). The membrane was incubated in a Hybaid hybridization oven at 60°C for 5 h in prehybridization/hybridization solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 5× Denhardt’s reagent, 0.5% SDS, 100 μg of denatured salmon sperm DNA per ml, 50 μg of yeast tRNA per ml, 50% formamide), followed by overnight incubation at 60°C in fresh hybridization solution supplemented with 2 × 107 cpm of labeled probe. The blot was then washed at 65°C three times for 30 min each with 1× SSPE–0.5% SDS and once for 30 min with 0.1× SSPE–0.5% SDS. Bands were visualized by X-ray autoradiography and quantified with a Molecular Imager (Bio-Rad Laboratories).
Metabolic labeling of viral RNA.
For [32P]orthophosphate incorporation, infected MDBK cells were cultured in phosphate-free DMEM supplemented with 2% heat-inactivated HS. Five hours postinfection, the cells were treated with dactinomycin (2 μg/ml) for 1 h prior to addition of [32P]orthophosphate (200 μCi/ml; ICN Pharmaceuticals, Inc.). Total RNA was harvested at 12 and 18 h postinfection, using TRIZOL reagent. RNA from 7 × 104 cells was denatured with glyoxal and separated by agarose gel electrophoresis as described above. The gel was then fixed with methanol and dried, and RNA was visualized and quantified as described above.
RESULTS
Construction of a full-length functional clone of BVDV NADL in low-copy-number plasmid pACNR1180.
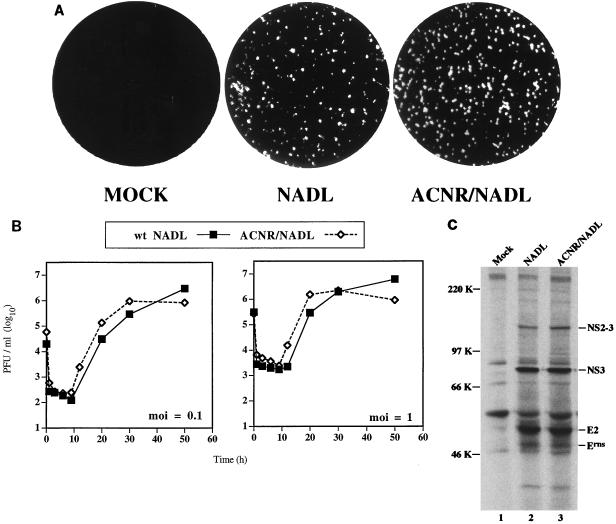
Initial attempts to assemble stable full-length BVDV NADL cDNA clones in high- or medium-copy-plasmid vectors failed. Finally, low-copy-number vector pACNR1180, which had been used for stable propagation of full-length CSFV cDNA clones (27), was successfully employed. pACNR/NADL contains a T7 promoter, the full-length BVDV NADL cDNA reconstructed from previously sequenced overlapping cDNA clones (6) or RT-PCR products, and a unique 3′ Sse8387I site for production of runoff RNA transcripts (see Materials and Methods) (Fig. 1). T7 polymerase transcription of Sse8387I-linearized pACNR/NADL template DNA produced RNA transcripts infectious for MDBK cells, as shown in Table 2. Cap analog was not included in transcription reactions since pestivirus RNAs are believed to be uncapped (4, 19, 27); in fact, capping of in vitro-transcribed CSFV RNA actually reduced specific infectivity about 10-fold (27). Optimized electroporation conditions yielded >105 PFU/μg of RNA transcript. Template DNA alone was not infectious, but intact template was required during transcription since DNase treatment abolished infectivity. After transcription, treatment with DNase had no effect whereas RNase treatment abolished infectivity of transcribed RNAs. These results establish that infectivity was derived by transcription of RNA from the full-length BVDV cDNA template. Typical virus yields harvested from the culture supernatant and cells (by freeze-thaw cycles) at 36 h were 3 × 106 to 107 PFU/ml. The resulting virus was neutralized by BVDV-specific antiserum, as demonstrated by both plaque and CPE reduction (data not shown).
TABLE 2.
