Abstract
The recent development and use of protease inhibitors have demonstrated the essential role that combination therapy will play in the treatment of individuals infected with the human immunodeficiency virus type 1 (HIV-1). Past clinical experience suggests that due to the appearance of resistant HIV-1 variants, additional therapeutics will be required in the future. To identify new options for combination therapy, it is of paramount importance to pursue novel targets for drug development. Ribosomal frameshifting is one potential target that has not been fully explored. Data presented here demonstrate that small molecules can stimulate frameshifting, leading to an imbalance in the ratio of Gag to Gag-Pol and inhibiting HIV-1 replication at what appears to be the point of viral particle assembly. Thus, we propose that frameshifting represents a new target for the identification of novel anti-HIV-1 therapeutics.
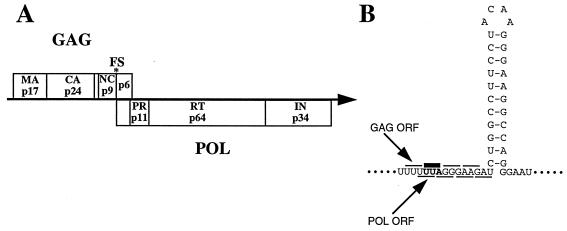
The full-length human immunodeficiency virus type 1 (HIV-1) mRNA encodes two polyprotein precursors, Gag and Pol (Fig. 1A). Although derived from the same mRNA, the precursor proteins are not produced with equal stoichiometry. Instead, 95% of the translating ribosomes produce Gag while 5% produce a Gag-Pol fusion protein (5, 14, 32). At the cell membrane, the Gag and Gag-Pol proteins assemble to form viral particles (33) and are subsequently processed into their individual protein components by the virally encoded protease (5, 32). Pol is always produced as a Gag-Pol fusion protein, even though the pol coding region partially overlaps and is in the −1 reading frame with respect to gag (Fig. 1A) (14, 27, 30). When ribosomes translating gag reach the start of the pol coding region, they must shift out of the gag reading frame by slipping back 1 nucleotide, to generate the Gag-Pol fusion protein (13). This is the process of −1 ribosomal frameshifting, and the structure within the gag-pol mRNA that causes frameshifting is shown in Fig. 1B (1, 8, 11, 13, 14). The frameshift signal is composed of two parts, the slippery sequence (UUUUUUA) and a stem-loop (a region of stable secondary structure in the mRNA). As ribosomes travel along the mRNA translating gag, they interact with the stem-loop and as a result are temporarily stalled on the slippery sequence (13, 15, 28). While stalled, a small percentage of the ribosomes slip back 1 nucleotide so that when the RNA stem-loop is unwound and the ribosomes continue, they now translate the pol open reading frame, producing the Gag-Pol fusion protein.
FIG. 1.
HIV-1 gene expression and frameshift signal. (A) The full-length HIV-1 mRNA encodes two precursor polyproteins, Gag and Pol, that are cleaved into their individual proteins during assembly of the mature viral particle. Abbreviations: MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; RT, reverse transcriptase; IN, integrase. (B) The HIV-1 frameshift signal is composed of two parts, the slippery sequence (UUUUUUA) and a stem-loop, a region of stable secondary structure in the mRNA. ORF, open reading frame.
Why has HIV-1, like many retroviruses, evolved the process of frameshifting for production of the Pol proteins? If the HIV-1 genome is modified (by the addition of 1 nucleotide at the frameshift site) so that gag and pol are in frame and Gag-Pol is produced 100% of the time, the resulting mutant viruses are unable to produce viral particles (16, 23). This result suggests that the ratio of Gag to Gag-Pol may be critical for particle assembly. The modified viruses, however, never produce p6 (at the carboxy terminus of Gag), so a role for p6 in particle formation cannot be eliminated. Studies using yeast retroviruses and retrotransposons that examine the relationship between the Gag/Gag-Pol ratio and viral particle assembly in more detail have been performed (7, 17, 34). These experiments used a variety of genetic mutations to demonstrate that either increasing or decreasing the level of frameshifting, by as little as twofold, was detrimental to replication of the yeast retroelements (7, 17, 34). Again it was hypothesized that by linking the synthesis of Gag and Gag-Pol through frameshifting, the production of Gag and Gag-Pol in the correct ratio for efficient viral particle assembly is ensured (7).
