Abstract
Expression of retroviral Gag polyproteins is sufficient for morphogenesis of virus-like particles with a spherical immature protein shell. Proteolytic cleavage of Gag into the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains (in the case of human immunodeficiency virus [HIV]) leads to condensation to the mature cone-shaped core. We have analyzed the formation of spherical or cylindrical particles on in vitro assembly of purified HIV proteins or inside Escherichia coli cells. CA protein alone yielded cylindrical particles, while all N-terminal extensions of CA abolished cylinder formation. Spherical particles with heterogeneous diameters or amorphous protein aggregates were observed instead. Extending CA by 5 amino acids was sufficient to convert the assembly phenotype to spherical particles. Sequences C-terminal of CA were not required for sphere formation. Proteolytic cleavage of N-terminally extended CA proteins prior to in vitro assembly led to the formation of cylindrical particles, while proteolysis of in vitro assembly products caused disruption of spheres but not formation of cylinders. In vitro assembly of CA and extended CA proteins in the presence of cyclophilin A (CypA) at a CA-to-CypA molar ratio of 10:1 yielded significantly longer cylinders and heterogeneous spheres, while higher concentrations of CypA completely disrupted particle formation. We conclude that the spherical shape of immature HIV particles is determined by the presence of an N-terminal extension on the CA domain and that core condensation during virion maturation requires the liberation of the N terminus of CA.
Retroviruses are released by budding from the plasma membrane of the infected host cell. Bud formation occurs either from preformed spherical particles (type B and D viruses, spumaviruses) or concomitant with assembly of the inner virion structure (type C viruses, lentiviruses [reviewed in references 16 and 59). Early immature retroviral particles always contain a spherical protein shell, approximately 80 to 100 nm in diameter, which is formed by assembly of the Gag- and Gag-Pol precursor polyproteins. Soon after budding, an extensive morphological rearrangement called maturation occurs, which is necessary for the virion to become infectious. Maturation requires proteolytic processing by the viral proteinase (PR) and leads to condensation of the spherical immature protein shell to a centrally located electron-dense structure, the mature core (reviewed in reference 55). Mature retroviral cores consist of a ribonucleoprotein (RNP) complex with two molecules of genomic RNA associated with the nucleocapsid (NC) protein and with the replication enzymes reverse transcriptase (RT) and integrase (IN), encased in a protein shell formed by the capsid (CA) protein (for nomenclature of retroviral proteins, see reference 36). In human immunodeficiency virus (HIV), additional viral and cellular proteins which are presumed to function during virus entry (e.g., cyclophilin A [CypA] [12, 38, 54]) are also packaged into virus particles.
The HIV Gag polyprotein (Pr55gag) is divided into the matrix (MA), CA, NC, and C-terminal p6 domain (Fig. 1). The N-terminal MA domain is cotranslationally myristoylated (6, 21) and closely apposed to the inner surface of the lipid envelope (17) and contains signals for intracellular transport of the polyprotein (33). A homomultimer of CA forms the cone-shaped capsid shell of the mature virus (17), while NC is involved in packaging of the genomic RNA (reviewed in reference 2) and p6 plays a role in release of the budding virus (20). In addition, there are short intervening sequences on the polyprotein called spacer peptides (23), which are released by PR-mediated cleavages at their N and C termini during maturation (41).
FIG. 1.
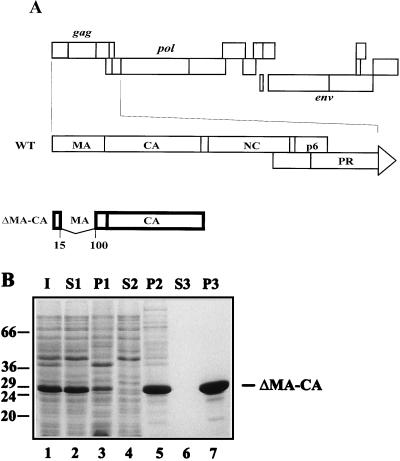
Expression and purification of the ΔMA-CA protein. (A) Schematic representation of the genetic structure of HIV-1 and of the region encoded in the expression plasmid. At the top, the coding region of the HIV-1 genome is shown, with the different open reading frames depicted as boxes. The gag reading frame and the 5′ terminal part of the pol reading frame are expanded below. The HIV-specific region encoded by plasmid pET ΔMA-CA is shown at the bottom. The numbers indicate the amino acids of MA flanking the deletion. (B) Coomassie blue-stained SDS-polyacrylamide gel representing purification of ΔMA-CA. Lanes: 1, lysate from induced bacteria; 2 and 3, soluble and insoluble material after high-speed centrifugation; 4 and 5, supernatant and precipitate following precipitation by addition of ammonium sulfate to 25% saturation; 6 and 7, supernatant and precipitate following a second ammonium sulfate precipitation after anion-exchange chromatography. Molecular mass standards (in kilodaltons) are indicated on the left; the position of ΔMA-CA is marked on the right.
Retroviral Gag proteins alone are sufficient to form immature spherical particles (48), which are released by budding but do not mature, do not contain viral glycoproteins, and are noninfectious. Accordingly, expression of Gag alone with recombinant viral expression systems (9, 18, 25, 28–30, 46, 49) or after transfection of expression plasmids (40, 50) consistently yielded the release of virus-like particles. The presence of RNA, heterogeneous in size and of both viral and cellular origins, within these particles has been reported (18, 49), but the role and significance of RNA in the assembly process are currently not clear. C-terminal deletions of Gag, removing p6 and most of NC, did not prohibit the formation of immature particles (25, 28, 29, 46), while further truncations extending into the spacer peptide at the C terminus of CA (SP1; Fig. 1) abolished particle formation (18, 25). Furthermore, large internal deletions within MA, removing most of its globular core (24, 39, 43), had no apparent effect on the morphology of immature or mature particles (11, 52). It appears likely, therefore, that the C-terminal segment of Gag and the globular domain of MA are not essential for assembly of immature HIV-like particles.
Assembly of retrovirus particles can occur at different sites within the cell, suggesting that no specific subcellular environment is required (44, 57). Formation of spherical nonenveloped HIV- or simian immunodeficiency virus (SIV)-like particles was observed in the cytoplasm or in the nucleus following baculovirus-mediated overexpression of the respective polyprotein (9, 46). Recent evidence suggests that assembly of retrovirus-related structures can even occur in a test tube following in vitro translation (47, 51) or bacterial expression of complete retroviral polyproteins or individual domains thereof. Klikova et al. (31) reported that Escherichia coli expression of Mason-Pfizer monkey virus Gag yielded spherical unenveloped particles similar to the immature virus, both inside bacterial cells and upon incubation of purified proteins. Formation of tubular structures has been observed following in vitro assembly of purified bacterially expressed fragments of Rous sarcoma virus (RSV) or HIV Gag polyproteins. In vitro assembly of RSV and HIV CA-NC led to RNA-dependent formation of hollow cylinders with a diameter of 55 nm and heterogeneous length (7). Morphologically identical structures were observed when purified HIV CA protein was used, but cylinder formation occurred at a 20-fold-higher molar concentration of protein, in high salt, and independent of RNA (22). These results suggested that at least for HIV, CA alone determines the shape of cylinders, which are stabilized by weak hydrophobic interactions. The NC domain probably aligns and concentrates the protein on the RNA and thereby facilitates efficient assembly at lower protein concentrations (7, 22). Campbell and Vogt (8) recently reported that in vitro assembly of larger fragments of RSV Gag yielded spherical particles if the p10 region (located upstream of CA on the RSV Gag polyprotein) was present. Gag fragments lacking p10 yielded cylindrical particles similar to CA-NC. These authors concluded that RSV p10 contains a shape-determining region which facilitates interactions required for spherical particle formation (8).