Specific infectivity of in vitro RNA transcripts generated from pACNR/NADLa
| Material used to transfect MDBK cells | Expt 1, yieldb (PFU) | Expt 2
|
|
|---|---|---|---|
| Yieldb (PFU) | Virus recoveredc (PFU/ml) | ||
| DNA linearized with Sse8387I | 0 | 0 | 0 |
| Transcription reaction | |||
| Complete | NDd | 2.3 × 105 | 6.7 × 106 |
| DNase during | 0 | 0 | 0 |
| DNase after | 2.5 × 105 | 2.9 × 105 | 3.9 × 106 |
| RNase after | 0 | 0 | 0 |
One microgram of pACNR/NADL linearized with Sse8387I was used for transcription either in the presence or in the absence of DNase I. Following synthesis, some transcription reactions were treated with DNase I or RNase A for 20 min at 37°C. After these treatments, samples were used to electroporate MDBK cells and infectious centers were determined as described in Materials and Methods.
Data are expressed in PFU per microgram of RNA or input DNA.
Viral titer harvested 36 h postelectroporation.
ND, not determined.
It should be noted that even in the pACNR1180 backbone, bacterial colonies harboring the full-length NADL cDNA were tiny, appearing on semisolid media only after 18 to 20 h at 37°C. The deleterious effects of long pestivirus cDNAs and full-length clones during propagation in E. coli have been noted previously (21, 27, 34). Since future genetic analyses depended on having a reliable NADL molecular clone for manipulation, we investigated the stability of pACNR/NADL in several bacterial hosts, including E. coli MC1061, ABLE-K, ABLE-C, XL1-Blue, and SURE cells. Plasmid DNA from our initial infectious clone was used to transform each of these strains. We monitored colony size, gross plasmid structure by restriction analysis, and the specific infectivity of transcribed RNAs. Among the host strains analyzed, MC1061, ABLE-K, and ABLE-C yielded heterogeneous mixtures of colony sizes. DNA from the larger colonies often showed evidence of deleted or rearranged sequences and no longer yielded infectious RNA transcripts. In contrast, transformation of XL1-Blue and SURE cells produced relatively uniform populations of small colonies, with no evidence of DNA rearrangement, and yielded transcribed RNAs with consistently high specific infectivities (data not shown). SURE cells proved slightly better (faster colony growth and higher specific infectivity RNA) and were used for all subsequent DNA manipulations.
Comparison of virus derived from pACNR/NADL to parental BVDV NADL.
As shown in Fig. 2A, plaques on MDBK cells produced by transfection with RNA transcribed from pACNR/NADL were homogeneous and similar to the BVDV NADL parental virus originally used for cDNA cloning. Similar results were obtained in plaque assays using virus harvested from cells transfected with pACNR/NADL transcript RNA (called ACNR/NADL) or infected with the NADL parent (data not shown). Growth properties of ACNR/NADL and the parent were compared after infection of MDBK cells at both low (0.1 PFU/cell) and high (1.0 PFU/cell) multiplicity of infection (MOI). As is apparent from the experiment shown in Fig. 2B, the kinetics of replication and the yield of infectious virus were similar for ACNR/NADL and the parental virus at both MOIs. The patterns of viral proteins were also compared by metabolic labeling between 20 and 24 h postinfection and immunoprecipitation with a BVDV-specific polyclonal antiserum (Fig. 2C). Identical patterns of virus-specific proteins were observed for both ACNR/NADL and the parent. Proteins indicated in Fig. 2C were identified not only by size but also by immunoreactivity with a panel of region-specific antisera (reference 36 and data not shown). Of note was the uncleaved NS2-3 species migrating at 125 kDa and the prominent 80-kDa NS3 cleavage product, which are characteristic of CP BVDV strains. The similar plaque morphology, cytopathogenicity, growth properties, and polyprotein processing patterns of ACNR/NADL and the BVDV NADL parent validated the use of pACNR/NADL for future molecular genetic studies.
FIG. 2.