We set out to investigate whether the ratio of Gag to Gag-Pol, as controlled by frameshifting, regulates HIV-1 particle assembly and, if so, would frameshifting be a suitable target for the development of novel anti-HIV-1 therapeutics. To this end, we looked for chemical agents that would affect the ratio of Gag to Gag-Pol (by either stimulating or inhibiting frameshifting) and then tested these agents for their effect on HIV-1 particle assembly and replication. Agents that affected frameshifting were identified, and it was determined that they did indeed inhibit HIV-1 replication.
MATERIALS AND METHODS
In vitro translations.
RNA encoding firefly luciferase was generated in vitro by using a T7 MegaScript kit (Ambion). Translation reactions contained 4 μl of RNA (stock concentration, 87 μg/ml), 3 μl of test sample (dissolved in 4% dimethyl sulfoxide [DMSO]), and 8 μl of rabbit reticulocyte lysate cocktail (Promega). After incubation at 30°C for 1 h, luciferase levels were measured by the addition of a luciferin reagent (Analytical Bioluminescence) and the light output in relative light units (RLUs) was detected with a luminometer (Dynatech ML3000).
Transfections.
COS cells (1.6 × 105) were transfected with 20 μg of DNA by a modified calcium phosphate technique (Stratagene). After 24 h at 37°C, the transfected cells were harvested and distributed into 12 wells (2-cm2 diameter) prior to incubation in the presence of compounds (final DMSO concentration, 0.5%) for 48 h. Plasmid-encoded secreted embryonic alkaline phosphatase (SEAP) levels were determined with a chemiluminescence SEAP kit (Tropix), and light output (in RLUs) was detected with a luminometer.
Acute infection assays.
CCRF-CEM cells and peripheral blood mononuclear cells were infected at a multiplicity of infection (MOI) of 0.1 with either HIV-1IIIB (27) or HIV-1RTMDR (19) for 1 h. The infected cells were incubated in the presence of a concentration range of RG501 until the untreated control demonstrated clear signs of infection (7 to 10 days). At the experimental end point, the levels of infectious virus were determined by using a p24 enzyme-linked immunosorbent assay (ELISA). Uninfected cells were also treated with a range of concentrations of RG501 for the same time period and then tested by using a tetrazolium assay (31) to determine RG501’s cellular toxicity profile. These assays were performed at ViroMed Inc.
Chronic infection experiments.
CH-1 cells (1.6 × 105) were incubated in the presence of compounds for 48 h, medium was removed, and the cells were washed. Fresh medium and compound were returned to the cells, and incubation was continued for a further 24 h. This medium exchange was performed to ensure that any particle formation that occurred did so in the presence of test compounds. At the experimental end point, viral particles in the medium were pelleted by centrifugation at 16,000 × g for 90 min and resuspended in Triton lysis buffer (100 μl). The amount of p24 within each of the pelleted viral particles was determined by using a standard p24 ELISA (Cellular Products). CH-1 cell extracts were prepared by removing the cells from the plate, washing with phosphate-buffered saline, and lysing in a standard Triton lysis buffer.
Immunoblots.
