Abstract
It has been previously shown that a proline substitution for any of the conserved leucine or isoleucine residues located in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 (HIV-1) gp41 renders viruses noninfectious and envelope (Env) protein unable to mediate membrane fusion (S. S.-L. Chen, C.-N. Lee, W.-R. Lee, K. McIntosh, and T.-M. Lee, J. Virol. 67:3615–3619, 1993; S. S.-L. Chen, J. Virol. 68:2002–2010, 1994). To understand whether these variants could act as trans-dominant inhibitory mutants, the ability of these mutants to inhibit wild-type (wt) virus infectivity was examined. Comparable amounts of cell- and virion-associated gag gene products as well as virion-associated gp41 were found in transfection with wt or mutant HIV-1 provirus. Viruses obtained from coexpression of wt provirus with mutant 566 or 580 provirus inhibited more potently the production of infectious virus than did viruses generated from cotransfection of wt provirus with other mutant proviruses. Nevertheless, all viruses produced from mixed transfection showed decreased infectivity compared with that of the wt virus when a multinuclear-activation β-galactosidase induction assay was performed. The ability of wt Env to induce cytopathic effects was inhibited by coexpression with mutant Env. Coexpression of mutants inhibited the ability of the wt protein to mediate virus-to-cell transmission, as demonstrated by an env trans-complementation assay with a defective HIV-1 proviral vector. These observations indicated that mutant Env, per se, interferes with wt Env function. Moreover, cotransfection of wt and mutant proviruses produced amounts of cell- and virion-associated gag gene products comparable to those produced by transfection of wt provirus. Similar amounts of gp41 were also found in virions generated from wt-mutant cotransfection as well as from wt transfection alone. These results indicated that the inhibitory effect conferred by mutants on the wt virus infectivity does not involve the late steps of Gag protein assembly and budding, but they suggest that the wt and mutant Env proteins form a dysfunctional hetero-oligomer which is impaired in an early step of the virus replication cycle. Our study demonstrates that mutations in the HIV-1 gp41 leucine zipper-like heptad repeat sequence dominantly inhibit infectious virus production.
The envelope (Env) glycoprotein of human immunodeficiency virus type 1 (HIV-1), forming as a gp120-gp41 heterodimer, plays a crucial role in viral infectivity and cytopathicity, as well as in viral transmission, pathogenesis, and cell and tissue tropism. HIV replication is initiated by attachment of the virus to the cell surface through the binding of viral extracellular Env glycoprotein gp120 to the cell surface primary receptor CD4 and a CXCR4 or CCR5 coreceptor, a member of the chemokine receptor family (for reviews, see references 2, 6, 20, and 55). Subsequently, the transmembrane (TM) protein gp41 mediates membrane fusion between viral and host cell membranes, thereby depositing the viral core into the cytoplasm (for reviews, see references 40 and 41). Nevertheless, the mechanism underlying gp41-mediated membrane fusion is still not well understood.
The best-characterized mechanism of membrane fusion is that of influenza virus mediated by hemagglutinin (HA) (7, 9, 64). Infection by influenza virus begins with the HA-mediated binding of the virus to a sialylated cellular receptor present on the surface of target cells, followed by internalization of virions into cellular endosomes by receptor-mediated endocytosis. HA consists of a trimer of HA1 and HA2 heterodimers linked by a disulfide bond. HA2 is a membrane-anchoring subunit forming the trimeric coiled-coil structure responsible for the oligomerization of the protein and membrane fusion. The HA1 subunit covers the inner core of the HA2 trimer. A change to an acidic pH in endosomes induces conformational changes of HA, including the displacement of HA1 from HA2 and formation of the B-loop region of HA2 into an extended coiled-coil structure, and exposes the N-terminal hydrophobic fusion peptide that is initially buried inside the molecule to the target membrane. Finally, membrane fusion between virus and target cells is triggered.
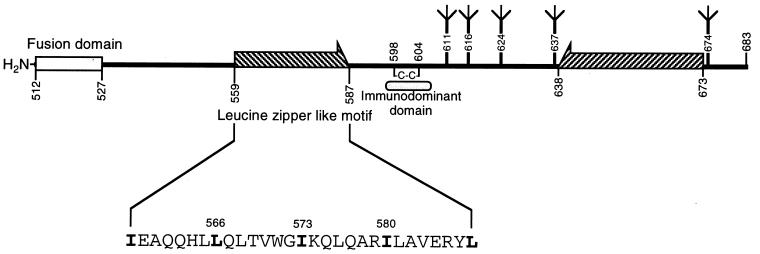
Topologically analogous to the trimeric stem region of HA2 in the HIV-1 TM protein is the leucine zipper-like motif, termed the zipper motif, located approximately 30 residues from the N-terminal fusion peptide sequence of gp41 (Fig. 1). This highly conserved motif, comprising a periodic repeat of leucine or isoleucine residues at every seventh position over eight helical turns, is explicit as a heptad repeat sequence, an extensive region containing nonpolar residues in all a positions and in most d positions when displayed on an α-helical wheel. Such an arrangement of hydrophobic residues within a putative α-helix was hypothesized to be structurally analogous to that of HA2, forming a coiled coil and playing a role in Env subunit assembly and virus fusion (18, 28).
FIG. 1.
Schematic representation of the HIV-1 gp41 ectodomain. Amino acid residues are numbered according to their positions in the HXB2 Env protein. The N-terminal leucine zipper-like heptad repeat sequence is shown as the hatched arrow directed to the right, and its amino acid sequence in single-letter code is indicated. The conserved leucine and isoleucine residues located in this motif, which are numbered and indicated by boldface, were each replaced by a proline residue and examined in this study. The C-terminal α-helical sequence is illustrated as the hatched arrow directed to the left. This domain forms a heterodimer with the N-terminal zipper motif α helix, and three molecules of heterodimers fold into a six-stranded helical bundle. Within the bundle the three N-terminal helices constitute a central, parallel, trimeric coiled-coil structure, whereas the three C-terminal helices pack antiparallelly into the hydrophobic groves on the surface of the N-terminal trimer. The N-glycosylation sites (Ψ) and intramolecular disulfide bond (C—C) are also shown.
Previously, to study the role of this hydrophobic heptad repeat sequence in the HIV-1 life cycle, we replaced each conserved leucine or isoleucine residue located in this region with a proline residue, which affected the α-helical structure more severely than any other amino acid. The results showed that all of the mutant viruses were severely impaired in virus infectivity and that mutant proteins were unable to mediate syncytium formation with CD4+ cells (14, 16). Nonetheless, all mutant proteins still formed oligomeric structures. Other investigators obtained similar findings when the middle isoleucine residue was replaced by amino acid residues that disrupted the α-helical structure less severely than the proline residue (21, 59). Studies on a peptide consisting of a segment corresponding to this heptad repeat region, DP107, and its proline substitution analog have shown that DP107 possesses anti-HIV activity and that the degree of the inhibitory effect correlates with the α-helix content in solution (62). Moreover, the destabilization effect of amino acid substitutions at Ile-573 on coiled-coil structures in peptide models correlates with the phenotype of virus infectivity and membrane fusion of mutants (59). These studies collectively indicate that this coiled-coil domain is important for virus infectivity and membrane fusion, although it is not involved in formation of a prefusogenic oligomer.