Comparison of virus derived from pACNR/NADL and wt BVDV NADL. (A) Plaques were visualized by crystal violet staining at 3 days postinfection (NADL) or post-RNA transfection (ACNR/NADL) as described in Materials and Methods. The mock-transfected monolayer was treated the same as the transfected monolayer except that no RNA was present in the transfection mixture. (B) MDBK cells were infected with either wt NADL or ACNR/NADL at an MOI (as determined on MDBK monolayers) of either 0.1 or 1 PFU/cell, washed, and harvested at the indicated times postinfection. Titers were determined by plaque assay on MDBK monolayers as described in Materials and Methods. (C) MDBK cells were mock infected or infected with wt NADL or ACNR/NADL at an MOI of 1 PFU/cell. At 20 h postinfection, proteins were labeled for 4 h with Expre35S35S label (NEN) and lysed, and BVDV-specific proteins were immunoprecipitated with a polyclonal anti-BVDV serum (α49). Proteins were separated by SDS-PAGE (8% gel) and visualized by fluorography. Molecular mass markers are indicated at the left; BVDV-specific proteins, identified by size and, in some cases, by immunoreactivity with region-specific antisera (data not shown), are indicated at the right.
Deletion of cIns abrogates processing at the 2/3 site and NS3 production, and produces replication-competent, non-CP BVDV.
Genome rearrangements and/or inserted sequences in CP isolates appear to be linked to processing at the 2/3 site, NS3 production, and cytopathogenicity. Although this hypothesis is supported by sequence comparisons of non-CP/CP pairs (17), it has been rigorously tested for only one CP isolate, CP7 (16, 30) (see Discussion). To address this for the NADL strain, we constructed pACNR/cIns−NADL in which the 270-base cIns was deleted. At the deletion breakpoint, two silent nucleotide changes were introduced to create a novel ApaI restriction site, which was used as an additional genetic marker for the deletion mutant (Fig. 1; see also Materials and Methods).
Transfection of MDBK cells with RNA transcripts from linearized pACNR/cIns−NADL template DNA did not induce CPE after 5 days at 37°C, and these cells looked similar to mock-transfected control monolayers. In contrast, RNA transcribed from pACNR/NADL induced CPE after 24 h (data not shown). We could, however, readily detect ACNR/cIns−NADL replication by immunostaining of foci by using a polyclonal anti-BVDV antiserum (Fig. 3A). Using an infectious center assay for electroporated MDBK cells (see Materials and Methods) and this immunostaining protocol, we determined that RNA transcripts from pACNR/cIns−NADL had a specific infectivity approaching that of pACNR/NADL (∼8 × 104 focus-forming units [FFU] per μg of RNA). Low- and high-multiplicity infection comparisons of ACNR/cIns−NADL and ACNR/NADL (Fig. 3B) revealed similar growth kinetics and virus yields, with the non-CP derivative showing slightly faster replication and higher cumulative virus titers, which approached 107 FFU/ml. As seen in Fig. 3C, ACNR/cIns−NADL did not induce CPE in cultures even at 50 h postinfection, when peak titers were reached, in contrast to ACNR/NADL and wt BVDV NADL, which had caused dramatic CPE by this time.
FIG. 3.
Virus lacking cIns is non-CP. (A) MDBK cells were transfected with RNA transcribed from linearized pACNR/cIns−NADL template DNA (ACNR/cIns−NADL) or were mock transfected (Mock), and then dilutions of transfected cells were assayed for infectious centers as described in Materials and Methods. Foci were visualized after 3 days by immunostaining with polyclonal α49 serum as the primary antibody. (B) Viral growth analyses after low (0.1 PFU or FFU per cell)- or high (1 PFU or FFU per cell)-MOI infection were conducted as described for Fig. 2B. Titers were determined by a standard plaque assay for CP ACNR/NADL (PFU/milliliter) or a focus forming assay for non-CP ACNR/cIns−NADL (FFU/milliliter). The data shown represent one of three independent experiments yielding similar results. (C) Phase-contrast photomicrographs of MDBK cells either mock infected or infected with the indicated viruses (MOI of 0.1). Pictures were taken at 50 h postinfection and correspond to the same cultures used for the growth analyses shown in panel B.