Protein samples were separated by gel electrophoresis (Novex) and transferred to nitrocellulose (Novex) in a Bio-Rad minigel transfer chamber. After transfer, 3% gelatin was used as a blocking reagent prior to incubation with a different primary antiserum. Antisera were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS Program, NIAID, NIH; these include antiserum to HIV-1 p25/24 Gag from K. Steimer, Chiron Corporation (29), human HIV-1 immune globulin from A. Prince, New York Blood Center (25, 26), and antiserum to HIV-1 RT from Division of AIDS, NIAID (24). Immunoreactive complexes were visualized by use of the enhanced chemiluminescence detection system (Amersham). Specific immunoreactive complexes were quantified with an Alphainnotech Imager 2000 documentation and analysis system.
RESULTS AND DISCUSSION
Assays to identify agents that affect frameshifting.
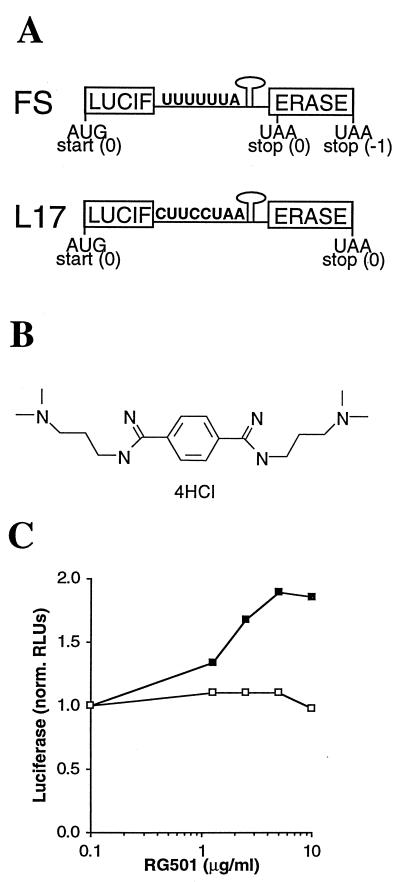
To study frameshifting in vitro, chimeric mRNAs that encode firefly luciferase were generated (Fig. 2A). In the FS construct (test RNA), DNA encoding the HIV-1 frameshift signal (Fig. 1B) was inserted into the luciferase-coding sequence (in place of nucleotides 676 to 682) so that the C-terminal portion of luciferase was in the −1 reading frame with respect to the luciferase start codon. Thus, the two sections of the luciferase-coding sequence represent the gag and pol coding regions and production of active luciferase from the FS construct is dependent on frameshifting. In the L17 construct (control RNA), DNA encoding a modified HIV-1 frameshift signal (CUUCCUAA instead of UUUUUUA at the slippery sequence) was inserted in the luciferase-coding sequence. The addition of the extra adenine residue leaves the two sections of the luciferase-coding sequence in the same reading frame, while changing the uracils to cytosines disrupts the slippery sequence but has no effect on the encoded amino acids. When translated, L17 mRNA generates a full-length protein identical to that produced from FS RNA but without the requirement for frameshifting. Thus, when the two RNAs are translated in parallel, the L17 translation serves as a control for nonspecific inhibitors of either luciferase activity or in vitro translation.
FIG. 2.
In vitro HIV-1 frameshifting assay. (A) Schematic representation of the RNA constructs used in the in vitro frameshifting assay. In the FS construct, functional luciferase is produced only when frameshifting occurs at the HIV-1 frameshift signal. The L17 construct contains a modified HIV-1 frameshift signal so when translated it produces the same protein as does the FS construct but without the requirement for frameshifting. (B) Chemical structure of RG501, which was provided by D. Boykin (Georgia State University). (C) RG501 stimulates frameshifting in vitro. Translations of either FS (▪) or L17 (□) RNA were performed in the presence of a range of RG501 concentrations (all at a final DMSO concentration of 0.8%). The amount of luciferase generated from each translation, as determined by light output measured in RLUs, was normalized to that from translations performed in the presence of DMSO alone.
