Abstract
The mouse mammary tumor virus (MMTV) encodes within the U3 region of the long terminal repeat (LTR) a protein known as the superantigen (Sag). Sag is needed for the efficient transmission of milk-borne virus from the gut to target tissue in the mammary gland. MMTV-infected B cells in the gut express Sag as a type II transmembrane protein that is recognized by the variable region of particular beta chains (Vβ) of the T-cell receptor (TCR) on the surface of T cells. Recognition of Sag by particular TCRs results in T-cell stimulation, release of cytokines, and amplification of MMTV infection in lymphoid cells that are needed for infection of adolescent mammary tissue. Because the C-terminal 30 to 40 amino acids of Sag are variable and correlate with recognition of particular TCR Vβ chains, we prepared a series of C-terminal Sag mutations that were introduced into a cloned infectious MMTV provirus. Virus-producing XC rat cells were used for injection of susceptible BALB/c mice, and these mice were monitored for functional Sag activity by the deletion of C3H MMTV Sag-reactive (CD4+ Vβ14+) T cells. Injected mice also were analyzed for mutant infection and tumor formation in mammary glands as well as milk-borne transmission of MMTV to offspring. Most mutations abrogated Sag function, although one mutation (HPA242) that changed the negative charge of the extreme C terminus to a positive charge created a weaker Sag that slowed the kinetics of Sag-mediated T-cell deletion. Despite the lack of Sag activity, many of the sag mutant viruses were capable of sporadic infections of the mammary glands of injected mice but not of offspring mice, indicating that functional Sag increases the probability of milk-borne MMTV infection. Furthermore, although most viruses encoding nonfunctional Sags were unable to cause mammary tumors, tumors were induced by such viruses carrying mutations in a negative regulatory element that overlaps the sag gene within the LTR, suggesting that loss of Sag function may be compensated, at least partially, by loss of transcriptional suppression in certain tissues. Together these results confirm the importance of Sag for efficient milk-borne transmission and indicate that the entire C-terminal region is needed for complete Sag function.
Mouse mammary tumor virus (MMTV) is transmitted through the germ line as integrated proviral DNA (endogenous viruses) or through maternal milk to susceptible offspring (exogenous or milk-borne virus) (10). Milk-borne MMTV infects B cells in gut-associated lymphoid tissue (29), where superantigen (Sag) is presented at the cell surface as a type II transmembrane glycoprotein in conjunction with major histocompatibility complex (MHC) class II protein (25, 32). The Sag-MHC complex interacts with particular variable regions of the beta chain (Vβ) of the T-cell receptor (TCR) on the surface of T cells, causing these cells to release cytokines and to proliferate (20, 25). The release of cytokines stimulates neighboring B and T cells to divide, creating additional target cells for MMTV integration and expanding the pool of cells that previously have been infected (20, 25). The infected lymphoid cells then act as a reservoir for infection of the mammary gland when this tissue begins development during puberty. Recent results have shown that both T cells and B cells are required for MMTV transmission from infected milk in the gut to the mammary gland (3, 14, 21). Other experiments have shown that injection of MMTV-infected CD4+ or CD8+ T cells as well as infected B cells will transfer viral infection to susceptible mice (60).
All known MMTVs encode Sag within the U3 region of the long terminal repeat (LTR) (5); this region also specifies many of the viral transcriptional regulatory sequences (6, 23, 36, 37, 42, 47). Expression of the endogenous MMTV Sag proteins results in the deletion of reactive T cells through the process of negative selection in the thymus (25), whereas expression of Sag protein from milk-borne virus is believed to result in stimulation and proliferation of cognate T cells followed by a gradual deletion of these cells (40). Thus, the complement of endogenous MMTV strains determines whether reactive T cells are available for exogenous MMTV infection (21). Indeed, previous experiments have shown that expression of the exogenous C3H MMTV sag gene from the germ line of transgenic animals is sufficient to prevent infection by milk-borne C3H virus (14).
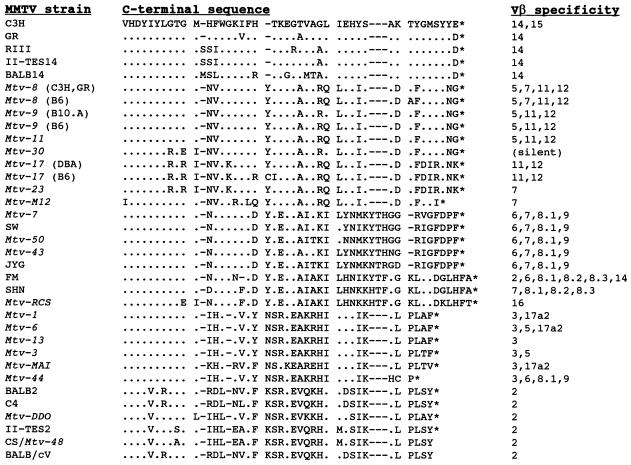
Sag is a type II transmembrane protein that contains a small N-terminal intracellular domain (32) and a large extracellular C terminus that interacts with the Vβ portion of the TCR (8). Sequence comparisons of the Sag proteins from several MMTV strains showed that there was a high degree of sequence identity between Sag molecules in two regions called polymorphic regions I (amino acids 164 to 198) and II (from amino acid 288 to the C terminus) (67). C-terminal variability in polymorphic regions I and II correlated well with observations that certain Sag molecules reacted with particular TCRs (67); e.g., the C3H and GR exogenous Sags reacted with T cells bearing Vβ14 chains (7). Moreover, experiments performed by Yazdanbakhsh et al. showed that substitution of the polymorphic region II of endogenous Mtv-1 Sag (Vβ3 reactive) for the polymorphic region II of Mtv-7 Sag (Vβ6 reactive) allowed the recombinant Sag to react with Vβ6+ T cells in stable transfection assays (67). The reciprocal experiment confirmed that polymorphic Sag region II (30 to 40 amino acids) at the C terminus is sufficient to specify interactions with certain TCR Vβ chains (67); however, the C-terminal half of Sag is insufficient to allow Sag function (34). Alignment of C-terminal Sag amino acids (Fig. 1) has been used to construct phylogenetic trees to predict relatedness among various MMTV strains (5).
FIG. 1.
Comparison of C-terminal sequences of MMTV Sag molecules and their TCR Vβ specificities. Amino acid identities are shown by dots compared to the C3H MMTV sequence. Sequence information was obtained from previous reports (1, 2, 17, 24, 26–28, 45, 48, 51, 62, 68, 69). Every effort was made to correct amino acid sequences from primary references. Gaps have been introduced (shown as dashes) to maximize amino acid identities. An asterisk indicates the position of the stop codon.
Although the C-terminal region of Sag has been shown to specify interactions with the TCR (67), little is known about the amino acid requirements within this region for Sag function. Therefore, a series of BglII linker substitutions and deletions were created within the region encoding the C terminus of Sag, and these substitution mutants were transferred into a cloned, infectious MMTV provirus (50) for in vivo analysis of C3H Sag function and effects on MMTV transmission and tumorigenicity. These experiments showed that virtually all C-terminal amino acid substitutions abolished Sag function and that sag mutant viruses failed to induce tumors in injected mice. However, mutants that lacked Sag function, but had overlapping mutations in a negative regulatory element (NRE) affecting MMTV transcription (4, 37), retained the ability to induce mammary tumors in mutant-injected animals. All mutants, except one that affected the C-terminal three amino acids and retained partial Sag function (HPA242), lost the ability to be transmitted through milk to susceptible offspring. Thus, virtually any mutation within the C-terminal region alters Sag function.
MATERIALS AND METHODS
Mice.
BALB/cJ mice were purchased from Jackson Laboratories (Bar Harbor, Maine). All animals were bred and maintained in our colony at The University of Texas at Austin Animal Resources Center. Animals were tested at intervals and were free of common bacterial and viral pathogens, including mouse hepatitis virus. Mice were injected with 2 × 106 XC cells expressing MMTV proviral constructs into each of five sites, four subcutaneous injections near the mammary glands proximal to each leg and one intraperitoneal injection (107 cells in total), as described by Shackleford and Varmus (50). All injected females were bred continuously to stimulate lactogenic hormones and MMTV production. Animals were palpated weekly for the appearance of mammary tumors.