To confirm the genomic structure of ACNR/cIns−NADL, we serially passaged the virus in MDBK cells, each time incubating the resulting virus with RNase and DNase to avoid carryover of input transcript RNA and plasmid template DNA. Total cellular RNA, isolated at each passage, was used for amplification of a NS2-3 subregion that included the cIns locus, and the resulting fragments were analyzed by agarose gel electrophoresis, either with or without digestion with ApaI (Fig. 4). As shown in Fig. 4, amplification of this region for the NADL parent and ACNR/NADL produced a fragment of 1,082 bp (Fig. 4A) that was resistant to digestion by ApaI (Fig. 4B, lanes 5 to 8). In contrast, amplification of both early (passage 1)- and late (passage 4)-passage RNA from ACNR/cIns−NADL yielded the expected smaller 812-bp fragment that was susceptible to digestion by ApaI (Fig. 4B, lanes 1 to 4).
FIG. 4.
Presence of the ApaI marker in virus derived from pACNR/cIns−NADL. (A) Diagram of the expected RT-PCR fragments of ACNR/NADL and ACNR/cIns−NADL. Indicated are the location of cIns (black box), the engineered ApaI site, the primers used for RT-PCR (arrows), and the NS2 and NS3 coding regions (hatched boxes). The expected sizes of the RT-PCR products are given, as are the sizes of the two ApaI digestion products for ACNR/cIns−NADL. (B) After infection (NADL) or transfection (ACNR/NADL and ACNR/cIns−NADL), virus was harvested at 20 to 26 h (when the CP derivatives caused demonstrable CPE) as described in Materials and Methods. At each passage, the resulting virus was treated with DNase I and RNase A for 30 min at 37°C before infection of new monolayers (0.3 ml of undiluted virus stock per 35-mm-diameter well). For the indicated passages, RNA from infected cells was used for RT-PCR and a portion of each amplification reaction was digested with ApaI. Products were separated by electrophoresis on a 1.5% agarose gel and visualized by staining with ethidium bromide. During this analysis, we consistently observed a small fraction of the RT-PCR product from ACNR/cIns−NADL which was resistant to digestion by ApaI (even in vast enzyme excess; lane 4). Control experiments demonstrated that this resistant fraction was generated not during virus propagation but rather during the T7 transcription or reverse transcription steps (data not shown).
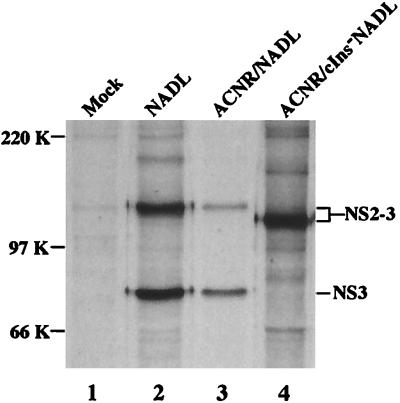
To examine protein processing in the NS2-3 region, MDBK cells were infected with the NADL parent (ACNR/NADL) or ACNR/cIns−NADL and metabolically radiolabeled, and the NS3-related proteins were immunoprecipitated with an NS3-specific polyclonal rabbit antiserum. As shown in Fig. 5, both NS2-3 and NS3 were present in cells infected with NADL and ACNR/NADL (lanes 2 and 3), whereas only NS2-3 was found in ACNR/cIns−NADL-infected cells (lane 4). NS2-3 produced by ACNR/cIns−NADL migrated faster than that produced by NADL and ACNR/NADL, presumably because of the cIns deletion that shortens NS2-3 by 90 amino acids (∼10 kDa).
FIG. 5.
Deletion of cIns abrogates NS3 production. MDBK cells were mock infected or infected with the indicated viruses at an MOI of 1 PFU/cell. At 20 h postinfection, monolayers were labeled for 4 h with Expre35S35S label (NEN) and lysed, and NS3-containing species were immunoprecipitated with a polyclonal anti-NS3 serum (G40 [5]). Proteins were separated by SDS-PAGE (8% gel) and visualized by fluorography. Molecular mass markers are indicated at the left; BVDV-specific proteins are indicated at the right.