The two chimeric mRNAs were utilized to find agents that had a specific effect on frameshifting. From 56,000 tests, several different samples with specific frameshifting activity were identified. To date, all the active samples identified have stimulated frameshifting. RG501 {1,4-bis-[N-(3-N,N-dimethylpropyl)amidino]benzene tetrahydrochloride} (Fig. 2B), as a representative of the active samples, increased translation of the luciferase-coding sequence from FS mRNA while having a minimal effect on translation of the luciferase-coding sequence from L17 RNA (Fig. 2C). From this result, it was hypothesized that RG501 was causing the ribosomes to pause above the slippery sequence for a longer period of time, thereby increasing their opportunity for slippage with a concomitant increase in the level of frameshifting. The most likely explanation for this extended pause was that RG501 interacted with and thereby stabilized the RNA stem-loop within the frameshift signal. A supporting result for this hypothesis was recently obtained in experiments where the HIV-1 frameshift signal stem-loop was replaced with the hairpin from the iron response element (IRE). In these in vitro experiments, addition of the IRE-binding protein (thereby stabilizing the stem-loop) significantly stimulated the levels of frameshifting (18).
To explore whether RG501 was indeed binding to the HIV-1 frameshift signal, a series of biophysical experiments were performed to determine the effect RG501 had on the thermal stability of the RNA stem-loop within the HIV-1 frameshift signal (20). The melting temperature (Tm) for the RNA stem-loop in the absence of RG501 was 78°C, but in the presence of RG501 (at a 1:1 molar ratio), the Tm was raised by 6.4°C (20). The ability to bind and stabilize RNA stem-loops was not due to a general RNA binding property of RG501 because this compound had little effect on the Tm for the RNA stem-loops within the REV response element (20). These data suggest that, as hypothesized, RG501 binds specifically to the stem-loop within the HIV-1 frameshift signal and stabilizes its structure, resulting in a concomitant increase in frameshifting.
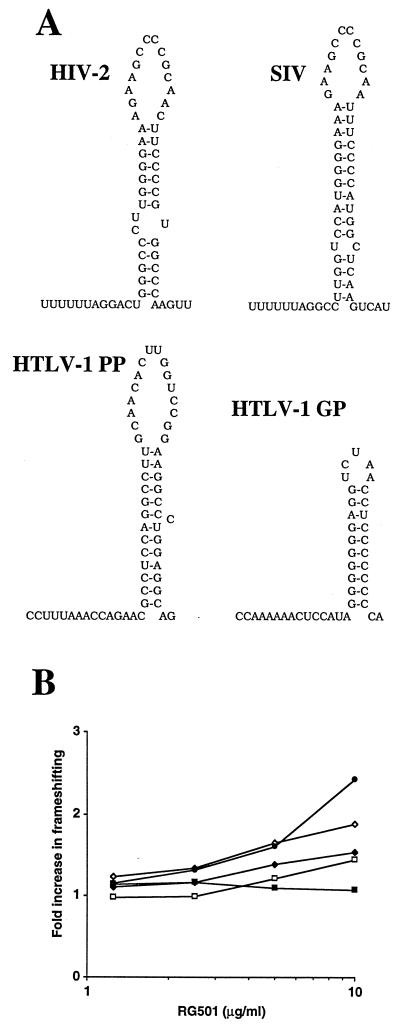
To explore further the effect of RG501 on frameshifting, constructs equivalent to FS and L17 were generated for four other −1 frameshift signals from other viruses (Fig. 3A). These signals occur at the gag-pol overlaps in HIV type 2 (HIV-2), in simian immunodeficiency virus (SIV) (9, 10), and at the two frameshift sites in human T-cell leukemia virus type 1 (HTLV-1) between gag-pro (GP) and pro-pol (PP) (12). In vitro assays were performed as described above, and the results are summarized in Fig. 3B. In Fig. 3B, the effect of RG501 on the different frameshift signals is presented as the fold increase in frameshifting; this value represents the percent frameshifting at a given concentration of RG501 divided by the percent frameshifting in the absence of RG501. RG501 enhances frameshifting at the different frameshift signals to quite different degrees. The HTLV-1 GP signal is the most enhanced, while there is no stimulation of frameshifting at the HTLV-1 PP signal. HIV-1 frameshifting is enhanced slightly more than that of HIV-2 or SIV, which are both stimulated to an equal extent. Other compounds have generated similar results as RG501, while additional compounds affect HIV-1, HIV-2, SIV, and HTLV-1 GP equally, but no compounds that enhance frameshifting at the HTLV-1 PP signal have been identified (data not shown). A recent publication (6) indicated that sparsomycin and anisomycin affect −1 frameshifting at the L-A double-stranded RNA virus pseudoknot but not at the Ty1 +1 frameshifting signal. Interestingly, neither of these compounds had frameshifting specific activity in our system against the HIV-1 frameshift stem-loop (data not shown). Clearly, different compounds affect frameshifting to different degrees depending on the individual frameshift signal. The physical properties (e.g., primary sequence, secondary structure, size, ΔG, or a combination of properties) that regulate the impact of RG501 on frameshifting still remain to be determined. A combination of RNA stability-structural analysis and mutational studies will be required to provide this information.
