Abstract
Purpose:
To predict the presence of angle dysgenesis on anterior-segment optical coherence tomography (ADoA) by using deep learning (DL) and to correlate ADoA with mutations in known glaucoma genes.
Participants:
In total, 800 high-definition anterior-segment optical coherence tomography (AS-OCT) images were included, of which 340 images were used to build the machine learning (ML) model. Images used to build the ML model included 170 scans of primary congenital glaucoma (16 patients), juvenile-onset open-angle glaucoma (62 patients), and adult-onset primary open-angle glaucoma eyes (37 patients); the rest were controls (n = 85). The genetic validation dataset consisted of another 393 images of patients with known mutations that were compared with 320 images of healthy controls.
Methods:
ADoA was defined as the absence of Schlemm’s canal, the presence of hyperreflectivity over the region of the trabecular meshwork, or a hyperreflective membrane. DL was used to classify a given AS-OCT image as either having angle dysgenesis or not. ADoA was then specifically looked for on AS-OCT images of patients with mutations in the known genes for glaucoma.
Results:
The final prediction, which was a consensus-based outcome from the three optimized DL models, had an accuracy of >95%, a specificity of >97%, and a sensitivity of >96% in detecting ADoA in the internal test dataset. Among the patients with known gene mutations, (MYOC, CYP1B1, FOXC1, and LTBP2) ADoA was observed among all the patients in the majority of the images, compared to only 5% of the healthy controls.
Conclusion:
ADoA can be objectively identified using models built with DL.
Keywords: Angle dysgenesis, artificial intelligence, AS-OCT, machine learning
Anterior-segment spectral-domain optical coherence tomography (SD-OCT) is being increasingly used in glaucoma patients, primarily to investigate the anterior chamber angle and visualize the trabecular meshwork (TM) and Schlemm’s canal (SC) in vivo.[1,2,3,4] This in vivo imaging of anterior chamber angle with AS-OCT has been used to detect gross features of angle dysgenesis in primary congenital glaucoma (PCG), juvenile-onset open-angle glaucoma (JOAG), and adult-onset primary open-angle glaucoma (POAG), which has been described either as an absence of SC and/or the presence of abnormal tissue or a hyperreflective membrane within angle recess.[5,6,7,8] These studies have shown that angle dysgenesis on AS-OCT (ADoA) can be observed even in eyes with gonioscopically normal appearing angles. Primary congenital glaucoma, JOAG, and adult-onset POAG form a spectrum in terms of severity of angle dysgenesis. Although most of the PCG eyes have features of ADoA, the same are present in 40% of JOAG eyes and in up to 35% of adult-onset POAG.[5,8] As there exists a wide spectrum of anatomical variability of the drainage angle, (the TM and SC morphology) in normal eyes[5,7,8] which can make it difficult to distinguish normal from abnormal; therefore, interpretation of AS-OCT images requires expertise and a deep understanding of the complexity involved in the developmental anomalies of the outflow pathways. Artificial intelligence (AI) has the potential to assist experts in disease diagnosis, progression, and management by performing rapid image classification, which otherwise is a difficult or ambiguous scenario for human experts. Deep learning (DL), a subtype of AI, uses the concept of biological neural networks and has demonstrated convincing results in ophthalmic diseases.[9,10,11]
While angle dysgenesis is associated with developmental immaturity of the outflow pathways regulated by genes, this could only be ascertained till now with histopathological studies. Mutations in some of the commonly associated genes with glaucoma, namely CYP1B1[12] FOXC1[13] PITX2,[14] and TEK,[15] have been shown to be associated with developmental abnormalities in the outflow pathways in experimental studies. The severity of angle dysgenesis has been correlated on histopathology with certain CYP1B1 gene mutations in PCG patients.[16] Although MYOC mutations are known to be associated with early-onset glaucoma of PCG[17,18,19] and JOAG,[20,21,22,23,24] no studies have shown the involvement of MYOC mutations in causing angle dysgenesis. Histopathological studies for angle dysgenesis in human glaucomatous eyes are difficult to perform and are inherently associated with tissue handling artifacts. Although grossly identifiable features of ADoA have been described before,[5,8] many subtle changes may also be present on AS-OCT scans that are challenging to detect or precisely quantify by human observers. Detecting ADoA could be especially important for non-invasively screening relatives of glaucoma patients to identify those at risk of developing glaucoma. Thus, in this study, we aimed to develop DL models that can identify ADoA among glaucoma patients in comparison to healthy controls and validate their performance in detecting angle dysgenesis in patients with known glaucoma-associated gene mutations.
Methods
Dataset details and study design
This observational case-control study was approved by the institutional ethics committee. It adhered to the tenets of the Declaration of Helsinki. Informed consent to participate was taken from all cases: parents of children less than 18 years of age and assent to participate from subjects <18 years of age. Patients coming to the institute from January 2020 to January 2021 were recruited. A detailed history was recorded, and all subjects underwent a thorough clinical examination.
Inclusion criteria
PCG: These were unrelated cases of PCG detected before 3 years of age with enlarged corneal diameters (>12 mm before 1 year of age) who had baseline IOP records of >22 mmHg and were now old enough (>10 years of age) to cooperate for anterior segment OCT scanning.
JOAG: These were unrelated primary open-angle glaucoma patients diagnosed between 10 and 40 years of age. These were patients without any acquired or non-acquired ocular conditions or syndromes, without signs of ocular enlargement (e.g., buphthalmos), and with a normal-appearing angle.
Adult-onset POAG: These were unrelated cases of POAG diagnosed after the age of 40 years with untreated IOP of >22 mmHg in one or both eyes on more than two occasions, open angle on gonioscopy in both eyes, and glaucomatous optic neuropathy in one or both eyes with visual field loss consistent with optic nerve damage.