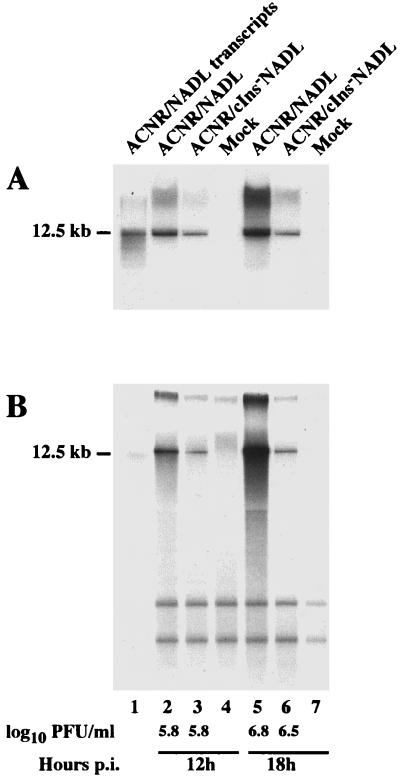
Parallel comparison of RNA, protein, and virus accumulation over time for ACNR/NADL and ACNR/cIns−NADL revealed significantly higher levels of RNA for the CP virus than for its non-CP derivative (Fig. 6), whereas the analyzed proteins NS2-3 or NS3 and E2 (Fig. 7) as well as the virus titers (Fig. 6 and 7) accumulated to similar levels. The RNA, proteins, and virus titers shown in Fig. 6 and 7 were obtained in parallel from one single experiment and are representative of three identical experiments repeated independently. For viral RNA, Northern blotting and metabolic labeling yielded similar results (Fig. 6). As quantified by Molecular Imager analysis, the calculated ratio of ACNR/NADL to ACNR/cIns−NADL RNA was 3 (Fig. 6A and B) at 12 h postinfection and 5 (Fig. 6A) or 8 (Fig. 6B) at 18 h postinfection.
FIG. 6.
Viral RNA accumulation in MDBK cells infected with ACNR/NADL and ACNR/cIns−NADL. MDBK cells were infected at an MOI of 2 with either ACNR/NADL (lanes 2 and 5) or ACNR/cIns−NADL (lanes 3 and 6) or were mock infected (lanes 4 and 7), and total RNA was harvested 12 h (lanes 2 to 4) and 18 h (lanes 5 to 7) postinfection (p.i.). (A) For each sample, RNA from 106 cells was analyzed by Northern blotting using a 32P-labeled RNA probe hybridizing to positive-sense viral RNA in the NS3 gene. (B) For direct analysis of total viral RNA accumulation, infected cells were metabolically labeled with [32P]orthophosphate between 6 and 18 h postinfection in the presence of dactinomycin. Glyoxal-denatured RNA from 7 × 104 cells harvested at 12 and 18 h postinfection was separated by agarose gel electrophoresis and visualized by X-ray autoradiography. Either unlabeled (A, lane 1) or 32P-labeled (B, lane 1) transcripts of pACNR/NADL served as size markers for full-length viral RNA (12.5 kb). The virus titers at the time of RNA harvest in the same experiment are indicated.
FIG. 7.
Levels of NS2-3, NS3, and E2 in MDBK cells infected with ACNR/NADL or ACNR/cIns−NADL. MDBK cells were infected at an MOI of 2 with either ACNR/NADL (lanes 1 and 4) or ACNR/cIns−NADL (lanes 2 and 5) or were mock infected (lanes 3 and 6), using the same inocula and cell densities described for the experiment shown in Fig. 6. The cells were lysed at 12 h (lanes 1 to 3) and 18 h (lanes 4 to 6) postinfection (p.i.), and lysates from 3 × 104 cells were separated by SDS-PAGE (10% gel) and analyzed by Western blotting using a low-dilution mix of rabbit antisera G40 and D31, specific for BVDV NS3 and E2, respectively. Molecular mass markers are indicated at the left. BVDV-specific proteins are indicated at the right.
These results demonstrate that cIns modulates cleavage at the 2/3 site, NS3 production, and cytopathogenicity but does not have dramatic effects on synthesis of virus-specific proteins or virus yield. Remarkably, however, deletion of cIns resulted in significantly lower levels of viral RNA synthesis and accumulation.
