Abstract
Translation of hepatitis C virus (HCV) and classical swine fever virus (CSFV) RNAs is initiated by cap-independent attachment (internal entry) of ribosomes to the ∼350-nucleotide internal ribosomal entry segment (IRES) at the 5′ end of both RNAs. Eukaryotic initiation factor 3 (eIF3) binds specifically to HCV and CSFV IRESs and plays an essential role in the initiation process on them. Here we report the results of chemical and enzymatic footprinting analyses of binary eIF3-IRES complexes, which have been used to identify the eIF3 binding sites on HCV and CSFV IRESs. eIF3 protected an internal bulge in the apical stem IIIb of domain III of the CSFV IRES from chemical modification and protected bonds in and adjacent to this bulge from cleavage by RNases ONE and V1. eIF3 protected an analagous region in domain III of the HCV IRES from cleavage by these enzymes. These results are consistent with the results of primer extension analyses and were supported by observations that deletion of stem-loop IIIb or of the adjacent hairpin IIIc from the HCV IRES abrogated the binding of eIF3 to this RNA. This is the first report that eIF3 is able to bind a eukaryotic mRNA in a sequence- or structure-specific manner. UV cross-linking of eIF3 to [32P]UTP-labelled HCV and CSFV IRES elements resulted in strong labelling of 4 (p170, p116, p66, and p47) of the 10 subunits of eIF3, 1 or more of which are likely to be determinants of these interactions. In the cytoplasm, eIF3 is stoichiometrically associated with free 40S ribosomal subunits. The results presented here are consistent with a model in which binding of these two translation components to separate, specific sites on both HCV and CSFV IRESs enhances the efficiency and accuracy of binding of these RNAs to 40S subunits in an orientation that promotes entry of the initiation codon into the ribosomal P site.
Hepatitis C virus (HCV) is the etiologic agent of most cases of posttransfusion hepatitis (1). It is a member of the genus Flaviviridae and is related to pestiviruses such as classical swine fever virus (CSFV) and bovine viral diarrhea virus. Members of this genus are enveloped positive-stranded RNA viruses. Their genomes encode a single polyprotein that is cleaved by virus-encoded and cellular proteinases to yield the various viral capsid and nonstructural proteins.
Initiation of translation of HCV and pestivirus polyproteins occurs as a result of ribosomal attachment to the ∼350-nucleotide (nt) internal ribosomal entry segment (IRES) (23, 28, 33). HCV and CSFV IRESs comprise almost the entire 5′ untranslated region and up to 30 nt of coding sequence. Although the sequences of these two IRESs differ at almost 50% of base positions, many of these nucleotide differences are covariant substitutions, indicative of conserved secondary and/or tertiary structures. HCV and CSFV IRESs contain four major structural domains, designated I to IV (see Fig. 1), and a pseudoknot upstream of the initiation codon (5, 11, 34). The structural integrity of the pseudoknot and of these domains is important for IRES function, but the roles that they play in internal initiation have not been defined (10, 11, 22, 25, 26, 28, 34, 35).
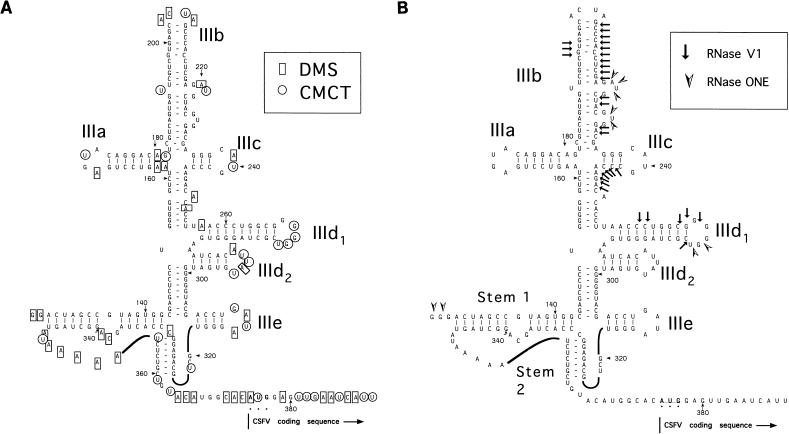
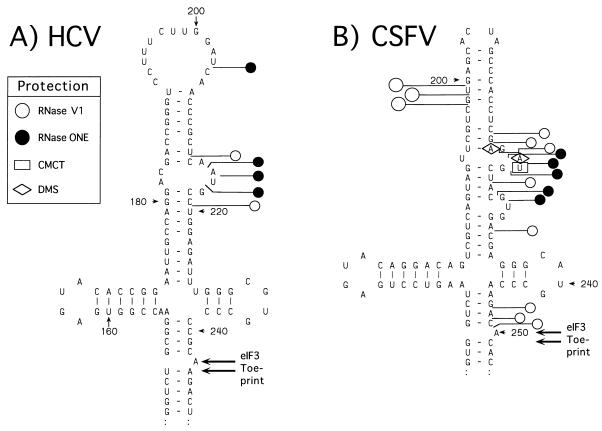
FIG. 1.
Summary of sites within nt 127 to 390 of the CSFV IRES that are modified by DMS and CMCT (A) or cleaved by RNase V1 and RNase ONE (B). These chemical and enzymatic probes are indicated by symbols at the upper right of each panel. The results are displayed on a structure that is based on previous proposals (5, 34) and that takes into account the results presented here. The nomenclature used to describe CSFV domains and hairpins is adapted from proposals (11) for the HCV IRES.
