Abstract
The small, 195-amino-acid form of the hepatitis delta virus (HDV) antigen (δAg-S) is essential for genome replication, i.e., for the transcription, processing, and accumulation of HDV RNAs. To better understand this requirement, we used purified recombinant δAg-S and HDV RNA synthesized in vitro to assemble high-molecular-weight ribonucleoprotein (RNP) structures. After transfection of these RNPs into human cells, we detected HDV genome replication, as assayed by Northern analysis or immunofluorescence microscopy. Our interpretation is that the input δAg-S is necessary for the RNA to undergo limited amounts of RNA-directed RNA synthesis, RNA processing, and mRNA formation, leading to de novo translation of δAg-S. It is this second source of δAg-S which then goes on to support genome replication. This assay made it possible to manipulate in vitro the composition of the RNP and then test in vivo the ability of the complex to initiate RNA-directed RNA synthesis and go on to achieve genome replication. For example, both genomic and antigenomic linear RNAs were acceptable. Substitution for δAg-S with truncated or modified forms of the δAg, and even with HIV nucleocapsid protein and polylysine, was unacceptable; the exception was a form of δAg-S with six histidines added at the C terminus. We expect that further in vitro modifications of these RNP complexes should help define the in vivo requirements for what we define as the initiation of HDV genome replication.
Human hepatitis delta virus (HDV) is a subviral satellite of hepatitis B virus, on which it is dependent for its envelope proteins (16). HDV particles contain a ribonucleoprotein (RNP) core composed of the circular 1.7-kb RNA genome and multiple copies of the delta antigen (δAg), the only HDV-encoded protein (25). There are two main forms of the δAg. The small 195-amino-acid (aa) form, δAg-S, is essential for genome replication (14). However, during replication, posttranscriptional RNA editing at a site corresponding to the termination codon for δAg-S allows the synthesis of an extra 19 aa to make the larger, 214-aa form, δAg-L (19).
Both δAg-S and δAg-L form a ribonucleoprotein complex with HDV RNA, and some of the functions of these proteins are probably mediated following incorporation into such complexes. Both proteins are found within the RNP core of mature virus (25).
δAg-L appears to have two roles late in virus replication. First, δAg-L can act as a dominant-negative inhibitor of genome replication (5). Thus, its natural appearance via editing could facilitate a suppression of RNA accumulation. Second, it is essential for particle assembly (4). There is evidence that this assembly is dependent on farnesylation of the cysteine 4 aa from the end of the 19-aa C-terminal extension (9).
The nature of the essential role(s) of δAg-S in virus replication is less clear. Attempts to achieve genome replication by transfection of cells with viral cDNAs have been successful, but only when the cDNA encodes a functional δAg-S protein (14). It is not known whether δAg-S has some cotranscriptional role in viral RNA synthesis in vivo. Transcription of HDV RNA in nuclear extracts was achieved by the host RNA polymerase II in the absence of δAg-S (8, 20); however, further studies are needed to confirm this and to directly test the effects of addition of δAg-S. It is clear that in vivo, δAg-S does have other roles. It stabilizes HDV RNA circles (18), and it facilitates ribozyme cleavage (13). Also, since δAg-S has both a nuclear localization signal and RNA-binding activity that is HDV RNA specific (12), it probably functions in both the initial transport of HDV RNP to the nucleus and the maintenance in this location during RNA-directed RNA synthesis.
While much valuable information can be gained about HDV genome replication by the use of cDNA transfections, there are obvious inadequacies when it comes to studying the early events of RNA-directed RNA synthesis. We and others have attempted transfections with HDV RNAs (10, 12). Consistent with the cDNA result, transfection of cells with HDV RNA isolated from virions or genomic RNA transcribed in vitro does not lead to initiation of genome replication; however, if the cells already contain δAg-S, then replication occurs (10). This requirement for δAg-S can also be met if cells are transfected with virions or viral RNP; under these conditions, the RNA presumably enters the cells already complexed with a mixture of δAg-S and δAg-L (1).
In the present study, we have used an RNP transfection approach to investigate the early role(s) of δAg-S in events leading to RNA-directed RNA synthesis. Our strategy was to assemble this RNP in vitro, using recombinant δAg-S and RNAs as synthesized in vitro. We found that after cells were transfected with such RNP, we could detect HDV genome replication. Application of this approach has allowed us to begin to define some of the protein and RNA requirements for the early events of HDV genome replication.