DISCUSSION
In this work, we succeeded in constructing a functional BVDV NADL cDNA clone in a low-copy-number plasmid. Full-length RNAs transcribed by T7 polymerase from this cDNA template have the authentic viral 5′- and 3′-terminal sequences, are highly infectious for MDBK cells (>105 PFU/μg), and yield a virus that has properties similar to those of the BVDV NADL parent. This plasmid clone is stable when propagated in the SURE strain of E. coli. Recently, assembly of a full-length BVDV NADL clone in a high-copy-number plasmid was reported by another group (34). Although infectious RNA could be transcribed from this template, the authors noted problems with plasmid transformation efficiency and stability and the production of full-length RNA transcripts. Some of these difficulties mimic our earlier unsuccessful attempts to construct such clones in high- or medium-copynumber plasmids. These problems were alleviated when the pACYC177 backbone was used. This plasmid had been used successfully for several other full-length pestivirus cDNAs (16, 19, 27). The reason(s) for the observed toxic effects of pestiviral cDNAs in some E. coli strains remains to be determined, but similar problems have also been encountered for several members of the flavivirus genus (22, 25).
The creation of a functional BVDV NADL cDNA clone allowed us to directly test the role of cIns in NS3 production and cytopathogenicity. Deletion of the 270-base cIns element produced a viable non-CP virus, in which detectable cleavage at the 2/3 site and NS3 production were abolished. Similar results were recently reported for CP BVDV strain CP7, which contains a 27-base duplication of viral sequences in the NS2 region (30). Using a vaccinia virus transient expression assay, deletion of this 27-base sequence eliminated cleavage at the 2/3 site (30). Further studies demonstrated that an isogenic derivative lacking this insertion was non-CP (16).
The mechanism(s) by which cIns (NADL) or the 27-base (CP7) insertions in NS2 promote cleavage at the 2/3 site is unknown. Although the NS3 N terminus has yet to be precisely determined for these strains, based on the similar apparent molecular masses of pestivirus NS3 proteins (10, 11, 30) and the conserved Ub-NS3, Npro-NS3, and Met-NS3 junctions observed for other CP isolates, Gly-1590 (SD-1 numbering [8, 9]) is the likely NS3 N-terminal residue. For NADL, this would imply that the 90-amino-acid cIns insertion, located 53 residues upstream of this Gly residue, somehow promotes cleavage at the 2/3 junction. In the case of CP7, the nine-residue insertion is located even further upstream of the putative 2/3 cleavage site. In addition to their different locations in NS2, there is no obvious sequence similarity between the NADL and CP7 inserts. Whether they activate a cryptic autoprotease present in the NS2-3 region or change the conformation of NS2-3 so as to render it susceptible to site-specific cleavage by a cellular enzyme remains to be determined (see references 29 and 36 for further discussion). Interestingly, in the absence of any inserted sequences or genome rearrangements, NS3 production occurs in cells infected with CSFV isolates (3, 33).
The strongest correlate of pestivirus cytopathogenicity is NS3 production, which is accomplished by myriad different strategies (17). Two groups have recently demonstrated that cell death induced by CP BVDV infection occurs via apoptosis (13, 37). It is possible that NS3 acts as a direct effector of apoptosis by somehow triggering cell death pathways. This is a plausible hypothesis given the obvious structural differences between NS2-3 and NS3, which could affect subcellular localization and interaction with host cell components, as previously discussed (35). Alternatively, cleavage at the 2/3 site (or NS3 production) could upregulate BVDV RNA replication to a level that is deleterious for host cells. In one model, viral RNA replication complexes might sequester cellular components present in limited quantities and required for maintaining homeostasis. In the case of BVDV NADL, increased numbers of replication complexes would then deplete such host factors to a level which triggers apoptosis. This model is consistent with our results, which demonstrate that RNA replication and accumulation are enhanced in ACNR/NADL-infected cells compared to ACNR/cIns−NADL-infected cells. It will be of interest to examine other isogenic non-CP/CP pairs to determine the generality of this observation and its possible correlation with cytopathogenicity.
In summary, genetic analyses of CP7 (16) and NADL (this report) have established that two distinct insertions in NS2 can regulate processing at the 2/3 site, NS3 production, and cytopathogenicity in cell culture. Such isogenic non-CP/CP pairs should be valuable for additional studies aimed at answering key questions in pestivirus biology. Examples include (i) defining the mechanism(s) of cleavage at the 2/3 site, including the responsible protease(s); (ii) establishing the pathway linking NS3 production to cytopathogenicity; and (iii) testing the hypothesis that CP strains with these insertions are sufficient to cause MD in animals persistently infected with the isogenic non-CP derivative.