Many other aspects of the mechanism of IRES-mediated initiation are also obscure. To identify the factors that are involved in initiation on HCV and CSFV IRESs and to characterize their functions in it, we reconstituted this process in vitro up to the stage of 48S preinitiation complex formation from individual purified translation components (mRNA, aminoacylated initiator tRNA, 40S subunits, and eukaryotic initiation factors [eIFs]) (22). These 48S complexes are competent to complete the remaining steps in initiation of translation, including joining with the large (60S) ribosomal subunits to form 80S complexes and formation of the first peptide bond. 40S ribosomal subunits bind directly to these IRESs without additional factors to form binary complexes and require only eIF2, GTP and aminoacylated initiator tRNA to assemble into a 48S complex at the authentic initiation codon. The process of 48S complex formation on HCV and CSFV IRESs is exceptional because it does not involve the canonical initiation factors eIF4A, eIF4B, or eIF4F. eIF3 enhances 48S complex formation and is absolutely required for 80S complex formation on these IRESs. Assembly of ribosomal complexes was assessed in these experiments by primer extension inhibition (toeprinting) and sucrose density gradient centrifugation. In the course of these experiments, we noted that eIF3 arrested primer extension at AA243–244 on the HCV IRES and at AC250–251 and U304 on the CSFV IRES. These stops are all within domain III. Toeprinting involves cDNA synthesis with reverse transcriptase (RT) on a template RNA to which a ribosome or protein is bound. cDNA synthesis is arrested either directly by the bound complex, yielding a stop or toeprint at its leading edge, or indirectly by stabilization of adjacent helices (3, 9). eIF3 therefore binds to HCV and CSFV IRESs, most probably in the large central domain III, but toeprinting did not permit the binding site to be determined precisely.
We have now investigated the interaction of eIF3 with HCV and CSFV IRES elements in detail by using UV cross-linking and chemical and enzymatic footprinting techniques. Our results show that eIF3 bound specifically and exclusively to the apical region of domain III of both HCV and CSFV, resulting in the formation of stable ribonucleoprotein complexes. UV cross-linking showed that this interaction involves the p170, p116, p66, and p47 subunits. This is the first time that a specific interaction of eIF3 with an eukaryotic mRNA has been described. The role of this and other interactions of HCV and CSFV IRESs with translation components in IRES-mediated initiation is discussed.
MATERIALS AND METHODS
Plasmids.
Plasmids pHCV(40–372).NS′ and pCSFV(1–442).NS′ contain HCV and CSFV sequences, respectively, linked to a truncated influenza virus NS′ reporter (22, 24). pΔB-CAT, pΔE-CAT, and pΔF-CAT were derived by deletion of HCV nt 26 to 67, 172 to 227, and 229 to 238, respectively, from plasmid pWT-CAT, which contains the full HCV 5′ untranslated region linked to a chloramphenicol acetyltransferase (CAT) reporter cistron (26).
Transcription of HCV and CSFV RNAs.
HCV and CSFV RNAs were transcribed in vitro with T7 RNA polymerase and either with or without [32P]UTP (∼3,000 Ci/mmol; ICN Radiochemicals, Irvine, Calif.) from plasmids that had been linearized at appropriate sites. These restriction sites included the EcoRI site after the NS′ cistron in the HCV-NS′ and CSFV-NS′ plasmids, the BamHI site at the junction of the HCV and NS′ sequences in the HCV-NS′ plasmid, and the HindIII site after the CAT cistron in the HCV-CAT plasmids. RNA was purified with Nuc-Trap columns (Stratagene, La Jolla, Calif.) as described previously (19). Radiolabeled RNAs had specific activities of ∼400,000 cpm/μg.
Purification of eIF3.
eIF3 was purified from rabbit reticulocyte lysate (RRL; Green Hectares, Oregon, Wis.) by established procedures (15). The purity and quality of this factor (see Fig. 6) were equal to or greater than those described by us previously (20–22).
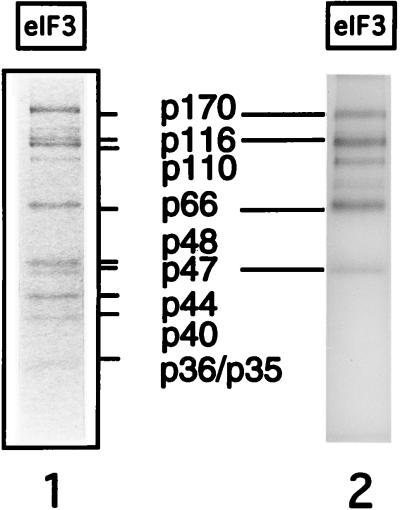
FIG. 6.
UV cross-linking in vitro of eIF3 and [32P]UTP-labelled HCV nt 40 to 373 RNA. Lanes: 1, Coomassie blue-stained gel of native eIF3; 2, autoradiograph of cross-linked eIF3. The cross-linked sample was digested with cobra venom nuclease and RNases A and T1. Polypeptides were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on an SDS–12% polyacrylamide gel. The designation of individual subunits is based on their electrophoretic mobility and that of known standards, except for p170, which was also identified by Western blotting.
Assembly and toeprint analysis of eIF3-HCV IRES complexes.