MATERIALS AND METHODS
Expression plasmids.
pR5δV5 was constructed for the high-level expression of δAg-S in Escherichia coli. The protein sequence of the American strain with the δAg-S (GenBank accession no. M28267) was back-translated with the program BACKTRANSLATE. (This program was from the Wisconsin Package, version 9.0 [Genetics Computer Group, Madison, Wis.], with E. coli codon frequencies obtained from gopher://weeds.mgh.harvard.edu:70/Oftp%3Aweeds.mgh.harvard.eduA@/pub/codon/eco.cod.) With the sequence obtained as shown in Fig. 1, the plasmid pR5δV5 was constructed by a two-step PCR method, as described previously (3), with the exception that Vent polymerase (New England BioLabs) was used instead of Taq polymerase. Eight overlapping synthetic primers were synthesized (Fig. 1). Changes in the back-translated sequence were made so that the overlaps of the PCR primers would have approximately the same melting temperature. Primers were electrophoresed into a 10% sequencing gel, visualized by UV shadowing, and excised from the gel. The primers were then purified with a Waters Sep-Pak column.
FIG. 1.
Sequence of synthetic gene for optimized expression of δAg-S in E. coli. The underlined sequences correspond to the eight primers used in the first round of PCR. The primers used in the second round are indicated with a dotted underline. The amino acid sequence is shown above the DNA sequence by the one-letter amino acid code. The restriction sites used in cloning are shown in italics.
The first PCR contained 4 pmol of each of the eight primers in a 100-μl reaction mixture. Ten microliters of the first PCR was added to a second reaction mixture that contained an upstream primer (5′-GGGCATATGAGCCGTAGCGA) and a downstream (5′-GCGCCATGGTTTACGGAAAG) primer designed to amplify the desired full-length product. Both reactions involved a hot start at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C, with a final 5-min extension at 72°C.
The PCR product from the second reaction was cloned into the vector pCR-Blunt (Invitrogen), which allows selection based on disruption of a toxic gene. Plasmids isolated from colonies were checked for the insert by restriction digest mapping. The open reading frame of δAg-S was subcloned into expression vector pRSETb (Invitrogen). The sequence of the resultant plasmid, pR5δV5, was verified by dye termination sequencing.
Plasmids pDL444 and pDL445 were constructed to express δAg-S and δAg-L, respectively, under the control of the simian virus 40 late promoter (17).
The plasmids described below were constructed to express proteins with a tag of six histidine residues. The following convention was adopted to indicate whether the tag was N or C terminal: His6δAg for an N-terminal tag and δAgHis6 for a C-terminal tag.
Plasmid pMB001 was constructed to express δAg-SHis6 in E. coli. Three DNA fragments were ligated: fragment 1 was cut from pDL670 (17) with AlwnI and NcoI, fragment 2 was cut from pGemex2 (Promega) with AlwnI and SalI, and fragment 3 was amplified by PCR from pDL444 and then cut with NcoI and SalI.
pTW203, which expresses δAg-SHis6 via the cytomegalovirus immediate-early promoter, was constructed as follows. The cDNA of δAg-S was amplified by PCR with primers that added six C-terminal histidines and then was cloned into vector pcDNA3.1/HisA (Invitrogen), which had been modified to allow initiation at the natural start of δAg-S.
pVB102 was constructed to express His6δAg-L under control of the immediate-early CMV promoter. The δAg-L open reading frame was amplified by PCR, cut with BamHI and XbaI, and ligated to the BamHI and XbaI sites of pcDNA3.1/His6A (Invitrogen). pVB103 was similarly constructed to express His6δAg-L, with cysteine 211 changed to alanine.
pδ1-84His6 was made to express a truncated form of δAg. It was made by cutting pR5δV5 with ApaI. Linkers were added to incorporate a histidine tag after residue 84, followed by a stop codon and an NcoI site. NcoI digestion was used to delete the remainder of the δAg-S gene, and the plasmid was religated.
pHIV-1NC, made for the expression in E. coli of the 71-aa form of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein (NC), with the sequence Met-(His)6 added at the N-terminal end, was obtained from Patrick Brown (28).
pSVL(D2M) (14) expresses a mutant dimer of the HDV genome with a 2-nucleotide deletion at position 1434 to 1435 which eliminates the ability to synthesize δAg.
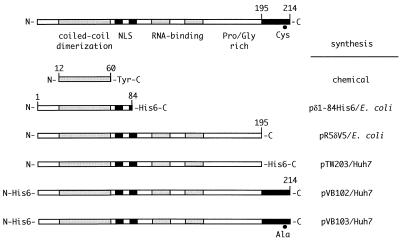
The various forms of δAg expressed by the plasmids described above are summarized in Fig. 2.
FIG. 2.
Various forms of δAg tested in RNP transfection assays. The domains indicated on δAg-L are as previously reviewed (16), but with the modification that the coiled-coil domain is actually larger (24). Also shown are δAg-S and five altered forms of it, along with an indication of how each protein was synthesized. Further details are in Materials and Methods.
Protein purification.