Interestingly, the zipper motif is conserved not only among HIV-1 isolates but also among the TM proteins of other retroviruses (18). The feature of heptad repeat sequences is also conserved in the fusion proteins of paramyxoviruses, influenza viruses, coronaviruses, and retroviruses (11, 42). Therefore, these conserved sequences may represent a class of structural motifs that can be targeted by similar antiviral therapeutic approaches.
Dominant negative inhibition is a phenomenon in which the functions of the wild-type (wt) gene products are inhibited or blocked by the coexpressed defective mutants of the same genes (33). trans-dominant negative inhibitors have been widely explored with various cellular and viral systems. A genetic intervention strategy based on trans-dominant negative mutants of HIV-1 proteins is an alternative to current vaccine development and pharmacological therapy for AIDS (26).
Numerous strategies to interfere with Env function have been proposed as potential anti-HIV therapies. One approach involves the use of trans-dominant mutant Env based on the oligomeric state of the Env proteins of primate immunodeficiency viruses (19, 22, 29, 31, 43, 49, 50). A polar substitution of Glu for Val at amino acid 2 of the hydrophobic fusion peptide sequence of gp41 inhibits production of infectious virus and syncytium formation induced by the wt Env (27). An HIV-2 Env mutant with an in-frame deletion in the CD4-binding site interferes with the wt virus infectivity (52). Recently, a gp41 cytoplasmic domain truncation Env variant was shown to inhibit wt virus infectivity (15). The mechanism responsible for the inhibitory effect conferred by the Env mutants involves interference with the functional assembly of oligomeric wt protein by coexpression with mutant Env, resulting in the formation of a nonfunctional Env protein complex (15).
In this study, we attempted to use the HIV-1 gp41 heptad repeat sequence as a model system to determine the feasibility of developing an antiviral strategy targeting this highly conserved sequence. Since the zipper motif Env mutants described previously are unable to mediate syncytium formation but still form oligomeric structures, the ability of these mutants to interfere with the wt virus infectivity was examined. The results showed that mutants with mutations in this region are able to dominantly inhibit the wt Env-mediated syncytia formation and virus entry into host cells. Also, the study may have implications for the design of trans-dominant negative Env mutants targeting the conserved heptad repeat sequences of fusion proteins of other membrane-enveloped viruses.
MATERIALS AND METHODS
Cells and antibodies.
HeLa-CD4-LTR-β-gal is a HeLa cell line that expresses a high level of CD4 and contains a single integrated copy of the β-galactosidase gene fused to the HIV-1 long terminal repeat (LTR) (35). HeLa-CD4-LTR-β-gal, CV-1, COS-1, and 293 cells were all cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum. SupT1 is a human CD4+ T-lymphoid cell line. PM1 is a derivative of Hut 78, which is a human cutaneous T-cell lymphoma cell line derived from the peripheral blood mononuclear cells of a patient with Sezary syndrome (39). Hybridoma Chessie 8 produces a murine monoclonal antibody (MAb) specific for gp160 and maps to amino acid residues 727 to 732 of HIV-1LAI (1). Hybridoma 183 (clone H12-5C) is a mouse MAb reactive with HIV-1 Gag p24 (17). SupT1, PM1, hybridoma Chessie 8, and hybridoma 183 were maintained in RPMI 1640 containing 10% fetal bovine serum. Hybridomas were injected intraperitoneally into BALB/c mice to produce ascitic fluids. HeLa-CD4-LTR-β-gal (from Michael Emerman), SupT1 (from James Hoxie), PM1 (from Marvin Reitz, Jr.), hybridoma 183 (from Bruce Chesebro), and Chessie 8 (from George K. Lewis) were obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Antisera obtained from the National Institutes of Health AIDS Research and Reference Reagent Program also included sheep anti-gp120 (strain IIIB) from Michael Phelan and rabbit antiserum to HIV-1 p25/p24 Gag from Kathelyn Steimer (53).
Plasmids.
The wt proviral DNA clone HXB2gpt (48) and mutant HXB2gpt proviruses that encode a proline substitution in the zipper motif of gp41 (Fig. 1) have been previously described (14, 16). The HXB2gpt provirus is abbreviated as HXB2 hereafter for simplicity. Vaccinia virus (VV)-based Env expression plasmids that encode the wt and zipper motif mutant Env proteins were previously described (14). pBaby is a simian virus 40 (SV40) late replacement expression vector and was used to generate pBSX for expression of the Env protein of strain HXB2. pSVE7 was cloned in an SV40-based vector that expresses the HXB2 strain Env driven by the HIV-1 LTR. pSVE7ΔKS, also known as pSVIIIenvΔKS (3), is a defective env expressor with a deletion at the KpnI (at nucleotide position 6351) to StuI (at nucleotide position 6834) sites of the env gene. pHXBCATΔBgl contains a defective HXB2 provirus with an in-frame deletion between the two BglII sites at nucleotides 7041 and 7621 in the env gene as well as a substitution of the chloramphenicol acetyltransferase (CAT) gene (cat) for the nef gene (32). pIIIextat expresses Tat under control of the HIV-1 LTR. All of these plasmids were previously described (15).
Construction of Env expression plasmids.
To construct env expression vectors that express zipper motif mutant proteins in the pSVE7 or pBSX backbone, the KpnI-BamHI fragments, i.e., the DNA sequence from nucleotide 6351 to 8475, of mutant HXB2 constructs were each substituted for the corresponding sequence in pSVE7 or pBSX. This version of pSVE7 contained the whole sequence of pSVE7 and a partial sequence of a neomycin resistance gene (neo) under control of an SV40 promoter. This partial sequence of the neo gene was inserted between the two EcoRI sites, separated by 440 bp, which was outside the LTR and env coding sequences. pSVE7ΔKS was similarly treated to contain a sequence of the neo gene and was used as a control. All constructed clones were sequenced to confirm mutations in the zipper motif by dideoxy chain termination with T7 Sequenase and the following oligonucleotides as primers: GGCGCAGCGTCAATGACG (at nucleotides 7814 to 7831 of the HXB2 sequence) for forward sequencing and GCTTGTGTAATTGTTAATTTCTCTGTCCCA (at nucleotides 8143 to 8114) for reverse sequencing.
Plasmid DNA transfection.
CV-1 cells grown in six-well plates were infected with wt VV (WR strain) and then transfected with wt or with wt and mutant VV env expression plasmids in the presence of Lipofectin reagent as previously described (14). HeLa-CD4-LTR-β-gal cells grown in six-well plates were transfected with pIIIextat and wt pSVE7 in the presence or absence of mutant pSVE7 plasmids by using Superfect transfection reagent according to the protocol provided by Qiagen (Valencia, Calif.). 293 cells grown in 100-mm-diameter petri dishes were transfected either with proviral DNA clones or with pSVE7-based expression plasmids and pIIIextat by a standard calcium phosphate coprecipitation protocol as previously described (15). COS-1 cells were transfected with pBSX-based expression plasmids by a DEAE-dextran method as previously described (16).
Western blot (immunoblot) analysis.
