Abstract
Glycoprotein gp150 is a highly glycosylated protein encoded by the BDLF3 open reading frame of Epstein-Barr virus (EBV). It does not have a homolog in the alpha- and betaherpesviruses, and its function is not known. To determine whether the protein is essential for replication of EBV in vitro, a recombinant virus which lacked its expression was made. The recombinant virus had no defects in assembly, egress, binding, or infectivity for B cells or epithelial cells. Infection of epithelial cells was, however, enhanced. The glycoprotein was sensitive to digestion with a glycoprotease that digests sialomucins, but no adhesion to cells that express selectins that bind to sialomucin ligands could be detected.
The envelope of Epstein-Barr virus (EBV), like that of all herpesviruses, includes multiple unique glycoprotein species. Among those mapped to their open reading frames (ORFs) in the virus genome (1) and characterized at least biochemically are gp350/220, the product of the BLLF1 ORF (2); glycoproteins gp85, gp42, and gp25, respective products of the BXLF2 (7, 21), BZLF2 (14), and BKRF2 ORFs (33), which make up the EBV gH-gL-gp42 complex; gp78, the product of the BILF2 ORF (16); gN, the product of the BLRF1 ORF (12); gM, the product of the BBRF3 ORF (12); and gp150, the product of the BDLF3 ORF (11, 20). Functions have been ascribed to gp350/220, which is the viral attachment protein that binds the virus to CR2 or CD21 (19, 30), and to the gH-gL-gp42 complex, which interacts with HLA class II molecules on B cells (27) and is involved in virus penetration through the membranes of both B cells and epithelial cells (6, 13, 17, 31, 32). Little, however, is yet known about the roles played by gN, gM, gp78, and gp150, including whether they are essential for virus replication.
Glycoprotein gp150, the largest of the four, is perhaps most remarkable for the extent of its posttranslational modification with sugars (11, 20). The backbone of the molecule has an apparent Mr of approximately 35,000, and the remaining mass is accounted for by O- and N-linked sugars. The protein is so heavily sialated that digestion with neuraminidase results in an increase in its apparent mass because of a decrease in the overall negative charge. Polyclonal antibodies made to gp150 expressed as a recombinant protein in vaccinia virus have no effect on transformation of B lymphocytes (11), implying that the protein probably does not play a major role in virus entry into this cell type. It remains possible, however, that the protein plays a role in virus assembly or egress or that it is uniquely required for infection of the other major target of EBV, the epithelial cell.
Generation of recombinant virus with a selectable marker in the BDLF3 ORF.
Over the last several years, a variety of strategies for abolishing the expression of proteins encoded by EBV have been developed. Most recently, we and others have begun to make use of the highly inducible Akata strain of virus as a background for mutation (26, 31). This has allowed us to produce recombinant virus in sufficient quantity to do an analysis of its behavior at different steps in the replication cycle and explore phenotypes that may have only subtle effects on virus behavior. To determine whether gp150 plays any essential role in replication of EBV in tissue culture, the BDLF3 ORF in the Akata strain of EBV was interrupted. Akata cells (a gift of John Sixbey, St. Jude Children’s Research Hospital, Memphis, Tenn.) were transfected with a 4.5-kb SacII fragment of Akata virus DNA which encompassed the BDLF3 ORF (Fig. 1) and included a neomycin resistance cassette inserted into a unique BsaBI site, 46 bp from the initiation codon of gp150, where it would interrupt the protein within its predicted signal peptide (11). Cells in which the fragment had recombined with viral episomes or host cell DNA were selected by growth in medium containing G418 as previously described (31). A total of 13 G418-resistant clones were derived from 7,488 wells of transfected cells, and each was examined by Southern blotting (Fig. 1) for evidence of homologous or illegitimate recombination with cellular or viral DNA (one clone contained a virus episome or episomes in which illegitimate recombination had occurred). One parental clone had a restriction pattern that was consistent with homologous recombination. This clone was maintained simultaneously in medium containing 1.5 mg of G418 per ml and 50 μM hydroxyurea to reduce the episome copy number while selecting for retention of those that had undergone recombination (4). Of 32 clones screened by Southern blotting for the presence of a mixture of wild-type and recombinant episomes or pure recombinant episomes, 31 contained recombinant episomes alone. Akata cells containing pure recombinant episomes were also obtained by the more conventional technique of inducing replication in the parental clone, rescuing released virus in EBV-negative Akata cells (a gift of Jeffrey Sample, St. Jude Children’s Research Hospital), and reselecting with G418. Twelve drug-resistant clones were obtained from the 384 wells seeded, and each contained only recombinant episomes. Restriction patterns typical of cells harboring only wild-type episomes, a mixture of both wild-type and recombinant episomes, or only recombinant episomes are shown in Fig. 2. Clones of cells carrying pure recombinant episomes were also evaluated by indirect immunofluorescence for the percentage of the population that could be induced to make late lytic cycle proteins after treatment with anti-human immunoglobulin. The percent inducibility varied extensively, from 0 to approximately 40%. Those with high levels of inducibility were studied further.