Recombinant δAg-S was expressed and purified as follows. Plasmid pR5δV5 was transformed into BL21(DE3)pLysS cells (Novagen). A single colony was used to inoculate a 100-ml overnight culture. Ten milliliters of this overnight culture was used to inoculate a 1-liter culture. At an optical density of between 0.4 and 0.6, the cells were induced with 3 ml of 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cell growth was continued for 3 h, and then cells were pelleted at 5,000 × g for 10 min. The cells were resuspended in 15 ml of 50 mM HEPES (pH 7.5)–250 mM NaCl–1 mM MgCl2 and stored at −20°C until needed.
The frozen cells (45 ml corresponding to three 1-liter cultures) were thawed, and one Complete Protease Inhibitor tablet (Boehringer Mannheim) was added, along with RNase A and DNase I, to a final concentration of 50 μg/ml. Cells were lysed by sonication and pelleted at 10,000 × g for 30 min. The lysate was diluted threefold with 50 mM HEPES buffer (pH 7.5) and then applied to a 10 × 1.5-cm Fast SP Sepharose column (Pharmacia) equilibrated with 50 mM HEPES buffer (pH 7.5) and eluted with a salt gradient from 0 to 1 M NaCl in 50 mM HEPES (pH 7.5). The fractions containing δAg-S were applied to a Superdex S-200 column (Pharmacia) equilibrated with 50 mM HEPES (pH 7.5), 500 mM NaCl, and 5% glycerol. The δAg-S obtained was >85% pure as judged by Coomassie blue staining of a sodium dodecyl sulfate gel.
Proteins with a histidine tag were purified as follows. Proteins expressed in E. coli were affinity purified with a Talon column according to the recommendations of the manufacturer (Clontech). Proteins expressed in mammalian cells were purified by the Invitrogen Xpress System. In both cases, the fractions containing the purified protein were identified by sodium dodecyl sulfate gel electrophoresis, pooled, dialyzed, and concentrated.
Other proteins.
The peptide δAg12-60Y was synthesized and purified as described by Rozzelle et al. (24). Poly-d-lysine (peak molecular mass, 513 kDa) was obtained from Sigma.
RNA transcription.
For RNP reconstitution experiments, we used genomic or antigenomic HDV RNA synthesized in vitro. A 1.2× unit-length cDNA was put under control of T7 promoter and terminator sequences in a modified pGEM4Z vector (Promega). The resulting plasmids, pTW101 for genomic RNA and pTW114 for antigenomic RNA, were used as the templates for RNA transcription (18).
RNP assembly.
Purified RNA and protein were combined in 10 μl of buffer (5 mM HEPES [pH 7.5], 50 mM NaCl, 0.5% glycerol) and incubated for 5 min at room temperature.
Nondenaturing gel electrophoresis of RNP.
Horizontal gels of 1.0% agarose were cast in Tris-borate-EDTA (TBE) buffer (90 mM boric acid, 10 mM Tris-base [pH 8.0], 1 mM EDTA). RNP samples were mixed with gel loading buffer (1× TBE, 30% glycerol, 0.2% bromophenol blue), and electrophoresis was performed at 120 V until the bromophenol blue dye front had migrated 8 cm.
Transfection.
Plastic 16-mm-diameter tissue culture wells (Costar) were seeded with approximately 0.1 × 106 Huh7 cells (21). For transfections with assembled RNP, 0.25 to 900 ng of δAg-S and 500 ng of genomic HDV RNA in 125 μl of Opti-MEM were combined with 2.7 μl of lipofectamine (2 mg/ml) in 125 μl of Opti-MEM, incubated for 30 min at room temperature, and applied to cells that had been washed with Opti-MEM (11). In control transfections, either δAg-S or HDV RNA was omitted. For cDNA transfections, 500 ng of plasmid DNA was used. At 5 h after transfection, the transfection mixture was changed to Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. At 4 days after transfection, cells were reseeded into a 30-mm-diameter dish containing a glass coverslip; at 8 days, the cells were examined by immunofluorescence microscopy. We picked 8 days for three reasons. (i) We needed to avoid detection of the transfected δAg-S. (ii) In the immunofluorescence assays, 8 days corresponded to the peak signal for a cell undergoing RNP-initiated replication. (iii) At 8 days, we could readily detect δAg-L, created as a consequence of both RNA editing and genome replication (19). In contrast, for Northern analyses, we were able to detect genome replication as early as 2 days (see Fig. 7).
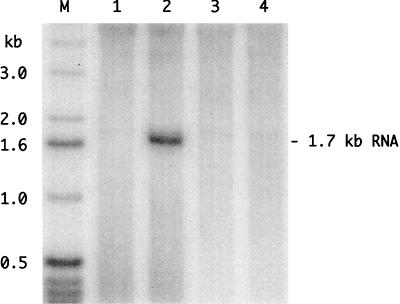
FIG. 7.