Only those patients who had been treated and had an IOP of <22 mmHg at the time of imaging were included.
Normal eyes: Age-matched healthy subjects of Indian ethnicity (age >10 years), who had IOP <21 mmHg, gonioscopically normal open angle, and no other ocular pathology on detailed ophthalmic evaluation.
Gonioscopy was performed by an experienced glaucoma specialist by using the Goldman two-mirror lens; the Schaffer system of gonioscopic grading was used.
Exclusion criteria
Patients excluded from the study: those with a history of steroid use; presence of any other retinal or neurologic pathology; evidence of secondary causes of raised IOP such as pigment dispersion, pseudoexfoliation, or trauma; those with any pathology detected on gonioscopy such as angle recession, pigmentation of the angle greater than grade 3 in any one or more quadrants, iridotrabecular contact, or peripheral anterior synechiae; those who had undergone angle surgery; and patients with nystagmus/and or poor fixation were excluded.
In patients who had undergone surgery and had either a superior trabeculectomy or a combined ab externo trabeculotomy with trabeculectomy, scans of AS-OCT from the nasal/temporal quadrant were selected per eye to exclude the superior quadrant.
SD-OCT examination
The AS-OCT examination was performed using the Spectralis OCT (software version 6.5; Heidelberg Engineering GmbH, Heidelberg, Germany). This machine uses an 880-nm wavelength and provides a resolution of 3.5 µm (digital) to 7 µm (optical) at 40 kHz. An anterior segment lens was used. Only those images that were considered good quality were included. AS-OCT B-scans from the nasal/temporal quadrant were selected per eye, and these images were analyzed by two blinded (blinded to the diagnosis) glaucoma experts for the presence or absence of ADoA, which were then subsequently used for machine learning (ML). A total of 800 AS-OCT B-scans were included, out of which 340 images (1 B-scan per eye) were used to build the ML model, and the rest were used for validation. Out of 340 images, 170 scans included PCG (n = 27), JOAG (n = 86), and POAG (n = 57) eyes, and the rest were healthy controls. ADoA was defined as the absence of Schlemm’s canal (SC), the presence of extensive hyperreflectivity over the region of the trabecular meshwork, or a hyperreflective membrane (HM) over the region of the trabecular meshwork.[7] As there are many channels seen on cross-sectional images of the angle,[25] it is essential to properly define the SC. We defined the SC as the first (nearest to the edge of the trabecular meshwork/angle recess) well-demarcated hypolucent space within a 350-µm perpendicular distance from the edge of the TM. It was considered present when observed on at least two consecutive B-scans (to differentiate it from artifacts). If SC/HM was identified as present in any one quadrant but absent in the other, it was considered as present for analysis.[8] Two glaucoma specialists masked to details of the patients evaluated the scans regarding the absence of SC and the presence of HM as indicative of ADoA. If there was discord between the two, a third adjudicated the case. Fig. 1 shows representative images of angles that are normal (a and b) and those with dysgenesis (c and d).
Figure 1.
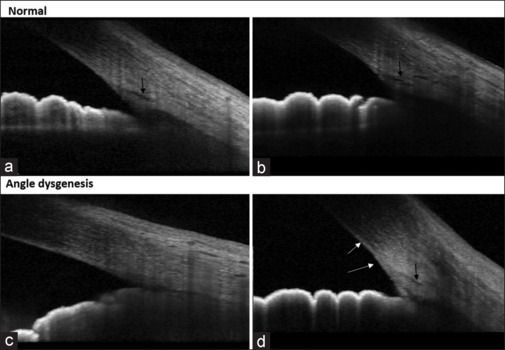
Representative high-definition AS-OCT images of normal healthy eyes (a and b) versus those with angle dysgenesis (c and d). Black arrows show the location of Schlemm’s canal, and the white arrows show the presence of a hyperreflective membrane at the angle
Data preprocessing
The images were encoded by removing patient details and giving unique reference numbers. Supplemental Fig. 1 (1.8MB, tif) summarizes the workflow used in the study. Each image was cropped manually in two ways to extract the iridocorneal angle (ICA) area and a trabecular meshwork (TM) area by a single observer. For this study, the ICA area was defined as 1100 × 900 pixels ± 10% (1210 × 990 µm) rectangular area including the region covering TM, SC, a part of cornea in continuation with a part of sclera, and iris. The images were further cropped to get a TM area defined as 600 × 400 pixels ± 10% (660 × 440 µm), including SC, scleral spur, and TM region [Fig. 2].
Figure 2.
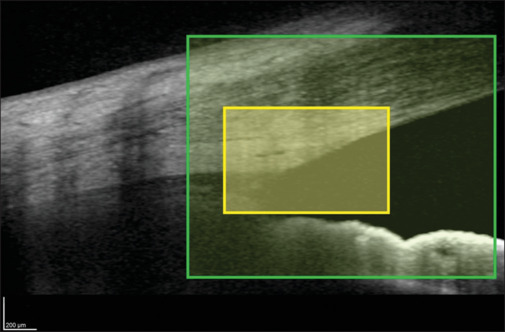
Anterior-segment SD-OCT image showing the iridocorneal angle area (green rectangle) and the trabecular meshwork area (yellow rectangle)
Model training and evaluation
The two datasets (ICA area and TM area images), each having 8160 images, were randomly split into training (n = 7996) and testing (n = 164) subsets with a ratio of 98:2 [Supplemental Fig. 1 (1.8MB, tif) ]. The applied split ratio was considered so that the maximum number of images could be used for model training. The test set was used only for the final evaluation of the model performance, and none of the images in the test set were used for training.