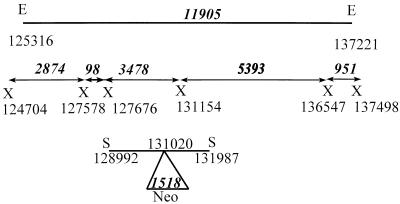
FIG. 1.
Diagram of the positions of the XhoI (X) restriction sites, numbered according to the B95-8 sequence, that surround the SacII (S) fragment shown at the bottom of the figure that was used for homologous recombination. The neomycin resistance cassette (Neo) was inserted in the BDLF3 ORF at position 131020 in the SacII fragment. Numbers in italics refer to the predicted sizes of the fragments resulting from digestion of wild-type virus DNA. The 11.9-kb EcoRI (E) fragment at the top of the figure was used as a probe of DNA digested with XhoI to determine whether homologous recombination had occurred and increased the size of the 3.5-kb XhoI fragment to 5.0 kb.
FIG. 2.
Southern blot analysis of DNA extracted from Akata cells harboring wild-type episomes (W), a parental clone of Akata harboring a mixture of wild-type and recombinant episomes (W/R), and two clones harboring pure recombinant episomes, derived either by infection of EBV-negative Akata cells (clone 2) (R) or by treatment of the parental clone with hydroxyurea (clone 26) (R/Hy). DNA was digested with XhoI, and the two identical halves of the membrane were cut apart and probed with either the EcoRI fragment (bp 125316 to 137221) of EBV (A) or the XmnI-HincII fragment containing the neomycin resistance gene (B). Sizes (in kilobases) are indicated by the arrows.
Lack of expression of gp150 in cells carrying recombinant virus episomes.
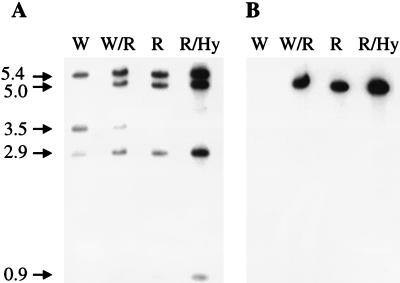
To confirm that expression of gp150 had been disrupted in cells that contained only recombinant episomes, the cells were induced with anti-human immunoglobulin, labeled with [3H]glucosamine, and immunoprecipitated, as previously described (31), with an antipeptide antibody, anti-gp150, which corresponded to the carboxyl-terminal residues 219 to 232 of gp150 (20), and with the following monoclonal antibodies (MAbs): 72A1 to gp350/220 (9), E2A5 to gp78 (16), and F-2-1 to gp42 within the gH-gL-gp42 complex (14). With the appropriate MAbs, the glycoproteins gp350 and gp78 and the gp85 (gH)-gp42-gp25 (gL) complex could be immunoprecipitated from cells expressing recombinant virus proteins. However, although anti-gp150 immunoprecipitated gp150 from cells expressing wild-type virus proteins, none of the protein was recovered from cells expressing recombinant virus (Fig. 3).
FIG. 3.