Complexes between eIF3 (7.2 pmol) and HCV RNA (2.4 pmol) were allowed to form during incubation for 5 min at 30°C in binding buffer (20 mM Tris-HCl [pH 7.4], 100 mM KCl, 2 mM magnesium acetate, 1 mM dithiothreitol). These complexes were analyzed by primer extension with primer 5′-CGCAAGCACCCTATC-3′ (complementary to HCV nt 295 to 309) and avian myeloblastosis RT (Promega, Madison, Wis.) as described previously (20), except that [α-32P]dATP was not present in the extension reaction mixtures and primers were instead first end labelled with [γ-32P]ATP (∼6,000 Ci/mmol; ICN Radiochemicals) by using T4 polynucleotide kinase.
Chemical and enzymatic footprint analysis of binary eIF3-IRES and ternary eIF3-40S subunit-IRES complexes.
RNP complexes were assembled from eIF3, 40S subunits, and HCV or CSFV RNAs as described above, with the amounts of these translation components described below. Free or protein-bound IRES-specific RNAs in binding buffer were digested enzymatically by incubation with either RNase V1 (Pharmacia, Piscataway, N.J.) or RNase ONE (Promega) or were modified chemically either at N-1 of adenines by incubation with dimethyl sulfate (DMS) or at N-3 of uracils and at N-1 of guanines by incubation with 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfate (CMCT), as described previously (13). Cleaved or chemically modified RNAs were hybridized to appropriate end-labelled DNA primers, and primer extension was done exactly as described previously (13). Primers 5′-CTCGTTTGCGGACATGCC-3′ and 5′-GGGATTTCTGATCTCGGCG-3′ (complementary to different parts of the NS′ coding sequence), 5′-GCAACTGACTGAAATGCC-3′ (complementary to part of the CAT coding sequence) and 5′-CGCAAGCACCCTATC-3′ (complementary to HCV nt 295 to 309) were used for analysis of complexes formed on CSFV-NS′, HCV-NS′, and HCV-CAT mRNAs, as appropriate. cDNA products were analyzed by electrophoresis on 10% polyacrylamide–8 M urea gels.
UV cross-linking.
UV cross-linking of binary IRES-eIF3 complexes, binary IRES-40S subunit complexes, and ternary IRES-eIF3-40S subunit complexes was done essentially as described previously (19, 22), with [32P]UTP-labelled CSFV nt 1 to 442 and HCV nt 40 to 372 RNAs, as appropriate.
RESULTS
Probing the structure of the CSFV IRES.
Models of the secondary and tertiary structures of the CSFV and HCV IRES have been proposed on the basis of minimum free-energy calculations, phylogenetic comparison, and genetic analysis (5, 11, 28, 30, 34). These models suggest that both RNAs are extensively base paired and consist of four major structural domains. Domain III consists of a basal pseudoknot and large irregular helix with several branching hairpins designated IIIa to IIIe. Domain IV of HCV contains the initiation codon within a single hairpin (11). The structural model of the HCV IRES has been experimentally verified and in part revised in light of the results of analysis with structure- and sequence-specific chemical and enzymatic probes (5, 10, 34). Models for the CSFV IRES have not been experimentally verified, and we therefore undertook a similar structural analysis of CSFV domains III and IV. In these experiments, we used RNase V1 from cobra venom, which cleaves double-stranded or other base-paired regions (6); RNase ONE from Escherichia coli, which cleaves single-stranded RNA without base specificity (14); DMS, which reacts with N-1 of adenine residues and to a lesser extent with N-3 of cytosine residues; and CMCT, which reacts with N-3 of uracil and N-1 of guanine residues (17). Chemical modification at these positions is inhibited by base pairing. Modification of these positions and strand scission both arrest cDNA elongation by RT and can therefore be analyzed by primer extension. Chemical modification results in arrest of primer extension at the nucleotide immediately 3′ to the modified position, and enzymatic cleavage results in arrest of primer extension at the nucleotide on the 3′ side of the cleaved bond. Numbering of the residues below indicates either the chemically modified base or the nucleotide on the 3′ side of the cleaved bond.
The CSFV IRES was extensively modified by DMS and CMCT (Fig. 1A). Residues that were modified by these chemicals were mostly clustered downstream of the pseudoknot, within the long interhelical strand of the pseudoknot, and at the apices of each of the proposed hairpins IIIa to IIIe in domain III. These results are almost wholly consistent with models of domain III and of the pseudoknot that have been proposed on the basis of phylogenetic comparisons (5, 34). However, they do not support the proposed structure of domain IV and instead indicate that this regions is almost entirely single stranded under ionic conditions that are used for efficient CSFV IRES-mediated translation in vitro.
The pseudoknot and the adjacent hairpins IIId2 and IIIe are resistant to cleavage by both RNase ONE and RNase V1 (Fig. 1B). The low reactivity of many DMS- and CMCT-insensitive residues in helices to RNase V1 and of DMS- and CMCT-sensitive regions in unpaired regions to RNase ONE suggests that the pseudoknot adopts a very compact structure and is thus sterically inaccessible to enzymatic cleavage. In contrast, the upper half of domain III was cleaved extensively by RNase V1 in a manner that is wholly consistent with the model proposed by Brown et al. (5). The small number of bonds that were cleaved by RNase ONE are clustered at the junction between domains II and III, in two internal bulges within the stem of hairpin IIIb, and in the apical loop of hairpin IIId1. The observation that these unpaired regions and some helical regions in the upper half of domain III are readily accessible to enzymatic cleavage indicates that they must be exposed.
Specific interaction of eIF3 with domain III of the CSFV IRES.
