Abstract
The differential use of CC chemokine receptor 5 (CCR5) and CXC chemokine receptor 4 (CXCR4) may be intimately involved in the transmission and progression of human immunodeficiency virus infection. Changes in coreceptor utilization have also been noted upon adaptation of primary isolates (PI) to growth in established T-cell lines. All of the T-cell line-adapted (TCLA) viruses studied to date utilize CXCR4 but not CCR5. This observation had been suggested as an explanation for the sensitivity of TCLA, but not PI, viruses to neutralization by recombinant gp120 antisera and V3-directed monoclonal antibodies, but recent studies have shown coreceptor utilization to be independent of neutralization sensitivity. Here we describe a newly isolated TCLA virus that is sensitive to neutralization but continues to utilize both CXCR4 and CCR5 for infection. This finding further divorces coreceptor specificity from neutralization sensitivity and from certain changes in cell tropism. That the TCLA virus can continue to utilize CCR5 despite the changes that occur upon adaptation and in the apparent absence of CCR5 expression in the FDA/H9 T-cell line suggests that the interaction between envelope protein and coreceptor may be mediated by multiple weak interactions along a diffuse surface.
The discovery of cellular molecules that act as coreceptors in conjunction with CD4 to mediate the binding and entry of human immunodeficiency virus type 1 (HIV-1) has provided a new perspective from which to approach questions of HIV-1 biology and pathogenesis. The differential use of CC chemokine receptor 5 (CCR5) and CXC chemokine receptor 4 (CXCR4) by primary isolates of HIV-1 throughout infection may have important implications for virus transmission and disease progression. HIV-1 infection first manifests as an acute viremic episode, typically involving a homogeneous outgrowth of monocytotropic, non-syncytium-inducing (NSI) viruses (53, 62) that utilize CCR5 as a coreceptor. Although the initial events in virus transmission are largely inaccessible to analysis, cells of the monocyte-macrophage lineage are believed to provide a portal for primary infection and a specific filter for monocytotropic NSI viruses (20). Persons lacking a functional CCR5 coreceptor are resistant to the establishment of HIV-1 infection (8, 28, 43).
Viruses that utilize the CXCR4 coreceptor evolve over the course of infection (61). These T-lymphocytotropic viruses no longer infect monocyte-derived macrophages (45, 46) but generally continue to utilize CCR5 in addition to CXCR4 (7). Endogenous production of CCR5-specific chemokines may provide the selective pressure for this broadening in coreceptor use (44). Importantly, the emergence of dual-coreceptor-utilizing syncytium-inducing (SI) viruses in a proportion of infected persons is prognostic for the development of clinical AIDS (50).
In contrast to primary isolate (PI) viruses, the commonly used laboratory isolates of HIV-1 utilize only CXCR4 as a coreceptor (1, 2, 6, 10, 12, 13, 27, 47). These isolates have been adapted to persistent growth in T-cell lines, and the loss of their ability to utilize CCR5 is perhaps understandable in that most T-cell lines express CXCR4 but not CCR5 (1, 15).
Coincident with changes in coreceptor utilization and cell tropism upon adaptation are changes in neutralization sensitivity. In contrast to PI viruses, T-cell line-adapted (TCLA) viruses are generally sensitive to neutralization by appropriate antibodies directed to the third variable loop (V3) of envelope surface protein gp120 (42, 55). In addition, PI viruses are entirely refractory to neutralization by recombinant HIV envelope protein gp120 (rgp120) antisera that potently neutralize related TCLA viruses (31, 55, 56). The unique ability of PI viruses to utilize CCR5 had been suggested as a basis for the ability of these viruses to escape neutralization, but recent reports have shown that PI viruses remain refractory to neutralization, regardless of the specific coreceptor utilized (27, 32, 52).
As part of our studies to define the relationship between changes in coreceptor utilization and virus phenotype, we isolated a TCLA derivative of molecularly cloned SI primary virus ACH320.2A.1.2 (21, 22). Although the TCLA virus was now able to infect T-cell lines and was sensitive to antibody-mediated virus neutralization, this virus continued to utilize both CCR5 and CXCR4 coreceptors. The intact capacity of this TCLA virus to utilize CCR5 suggests that changes in coreceptor utilization are neither associated with changes in neutralization sensitivity nor required for changes in cell tropism.
Adaptation of a molecularly cloned SI primary virus.