Electrophoretic analysis of proteins immunoprecipitated from Akata cells harboring wild-type or recombinant episomes by MAb 72A1 to gp350, MAb F-2-1 to gp42 and the associated proteins gp85 and gp25, MAb E2A5 to gp78, and rabbit antibody to gp150. The cells were induced with anti-human immunoglobulin and labeled with [3H]glucosamine. The numbers and arrows at the right indicate the masses of the immunoprecipitated proteins in kilodaltons.
Recombinant virus is not impaired in egress.
Virus glycoproteins may be involved in the assembly of virus and its exit from cells. To determine whether gp150 plays any critical role in either of these processes, a slot blot assay (31) was used to compare, for wild-type and recombinant viruses, the amounts of virus DNA within the induced cell and the amounts released extracellularly as encapsidated virus. The measurements were made at 120 h postinduction for clones 2 and 26, derived by infection with EBV-negative Akata cells and by treatment of the parental clone with hydroxyurea, respectively, and for wild-type Akata cells. The ratio of external virion DNA, expressed as a percentage of that which remained cell associated, varied significantly between inductions for each cell line. However, with repeated analysis, no significant trends could be seen for any, indicating that the recombinant viruses had no significant lesions in egress (Table 1).
TABLE 1.
Slot blot analysis of the ratio of extracellular virion DNA to cell-associated virus DNA after induction of wild-type or recombinant virus
| Expt no. | Ratio of extracellular virion DNA to cell-associated virus DNA (102) for:
|
||
|---|---|---|---|
| Wild-type virusa | Recombinant clone 26 | Recombinant clone 2 | |
| 1 | 3.6 | 1.7 | 1.7 |
| 2 | 5.2 | 4.2 | 6.2 |
| 3 | 10.2 | 12.8 | 13.6 |
| 4 | 7.0 | 4.0 | NDb |
| Avg | 6.5 | 5.7 | 7.2 |
The relative amounts of virion DNA released from the wild type and recombinant clone 26 were similar for experiments 1 and 2 and were approximately half for recombinant virus in experiments 3 and 4. Blots of recombinant clone 2 were probed at a different time and cannot be directly compared.
ND, not determined.
Recombinant virus binds and infects B cells normally.
To determine whether recombinant virus could bind to B cells normally, virus was labeled intrinsically with [3H]thymidine and incubated with receptor-positive Raji cells (23) or receptor-negative P3HR1-C15 cells (8) (a gift of George Miller, Yale University, New Haven, Conn.) in the presence or absence of MAb 72A1 to gp350 as previously described (31). The amount of acid-precipitable radioactivity bound to Raji cells was similar for wild-type and recombinant viruses and in each case, by preincubation of virus with MAb 72A1, could be reduced to levels similar to those bound to receptor-negative P3HR1-C15 cells (Table 2). To determine whether recombinant virus could also infect B cells normally, the abilities of the wild-type and recombinant viruses to infect EBV-negative Akata were compared. Initial virus concentrations were adjusted to be equal as judged by slot blot analysis of virus DNA. The amounts of EBNA induced in EBV-negative Akata cells were compared by Western blotting as previously described (31). No reproducible differences in the abilities of the wild-type virus and the two clones derived by hydroxyurea treatment to induce EBNA were found (Fig. 4). The relative abilities of the wild-type and recombinant viruses to transform peripheral blood mononuclear cells were also tested as previously described (31). When adjusted to contain equal amounts of virus, both stocks had a titration endpoint of 1/128.
TABLE 2.
Binding of [3H]thymidine-labeled recombinant or wild-type virus to receptor-positive and -negative cells
| Virus | Acid-precipitable radioactivity (cpm) bound to:
|
|||
|---|---|---|---|---|
| Rajia
|
P3HR1-C15b
|
|||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | |
| Wild type | ||||
| Without MAb | 3,152 | 4,781 | 319 | 283 |
| With MAb 72A1 | 318 | 343 | NDc | ND |
| Recombinant clone 2 | ||||
| Without MAb | 2,127 | 3,532 | 467 | 636 |
| With MAb 72A1 | 532 | 491 | ND | ND |
| Recombinant clone 26 | ||||
| Without MAb | 2,822 | 2,912 | 285 | 591 |
| With MAb 72A1 | 374 | 391 | ND | ND |
Receptor-positive cells.