Antibodies and flow cytometry analysis.
Injected mice and their progeny were bled from the retro-orbital sinus at 1-month intervals. The peripheral lymphocytes were purified on a Histopaque cushion (Sigma Chemical Co., St. Louis, Mo.) and then stained with phycoerythrin-conjugated CD4 and fluorescein-conjugated Vβ14 antibodies (PharMingen, San Diego, Calif.) as described previously (61). In some cases, when mice were sacrificed for mammary gland analysis, lymph node cells were obtained and subjected to dual staining with CD4 antibody and a panel of fluorescein-conjugated antibodies specific for Vβ2, -5, -6, -7, -8, -9, and -14 (all obtained from PharMingen).
Construction and growth of plasmid constructs.
Construction of the BglII substitution mutants p852, p867, p899, p909, p924, and p907/924 in the p19-LUC vector (57) was as described previously (4) except that the 907/924 mutation was CTAGATCTTAGAACATTCAGATCTG instead of GAGATCTGTAGAACATTCAGATCTG. The pA series of deletion constructs were prepared by digesting the wild-type C3H LTR in pLC1 (43) at the AflII site (−201 relative to the start of MMTV transcription at the U3/R junction) with Bal 31 (New England Biolabs, Beverly, Mass.). Digestion was terminated at intervals by the addition of EGTA (final concentration of 20 mM), and the Bal 31 nuclease was removed by digestion with proteinase K, phenol extraction, and ethanol precipitation. The linear ends of the DNA were filled with Klenow fragment of DNA polymerase (GIBCO BRL, Gaithersburg, Md.), and the DNA again was phenol extracted, precipitated with ethanol, and cleaved with ClaI. The pLC1 vector was cleaved with AflII, the ends were filled, and the DNA then was digested with ClaI. The large vector fragment was purified, ligated to the Bal 31-digested fragment, and used to transform Escherichia coli to ampicillin resistance. Plasmids from individual transformants were recovered and sequenced to determine the exact ends of the deletion. Mutant LTRs were substituted into the 3′ LTR of the cloned infectious provirus of Shackleford and Varmus (50), using mutant LTRs subcloned into pUC19 (59) as an intermediate; the infectious provirus contains the 5′ LTR and gag-pol region of Mtv-1 and the envelope region and 3′ LTR of C3H MMTV.
Transfections.
Supercoiled wild-type or mutant DNAs (1.8 μg) and 0.2 μg of selectable pSV2neo DNA (54) were transfected into rat XC cells (31), using Lipofectin (GIBCO BRL) as recommended by the manufacturer. Cells were selected in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah), streptomycin (50 μg/ml), penicillin (100 U/ml), and G418 (1 mg/ml; GIBCO BRL) until the appearance of discrete colonies. Colonies were pooled, and a portion was extracted for RNA to compare expression levels with those of wild-type plasmid transfectants. We also measured the amount of virus produced by Western blotting, by RNase protection experiments, and by reverse transcriptase assays, but the amount produced was too low to measure accurately, whereas the RNase protection assays (RPAs) described here were very quantitative, and the results did not depend on an enzymatic assay. We have observed that MMTV infection by inoculation of wild-type-transfected XC cells is faster when the XC cells are producing more intracellular MMTV RNA as measured by the RPAs (data not shown), which suggests that virion production is proportional to intracellular RNA levels.
RNA extractions and RPAs.
RNA extractions were performed essentially as described by Xu et al. (63) by the single-step guanidinium method (30) except that an ethanol precipitation was used instead of an isopropanol precipitation. DNA and low-molecular-weight RNA were removed from samples by precipitation in 3 M sodium acetate, pH 6.0 (46). RNA was extracted from mouse milk by the method of Golovkina et al. (16) except that virus was purified over a 30% sucrose cushion prior to extraction. RNA levels were determined by absorbance readings at 260 nm. RPAs were performed as described by Yang and Dudley (66) except that hybridizations were performed at 56°C. The riboprobe contained the Sau3A fragment (−455 to −116 relative to the start of MMTV transcription) that includes the sag gene polymorphic region II (50) and the promoter proximal NRE (4).
DNA extractions and Southern blotting.
High-molecular-weight DNA was extracted from the HP907/924 tumor and subjected to blotting by the method of Southern (53) as described previously (11).
RT-PCRs and cloning of PCR products.
Reverse transcription-PCRs (RT-PCRs) were performed essentially as described by Xu et al. (63) except that a random hexamer was used instead of oligo(dT) for priming cDNA synthesis. RNAs were obtained from the mammary glands of HPA242-injected mice and progeny of injected mice. The primers used for PCR of the MMTV LTR U3 region were 5′ GGCATAGCTCTGCTTTGC 3′ and 5′ AACACTCAGAGCTCAGATCAGAACC 3′. RT-PCR products were cloned by using the pGEM-T vector (Promega, Madison, Wis.). After selection of independent colonies, plasmid DNA was extracted, purified by using a JETstar 2.0 Plasmid MIDI Kit 25 (PGC Scientific, Gaithersburg, Md.), and sequenced by the DNA Sequencing Facility (Institute for Cellular and Molecular Biology, The University of Texas at Austin), using fluorescently tagged dideoxynucleotides and an automated ABI Prism 377 DNA Sequencer (Perkin-Elmer, Foster City, Calif.).
RESULTS
In vivo Sag activity of Sag mutant proviral constructs.
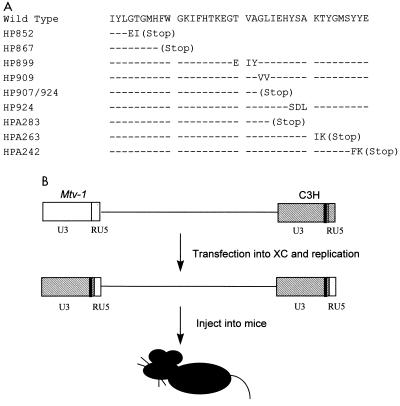
Because the C-terminal 30 to 40 amino acids of Sag correlates with recognition of particular TCR Vβ chains (Fig. 1) (5) and because this region directly has been shown by recombinant DNA switching experiments to confer Vβ specificity in in vitro transfection experiments (67), we performed deletion and linker substitution mutagenesis on the C-terminal 35 amino acids of Sag (Fig. 2A). Although some mutations truncated Sag due to the formation of a stop codon, other mutations consisted of a two- to three-amino-acid substitution. These mutations were inserted into the 3′ LTR of the infectious MMTV provirus described by Shackleford and Varmus (50) and transfected into rat XC cells, a cell line lacking endogenous MMTVs but permissive for virus replication (12). Since sag is encoded within the U3 region of the LTR, transcription of the integrated mutant virus, followed by proviral replication, will result in the duplication of the mutation within the 5′ LTR (58) (Fig. 2B). Although this strategy also may produce effects on the transcription of the integrated provirus, a number of these mutations previously have been shown to have no effect on basal or glucocorticoid-inducible expression of the virus (4). This was substantiated by using RPAs and RNA extracted from pools of XC cell transfectants (Fig. 3). Because of differences with the C3H LTR riboprobe, RPAs generated readily distinguishable RNase protection patterns (Fig. 3; compare lanes 1 and 3). Levels of wild-type MMTV (lane 1) and mutant expression (lanes 3 to 7, 9, 10, and 12) in XC cells were comparable. sag expression should not be affected directly by the LTR mutations since the cloned infectious provirus and C3H MMTV have been shown to express spliced sag mRNA only from an intragenic envelope promoter (13, 41) and not from the LTR (64). Also, because Sag is not believed to be a structural component of MMTV virions, these mutations should not affect production of virus particles.
FIG. 2.