FIG. 3.
RG501 affects frameshifting at multiple viral frameshift signals. (A) Proposed frameshifting signals from other retroviruses. The ribosomal frameshift signals are shown for HIV-2 gag-pol, SIV gag-pol, HTLV-I GP, and HTLV-I PP. (B) RG501 stimulates frameshifting at multiple frameshift signals. Translations were performed in the presence of RG501 as described in Materials and Methods. For each pair of RNAs representing the different viral frameshift signals, at a given RG501 concentration, the percent frameshifting was determined by comparing the light output derived from the FS RNA with the light output derived from the L17 RNA. The percent frameshifting for each RG501 concentration was then compared to the percent frameshifting for the same viral constructs in the absence of RG501 and expressed as a fold increase in frameshifting HIV-1 (◊), HIV-2 (□), SIV (⧫), HTLV-I GP (•), and HTLV-I PP (▪).
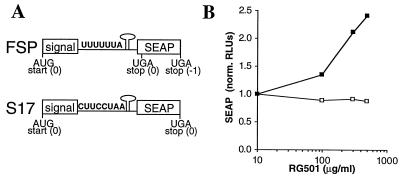
Having identified samples that affect frameshifting in vitro, it was next determined whether these samples also affected ribosomal frameshifting in cell-based experiments. An assay analogous to the in vitro translation assay was developed (Fig. 4A) based on the transfection of COS cells with vectors that encode SEAP (4). In these constructs, the HIV-1 frameshift signal was inserted between nucleotides 75 and 76 of the SEAP-coding sequence. This is after the secretion signal and in a region of the coding region that is not required for phosphatase activity (21). As expected from the in vitro translation results, RG501 caused a preferential stimulation of SEAP expression from the FSP construct compared to the S17 construct (Fig. 4B).
FIG. 4.
Cell-based HIV-1 frameshifting assay. (A) Schematic representation of the RNA constructs used in the cell-based frameshifting assay. The constructs were designed along similar lines as the in vitro constructs. In the FSP construct, functional SEAP (4) is produced only when frameshifting occurs. The S17 construct contains a modified HIV-1 frameshift signal so that when translated it produces the same protein as the FSP construct but without the requirement for frameshifting. (B) RG501 stimulates frameshifting in cells. COS cells were transfected with either FSP (▪) or S17 (□) expression vectors. After incubation in the presence of RG501 (final DMSO concentration, 0.5%) for 48 h, SEAP levels were determined with a chemiluminescence SEAP kit (Tropix). The RLUs for each concentration of RG501 were normalized to those from cells incubated in medium supplemented with 0.5% DMSO.