To define the requirements for assembly of immature and mature HIV-like particles, we constructed bacterial expression vectors for segments of the HIV Gag polyprotein. Here, we report that CA alone formed cylindrical particles in vitro and inside bacterial cells while N-terminal extensions abolished cylinder formation. Spherical particles with heterogeneous diameter or amorphous protein aggregates were observed instead. Cylinder formation was restored if proteins were cleaved by HIV PR before in vitro assembly. These results show that N-terminal extensions of CA convert its assembly products from tubular to spherical particles and suggest that liberation of the CA N terminus is required for core condensation during maturation.
MATERIALS AND METHODS
Expression plasmids.
DNA manipulations were carried out by standard methods. All plasmids were derived from the prokaryotic expression vector pET11 3xc (Novagen, Madison, Wis.), which carries a T7 expression cassette. HIV-1 coding sequences were amplified from the proviral clone pNL4-3 (1) by PCR. For construction of the CypA expression vector, plasmid pGEX-2T/hu–cyclophilin A (3) was used as a template. By choosing appropriate primer sequences, we introduced a translational start codon at the 5′ end (part of the newly introduced NdeI site) and two stop codons (TGA/TAG) at the 3′ end of the coding sequences and also introduced novel restriction sites at the 5′ end (NdeI) and at the 3′ end (BamHI).
The amplified fragments were cleaved with NdeI and BamHI and ligated into pET11 3xc that was also cleaved with NdeI and BamHI, resulting in the expression plasmids pET MA-CA, pET ΔMA-CA, pET N15MA-CA, pET MA100CA, pET MA115CA, pET MA120CA, pET MA128CA, pET CA, pET CAΔ13 (see Fig. 5A), and pET-CypA. These plasmids encode recombinant proteins with predicted molecular masses of 40.5 kDa (MA-CA), 31 kDa (ΔMA-CA), 27 kDa (N15MA-CA), 29 kDa (MA100CA), 27.5 kDa (MA115CA), 27 kDa (MA120CA), 26 kDa (MA128CA), 25.6 kDa (CA), and 24 kDa (CAΔ13), all containing an N-terminal Met residue in addition to the sequence of the respective viral protein. In the case of CA and ΔMA-CA, N-terminal sequence analysis of purified proteins indicated removal of the N-terminal methionine.
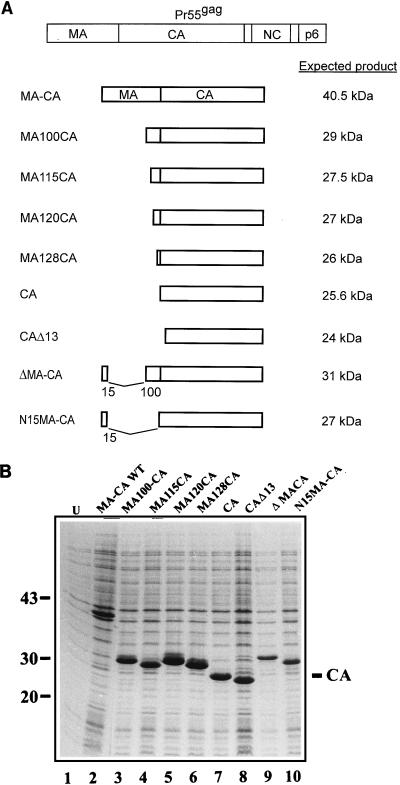
FIG. 5.
(A) Schematic representation of the various expression vectors. The Gag polyprotein is shown at the top, and the regions encoded in the expression vectors and the expected products are indicated below. The numbers correspond to the amino acids of MA or CA (CAΔ13) flanking the deletion. (B) Coomassie blue-stained SDS-polyacrylamide gel representing induced bacterial cells harboring various expression vectors. U, uninduced bacteria. Molecular mass standards (in kilodaltons) are indicated on the left, and the position of CA is marked on the right.
Expression and purification of recombinant proteins.
Induction of Escherichia coli BL21 DE3 cells was performed essentially as described previously (22). CypA was purified by a published procedure (37). Purification of CA-derived proteins was modified from our published procedure (22). Bacterial cells were resuspended in cold lysis buffer (50 mM morpholineethanesulfonic acid [MES; pH 6.5], 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 100 mM NaCl) and disrupted by cell disintegration (glass beads; Biomatik) and subsequent sonication (Branson sonifier B12). After consecutive centrifugation at 3,000 × g for 5 min, 27,000 × g for 15 min, and 246,000 × g for 60 min, CA proteins were precipitated from the soluble fraction by the addition of ammonium sulfate to 25% saturation, redissolved in buffer containing 50 mM Tris-HCl (pH 8.0), 30 mM NaCl, 1 mM EDTA, and 1 mM DTT, and applied to DEAE-cellulose (Whatman DE52) equilibrated with the same buffer in a batch procedure. CA proteins were collected from the unbound material by the addition of ammonium sulfate to 50% saturation followed by centrifugation. In some cases, proteins were further purified by cation-exchange chromatography with a POROS SP 20/M column on a BioCAD Sprint Perfusion Chromatography System (PerSeptive Biosystems). Proteins were applied in 50 mM MES (pH 6.0)–50 mM NaCl–1 mM EDTA–1 mM DTT and eluted with a salt gradient. For the MA120CA and MA128CA proteins, lysis was performed in 50 mM Tris-HCl (pH 8.9)–1 mM EDTA–1 mM DTT–1 M NaCl, high-speed centrifugation and anion-exchange chromatography were omitted, and the ammonium sulfate precipitates were redissolved in 1 M salt buffer (pH 6.0) and diluted to 50 mM NaCl before undergoing cation-exchange chromatography. All purified proteins were collected by ammonium sulfate precipitation, redissolved to a concentration of approximately 3 mg/ml in 30 mM MES (pH 6.0)–1 mM EDTA–1 mM DTT–100 mM NaCl (or lacking NaCl in the case of CA and ΔMA-CA), and stored frozen at −70°C.
Analysis of expression products.
Protein samples were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels containing 15% polyacrylamide (30:1 ratio of acrylamide to N,N-methylenebisacrylamide) and stained with Coomassie blue. The protein concentration was determined by the method of Layne (34). Purity was assessed by scanning of Coomassie blue-stained gels with a Desaga CD50 densitometer.
In vitro assembly.
Protein stock solutions were diluted to the appropriate concentration with storage buffer (30 mM MES [pH 6.0], 1 mM EDTA, 1 mM DTT, 100 mM NaCl) and dialyzed overnight at 4°C against assembly buffer (50 mM Tris-HCl [pH 8.0], 1 M NaCl) unless otherwise indicated. For PR treatment before assembly, ΔMA-CA was diluted to 3 mg/ml with storage buffer and active-site titrated HIV-1 PR (32) was added at a molar ratio of 1:10 (enzyme:substrate). After a 6-h incubation, the specific PR inhibitor Ro31-8959 (45) was added to a concentration of 5 μM and samples were dialyzed against assembly buffer and analyzed by negative-stain electron microscopy. For PR-treatment after in vitro assembly, ΔMA-CA was first dialyzed overnight against 50 mM Tris-HCl (pH 7.0)–0.65 M NaCl and PR was added subsequently at a molar ratio of 1:10 (E:S). Incubation was performed at 4°C on the grid, and PR treatment was stopped at various time points by the addition of Ro31-8959 to a final concentration of 5 μM and subsequent preparation for negative staining. For analysis of the effects of CypA on in vitro assembly, purified CA or ΔMA-CA was mixed with purified CypA at molar ratios between 1:1 and 10:1 and dialyzed against assembly buffer as indicated above.