Sequences and positions of C-terminal Sag mutants expressed from an infectious MMTV provirus. (A) Sequence of the final 40 amino acids of the C3H MMTV Sag (wild type) compared to the predicted sequence of each mutant. Dashes correspond to identity between the wild-type and mutant Sag molecules. The numbers for each mutant refer to the 5′ base at the beginning of the introduced substitution mutation relative to base 1 of the C3H LTR (5), whereas the deletion mutations (A series) are numbered in negative numbers relative to the start of transcription; i.e., HPA242 has a deletion from the AflII site to −242. (B) The 5′ half of the hybrid infectious provirus is composed of the 5′ LTR and gag-pol genes from the endogenous Mtv-1 provirus, whereas the 3′ end is composed of the env and 3′ LTR from a C3H MMTV provirus (50). Mutations are depicted as black bars within the U3 region of the LTR. Transcription from the 5′ LTR produces a genomic-length transcript that is packaged into virions, used for translation of the Gag, Gag-Pro, or Gag-Pro-Pol proteins, or spliced to give mRNAs for other viral proteins. After integration of the mutant plasmid DNA, genomic transcripts packaged into virions will enter new cells and be reverse transcribed into proviral DNA so that the unique regions U5 and U3 (and the U3-associated mutation) are duplicated.
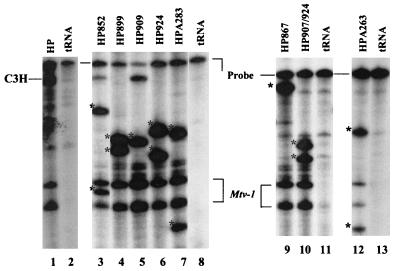
FIG. 3.
Equivalent transcription of mutant proviruses in stably transfected XC cells. Total RNA (20 μg) from transfected XC cells was subjected to an RPA using a riboprobe specific for the C3H LTR. Protection of the probe by sag mutant RNAs is indicated by asterisks. Yeast RNA (50 μg) was used as a negative control (lanes 2, 8, 11, and 13). Mtv-1 expression may be due to readthrough of the 5′ LTR in tandemly integrated proviruses or the initiation of transcription within the U3 region of the Mtv-1 LTR (19) of the hybrid provirus. RNA from the hybrid provirus and mutant HP867 gives full-length protection of the riboprobe. The appearance of full-length protection for HP852 and HP909 appears to be an artifact of this particular assay. XC rat cells lack endogenous MMTVs.
To test the activity of Sag mutant viruses in vivo, we used the strategy initially described by Shackleford and Varmus (50) to show the infectivity and tumorigenicity of their cloned MMTV provirus. Injection of purified virus was not considered for these experiments because of the unstable nature of MMTV and the loss of infectivity encountered during viral concentration steps (50). Therefore, transfected XC cells were inoculated subcutaneously and intraperitoneally into BALB/c weanling mice that were monitored for the deletion of C3H MMTV Sag-reactive (Vβ14+) T cells at different intervals after injection. Although the deletion of Sag-reactive cells had not been shown previously by this method, as expected, mice injected with XC cells transfected with the wild-type infectious hybrid provirus (HP/XC) showed a 37% reduction in the percentage of CD4+ Vβ14+ T cells compared to mice injected with untransfected XC cells at 12 weeks and a 52% reduction of cognate T cells compared to XC-injected controls at 24 weeks (data not shown). Therefore, Sag activity can be determined by injection of MMTV-expressing XC cells into adult BALB/c mice.
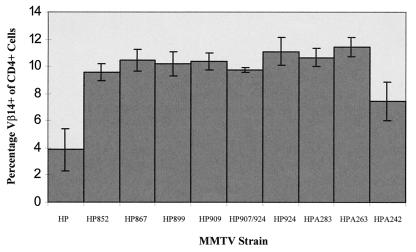
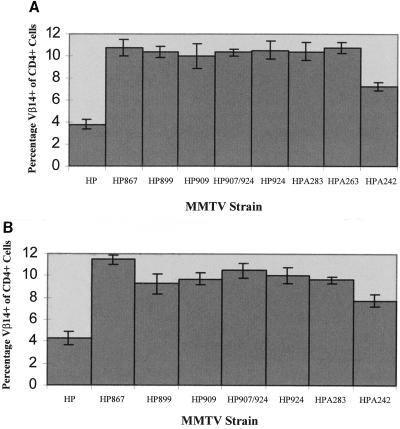
To maximize our ability to detect mutant Sag activity, mice inoculated with XC cells expressing sag mutant MMTVs were analyzed for deletion of Sag-reactive T cells (Fig. 4). With the exception of mutant HPA242, all mutants were unable to mediate the deletion of Vβ14+ T cells. Furthermore, only three of four mice injected with XC cells transfected with the HPA242 mutant deleted cognate T cells, whereas all mice of eight injected with transfectants expressing the wild-type provirus showed T-cell deletion with similar kinetics (data not shown). In addition, the deletion of Vβ14+ T cells in HPA242-injected mice was much less dramatic (6.8% ± 0.6% in infected mice) than that observed in mice injected with wild-type provirus transfectants (4.4% ± 0.8%). Therefore, it appears that alteration of almost any amino acid within the C-terminal 35 amino acids is sufficient to alter Sag function as measured by deletion of cognate T cells. Interestingly, the one mutation tested that appears to retain some Sag activity (HPA242) completely changed the charge at the end of Sag from a negatively charged glutamic acid to a positively charged lysine (Fig. 2A).
FIG. 4.
Deletion of Vβ14+ T cells in mice injected with XC transfectants or wild-type and sag mutant constructs. Peripheral blood lymphocytes were obtained from three to five mice at least 7 months postinjection to maximize the detection of potential Sag activity. Cells were stained with antibodies to CD4 and Vβ14 and analyzed by flow cytometry. Each number shown on the y axis is a percentage obtained by dividing the number of double-positive cells (CD4+ Vβ14+) cells by the number of CD4+ cells and then multiplying by 100. Standard deviations from the mean are given by error bars. Age-matched uninfected mice had 10.3% ± 0.4% Vβ14+ cells of CD4+ cells. The value given for HPA242 (7.45% ± 1.4% Vβ14+ T cells) includes one uninfected animal of four injected animals; exclusion of this animal gives 6.8% ± 0.6% Vβ14+ T cells. Only the wild-type- and mutant A242-injected animals had statistically different levels of Vβ14+ T cells compared to uninfected BALB/c controls.
Using C3H/HeN transgenic mice expressing the infectious hybrid provirus with a frameshift mutation in the first one-third of the sag gene (HYB PRO/Cla), Golovkina et al. showed that the HYB PRO/Cla did not delete C3H MMTV-cognate T cells, yet the offspring of these mice deleted Vβ14+ T cells (15). When virus from the C3H/HeN offspring was recovered and sequenced, the region containing the frameshift had reverted by recombination with endogenous Mtv-1 (15). Although BALB/c mice do not contain endogenous Mtv-1 (33), we analyzed the first and third litters of Sag mutant-injected mice for deletion of C3H Sag-reactive T cells to determine if recombination with endogenous MMTVs would generate functional Sags. Reversion was not observed in first- and third-litter offspring of mutant-injected mice (Fig. 5A and B, respectively), since only the wild-type- and HPA242-injected animals showed deletion of Vβ14+ T cells. Again, the Sag encoded by HPA242 was not as effective for cognate T-cell deletion as the wild-type C3H Sag, indicating that the mutant Sag had not recovered wild-type function by recombination.
FIG. 5.