Two days after transfection, cell lysates were prepared as previously described (15). Briefly, culture media were spun at 1,200 rpm in a Beckman GS-6R centrifuge for 5 min followed by centrifugation at 2,400 rpm in the same centrifuge for 20 min. Supernatants were layered over a 20% sucrose cushion prepared in phosphate-buffered saline and centrifuged at 27,000 rpm in a Beckman SW41 rotor for 2 h at 4°C, and the viral pellets were lysed with phosphate-buffered saline containing 1% Nonidet P-40 and 1% sodium deoxycholate. Equal volumes of cell or virion lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto nitrocellulose membranes (0.45-μm pore size), and analyzed by using a biotin-streptavidin-amplified horseradish peroxidase system coupled with an enhanced chemiluminescence assay (Amersham, Arlington Heights, Ill.) as previously described (15).
Virus infection, RT assay, and virus titration.
Two days after transfection, culture media from cells transfected with HXB2 proviruses or with pHXBCATΔBgl and env expression plasmids were spun at 2,000 rpm in a Beckman GS-6R centrifuge for 5 min, and supernatants were filtered through a 0.45-μm-pore-size membrane. One-milliliter aliquots of filtered supernatants were concentrated by use of polyethylene glycol and assayed for virion-associated reverse transcriptase (RT) activity (16). Virus infectivity was assayed in SupT1 or PM1 cells by using aliquots of virus containing 105 cpm (or as specifically indicated for each experiment) of RT activity. SupT1 culture supernatants at different days postinfection were then monitored for RT activity. Virus infectivity was also titrated on HeLa-CD4-LTR-β-gal indicator cells by the MAGI (multinuclear activation of a galactosidase indicator) assay as previously described (35).
CAT assay.
CAT activity was determined 3 days after infection of HeLa-CD4-LTR-β-gal or PM1 cells as previously described, using aliquots of cell lysates containing equal amounts of proteins (15). The percentage of acetylation of radioactive chloramphenicol was quantitated by determining the relative signals of acetylated products and the unreacted chloramphenicol with an Instant Imager (Packard Instrument Company, Meriden, Conn.). The degree of entry of defective virus into CD4+ cells mediated by Env proteins supplied in trans was calculated by subtracting the percentage of acetylation of the defective virus in the absence of Env expression from that of the defective virus pseudotyped with wt or with wt and mutant Env proteins.
RESULTS
Analysis of viral proteins expressed in cells transfected with mutant HXB2 proviruses.
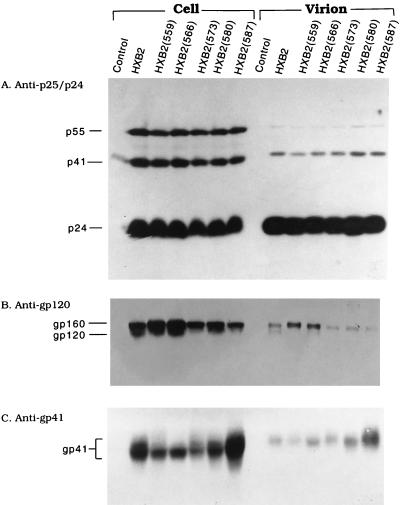
To analyze viral proteins expressed by zipper motif mutants in the context of replication of a whole virus, 293 cells were transfected with equal amounts of wt or mutant HXB2 proviruses by a calcium phosphate coprecipitation method. Two days after transfection, equal volumes of cell lysates were analyzed by SDS-PAGE followed by Western blotting with rabbit anti-p25/p24. Similar levels of gag gene products were produced by the wt provirus and all of the mutant proviruses (Fig. 2A). When lysates were analyzed with anti-gp120, all transfections produced similar levels of cell-associated gp160 (Fig. 2B). Intracellular gp120 was detected in the wt and some of the mutants (Fig. 2B). When lysates of transfected cells were analyzed with the Chessie 8 MAb, which is reactive with an epitope located in the gp41 cytoplasmic domain, all mutant proviruses except mutant 587 apparently produced less cell-associated gp41 than the wt provirus (Fig. 2C). To examine viral proteins assembled into mutant virions, virions were isolated by a standard sucrose cushion centrifugation as previously reported (30). Equal aliquots of virus lysates were then analyzed with these three antibodies. Similar amounts of p24 were detected in the wt and mutant virions (Fig. 2A). gp120 was found associated with wt and mutant 566 virions but could not be detected with other mutant virions (Fig. 2B). Amounts of gp41 similar to or even greater than that detected in the wt virus were detected in all mutant virions (Fig. 2C).
FIG. 2.
Expression of viral proteins encoded by wt and zipper motif mutant HXB2 proviruses. 293 monolayers were transfected by a standard calcium phosphate coprecipitation method with 10 μg each of wt or mutant HXB2 proviruses as indicated. The numbers shown in parentheses indicate the locations where the conserved leucine or isoleucine residues in the gp41 zipper motif were replaced by a proline residue. Mock-transfected cells were included as a negative control (lane 1). Two days after transfection, lysates of cells and virions were prepared as described in Materials and Methods. Equal aliquots of cell lysates and virus fractions were separated by SDS–10% PAGE, and proteins were visualized by Western immunoblotting analysis with rabbit anti-p25/p24 (strain SF2) (A), sheep anti-gp120 (B), or anti-gp41-specific Chessie 8 MAb (C).
Interference with infectious virus production by cotransfection with the mutant HXB2 proviruses.
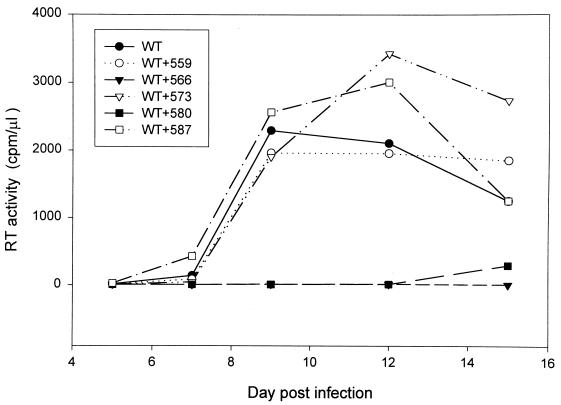
To examine whether mutant HXB2 could interfere with the wt virus infectivity, virus stocks were generated from 293 cells transfected with wt or with wt and mutant proviruses at a wt/mutant DNA ratio of 1:2. In each transfection the amount of wt HXB2 DNA was held constant and pHXBCATΔBgl DNA was added to the wt provirus transfection to maintain a constant final DNA concentration. Equal amounts of cell-free virus, as normalized by RT, were used to challenge SupT1 cells, and RT activity was monitored after infection. SupT1 cells infected with virus produced from wt HXB2-transfected 293 cells began to produce RT 7 days after infection, and the RT activity reached its peak 9 days after infection (Fig. 3). When viruses generated from cotransfection with wt and mutant 566 proviruses or with wt and mutant 580 proviruses were used for SupT1 inoculation, there was a substantial delay in the appearance of RT activity (Fig. 3). When viruses generated from cotransfection with wt and mutant 559, wt and mutant 573, or wt and mutant 587 were examined, there appeared to be no significant delay in virus production (Fig. 3). This study indicated that mutants 566 and 580 exert an inhibitory effect on the production of infectious virus. The other mutants did not seem to interfere with the production of infectious virus in this continuous virus growth study.