Receptor-negative cells.
ND, not done.
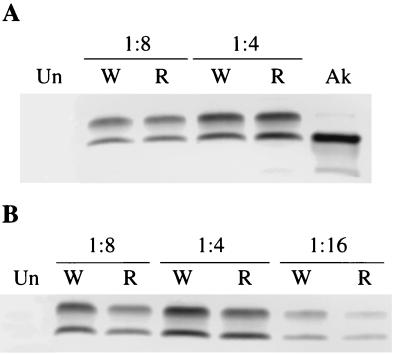
FIG. 4.
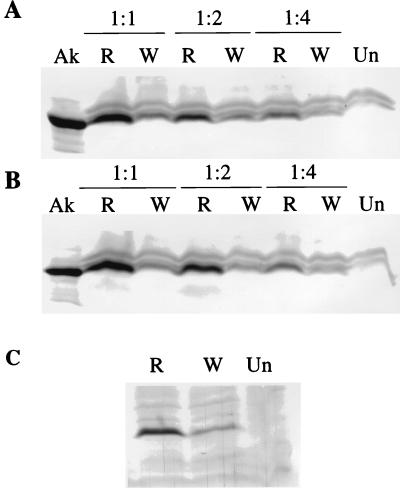
Induction of EBNA1 in EBV-negative Akata cells. Cells were mock infected (Un) or infected with wild-type virus (W) or with one of two different clones (one in panel A and the other in panel B) of recombinant virus (R) at the dilutions shown above the gels, and 5 days later they were harvested for Western blotting with human serum containing antibodies to EBNA1. The starting stocks of the viruses were adjusted so that they contained equal amounts of virus DNA. A sample of uninduced Akata cells (Ak) was included in panel A to demonstrate the mobility of EBNA1 in this strain.
Recombinant virus can infect epithelial cells more efficiently than wild-type virus.
Despite both the presence of EBV in all nasopharyngeal carcinomas (5, 24) and the high levels of virus replication seen in the epithelial lesions of oral hairy leukoplakia (25), there are few cultured epithelial cell lines that can be readily infected with EBV and none that consistently support the lytic cycle of virus replication. This is particularly unfortunate in that the BDLF3 transcript has been reported to be expressed at a high frequency in oral hairy leukoplakia biopsy specimens (22). However, the SVKCR2 cell line (15) (a gift of Alan Rickinson, University of Birmingham, Birmingham, England), derived from keratinocytes transformed with simian virus 40 and stably transfected with a construct expressing the B-cell receptor for EBV, CR2, does at least allow us to examine virus entry, since SVKCR2 cells express latent proteins after infection with wild-type virus. Equal amounts of wild-type and recombinant viruses were repeatedly tested at different dilutions for the ability to induce EBNA1 in SVKCR2 cells. The recombinant virus was slightly, but reproducibly, more efficient at inducing an EBNA1 signal than was wild-type virus (Fig. 5A and B). To determine whether this reflected a difference in the amounts of virus that could bind to cells or a subsequent event in infection, the relative amounts of the radiolabeled wild-type and recombinant viruses that could bind to the B-cell line Raji or the epithelial cell line SVKCR2 were evaluated. Although the ratios varied among experiments because of fluctuations in expression of CR2 on the transfected SVKCR2 cell line, no significant, reproducible difference in the relative abilities of the two virus types to bind to epithelial cells could be detected (Table 3).
FIG. 5.
Induction of EBNA1 in SVKCR2 cells and the AGS gastric carcinoma cell line. (A and B) SVKCR2 cells were mock infected (Un) or infected with either wild-type virus (W) or one of two different clones (one in panel A and the other in panel B) of recombinant virus lacking gp150 (R) at the dilutions shown above the gels. The starting stocks of the viruses were adjusted so that they contained equal amounts of virus DNA. Samples of uninduced Akata cells (Ak) were included to demonstrate the mobility of EBNA1 in this strain. (C) AGS cells were mock infected (Un) or infected with equal amounts of recombinant virus (R) or wild-type virus (W). In all cases, 5 days after infection, cells were harvested for Western blotting with human serum containing antibodies to EBNA1.
