Abstract
The G protein of 23 strains of human respiratory syncytial virus isolated in Havana, Cuba, between October 1994 and January 1995 was analyzed at the antigenic and genetic level. All viruses reacted with 10 of 11 antibodies specific for the Long strain. Moreover, the G protein gene of the Cuban isolates had only five nucleotide differences from the sequence of the Long gene. The homogeneity of the Cuban isolates and their resemblance to an ancient strain, such as Long, are at odds with previous findings for viruses isolated in countries with a temperate climate and different socioeconomic status. The G proteins of three of four other viruses isolated in Havana 2 years later (1996) were also identical to those of the 1994-to-1995 isolates, and the fourth virus had a single extra nucleotide difference. This, again, is unusual, since no identical viruses had been isolated in different epidemics previously. The singular characteristics of the Cuban isolates reported here are discussed in terms of the epidemiological, climatic, and socioeconomic characteristics of Cuba.
Human respiratory syncytial virus (HRSV) isolates have been classified into two antigenic groups (A and B) by their reactivities with different panels of monoclonal antibodies (1, 15). Sequence comparison of reference strains showed that the attachment (G) protein is less conserved between viruses of the two antigenic groups than other gene products (12), including the other surface glycoprotein of virions (the fusion protein, F). For each group, the G protein also showed the highest antigenic and genetic divergence among viral isolates (reviewed in reference 14). Thus, recent studies of the molecular evolution of HRSV have focused on the G protein because it has the highest capacity to differentiate strains that may be identical in terms of other gene products. In addition, the G protein is one of the targets of neutralizing and protective antibodies; thus, sequence changes corresponding to antibody binding sites of the G protein may be related to immune selection of variants during natural propagation of the virus.
The most extensive analysis of HRSV G protein evolution has been done with isolates from antigenic group A (2, 6). The emerging idea from those studies is that viruses from different evolutionary lineages cocirculate in each epidemic and that there is a progressive accumulation with time of antigenic and genetic changes in the G protein of viruses belonging to the same lineage. However, the viruses used in those studies were isolated mainly in places with a temperate climate—where annual outbreaks of HRSV occur in winter months—and, in most cases, in developed countries (14). Very limited epidemiological data are available from tropical and developing countries, where HRSV infections may follow a different pattern (11).
Cuba has unique geographical, climatic, and socioeconomic characteristics that might influence the dissemination and evolution of HRSV (10). Thus, 23 viruses isolated during the period October 1994 to January 1995 and 4 viruses isolated in 1996 were included in an antigenic and genetic analysis of the G protein (Table 1). All viruses were isolated at the Instituto Pedro Kouri (Havana, Cuba) from specimens collected in three hospitals of the same region. The majority of isolates were from children under 1 year of age admitted to the hospital because of severe respiratory infections. Bronchiolitis was the most common disease in those children.
TABLE 1.