Deletion of Vβ14+ T cells in first- and third-litter progeny of mice injected with XC cell transfectants. (A) The percentage of Vβ14+ CD4+ T cells was determined in first litters of mice injected with transfectants of wild-type or sag mutant constructs. All animals analyzed were at least 7 months old. Errors bars indicate standard deviation from the mean. Offspring of HP852 mutant-injected mice were not available for analysis. Age-matched uninfected mice had 10.3% ± 0.4% Vβ14+ cells of CD4+ cells. Only wild-type- and HPA242-infected animals had statistically different levels of Vβ14+ T cells compared to uninfected BALB/c controls. (B) The percentage of Vβ14+ CD4+ cells was determined in third litters of mice injected with transfectants of wild-type or sag mutant constructs. All animals analyzed were at least 7 months old, but offspring of HP852 and HPA263 mutant-injected mice were not available for analysis. Only wild-type- and HPA242-infected animals had statistically different levels of Vβ14+ T cells compared to uninfected BALB/c controls.
Infection of the mammary gland by Sag mutant MMTVs.
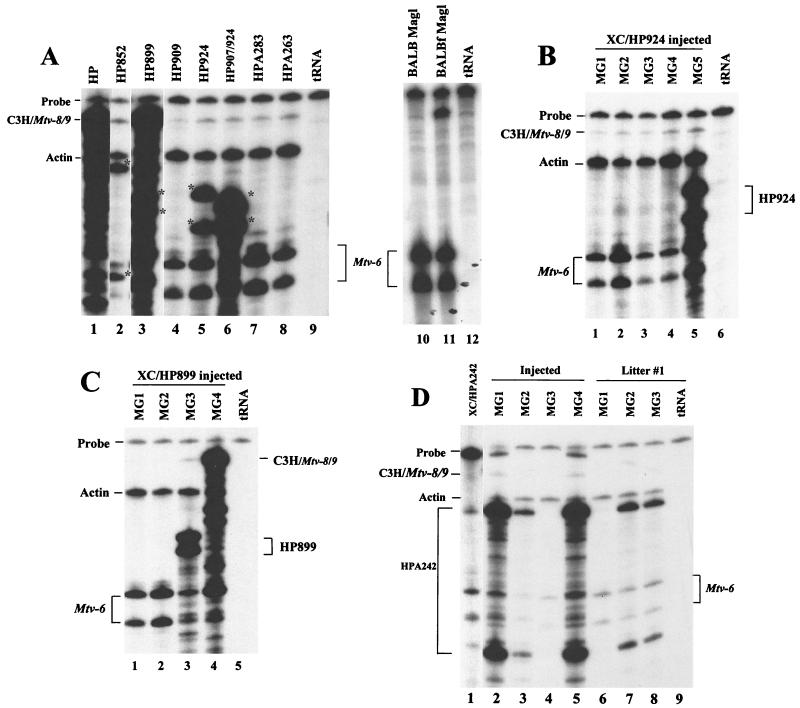
A considerable amount of evidence documents the need for Sag in the MMTV infectious cycle (3, 14, 15, 21). However, it is unknown whether MMTVs lacking functional Sag will infect after injection into adult mice. Therefore, mammary glands from sag mutant-injected mice were removed from adult (approximately 1-year-old) females, and total RNA was extracted. Subsequently, RNA was subjected to an RPA using a riboprobe that spans the LTR region containing sag mutations. Because RNase T1 cleaves 3′ to unpaired G residues, the riboprobe will be cleaved at regions that have a mismatch between the wild-type and mutant sequences. Mutant infection of the mammary gland was demonstrable by the appearance of protected fragments that migrated differently than those protected by actin and Mtv-6 RNA, e.g., HP852 (asterisks in Fig. 6A, lane 2); such fragments were characteristic of each mutant and were not observed in normal BALB/c mammary gland RNA (lane 10). Thus, results from RPAs showed that most sag mutant viruses were capable of mammary gland infection, despite their inability to delete cognate T cells.
FIG. 6.
RPAs of sag mutant expression in the mammary gland. (A) Mammary gland total RNA was extracted from mice injected with XC transfectants of sag mutant constructs. RPAs were performed with 10 to 40 μg of total RNA and a riboprobe specific for the C3H MMTV polymorphic region II. RNA pools were prepared from one to four injected animals (10 μg/animal). All samples were adjusted to a total of 50 μg by using yeast RNA. Partial protection of the probe by sag mutant RNAs is shown by asterisks, whereas the wild-type RNA gives full-length protection of the probe (lane 1). Other RNA samples showed small amounts of full-length protection of the riboprobe compared to uninfected BALB/c mammary gland (lane 10), perhaps due to the presence of recombinant MMTVs. RPAs from uninfected BALB/c and MMTV-infected (BALB) (4-month-old) (lane 11) mammary glands were shown previously (63); they are shown here for comparative purposes. Lanes 9 and 12 contained only yeast RNA. Actin riboprobe was included in each hybridization reaction as an internal control for RNA quality. Partial protection of the C3H riboprobe by endogenous Mtv-6 RNA also serves as an internal control; the position of the Mtv-6-specific fragments varies because lanes were derived from different gels. (B) Total RNA was prepared from the mammary glands of mice injected with XC transfectants with pHP924. RNA (20 μg) from individual mice was analyzed (lanes 1 to 5) as described for panel A. The partial protection of the riboprobe expected of the HP924 mutation is indicated at the right; the protection pattern expected of endogenous Mtv-6 is shown at the left. (C) Total RNA was prepared from the mammary glands of mice injected with XC transfectants with pHP899. RNA (20 μg) from individual mice was analyzed (lanes 1 to 4) as described for panel A. The partial protection of the probe expected of the HP899 mutations is shown at the right; protected fragments expected of endogenous Mtv-6 RNA are shown at the left. High-level expression of a recombinant virus can be observed in lane 4. (D) Total RNA was prepared from the mammary glands of mice injected with XC transfectants of pHPA242 (lanes 2 to 5) or three first-litter offspring of these injected mice (lanes 6 to 8). RNA (20 μg) from individual mice was used in an RPA with C3H MMTV-specific riboprobe (lanes 2 to 8). An RPA using 10 μg of total RNA from XC transfectants is shown as a control in lane 1; because the results of this RPA were analyzed on a separate gel, the HPA242-specific bands in lane 1 and those in lanes 2 to 8 migrate differently. Bands shown just below the probe in lanes 2 and 5 are an artifact. A longer exposure of the gel revealed expression of HPA242 in the mammary gland of mouse 1 of the first litter (not shown).
Although analysis of Vβ14+ T cells in injected mice indicated that sag mutants did not revert to give wild-type C3H Sag function, most mutants protected RNA fragments, consistent with the expression of endogenous Mtv-8 and/or Mtv-9 RNAs. Since expression of Mtv-8 and Mtv-9 RNAs is not detectable in normal BALB/c mammary glands (63), these results suggested that Mtv-8 or Mtv-9 recombined with the sag mutant viruses to produce novel MMTVs. Such novel MMTVs must lack Sags with reactivity for Vβ14+ TCRs.
Because mammary glands from several mice injected with the same mutant were pooled and analyzed (Fig. 6A), it was unclear whether all of the injected mice were infected. When mammary glands from mice injected with HP924 transfectants were analyzed individually, RPAs showed that only one of the five injected mice was infected by the mutant virus (Fig. 6B, lane 5). Similarly, only one of the four mice injected with HP899 transfectants was infected with predominantly mutant virus (Fig. 6C, lane 3), and another of the four mice appeared to be infected with high levels of a recombinant virus that protected a fragment typical of endogenous Mtv-8 or Mtv-9 (Fig. 6C, lane 4). Thus, the ability of MMTV to infect the mammary gland under these conditions is enhanced by, but does not require, Sag activity.