We applied the transfer learning (TL) method to classify a given SD-OCT image as either having a normal angle or angle dysgenesis. In MATLAB, all the available 19 pretrained convolutional neural network (CNN) models, including SqueezeNet, ResNet-18, GoogleNet, ResNet-50, DarkNet-53, DarkNet-19, ShuffleNet, NasnetMobile, Nasnet Large, Xception, Place-365-Google, MobileNet V2, DenseNet-201, Inception-ResNet V2, Inception-V3, ResNet-101, VGG-19, VGG-16, and AlexNet, were trained using our datasets. All the images were resized to the required pixels depending on the CNN model being trained. First, all the models were trained using the default parameters, and the most efficient ones were prioritized. The hyperparameters of the prioritized models were further tuned in a stochastic gradient descent manner (SGDM) based on the minimization of mean squared error with the combinations of different batch sizes, epochs, learning rates, momentum, and drop factor.
To obtain robust models, different groupings of 23 augmented images were also evaluated along with models with single augmented images and models with all combined augmented images. Finally, 74 models were developed using varied hyperparameters and augmented image combinations. Prediction quality was assessed by overall accuracy, specificity, sensitivity, area under the ROC curve, and comparison with the image annotations of two glaucoma experts (glaucoma specialists with 10 years of specialization). Whenever there was a lack of consensus between the two glaucoma experts, a third glaucoma specialist adjudicated the case.
Genetic correlation with ADoA
In total, 393 AS-OCT angle images of 27 unrelated JOAG patients with known mutations were compared with 320 images of healthy controls. These JOAG patients had undergone whole exome sequencing (WES) followed by bioinformatics analysis and had been found to harbor a mutation (that was either of uncertain significance, likely pathogenic or pathogenic as per the ACMG criteria or known to be associated with glaucoma) in a known glaucoma gene. WES capture was performed using the Sure Select Clinical Research Exome V2 kit (Agilent Technologies, Santa Clara, CA). Variant analysis was performed using the GenomeAnalysisTK-3.6 toolkit. The variant call files (VCFs) generated were analyzed using Golden Helix VarSeq Software v. 1.2.1 (Bozeman, MT). VarSeq variants with read depth <15 and genotype quality score <20 were excluded. To identify rare mutations, variant frequency databases were used to remove variants that were present at high frequencies among large population groups. The remaining variants were filtered according to minor allele frequency (MAF) <0.001 in multiple databases, including Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/), 1000 Genomes Project (http://browser.1000genomes.org), and gnomAD (http://gnomad.broadinstitute.org/). Variants among the known glaucoma genes were filtered based on only those exonic variants that were non-synonymous missense variants, frameshift and indels, splice region variants, and were absent among ethnically and geographically matched healthy controls.
The DL models were tested with these patients’ scans to detect ADoA.
Validation of DL predictions
Validations of the final DL models were performed using the following types of independent datasets: a) An independent external validation dataset of 67 images; b) A comparative validation between the model prediction and human experts where the two glaucoma experts masked to the details of the patients evaluated the SD-OCT scans (n = 73), and their results were compared with the final model prediction; and c) A genetics validation dataset consisted of 393 images of patients with known mutations and 320 images of healthy controls without any glaucoma gene mutation. This was a blind check validation where the results of the molecular analysis were blinded from the AI experts.
Statistical analysis
To compare the outcomes from the validation studies, statistical analyses were performed using a statistical software package (SPSS v. 26.0; SPSS, Inc., Chicago, IL, USA). To determine the agreement between the experts and DL prediction, Cohen’s κ test was applied. The receiver operating characteristic (ROC) analysis was performed for the external validation dataset, and the area under the ROC (AUROC) curve was determined for comparisons.
Results
Table 1 shows the clinical and demographic characteristics of the study subjects. Using TL on two approaches as mentioned below, we built a consensus-based algorithm consisting of the three best models for differentiating angle dysgenesis from the normal angle [Fig. 1]. The performance measures of these models are given in Supplemental Table 1.
Table 1.
Demographic and clinical details of subjects whose AS-OCT B-scans (1 B-scan image per eye, n=340) were used for machine learning model preparation
| Characteristics | PCG | JOAG | POAG | Normal (Control) |
|---|---|---|---|---|
| Number of subjects | 16 | 62 | 37 | 85 |
| Number of eyes | 27 | 86 | 57 | 170 |
| Laterality | ||||
| • Bilateral | 11 | 24 | 20 | 85 |
| • Unilateral | 5 | 38 | 17 | 0 |
| Age (years) at the time of study (Mean±SD) | 14.9±3.7 | 27±7.1 | 62.9±9.24 | 38.4±8.6 |
| Gender | ||||
| • Male | 5 (31.25%) | 44 (71%) | 25 (67.5%) | 45 (53%) |
| • Female | 11 (68.75%) | 18 (29%) | 12 (32.5%) | 40 (47%) |
| IOP (mmHg) at the time of the study (Mean±SD) | 14.2±1.4 | 13.8±1.2 | 15.1±1.3 | 16.2±0.8 |
PCG=Primary congenital glaucoma, JOAG=Juvenile open-angle glaucoma, POAG=Primary open-angle glaucoma. AS-OCT=Anterior Segment Optical Coherence tomography, IOP=Intraocular pressure, SD=Standard deviation
Supplemental Table 1.