Cuban isolates of HRSV used in the present study
| Virusa | Date of isolation (day/mo/yr) | Age of patient | Disease |
|---|---|---|---|
| Cub/52/94 | 18/10/94 | 7 mo | Bronchiolitis |
| Cub/54/94 | 18/10/94 | 6 yr | URI (asthma)b |
| Cub/60/94 | 20/10/94 | 2 mo | URI |
| Cub/67/94 | 25/10/94 | 2 yr | Laryngitis |
| Cub/69/94 | 25/10/94 | 1 mo | URI |
| Cub/81/94 | 01/11/94 | 3 mo | Bronchiolitis |
| Cub/82/94 | 01/11/94 | 4 mo | Bronchiolitis |
| Cub/83/94 | 01/11/94 | 6 mo | Bronchiolitis |
| Cub/97/94 | 08/11/94 | 6 mo | Bronchiolitis |
| Cub/105/94 | 15/11/94 | 2 mo | Bronchiolitis |
| Cub/106/94 | 15/11/94 | 5 mo | Bronchiolitis |
| Cub/107/94 | 15/11/94 | 5 mo | Bronchiolitis |
| Cub/111/94 | 15/11/94 | 5 mo | Bronchiolitis |
| Cub/115/94 | 17/11/94 | Not known | URI |
| Cub/128/94 | 01/12/94 | 10 mo | Bronchiolitis |
| Cub/134/94 | 06/12/94 | 3 mo | URI |
| Cub/140/94 | 13/12/94 | 22 days | URI |
| Cub/141/94 | 13/12/94 | 4 mo | Bronchiolitis |
| Cub/151/94 | 20/12/94 | 8 mo | Bronchiolitis |
| Cub/5/95 | 17/01/95 | 5 mo | URI |
| Cub/8/95 | 17/01/95 | 9 mo | Bronchiolitis |
| Cub/10/95 | 19/01/95 | 5 mo | Bronchiolitis |
| Cub/11/95 | 19/01/95 | 6 mo | Bronchiolitis |
| Cub/104/96 | 02/04/96 | 5 yr | URI |
| Cub/195/96 | 08/10/96 | 2 yr | Bronchiolitis |
| Cub/201/96 | 11/10/96 | 10 mo | Bronchiolitis |
| Cub/220/96 | 29/10/96 | 3 mo | Bronchiolitis |
Viruses are designated by country (Cuba)/number/year of isolation.
URI, upper respiratory infection.
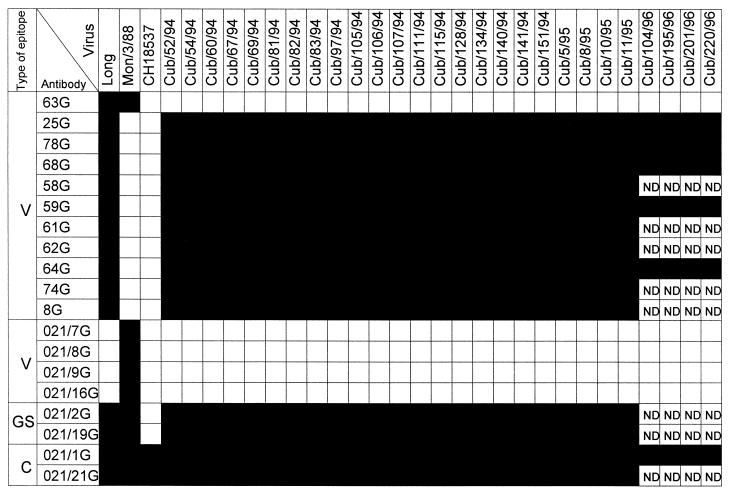
The reactivity of Cuban isolates with monoclonal antibodies specific for the G protein was assayed with a dot test (Fig. 1). Two reference strains of group A (Long and Montevideo/3/88) and one of group B (CH18537) were included in the same assay. All viruses reacted with antibodies that recognized conserved epitopes of the G protein shared by all human isolates. The Cuban isolates reacted with two antibodies (021/2G and 021/19G) that recognized different group-specific epitopes common to all viruses of antigenic group A. (Note that CH18537 did not react with these two antibodies.) Finally, the Cuban isolates reacted with 10 of 11 antibodies specific for the Long strain (except antibody 63G, whose epitope is shared by Mon/3/88 virus) and did not react with 4 other antibodies specific for the Mon/3/88 strain.
FIG. 1.
Reactivity of Cuban isolates with monoclonal antibodies specific for the G protein. HEp-2 cells were infected with the viruses indicated at the top. Cells were collected and extracts were made as previously described (13). Five microliters of extract dilutions was applied to Immobilon-P strips. These were saturated with 10% nonfat milk in phosphate-buffered saline and incubated with culture supernatant from the hybridomas indicated to the left. The bound antibody was visualized with biotinylated antimouse serum, streptavidin-conjugated peroxidase, and 4-chloro-1-naphthol. The isolation and characterization of the monoclonal antibodies used in this study have been described previously (7, 13). The epitopes recognized by these antibodies have been classified as conserved (C), group specific (GS), and strain specific or variable (V). ■, full reactivity; □, no reactivity. ND, not done.