Although this was not a statistically significant difference, only three of the four mice injected with HPA242 transfectants showed deletion of Vβ14+ cells, whereas all mice injected with wild-type transfectants showed deletion of these cells (Fig. 4). Because of the observed variability of HPA242 infection and because infection by this mutant caused lower levels of deletion than that observed with the wild-type virus, it was possible that HPA242 virus, but not other viruses, gained Sag activity by recombination with endogenous MMTVs. Because HPA242 encodes a deletion of ∼40 bp, reversion of this mutation likely would change the RNase protection pattern of the expressed virus in the mammary gland relative to that observed in the HPA242-transfected cells. When the mammary glands of mice injected with HPA242 transfectants were analyzed individually by RPA, the protection pattern matched that of the virus expressed in the XC cell transfectants (Fig. 6D). Additionally, lack of revertant virus in the mammary glands of mice injected with HPA242 transfectants was confirmed by sequencing of RT-PCR products obtained using LTR primers and RNA from mammary glands of injected mice or progeny of these mice (data not shown). The data suggest that HPA242 encodes a functional Sag, since all mice infected by the HPA242 virus deleted Vβ14+ T cells, whereas an injected mouse that remained uninfected showed no Vβ14+ deletion. This conclusion is supported by our failure to detect reversion of the HPA242 mutation as well as by the similar low level of deletion observed in first- and third-litter offspring of HPA242-injected animals (Fig. 5).
Requirement for Sag activity during milk-borne MMTV transmission.
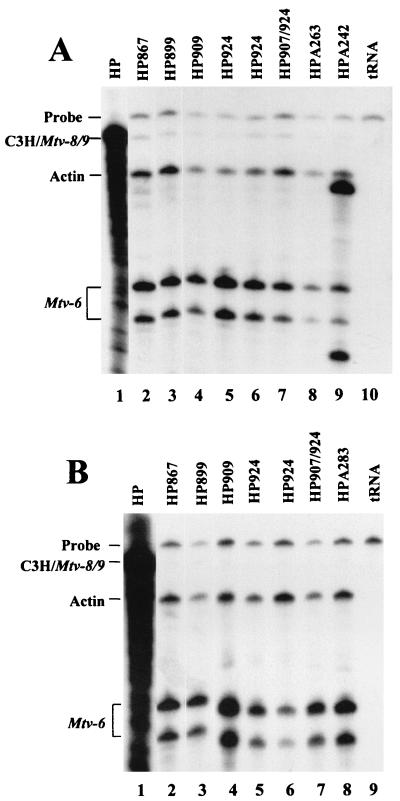
Because injection of mice with MMTV-infected cells may have bypassed the need for Sag, the offspring of injected mice were monitored for mammary gland infection. If Sag is needed to establish infection in gut-associated lymphocytes, neonates receiving virus lacking Sag activity will remain uninfected. Thus, mammary glands from first and third litters of mutant-injected mice were analyzed by RPA (Fig. 7A and B, respectively). Although some mutant-injected mice did not have offspring that could be examined, the available results indicated that MMTV infection occurred only in progeny of wild-type- and HPA242-injected animals (Fig. 7A, lanes 1 and 9, respectively; Fig. 7B, lane 1). Also HP867-injected mice showed mutant virus in the milk, but this virus was not transmitted to offspring (data not shown). Because progeny of wild-type- and HPA242-injected animals were the only offspring of injected animals to show Vβ14+ T-cell deletion, these results suggested that Sag is needed to establish infection in gut-associated lymphoid tissue. Although it is possible that the mammary glands of mice injected with virus lacking Sag activity had produced insufficient virus to establish viral infection in their offspring, our previous results with a sag frameshift virus argue against this explanation (15).
FIG. 7.
Mammary gland expression of sag mutants in first and third litters of mice injected with XC transfectants. (A) Mammary gland total RNA (10 to 40 μg) of first-litter offspring of injected mice was used for an RPA with a C3H MMTV-specific riboprobe. RNA from one to four females was pooled (10 μg/animal) and adjusted to a total of 50 μg by using yeast tRNA. Actin riboprobe was added to all samples as an internal control. Yeast tRNA was used as a negative control (lane 10). Pools of two different litters of HP924-injected mice are shown. (B) Mammary gland total RNA (10 to 40 μg) of third-litter offspring of injected mice was used for an RPA with a C3H MMTV-specific riboprobe. Other parameters were the same as those described for panel A.
Tumorigenesis in injected animals.
MMTV induces mammary tumors by insertion of proviral DNA near proto-oncogenes that are silent transcriptionally in mature mammary glands (35, 38, 39). Enhancers within the viral LTR apparently activate proto-oncogene transcription, leading to aberrant growth of mammary epithelial tissues. Because tumorigenesis is believed to be a multistep process (38), MMTV integration at other chromosomal sites increases the likelihood that initial growth stimulation will lead to the formation of tumor cells. Since integration near any given gene is a relatively random event, increased levels of retroviral replication and integration will increase the chance of tumor formation.
Because the mammary glands of injected mice were infected to different extents, animals were subjected to continuous breeding and monitored for the formation of mammary tumors. Approximately 63% (five of eight) of wild-type virus-injected animals developed mammary cancers with an average latency of 9 months (Table 1). This is similar to the average latency of MMTV-induced mammary tumors in C3H/HeN Mtv+ mice (9 months) (18) or BALB/c mice injected by XC cells producing the hybrid infectious MMTV provirus (57% of injected females had tumors with an average latency of 7.3 months) (65). Although Shackleford et al. observed a higher frequency of mammary tumors induced by injection of hybrid MMTV-producing XC cells (89% with an average latency of 6.5 months) (49), the increased tumor frequency may be due to inoculation of XC cells producing higher levels of MMTV (our unpublished results).
TABLE 1.
Mammary tumor incidence in sag mutant-injected mice
| Virus | Sag type | Tumor incidencea | Tumor latencyb |
|---|---|---|---|
| HP | Functional | 5/8 (63) | 9 (6–15) |
| HPA242 | Functional | 2/4 (50) | 12 |
| HP924 | Nonfunctional | 1/8 (13)c | 11 |
| HP907/924 | Nonfunctional | 2/3 (67) | 12 (11–13) |
| All others | Nonfunctional | 0/22 (0) | NA |
| Uninjected | None | <0.1%d |
Number of female mice developing mammary tumors divided by the number of injected females. Percentage of tumors is shown in parentheses. NA, not applicable.
Average latency of mammary tumors in months. Range for appearance of mammary tumors is shown in parentheses.
Only two of the injected females were confirmed to be infected; thus, tumor incidence in confirmed infected females was 50%.
Incidence of spontaneous mammary tumors in breeding BALB/c females.
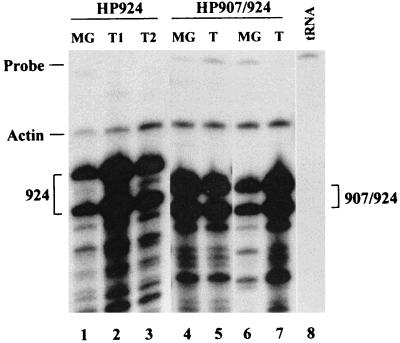
HPA242, the only sag mutant shown to have Sag activity, caused tumors in two of four injected mice (50%) with an average latency of 12 months (Table 1); however, only three of four injected mice had detectable MMTV infection. Most of the other mutants were unable to cause tumors, despite the ability of these mutants to infect the mammary glands following injection. Interestingly, mammary tumors were produced by viruses containing mutations in an LTR region shown to be important for negative regulation of MMTV expression in lymphoid tissues (37). Despite its lack of Sag activity, HP924 caused tumors in one of eight injected females (13%) with a latency of 11 months (Table 1); this tumor frequency is higher (one of two, or 50%) among females shown to be infected by RPAs. The HP924 mutation is located in the 3′ half of an imperfect inverted repeat within a promoter-proximal NRE (37). Mice injected with virus containing mutations in both the 5′ and 3′ halves of the inverted repeat (HP907/924) developed tumors at a frequency equivalent to that of viruses encoding functional Sags (67%) despite a longer latency (12 months) (Table 1). Mammary tumors induced by mutant viruses showed expression of the injected virus as detected by RPAs (Fig. 8, lanes 1 to 7), and integration of the mutant provirus was detectable by using a BglII restriction enzyme polymorphism and Southern blotting (data not shown). The tumors induced by mutant viruses were type B adenocarcinomas, typical of mammary tumors induced by wild-type MMTV (44). All viruses lacking Sag activity, except those containing mutations at the 924 site in the proximal MMTV NRE (4, 37), were unable to cause mammary tumors (0 of 22 injected mice).