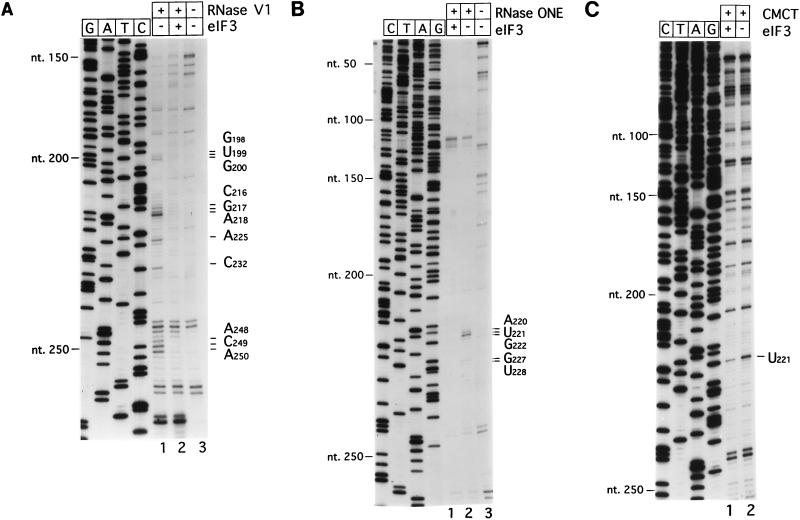
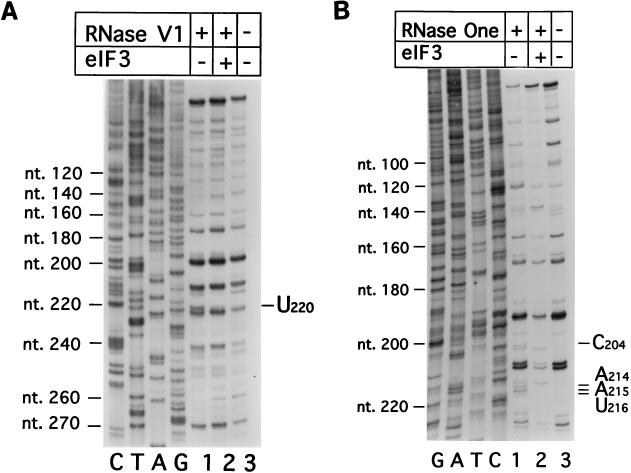
To localize eIF3 binding sites on the CSFV IRES, ribonucleoprotein complexes were assembled from these two moieties under buffer conditions and at a temperature similar to those that are used normally for translation of CSFV RNA (22). eIF3 was present in a threefold molar excess over IRES-containing RNA. We first used RNase V1 to identify cleavage sites within the CSFV IRES that are occluded by the binding of eIF3, because this enzyme cleaves the IRES at numerous sites in close proximity to the toeprint at AC250–251 that is caused by binding of eIF3 (22). eIF3 protected residues GUG198–200; CGA216–218, A225, C232, and ACA248–250 from cleavage by RNase V1 (Fig. 2A, lane 2). These protected regions flank two internal bulges in hairpin IIIb that are sensitive to cleavage by RNase ONE, and we therefore next assayed the protection of this region of the CSFV IRES by eIF3 against cleavage by this enzyme. These two bulges were bound by eIF3: residues GU227–228 and, particularly, AUG220–222 were protected by eIF3 from cleavage by RNase ONE (Fig. 2B, lane 1).
FIG. 2.
Chemical and enzymatic footprinting of the eIF3-CSFV IRES complex. Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension shows the sensitivity of CSFV RNA upstream of nt 265 to cleavage by RNase V1 (lanes 1 and 2) either alone (lane 1) or complexed with eIF3 (lane 2) (A), the sensitivity of CSFV RNA upstream of nt 259 to cleavage by RNase ONE (lanes 1 and 2) either alone (lane 2) or complexed with eIF3 (lane 1) (B), and the reactivity of CSFV RNA upstream of nt 252 to modification by CMCT (lanes 1 and 2) either alone (lane 2) or complexed with eIF3 (lane 1) (C). cDNA products obtained after primer extension of untreated CSFV RNA are shown in lanes 3 of panels A and B. A dideoxynucleotide sequence generated with the same primer was run in parallel on each gel. The positions of protected residues are indicated to the right of each panel, and the positions of CSFV nucleotides at 50-nt intervals are indicated to the left of each panel.
Binary CSFV IRES-eIF3 complexes were next footprinted with the base-specific chemical probes CMCT and DMS. Binding of eIF3 to CSFV RNA protected U221 from modification by CMCT (Fig. 2C, lane 1) and A218 and A220 from modification by DMS (data not shown). Residues within the apical loops of hairpins IIIa, IIIb, IIIc, IIId1, and IIId2 that are susceptible to chemical modification were not protected from such modification by eIF3. Bound eIF3 did not protect residues in CSFV domains I, II, or IV from enzymatic cleavage or chemical modification (data not shown).
The results of chemical and enzymatic footprinting are summarized in Fig. 5B. They indicate that eIF3 binds strongly and specifically to the apical region of domain III of the CSFV IRES. These results strongly support the results of primer extension analysis; the toeprint at AC250–251 caused by bound eIF3 probably corresponds to the leading edge of this factor, whereas the toeprint at U304 that is strengthened by the binding of eIF3 occurs within an adjacent helix that is stabilized by this interaction.