Electron microscopy analysis.
The procedure for analysis of assembly products within induced bacteria will be described elsewhere (26). Briefly, induced E. coli BL21 DE3 cells were collected by brief centrifugation, resuspended and fixed in 4% paraformaldehyde–2% glutaraldehyde in phosphate-buffered saline (PBS), incubated for 5 min on ice, washed with PBS, and collected by brief centrifugation. The wet bacterial soft pellet was drawn into cellulose capillary tubes by capillary action as described previously (27). Subsequently, bacterial cells were postfixed within the capillaries for 30 min with 1% OsO4 in PBS, washed with water, stained for 30 min in 1% uranyl acetate in water, and dehydrated in a graded series of ethanol. The capillary tubes were embedded in ERL resin for sectioning. Ultrathin sections were counterstained with 2% uranyl acetate and lead citrate. Micrographs were obtained with a Philips CM120 transmission electron microscope at 80 kV.
For negative staining of in vitro assembly products, 5-μl samples of dialyzed protein solutions were applied to Parafilm and covered with a UV-irradiated Formvar/carbon-coated grid (200-mesh size) for 5 min. In some cases, purified antibodies to HIV-1 CA were bound to the grid to immobilize spherical particles. An immunoglobulin G fraction was prepared from rabbit polyclonal antiserum against CA (40) by successive precipitation with caprylic acid and ammonium sulfate as described previously (53). Purified antibodies were redissolved in PBS and incubated with UV-irradiated grids for 30 min at room temperature (protein concentration, 0.9 mg/ml). Subsequently, the grids were washed three times with 50 mM Tris-HCl (pH 7.2) and immediately used for binding of in vitro assembly products (1 h at room temperature). After binding, the grids were washed three times with Tris-HCl (pH 7.2) and three times with water and stained with 1% uranyl acetate for 5 min. Excess stain was removed by touching the grid to a filter paper. After being stained, the grid was air dried for 5 min and analyzed with a Philips CM 120 transmission electron microscope.
RESULTS
Purification of N-terminally extended CA proteins and in vitro assembly of spherical particles.
Previously, we had shown that in vitro assembly of purified HIV-1 CA protein with or without the C-terminally adjacent NC domain led to the formation of tubular particles (22). These hollow cylinders are likely to share structural features with mature HIV capsids, which exhibit a cone-shaped morphology in thin-section electron microscopy (16, 17) and cryoelectron microscopy (13). To extend the in vitro assembly system to the analysis of spherical HIV core-like particles corresponding to the immature virion, we constructed expression vectors for extended CA proteins. Previous studies with recombinant baculoviruses for expression had shown that the C-terminal p6 domain of Gag and most of the NC domain are not essential for assembly and extracellular release of spherical particles (25, 46). Since the presence or absence of NC also did not alter the in vitro assembly phenotype (22), we reasoned that the shape of assembly products may be determined by the N-terminal MA domain. Furthermore, a large internal deletion within the MA domain of HIV Gag (amino acids 16 to 99) did not change the morphology of the immature spherical protein shell (11). We therefore made a construct (pET ΔMA-CA) for expression of a protein containing the first 15 amino acids and the last 33 amino acids of the MA domain fused to CA (ΔMA-CA, containing a deletion from amino acids 16 to 99 of MA [Fig. 1A]). Following expression in E. coli BL21 DE3 cells, the specific product (31 kDa) accumulated to about 10% of the total E. coli protein after induction (Fig. 1B, lane 1) and reacted with specific antisera against CA in immunoblot analysis (data not shown).
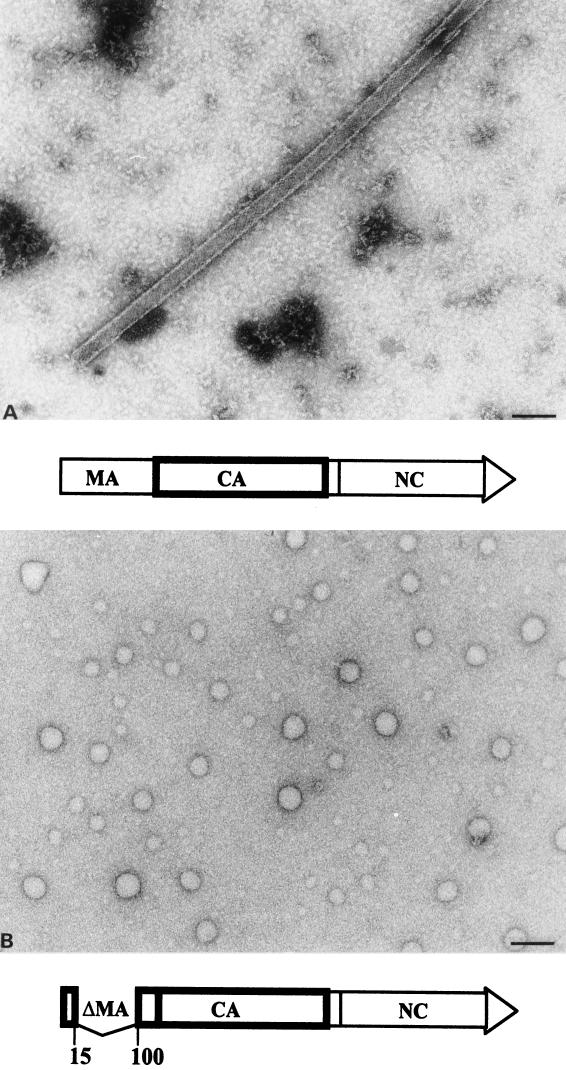
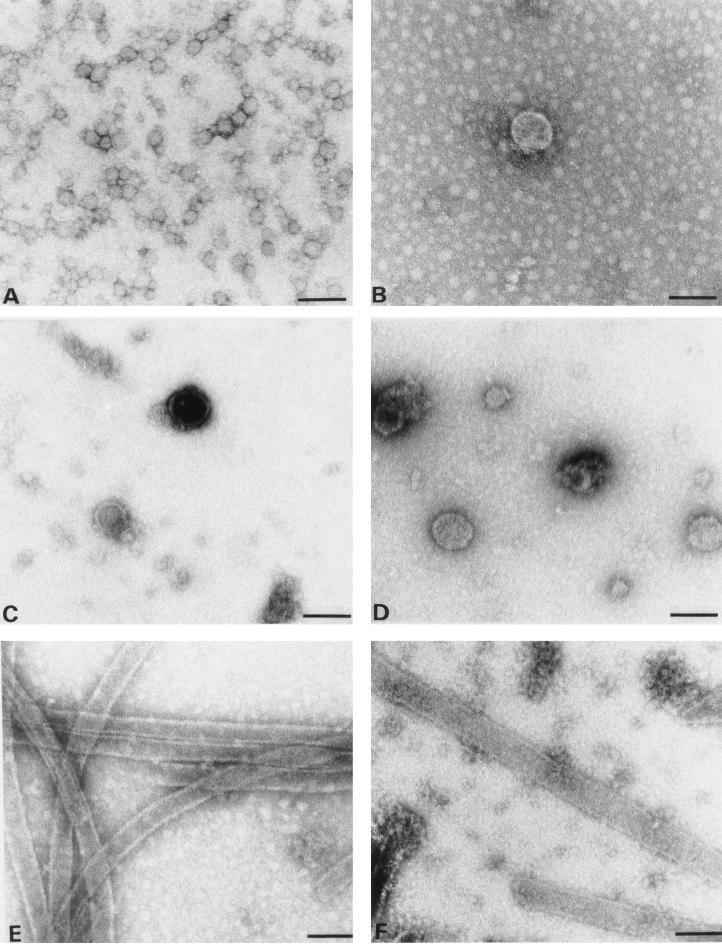
The ΔMA-CA protein was purified essentially as described for HIV-1 CA (22). Following high-speed centrifugation (Fig. 1B, lane 2), precipitation with ammonium sulfate (lane 5), and anion-exchange chromatography, approximately 20 mg of ΔMA-CA per liter of induced bacterial culture was obtained at a purity of >95% (lane 7). For in vitro assembly, purified CA and ΔMA-CA proteins were diluted to a concentration of 3 mg/ml and dialyzed overnight against assembly buffer (1 M NaCl [pH 8]). Samples from both reactions were analyzed by negative-stain electron microscopy. Assembly into long and hollow tubular structures with an external diameter of 50 nm was observed for CA (Fig. 2A), while in vitro assembly of ΔMA-CA yielded spherical particles of heterogeneous size but no cylinders (Fig. 2B). Both CA-derived cylinders and ΔMA-CA-derived spheres exhibited a regular substructure. The diameter of spherical particles ranged from 20 to 100 nm, with approximately 50% of particles showing a diameter of about 60 nm. Some incomplete and broken spheres were observed, with stain penetrating into the interior of the particle, which indicated that they were hollow with a wall thickness of 5 to 6 nm (data not shown; see below). The ΔMA-CA-derived particles appeared mostly circular with occasional flat or angular facets but without evidence for icosahedral symmetry (Fig. 2B). Approximately threefold fewer spherical particles were observed compared to CA-derived tubular structures, and we therefore used antibody-coated grids for immobilization of in vitro assembly products in most experiments. No difference in the shape or morphology of spheres was observed when grids with or without antibodies were analyzed in parallel (data not shown). Spherical particles were first observed after dialysis for 15 min. Large precipitates but no spherical or tubular particles were detected when ΔMA-CA or CA was not dialyzed but immediately adjusted to 1 M salt.