RG501 stimulated frameshifting approximately twofold in both the luciferase and SEAP assays, although presumably because of permeability issues, the stimulation of frameshifting in the cell-based SEAP assay required a higher concentration of RG501. Although the effect on frameshifting appeared modest, a two- to threefold increase (or decrease) in the level of frameshifting was enough to block replication of yeast retroelements (7, 17, 34); therefore, experiments were performed to determine whether RG501 could inhibit HIV-1 replication.
Agents that stimulate frameshifting block acute HIV-1 infection in culture.
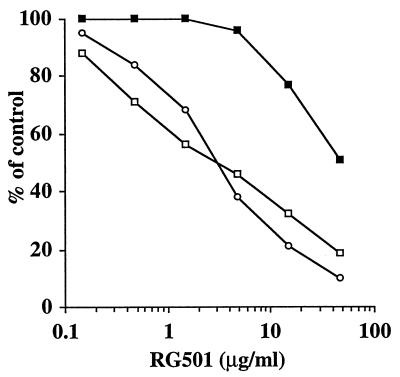
To determine whether stimulating frameshifting affected HIV-1 replication, the activity of RG501 in acute HIV-1 infection assays was examined. Infected cells (MOI = 0.1) were incubated in the presence or absence of RG501 to determine its ability to inhibit viral spread. At the same time, uninfected cells were incubated with or without RG501 to examine its cellular toxicity. From the results of two different virus strains, it can be seen that RG501 specifically inhibited the spread of HIV-1 from acutely infected cells (Fig. 5). RG501 inhibited both viral and cellular replication; however, in each case viral replication was inhibited by 50% at a concentration (IC50) (2.89 μg/ml) that was at least 10-fold lower than the concentration required to inhibit cellular replication by 50% (i.e., the therapeutic index of RG501 was >10). In addition, RG501 had a therapeutic index of >10 for HIV-1IIIB infection of peripheral blood mononuclear cells but in this case the IC50 was closer to 11 μg/ml (data not shown). It should be noted that RG501 appeared both active and toxic at lower concentrations in T cells (used for infection assays) than in COS cells (used for SEAP assays). This was assumed to be due to cell type variation in cellular permeability by the tetracation, RG501. To confirm that RG501’s activity was not due to some unexpected activity, in vitro assays which demonstrated that RG501 has no activity against HIV-1 reverse transcriptase (RT), integrase, or protease were performed (data not shown).
FIG. 5.
RG501 inhibits acute HIV-1 replication. CCRF-CEM cells were infected (MOI = 0.1) with either HIV-1IIIB (○) or HIV-1RTMDR (□) for 1 h. Infected and uninfected cells were incubated in the presence of a range of concentrations of RG501 for 7 to 10 days. The levels of infectious virus (○ and □) were determined with a p24 ELISA and the cellular toxicity profile of RG501 (▪) was determined with a tetrazolium assay.
Agents that stimulate frameshifting inhibit HIV-1 replication from chronically infected cells.
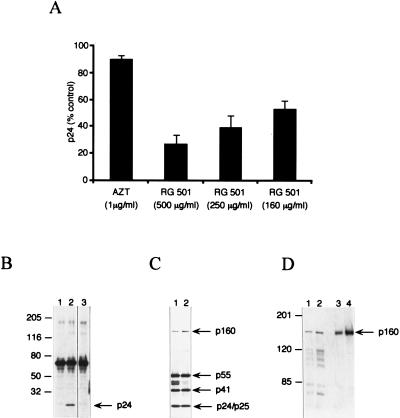
If the role of frameshifting is to control the precise Gag/Gag-Pol ratio required for particle assembly, then compounds that affect frameshifting should inhibit particle assembly and therefore block viral replication in cells chronically infected with HIV-1. CH-1 cells are COS cells stably transfected with HIV-gpt (2). This variant of HIV-1 has most of the ENV gene replaced by the selectable marker gpt, but all other viral genes are intact (22). Thus, CH-1 cells are chronically infected and produce normal viral particles, except that the particles are nonenveloped. CH-1 cells were incubated in the presence of compound for 3 days, at which point the medium was harvested and the number of viral particles was quantified with a p24 ELISA kit. During the formation of mature viral particles, the Gag and Gag-Pol polyproteins are cleaved by the viral protease into their individual components. Thus, the detection of p24 in medium indicates the presence of mature viral particles. RG501 significantly reduced the level of viral particles in the CH-1 cell medium (Fig. 6A) in a concentration-dependent manner and at concentrations that correspond to those having an effect on frameshifting in the COS-cell-based SEAP experiments (Fig. 4B). Zidovudine (AZT), on the other hand, had little effect on particle production from the CH-1 cells (Fig. 6A), even at a concentration >1,000-fold higher than its IC50 in acute infections. This was expected since AZT inhibits the activity of RT, which has no role in either viral replication or particle production from chronically infected cells.