ACKNOWLEDGMENTS
We thank Carol Read for expert technical assistance. We are also grateful to many colleagues for helpful discussions during the course of this work and to M. Scott McBride, Tina Myers, and Karen Reed for critical reading of the manuscript.
E.M. was supported by a fellowship from the Human Frontiers of Science Program Organization and by the Universidad Nacional Autónoma de México. N.R. was supported by fellowships from the Swiss National Science Foundation and from the Swiss Foundation for Biomedical Stipends (SSMBS).
REFERENCES
- 1.Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. [PubMed] [Google Scholar]
- 2.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher P, Shannon A D, Tautz N, Thiel H-J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 4.Brock K V, Deng R, Riblet S M. Nucleotide sequencing of 5′ and 3′ termini of bovine viral diarrhea virus by RNA ligation and PCR. J Virol Methods. 1992;38:39–46. doi: 10.1016/0166-0934(92)90167-c. [DOI] [PubMed] [Google Scholar]
- 5.Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 6.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 7.Collett M S, Wiskerchen M A, Welniak E, Belzer S K. Bovine viral diarrhea virus genomic organization. Arch Virol Suppl. 1991;3:19–27. doi: 10.1007/978-3-7091-9153-8_3. [DOI] [PubMed] [Google Scholar]
- 8.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–869. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 9.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genomes: primary and secondary structure analyses. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donis R O, Dubovi E J. Characterization of bovine viral diarrhoea-mucosal disease virus-specific proteins in bovine cells. J Gen Virol. 1987;68:1597–1605. doi: 10.1099/0022-1317-68-6-1597. [DOI] [PubMed] [Google Scholar]
- 11.Greiser-Wilke I, Dittmar K E, Liess B, Moennig V. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J Gen Virol. 1992;73:47–52. doi: 10.1099/0022-1317-73-1-47. [DOI] [PubMed] [Google Scholar]
- 12.Hertz J M, Huang H V. Evolution of the Sindbis virus subgenomic mRNA promoter in cultured cells. J Virol. 1995;69:7768–7774. doi: 10.1128/jvi.69.12.7768-7774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoff H S, Donis R O. Induction of apoptosis and cleavage of poly(ADP-ribose) polymerase by cytopathic bovine viral diarrhea virus infection. Virus Res. 1997;49:101–113. doi: 10.1016/s0168-1702(97)01460-3. [DOI] [PubMed] [Google Scholar]
- 14.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laskey R A, Mills A D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 16.Meyers G, Tautz N, Becher P, Thiel H J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 18.Meyers G, Thiel H J. Cytopathogenicity of classical swine fever virus caused by defective interfering particles. J Virol. 1995;69:3683–3689. doi: 10.1128/jvi.69.6.3683-3689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyers G, Thiel H J, Rumenapf T. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol. 1996;70:1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittelholzer C, Moser C, Tratschin J-D, Hoffman M A. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell culture. Virus Res. 1997;51:125–137. doi: 10.1016/S0168-1702(97)00081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moormann R J, van Gennip H G, Miedema G K, Hulst M M, van Rijn P A. Infectious RNA transcribed from an engineered full-length cDNA template of the genome of a pestivirus. J Virol. 1996;70:763–770. doi: 10.1128/jvi.70.2.763-770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J Virol. 1997;71:5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole T L, Wang C-Y, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 24.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–960. [Google Scholar]
- 25.Rice C M, Grakoui A, Galler R, Chambers T J. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- 26.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruggli N, Tratschin J D, Mittelholzer C, Hofmann M A. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tautz N, Thiel H J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Raven Press; 1996. pp. 1059–1073. [Google Scholar]
- 33.Thiel H-J, Stark R, Weiland E, Rümenapf T, Meyers G. Hog cholera virus: molecular composition of virions from a pestivirus. J Virol. 1991;65:4705–4712. doi: 10.1128/jvi.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, Aldridge S, Clarke M C, McCauley J W. Cell death induced by cytopathic bovine viral diarrhea virus is mediated by apoptosis. J Gen Virol. 1997;77:1677–1681. doi: 10.1099/0022-1317-77-8-1677. [DOI] [PubMed] [Google Scholar]