Northern analysis to detect genome replication following transfection of cells with assembled RNPs. RNPs were assembled with 500 ng of genomic RNA that was either wild type (lanes 1 and 2) or mutated to block the open reading frame for δAg-S (lanes 3 and 4). RNPs were assembled either without (lanes 1 and 3) or with (lanes 2 and 4) 200 ng of recombinant δAg-S. At 2 days after transfection, RNA was isolated and subjected to Northern analysis to detect antigenomic RNA. Lane M shows end-labeled single-stranded DNA size markers. To the right is indicated the position of unit-length antigenomic RNA.
Immunofluorescence.
Cell monolayers on glass coverslips were fixed and stained as previously described (1). A human anti-δAg antibody, conjugated with fluorescein (a gift of John Gerin), was used to detect total δAg. For the detection of δAg-L, a rabbit antipeptide antibody rab LP3 (a gift of Stan Lemon [29]) was used in a combination with goat anti-rabbit immunoglobulin G polyclonal antibody conjugated with rhodamine (Sigma). To stain the cellular DNA, 2 μg of 4′,6-diamidino-2-phenylindole (DAPI [Sigma]) was added to secondary antibody solutions. After mounting, the samples were viewed with a Zeiss Axiophot microscope with a ×40 or ×100 objective and specific filter blocks. Images were acquired with a charge-coupled device camera (Photometrics, model CH 250) and processed with Photoshop and Canvas software.
Northern blot analysis.
For studies of the initiation of genome replication in transfected cells, total cellular RNA was extracted with Tri Reagent (Molecular Research Center), glyoxalated, and analyzed by electrophoresis as previously described (1). RNA was transferred electrophoretically to a nylon membrane (Zeta-probe GT; Bio-Rad) and immobilized by UV cross-linking. Hybridization was performed with 32P-labeled RNA probes specific for genomic HDV RNA, as described previously (1). Probe was synthesized in vitro in the presence of [α-32P]UTP (DuPont). Detection and quantitation of radioactivity were performed with a phosphoimager (Fuji Bio-Imaging system).
Immunoblot analysis.
Total cell protein was analyzed under denaturing conditions as described by Laemmli (15). After electrotransfer to nitrocellulose, δAg was detected with a rabbit polyclonal antibody followed by enhanced chemiluminescence (ECL; Amersham).
Nucleotide sequence accession number.
The sequence of plasmid pR5δV5 has been assigned GenBank accession no. U88619.
RESULTS
Synthesis of δAg-S in E. coli.
Initial studies with E. coli demonstrated poor expression of δAg-S from the wild-type sequence. We noted that about 18% of the codons in the natural δAg sequence are rarely used by E. coli. Others have shown that attempted overexpression of codons that are rare in E. coli not only can inhibit expression but also can lead to misincorporation (7). Therefore, we designed a nucleotide sequence which maintained the amino acid sequence, but increased the percentage of codons that were most favored for expression in E. coli from 26% to 85%. This optimized sequence (Fig. 1) was used to construct expression plasmid pR5δV5. We thus obtained a 40-fold increase in expression and went on to purify the recombinant protein to >85% homogeneity.
Recombinant δAg-S forms RNP with HDV RNA.
Experiments were performed to determine whether the purified δAg-S could interact in vitro with HDV RNA. Aliquots of 32P-labeled linear genomic HDV RNA were incubated with increasing amounts of δAg-S. RNP assembly was assayed by electrophoresis into agarose gels under nondenaturing conditions, after which the gels were dried and the radioactivity was quantitated with a phosphoimager.
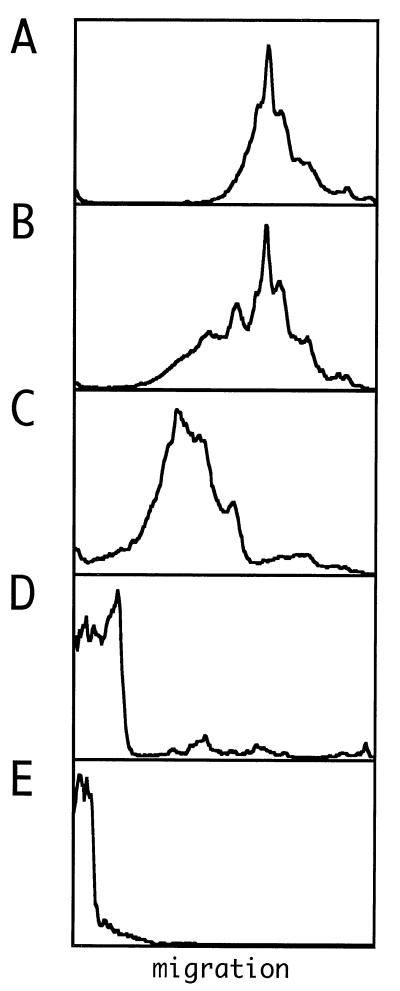
We observed that for concentrations of δAg-S of ≤100 nM (22 ng in 10 μl), the migration of the RNA was not significantly reduced relative to that of free RNA (Fig. 3A). At 300 nM, the RNA migration was reduced (Fig. 3B). Also, as the δAg-S concentration was increased, the migration of the complexes was reduced even further, and ultimately the complexes were barely able to enter the gel (Fig. 3C to E).