The infectious molecularly cloned provirus ACH320.2A.1.2 was isolated from a biologically cloned SI PI obtained from a member of the Amsterdam Cohort 9 weeks after seroconversion (21, 22). The ACH320.2A.1.2 plasmid was obtained from Hanneke Schuitemaker (Central Laboratory of the Netherlands Red Cross) through the NIBSC AIDS Reagent Project (United Kingdom). Virus (320SI) was regenerated by electroporation and expansion in phytohemagglutinin-activated peripheral blood lymphocytes (PBLs). Supernatants containing high levels of 320SI were used in efforts to adapt this PI virus to growth in an established T-cell line. As in previous studies (55), we utilized the Food and Drug Administration (FDA) isolate of the H9 cell line (37) because of its increased ability, relative to other H9 cell lines, to manifest a cytopathic effect upon infection. By other measures, FDA/H9 cells behave similarly to other H9 cell lines (unpublished data).
In contrast to our previous experience obtained by using a biological isolate of another primary SI virus, ACH168.10 (50, 55), several long-term attempts to obtain infection of FDA/H9 cells by 320SI were without success. In an effort to increase the genetic complexity of the virus population, 320SI virus infection was first established in the permissive human T-cell leukemia virus type 1 (HTLV-1)-transformed MT4 T-cell line (25). Continued cocultivation of these 320SI-infected MT4 cell cultures with FDA/H9 cells yielded immunofluorescence evidence of infection of the FDA/H9 cells after 4 weeks, and a low level of FDA/H9 cell infection was subsequently obtained on passage of the culture supernatant onto naive FDA/H9 cells. Complete and persistent infection of this culture was obtained in an additional 3 weeks, and the TCLA virus produced was designated 320SI-C3. Subsequent low-multiplicity passages onto fresh FDA/H9 cells yielded virus populations 320SI-C3.1 through -C3.3 over the course of 5 months. These TCLA viruses readily infect FDA/H9 and H9 cells and yield virus titers comparable to those of prototypic TCLA viruses.
Coreceptor utilization by the pedigreed PI and TCLA viruses.
Based on the SI phenotype of the parental 320SI virus, we anticipated that this virus would utilize both CCR5 and CXCR4 as coreceptors for infection. Similarly, we anticipated that the TCLA 320SI-C3 virus would have lost the ability to utilize CCR5, as all of the TCLA isolates analyzed to date utilize CXCR4 but not CCR5 (1, 2, 6, 10, 12, 13, 27, 47). To assess coreceptor utilization, cell culture supernatants of infected PBLs (320SI) or FDA/H9 cells (320SI-C3) were titrated for infectivity in U87 human glioma cells expressing CD4 and either CCR5 or CXCR4 (23, 27). After 2 days of incubation, cell monolayers were fixed with methanol-acetone and immunochemically stained by using HIV/IG (38), an anti-human ABC kit (Biomeda Corp.), and a diaminobenzamidine substrate.
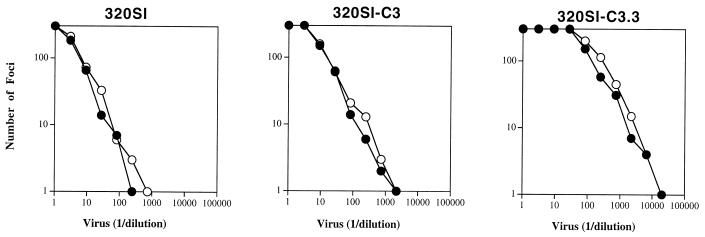
Contrary to expectations based on other TCLA viruses, relative utilization of CCR5 and CXCR4 remained unchanged upon adaptation (Fig. 1). The TCLA 320SI-C3 virus continued to utilize CCR5, despite adaptation and growth in a T-cell line that nominally does not express CCR5. Flow cytometric analysis using CCR5-directed monoclonal antibody (MAb) 2D7 (58) was able to detect CCR5 expression on PBLs and on the PM-1 T-cell line (29) but was unable to detect its expression on FDA/H9 cells (data not shown).
FIG. 1.
Coreceptor utilization by pedigreed 320SI viruses. U87-CD4 cells expressing CCR5 (○) or CXCR4 (•) were infected with serial dilutions of the following viruses: 320SI from PBL culture supernatant and 320SI-C3 and 320SI-C3.3 from FDA/H9 cell culture supernatant. Foci were quantitated 2 days later as described in the text. Relative utilization of CCR5 and CXCR4 was judged by the relative titer on the respective coreceptor-expressing cells. Coreceptor-specific titer was determined on the linear portion of the virus titration curve.