The antigenic properties of the Cuban isolates were rather surprising, because none of the recent isolates from other countries tested to date reacted with antibodies 58G, 59G, 61G, 62G, 64G, 74G, and 8G (6). All of these antibodies recognize epitopes which are influenced by changes in the C-terminal two to three amino acids of the G glycoprotein (16). The sequence of the G protein of the Long strain has a unique stretch of amino acids at the C-terminal end (6), explaining the lack of reactivity of the antibodies described above with other HRSV strains. Another unique feature of the Cuban isolates was the lack of reactivity with antibody 63G. This epitope is conserved in most group A viruses, including Long and Mon/3/88 (Fig. 1), although occasionally strains of antigenic group A lacking reactivity with antibody 63G have been observed (6). The different reactivities of Cuban isolates and the Long strain with antibody 63G suggested that the former were not contaminated with the Long strain in the course of laboratory manipulations (however, see the results presented below).
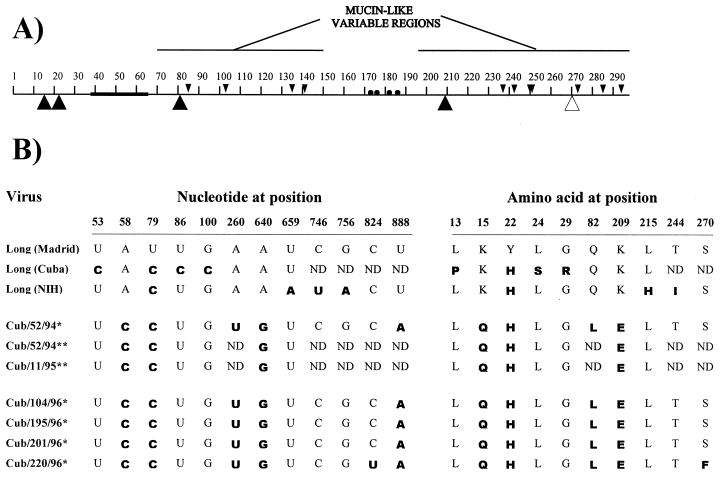
To understand the genetic basis of the unusual antigenic characteristics of the G protein from Cuban isolates, this gene was amplified by reverse transcription (RT)-PCR and sequenced for the virus Cuba/52/94 (Cub/52/94). There was a high degree of identity with the published sequence of the G protein of the Long strain available in Madrid, Spain (Fig. 2) (8). Only five nucleotide differences were found (positions 58, 79, 260, 640, and 888), and four were translated into amino acid changes, K15Q, Y22H, Q82L, and K209E (the change at position 888 was silent). The changes K15Q and Y22H were within the cytoplasmic domain. The change K209E is unique to the Cuban isolates, and it has not been found in the G protein of more than 30 group A isolates sequenced in our laboratory (6). Amino acid 209K is included in the sequence of a synthetic peptide that reproduced epitope 63G (9), explaining the lack of reactivity of the Cuban isolates with antibody 63G (Fig. 1).
FIG. 2.
G protein primary structure and sequence changes observed in Cuban isolates compared with those in Long virus. (A) G protein primary structure of the Long strain denoting the transmembrane region (thick line), the potential sites for N glycosylation (▾), the cysteine residues (•), the amino acid changes in Cuban isolates (▴), the unique change in isolate Cub/220/96 (▵), and the variable regions with sequence similarity to mucins (14). (B) Nucleotide and amino acid changes (boldface) of the G protein of indicated viruses compared with that of Long virus from the Madrid laboratory (see text for details). ∗, viral isolates; ∗∗, clinical samples. NIH, National Institutes of Health; ND, not determined.
The G protein gene of all Cuban viruses isolated during the 1994-to-1995 outbreak was amplified by RT-PCR and sequenced from nucleotide 533 to the end by using only dideoxyadenosine. The loss of A at nucleotide 640 and the gain of A at nucleotide 888 were detected in all isolates, and no other sequence changes were observed. The genetic homogeneity of 23 viruses from the same Cuban outbreak is in contrast with the results obtained with viruses from other places where strains belonging to different lineages are commonly isolated during the same epidemic.