FIG. 3.
Electrophoretic analysis of RNPs assembled in vitro with recombinant δAg-S and in vitro-transcribed genomic HDV RNA. (A) Profile of 500 ng of genomic RNA synthesized in vitro. (B to E) Aliquots of 500 ng of RNA incubated with 67, 200, 600, and 900 ng of δAg-S, respectively. A trace amount of 32P-labeled genomic RNA was included to allow detection following electrophoresis. Quantitation was via a phosphoimager (Fuji). Images were further processed with Canvas software. In each panel, electrophoresis was from left to right, with the gel origin shown at the left side.
Transfection of cells with RNP assembled in vitro leads to genome replication.
Each of the RNPs shown in Fig. 3 was tested in a transfection assay. After 8 days, the total RNA was extracted and subjected to Northern analysis to detect antigenomic (Fig. 4A) and genomic (Fig. 4B) RNA. At levels of δAg-S in which no RNP formation had been previously observed (Fig. 3A), no replication was detected (Fig. 4, lanes 1 to 4). However, when the level of δAg-S was sufficient for RNP formation (Fig. 3B to E), there was significant accumulation of δAg and unit-length HDV RNAs in the transfected cells (Fig. 4, lanes 5 to 9). No replication was detected when genomic RNA was transfected alone (Fig. 4, lanes 10).
FIG. 4.
Northern analysis to detect genome replication following transfection of cells with assembled RNPs. RNPs were assembled with 500 ng of genomic HDV RNA and a range of amounts of δAg-S. At day 8 after transfection, RNA was isolated and assayed by Northern analysis for antigenomic (A) and genomic (B) RNA. In lanes 1 to 10, the amounts of δAg-S used were 0.26, 0.8, 2.4, 7.4, 22, 67, 200, 600, 900, and 0 ng, respectively. These correspond to molar ratios of δAg-S per RNA of 0.016, 0.048, 0.14, 0.44, 1.3, 4.0, 12, 36, 54, and 0, respectively.
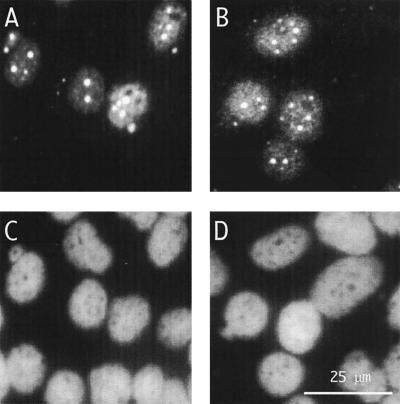
To obtain additional evidence for genome replication, the transfected cells were examined by immunofluorescence microscopy for δAg-S. This was detected in the nucleus (Fig. 5A), and the pattern was characteristic of HDV replication; that is, there was a diffuse nucleoplasmic staining as well as an accumulation of small granules and larger discrete speckles, as observed previously (2). No signal was detected in cells transfected with either genomic RNA or δAg-S alone (data not shown).
FIG. 5.
Immunofluorescence microscopy to detect genome replication following transfection of cells with assembled RNPs. RNPs were assembled as described in the legend to Fig. 3C. At day 8 after transfection, immunofluorescence was used to detect either total δAg (A) or, specifically, δAg-L (B). (C and D) Same fields as panels A and B, but with detection of cellular DNA by DAPI staining. The scale is indicated in panel D.
Immunofluorescence microscopy was also performed with an antibody specific for δAg-L. This assay represented an additional control, because δAg-L was not present in the input RNP and could only be produced by genome replication coupled with posttranscriptional RNA editing (19). As shown in Fig. 5B, this antibody detected the characteristic nucleoplasmic signal, further confirming our interpretation that HDV genome replication was occurring.
In summary, these studies provided three lines of evidence that recombinant δAg-S was necessary for the RNP, as assembled in vitro, to be able to lead to genome replication when transfected into cells.
Are the events which lead to genome replication specific for δAg-S?
It needs to be pointed out that the assay of transfection with RNP as described above, involves not one but two “sources” of functional δAg-S. The “early” source is that which is present on the RNA as it is transfected into the cell. The “late” source is de novo, in that it depends upon the ensuing RNA-directed RNA replication, together with mRNA synthesis and translation. Experiments were next carried out to determine whether the initial source, the recombinant δAg-S, was specific, or whether other RNA-binding proteins, including mutant forms of δAg-S, could substitute.