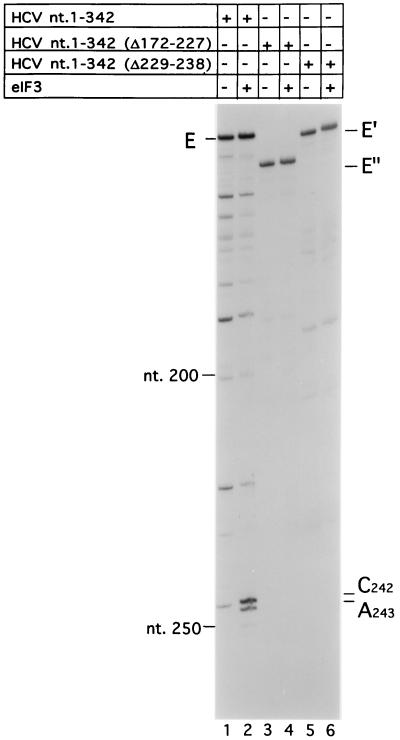
FIG. 5.
Primer extension analysis of the dependence of eIF3-HCV IRES ribonucleoprotein complex formation on the presence of hairpins IIIb and IIIc in domain III of the IRES. HCV nt 1 to 342 CAT mRNA (lanes 1 and 2), HCV nt 1 to 342(Δ172–227) CAT mRNA (lanes 3 and 4), and HCV nt 1 to 342(Δ229–238) CAT mRNA (lanes 5 and 6) were incubated with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) eIF3 under standard conditions. Primer 5′-CGCAAGCACCCTATC-3′ was annealed to HCV nt 295 to 309 of these mRNAs and extended with avian myeloblastosis virus RT. The full-length cDNA extension products derived from reverse transcription of HCV nt 1 to 342 CAT, nt 1 to 342(Δ229–238) CAT, and nt 1 to 342(Δ172–227)- CAT mRNAs are marked E, E′, and E", respectively. The cDNA products labelled A243 and A244 on the right terminated at these nucleotides. The positions of HCV nucleotides are indicated on the left.
Specific interaction of eIF3 with domain III of the HCV IRES.
To localize the eIF3 binding site on the HCV IRES, we assembled RNP complexes from these two moieties as described above for CSFV and then used RNases V1 and ONE as probes to identify sites in the IRES that are occluded from cleavage by the binding of eIF3. A single prominent RNase V1 cleavage site at U220 was occluded by binding of eIF3 (Fig. 3A, lane 2; also see Fig. 7A, lane 2). A number of other sites in the HCV IRES that appear to be protected by eIF3 from cleavage coincide with strong stops formed during primer extension on this highly structured RNA (Fig. 3, lanes 3). Protection of all but one of these cleavage sites is therefore equivocal and is not discussed below. The exception is the cleavage site at U212, which in Fig. 7A (lane 2) was clearly occluded by eIF3. In addition, RNase ONE cleavage at C204, A214, A215, and U216 was impaired by the binding of eIF3 (Fig. 3B, lane 2). These five protected sites are located close together within the irregular apical hairpin IIIb (Fig. 4A). The identification of this region of the HCV IRES as the eIF3 binding site strongly supports our earlier identification by toeprinting of an interaction between eIF3 and the apical half of domain III of this RNA (22). Bound eIF3 did not protect residues elsewhere in the HCV IRES from enzymatic cleavage (data not shown).
FIG. 3.
Enzymatic footprinting of the eIF3-HCV IRES complex. Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension shows the sensitivity of HCV RNA upstream of nt 225 to cleavage by RNase V1 (lanes 1 and 2) either alone (lane 1) or complexed with eIF3 (lane 2) (A) and the sensitivity of HCV RNA upstream of nt 277 to cleavage by RNase ONE (lanes 1 and 2) either alone (lane 1) or complexed with eIF3 (lane 2) (B). cDNA products obtained after primer extension of untreated HCV RNA are shown in lanes 3. A dideoxynucleotide sequence generated with the same primer was run in parallel on each gel. The positions of protected residues are indicated to the right of each panel, and those of HCV nucleotides are indicated to the left of each panel.
FIG. 7.