Architecture, optimized hyperparameters and performance of the three final classification models
| Model-1 (inceptionresnet-v2) | Model-2 (inceptionresnet-v2) | Model-3 (mobilenet-v2) | |
|---|---|---|---|
|
| |||
| Internal testing dataset | |||
| Total images | 8160 | 8160 | 8160 |
| Training dataset (98%) | 7996 | 7996 | 7996 |
| Testing dataset (2%) | 164 | 164 | 164 |
| Accuracy % | 97.56 | 98.17 | 98.78 |
| Sensitivity | 0.964 | 0.97 | 0.987 |
| Specificity | 0.987 | 0.987 | 0.987 |
| Precision | 0.987 | 0.98 | 0.987 |
| Recall | 0.964 | 0.97 | 0.987 |
|
External independent validation dataset | |||
| External dataset (n) | 67 | 67 | 67 |
| Accuracy % | 83.58 | 74.6 | 80.6 |
| Sensitivity | 0.68 | 0.64 | 0.80 |
| Specificity | 0.92 | 0.78 | 0.80 |
In the first approach, the iridocorneal angle area dataset was used to train all the 19 CNN models [Fig. 2]. The most efficient model was built using Inception-ResNetV2, a 164-layer-deep convolutional neural network previously trained on more than a million images.[26] All the images were rescaled to 299 × 299 pixels as the input image prerequisite of Inception-ResNetV2 and finally utilized SGDM optimizer with a learning rate of 0.005, 45 epochs, and mini-batch size of 64 after hyperparameter optimization [Supplemental Table 1]. The model achieved the accuracy, sensitivity, and specificity of 97.56%, 96.4%, and 98.7%, respectively, on the internal test dataset.
In the second approach, the TM area was used, and the two best models were trained using Inception-ResNetV2[26] and MobileNetV2[27] neural networks. MobileNetV2 is a convolutional neural network that requires an input image of 229 × 229 pixels. Using the TM area test images, the models achieved an accuracy of 98.17% and 98.78%, with a sensitivity of 97% and 98.7%, and a specificity of 98.7% in each of the cases, respectively [Supplemental Table 1]. The consensus-based outcome from the three CNN TL models is the final predicted classification, which could recognize pixel patterns corresponding to the abnormalities at the angle, helping in better classification among the glaucoma group and controls.
External validation dataset
To further evaluate the accuracy and reproducibility of our models, we tested them on an independent external validation dataset consisting of 67 images. The models trained with the combined augmented and the original images exhibited lower accuracy than those trained on original images alone. Thus, we did not proceed with the augmented images, and all the validation studies were carried out using original images only. Model 1 had the best accuracy and specificity but the lowest sensitivity, whereas the other two models showed good sensitivity and comparable accuracies [Supplemental Table 1]. The consensus-based outcome ensures inclusiveness of the mandatory training features after the tradeoff and reaching one outcome. The area under the ROC curves was >0.80 for all three models, indicating good performances of the models in detecting ADoA [Fig. 3].
Figure 3.
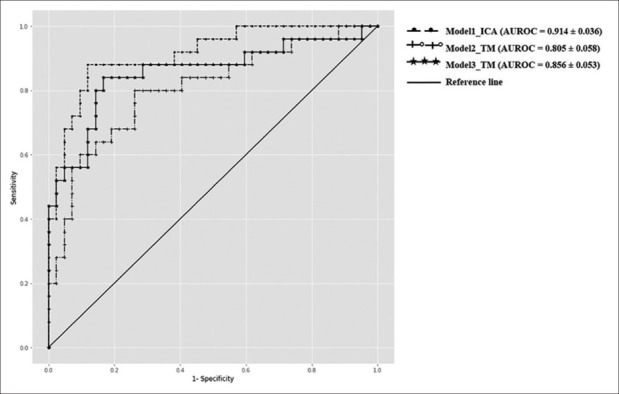
Receiver operating characteristic (ROC) curves for the three deep learning models using an external validation dataset. *AUROC = Area under the ROC curve, TM = Trabecular meshwork, ICA = Iridocorneal angle
Comparison of the model’s performance with human experts
The comparative prediction analysis is summarized in Table 2. The consensus-based result achieved a maximum accuracy of 83%, reiterating the importance of consensus-based decision-making in clinical settings. To determine the agreement between the expert’s decision and consensus-based prediction, Cohen’s Kappa test was carried out between expert1-model prediction and expert2-model prediction. There was a good agreement between the expert1-model prediction (κ = 0.619, P < 0.05) and expert2-model prediction (κ = 0.60, P < 0.05) suggesting a high degree of similarity between the expert’s decision and the model prediction.
Table 2.
Comparison between the models (M1, M2, and M3) prediction and expert’s decision
| M1 | M2 | M3 | Final consensus-based prediction | |
|---|---|---|---|---|
| Expert1 | 80.82% | 79.45% | 80.82% | 83.56% |
| Expert2 | 67.12% | 79.45% | 78% | 72.6% |
MI: Model1. M2: Model2. M3: Model3
Genetic validation dataset
Out of 27 (unrelated) patients who had known gene mutations, 15 had MYOC mutations, eight had CYP1B1, two had FOXC1, and two had LTBP2 mutation. The detailed genotype of these patients is provided in Table 3. These patients had 12 different mutations, and all except three (that were frameshift) were missense. All mutations except one in the CYP1B1 gene (p.Arg368His) were heterozygous.
Table 3.