FIG. 2.
Negative-stain electron micrograph of in vitro assembly products. (A) CA. (B) ΔMA-CA. Protein solutions (3 mg/ml) were dialyzed overnight against 50 mM Tris HCl (pH 8.0) containing 1 M NaCl, stained with 1% uranyl acetate, and analyzed by electron microscopy. To immobilize ΔMA-CA-derived particles, the grid had been coated with purified CA-specific antibodies. No difference in size or morphology of particles was observed when grids without antibodies were used instead. Bar, 100 nm.
Optimal in vitro assembly of spherical structures from purified ΔMA-CA protein occurred in buffer of neutral to alkaline pH, at high ionic strength, and at a protein concentration of 3 mg/ml or more (100 μM), similar to the conditions determined for CA (22). Virtually no particulate structures were observed at pH 8 and 0.2 M salt, and the efficiency of sphere formation was only 5 to 10% at 0.5 M salt compared to 1 M salt. No ordered structures were observed if ΔMA-CA protein was incubated at pH 6 and 0.1 M salt, while assembly of spherical particles occurred when the salt concentration was raised to 1 M, albeit at a threefold-lower efficiency than that at pH 8 and 1 M salt. Thus, in vitro assembly of ΔMA-CA-derived spheres appeared less sensitive to pH and more sensitive to salt concentration than did assembly of CA-derived tubular particles (22). No particulate structures were observed when assembly reactions (in 1 M salt buffer [pH 8]) were performed at a protein concentration of 0.2 mg/ml ΔMA-CA. Increasing numbers of particles were detected when the protein concentration was raised above 1 mg/ml. Addition of 5 μM Ca2+ had no effect on the efficiency of assembly or on the morphology of spherical particles. Similarly, addition of RNA at 3% (by weight) of protein or pretreatment with RNase A (0.2 mg/ml for 1 or 2 h) did not alter the yield or structure of assembly products. Spherical particles were unaltered following incubation in 0.5% Triton X-100 for 150 min.
Effect of PR treatment and of cyclophilin A on in vitro assembly.
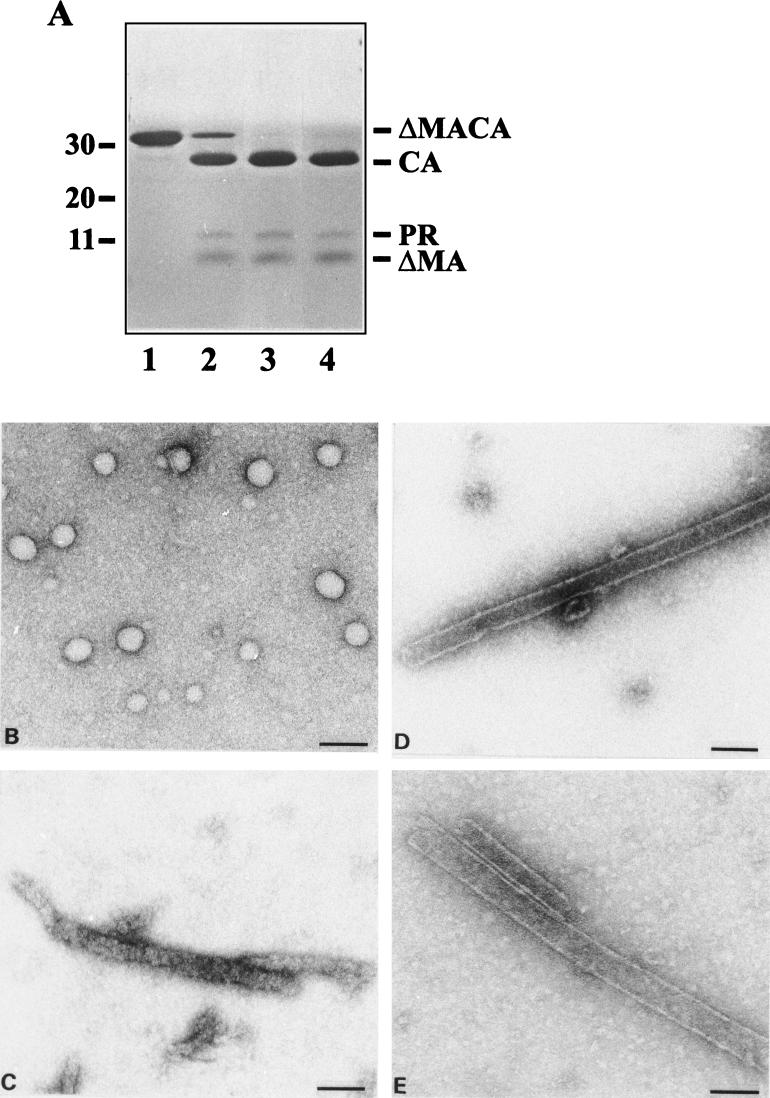
To analyze the effect of PR treatment on in vitro assembly, we incubated ΔMA-CA with purified HIV-1 PR at a molar ratio of 1:10 (E:S) either before or after performing the assembly reaction. Incubation of in vitro-assembled spherical particles (Fig. 3B) with PR for 10 min led to cleavage of most of the ΔMA-CA protein to ΔMA and CA (Fig. 3A, lane 2). Spherical particles were completely disrupted under these conditions, and no ordered structures were observed on the grid (data not shown). Incubation of assembly products with PR for 1 h or longer resulted in complete cleavage of ΔMA-CA (lane 3) and formation of long and irregular rod-like protein conglomerates with similar overall dimensions to those of CA-derived cylinders (Fig. 3C). However, these structures were not cylindrical and did not exhibit regular substructures. Cleavage of ΔMA-CA before in vitro assembly, on the other hand, resulted in efficient formation of cylindrical structures which were morphologically identical to CA-derived tubes (Fig. 3D). In a separate experiment, ΔMA-CA and PR were mixed and the entire sample was dialyzed against assembly buffer during the cleavage reaction. Dialysis for 6 h led to the formation of tubular structures which were slightly more heterogeneous in diameter than CA-derived cylinders (external diameters ranging from 40 to 60 nm [Fig. 3E]).