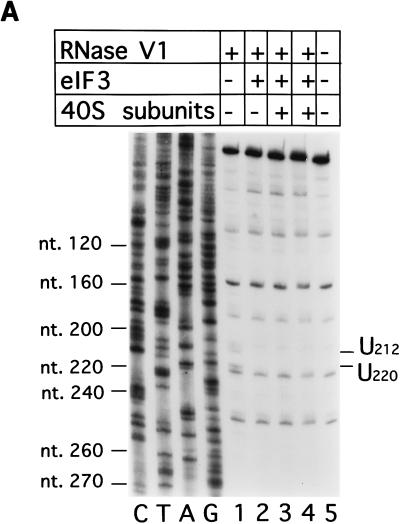
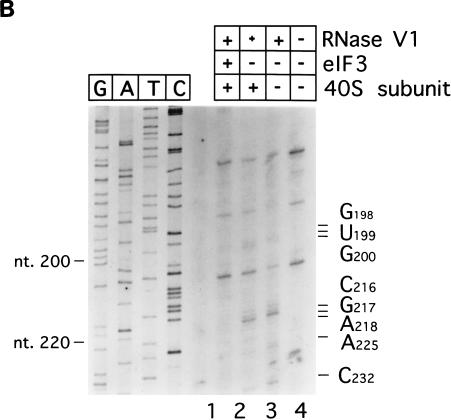
Enzymatic footprinting and UV cross-linking analysis of ternary eIF3-40S subunit-IRES complexes. (A) Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension showing the sensitivity of HCV RNA (1 pmol) upstream of nt 273 to cleavage by RNase V1 (lanes 1 to 4) either alone (lane 1), with 3 pmol of eIF3 (lane 2), with 3pmol of eIF3 and 3pmol of 40S subunits (lane 3), or with 3 pmol of eIF3 and 9 pmol of 40S subunits (lane 4). (B) Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension showing the sensitivity of CSFV RNA (1 pmol) upstream of nt 232 to cleavage by RNase V1 (lanes 1 to 3) either alone (lane 3), with 9 pmol of 40S subunits (lane 2), or with 3 pmol of eIF3 and 9 pmol of 40S subunits (lane 1). cDNA products obtained after primer extension of untreated HCV and CSFV RNA are shown in lane 5 of panel A in lane 4 of panel B, respectively. A dideoxynucleotide sequence generated with the same primer was run in parallel on each gel. The positions of protected residues are indicated to the right of each panel, and those of HCV or CSFV nucleotides are indicated to the left of each panel. (C) Autoradiograph of [32P]UTP-labelled CSFV nt 1 to 442 RNA (1 pmol) after UV cross-linking to 3 pmol of eIF3 (lanes 1 to 4), 9 pmol of eIF3 (lane 6), or 15 pmol of eIF3 (lane 7) and either 3 pmol of 40S subunits (lanes 2 and 5 to 7), 9 pmol of 40S subunits (lane 3), or 15 pmol of 40S subunits (lane 4), followed by RNase digestion and electrophoretic separation of polypeptides by SDS-polyacrylamide gel electrophoresis on an SDS–12% polyacrylamide gel.
FIG. 4.
Summary of the sites within the HCV (A) and CSFV (B) IRESs that are protected from modification by CMCT and DMS and from cleavage by the single-strand-specific RNase ONE and the double-strand-specific RNase V1. These chemical and enzymatic probes are indicated by symbols at the upper left. The results are displayed on secondary-structure models of the upper half of domain III that are based on previous proposals (5) (A) and on the results shown in Fig. 1 (B). Domains and subdomains are described according to the nomenclature in reference 11.
Abrogation of interaction of eIF3 with the HCV IRES by deletion of hairpins IIIb and IIIc.
The results of chemical and enzymatic footprinting analysis described above indicate that eIF3 binds to the apical stem of a cloverleaf-like structure that constitutes the apical half of domain III of the HCV IRES. To confirm that elements of this structure are required for this interaction to occur, we used toeprinting to assess the binding of eIF3 to the wild-type (wt) HCV IRES and to mutant HCV IRES transcripts that lack either the IIIb or IIIc hairpin (26). eIF3 strongly enhanced the arrest of primer extension at A243 and yielded a new stop at A244 on wt HCV nt 1 to 341 CAT RNA (Fig. 5, lane 2), as reported previously (22). However, eIF3 did not arrest primer extension on corresponding mRNAs that lacked either the IIIb hairpin or the IIIc hairpin (lanes 4 and 6). These results show that eIF3 did not bind stably to these two mutant IRES elements. They confirm that the interaction of eIF3 with the wt HCV IRES is highly specific and indicate that the IIIb and IIIc hairpins are important determinants of this interaction.
Identification of eIF3 subunits that bind to CSFV and HCV IRES elements.
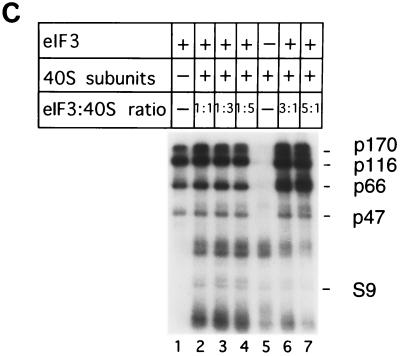
The mammalian eIF3 complex consists of at least 10 subunits: p170, p116, p110, p66, p48, p47, p44, p40, p36, and p35 (16). These subunits are shown in Fig. 6 (lane 1). Binary eIF3-HCV IRES complexes were assembled with [32P]UTP-labelled HCV nt 40 to 372 RNA, and UV cross-linking was then used to identify which of these subunits are in direct contact with the HCV IRES. The p116 and p66 subunits became prominently labelled after UV irradiation of this complex; strong labelling of p170 and p47 was also apparent (lane 2). The other labelled bands shown in this lane are likely to be degradation products of p170 and p116. None of the bands described above was detected if eIF3 was omitted from the reaction mixtures (data not shown).
In a parallel series of experiments, UV cross-linking of binary complexes formed by binding eIF3 to [32P]UTP-labelled CSFV nt 1 to 442 RNA transcripts also resulted in strong labelling of the p170, p116, p66, and p47 subunits of eIF3 (see Fig. 7C, lane 1).
Simultaneous binding of eIF3 and 40S ribosomal subunits to CSFV and HCV IRES elements.
Initiation factor eIF3 is stoichiometrically associated with native 40S ribosomal subunits in the cytoplasm (31). We have previously reported that 40S subunits can bind directly to HCV and CSFV IRESs to form stable binary complexes (22). In light of the results presented above, which show that eIF3 also binds specifically to these IRESs, it was interesting to examine whether eIF3 and 40S subunits could bind simultaneously to the same IRES or whether binding of one to the IRES precluded binding of the other.