Genetic mutations among 27 patients and angle dysgenesis on AS-OCT as determined by AI consensus
| Gene (number of patients) | Mutation | South Asian MAF | Age of onset (y) | Percentage scans with angle dysgenesis predicted by AI |
|---|---|---|---|---|
| MYOC(1) | p.Pro481Thr | NA | 12 | 88 |
| MYOC(2) | p.Lys423Gln | NA | 15±2.8 | 90 |
| MYOC(2) | p.Thr377Lys | NA | 22±1.4 | 68 |
| MYOC(3) | p.Pro370Leuc | 0 | 14.3±3 | 95 |
| MYOC(4) | p.Gly367Arg | 0 | 28.3±6.5 | 60 |
| MYOC(3) | p.Gln337Arg | 0 | 31±6.2 | 70 |
| CYP1B1(3) | p.Arg368His | 0.01 | 30.7±5.3 | 95 |
| CYP1B1(1) | Frameshift (p.Pro321Serf*104) | NA | 31 | 90 |
| CYP1B1(4) | p.Pro193Leu | 0.01 | 19.2±6.2 | 75 |
| FOXC1(2) | Frameshift( p.Gly418Alafs*27) | NA | 20±1.4 | 80 |
| LTBP2(1) | Frameshift (p.Val801Hisfs*18) | NA | 36 | 100 |
| LTBP2(1) | p.Pro229Thr | 0 | 30 | 100 |
MAF=Minor allele frequency, AS-OCT=Anterior segment optical coherence tomography, AI=Artificial intelligence
Among these patients, ADoA was observed as predicted by AI among all patients with known gene mutations. The maximum number of scans showing ADoA were observed with MYOC p.Pro370Leuc, CYP1B1 p.Arg368His, and with LTBP2 frameshift (p.Val801Hisfs*18) and p.Pro229Thr mutation. Gonioscopically angle dysgenesis was not seen among any of the MYOC patients [Fig. 4]. However, features of angle dysgenesis were seen both on gonioscopy and on AS-OCT among those with CYP1B1, FOXC1, and LTBP2 mutation.
Figure 4.
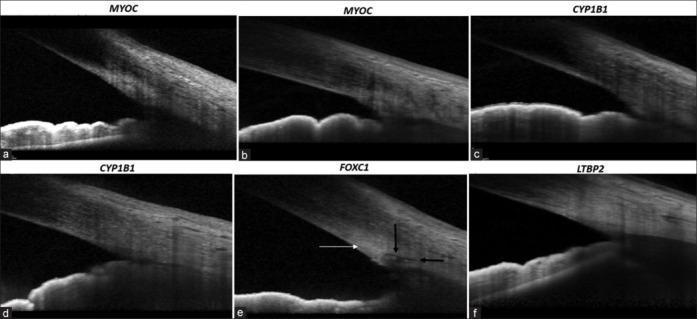
Anterior-segment SD-OCT images of patients with (a) MYOC p.Gly367Arg showing intense hyperreflectivity at the TM, (b) MYOC p.Pro370Leuc showing intense hyperreflectivity at the TM, (c) CYP1B1 p.Arg368His showing intense hyperreflectivity at the TM with non-discernible SC, (d) CYP1B1 p.Pro193leu with non-discernible SC, (e) FOXC1 p.Gly418Alafs*27 showing intense hyperreflectivity at the TM (white arrow) with the presence of SC (black arrows), (f) LTBP2 p.Pro229Thr showing intense hyperreflectivity at the TM with absent SC
Overall, AI was predictive of angle dysgenesis in 81% of scans among MYOC-positive patients, 89% of CYP1B1 patients, 85% of FOXC1, and 96% among those with LTBP2 mutation on average compared to only 5% of the healthy controls. Although CYP1B1 and LTBP2 mutations were seen to primarily affect SC morphology, the MYOC and FOXC1 mutations were found to be associated with morphological variations in the TM because the SC was visualized on most scans. The age of onset of glaucoma did not correlate with the extent of angle dysgenesis.
Discussion
This study used DL to build models that identify ADoA among open-angle glaucoma patients. We hypothesized that the angle dysgenesis exhibited as disturbances in the extracellular matrix of TM, SC, and adjoining regions in open-angle glaucoma patients could be reflected as changes (as they occur on histopathological sections), and the DL-based models can identify these variations by classifying the iridocorneal angle as having dysgenesis or not based on SD-OCT scans.
High-definition AS-OCT can pick up anatomical variations in SC and TM morphology, which are not visible on enface gonioscopy.[7] Gonioscopy and goniophotography, in contrast, require much expertise and remain observer-dependent. Moreover, sometimes what appears to be a normal open angle on gonioscopy, may be harboring dysgenesis in the form of an impermeable hyperreflective membrane that may be visible only on an AS-OCT.[7] Hence, AS-OCT,[1] which has been used primarily for identifying angle closure, would be of use even in patients with POAG and their relatives in identifying ADoA, which could be important in deciding the role of angle-based surgeries.[8]
In the present study, three DL-based models were developed for a consensus-based outcome to predict the presence of ADoA among open-angle glaucoma patients. Out of all the 19 TL algorithms used in model building, Inception ResNetV2 and MobileNetV2 achieved superior performances on the ICA and TM areas. Whether an eye has angle dysgenesis was predicted with >95% accuracy in the internal dataset and >80% accuracy in the two different external validation datasets used in the present study.
Considering the phenotypic, genotypic, and histopathological complexity in open-angle glaucoma, DL has been implemented using different approaches.[28,29,30,31,32] Studies have evaluated the potential of implementing DL in primary angle closure disease and have shown promising results.[33,34] We have shown that SD-OCT scans can successfully capture the anterior angle at high resolution and may help in identifying angle dysgenesis in cases where angle anomalies are subtle enough to go unnoticed. Although identifying gross dysgenesis of the angle, as in PCG may be easier,[35] subtle angle anomalies as in JOAG or POAG are more challenging to identify. The biological changes in the extracellular matrix (ECM) comprising the trabecular drainage pathways that lead to IOP elevation have been identified in histopathological studies[36,37] in POAG. With the DL models used in the present study, we could identify these ECM changes in the TM along with abnormalities in the SC morphology in vivo.