FIG. 6.
Stimulation of frameshifting is detrimental to HIV-1 replication. (A) Increased frameshifting inhibits viral production from chronically infected cells. After CH-1 cells were incubated in the presence of RG501 for 72 h, the levels of viral particles were determined with a standard p24 ELISA (Cellular Products). In these experiments, neither RG501 nor AZT had any discernible cellular toxicity and each data point represents the average of at least four samples. (B) Immunoblot analysis of Gag p24 immunoreactive products present in viral pellets from chronically infected cells. Proteins (5 μl of viral pellet) were separated by electrophoresis in 4 to 12% gradient polyacrylamide–sodium dodecyl sulfate (SDS) gels and immunoblotted with a Gag p24 polyclonal antibody. Lanes: 1, untreated COS cells; 2, untreated CH-1 cells; 3, RG501 (500 μg/ml)-treated CH-1 cells. (C) Immunoblot analysis of human HIV-1 immunoglobulin-reactive products present in CH-1 cell lysates. After incubation in the presence of RG501, CH-1 cell lysates were prepared by using a Triton buffer. CH-1 cell lysates (5 μg of protein) from either untreated (lane 1) or RG501 (500 μg/ml)-treated (lane 2) cells were separated by electrophoresis in 4 to 12% gradient polyacrylamide–SDS gels and immunoblotted with human HIV-1 immunoglobulin. (D) Immunoblot analysis of RT immunoreactive products present in cell lysates. Lysates (5 μg of protein) prepared as for panel C were separated in 6% polyacrylamide–SDS gels and immunoblotted with a polyclonal antibody raised against RT. Samples represent lysates of untreated (lane 1) or RG501 (500 μg/ml)-treated (lane 2) CH-1 cells and of untreated (lane 3) or RG501 (500 μg/ml)-treated (lane 4) 12A2 cells. The numbers to the left of panels B and D represent the relative positions of molecular size markers (in kilodaltons), and the numbers to the right of panels B to D indicate the positions of viral proteins p55 (Gag), p160 (Gag-Pol), p50s (MA, CA, and NC with or without p1 spacer), p41 (MA, CA), and p24 (CA).
As an alternative method of examining the effect of RG501 on viral replication, medium samples were also subjected to immunoblot analysis with a Gag p24 polyclonal antibody. In the medium from untreated CH-1 cells, p24 was the only specific viral protein detected by the p24 antibody (Fig. 6B, lane 2). A 68-kDa protein was detected, but this was also detected in the COS cell control (Fig. 6B, lane 1). No p24 or p24-containing polyproteins were detected in the medium from RG501-treated CH-1 cells (Fig. 6B, lane 3), although upon significantly longer exposure, a faint p24 band did start to appear. These results show that RG501 inhibits HIV-1 particle formation, presumably by its ability to stimulate frameshifting.