We wanted to confirm that this unique pattern of dual coreceptor use by a TCLA virus was stable through additional passage in FDA/H9 cells. We examined coreceptor utilization by the repeatedly passaged 320SI-C3.3 virus. This virus was also indistinguishable from the original PI virus 320SI in coreceptor specificity (Fig. 1). Thus, dual use of CCR5 and CXCR4 appears to be a stable property of the TCLA 320SI-C3 virus.
The adaptation procedure used here to obtain 320SI-C3 differs from that used previously to isolate a TCLA derivative of another SI primary virus, ACH168.10 (50, 55). This biological isolate was able to directly establish infection in the FDA/H9 cell line, albeit over the course of 4 to 12 weeks, without the need for expansion in MT4 cells. Although expansion in permissive MT4 cells may have been important in generating sequence diversity in the molecularly cloned 320SI virus population, it was also conceivable that this passage history affected the adaptation process and coreceptor preference of the ultimate TCLA virus.
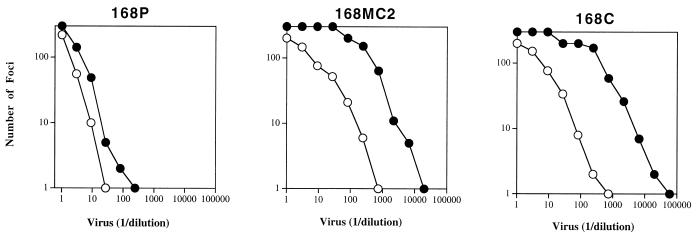
To address this possibility, we repeated the adaptation of ACH168.10 (168P) by using an MT4 cell line intermediate. The 168M virus derived from the MT4 cell culture retained PI virus phenotypes, including dual-coreceptor utilization and the lack of measurable FDA/H9 cell tropism, as did the MT4 cell line-expanded 320SI virus (data not shown). This observation supports the contention that MT4 cells are permissive and nonselective towards SI viruses, serving here simply to broaden the quasispecies distribution of the molecularly cloned 320SI virus. Following cocultivation and adaptation of 168M to persistent growth in FDA/H9 cells, the newly derived TCLA virus 168MC2 lost the ability to utilize CCR5 (Fig. 2), as had the original TCLA 168C virus that was directly adapted to growth in FDA/H9 cells (27, 55). Thus, the retention of dual-coreceptor use by the TCLA 320SI-C3 virus is not a reflection of its passage through MT4 cells prior to adaptation to growth in the FDA/H9 cell line. Rather, different viruses appear to respond differently to selection for growth in T-cell lines.
FIG. 2.
Coreceptor utilization by TCLA virus 168MC2 adapted through an MT4 cell line intermediate. U87-CD4 cells expressing CCR5 (○) or CXCR4 (•) were infected with serial dilutions of the following viruses: 168P from PBL culture supernatant (27, 55), 168MC2 from FDA/H9 cell culture supernatant following expansion in an MT4 cell culture, and 168C from FDA/H9 cell culture supernatant following direct adaptation to FDA/H9 cells (27, 55).
Neutralization sensitivity of the TCLA 320SI-C3 virus.
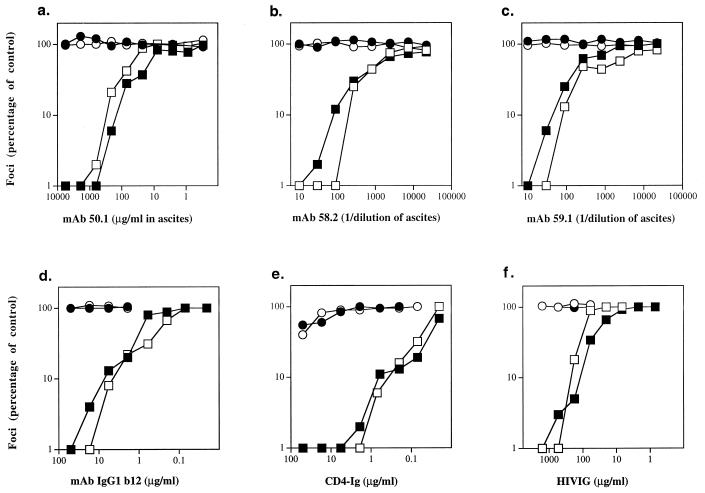
Previous studies had demonstrated that adaptation to persistent growth in established T-cell lines renders the TCLA virus sensitive to neutralization by a broad range of reagents (33, 55). Similarly, TCLA virus 320SI-C3 demonstrated de novo sensitivity to V3-directed MAbs 50.1, 58.2, and 59.1 (Fig. 3a to c, respectively) (54). The core epitopes of the latter two MAbs have been mapped and are present in the known amino acid sequence of 320SI (IGPGRAF and GPGRAF, respectively [19, 54]); the core epitope of MAb 50.1 has also been determined crystallographically (RIHIG [39]) and differs from that present in 320SI (GIHIG). The 320SI-C3 virus was also newly sensitive to CD4 binding domain-directed MAb IgG1 b12 (5), to CD4 immunoadhesin (CD4-Ig) (48), and to HIVIG (Fig. 3d to f, respectively). Thus, as in previous studies, increased sensitivity to virus neutralization appeared coordinately with adaptation to growth in established T-cell lines.