The four viruses isolated in Cuba during 1996 (one in April and three in October) showed the same reactivity with monoclonal antibodies as the viruses from the 1994-to-1995 outbreak (Fig. 1). The entire G protein gene sequence of the four 1996 isolates was determined directly from the RT-PCR product. The four viruses have the same nucleotide changes that distinguished the 1994-to-1995 isolates from Long virus (Fig. 2). One of the 1996 viruses (Cub/220/96) had, in addition, the nucleotide change C824U, which led to the amino acid replacement S270F. This is also an unusual finding, since until now, viruses with identical G protein sequences have been isolated only during the same outbreak and in the same place. The genetic identity of the G protein from viruses isolated in Cuba 2 years apart is thus surprising.
Four clinical samples from the 1994-to-1995 epidemic were still available at the time these studies were being done. Amplification of the G protein gene directly from the clinical specimen was attempted with different primer pairs. Two overlapping gene segments, encompassing nucleotides 1 to 316 and 295 to 695, could be amplified from samples corresponding to isolates Cub/52/94 and Cub/11/95. Direct sequencing of the PCR products identified the diagnostic nucleotide changes at positions 58, 79, and 640 that were found in isolated viruses (Fig. 2). This result ruled out the possibility that viruses with singular sequence changes were selected during adaptation to grow in tissue culture.
The possibility of laboratory contamination of Cuban isolates by Long virus was also thoroughly investigated. (i) As mentioned before, partial cDNA sequences of clinical samples contained the diagnostic nucleotide changes that were found in the corresponding viruses. (ii) The Long strains available in Madrid (where antigenic and genetic analysis were carried out) and Havana (where virus isolation was done) were tested for reactivity with the monoclonal antibodies shown in Fig. 1. The Havana virus reacted with all of the antibodies specific for the Long strain, including 63G, that did not react with the Cuban isolates (data not shown). (iii) The G genes of the Long strains from Madrid and Havana were amplified by RT-PCR. Sequencing of the regions where nucleotide changes were observed in the Cuban isolates was done for the Long viruses from Madrid and Havana side by side. The sequence obtained for the G gene of the Long strain from Madrid was identical to that reported previously (8). The sequence of the G gene corresponding to the Long virus from Havana had four nucleotide differences (positions 53, 79, 86, and 100) compared with the Long virus from Madrid (Fig. 2). That the same viral strain from different laboratories contained minor sequence differences might reflect a different passage history since the isolation of Long virus in Baltimore, Md., in 1956 (4). In fact, we have noticed previously that there were four nucleotide differences between the G gene of Long virus from Madrid and that published by the National Institutes of Health group (12) (Fig. 2). However, the most important conclusion from this analysis is that none of the Long virus sequences were identical to those of the Cuban isolates. Thus, the possibility of a laboratory contamination of the Cuban isolates by Long virus was negligible by the three criteria presented above.
It is difficult, of course, to envisage the reasons for the singular properties of the Cuban isolates. The seasonality of HRSV infections differs between temperate zones and tropical countries (reviewed in reference 5). Whereas severe HRSV outbreaks are circumscribed to the winter months in places with a temperate climate (December to February in the Northern Hemisphere), HRSV infections are widely distributed throughout the year in Cuba, and virus circulation is observed from September to January and, to some extent, in May and June (10). The influence of seasonality on HRSV evolution is not known, but it is feasible that severe outbreaks of virus infections may contribute to generation of virus diversity through bottleneck selection imposed by large fluctuations in the size of the virus population. Interestingly, Cane has found that some viruses isolated in Gambia in 1994 were very similar to viruses isolated in Madrid 10 years before (1a, 3, 14).
Another factor that may contribute to the genetic stability of HRSV in Cuba is restriction of travel both to and outside of the island and within the country itself. In other studies, it has been observed that very similar viruses could be isolated in distant places (2, 6), indicating the capacity of HRSV to disseminate worldwide. However, limitations on travelling, as found in Cuba, may favor the stability of viruses.