TABLE 3.
Relative amounts of [3H]thymidine-labeled wild-type and recombinant virus bound to receptor-positive Raji cells or SVKCR2 cells
| Virus | Acid-precipitable radioactivity (cpm) bound to Raji cells/acid-precipitable radioactivity bound to SVKCR2 cells in expt:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Wild type | 0.54 | 2.00 | 0.98 | 0.77 |
| Recombinant | 0.85 | 1.22 | 0.96 | 0.92 |
Very recently, certain gastric carcinoma cell lines have also been shown to be infectable by EBV (10, 34). The technique used has been to infect cells with a virus containing a neomycin resistance cassette in the nonessential thymidine kinase gene and to select for cells that become resistant to G418. To determine whether our findings with the SVKCR2 cell line were more generally applicable to epithelial cells and to examine epithelial cell infection in a more readily quantifiable manner, we infected six-well plates of 70% confluent AGS cells (a gift of Shosuke Imai, Hokkaido University School of Medicine, Sapporo, Japan) with equal amounts of recombinant virus lacking gp150 or a recombinant lacking gp42. The gp42-negative virus also contains a neomycin resistance cassette and has the same ability to infect SVKCR2 cells as does the wild-type virus (32). Infected cells were seeded in 96-well tissue culture plates and grown in medium containing G418. The number of wells containing G418-resistant cells was counted after 2 weeks. Approximately 10 times as many resistant cultures were obtained with the virus that lacked gp150 than resulted with the virus that lacked gp42 (Table 4). This finding was confirmed with wild-type virus by examining EBNA1 expression by Western blotting of cells that had been infected 5 days previously with equal amounts of wild-type virus or recombinant virus lacking gp150 (Fig. 5C). Again, the gp150-negative virus infected the cells more efficiently.
TABLE 4.
Comparison of the abilities of recombinant virus lacking gp150 and recombinant virus lacking gp42 to infect the AGS gastric carcinoma cell line
| Expt | Recombinant virus type | No. of G418-resistant cultures in three 96-well plates at a virus dilution ofa:
|
||
|---|---|---|---|---|
| 1/1 | 1/2 | 1/4 | ||
| 1 | gp150 negative | 48 | 24 | 11 |
| gp42 negative | 4 | 2 | NDb | |
| 2 | gp150 negative | 48 | 23 | ND |
| gp42 negative | 3 | 1 | ND | |
Starting stocks of virus were adjusted so that each contained the same amount of virus DNA.
ND, not done.
Sensitivity of gp150 to glycoprotease digestion.