The HIV-1 nucleocapsid protein (NC) was first tested since it shares many properties with the δAg. These include multimerization ability, high basicity, and a strong affinity for nucleic acids (6). However, although the NC was able to form RNP with HDV RNA, as assayed by gel electrophoresis (data not shown), transfection of cells with these RNP complexes was unable to lead to detectable genome replication, as assayed after 8 days, by immunofluorescence microscopy, which is more sensitive than Northern analysis (data not shown). Polylysine was also tested, because it is even more basic than δAg and NC, and it also formed RNP complexes with HDV RNA, just as for δAg-S in Fig. 3E (data not shown). Transfection with complexes made with polylysine did not achieve detectable genome replication (data not shown).
We next tested various truncated and modified forms of δAg (Fig. 2) for their capacity to form RNP. Four were able to form RNP, as assayed by gel electrophoresis, but on transfection failed to achieve genome replication (data not shown): (i) a peptide consisting of the δAg sequence aa 12 to 60 plus a C-terminal tyrosine (12-60Y) (17, 24, 30); (ii) another truncated version of δAg-S, this time with aa 1 to 84His6; (iii) a protein, His6δAg-L; (iv) and a protein closely related to that of (iii) with the cysteine at position 211 replaced by alanine. (In contrast to the wild-type δAg-L, this species could not be farneslyated, a modification which is considered to enable packaging [22].)
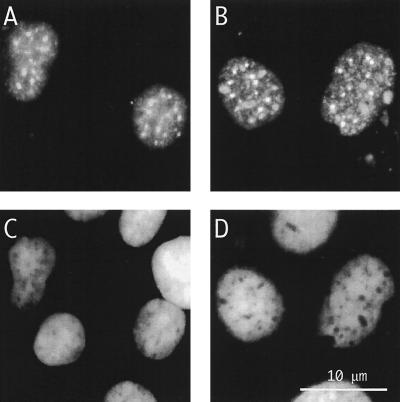
A modified form of δAg-S whose transfection led to genome replication was one with a C-terminal histidine tag. As detected by immunofluorescence microscopy, the resultant replication level in transfected cells (Fig. 6A) was comparable to that obtained with wild-type δAg-S (Fig. 6B). Comparable results were obtained with protein purified from Huh7 cells rather than from E. coli; also functional was a δAg-S modified with an N-terminal tag (data not shown).
FIG. 6.
Immunofluorescence microscopy to detect genome replication following transfection of cells with assembled RNPs. (B and D) Cells were transfected with the RNP, assembled as described in the legends to Fig. 3C and 5. In panels A and C, the δAg-S was replaced by δAg-SHis6. At day 8 after transfection, immunofluorescence was used to detect total δAg (A and B). (C and D) Same fields as panels A and B, but with detection of cellular DNA by DAPI staining. The scale is indicated in panel D.
These studies showed that δAg-S, modified with either a C- or N-terminal tag, in contrast to more extensively mutated forms, was able to supply what we have defined as the early source of δAg-S. The data support the interpretation that RNP formed by δAg-S and HDV RNA has unique properties that, following transfection, lead to genome replication.
Is de novo synthesis of δAg-S necessary for genome replication initiated by transfection with RNP?
The studies mentioned above showed that the early source of δAg-S, as delivered by transfection, was necessary for achieving genome replication. We next asked whether the same source might be sufficient for at least a limited amount of genome replication. To remove the late or de novo source of δAg-S, we replaced the linear multimeric HDV genomic RNA with a form that was identical, except for a modification to block the open reading frame of δAg-S.
By immunofluorescence microscopy it was clear that much of the transfected δAg-S was still present at 2 days after transfection (data not shown). Therefore, cultures were transfected with either the wild-type or mutated form of the HDV RNA, with or without prior complexing with δAg-S. At 2 days the cell RNA was examined by Northern analysis to detect antigenomic HDV RNA. As shown in Fig. 7 (lane 2), we only detected unit-length antigenomic RNA in cells transfected with the wild-type RNA, as delivered in a complex with δAg-S.
These studies show that de novo synthesis of δAg-S is necessary for readily detectable levels of genome replication. However, they do not exclude that with assays more sensitive than Northern analysis, some levels of RNA-directed RNA synthesis might be detected.
Can the modified form of δAg-S replace the wild type as a late source of δAg-S?
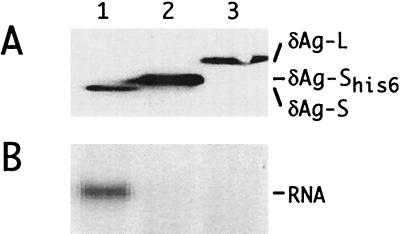
The studies described above showed that δAg-S with a C-terminal histidine tag could replace the wild-type protein as the early source of δAg-S. We next asked whether the same protein could also provide the late de novo source. To answer this, we could not use a genome that encoded such a protein, because the associated genomic alterations would most likely have additional inhibitory consequences. Therefore, we made use of a cDNA cotransfection assay. Cultures were transfected with a vector that expressed multimers of a genome that was modified to block the open reading frame for δAg-S. Huh7 cells were cotransfected with additional vectors, expressing either the modified δAg-S protein, wild-type δAg-S as a positive control, or δAg-L as a negative control. By immunoblotting, we found that all three proteins were expressed (Fig. 8A). By Northern analysis, we found that only the positive control supported genome replication; modified δAg-S did not (Fig. 8B).