Inclusion of eIF3 in reaction mixtures in a threefold molar excess over the HCV IRES resulted in protection of U212 and U220 from cleavage by RNase V1 (Fig. 7A, lanes 1 and 2). Protection of U212 and U220 by eIF3 was not affected by inclusion of 40S subunits in reaction mixtures in equimolar amounts with eIF3 or in a threefold molar excess over eIF3 (lanes 2 to 4). 40S subunits bound to the HCV IRES in these conditions as shown by sucrose density gradient centrifugation analysis (22).
In similar footprinting experiments, inclusion of 40S subunits in a threefold molar excess over eIF3 did not prevent the binding of eIF3 to the CSFV IRES: under these conditions, the presence of eIF3 in threefold molar excess over the RNA resulted in protection of nucleotides GUG198–200, CGA216–218, A225, and C232 from cleavage by RNase V1 (Fig. 7B, lanes 1 to 3), exactly as described above for eIF3 alone (Fig. 2A). We used UV cross-linking to complement enzymatic footprinting in experiments to assess whether eIF3 and 40S subunits bind simultaneously to the CSFV IRES. UV cross-linking of eIF3 to CSFV nt 1 to 442 resulted in labelling of the p170, p116, p66, and p47 subunits of this factor, and this was not altered by inclusion of increasing amounts of 40S subunits in reaction mixtures (Fig. 7C, lanes 2 to 4). We have previously reported that UV cross-linking of binary 40S subunit-CSFV IRES complexes resulted in specific radiolabelling of ribosomal protein S9 (22). Radiolabelling of this ribosomal protein was apparent in reactions with various different molar ratios of 40S subunits and eIF3 (lanes 2 to 7). In these experiments, the translation component present at the lower concentration was always present in a threefold molar excess over the RNA. eIF3 and 40S subunits can therefore both bind simultaneously to both the CSFV IRES and to the HCV IRES.
DISCUSSION
We have used chemical and enzymatic footprinting techniques to define the binding site for the translation initiation factor eIF3 on the IRES elements of HCV and CSFV RNAs. This factor binds specifically to the apical half of domain III in both IRES elements and protects both unpaired and base-paired residues in the irregular central helix of this domain from chemical modification and enzymatic cleavage. The binding site on both RNAs consists of a large cloverleaf-like structure composed of the central helix of domain III and hairpins IIIa, IIIb, and IIIc. These results are consistent with the results of our earlier toeprinting analysis (22) and are strongly supported by the observation that deletion of hairpin IIIb or the adjacent hairpin IIIc from the HCV IRES abrogates the binding of eIF3 to this RNA (Fig. 5).
Initiation factors and mRNAs commonly form RNP complexes that are able to withstand sucrose density gradient centrifugation, but these complexes do not yield distinct toeprints on primer extension analysis (2), indicating that these RNA-protein interactions are weak and not sequence specific. eIF3 is known to have RNA binding properties (7, 18, 36). However, this is the first report that eIF3 is able to bind mRNAs in a sequence- or structure-specific manner. We have found that many of the residues in the HCV and CSFV IRESs that are bound by eIF3 are centered on an internal bulge in the irregular apical helix of domain III, but we have not yet identified sequence or structural determinants of this interaction. Hairpins IIIb and IIIc are essential for binding of eIF3 to the HCV IRES (Fig. 5), but the extensive covariant substitutions in this region (30) and the differences in sequence between these regions in HCV and CSFV IRESs suggests that their structure is likely to be at least as important a determinant of their binding as their sequence.
We have previously found that the EMCV IRES contains specific binding sites for eIF4G and the pyrimidine tract binding protein (13, 20, 21). These observations have led us to propose a model for IRES function in which these large complex RNAs contain (i) specific binding sites for incoming 40S subunits and factors associated with them in 43S preinitiation complexes and (ii) structural elements that orient these binding sites in such a way that their interaction with components of the 43S complex correctly places the initiation codon of the mRNA at or in the immediate vicinity of the ribosomal P site. The results presented here and previous observations that HCV and CSFV IRESs interact directly with 40S ribosomal subunits (22) are consistent with this model.
UV cross-linking to [32P]UTP-labelled CSFV and HCV IRES-specific RNAs revealed that four subunits of eIF3 (p170, p116, p66, and p47) make contacts with these IRESs (Fig. 6 and 7C). The interactions of eIF3 with these two IRESs are therefore similar and are more extensive than its interactions with either Semliki Forest virus mRNA (29) or rabbit globin mRNA (36), which appear to involve only the p66 and p116 subunits. These two subunits both contain an RNA recognition motif (7, 16), whereas p170 does not contain this or any other RNA binding motifs (12). The p116 and p66 subunits are therefore likely to be determinants of the interaction of eIF with HCV and CSFV IRES elements.