FIG. 8.
Detection of sag mutants in mammary tumors of injected mice. Mice that developed tumors were sacrificed, and total RNA was extracted from the tumors and normal mammary gland. The RNA (20 μg) was subjected to an RPA using the C3H MMTV-specific riboprobe. One mouse injected with XC cells transfected with pHP924 developed two tumors (T1 and T2; lanes 2 and 3, respectively). Two tumors from individual mice injected with XC cells transfected with pHP907/924 were analyzed (lanes 5 and 7). Normal mammary gland (MG) RNA from each mouse also is shown (lanes 1, 4, and 6). Actin riboprobe was added to hybridizations as a control for RNA integrity. Yeast RNA was used as a negative control (lane 8).
DISCUSSION
Abrogation of Sag function by most C-terminal mutations.
All known MMTVs encode functional Sags, and comparison of the primary sequences of MMTV Sags shows high amino acid conservation in all but two polymorphic regions (5, 67). Only the C-terminal polymorphic 30 to 40 amino acids (region II) has been shown to determine the specificity of interaction with the variable portion of the TCR β chain (67). The viruses used in this study encoded mutations that spanned the C-terminal 35 amino acids. Since most mutations led to loss of MMTV Sag activity, including one mutant (HP909) encoding a two-amino-acid substitution, the entire C-terminal region appears to be important for Sag function, including TCR interactions. Nevertheless, we cannot exclude the possibility that the majority of C-terminal mutations tested prevent Sag presentation, since no reagents are available for the detection of C3H-specific Sag protein. Interestingly, the HPA242 mutation, encoding a basic lysine residue at the extreme C terminus of Sag, was functional, despite the fact that all other Vβ14-reactive Sags terminate with acidic residues (Fig. 1). However, this change impaired the ability of the HPA242 virus to cause deletion (Fig. 4 and 5).
All mice infected with HPA242 had similar residual populations of Vβ14+ T cells in their immune repertoire at 7 months postinfection (Fig. 4 and 5). Either the remaining population was unreactive with the HPA242 Sag, or these cells were reactive but their kinetics of deletion were very slow. What might distinguish the reactive versus the nonreactive Vβ14+ cells? Some Sags are unreactive with normally reactive TCR β chains when they are paired with particular α chains (55). If the HPA242 Sag stimulates only Vβ14+ T cells that expressed particular variable regions on their α chains, then deletion of Vβ14+ T cells would cease when the reactive cells have been depleted. This latter scenario did not occur since deletion of Vβ14+ cells in HPA242-infected mice never appeared to stop. However, the rate of deletion was twice as slow in these mice as in mice infected with the wild-type virus (data not shown). Therefore, it is likely that levels of Vβ14+ T cells in HPA242-infected mice eventually will equal that of the wild-type-infected mice with time.
Infection of the mammary gland by viruses lacking Sag activity.
The importance of Sag in the MMTV infectious cycle is well established. Golovkina et al. demonstrated the requirement of Sag-reactive T cells for MMTV infection (14), and subsequently the necessity of Sag-presenting B cells for MMTV infectivity was shown (3). Selection for functional Sags during milk-borne infection was observed following isolation of MMTVs encoding functional Sags from mammary glands of mice receiving MMTVs encoding a nonfunctional Sag (15). Although all evidence suggests that there is a strong selection for MMTVs encoding active Sag proteins, it is not known whether Sag is necessary for infection of mammary glands by MMTV. This study indicates that Sag is not required for mammary gland infection if virus-producing cells are introduced intraperitoneally and subcutaneously into susceptible mice. Clearly, MMTV infection of mammary glands resulted from injection of mice with XC cells expressing MMTVs encoding nonfunctional Sag proteins (Fig. 6). However, there was a direct correlation between functional Sag activity and the likelihood of mammary gland infection. All mice injected with wild-type transfectants showed mammary gland infection, whereas only three of four mice injected with HPA242 transfectants had infected mammary glands. Furthermore, the mammary glands of mice injected with transfectants expressing nonfunctional sag genes were infected sporadically (Fig. 6). Therefore, Sag activity greatly increases the probability that MMTV will infect its host.
It may be argued that sag mutant viruses infecting mammary glands sustained further mutations to restore Sag function and that this activity was overlooked by analyzing the Vβ14+ population. However, when the percentages of several other T-cell subsets expressing different Vβ chains were compared in infected versus uninfected animals, no significant differences were observed for the deletion of the particular T-cell subsets tested (Vβ2, -5, -6, -7, -8, -9, and -14), nor did we observe in any of the subsets analyzed a compensatory increase that would indicate deletion of a T-cell subset not tested (data not shown). Also, all viruses maintained their characteristic riboprobe protection pattern in RPAs with RNA extracted from mammary glands of injected animals (Fig. 6). Finally, if the infecting mutant viruses contained compensatory mutations or if the original mutation encoded a Sag of unknown specificity, it is likely that these viruses would be infectious for susceptible offspring. However, only the offspring of mice injected with MMTVs causing deletion of Vβ14+ T cells showed infection of the mammary glands by milk-borne MMTV (Fig. 7).
The lack of MMTV infection of the first and third litters of mice injected with Sag mutant-expressing cells suggested that Sag was necessary for infection of lymphoid cells in the gut. Alternatively, the newborns received a very limited amount of virus from the sag mutant-injected mothers. The limited amount of virus in maternal milk combined with the lack of functional Sag activity resulted in failure to transmit MMTV to offspring of most mutant-injected mice. This implies that Sag greatly increases the probability of MMTV infection, and without functional Sag, MMTV infection is sporadic and inefficient. Given the selection against MMTV-infected individuals due to the development of mammary tumors, transmission of virus to all members of a population eliminates the selective disadvantage of MMTV infection. Indeed, the loss of MHC class II I-E expression by some mouse strains (1) suggests that such a selective pressure (either MMTV or another pathogen encoding Sag) may have existed in the past.
Similarities in the N- and C-terminal portions of Sag have been used to deduce phylogenetic relationships among MMTV species (5). Given the sensitivity of the Sag C terminus to amino acid substitutions, our study indicates that C-terminal comparisons of MMTV Sags for phylogenetic studies probably are misleading. There is substantial pressure on MMTV Sag to encode a functional Sag that allows efficient infection of the mammary gland (reference 15 and this study), and Sag must react with a TCR displaying a Vβ specificity different from that of endogenous MMTV Sags expressed in the host (14, 21). This pressure combined with very limited combinations of amino acids that form a functional Sag would allow chance mutations in very divergent viruses to appear highly related. Therefore, similarities in the carboxyl terminus of MMTV Sags may be the result of convergent evolution. Phylogenetic comparisons of MMTV strains are more meaningful when the highly conserved regions of Sag or the viral structural genes are compared.
Tumor induction by MMTVs with NRE mutations.
Surprisingly, some MMTV mutants were able to cause tumors despite their lack of Sag activity. These mutants contained alterations in a region of the LTR (NRE) that is important for the negative regulation of MMTV transcription (4, 22). The NRE region is composed of promoter-proximal and promoter-distal elements that have binding sites for the nuclear matrix-associated region-binding proteins SATB1 and Cux/CDP. Both proteins are homeodomain-containing transcription factors that have been associated with tissue-specific expression of promoters to which they bind (9, 37, 52, 56). A mutation in the 3′ half of an imperfect inverted repeat in the proximal NRE (called 924) resulted in high-level expression from the C3H MMTV LTR in lymphoid tissue (37) and increased LTR-directed reporter gene expression in transient transfection assays (4). The 924 mutation and a mutation in both halves of the inverted repeat in the proximal NRE (907/924) were introduced into the wild-type infectious provirus and transfected into XC cells. Injection of XC cells producing the 924 and 907/924 viruses each caused mammary tumors (Table 1), despite the lack of detectable Sag activity encoded by these viruses.