FIG. 3.
Effect of cotransfection with wt and mutant HXB2 proviruses on production of infectious virus. 293 cells were transfected with 5 μg of wt HXB2 or 5 μg of wt HXB2 plus 10 μg of mutant HXB2 proviruses as indicated. The total amount of DNA in all transfections was kept constant at 15 μg by adding pHXBCATΔBgl plasmid DNA. Two days after transfection, culture supernatants were collected, centrifuged, and passed through 0.45-μm-pore-size membranes. Virions containing 5 × 104 cpm of RT activity from each virus stock were used to challenge 2 × 106 SupT1 cells. The cultures were monitored for virion-associated RT production at different times postinfection.
Quantitation of infectivity of viruses generated from cotransfection with wt and mutant proviruses.
To quantitate the infectivity of these mixed viruses, virus from each stock containing equal amounts of RT activity was measured by MAGI assay with HeLa-CD4-LTR-β-gal indicator cells (35). This method can detect productive infection of a single viral particle by the ability of viral Tat protein upon HIV-1 infection to transactivate the HIV-1 LTR-linked β-galactosidase gene. Virus infection was then monitored by the appearance of foci of the target cells by using X-Gal (5-bromo-4-chloro-3-indolyl-d-galactopyranoside) staining under a light microscope 3 days postinfection. All viruses generated from cotransfection with wt and mutant HXB2 showed a 64 to 99% reduction in titer compared to that of the wt virus (Table 1). Consistent with Fig. 3, cotransfection of wt provirus with mutant 559, 573, or 587 provirus showed a lower degree of inhibition of virus infectivity than cotransfection of wt provirus with mutant 566 or 580 provirus. Collectively, these results demonstrated that coexpression of wt and zipper motif mutant proviruses inhibits wt virus infectivity.
TABLE 1.
Titration of viruses produced by wt-mutant coexpression
| Expt no. and transfection | No. of blue cellsa | Relative infectivity (%)b |
|---|---|---|
| 1c | ||
| HXB2 | 15,772 | 100 |
| HXB2 + HXB2(559) | 5,568 | 35.3 |
| HXB2 + HXB2(566) | 154 | 1.0 |
| HXB2 + HXB2(573) | 1,908 | 12.1 |
| HXB2 + HXB2(580) | 225 | 1.4 |
| HXB2 + HXB2(587) | 5,724 | 36.3 |
| 2d | ||
| pSVE7 | 2,369 | 100 |
| pSVE7 + pSVE7(566) | 1,064 | 44.9 |
| pSVE7 + pSVE7(573) | 905 | 38.2 |
| 3e | ||
| pSVE7 | 1,100 | 100 |
| pSVE7 + pSVE7(566) (2:1) | 362 | 32.9 |
| pSVE7 + pSVE7(566) (1:1) | 225 | 20.5 |
| pSVE7 + pSVE7(573) (2:1) | 525 | 47.7 |
| pSVE7 + pSVE7(573) (1:1) | 395 | 35.9 |
Infectivities of viruses were measured by the MAGI assay with HeLa-CD4-LTR-β-gal indicator cells.
Expressed as (number of blue cells found in wt-mutant cotransfection/number of blue cells found in wt transfection) × 100%.
Viruses were produced in 293 cells by transfection with 5 μg of wt HXB2 or with 5 μg of wt HXB2 and 10 μg of mutant HXB2 as indicated. DNA of pHXBCATΔBgl was added to make the amounts of total DNA in transfections the same. Two days after transfection, cell-free viruses were prepared, and viruses with 2 × 105 cpm of RT activity were assayed on HeLa-CD4-LTR-β-gal cells as described in Materials and Methods.
Recombinant viruses were produced in 293 cells by transfection with 10 μg of pHXBCATΔBgl together with 10 μg of wt pSVE7 or with 10 μg each of wt and mutant pSVE7. Transfection with pHXBCATΔBgl only was used as a negative control. The amount of total DNA was kept constant by adding pSVE7ΔKS. Two days after transfection, cell-free virus supernatants were prepared and assayed for RT activity. Viruses containing 2 × 105 cpm of RT activity were assayed on HeLa-CD4-LTR-β-gal cells.
293 cells were transfected with 10 μg of pHXBCATΔBgl together with 10 μg of wt pSVE7 or with 10 μg of wt pSVE7 in the presence of mutant pSVE7 at different wt/mutant DNA ratios as indicated in parentheses following the plasmid names. Two days after transfection, cell-free viruses were prepared, and viruses containing 105 cpm of RT activity were assayed on indicator cells.
Interference with wt Env-induced cytopathic effects by cotransfection with mutant proteins.
To understand whether mutant proteins were responsible for the phenotype of inhibition of infectivity of viruses obtained from cotransfection, the ability of mutant proteins to inhibit wt Env-induced cytopathic effects was examined. wt VV-infected CV-1 cells were transfected by a liposome-mediated method with wt or with wt and mutant env plasmids cloned in a VV vector (14) at different wt/mutant DNA ratios. Transfected cells were mixed with SupT1 cells, and syncytium formation was examined under a light microscope 18 h after cocultivation. As summarized in Table 2, syncytium formation induced by the wt Env was not detected when the wt/mutant plasmid DNA ratio was 1:3. At a 1:2 ratio of wt to mutant plasmid DNA, wt protein-mediated syncytium formation was also significantly inhibited. When the wt/mutant plasmid DNA ratio was set at 1:1, many fewer or even no syncytia were observed in the wt-mutant cotransfection compared to the amount found in the wt transfection.
TABLE 2.
Inhibition of wt Env-mediated syncytium formation by coexpression with mutant proteinsa
| Plasmid(s) transfected | Relative degree of syncytium formationb at the following wt/mutant plasmid DNA ratio:
|
||
|---|---|---|---|
| 1:1c | 1:2d | 1:3d | |
| wt | ++++ | ++++ | ++++ |
| wt + 559 | −/+ | − | − |
| wt + 566 | − | − | − |
| wt + 573 | −/+ | − | − |
| wt + 580 | − | − | − |
| wt + 587 | − | − | − |
VV-infected CV-1 cells were transfected with a VV-based wt env plasmid or with wt and mutant plasmids at different wt/mutant DNA ratios by the Lipofectin-mediated DNA transfer technique, according to the procedure provided by the manufacturer (Bethesda Research Laboratories, Gaithersburg, Md.). The total DNA amount was kept constant by the addition of 3PrV DNA, a VV vector without an env insert. Six hours after transfection, SupT1 cells were added to each transfected culture, and syncytium formation was scored under a light microscope 18 h after transfection.
++++, more than 250 syncytia were observed in a well; −/+, fewer than 20 syncytia were observed; −, no apparent syncytia were observed.
Five micrograms each of wt and mutant plasmid DNA were used to transfect VV-infected CV-1 cells grown in six-well plates. The experiment was performed more than three times, and similar results were obtained. In each experiment, transfection with wt plasmid DNA resulted in significant syncytium formation (more than 250 syncytia).