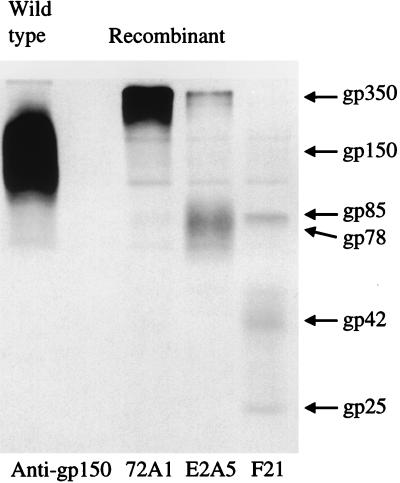
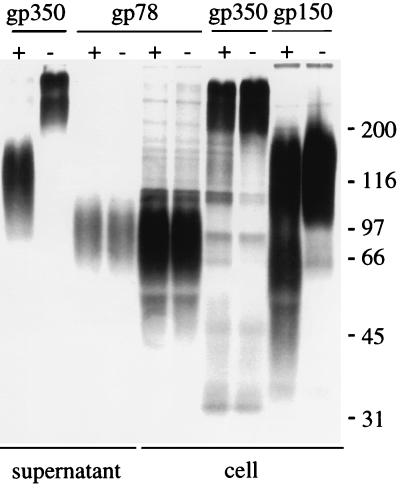
The resemblance of gp150 to a mucin has led to speculation that it may play a role in localization of virus or virus-infected cells to mucosal surfaces (11, 20). Mucin-like glycoproteins are the ligands for selectins, molecules that mediate low-affinity interactions with endothelial surfaces and begin the process of diapedesis. We previously hypothesized that a rare circulating B cell, shifting from the latent to the lytic cycle, might be induced by the expression of such a glycoprotein on its surface to traffic out of circulation into tissues such as the submucosal epithelium in the oropharynx and facilitate the shedding of virus in saliva (11). Since no critical role for gp150 in replication of virus in vitro could be demonstrated, the hypothesis that the glycoprotein might influence B-cell trafficking was evaluated further. Certain selectin ligands are susceptible to digestion with a unique O-sialoglycoprotein endopeptidase (glycoprotease) made by Pasteurella haemolytica which digests only cell surface, not soluble, mucin-like molecules (28, 29). The sensitivities of three EBV glycoproteins—gp350, which carries both N- and O-linked sugars; gp78, which carries only N-linked sugars; and gp150—were therefore compared by exposing induced Akata cells that had been labeled with [3H]glucosamine to this glycoprotease (a gift of Alan Mellors, University of Guelph) for 1 h at 37°C. The cells were pelleted, and medium supernatant and solubilized cells were subjected to immunoprecipitation with MAbs to gp350/220 and gp78, since the potential sites of cleavage relative to the antibody epitopes were unknown for these proteins. Solubilized cells alone were subjected to immunoprecipitation with anti-gp150, since the epitope recognized by this antibody is known to be on the intracellular cytoplasmic tail of gp150 (20). No cleavage of gp78 was detected in either cells or medium supernatant (Fig. 6). In contrast, both gp350 and gp150 were cleaved. The cleavage site on gp350 was carboxyl terminal to the 72A1 epitope, which has been tentatively mapped to the amino-terminal 162 residues, and was preserved in gp220, which is missing residues 500 to 757.
FIG. 6.
Electrophoretic analysis of proteins immunoprecipitated from Akata cells induced with anti-human immunoglobulin, radiolabeled with [3H]glucosamine, and treated with glycoprotease (+) or buffer alone (−). Cells were separated from the supernatant medium by centrifugation, and samples of supernatant and lysed cells were immunoprecipitated with antibodies to gp350, gp78, or gp150 as indicated. Sizes are indicated in kilodaltons.
gp150 does not bind E-selectin or P-selectin.
To test more directly whether the ability of cells expressing EBV glycoproteins to bind to cells expressing a potential homing receptor, such as E-selectin or P-selectin, had been altered, Akata cells were induced with anti-human immunoglobulin for 24 h and then, along with uninduced cells, tested for their ability to adhere to monolayers of CHO cells (a gift of Karen Bame, University of Missouri—Kansas City), CHO cells that stably expressed E-selectin or P-selectin (a gift of the Genetics Institute, Cambridge, Mass.), or freshly isolated human platelets prepared according to standard protocols (3). No difference in adherence of induced and uninduced Akata cells was seen, with few cells of either type adhering to monolayers. In contrast, HL60 cells (American Type Culture Collection), used as a positive control, demonstrated marked adherence to platelets as well as to CHO cells expressing E-selectin or P-selectin and did not adhere to untransfected CHO cells.
Conclusion.
The observations reported here establish the important fact that gp150 is not essential for replication in the B lymphocyte, despite the fact that this cell type could well be considered the most virus-specific niche of infection. Glycoprotein gp150 is also clearly not needed for infection of epithelial cells either, although there its behavior could only be studied in a more limited way. The finding that virus lacking gp150 could consistently infect epithelial cells more efficiently than wild-type virus was unexpected and curious. The relative amounts of both wild-type and recombinant viruses that bound to B cells and SVKCR2 cells were not significantly different, and although it is possible that subtle changes in the affinity for binding would not be picked up in the assay that was used, the fact that the finding was reproduced in a cell line to which EBV attaches in a CR2-independent manner suggests that a postbinding event is affected. Indeed, it is already known that there are significant differences in the postbinding events that occur on B lymphocytes and epithelial cells (32). The charge of a virus particle that lacks gp150, and thus lacks the large amount of sugar carried by that glycoprotein, is presumably very different from that of the wild-type virion and perhaps alters constraints on conformational changes in other proteins that are required for penetration.