FIG. 3.
Effect of PR treatment on in vitro assembly. (A) Coomassie blue-stained SDS-polyacrylamide gel of ΔMA-CA and cleavage products. In vitro assembly of ΔMA-CA was performed as described in the legend to Fig. 2, and assembly products were analyzed either directly (lane 1) or after incubation with purified HIV-1 PR (E:S, 1:10) for 10 min (lane 2) or 1 h (lane 3). Alternatively, ΔMA-CA was first cleaved with PR for 6 h (lane 4) and subsequently used for in vitro assembly. Molecular mass standards (in kilodaltons) are depicted on the left; HIV-specific proteins are identified on the right. (B to E) Negative-stain electron micrographs showing the following reaction products: (B) ΔMA-CA in vitro assembly without PR incubation; (C) products after PR cleavage of ΔMA-CA-derived spherical particles for 1 h; and (D) products obtained when ΔMA-CA was first cleaved with PR for 6 h and cleavage products were subsequently used for in vitro assembly. (E) ΔMA-CA and PR were mixed and the entire reaction was used for in vitro assembly as described in Materials and Methods. Bar, 100 nm.
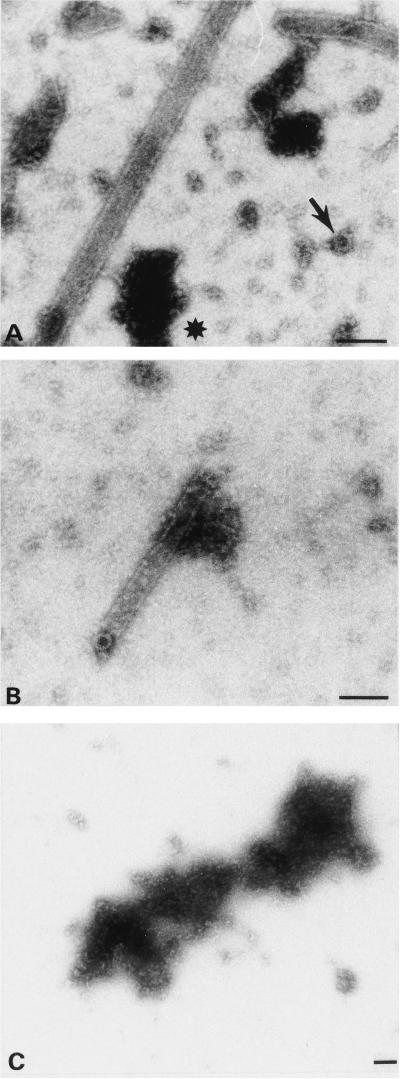
CypA, a cellular peptidylprolyl isomerase, is incorporated into HIV-1 particles by directly binding to the CA domain of Gag and is required for viral infectivity at an early step of replication (12, 38, 54). Virions lacking CypA appear to have no defect in assembly or release, and it has been suggested that CypA plays a role in virus entry, destabilizing the mature core for disassembly (5, 14). To analyze the effect of CypA on in vitro assembly of tubular and spherical particles, we performed assembly reactions in the presence of increasing concentrations of purified CypA. In the virion, the ratio of CA to CypA is approximately 10:1. In vitro assembly of CA in the presence of CypA at the same molar ratio of 10:1 did not alter the morphology, diameter, or wall thickness of the resulting cylinders (Fig. 4A). However, cylinders formed in the presence of CypA were significantly longer (average length, >3 μm) than those obtained in the absence of CypA (average length, 600 to 700 nm). Furthermore, assembly in the presence of CypA prevented aggregation of individual tubes, which was common in the absence of CypA. Besides hollow cylinders, we also observed small (diameter, approximately 30 nm) ringlike structures (Fig. 4A) and accumulation of unstructured protein aggregates (Fig. 4A) in the presence of CypA. At a molar ratio of 3:1 (CA to CypA), approximately 100-fold fewer cylinders were observed than in the absence of CypA. Individual cylinders were morphologically similar and had the same diameter of 50 nm but were shorter (Fig. 4B; average length, 300 nm). Small ring-like structures attached to the end of a cylinder were also observed (Fig. 4B). At a molar ratio of 1:1, neither cylinders nor any other organized structures were observed on the grid (Fig. 4C). In vitro assembly of ΔMA-CA in the presence of CypA at a molar ratio of 1:1 also completely disrupted particle formation (data not shown). Virtually no spherical particles but small unstructured protein aggregates of variable sizes were observed at a molar ratio of 3:1 (ΔMA-CA to CypA [Fig. 4E]), while assembly efficiency was not significantly altered when CypA was added at a molar ratio of 10:1. However, assembly products were considerably more heterogeneous in size and shape (Fig. 4D). Larger particles with a diameter up to 200 nm, as well as very small particles, were observed. Furthermore, only 50% of particles appeared spherical and various altered morphologies (elliptical, indented, teardrop shaped [Fig. 4D]) were observed. Very rarely, cylinders with similar dimensions to those of CA-derived particles, which had never been observed in the absence of CypA, were also detected. We conclude that CypA affects the in vitro assembly of both spherical and tubular particles, leading to a complete loss of particle formation if equimolar amounts of CypA are used and to altered assembly properties at substoichiometric concentrations. The effects of CypA can be blocked by the immunosuppressive drug cyclosporine, and assembly of both tubular and spherical particles was restored if 10 μM cyclosporine was added to an assembly reaction mixture containing equimolar amounts of CypA and CA or ΔMA-CA (data not shown).
FIG. 4.
Effect of CypA on in vitro assembly. Negative-stain electron micrographs corresponding to in vitro assembly of CA (A to C) or ΔMA-CA (D and E) in the presence of purified CypA at a CA-to-CypA molar ratio of 10:1 (A and D), 3:1 (B and E), or 1:1 (C). (D and E). Grids had been coated with purified CA-specific antibodies. The arrow in panel A identifies a ring-like structure approximately 30 nm in diameter; the asterisk denotes a large, nonstructured protein aggregate. Bar, 100 nm.
In vitro assembly of CA proteins with short N-terminal extensions.
To determine whether the ΔMA-CA protein contains a specific shape-determining region giving rise to assembly of spherical particles, we constructed additional bacterial expression vectors for N-terminally extended CA proteins. Besides MA-CA, containing the entire MA and CA sequence, the following extended CA proteins were expressed in E. coli: N15MA-CA (containing the first 15 amino acids of MA fused to the N terminus of CA), MA100CA (from amino acid 100 of MA to the C terminus of CA), MA115CA, MA120CA, and MA128CA. N15MA-CA and MA100CA contain the N-terminal or C-terminal segment of MA that is also present in the ΔMA-CA protein. In parallel experiments, we also analyzed the in vitro assembly of CA and of a truncated CA protein lacking the N-terminal 13 amino acids of CA (CAΔ13), which correspond to a tightly folded β-hairpin in the three-dimensional structure model of CA (14, 19). A schematic representation of the HIV-specific regions encoded by the various constructs and of the expected expression products is shown in Fig. 5A. Following induction of E. coli BL21 DE3, all proteins were produced equally well and accumulated to about 10% of total E. coli protein (Fig. 5B). Full-length MA-CA protein was poorly soluble after bacterial lysis and was not further analyzed. N15MA-CA, MA100CA, MA120CA, MA128CA, CA, and CAΔ13 were purified as described above and were all obtained at a purity of 90% or better.