VV-infected CV-1 cells were transfected with 1 μg of wt plasmid together with the various mutant plasmids at the specified wt/mutant DNA ratio.
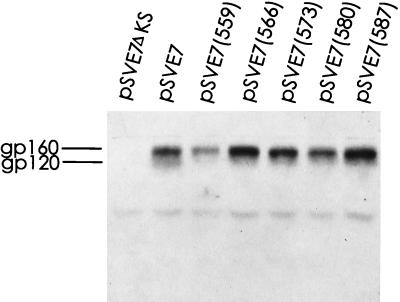
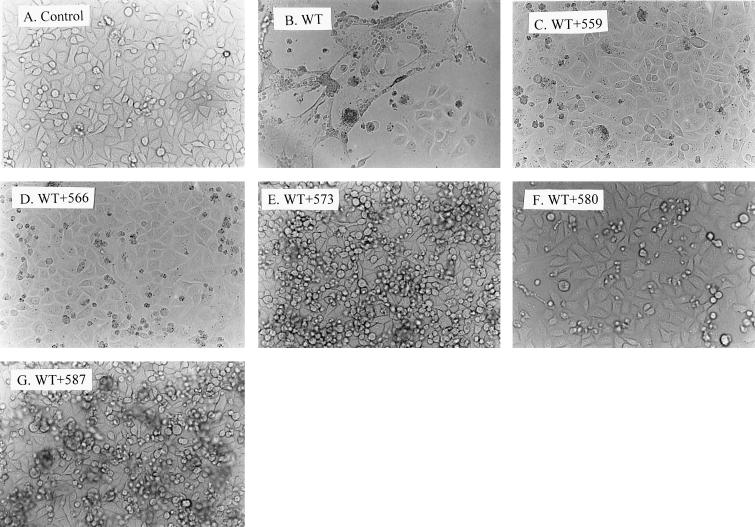
To confirm that the zipper motif mutant proteins interfered with wt Env-induced cytopathic effects, the pSVE7 expression system was also employed. The 2.1-kb KpnI-BamHI fragments isolated from the mutant HXB2 proviruses were used to substitute for the corresponding sequence in a modified version of pSVE7 to yield mutant pSVE7 plasmids. These mutants were examined for Env protein expression by Western blotting analysis. Except for mutant 559, which showed a slightly lower level, all mutant plasmids produced Env proteins at a level similar to or even slightly greater than that produced by the wt plasmid in the presence of pIIIextat during transfection (Fig. 4). HeLa-CD4-LTR-β-gal cells were then cotransfected with pIIIextat and pSVE7 in the presence or absence of mutant plasmids by the Superfect transfection method. Two days after transfection, cell cultures were examined under a light microscope. No syncytia or cytopathic effects were observed in the culture that was transfected with the tat expression plasmid alone (Fig. 5A). Transfection with wt pSVE7 together with pIIIextat produced striking cytopathic effects, including syncytium formation and floating and dead cells (Fig. 5B). In addition, a large number of cells had undergone fusion and lysis. In contrast, cotransfection of wt and mutant plasmids strikingly inhibited or delayed the wt Env-induced cytopathic effects (Fig. 5C to G). Although a few syncytia were about to form in cells cotransfected with mutant 573 or 587, the cytopathic effects in these cultures were much delayed compared to that found with the wt transfection. Taken together, these observations indicated that zipper motif mutant proteins are able to interfere with cytopathic effects mediated by the wt protein.
FIG. 4.
Env proteins encoded by wt and mutant pSVE7 plasmids. COS-1 cells were cotransfected by the DEAE-dextran method with 5 μg each of wt or mutant pSVE7 plasmid in the presence of 2 μg of pIIIextat. Two days after transfection, cell lysates were prepared and equal amounts of cell lysates were subjected to SDS-PAGE followed by Western blotting with sheep anti-gp120.
FIG. 5.
Inhibition of the wt Env-mediated cytopathic effects by coexpression with wt and mutant Env proteins. HeLa-CD4-LTR-β-gal cells grown in six-well plates were transfected with 1 μg of pIIIextat and 2 μg of wt pSVE7 in the presence or absence of 4 μg of mutant pSVE7 plasmids as indicated, using 10 μl of Superfect transfection reagent according to the Qiagen protocol. Transfection with pIIIextat was used as a negative control (A). DNA of pSVE7ΔKS was added to transfection mixtures to keep the total DNA amount in each transfection constant. Two days after transfection, cell cultures were photographed under a light microscope. Magnification, ×200.
Interference with wt Env-mediated viral transmission by mutant Env proteins.
To confirm that zipper motif mutant Env proteins conferred interference with wt virus infectivity, an env trans-complementation assay utilizing a defective HIV-1 proviral vector (32) was performed to study virus replication in a context of virus-to-cell transmission. This assay determines the ability of Env proteins to complement a defective pHXBCATΔBgl provirus for a single round of virus replication. 293 cells were transfected with pHXBCATΔBgl together with wt pSVE7 in the presence or absence of mutant pSVE7 plasmids. The wt/mutant plasmid DNA ratio was set to 1:1. Two days after transfection, culture supernatants were filtered and their RT activity was determined. Viruses containing equal amounts of RT activity from each transfection were used to infect HeLa-CD4-LTR-β-gal cells. Three days after infection, cell lysates were prepared and assayed for CAT activity. Viruses derived from defective provirus transfection alone did not show entry into CD4+ cells, as CAT activity was undetected (Fig. 6A, lane 1). wt Env successfully mediated virus-to-cell transmission as evidenced by the increased CAT activity compared with the Env-negative control (Fig. 6A, lane 2). In contrast, mutant Env proteins did not support entry of the defective virus into CD4+ cells (data not shown). On the other hand, wt-mutant cotransfection decreased CAT activity by 49 to 71% compared with that with wt transfection alone (Fig. 6A, lanes 3 to 7).
FIG. 6.
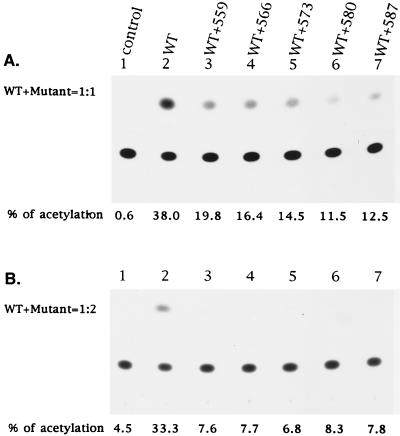
Virus-to-cell transmission of a defective provirus mediated by wt and mutant Env coexpression. (A) wt/mutant pSVE7 DNA ratio of 1:1. 293 cells were cotransfected with 10 μg of pHXBCATΔBgl and 10 μg of wt pSVE7 or with 10 μg each of pHXBCATΔBgl, wt, and mutant pSVE7 plasmids as indicated. Transfection with the defective provirus alone was used as a control (lane 1). Plasmid SVE7ΔKS was added to transfection mixtures to make the total amount of DNA in each transfection constant. Two days after transfection, cell-free viruses were prepared, and virus from each stock containing 5 × 104 cpm of RT activity was applied to subconfluent HeLa-CD4-LTR-β-gal cells grown in 60-mm-diameter petri dishes. Unbound viruses were removed after overnight incubation at 37°C, and fresh media were added to cultures. Three days after transfection, cell lysates were prepared and assayed for CAT activity. (B) wt/mutant pSVE7 DNA ratio of 1:2. Transfection was performed as described for panel A except that 7.5 μg of pHXBCATΔBgl, 5 μg of wt pSVE7, and 10 μg of various pSVE7 mutants were used in transfection. Viruses with 2 × 105 cpm of RT activity were applied to HeLa-CD4-LTR-β-gal cells, and CAT activity was assayed 3 days after infection.