FIG. 3.
Neutralization sensitivity of 320SI (PI) and 320SI-C3 (TCLA) in U87-CD4 cell lines expressing the CCR5 or CXCR4 coreceptor. Virus stocks of 320SI (○, •) and 320SI-C3 (□, ■) comprising cell culture supernatants (from PBLs and FDA/H9 cells, respectively) were standardized to yield a submaximal number of foci of infection on U87-CD4-CCR5 (○, □) or U87-CD4-CXCR4 (•, ■) cells. The virus was incubated with the indicated reagents for 1 h prior to infection (a to f).
Because of the dual-coreceptor use by these PI and TCLA viruses, we were able to investigate neutralization sensitivity as a function of coreceptor use (Fig. 3). In all cases, neutralization sensitivity was independent of the coreceptor utilized for infection of U87-CD4 cells. We had previously demonstrated this for PI virus ACH168.10 (27); we can now extend this conclusion to a TCLA virus. Furthermore, not only is neutralization sensitivity independent of the process of utilizing a specific coreceptor, it is also independent of the changes in coreceptor specificity that often accompany adaptation to growth in T-cell lines.
DNA sequence analysis of the 320SI-C3 envelope gene.
In our previous analysis of the adaptation of ACH168.10 (168P), we defined three adaptation-associated amino acid changes within the envelope protein of TCLA virus 168C: I166R in V2, I282N in C2, and G318R in V3 (55). For the present study, we wished to determine whether similar amino acid changes had arisen during adaptation of the 320SI virus. Proviral DNA from FDA/H9 cells infected with 320SI-C3 was amplified by using envelope-specific primers (envA and envN [18]) and high-fidelity XL PCR (PE Applied Biosystems) (27), and the DNA sequence of the envelope gene was compared with those of ACH320.2A.1.2 (22) and 320SI. Two amino acid changes in gp120 were noted (I166K and H317R (GenBank accession no. AF069524). The I166K change in the V2 region is similar to the I166R change in the unrelated 168C envelope. We do not know whether the change is fortuitous or whether this common amino acid change points to a common function. The H317R change is located within the crown of the V3 loop (KGIHIGPGRAF to KGIRIGPGRAF) but distant from the G318R change in the 168C envelope (GRAFYTTRQII). The increased positive charge in the V3 loop is consistent with similar trends in other PI and TCLA viruses (9, 17, 55). In the case of the 320SI-C3 virus, however, the increased positive charge may be associated with adaptation and neutralization sensitivity but is not associated with altered coreceptor use.
In summary, our finding of this unique TCLA virus that is now sensitive to neutralization but continues to utilize both CCR5 and CXCR4 extends our earlier observations divorcing coreceptor utilization and sensitivity to neutralization. Not only are PI viruses resistant to neutralization regardless of whether CCR5 or CXCR4 is used in infection, but changes in the ability to utilize these coreceptors are not required for the acquisition of neutralization sensitivity in the TCLA virus. Furthermore, the TCLA virus remains sensitive to neutralization regardless of the coreceptor utilized for infection.