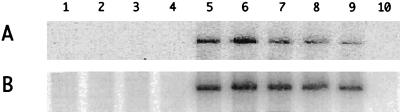
FIG. 8.
Immunoblotting and Northern analyses following transfection of cells with cDNAs. Cells were transfected with pSVL(D2m) together with one of the following vectors: lane 1, pDL444 (δAg-S); lane 2, pTW203 (δAg-SHis6); and lane 3, pDL445 (δAg-L). At 6 days, δAg expression was examined by immunoblotting (A) and HDV genomic RNA was examined by Northern analysis (B). To the right are indicated the different δAg species detected by immunoblotting and the unit-length HDV RNA detected by Northern hybridization.
This result—that the modified δAg-S is sufficient early but not sufficient late—raises the question of whether there is a difference in the roles of δAg-S between the early and late effects. One model is that there is a major difference; e.g., the early role may be simply binding HDV RNA and transporting it to the nucleus. However, an opposing model is that there is no qualitative difference; e.g., genome replication involves the compound effect of many cycles, each with many roles for δAg-S, so that even a minor deficiency in one or more roles is ultimately compounded into a major effect on genome replication. Further experiments are needed to distinguish between these models.
DISCUSSION
In a natural HDV infection, we expect that the virus binds to a susceptible cell via an as yet unidentified receptor, is internalized, uncoated, and transported to the nucleus, and then RNA-directed RNA synthesis is somehow initiated. These events take place as infection is established in a susceptible animal (23) and could be studied, not without difficulty, with cultures of primary human or woodchuck hepatocytes (26, 27). We hope that our more convenient receptor-independent transfection assay as described here, proves to be a useful model for studying what we have defined as the “initiation of genome replication”. This assay has some unique advantages relative to initiation of genome replication by cDNA transfection (14). It may even have a major advantage over natural infections, in that we can readily manipulate in vitro, the nature and composition of the RNP prior to the assay in vivo, of genome replication. In the present study, we have begun to test some variations for the RNP. For example, linear multimers of genomic RNA could be replaced with antigenomic RNA (data not shown). Also successful was a range of stoichiometric amounts of δAg-S per RNA (Fig. 4). The δAg-S was not only necessary but also specific, in that several deleted forms of δAg-S or other RNA-binding proteins, were not acceptable. In future studies, it will be possible to test other variables, such as posttranslational modifications of the δAg-S, different protein combinations, or modifications of the RNA, such as by binding of oligonucleotides to specific regions that might be important for the initiation of RNA-directed RNA synthesis.
In our transfection assay, if genome replication is ultimately detected (either at 8 days by immunofluorescence microscopy [Fig. 5] and Northern analysis [Fig. 4], or even at 2 days by Northern analysis [Fig. 7]), then we can conclude that earlier, the transfected RNP was used for RNA-directed RNA synthesis, leading to RNA processing and mRNA synthesis. We say this because the genome replication, as defined, depended not only an early source of δAg-S, as directly provided by the transfection, but also on a de novo or late source of δAg-S (Fig. 7). Further experiments are needed to clarify the manner by which the early source of δAg-S facilitates the initiation of genome replication. Also, we need to know the extent to which these early transcription and processing events taking place after RNP transfection are a valid model of what happens in a natural infection. Currently, we are examining the early RNA transcripts with procedures such as 5′ rapid amplification of cDNA ends, which are both more sensitive and specific than Northern analyses, and we have begun to address such important questions as defining the early role of δAg-S and determining the importance of the conformation (linear versus circular) and local structure of the RNA template.
ACKNOWLEDGMENTS
J.T. was supported by grants AI-26522 and CA-06927 from the NIH and by an appropriation from the Commonwealth of Pennsylvania. J.H. was supported by grant AI-32480 from the NIH.
Constructive comments on the manuscript were given by William Mason, Glenn Rall, and Ting-Ting Wu. Antibodies essential to this work were provided by John Gerin and Stan Lemon. The synthetic peptide 12-60Y was also obtained from Stan Lemon. Patrick Brown provided the expression plasmid for HIV-1 nucleocapsid protein. Vadim Bichko constructed pVB101, -102, and -103; Matt Bockol constructed pMB001; and Ting-Ting Wu constructed pTW203. The proteins HIV-1NCHis6 and δAg-SHis6 were purified by Gloria Moraleda. Daphne Bell and Binaifer Balsara assisted in the collection of charge-coupled device images.