To further address the interference with wt Env-mediated virus entry conferred by mutant Env, recombinant viruses were generated from 293 cells cotransfected either with the cat-containing defective provirus along with wt pSVE7 or with wt and mutant pSVE7 at a wt/mutant plasmid DNA ratio of 1:2. Defective viruses pseudotyped with wt and zipper motif Env mutants showed significant reductions, ranging from 87 to 92%, in CAT activity compared with the defective virus supplemented with the wt protein alone (Fig. 6B). By comparing Fig. 6A and B, it was also noted that the interference with wt Env-mediated virus entry conferred by zipper motif mutants was dependent on the amounts of mutant plasmids used in cotransfection.
To examine whether wt Env-mediated virus entry measured by CAT activity correlated with virus infectivity, mutants 566 and 573 were chosen for more characterization. Mutants 566 and 573 represented the group of mutants that showed a more pronounced interference effect or a lesser interference effect, respectively, on infectious virus production (Fig. 3). Infectivity of recombinant viruses generated from pHXBCATΔBgl cotransfection with wt pSVE7 or with wt and mutant pSVE7 at a wt/mutant DNA ratio of 1:1 was determined by MAGI assay. Virus derived from cotransfection with wt and mutant 566 showed a 55% reduction in CAT activity, and virus from cotransfection with wt and mutant 573 showed a 62% reduction in infectivity, compared with that of virus obtained from wt pSVE7 transfection alone (Table 1). In a separate experiment, defective recombinant viruses were generated from cotransfection of wt and mutant 566 and of wt and mutant 573 at different wt/mutant DNA ratios. The degree of inhibition of virus infectivity was dependent on the amount of mutant plasmid DNA used in cotransfection (Table 1).
Interference with wt Env-mediated virus entry conferred by mutant proteins expressed by another expression system.
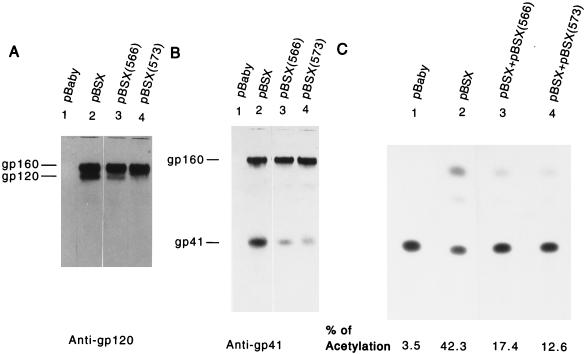
To determine whether the interference effect conferred by the zipper motif mutants was independent of the expression vectors used, mutants 566 and 573 were analyzed by utilizing the pBSX expression system, in which Env expression is independent of Tat protein. A smaller amount of cell-associated gp120 was detected for mutant 566 or 573 transfection than for wt pBSX transfection (Fig. 7A). Transfection with mutant 566 or mutant 573 also produced a smaller amount of cell-associated gp41 than transfection with wt plasmid (Fig. 7B). Moreover, defective recombinant viruses generated from cotransfection with wt and mutant 566 at a plasmid DNA ratio of 1:1 showed a 64% reduction in CAT activity, and those from cotransfection with wt and mutant 573 showed a 77% reduction in CAT activity, compared with viruses derived from wt transfection alone (Fig. 7C).
FIG. 7.
Effect of coexpression of wt and zipper motif mutant proteins on the wt Env-mediated virus-to-cell transmission. (A and B) Env protein expression. COS-1 cells were transfected with 5 μg each of pBaby, wt pBSX, or mutant pBSX, as indicated, by the DEAE-dextran method. Two days after transfection, equal amounts of cells lysates from each transfection were analyzed by Western blotting with anti-gp120 (A) or Chessie 8 anti-gp41 MAb (B). (C) Ability of mutant proteins to inhibit wt Env-induced virus entry into CD4+ cells. COS-1 cells were transfected with 5 μg of pHXBCATΔBgl and 2 μg of wt pBSX in the presence or absence of 2 μg of mutant pBSX as indicated. Transfection with pHXBCATΔBgl and pBaby was used as a negative control (lane 1). The total DNA amount in all transfections was kept constant by adding pBaby DNA. Two days following transfection, cell-free viruses were prepared, and 105 cpm of RT activity from each virus stock was then applied to HeLa-CD4-LTR-β-gal cells. Three days after infection, cell lysates were prepared and assayed for CAT activity.
Interference conferred by zipper motif Env mutants assayed in a T-cell line.
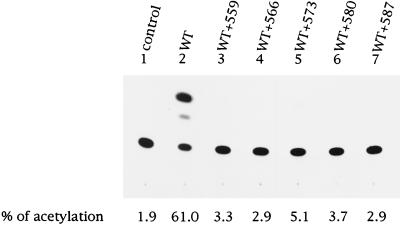
To determine whether the interference effect conferred by the zipper motif mutants was independent of the CD4+ cells used in the env trans-complementation assay, PM1, a CD4+-T-cell line derived from Hut78, was utilized to assess virus entry of a defective proviral vector pseudotyped with the wt Env or with the wt and mutant coexpressed proteins at a wt/mutant molar ratio of 1:2. wt Env effectively mediated virus entry into PM1 cells (Fig. 8, lane 2). Again, all of the zipper motif mutants significantly inhibited the ability of the wt Env to mediate virus entry when they were coexpressed with the wt Env (Fig. 8, lanes 3 to 7). Taken together, interference studies using HeLa-CD4-LTR-β-gal and PM1 as host cells clearly demonstrated that all zipper motif mutant Env proteins confer an interference effect on the wt Env-mediated virus entry into CD4+ cells.
FIG. 8.
Interference conferred by zipper motif Env mutants with virus-to-cell transmission assayed in PM1 cells. Recombinant viruses were generated from 293 cells cotransfected with the cat-containing defective provirus along with the wt pSVE7 or with the wt and mutant pSVE7 at a wt/mutant DNA ratio of 1:2 as described in the legend to Fig. 6B. Viruses containing 8 × 104 cpm of RT activity were used to challenge PM1 cells. Three days after infection, cell lysates were prepared and assayed for CAT activity.
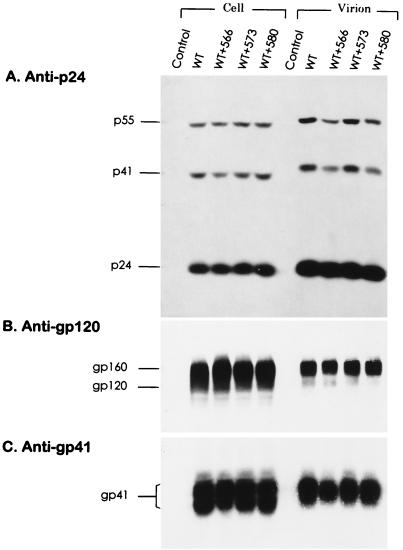
Incorporation of viral proteins into virions after cotransfection with wt and mutant HXB2 proviruses.