Our findings also divorce changes in coreceptor utilization and adaptation to growth in established T-cell lines. Clearly, the often-observed loss of the ability of TCLA viruses to utilize CCR5 is not necessary for adaptation. It is possible, however, that adaptation might derive from more subtle changes in the virus’s interaction with CXCR4. Initial studies to examine the sensitivity to inhibition by the CXCR4 ligand SDF-1 (4, 34), however, did not yield a consistent pattern: whereas 320SI-C3 showed increased sensitivity to inhibition by SDF-1, 168C showed decreased sensitivity (data not shown). Thus, the relationship between coreceptor utilization and adaptation to growth in T-cell lines remains elusive. Appropriate coreceptor expression may be necessary for infection, but it is by no means sufficient, as restrictions to productive infection can exist at multiple levels in the viral life cycle (16, 59, 60).
Our observations that these phenotypically distinct viruses can continue to utilize both coreceptors and that the capacity to utilize CCR5 is retained despite the apparent lack of the CCR5 coreceptor on FDA/H9 cells suggest that the structural requirements for specific coreceptor binding are relatively minimal. In fact, multiple studies to define critical sites for envelope-coreceptor interaction have not yielded entirely consistent generalizations (3, 11, 26, 35, 36, 40, 41, 49). The variable and diffuse nature of functional envelope-coreceptor pairings is consistent with an interaction surface comprising multiple weak contacts. We suggest that the free energy of coreceptor binding may derive in part from the entropic contribution of the initial binding to CD4 in localizing the envelope-coreceptor interaction to two dimensions. Subsequent interactions are driven both by conformational changes induced within the envelope protein complex upon CD4 binding (51, 57) and by proximity on the membrane but ultimately involve multiple weak contacts over a variable surface of envelope-coreceptor interaction. The observed relationship between infectivity by PI viruses and cell surface CD4 density is consistent with this model (24).
In certain instances, the entropic contributions of CD4 binding may be less significant to the overall free energy of envelope protein binding, as in certain human and simian immunodeficiency virus isolates that are able to interact directly with a coreceptor to allow CD4-independent infection (14, 15, 30). This capability may derive from an envelope protein conformation that more closely approximates that induced by CD4 binding, or perhaps from other enhancements in the envelope-coreceptor interaction.
To the extent that the envelope-coreceptor interaction is diffuse and that specificity is determined by the sum of many weak contacts, the effects of specific changes in either protein on overall binding may not be readily predictable. The changes in the 320SI-C3 envelope protein that determine cell tropism appear not to perturb significantly the envelope-CCR5 binding surface, whereas those in the 168C envelope protein preclude effective interaction. Perhaps subtle changes in envelope-coreceptor interaction underlie the observed changes in cell tropism and neutralization sensitivity that define adaptation. At the current level of analysis, however, these fundamental viral phenotypes appear to be independent of specific coreceptor utilization.
Acknowledgments
This work was supported by NIH AREA grant AI41165 to J.H.N.
We thank Jim Rusche and Carolyn Muermann (Repligen Corp.) for gifts of V3-directed MAbs 50.1, 58.2, and 59.1. Additional reagents were kindly provided by Steve Chamow (Genentech, Inc.), Dan Littman (HHMI, NYU Medical Center), Ian Clark-Lewis (University of British Columbia), Susan Zolla-Pazner (NYU Medical Center), and Fred Prince (NY Blood Center). The following MAbs were obtained through the AIDS Research and Reference Reagent Program, NIH, NIAID: 2D7 from LeukoSite, Inc., and IgG1 b12 from Dennis Burton and Carlos Barbas. The infectious molecularly cloned provirus ACH320.2A.1.2 was obtained through the NIBSC AIDS Reagent Project (United Kingdom) from Hanneke Schuitemaker. We thank Dave Holley for technical assistance, Joan Strange for DNA sequencing services (The University of Montana M. J. Murdock Molecular Biology Facility), Richard Field (Department of Chemistry) for valuable discussions, and Ed Walker and Linda Griggs (Ribi ImmunoChem Research, Inc.) for flow cytometry.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions with both the viral envelope and the CCR5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N L. Changes in coreceptor use correlate with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.de Jong J-J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Lu Z-H, Rucker J, Zhang T-Y, Sharron M, Cen Y-H, Wang Z-X, Guo H-H, Du J-G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 13.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 14.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 16.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Invest. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H The WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghiara J B, Stura E A, Stanfield R L, Profy A T, Wilson I A. Crystal structure of the principal neutralization site of HIV-1. Science. 1994;264:82–85. doi: 10.1126/science.7511253. [DOI] [PubMed] [Google Scholar]