DL requires an enormous amount of annotated training data, which is challenging to obtain in rare disorders.[38] However, TL and augmentation techniques are effective strategies to be used in cases with a limited dataset size.[39] TL demonstrates compelling results, particularly in cases where the data available for building the models is limited.[11,39] In the present study, 19 types of CNN algorithms were trained, with each image in the training dataset augmented in 23 different ways. This increased the number of images in the training dataset and ensured that the model was trained on various images, making it more robust and reliable to be used in clinical settings. The robustness was also evident because the prediction for external dataset images displayed better results in their original form than with augmentations. This indicates that in natural settings, apart from pixel changes, no query image augmentation is required.
In addition, we looked for any pattern between the DL predictions of ADoA and specific gene mutations. Interestingly in all the mutation-positive patients, ADoA was predicted in over 80% of images. This highlights the fact that the model efficiently captures the differentiating extracellular matrix features in normal and mutation-positive AS-OCT scans. Most gene mutation studies on animal models of glaucoma have provided insights into the pathogenesis of outflow channels in controlled experiments. In contrast, in-vivo analyses of human eyes with rare disease-causing mutations provide a better understanding of the anatomical effects of these mutations. Although the mutations in the MYOC gene are known to cause aggregation of the misfolded myocilin protein, leading to TM cell toxicity and eventually death, there is no evidence in the literature to suggest the role of the MYOC gene mutations in alterations in angle morphology. There is only one histopathological report of a JOAG patient with MYOC Tyr453His mutation, where no apparent changes of the TM or SC were noted, though intense MYOC immune reactivity was observed at the TM.[40] Nevertheless, there is evidence to suggest that MYOC mutations are associated with goniodysgenesis. Chen X et al.[41] reported a 3-generation JOAG family with Pro370Leu mutation in the MYOC gene in all affected members who also had goniodysgenesis. This evidence is further strengthened by the reports of the association of MYOC gene mutations with congenital glaucoma.[17,41] In contrast, CYP1B1-related cases of PCG have been shown to have histopathological evidence of goniodysgenesis, involving not only the TM and SC but also the collector channels.[42]
In our study too, the subset of JOAG patients with CYP1B1 gene mutations showed ADoA, as predicted by DL models. We also found FOXC1 and LTBP2 mutations among our patients with no other features of either Axenfeld–Reiger Syndrome (ARS) or zonular abnormalities classically associated with these gene mutations. Two cases of JOAG with LTBP2 mutations have been described,[43,44] one by Saeedi et al.[43] and the other by our group.[44] LTBP2 gene mutations are known to express a wide variety of ocular phenotypes (as with other monogenic disorders), ranging from primary trabecular meshwork dysgenesis to a Marfans-like zonular disease. Although FOXC1 mutations have been commonly associated with ARS, they are also known to occur in adult-onset POAG and JOAG.[45] In our study, two unrelated patients harbored the same FOXC1 frameshift mutation, which is novel. Our findings demonstrate that probably different gene mutations affect different parts of the proximal outflow pathways. CYP1B1 and LTBP2 were found to affect primarily the SC morphology; in contrast, MYOC and FOXC1 were found to be associated with morphological variations in the TM because the SC in the latter was normally developed.
The strength of the study is that the final prediction was based on three distinct models, each utilizing unique regions of ICA or TM, to ensure there was no repeat sampling bias. The study’s limitation was the small and heterogeneous sample size in the training dataset (n = 340) used for model building. However, we enhanced the input data by using augmentation techniques. Another drawback of the limited data set was our inability to correlate the gene mutations with the clinical severity of the disease, which was not within the ambit of our research, as our study was focused on evaluating the association of gene mutations with DL-predicted angle dysgenesis. There is also the possibility that surgical intervention would influence the changes occurring at the angle, though we took only images of the nasal and temporal quadrants, away from the superior site of surgery, and excluded eyes that had had angle surgery. Many images on AS-OCT have to be discarded due to poor quality and image artifacts at the ICA area due to the reflectance from the superficial vessels. This would be taken care of, hopefully, in the newer generation machines, which would have better resolution too. Moreover, the results of this study need to be replicated in other populations.
Conclusion
Notwithstanding these limitations, the importance of the study lies in having addressed a crucial as well as unique issue of in vivo identification of angle dysgenesis. In conclusion, the DL models in this study were effective in identifying angle dysgenesis on AS-OCT images and could be correlated with gene mutations in a subset of patients.
Financial support and sponsorship:
Funding for this study was received from Indian Council of Medical Research, New Delhi (Grant no. ISRM/12 (58)/2019). DG acknowledges the funding by the Department of Biotechnology, Government of India (Grant No. BT/PR40151/BTIS/137/5/2021).
Conflicts of interest:
There are no conflicts of interest.