The ability of NRE mutant viruses to cause tumors may be explained in at least two nonmutually exclusive ways. First, the increased expression of such viruses in lymphoid cells may lead to greater numbers of infected mammary cells and random mutagenic events leading to tumor formation. Second, MMTV expression in mammary tissue may be restricted to periods of pregnancy and lactation, since the NRE binding activities of SATB1 and Cux/CDP are detectable in virgin mammary gland (36a), but these activities are undetectable in the lactating mammary gland (37). Thus, NRE mutation may allow viral integration and expression at all stages of mammary development, whereas the presence of SATB1 and Cux in developing mammary gland may diminish wild-type MMTV expression in developing breast tissue. Loss of transcriptional suppression may result in increased MMTV integrations and increased tumorigenesis by NRE mutants; however, loss of Sag activity by such mutants partially may obscure the transcriptional advantage of NRE mutants over wild-type MMTV. Thus, we predict that NRE mutant viruses with functional Sag will develop tumors very rapidly. However, not all NRE mutations have the same effect, since the HP909 virus (also carrying a mutation in the proximal NRE) does not appear to induce mammary tumors (Table 1).
Why would MMTV encode elements, like the NRE, that suppress viral spread? While transcriptional suppression of MMTV may be a means to minimize immune system detection, it also is possible that loss of the NRE in the presence of a functional Sag yields MMTVs that cause tumors at a very young age (within a few litters). Since early tumorigenesis will limit MMTV spread by decreasing the number of offspring produced by an infected mouse, early-appearing mammary tumors likely will be an unwanted by-product of efficient MMTV replication. However, by suppressing MMTV expression until viral particles are needed for transmission during lactation, the presence of the NRE will decrease mortality compared to MMTV strains lacking this element and thereby increase the number of offspring receiving virus.
ACKNOWLEDGMENTS
We thank Susan Ross and Tatyana Golovkina for useful discussions and Lakshmi Rajan for the Southern blot analysis.
This work was supported by grants R01 CA34780 and R01 CA52646 from the National Institutes of Health. T.J.W. was supported by NIH training grant T32 CA09583.
REFERENCES
- 1.Acha-Orbea H, MacDonald H R. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 2.Ando Y, Wajjwalku W, Niimi N, Hiromatsu K, Morishima T, Yoshikai Y. Concomitant infection with exogenous mouse mammary tumor virus encoding I-E-dependent superantigen in I-E-negative mouse strain. J Immunol. 1995;154:6219–6226. [PubMed] [Google Scholar]
- 3.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bramblett D, Hsu C-L L, Lozano M, Earnest K, Fabritius C, Dudley J. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt-Carlson C, Butel J S, Wheeler D. Phylogenetic and structural analyses of MMTV LTR ORF sequences of exogenous and endogenous origins. Virology. 1993;193:171–185. doi: 10.1006/viro.1993.1113. [DOI] [PubMed] [Google Scholar]
- 6.Cavin C, Buetti E. Tissue-specific and ubitquitous factors binding next to the glucocorticoid receptor modulate transcription from the mouse mammary tumor virus promoter. J Virol. 1995;69:3759–3770. doi: 10.1128/jvi.69.6.3759-3770.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi Y, Kappler J W, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of the mouse mammary tumour virus. Nature. 1991;350:203–207. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 8.Choi Y, Marrack P, Kappler J W. Structural analysis of a mouse mammary tumor virus superantigen. J Exp Med. 1992;175:847–852. doi: 10.1084/jem.175.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickinson L A, Dickinson C D, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 10.Dudley J. Mouse mammary tumor virus. In: Webster R G, Granoff A, editors. Encyclopedia of virology. New York, N.Y: Academic Press; 1994. pp. 853–861. [Google Scholar]
- 11.Dudley J, Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984;49:92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudley J P, Varmus H E. Purification and translation of murine mammary tumor virus mRNA’s. J Virol. 1981;39:207–218. doi: 10.1128/jvi.39.1.207-218.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott J F, Pohajdak B, Talbot D J, Shaw J, Paetkau V. Phorbol diester-inducible, cyclosporine-suppressible transcription from a novel promoter within the mouse mammary tumor virus env gene. J Virol. 1988;62:1373–1380. doi: 10.1128/jvi.62.4.1373-1380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 15.Golovkina T V, Dudley J P, Jaffe A B, Ross S R. Mouse mammary tumor viruses with functional superantigen genes are selected during in vivo infection. Proc Natl Acad Sci USA. 1995;92:4828–4832. doi: 10.1073/pnas.92.11.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovkina T V, Jaffe A B, Ross S R. Coexpression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J Virol. 1994;68:5019–5026. doi: 10.1128/jvi.68.8.5019-5026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovkina T V, Piazzon I, Nepomnaschy I, Buggiano V, de Olano Vela M, Ross S R. Generation of a tumorigenic milk-borne mouse mammary tumor virus by recombination between endogenous and exogenous viruses. J Virol. 1997;71:3895–3903. doi: 10.1128/jvi.71.5.3895-3903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovkina T V, Prescott J A, Ross S R. Mouse mammary tumor virus-induced tumorigenesis in sag transgenic mice: a laboratory model of natural selection. J Virol. 1993;67:7690–7694. doi: 10.1128/jvi.67.12.7690-7694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunzburg W H, Heinemann F, Wintersperger S, Miethke T, Wagner H, Erfle V, Salmons B. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature. 1993;364:154–158. doi: 10.1038/364154a0. [DOI] [PubMed] [Google Scholar]
- 20.Held W H, Acha-Orbea H, MacDonald H R, Waanders G A. Superantigens and retroviral infection: insights from mouse mammary tumor virus. Immunol Today. 1994;15:184–190. doi: 10.1016/0167-5699(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 21.Held W, Waanders G A, Shakhov A N, Scarpellino L, Acha-Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 22.Hsu C L, Fabritius C, Dudley J. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J Virol. 1988;62:4644–4652. doi: 10.1128/jvi.62.12.4644-4652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang M, Lee J W, Peterson D O. Functional redundancy of octamer elements in the mouse mammary tumor virus promoter. Nucleic Acids Res. 1993;21:5235–5241. doi: 10.1093/nar/21.22.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatowicz L, Kappler J W, Marrack P, Scherer M T. Identification of two Vβ7-specific viral superantigens. J Immunol. 1994;152:65–71. [PubMed] [Google Scholar]
- 25.Janeway C A. Mls: makes a little sense. Nature. 1991;349:459–461. doi: 10.1038/349459a0. [DOI] [PubMed] [Google Scholar]
- 26.Jouvin-Marche E, Cazenave P A, Voegtle D, Marche P N. Vβ17 T-cell deletion by endogenous mammary tumor virus in wild-type-derived mouse strain. Proc Natl Acad Sci USA. 1992;89:3232–3235. doi: 10.1073/pnas.89.8.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jouvin-Marche E, Marche P N, Six A, Liebe-Gris C, Voegtle D, Cazenave P A. Identification of an endogenous mammary tumor virus involved in the clonal deletion of Vβ2 T cells. Eur J Immunol. 1993;23:2758–2764. doi: 10.1002/eji.1830231106. [DOI] [PubMed] [Google Scholar]
- 28.Kang J J, Schwegel T, Knepper J E. Sequence similarity between the long terminal repeat coding regions of mammary-tumorigenic BALB/cV and renal-tumorigenic C3H-K strains of mouse mammary tumor virus. Virology. 1993;196:303–308. doi: 10.1006/viro.1993.1480. [DOI] [PubMed] [Google Scholar]
- 29.Karapetian O, Shakhov A N, Kraehenbuhl J-P, Acha-Orbea H. Retroviral infection of neonatal Peyer’s patch lymphocytes: the mouse mammary tumor virus model. J Exp Med. 1994;180:1511–1516. doi: 10.1084/jem.180.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston R E, Chomczynski P, Sacchi N. Guanidinium methods for total RNA preparation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 4.2.1–4.2.9. [Google Scholar]
- 31.Klement V, Rowe W P, Hartley J W, Pugh W E. Mixed culture cytopathogenicity: a new test for growth of murine leukemia viruses in tissue culture. Proc Natl Acad Sci USA. 1969;63:753–758. doi: 10.1073/pnas.63.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korman A J, Bourgarel P, Meo T, Rieckhof G E. The mouse mammary tumour virus long terminal repeat encodes a type II transmembrane glycoprotein. EMBO J. 1992;11:1901–1905. doi: 10.1002/j.1460-2075.1992.tb05242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak C, Peters G, Pauley R, Morris V, Michalides R, Dudley J, Green M, Davisson M, Prakash O, Vaidya A, Hilgers J, Verstraeten A, Hynes N, Diggelmann H, Peterson D, Cohen J C, Dickson C, Sarkar N, Nusse R, Varmus H, Callahan R. A standardized nomenclature for endogenous mouse mammary tumor viruses. J Virol. 1987;61:1651–1654. doi: 10.1128/jvi.61.5.1651-1654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert J F, Acha-Orbea H, Kolb E, Diggelmann H. The 3′ half of the mouse mammary tumor virus orf gene is not sufficient for its superantigen function in transgenic mice. Mol Immunol. 1993;30:1399–1404. doi: 10.1016/0161-5890(93)90101-g. [DOI] [PubMed] [Google Scholar]
- 35.Lee F S, Lane T F, Kuo A, Shackleford G, Leder P. Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 mice. Proc Natl Acad Sci USA. 1995;92:2268–2272. doi: 10.1073/pnas.92.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J W, Moffitt P G, Morley K L, Peterson D O. Multipartite structure of a negative regulatory element associated with a steroid hormone-inducible promoter. J Biol Chem. 1991;266:24101–24108. [PubMed] [Google Scholar]
- 36a.Liu, J. Unpublished data.