REFERENCES
- 1.Bichko V, Netter H J, Taylor J. Introduction of hepatitis delta virus into animal cell lines via cationic liposomes. J Virol. 1994;68:5247–5252. doi: 10.1128/jvi.68.8.5247-5252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bichko V V, Taylor J M. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J Virol. 1996;70:8064–8070. doi: 10.1128/jvi.70.11.8064-8070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casimiro D R, Toy-Palmer A, Blake II R C, Dyson H J. Gene synthesis, high-level expression, and mutagenesis of Thiobacillus ferrooxidans rusticyanin: his 85 is a ligand to the blue copper center. Biochemistry. 1995;26:6640–6648. doi: 10.1021/bi00020a009. [DOI] [PubMed] [Google Scholar]
- 4.Chang F L, Chen P J, Tu S J, Chiu M N, Wang C J, Chen D S. The large form of hepatitis δ antigen is crucial for the assembly of hepatitis δ virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao M, Hsieh S-Y, Taylor J. Role of two forms of the hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. The first glimpses at structure-function relationships of nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 7.Del Tito B J, Jr, Ward J M, Hodgson J, Gershater C J L, Edwards H, Wysocki L A, Watson F A, Sathe G, Kane J F. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu T-B, Taylor J. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J Virol. 1993;67:6965–6972. doi: 10.1128/jvi.67.12.6965-6972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glenn J S. Prenylation and virion morphogenesis. In: Dinter-Gottlieb G, editor. The unique hepatitis delta virus. R. G. Austin, Tex: Landes, Co.; 1995. pp. 83–94. [Google Scholar]
- 10.Glenn J S, Taylor J M, White J M. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J Virol. 1990;64:3104–3107. doi: 10.1128/jvi.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawley-Nelson P, Ciccaroni V, Gebeyehu G, Jessee J. Lipofectamine reagent: a new, higher efficiency polycationic liposome transfection reagent. Focus. 1993;15:73–79. [Google Scholar]
- 12.Hwang S B, Jeng K-S, Lai M M C. Studies of functional roles of hepatitis delta antigen in delta virus RNA replication. In: Dinter-Gottlieb G, editor. The unique hepatitis delta virus. R. G. Austin, Tex: Landes, Co.; 1995. pp. 95–110. [Google Scholar]
- 13.Jeng K-S, Su P-Y, Lai M M C. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J Virol. 1996;70:4205–4209. doi: 10.1128/jvi.70.7.4205-4209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 17.Lazinski D W, Taylor J M. Relating structure to function in the hepatitis delta virus antigen. J Virol. 1993;67:2672–2680. doi: 10.1128/jvi.67.5.2672-2680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazinski D W, Taylor J M. Intracellular cleavage and ligation of hepatitis delta virus genomic RNA: regulation of ribozyme activity by cis-acting sequences and host factors. J Virol. 1995;69:1190–1200. doi: 10.1128/jvi.69.2.1190-1200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo G, Chao M, Hsieh S-Y, Sureau C, Nishikura K, Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990;64:1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macnaughton T B, Gowans E J, McNamara S P, Burrell C J. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology. 1991;184:387–390. doi: 10.1016/0042-6822(91)90855-6. [DOI] [PubMed] [Google Scholar]
- 21.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 22.Otto J C, Casey P J. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J Biol Chem. 1996;271:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 23.Ponzetto A, Cote P J, Popper H, Hoyer B H, London W T, Ford E C, Bonino F, Purcell R H, Gerin J L. Transmission of the hepatitis B virus-associated δ agent to the eastern woodchuck. Proc Natl Acad Sci USA. 1984;81:2208–2212. doi: 10.1073/pnas.81.7.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozzelle J, Wang J-G, Wagner D, Erickson B, Lemon S. Self-association of a synthetic peptide from the N terminus of the hepatitis delta virus protein into an immunoreactive alpha-helical multimer. Proc Natl Acad Sci USA. 1995;92:382–386. doi: 10.1073/pnas.92.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu W-S, Netter H J, Bayer M, Taylor J. Ribonucleoprotein complexes of hepatitis delta virus. J Virol. 1993;67:3281–3287. doi: 10.1128/jvi.67.6.3281-3287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sureau C, Jacob J R, Eichberg J W, Lanford R E. Tissue culture system for infection with human hepatitis delta virus. J Virol. 1991;65:3443–3450. doi: 10.1128/jvi.65.7.3443-3450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor J, Mason W, Summers J, Goldberg J, Aldrich C, Coates L, Gerin J, Gowans E. Replication of human hepatitis delta virus in primary cultures of woodchuck hepatocytes. J Virol. 1987;61:2891–2895. doi: 10.1128/jvi.61.9.2891-2895.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J G, Cullen J, Lemon S M. Immunoblot analysis demonstrates that the large and the small forms of hepatitis delta virus antigen have different C-terminal amino acid sequences. J Gen Virol. 1992;73:183–188. doi: 10.1099/0022-1317-73-1-183. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y-P, Lai M M C. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J Virol. 1992;66:6641–6648. doi: 10.1128/jvi.66.11.6641-6648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]