To determine whether the impaired infectivity in virus generated from wt-mutant cotransfection was due to impairment in the late steps of the virus life cycle, 293 cells were transfected with wt or with wt and mutant 566, 573, or 580 HXB2 proviral DNA in a 1:1 ratio. Cotransfection of wt and mutant proviruses produced levels of cell-associated and virion-associated gag gene products comparable to those produced by wt provirus transfection (Fig. 9A). Virion-associated gp120 was also detected in wt-mutant cotransfection as well as in wt transfection alone (Fig. 9B). Comparable amounts of cell-associated and virion-associated gp41 were found in wt-mutant cotransfection as well as in wt transfection alone (Fig. 9C).
FIG. 9.
Viral protein expression in cells cotransfected with wt and mutant HXB2 proviruses. 293 cells were transfected either with 10 μg of wt HXB2 or with 5 μg each of wt and mutant HXB2 proviruses as indicated. Two days after transfection, equivalent portions of cell lysates and virion fractions were analyzed with mouse anti-p24 MAb (A), sheep anti-gp120 (B), or anti-gp41 MAb (C).
DISCUSSION
In this study we further characterized the zipper motif mutants that we have previously described (14, 16) and found that gp41 zipper motif mutants are able to dominantly inhibit the wt Env-mediated function in an early step of the virus life cycle. Mutants 566 and 580 dominantly interfere with infectious virus production more significantly than other mutants (Fig. 3). Although other mutants do not exhibit a significant inhibitory effect on production of infectious virus in the SupT1 infection study (Fig. 3), these mutants also show significant reduction in virus infectivity compared to the wt virus when the infectivity of mixed viruses is assessed by MAGI assay (Table 1). In addition, all mutants are able to interfere with wt Env-induced cytopathicity (Table 2; Fig. 5) and virus entry, assayed in either the HeLa-CD4-LTR-β-gal indicator cell line (Fig. 6 and 7; Table 1) or a CD4+ T-cell line, PM1 (Fig. 8). Thus, the observation that viruses derived from mixed cotransfection eventually lead to productive virus replication in SupT1 cells after a long period of culture (Fig. 3) may result from multiple infections of the residual virus infectivity which is not completely knocked out by coexpression with mutant proviruses.
From comparison of the abilities of mutant Env proteins to complement virus entry and to mediate syncytium formation, it was previously reported that virus-cell and cell-cell fusion may require different numbers of successful Env-CD4 interactions (8, 32, 36). These studies suggested that virus-cell fusion and syncytium formation may have different requirements for the Env structures. Since mutants 566 and 573 show different degrees of precursor processing and gp120-gp41 association (14), it is plausible that the hetero-oligomers formed by wt and mutant 566 and by wt and mutant 573 may have qualitative differences in their structures. It is likely that the altered structure of the wt-mutant 573 hetero-oligomer may still be sufficient to mediate a low level of cell-cell fusion, as shown in the syncytium formation assay (Table 2; Fig. 5) and the SupT1 infection study (Fig. 3), in which cell-cell fusion is also encountered. However, such a structural alteration may be restricted for its function in virus-cell fusion (Table 1; Fig. 6, 7, and 8). In contrast, the altered structure formed by the wt and mutant 566 proteins may be insufficient for mediating both syncytium formation and virus entry.
Evidence supporting that this motif contributes to the Env oligomeric structure has been documented. Sequences or determinants that are involved in Env oligomerization of immunodeficiency viruses have been mapped to the ectodomain of the TM protein, in particular, to the zipper motif (10, 22, 23, 44, 45, 54). Moreover, the gp41 zipper motif is sufficient to promote monomeric proteins to become tetrameric or trimeric structures when it is fused to otherwise monomeric proteins (4, 51, 58). Also, synthetic peptides or recombinant proteins containing sequences corresponding to this motif form a rod-like molecule with a high degree of α-helical structure (37, 46, 47, 56, 58, 59, 62). Two peptides corresponding to the heptad repeat sequence and a segment close to the TM region expressed as recombinant proteins or approached by peptide modeling form a stable, coiled-coil trimer that consists of two peptides packed as an antiparallel heterodimer (38, 47). Recent crystallographic data on a complex formed by the peptides containing the N- and C-terminal sequences of the gp41 ectodomain and of a GCN4-gp41 ectodomain chimera also demonstrate the conformation of the gp41 ectodomain core as being an interior, parallel coiled-coil trimer with the N terminus at its tip and the other three helices packed in the reverse direction against the outside of the inner trimer (12, 57). Moreover, the TM protein ectodomains of Moloney murine leukemia virus and simian immunodeficiency virus also show trimeric conformations formed by the dimeric N- and C-terminal segments that pack antiparallelly (5, 24, 25). These extended coiled coils exhibit striking structural similarity to the low-pH-converted conformation of the fusogenic form of HA2, suggesting a similar mode of function of these coiled-coil domains in membrane fusion (40, 59).
The inhibitory effect on the wt virus infectivity conferred by these mutants does not seem to involve the assembly and budding of Gag proteins, since cotransfection with wt and mutant proviruses produces levels of cell-associated and virion-associated gag gene products comparable to those produced by the wt transfection alone (Fig. 9A). In addition, virions derived from wt-mutant cotransfection contain amounts of gp41 comparable to that found in the wt virus (Fig. 9C). The interference conferred by mutant coexpression appears to be attributable to the ability of mutant Env to form a hetero-oligomer with the wt protein which is impaired in an early step of the virus replication cycle. The possibility that the interference is due to competition between wt and mutant homo-oligomers for the same receptor molecule is less likely. If a 1:1 ratio of wt to mutant plasmid DNA is used in cotransfection and if wt and mutant proteins are present as separate homo-oligomeric entities, at most a 50% inhibitory effect would be observed in viruses obtained from wt-mutant coexpression. However, as much as 80% inhibition of virus entry is detected when mixed virus is measured in a trans-complementation assay (Table 1). Nevertheless, the detailed molecular mechanism underlying the interference effects conferred by these mutants remains to be determined.
Synthetic peptides that contain sequences corresponding to the heptad repeat sequence or the C-terminal segment of the gp41 ectodomain have been found to be effective inhibitors in blocking virus infectivity and membrane fusion (13, 34, 38, 60–63). It has been proposed that the inhibitory activity of the N-terminal and C-terminal peptide inhibitors can be attributable to their interactions with the C-terminal and N-terminal sequences, respectively, within endogenous gp41 (13, 40, 61). In this study we describe an anti-Env approach, different from the use of peptide inhibitors, that targets the heptad repeat region of gp41 ectodomain by using a trans-dominant inhibitory mutant. Since heptad repeat sequences are highly conserved among the TM proteins of various membrane-enveloped viruses, the approach described here may be applied to other viral systems in order to develop dominant-negative mutant-based antiviral therapeutics against other viral infections.