- 20.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman R M. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groenink M, Fouchier R A M, de Goede R E Y, de Wolf F, Gruters R A, Cuypers H T M, Huisman H G, Tersmette M. Phenotypic heterogeneity in a panel of infectious molecular human immunodeficiency virus type 1 clones derived from a single individual. J Virol. 1991;65:1968–1975. doi: 10.1128/jvi.65.4.1968-1975.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillon C, Bedin F, Fouchier R A M, Schuitemaker H, Gruters R A. Completion of nucleotide sequences of non-syncytium-inducing and syncytium-inducing HIV type 1 variants isolated from the same patient. AIDS Res Hum Retroviruses. 1995;11:1537–1538. doi: 10.1089/aid.1995.11.1537. [DOI] [PubMed] [Google Scholar]
- 23.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabat D, Kozak S L, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koot M, Vos A H V, Keet R P M, de Goede R E Y, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 coculture assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infection by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaCasse R A, Follis K E, Moudgil T, Trahey M, Binley J M, Planelles V, Zolla-Pazner S, Nunberg J H. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J Virol. 1998;72:2491–2495. doi: 10.1128/jvi.72.3.2491-2495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 29.Lusso P, Cocchi F, Balotta C, Markham P D, Louie A, Farci P, Pal R, Gallo R C, Reitz M S., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 31.Mascola J R, Snyder S W, Weislow O S, Belay S M, Belshe R B, Schwartz D H, Clements M L, Dolin R, Graham B S, Gorse G J, Keefer M C, McElrath M J, Walker M C, Wagner K F, McNeil J G, McCutchan F E, Burke D S NIAID-AVEG. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 32.Montefiori D C, Collman R G, Fouts T R, Zhou J Y, Bilska M, Hoxie J A, Moore J P, Bolognesi D P. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J Virol. 1998;72:1886–1893. doi: 10.1128/jvi.72.3.1886-1893.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J P, Burkly L C, Connor R I, Cao Y, Tizard R, Ho D D, Fisher R A. Adaptation of two primary human immunodeficiency virus type 1 isolates to growth in transformed T cell lines correlates with alterations in the response of their envelope glycoproteins to soluble CD4. AIDS Res Hum Retroviruses. 1993;9:529–539. doi: 10.1089/aid.1993.9.529. [DOI] [PubMed] [Google Scholar]
- 34.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 35.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleskoff O, Sol N, LaBrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popovic M, Sarnagadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 38.Prince A M, Reesink H, Pascual D, Horowitz B, Hewlett I, Murthy K K, Cobb K E, Eichberg J W. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res Hum Retroviruses. 1991;7:971–973. doi: 10.1089/aid.1991.7.971. [DOI] [PubMed] [Google Scholar]
- 39.Rini J M, Stanfield R L, Stura E A, Salinas P A, Profy A T, Wilson I A. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci USA. 1993;90:6325–6329. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross T M, Bieniasz P D, Cullen B R. Multiple residues contribute to the inability of murine CCR5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rucker J, Samson M, Doranz B J, Liebert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions of β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 42.Rusche J R, Javaherian K, McDanal C, Petro I, Lynn D L, Grimalia R, Langlois A, Gallo R C, Arthur L O, Fischinger P J, Bolognesi D P, Putney S D, Matthews T J. Antibodies that inhibit fusion of HIV-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci USA. 1988;85:3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapouméroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 44.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 45.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuitemaker H, Kootstra N A, de Goede R E Y, de Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith D H, Byrn R A, Marsters S A, Gregory T, Groopman J E, Capon D J. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238:1704–1707. doi: 10.1126/science.3500514. [DOI] [PubMed] [Google Scholar]
- 49.Strizki J M, Turner J d, Collman R G, Hoxie J, Gonzalez-Scarano F. A monoclonal antibody (12G5) directed against CXCR4 inhibits infection with a dual-tropic human immunodeficiency virus type 1 isolate but not the T-tropic isolate HIV-1 HxB. J Virol. 1997;71:5678–5683. doi: 10.1128/jvi.71.7.5678-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tersmette M, Gruters R A, deWolf F, de Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 52.Trkola A, Ketas T, KewalRamani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibody and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van’t Wout A B, Koostra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema R A, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White-Scharf M E, Potts B J, Smith L M, Sokolowski K A, Rusche J R, Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993;192:197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- 55.Wrin T, Loh T P, Charron-Vennari J, Schuitemaker H, Nunberg J H. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wrin T, Nunberg J H. HIV-1 MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS. 1994;8:1622–1623. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- 57.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardosa A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 58.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 62.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]