While it remains possible that gp150 is critical to virus assembly or egress in epithelial tissues, it seems more likely at this point that the protein is nonessential. Its susceptibility to an enzyme which is known to cleave sialomucins was at least consistent with the hypothesis that expression of this molecule on lymphocytes in which the virus had moved into the lytic cycle alters interactions with selectins. However, no binding of cells expressing wild-type virus proteins to either E-selectin or P-selectin could be detected. While there may be as-yet-undiscovered adhesins for which gp150 can serve as a ligand, it also seems more likely that the molecule is not functioning to modify homing of infected cells either. The possibilities that remain for in vitro exploration of the role of that protein in EBV infections are limited. However, the conservation and expression of the BDLF3 gene in all isolates of EBV so far examined imply that it serves the virus in some function important to its persistence or spread within or between hosts. Examination of its homologs in very closely related viruses, such as the lymphocryptovirus of rhesus monkeys (18), may prove informative in this regard.
Acknowledgments
This research was supported by Public Health Service grant DE11116 from the National Institute of Dental Research.
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Beisel C, Tanner J, Matsuo T, Thorley-Lawson D, Kezdy F, Kieff E. Two major outer envelope glycoproteins of Epstein-Barr virus are encoded by the same gene. J Virol. 1985;54:665–674. doi: 10.1128/jvi.54.3.665-674.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalfamo J L, Dodds W J. Isolation of platelets from laboratory animals. Methods Enzymol. 1989;169:27–33. doi: 10.1016/0076-6879(89)69048-9. [DOI] [PubMed] [Google Scholar]
- 4.Chodosh J, Holder V P, Gan Y J, Belgaumi A, Sample J, Sixbey J W. Eradication of latent Epstein-Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J Infect Dis. 1998;177:1194–1201. doi: 10.1086/515290. [DOI] [PubMed] [Google Scholar]
- 5.Desgranges C, Wolf H, de The G, Shanmugaratnam K, Cammoun N, Ellouz R, Klein G, Lennert K, Munoz N, zur Hausen H. Nasopharyngeal carcinoma. X. Presence of Epstein-Barr virus genomes in separated epithelial cells of tumors in patients from Singapore, Tunisia and Kenya. Int J Cancer. 1975;16:7–15. doi: 10.1002/ijc.2910160103. [DOI] [PubMed] [Google Scholar]
- 6.Haddad R S, Hutt-Fletcher L M. Depletion of glycoprotein gp85 from virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J Virol. 1989;63:4998–5005. doi: 10.1128/jvi.63.12.4998-5005.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heineman T, Gong M, Sample J, Kieff E. Identification of the Epstein-Barr virus gp85 gene. J Virol. 1988;62:1101–1107. doi: 10.1128/jvi.62.4.1101-1107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heston L, Rabson M, Brown N, Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982;295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman G J, Lazarowitz S G, Hayward S D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc Natl Acad Sci USA. 1980;77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurilla M G, Heineman T, Davenport L C, Kieff E, Hutt-Fletcher L M. A novel Epstein-Barr virus glycoprotein, gp150, expressed from the BDLF3 open reading frame. Virology. 1995;209:108–121. doi: 10.1006/viro.1995.1235. [DOI] [PubMed] [Google Scholar]