ACKNOWLEDGMENTS
This work was supported by grants from the National Sciences Council (NSC 85-2331-B-001-010 and NSC 86-2314-B-001-017) and from the Institute of Biomedical Sciences at Academia Sinica, Taipei, Taiwan, Republic of China.
We thank Hong-Huat Loh for technical help and Chien-Ting Lin for figure labeling. We are also indebted to Douglas Platt for text editing.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claasen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Bates P. Chemokine receptors and HIV-1: an attractive pair? Cell. 1996;86:1–3. doi: 10.1016/s0092-8674(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron L, Sullivan N, Sodroski J. Target cell-specific determinants of membrane fusion within the human immunodeficiency virus type 1 gp120 third variable region and gp41 amino terminus. J Virol. 1992;66:2389–2397. doi: 10.1128/jvi.66.4.2389-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein H B, Tucker S P, Kar S R, McPherson S A, McPherson D T, Dubay J W, Lebowitz J, Compans R W, Hunter E. Oligomerization of the hydrophobic heptad repeat of gp41. J Virol. 1995;69:2745–2750. doi: 10.1128/jvi.69.5.2745-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blacklow S C, Lu M, Kim P S. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 6.Broder C C, Dimitrov D S. HIV and the 7-transmembrane domain receptors. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 7.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 10.Center R J, Kemp B E, Poumbourios P. Human immunodeficiency virus type 1 and 2 envelope glycoproteins oligomerize through conserved sequences. J Virol. 1997;71:5706–5711. doi: 10.1128/jvi.71.7.5706-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 12.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 13.Chen C-H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral function. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S S-L. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1994;68:2002–2010. doi: 10.1128/jvi.68.3.2002-2010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S S-L, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 16.Chen S S-L, Lee C-N, Lee W-R, McIntosh K, Lee T-H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delwart E L, Mosialos G, Gilmore T. Retroviral envelope glycoproteins contain a “leucine zipper”-like repeat. AIDS Res Hum Retroviruses. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 19.Doms R W, Earl P L, Chakrabarti S, Moss B. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus env proteins possess a functionally conserved assembly domain. J Virol. 1990;64:3537–3540. doi: 10.1128/jvi.64.7.3537-3540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Souza M P, Harden V A. Chemokines and HIV-1 second receptors. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 21.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl P L, Doms R W, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl P L, Moss B. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res Hum Retroviruses. 1993;9:589–594. doi: 10.1089/aid.1993.9.589. [DOI] [PubMed] [Google Scholar]
- 24.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 25.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg M B, Trono D. Intracellular immunization: trans-dominant mutants of HIV gene products as tools for the study and interruption of viral replication. AIDS Res Hum Retroviruses. 1992;8:1013–1022. doi: 10.1089/aid.1992.8.1013. [DOI] [PubMed] [Google Scholar]
- 27.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 29.Gelderblom H R, Hausmann E H, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1988;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 30.Göttlinger H G, Dorfman T, Cohen E A, Haseltine W A. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci USA. 1993;90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grief C, Hockley D J, Fromholc C E, Kitchin P A. The morphology of simian immunodeficiency virus as shown by negative staining electron microscopy. J Gen Virol. 1989;70:2215–2219. doi: 10.1099/0022-1317-70-8-2215. [DOI] [PubMed] [Google Scholar]
- 32.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 34.Jiang S, Lin K, Strick N, Neurath A R. HIV-1 inhibition by a peptide. Nature (London) 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 35.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalski M, Bergeron L, Dorfman T, Haseltine W, Sodroski J. Attenuation of human immunodeficiency virus type 1 cytopathic effect by a mutation affecting the transmembrane envelope glycoprotein. J Virol. 1991;65:281–291. doi: 10.1128/jvi.65.1.281-291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawless M K, Barney S, Guthrie K L, Bucy T B, Petteway S R, Jr, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 38.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 39.Lusso P, Cocchi F, Balotta C, Markham P D, Louie A, Farci P, Pai R, Gallo R C, Reitz M S., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews T J, Wild C, Chen C-H, Bolognesi D P, Greenberg M L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 41.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. The HIV-cell fusion reaction. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 233–289. [Google Scholar]
- 42.Patarca R, Haseltine W A. Similarities among retrovirus proteins. Nature (London) 1984;312:496. doi: 10.1038/312496a0. [DOI] [PubMed] [Google Scholar]
- 43.Pinter A, Honnen W J, Tilley S A, Bona C, Zaghouani H, Gorny M K, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poumbourios P, Ahmar W E, McPhee D A, Kemp B E. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J Virol. 1995;69:1209–1218. doi: 10.1128/jvi.69.2.1209-1218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poumbourios P, Wilson K A, Center R J, Ahmar W E, Kemp B E. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic α-helical/leucine zipper-like sequence. J Virol. 1997;71:2041–2049. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabenstein M, Shin Y-K. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 47.Rabenstein M D, Shin Y-K. HIV-1 gp41 tertiary structure studied by EPR spectroscopy. Biochemistry. 1996;35:13922–13928. doi: 10.1021/bi961743t. [DOI] [PubMed] [Google Scholar]
- 48.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R-S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 49.Rey M-A, Krust B, Laurent A G, Montagnier L, Hovanessian A G. Characterization of human immunodeficiency virus type 2 envelope glycoproteins: dimerization of the glycoprotein precursor during processing. J Virol. 1989;63:647–658. doi: 10.1128/jvi.63.2.647-658.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rey M-A, Laurent A G, McClure J, Krust B, Montagnier L, Hovanessian A G. Transmembrane envelope glycoproteins of human immunodeficiency virus type 2 and simian immunodeficiency virus SIV-mac exist as homodimers. J Virol. 1990;64:922–926. doi: 10.1128/jvi.64.2.922-926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shugars D C, Wild C T, Greenwell T K, Matthews T J. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70:2982–2991. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steffy K R, Wong-Staal F. Transdominant inhibition of wild-type human immunodeficiency virus type 2 replication by an envelope deletion mutant. J Virol. 1993;67:1854–1859. doi: 10.1128/jvi.67.4.1854-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P, Barr P J, Hallewell R A. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral antigen p25 gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 54.Thomas D J, Wall J S, Hainfeld J F, Kaczorek M, Booy F P, Trus B L, Eiserling F A, Steven A C. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J Virol. 1991;65:3797–3803. doi: 10.1128/jvi.65.7.3797-3803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unutmaz D, Littman D R. Expression pattern of HIV-1 coreceptors on T cells: implications for viral transmission and lymphocyte homing. Proc Natl Acad Sci USA. 1997;94:1615–1618. doi: 10.1073/pnas.94.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissenhorn W, Calder L, Dessen A, Laue T, Skehel J J, Wiley D C. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Willey D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 58.Weissenhorn W, Wharton S A, Calder L J, Earl P L, Moss B, Aliprandis E, Skehel J J, Wiley D C. The ectodomain of HIV-1 env subunit gp41 forms a soluble, α-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 1996;15:1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 59.Wild C, Dubay J W, Greenwell T, Baird T, Jr, Oas T G, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 61.Wild C, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res Hum Retroviruses. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 62.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wild C, Shugars D C, Greenwell T K, McDonal C B, Matthews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson I A, Skehel J J, Wiley D C. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]