Workflow used in deep learning of anterior-segment SD-OCT images
References
- 1.Ang M, Baskaran M, Werkmeister RM, Chua J, Schmidl D, Aranha Dos Santos V, et al. Anterior segment optical coherence tomography. Prog Retin Eye Res. 2018;66:132–56. doi: 10.1016/j.preteyeres.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Hong J, Xu J, Wei A, Wen W, Chen J, Yu X, et al. Spectral-domain optical coherence tomographic assessment of schlemm’s canal in chinese subjects with primary open-angle glaucoma. Ophthalmology. 2013;120:709–15. doi: 10.1016/j.ophtha.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Kagemann L, Wollstein G, Ishikawa H, Sigal IA, Folio LS, Xu J, et al. 3D visualization of aqueous humor outflow structures in-situ in humans. Exp Eye Res. 2011;93:308–15. doi: 10.1016/j.exer.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Usui T, Tomidokoro A, Mishima K, Mataki N, Mayama C, Honda N, et al. Identification of Schlemm’s canal and its surrounding tissues by anterior segment fourier domain optical coherence tomography. Invest Opthalmol Vis Sci. 2011;52:6934–9. doi: 10.1167/iovs.10-7009. [DOI] [PubMed] [Google Scholar]
- 5.Buller C, Johnson D. Segmental variability of the trabecular meshwork in normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 1994;35:3841–51. [PubMed] [Google Scholar]
- 6.Gould DB, Smith RS, John SW. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48:1015–29. doi: 10.1387/ijdb.041865dg. [DOI] [PubMed] [Google Scholar]
- 7.Gupta V, Chaurasia AK, Gupta S, Gorimanipalli B, Sharma A, Gupta A. In vivo analysis of angle dysgenesis in primary congenital, juvenile, and adult-onset open angle glaucoma. Invest Opthalmol Vis Sci. 2017;58:6000.. doi: 10.1167/iovs.17-22695. [DOI] [PubMed] [Google Scholar]
- 8.Varshney T, Azmira K, Gupta S, Mahalingam K, Singh A, Angmo D, et al. In vivo imaging of the schlemm’s canal and the response to selective laser trabeculoplasty. Am J Ophthalmol. 2022;234:126–37. doi: 10.1016/j.ajo.2021.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Fauw Jd, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–50. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 10.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 11.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–31.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Bayat B, Yazdani S, Alavi A, Chiani M, Chitsazian F, Tusi BK, Suri F, et al. Contributions of MYOC and CYP1B1 mutations to JOAG. Mol Vis. 2008;14:508–17. [PMC free article] [PubMed] [Google Scholar]
- 13.van der Merwe EL, Kidson SH. Wholemount imaging reveals abnormalities of the aqueous outflow pathway and corneal vascularity in Foxc1 and Bmp4 heterozygous mice. Exp Eye Res. 2016;146:293–303. doi: 10.1016/j.exer.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Ji Y, Buel SM, Amack JD. Mutations in zebrafish pitx2 model congenital malformations in Axenfeld-Rieger syndrome but do not disrupt left-right placement of visceral organs. Deve Biol. 2016;416:69–81. doi: 10.1016/j.ydbio.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souma T, Tompson SW, Thomson BR, Siggs OM, Kizhatil K, Yamaguchi S, et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest. 2016;126:2575–87. doi: 10.1172/JCI85830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollander DA, Sarfarazi M, Stoilov I, Wood IS, Fredrick DR, Alvarado JA. Genotype and phenotype correlations in congenital glaucoma: CYP1B1 mutations, goniodysgenesis, and clinical characteristics. Am J Ophthalmol. 2006;142:993–1004. doi: 10.1016/j.ajo.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti S, Kaur K, Komatireddy S, Acharya M, Devi KR, Mukhopadhyay A, et al. Gln48His is the prevalent myocilin mutation in primary open angle and primary congenital glaucoma phenotypes in India. Mol Vis. 2005;11:111–3. [PubMed] [Google Scholar]
- 18.Chen Y, Jiang D, Yu L, Katz B, Zhang K, Wan B, et al. CYP1B1 and MYOC mutations in 116 Chinese patients with primary congenital glaucoma. Arch Ophthalmol. 2008;126:1443–7. doi: 10.1001/archopht.126.10.1443. [DOI] [PubMed] [Google Scholar]
- 19.Kaur K, Reddy AB, Mukhopadhyay A, Mandal AK, Hasnain SE, Ray K, et al. Myocilin gene implicated in primary congenital glaucoma. Clin Genet. 2005;67:335–40. doi: 10.1111/j.1399-0004.2005.00411.x. [DOI] [PubMed] [Google Scholar]
- 20.Braghini CA, Neshich IA, Neshich G, Soardi FC, de Mello MP, Costa VP, et al. New mutation in the myocilin gene segregates with juvenile-onset open-angle glaucoma in a Brazilian family. Gene. 2013;523:50–7. doi: 10.1016/j.gene.2013.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Bruttini M, Longo I, Frezzotti P, Ciappetta R, Randazzo A, Orzalesi N, et al. Mutations in the myocilin gene in families with primary open-angle glaucoma and juvenile open-angle glaucoma. Arch Ophthalmol. 2003;121:1034–8. doi: 10.1001/archopht.121.7.1034. [DOI] [PubMed] [Google Scholar]
- 22.Wiggs JL, Allingham RR, Vollrath D, Jones KH, De La Paz M, Kern J, et al. Prevalence of mutations in TIGR/Myocilin in patients with adult and juvenile primary open-angle glaucoma. Am J Hum Genet. 1998;63:1549–52. doi: 10.1086/302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao YH, Wang YQ, Fang WF, Zhang L, Yang JH, Zhu YH. A recurrent G367R mutation in MYOC associated with juvenile open angle glaucoma in a large Chinese family. Int J Ophthalmol. 2018;11:369–74. doi: 10.18240/ijo.2018.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoilova D, Child A, Brice G, Desai T, Barsoum-Homsy M, Ozdemir N, et al. Novel TIGR/MYOC mutations in families with juvenile onset primary open angle glaucoma. J Med Genet. 1998;35:989–92. doi: 10.1136/jmg.35.12.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Butt A, Chien JL, Ghassibi MP, Furlanetto RL, Netto CF, et al. Characteristics and variations of in vivo Schlemm’s canal and collector channel microstructures in enhanced-depth imaging optical coherence tomography. Br J Ophthalmol. 2117;101:808–13. doi: 10.1136/bjophthalmol-2016-309295. [DOI] [PubMed] [Google Scholar]