- 37.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S R, Dudley J P. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacArthur C A, Shankar D B, Shackleford G M. Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol. 1995;69:2501–2507. doi: 10.1128/jvi.69.4.2501-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith G H, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrack P, Kushnir E, Kappler J. A maternally inherited superantigen encoded by a mammary tumour virus. Nature. 1991;349:524–526. doi: 10.1038/349524a0. [DOI] [PubMed] [Google Scholar]
- 41.Miller C L, Garner R, Paetkau V. An activation-dependent, T-lymphocyte-specific transcriptional activator in the mouse mammary tumor virus env gene. Mol Cell Biol. 1992;12:3262–3272. doi: 10.1128/mcb.12.7.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mink S, Hartig E, Jennewein P, Doppler W, Cato A C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morley K L, Toohey M G, Peterson D O. Transcriptional repression of a hormone-responsive promoter. Nucleic Acids Res. 1987;15:6973–6989. doi: 10.1093/nar/15.17.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nandi S, McGrath C M. Mammary neoplasia in mice. Adv Cancer Res. 1973;17:353–414. [Google Scholar]
- 45.Niimi N, Wajjwalku W, Ando Y, Nakamura N, Ueda M, Yoshikai Y. A novel Vβ2-specific endogenous mouse mammary tumor virus which is capable of producing a milk-borne exogenous virus. J Virol. 1995;69:7269–7273. doi: 10.1128/jvi.69.11.7269-7273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmiter R D. Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undegraded polysomes and messenger ribonucleic acid. Biochemistry. 1974;13:3606–3615. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- 47.Payvar F, DeFranco D, Firestone G L, Edgar B, Wrange O, Okret S, Gustafsson J A, Yamamoto K R. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983;35:381–392. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- 48.Rosenwasser O A, Fairchild S, Tomonari K. New superantigen specificity created by two amino acid replacements. Immunogenetics. 1993;38:367–369. doi: 10.1007/BF00210480. [DOI] [PubMed] [Google Scholar]
- 49.Shackleford G M, MacArthur C A, Kwan H C, Varmus H E. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 1993;90:740–744. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shackleford G M, Varmus H E. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherer M T, Ignatowicz L, Pullen A, Kappler J, Marrack P. The use of mammary tumor virus (Mtv)-negative and single-Mtv mice to evaluate the effects of endogenous viral superantigens on the T cell repertoire. J Exp Med. 1995;182:1493–1504. doi: 10.1084/jem.182.5.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skalnik D G, Strauss E C, Orkin S H. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 53.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 54.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 55.Vacchio M S, Kanagawa O, Tomonari K, Hodes R J. Influence of T cell receptor Vα expression on Mlsa superantigen-specific T cell responses. J Exp Med. 1992;175:1405–1408. doi: 10.1084/jem.175.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valarche I, Tissier-Seta J-P, Hirsch M-R, Martinez S, Goridis C, Brunet J-F. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 57.van Zonneveld A-J, Curriden S A, Loskutoff D J. Type 1 plasminogen activator inhibitor gene: functional analysis and glucocorticoid regulation of its promoter. Proc Natl Acad Sci USA. 1988;85:5525–5529. doi: 10.1073/pnas.85.15.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varmus H, Swanstrom R. Replication of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. Molecular biology of tumor viruses: RNA tumor viruses. 2nd ed. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory; 1985. pp. 75–134. [Google Scholar]
- 59.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 60.Waanders G A, Shakhov A N, Held W, Karapetian O, Acha-Orbea H, MacDonald H R. Peripheral T cell activation and deletion induced by transfer of lymphocyte subsets expressing endogenous or exogenous mouse mammary tumor virus. J Exp Med. 1993;177:1359–1366. doi: 10.1084/jem.177.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrona T, Dudley J P. Major histocompatibility complex class II I-E-independent transmission of C3H mouse mammary tumor virus. J Virol. 1996;70:1246–1249. doi: 10.1128/jvi.70.2.1246-1249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu L, Haga S, Imai S, Sarkar N H. Cloning in a plasmid of an MMTV from a wild Chinese mouse: sequencing of the viral LTR. Virus Res. 1994;33:167–178. doi: 10.1016/0168-1702(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 63.Xu L, Wrona T J, Dudley J P. Exogenous mouse mammary tumor virus (MMTV) infection induces endogenous MMTV sag expression. Virology. 1996;215:113–123. doi: 10.1006/viro.1996.0014. [DOI] [PubMed] [Google Scholar]
- 64.Xu L, Wrona T J, Dudley J P. Strain-specific expression of spliced MMTV RNAs containing the superantigen gene. Virology. 1997;236:54–65. doi: 10.1006/viro.1997.8717. [DOI] [PubMed] [Google Scholar]
- 65.Yanagawa S-I, Kakimi K, Tanaka H, Murakami A, Nakagawa Y, Kubo Y, Yamada Y, Hiai H, Kuribayashi K, Masuda T, Ishimoto A. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J Virol. 1993;67:112–118. doi: 10.1128/jvi.67.1.112-118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J N, Dudley J. Endogenous Mtv-8 or a closely linked sequence stimulates rearrangement of the downstream Vκ9 gene. J Immunol. 1992;149:1242–1251. [PubMed] [Google Scholar]
- 67.Yazdanbakhsh K, Park C G, Winslow G M, Choi Y. Direct evidence for the role of COOH terminus of mouse mammary tumor virus superantigen in determining T cell receptor Vβ specificity. J Exp Med. 1993;178:737–741. doi: 10.1084/jem.178.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshimoto T, Nagase H, Nakano H, Matsuzawa A, Nariuchi H. A Vβ8.2-specific superantigen from exogenous mouse mammary tumor virus carried by FM mice. Eur J Immunol. 1994;24:1612–1619. doi: 10.1002/eji.1830240724. [DOI] [PubMed] [Google Scholar]
- 69.Yoshimoto T, Nagase H, Nakano H, Matsuzawa A, Nariuchi H. Deletion of CD4+ T cells by mouse mammary tumor virus (FM) superantigen with broad specificity of T cell receptor β-chain variable region. Virology. 1996;223:387–391. doi: 10.1006/viro.1996.0492. [DOI] [PubMed] [Google Scholar]