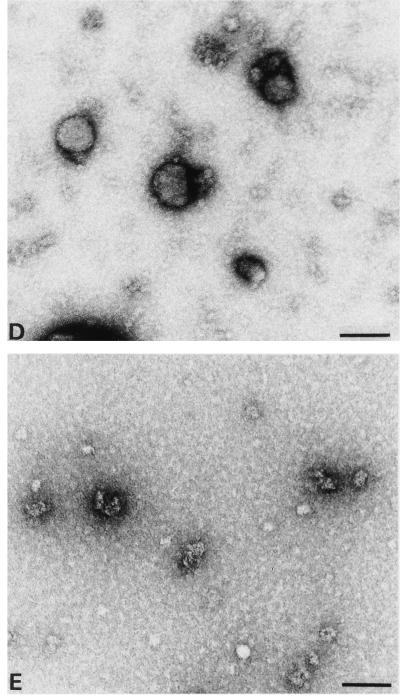
Dialysis of N15MA-CA and MA100CA against assembly buffer (1 M NaCl [pH 8]) yielded different results. With N15MA-CA, a large number of small, heterogeneous, spherical particles (diameter, 25 to 35 nm) with a strong tendency for aggregation into strings and grape-like structures was observed (Fig. 6A). In contrast to ΔMA-CA-derived particles, these smaller spheres did not exhibit any regular substructure. MA100CA, on the other hand, produced particles very similar to those observed for ΔMA-CA, albeit at more than a 100-fold-lower efficiency. Occasional particles with an average diameter of 60 nm, a circular appearance, and a regular substructure were detected (Fig. 6B). These results indicate that N-terminal extensions of CA by heterologous (N15MA-CA) or homologous (MA100CA) MA-derived sequences can block in vitro assembly of cylinders and convert the assembly phenotype to spherical particles.
FIG. 6.
Electron micrographs showing in vitro assembly products of N15MA-CA (A), MA100CA (B), MA120CA (C), MA128CA (D), CA (E), and CAΔ13 (F). Proteins (3 mg/ml) were dialyzed overnight against 50 mM Tris-HCl (pH 8.0) containing 1 M NaCl and analyzed by negative staining with 1% uranyl acetate. All the grids had been coated with purified CA-specific antibodies. No difference in the size or morphology of particles was observed when grids without antibodies were used instead. Bar, 100 nm.
To delineate a minimal MA-derived sequence required for production of spherical particles, we analyzed the in vitro assembly properties of MA120CA and MA128CA containing the C-terminal 13 and 5 amino acids of MA, respectively. Both proteins yielded spherical particles at comparable efficiency to that of ΔMA-CA. However, these particles were considerably more heterogeneous in morphology, and many broken or incomplete particles were observed (Fig. 6C and D), in contrast to the regular assembly products of ΔMA-CA (Fig. 2B). Many half-shells filled with stain were observed on the grid (Fig. 6C), which permitted accurate determination of their wall thickness. Similar to the tubular particles analyzed previously (22), the walls of these spheres were approximately 6 nm thick. No tubular particles typical of in vitro assembly of CA (Fig. 6E) were ever detected in MA120CA or MA128CA. The shortest protein analyzed in the in vitro assembly system contained a deletion of the first 13 amino acids of CA (CAΔ13). This protein yielded tubular particles at similar efficiency and of similar morphology to CA itself (Fig. 6F). CAΔ13-derived particles were slightly more heterogeneous in diameter but otherwise indistinguishable from CA-derived particles. However, more unstructured protein aggregates were observed with CAΔ13.
Assembly of N-terminally extended CA proteins in E. coli.
The Gag protein of Mason-Pfizer monkey virus and fragments of the RSV Gag protein assemble into either spherical or tubular particles inside bacterial cells, and their assembly properties in E. coli correlate with the in vitro assembly properties of the respective purified proteins (8, 31). However, these proteins contain the RNA-binding NC domain and in vitro assembly occurs in an RNA-dependent manner in low-salt buffer. To analyze whether CA and N-terminally extended CA proteins also assembled into particulate structures inside E. coli cells, the respective expression vectors were transformed into E. coli and bacterial cells were prepared for thin sectioning 3 h after induction. Fixation and further preparation were performed on bacteria drawn into cellulose capillary tubes, which obviated the need for repeated centrifugations and facilitated the handling and preparation of specimens.
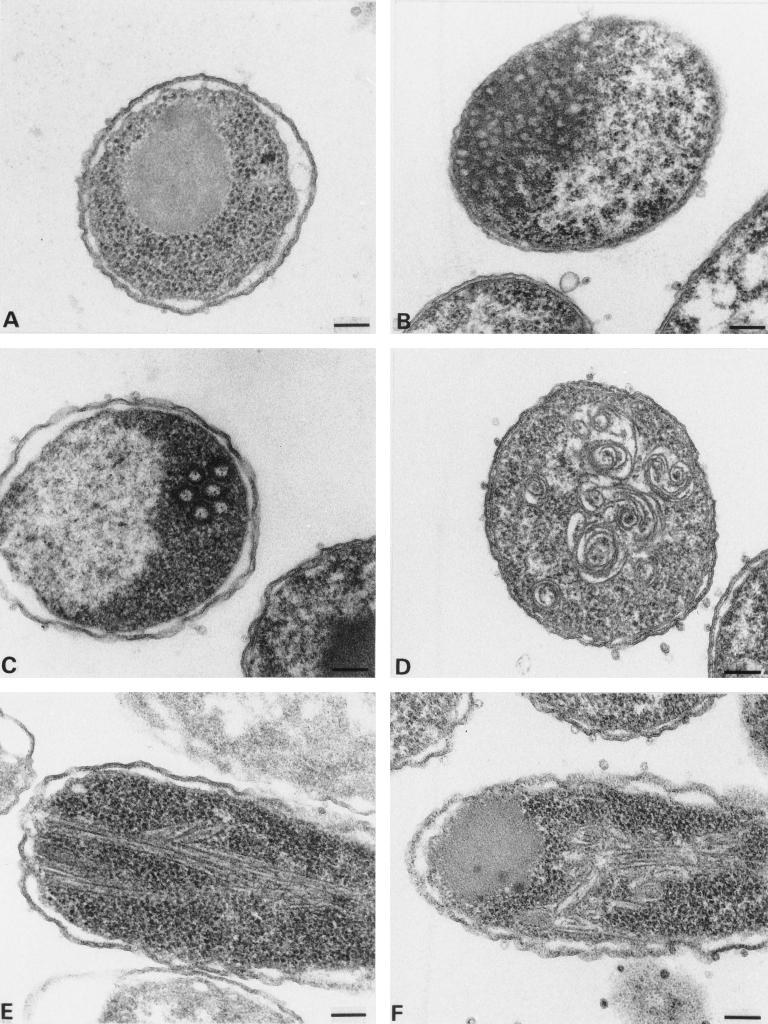
Induced bacteria harboring the CA expression vector readily showed the accumulation of intracellular cylinders (Fig. 7E). These cylinders were smaller in diameter than those observed in vitro (30 nm compared to 50 nm) and were considerably more branched. Furthermore, the cylinder lumen was difficult to detect in most cases, indicating that many particles had collapsed. MA-CA, containing the entire MA and CA regions of HIV-1, did not yield any ordered structures inside E. coli cells, and only large amorphous protein aggregates were observed (Fig. 7A). Similar results were obtained for MA120CA and MA128CA (data not shown), both of which had been shown to assemble into spherical particles in vitro (Fig. 6C and D). N15MA-CA gave rise to large arrays of small irregular particles of variable sizes very similar to the in vitro assembly products (Fig. 7B). MA100CA produced predominantly unstructured protein aggregates similar to MA-CA, but occasional regular spherical particles similar to those observed in vitro were also detected (Fig. 7C). Unexpectedly, ΔMA-CA which assembled into regular spherical particles with much higher efficiency in vitro gave a similar result to MA100CA in induced bacteria. Only rare spherical particles but predominantly unstructured protein aggregates were detected (data not shown). Induced bacteria harboring pET MA115CA (containing the last 18 amino acids of MA upstream of CA) showed large spiral structures (Fig. 7D). These structures most probably correspond to two-dimensional sheet-like arrays of protein. Similar spirals of slightly different morphology as well as unstructured protein aggregates but no regular cylinders were observed for CAΔ13 inside bacterial cells (Fig. 7F).