It would be interesting to analyze isolates from other tropical countries with different socioeconomic characteristics. Of course, it would be also interesting to continue studies of molecular epidemiology of HRSV in Cuba to find out whether the viruses described here are maintained in future epidemics or are replaced by new variants.
Acknowledgments
O. Valdés and I. Martinez contributed equally to this work.
We thank Juan Ortín and Agustín Portela for critical reading of the manuscript.
This work was funded in part by grant BIO95-2066-E from Comisión Interministerial de Ciencia y Tecnología for Spanish-Cuban cooperation and grant CI1*-CT94-0012 from the European Union (DG12 HSMU).
REFERENCES
- 1.Anderson L J, Heirholzer J C, Tson C, Hendry R M, Fernie B N, Stone Y, McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 1a.Cane, P. A. Personal communication.
- 2.Cane P A, Pringle C R. Evolution of subgroup A respiratory syncytial virus: evidence of progressive accumulation of amino acid changes in the attachment protein. J Virol. 1995;69:2918–2925. doi: 10.1128/jvi.69.5.2918-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cane P A, Pringle C R. Molecular epidemiology of human respiratory syncytial virus. Semin Virol. 1995;6:371–378. [Google Scholar]
- 4.Chanock R M, Roizman B, Myers R. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent. I. Isolation, properties and characterization. Am J Hyg. 1957;66:281–290. doi: 10.1093/oxfordjournals.aje.a119901. [DOI] [PubMed] [Google Scholar]
- 5.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields’ virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1351. [Google Scholar]
- 6.García O, Martín M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Breña P, Martínez I, García-Barreno B, Melero J A. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol. 1994;68:5448–5459. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Barreno B, Palomo C, Peñas C, Delgado T, Perez-Breña P, Melero J A. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J Virol. 1989;63:925–932. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Barreno B, Portela A, Delgado T, López J A, Melero J A. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO J. 1990;9:4181–4187. doi: 10.1002/j.1460-2075.1990.tb07642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Barreno B, Delgado T, Akerlind-Stopner B, Norrby E, Melero J A. Location of the epitope recognized by monoclonal antibody 63G on the primary structure of human respiratory syncytial virus G glycoprotein and the ability of synthetic peptides containing this epitope to induce neutralizing antibodies. J Gen Virol. 1992;73:2625–2630. doi: 10.1099/0022-1317-73-10-2625. [DOI] [PubMed] [Google Scholar]
- 10.Goyenechea A, Bello M, Clua A, Savon C, Valdivia A, Oropesa S, Díaz O, Hernandez B. Determinación de anticuerpos fijadores de complemento al virus sincitial respiratorio. Estudio longitudinal en una población meno de 1 año en Ciudad de La Havana. Rev Cuba Med Trop. 1994;46:79–85. [PubMed] [Google Scholar]
- 11.Heirholzer J C, Tannock G A, Heirholzer C M, Coombs R A, Kennett M L, Phillips P A, Gust I D. Subgrouping of respiratory syncytial virus strains from Australia and Papua New Guinea by biological and antigenic characteristics. Arch Virol. 1994;136:133–147. doi: 10.1007/BF01538823. [DOI] [PubMed] [Google Scholar]
- 12.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez I, Dopazo J, Melero J A. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol. 1997;78:2419–2429. doi: 10.1099/0022-1317-78-10-2419. [DOI] [PubMed] [Google Scholar]
- 14.Melero J A, García-Barreno B, Martínez I, Pringle C R, Cane P A. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78:2411–2418. doi: 10.1099/0022-1317-78-10-2411. [DOI] [PubMed] [Google Scholar]
- 15.Mufson M A, Örvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 16.Rueda P, Palomo C, García-Barreno B, Melero J A. The three C-terminal residues of human respiratory syncytial virus G glycoprotein (Long strain) are essential for integrity of multiple epitopes distinguishable by antiidiotypic antibodies. Viral Immunol. 1995;8:37–46. doi: 10.1089/vim.1995.8.37. [DOI] [PubMed] [Google Scholar]