- 12.Lake C M, Molesworth S J, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J Virol. 1998;72:5559–5564. doi: 10.1128/jvi.72.7.5559-5564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Spriggs M K, Kovats S, Turk S M, Comeau M R, Nepom B, Hutt-Fletcher L M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 16.Mackett M, Conway M J, Arrand J R, Haddad R S, Hutt-Fletcher L M. Characterization and expression of a glycoprotein encoded by the Epstein-Barr virus BamHI I fragment. J Virol. 1990;64:2545–2552. doi: 10.1128/jvi.64.6.2545-2552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller N, Hutt-Fletcher L M. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment of Epstein-Barr virus. J Virol. 1988;62:2366–2372. doi: 10.1128/jvi.62.7.2366-2372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moghaddam A, Rosenzweig M, Lee-Parrits D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 19.Nemerow G R, Mold C, Keivens Schwend V, Tollefson V, Cooper N R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolan L A, Morgan A J. The Epstein-Barr virus open reading frame BDLF3 codes for a 100-150 kDa glycoprotein. J Gen Virol. 1995;76:1381–1392. doi: 10.1099/0022-1317-76-6-1381. [DOI] [PubMed] [Google Scholar]
- 21.Oba D E, Hutt-Fletcher L M. Induction of antibodies to the Epstein-Barr virus glycoprotein gp85 with a synthetic peptide corresponding to a sequence in the BXLF2 open reading frame. J Virol. 1988;62:1108–1114. doi: 10.1128/jvi.62.4.1108-1114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penaranda M E, Lagenaur L A, Pierek L T, Berline J W, MacPhail L A, Greenspan D, Greenspan J, Palefsky J M. Expression of Epstein-Barr virus BMRF-2 and BDLF-3 genes in hairy leukoplakia. J Gen Virol. 1997;78:3361–3370. doi: 10.1099/0022-1317-78-12-3361. [DOI] [PubMed] [Google Scholar]
- 23.Pulvertaft R J V. Cytology of Burkitt’s tumor (African lymphoma) Lancet. 1964;i:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 24.Raab-Traub N, Flynn K, Pearson G, Huang A, Levine P, Lanier A, Pagano J. The differentiated form of nasopharyngeal carcinoma contains Epstein-Barr virus DNA. Int J Cancer. 1987;39:25–29. doi: 10.1002/ijc.2910390106. [DOI] [PubMed] [Google Scholar]
- 25.Rabanus J P, Greenspan D, Petersen V, Leser U, Wolf H, Greenspan J S. Subcellular distribution and life cycle of Epstein-Barr virus in keratinocytes of oral hairy leukoplakia. Am J Pathol. 1991;139:185–197. [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu N, Yoshiyama H, Takada K. Clonal propagation of Epstein-Barr virus (EBV) recombinants in EBV-negative Akata cells. J Virol. 1996;70:7260–7263. doi: 10.1128/jvi.70.10.7260-7263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spriggs M K, Armitage R J, Comeau M R, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson M R, Müllberg J, Cohen J I. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR β chain and inhibits antigen presentation. J Virol. 1996;70:5557–5563. doi: 10.1128/jvi.70.8.5557-5563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steininger C N, Eddy C A, Leimgruber R M, Mellors A, Welpy J K. The glycoprotease of Pasteurella haemolytica A1 eliminates binding of myeloid cells to P-selectin but not to E-selectin. Biochem Biophys Res Commun. 1992;188:760–766. doi: 10.1016/0006-291x(92)91121-6. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland D R, Abdullah K M, Cyopick P, Mellors A. Cleavage of the cell-surface O-sialoglycoproteins CD34, CD43, CD44 and CD45 by a novel glycoprotease from Pasteurella haemolytica. J Immunol. 1992;148:1458–1464. [PubMed] [Google Scholar]
- 30.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Hutt-Fletcher L M. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Kenyon W J, Li Q, Müllberg J, Hutt-Fletcher L M. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J Virol. 1998;72:5552–5558. doi: 10.1128/jvi.72.7.5552-5558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaswen L R, Stephens E B, Davenport L C, Hutt-Fletcher L M. Epstein-Barr virus glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology. 1993;195:387–396. doi: 10.1006/viro.1993.1388. [DOI] [PubMed] [Google Scholar]
- 34.Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688–5691. doi: 10.1128/jvi.71.7.5688-5691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]