FIG. 7.
Thin-section electron microscopy analysis of induced bacterial cells harboring various expression vectors. (A) pET MA-CA; (B) pET N15MA-CA; (C) pET MA100CA; (D) pET MA115CA; (E) pET CA; (F) pET CAΔ13. Bar, 100 nm.
DISCUSSION
In this report, we have shown that fragments of the HIV-1 Gag polyprotein can assemble into hollow spherical particles in vitro provided that an N-terminally extended form of CA is used for assembly. Cylindrical particles were produced, on the other hand, if CA protein with its native N terminus or with a short N-terminal deletion (CAΔ13) was used. Formation of spherical particles did not require any other viral or cellular protein or nucleic acid. In vitro assembly of spheres occurred at neutral pH and high protein and salt concentrations, similar to the previously described in vitro assembly of cylindrical particles from purified CA (22). These results indicate that sphere formation is also mediated by weak hydrophobic interactions, most probably contributed by the CA domain. C-terminal segments of Gag were not required for in vitro assembly of spherical particles.
The observation that the presence of the N-terminal MA domain or a segment thereof determines the shape of the HIV core particle is in agreement with studies reporting the assembly phenotype of gag deletion mutants in tissue culture. Deletion of the entire p6 domain (28, 29, 46) and of most or all of the NC domain (25, 29, 46) did not prevent the release of HIV-like particles containing a spherical protein shell following baculovirus-mediated expression. Furthermore, large internal deletions within the HIV MA domain also had no apparent effect on the morphology of the resulting particles. A Gag protein lacking amino acids 16 to 99 of MA directed the budding of morphologically normal immature particles into the cisternae of the endoplasmic reticulum (11). A longer deletion mutant (amino acids 16 to 120 of MA) permitted extracellular release of particles (56), while deletion of almost the entire MA domain (amino acids 16 to 132, retaining only the first 15 amino acids of MA upstream of CA) led to accumulation of nonenveloped spherical particles with a similar morphology to authentic HIV (52). Lee and Linial (35) reported that replacing the entire MA domain of HIV-1 by a short signal for N-terminal myristoylation also permitted the assembly and release of spherical HIV-like particles, albeit at lower efficiency. In this case, the deletion included the first 10 amino acids of CA, indicating that the CA N-terminal region may not be essential for assembly of immature HIV particles. A similar result was obtained following baculovirus-mediated expression of a deletion mutant lacking codons 41 to 143 of the HIV gag reading frame (including the first 10 codons for CA), which led to the efficient release of immature particles with a morphology very similar to that of wild-type particles (4). While the expression products were quite different in these various studies, they all had in common that upstream sequences were present at the N terminus of CA. A similar phenotype was observed in the in vitro assembly system: spherical particles were formed when N-terminally extended CA proteins were used. These results suggest that morphogenesis of spherical particles may be controlled by the presence of additional sequences upstream of the CA domain on the polyprotein.
Only cylindrical particles, morphologically identical to CA-derived cylinders, were obtained when the ΔMA-CA protein was cleaved with HIV-1 PR prior to the in vitro assembly reaction. This result indicates that neither the synthesis as a polyprotein before cleavage nor the presence of the ΔMA peptide in the assembly reaction was responsible for the assembly phenotype, but only the presence of additional sequences at the N terminus of CA. Proteolytic separation of MA and CA should therefore also be required for condensation of the mature cone-shaped capsid shell. Mutation of the MA-CA cleavage site in the context of an HIV-1 proviral clone led to noninfectious particles containing an electron-dense RNP core and a thickening of the viral membrane but no conical capsid shell (21). The additional density at the membrane was most probably due to the CA domain, which had not been removed from the membrane-attached MA domain. Recent experiments showed that cleavage at both sides of SP1, separating NC and SP1 from the CA domain, is also essential for virion maturation, and it has been suggested that HIV-1 maturation is a sequential process governed by the rate of processing at individual cleavage sites (58).
The three-dimensional structure model of the N-terminal segment of HIV-1 CA, obtained by nuclear magnetic resonance spectroscopy (19) and X-ray analysis (14, 42), provides a likely explanation for the role of N-terminal extensions of CA during assembly. The structural studies indicated that CA is a largely helical protein with several putative interfaces for intermolecular interaction. The two most prominent interfaces are the C-terminal dimerization domain (15) and the N-terminal part of CA including helices 1 and 2 (amino acids 17 to 43 of CA) and a N-terminal β-hairpin structure from amino acids 1 to 13 of CA (14, 19). In the cleaved CA protein, the N-terminal NH2+ group of Pro1 of CA is oriented toward helix 3 and forms a salt bridge with Asp51, which is presumed to stabilize the β-hairpin (19). Preventing the formation of this salt bridge by substitution of the Asp codon in a proviral clone abolished condensation of the mature cone-shaped core, and the same substitution also prevented the formation of tubular particles in the in vitro assembly system (55a). N-terminal extensions of CA are predicted to block formation of the β-hairpin, because additional residues would sterically clash with the first helices and no positively charged group would be available for interaction with Asp51 (19). Presumably, N-terminal extensions cause unfolding of the β-hairpin structure into an extended conformation, making the MA-CA bond accessible for proteolytic cleavage. This alteration may prevent cylinder formation, either because the β-hairpin structure is an essential determinant of the CA interface in the cylinder or because the flexible extended N terminus interferes with cylinder formation. In either case, the assembly of spherical particles would be determined primarily by CA-mediated interactions and not by additional protein-protein interactions provided by the N-terminally fused sequences. This model can also explain why short N-terminal extensions of only 5 amino acids (MA128CA), as well as heterologous MA-derived sequences (N15MA-CA), can convert the assembly phenotype from cylinders to spheres. It should be noted, however, that particles derived from ΔMA-CA appeared more regular and were mostly intact whereas MA128CA and other shorter proteins yielded heterogeneous particles which were mostly incomplete. These results indicate that MA sequences may contribute to the morphogenesis of the spherical protein shell in additional ways besides anchoring the Gag protein to the membrane and keeping the N terminus of the CA domain in an unfolded state. All particles obtained in the in vitro system were smaller than the Gag protein shells of immature HIV particles, and additional segments of Gag and/or nucleic acids or cellular proteins may be required for the assembly of particles of regular size.