- 26.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence. San Francisco, California, USA: AAAI Press; 2017. Inception-v4, inception-ResNet and the Impact of Residual Connections on Learning; pp. 4278–84. [Google Scholar]
- 27.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition: IEEE. 2018:4510–20. [Google Scholar]
- 28.Medeiros FA, Jammal AA, Thompson AC. From machine to machine: An OCT-trained deep learning algorithm for objective quantification of glaucomatous damage in fundus photographs. Ophthalmology. 2019;126:513–21. doi: 10.1016/j.ophtha.2018.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ran AR, Cheung CY, Wang X, Chen H, Luo LY, Chan PP, et al. Detection of glaucomatous optic neuropathy with spectral-domain optical coherence tomography: A retrospective training and validation deep-learning analysis. Lancet Digit Health. 2019;1:e172–e82. doi: 10.1016/S2589-7500(19)30085-8. [DOI] [PubMed] [Google Scholar]
- 30.Shuldiner SR, Boland MV, Ramulu PY, De Moraes CG, Elze T, Myers J, et al. Predicting eyes at risk for rapid glaucoma progression based on an initial visual field test using machine learning. PLoS One. 2021;16:e0249856.. doi: 10.1371/journal.pone.0249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson AC, Jammal AA, Medeiros FA. A deep learning algorithm to quantify neuroretinal rim loss from optic disc photographs. Am J Ophthalmol. 2019;201:9–18. doi: 10.1016/j.ajo.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefi S, Gupta K, Sun J, Huang X, Pasquale L, Lu L, et al. Novel genetic factors associated with primary open-angle glaucoma identified using artificial intelligence. Invest Ophthalmol Vis Sci. 2021;62:1491.. [Google Scholar]
- 33.Fu H, Xu Y, Lin S, Wong DWK, Baskaran M, Mahesh M, et al. Angle-closure detection in anterior segment OCT based on multilevel deep network. IEEE Trans Cybern. 2020;50:3358–66. doi: 10.1109/TCYB.2019.2897162. [DOI] [PubMed] [Google Scholar]
- 34.Xu BY, Chiang M, Chaudhary S, Kulkarni S, Pardeshi AA, Varma R. Deep learning classifiers for automated detection of gonioscopic angle closure based on anterior segment OCT images. Am J Ophthalmol. 2019;208:273–80. doi: 10.1016/j.ajo.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta V, Singh A, Pandya I, Sofi R, Sen S, Somarajan BI, et al. Differences in outflow channels between two eyes of unilateral primary congenital glaucoma. Acta Ophthalmol. 2021;99:187–94. doi: 10.1111/aos.14540. [DOI] [PubMed] [Google Scholar]
- 36.Alvarado J, Murphy C, Juster R. Trabecular meshwork cellularity in primary open-angle glaucoma and nonglaucomatous normals. Ophthalmology. 1984;91:564–79. doi: 10.1016/s0161-6420(84)34248-8. [DOI] [PubMed] [Google Scholar]
- 37.Alvarado J, Murphy C, Polansky J, Juster R. Age-related changes in trabecular meshwork cellularity. Invest Ophthalmol Vis Sci. 1981;21:714–27. [PubMed] [Google Scholar]
- 38.Altaf F, Islam SMS, Janjua NK. A novel augmented deep transfer learning for classification of COVID-19 and other thoracic diseases from X-rays. Neural Comput Appl. 2021;33:14037–48. doi: 10.1007/s00521-021-06044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imai S, Kawai S, Nobuhara H. Stepwise PathNet: A layer-by-layer knowledge-selection-based transfer learning algorithm. Sci Rep. 2020;10:8132.. doi: 10.1038/s41598-020-64165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Heide CJ, Alward WLM, Flamme-Wiese M, Riker M, Syed NA, Anderson MG, et al. Histochemical analysis of glaucoma caused by a myocilin mutation in a human donor eye. Ophthalmol Glaucoma. 2018;1:132–8. doi: 10.1016/j.ogla.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Yan N, Yun H, Sun J, Yu M, Zhou J, et al. Sequence analysis of MYOC and CYP1B1 in a Chinese pedigree of juvenile glaucoma with goniodysgenesis. Mol Vis. 2009;15:1530–6. [PMC free article] [PubMed] [Google Scholar]
- 42.García-Antón MT, Salazar JJ, Hoz Rd, Rojas B, Ramírez AI, Triviño A, et al. Goniodysgenesis variability and activity of CYP1B1 genotypes in primary congenital glaucoma. PLoS One. 2017;12:e0176386.. doi: 10.1371/journal.pone.0176386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeedi O, Yousaf S, Tsai J, Palmer K, Riazuddin S, Ahmed ZM. Delineation of novel compound heterozygous variants in LTBP2 associated with juvenile open angle glaucoma. Genes. 2018;9:527.. doi: 10.3390/genes9110527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somarajan BI, Gupta S, Mahalingam K, Azmira K, Gupta V. Digenic inheritance in juvenile open-angle glaucoma. J Pediatr Gene. 2021;12:150–4. doi: 10.1055/s-0040-1722213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina-Trillo C, Sanchez-Sanchez F, Aroca-Aguilar JD, Ferre-Fernández JJ, Morales L, Méndez-Hernández CD, et al. Hypo- and hypermorphic FOXC1 mutations in dominant glaucoma: Transactivation and phenotypic variability. PLoS One. 2015;10:e0119272.. doi: 10.1371/journal.pone.0119272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow used in deep learning of anterior-segment SD-OCT images