Our previous studies of initiation mediated by HCV and CSFV IRESs involved reconstitution of this process in vitro from fully fractionated translation components. This analysis suggested a model for the initiation of HCV and CSFV translation that involves direct binding of the 40S subunit to the IRES and otherwise requires only the eIF2-GTP-Met-tRNAiMet ternary complex for a 48S preinitiation complex to assemble at the initiation codon (22). eIF3 enhanced the process of 48S complex formation on the CSFV IRES in vitro and was found to be essential for the formation of active 80S ribosomal complexes. The molecular basis for the second of these activities has not yet been elucidated; a model for the first is discussed below. In addition to these roles, previous biochemical studies have implicated eIF3 in dissociation of 80S ribosomes into 40S and 60S subunits and in stabilization of the binding of mRNA and the Met-tRNAiMet ternary complex to the 40S subunit (4, 8, 32). These activities would contribute to the efficient translation of all mRNAs, including those of HCV and CSFV. The observations reported here suggest an additional specific role for eIF3 in initiation on these viral RNAs. eIF3 is stoichiometrically associated with free 40S ribosomal subunits in the cytoplasm and can there be considered to effectively be a constitutive component of native 40S subunits (31). The affinity of the 40S subunit (22) and of eIF3 (see above) for distinct structural elements within HCV and CSFV IRESs could therefore result in selective binding of native 40S subunits to these viral RNAs in the cytoplasm when they compete with cellular mRNAs for the translation apparatus. This model is strongly supported by the results reported here (Fig. 7), which indicate that these two IRES elements are able to bind to eIF3 and to a 40S subunit simultaneously. The presence of high-affinity binding sites for two different components of the native 40S subunit on HCV and CSFV IRESs may therefore enhance the efficiency and accuracy of binding of these RNAs to the 40S subunit in an orientation that promotes entry of the initiation codon into the ribosomal P site.
ACKNOWLEDGMENTS
This work was supported by grants from the Council for Tobacco Research, Inc. (to C.U.T.H.), from the Russian Foundation for Basic Investigations (to I.N.S.), and from NATO and the Howard Hughes Medical Institute (to C.U.T.H. and I.N.S.).
We thank Dmitry Kolkevich and Rodney Romain for technical assistance, Ernest Cuni for photography, and P. Bredenbeek, S. Fletcher, and R. Jackson for the CSFV and HCV plasmids.
REFERENCES
- 1.Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 2.Anthony D D, Merrick W C. Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 3.Baker A-M, Draper D E. Messenger RNA recognition by fragments of ribosomal protein S4. J Biol Chem. 1995;270:22939–22945. doi: 10.1074/jbc.270.39.22939. [DOI] [PubMed] [Google Scholar]
- 4.Benne R, Hershey J W B. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 5.Brown E A, Zhang H, Ping L-H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J P, Ehresmann B. Probing the structure of RNA in solution. Nucleic Acids Res. 1987;19:665–671. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Barrio M T, Naranda T, Vasquez de Aldana C R, Cuesta R, Hinnebusch A G, Hershey J W B, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 8.Hannig E M. Protein synthesis in eukaryotic organisms: new insights into the function of translation initiation factor eIF-3. Bioessays. 1995;17:915–919. doi: 10.1002/bies.950171103. [DOI] [PubMed] [Google Scholar]
- 9.Hartz D, McPheeters D S, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 10.Honda M, Ping L H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 11.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson K R, Merrick W C, Zoll W L, Zhu Y. Identification of cDNA clones for the large subunit of eukaryotic translation initiation factor 3. Comparison of homologues from human, Nicotiana tabacum, Caenorhabditis elegans and Saccharomyces cerevisiae. J Biol Chem. 1997;272:7106–7113. doi: 10.1074/jbc.272.11.7106. [DOI] [PubMed] [Google Scholar]
- 13.Kolupaeva V G, Hellen C U T, Shatsky I N. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 14.Meador J, Cannon B, Cannistraro V J, Kennel D. Purification and characterization of Escherichia coli RNase I. Comparison with RNase M. Eur J Biochem. 1990;187:549–553. doi: 10.1111/j.1432-1033.1990.tb15336.x. [DOI] [PubMed] [Google Scholar]
- 15.Merrick W C. Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 1979;60:101–108. doi: 10.1016/s0076-6879(79)60010-1. [DOI] [PubMed] [Google Scholar]
- 16.Methot N, Rom E, Olsen H, Sonenberg N. The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J Biol Chem. 1997;272:1110–1116. doi: 10.1074/jbc.272.2.1110. [DOI] [PubMed] [Google Scholar]
- 17.Moazed D, Stern S, Noller H F. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 18.Naranda T, McMillan S E, Hershey J W B. Purified yeast translation initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 19.Pestova T V, Hellen C U T, Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′-nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of hepatitis C virus and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds J E, Kaminski A, Carroll A R, Clarke B E, Rowlands D J, Jackson R J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 26.Rijnbrand R, Bredenbeek P J, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 27.Rijnbrand R, Abbink T E M, Haasnoot P C J, Spaan W J M, Bredenbeek P J. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 28.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setyono B, Van Steeg H, Voorma H O. Ultraviolet-crosslinking reveals specific affinity of eukaryotic initiation factors for Semliki forest virus mRNA. Biochim Biophys Acta. 1984;782:242–246. doi: 10.1016/0167-4781(84)90058-7. [DOI] [PubMed] [Google Scholar]
- 30.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P-L, Simmonds P The International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 31.Sundkvist I C, Staehelin T. Structure and function of free 40S ribosome subunits: characterization of initiation factors. J Mol Biol. 1975;99:401–418. doi: 10.1016/s0022-2836(75)80135-5. [DOI] [PubMed] [Google Scholar]
- 32.Trachsel H, Staehelin T. Initiation of mammalian protein synthesis: the multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 33.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosomal entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Le S Y, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westermann P, Nygard O. Cross-linking of mRNA to initiation factor eIF-3, 24kDa cap binding protein and ribosomal proteins S1, S3/3a, S6 and S11 within the 48S pre-initiation complex. Nucleic Acids Res. 1984;12:8887–8897. doi: 10.1093/nar/12.23.8887. [DOI] [PMC free article] [PubMed] [Google Scholar]