The N-terminal sequence of CA corresponding to the β-hairpin is not required for the assembly of spherical protein shells (4, 35). Our in vitro assembly experiments have shown that it is also not required for the formation of tubular particles of similar morphology to CA-derived cylinders. Thus, the N-terminal region of CA appears to have a decisive influence on particle morphogenesis when it is present in the protein, although it is not required for either cylinder or sphere formation. Most probably, this part of the protein serves as a molecular switch which is triggered by PR-mediated proteolytic cleavage during maturation. Gamble et al. (14) reported that the N-terminal interface of CA occludes 800 Å2 of accessible surface area and the β-hairpin structure contributes 230 Å2 to this value. The residual interactions mediated by the two N-terminal helices of CA may be sufficient for cylinder formation in the in vitro system, where only a single protein (CAΔ13) is present. However, no cylinders were detected for CAΔ13 in the more complex environment inside bacterial cells, suggesting that the β-hairpin contributes to cylinder formation. Full-length CA protein, on the other hand, also gave rise to cylinders inside bacterial cells, although these structures appeared less ordered and were mostly collapsed.
CypA is a cellular protein that is incorporated into HIV-1 particles at a ratio of 1:10 (relative to CA) and is important for virus replication (12, 38, 54). It binds to a flexible loop in CA (14) and is incorporated into the particle via its interaction with the CA domain of Gag (38). Based on analysis of individual steps in viral replication, it has been suggested that CypA plays a role in viral entry, at an early step preceding reverse transcription (5). Structural analysis of CA-CypA cocrystals (at a CA-to-CypA ratio of 1:1) showed that CypA binding may inhibit interactions between planar CA strips, thereby preventing the formation of higher-order structures (14). Given the 10-fold-lower CypA-CA stoichiometry in the virion, this inhibition may have a destabilizing effect preparing the core for disassembly (14). In vitro assembly of CA or ΔMA-CA in the presence of CypA at a molar ratio of 1:1 prevented formation of ordered structures, as suggested by the structural studies. Furthermore, a severe reduction in assembly efficiency and formation of significantly shorter cylinders was also observed at a CypA-to-CA molar ratio of 1:3 and no particles were detected for ΔMA-CA at this ratio. When CypA was added at a molar ratio of 1:10, as found in the virion, on the other hand, there was no significant difference in the assembly efficiency for either CA or ΔMA-CA compared to experiments in the absence of CypA. Significantly longer cylinders which had lost their tendency to aggregate were observed in the case of CA. Thus, at least in our in vitro system (1 M NaCl [pH 8]), the presence of CypA at a concentration of 10% of the CA level does not interfere with cylinder formation and the observed phenotype could be explained by a chaperone function of CypA which may enhance CA reorganization, resulting in longer cylinders in the in vitro system. It is interesting that the same concentration of CypA altered the shape of ΔMA-CA-derived particles from spheres to polymorphic structures with occasional cylinders with diameters of 50 nm, which had never been observed in the absence of CypA. Conceivably, CypA may also destabilize the spherical protein shell during virion maturation and facilitate the formation of the cone-shaped core. Even minor alterations in the ordered reorganization of the mature core may produce subsequent defects in virus entry.
ACKNOWLEDGMENTS
We are grateful to A. Billich for the CypA vector and to J. Konvalinka for ETG. We thank C. Hegert for preparation of recombinant proteins and for sequence analysis, B. Müller for critically reading the manuscript, and I. Ellhof for expert assistance and photography. We are grateful to W. Sundquist, S. Campbell, and V. Vogt for communicating their experimental results on in vitro assembly prior to publication.
This work was supported in part by a grant from the German ministry for education and research to H.-G.K.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Billich A, Hammerschmid F, Peichl P, Wenger R, Zenke G, Queshian V, Rosenwirth B. Mode of action of SDZ NIM811, a non immunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J Virol. 1995;69:2451–2461. doi: 10.1128/jvi.69.4.2451-2461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulanger, P. Personal communication.
- 5.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the iniziation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The Gag precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, III Hutchison C A, Loeb D D, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 11.Fäcke M, Janetzko A, Shoeman R L, Kräusslich H-G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 13.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 14.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 15.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;273:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 16.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 17.Gelderblom H R, Hausmann E H S, Özel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 18.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55 gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 19.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 20.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation on infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross I, Hohenberg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 23.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structure of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 26.Hohenberg, H., C. Huckhagel, I. Gross, and H.-G. Kräusslich. Unpublished data. [DOI] [PMC free article] [PubMed]
- 27.Hohenberg H, Mannweiler K, Müller M. High-pressure freezing of cell suspensions in cellulose capillary tubes. J Microsc. 1994;175:34–43. doi: 10.1111/j.1365-2818.1994.tb04785.x. [DOI] [PubMed] [Google Scholar]
- 28.Hoshikawa N, Kojima A, Yasuda A, Takayashiki E, Masuko S, Chiba J, Sata T, Kurata T. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J Gen Virol. 1991;72:2509–2517. doi: 10.1099/0022-1317-72-10-2509. [DOI] [PubMed] [Google Scholar]
- 29.Jowett J B M, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 30.Karacostas V, Najashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression system. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klikova N, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konvalinka J, Heuser A-M, Hruskova-Heidingsfeldova O, Vogt V M, Sedlacek J, Strop P, Kräusslich H-G. Proteolytic processing of particle-associated retroviral polyproteins by homologous and heterologous viral proteinases. Eur J Biochem. 1995;228:191–198. doi: 10.1111/j.1432-1033.1995.tb20249.x. [DOI] [PubMed] [Google Scholar]
- 33.Kräusslich H-G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 34.Layne E. Spectrophotometric and turbidometric methods for measuring proteins. Methods Enzymol. 1957;3:447–454. [Google Scholar]
- 35.Lee P P, Linial M. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common for all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Albers M W, Chen Q-M, Schreiber S L, Walsh C T. Cloning, expression, and purification of human cyclophilin A in Escherichia coli and assessment of the catalytic role of cysteins by site-directed mutagenesis. Proc Natl Acad Sci USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 gag protein binds to cyclophilins A and B. Cell. 1993;73:1076–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 39.Massiah M A, Starich M R, Paschal C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 40.Mergener K, Fäcke M, Welker R, Brinkmann V, Gelderblom H R, Kräusslich H-G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 41.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 43.Rao Z, Balyaev A S, Fry E, Jones I M, Stuart D I. Crystal structure of the SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 44.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 45.Roberts N A, Martin J A, Kinchington D, Broadhurst A V, Craig J C, Duncan I B, Galpin S A, Handa B K, Kay J, Kröhn A, Lambert R W, Merrett J H, Mills J S, Parkes K E B, Redshaw S, Ritchie A J, Taylor D L, Thomas G J, Machin P J. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 46.Royer M, Cerutti M, Gay B, Hong S S, Devauchelle G, Boulanger P. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 47.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsid in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shields A, Witte O N, Rothenberg R, Baltimore D. High frequency of aberrant expression of Moloney leukemia virus in clonal infections. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 49.Shioda T, Shibuta H. Production of human immunodeficiency virus (HIV)-like particles from cells infected with recombinant vaccinia viruses carrying the gag gene of HIV. Virology. 1990;175:139–148. doi: 10.1016/0042-6822(90)90194-v. [DOI] [PubMed] [Google Scholar]
- 50.Smith A J, Cho M, Hammarskjöld M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55 gag and Pr160 gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spearman P, Wang J J, Van der Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinbuch M, Andran R. The isolation of IgG from mammalian sera with the aid of caprylic acid. Arch Biochem Biophys. 1969;134:279–284. doi: 10.1016/0003-9861(69)90285-9. [DOI] [PubMed] [Google Scholar]
- 54.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C, Sodroski J, Göttlinger H. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 55.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–132. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 55a.Von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C-T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welker R, Janetzko A, Kräusslich H-G. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J Virol. 1997;71:5209–5217. doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]