There is some discrepancy between the results of the ELISA and those of the Western blot analysis: the ELISA predicts a fourfold decrease in the level of p24, whereas in the Western blot analysis the decrease in p24 appears more dramatic. This difference was observed consistently. Likewise, experiments with a protease inhibitor also showed such a discrepancy, so that concentrations of protease inhibitor that blocked p24 production completely according to Western blot analysis caused only an apparent five- to sixfold reduction in p24 according to the ELISA (data not shown). These discrepancies most likely reflect the differential sensitivities of the two detection systems, but they could be accounted for by contamination of the particle preparations with p24 degradation products that register in the ELISA but that are too small to be retained by the polyacrylamide gel.
Direct correlation between inhibiting HIV-1 replication and stimulating frameshifting.
The fact that RG501 blocked production of both mature and immature viral particles from the chronically infected cells eliminates the possibility that this compound affects RT, integrase, or protease. As yet, however, no evidence has been shown that RG501 affects frameshifting in virally infected cells; indeed, if RG501 blocked the activity of TAT or REV, a result similar to that shown in Fig. 4B would be expected. If the inhibition of particle production by RG501 were due to stimulation of frameshifting, then the intracellular levels of Gag-Pol p160 should be higher in RG501-treated cells than in untreated controls.
Immunoblots were used to analyze the levels of viral proteins in lysates prepared from CH-1 cells. In untreated cell lysates, human anti-HIV-1 immunoglobulin reacted with virus-specific proteins that corresponded to Gag p55, Gag-Pol p160, and cleavage products from the Gag polyprotein including p24, p25, and p41 (Fig. 6C, lane 1). Cytoplasmic proteolytic processing also resulted in the appearance of unusual intermediate cleavage products around 50 kDa (Fig. 6C, lane 1). In the RG501-treated cells, the human anti-HIV-1 immunoglobulin detected many viral proteins, in particular Gag p55 and Gag-Pol p160, indicating that normal viral gene expression was occurring (Fig. 6C, lane 2). The continued expression of Gag and Gag-Pol eliminates the possibility that RG501 was having any effect on either of the viral regulatory proteins, TAT or REV, since this would dramatically reduce the production of both Gag and Gag-Pol. There did appear to be a small increase in the amount of p160 in the RG501-treated cell lysates relative to the levels of p55 and p24 (Fig. 6C, lane 2), suggesting that, indeed, RG501 had specifically stimulated frameshifting and therefore the production of Gag-Pol p160. It should be noted that the intensity of the 50-kDa bands decreased in the RG501-treated cells. A possible explanation for this result is that these intermediate proteolytic products may be hypersensitive to the levels of protease. If frameshifting were stimulated by RG501, the levels of protease would increase, and as a result, increased intracellular proteolysis would reduce the amount of the more sensitive 50-kDa intermediates.
Since the human anti-HIV-1 immunoglobulin reacted only weakly with Gag-Pol p160, it was difficult to determine reliably the magnitude of the observed stimulation of frameshifting. To examine this question further, immunoblots were probed with an HIV-1 RT polyclonal antibody. In addition to the CH-1 cell lysates used previously (Fig. 6C), lysates were also prepared from a second cell line, 12A2 (3). 12A2 cells are equivalent to CH-1 cells except that the integrated HIV-gpt is protease deficient; therefore, in these cells, an accurate determination of the levels of Gag-Pol p160 is not compromised by proteolysis. Lysates prepared from both cell lines after treatment with RG501 showed a marked increase in the amount of Gag-Pol p160 compared to the untreated cell lysates (Fig. 6D). The increase in Gag-Pol p160 was especially pronounced in the 12A2 cells, where intracellular proteolytic processing is impaired (Fig. 6D, lanes 3 and 4). Quantification of these results indicated that RG501 treatment resulted in a 2.2-fold increase in the levels of Gag-Pol p160 in CH-1 cells and a 2.8-fold increase in the 12A2 cells. The RG501-treated CH-1 cell lysates showed an increase in the levels of all the RT-containing p160 proteolytic processing products, implying that the increased level of protease within Gag-Pol p160 resulted in an increase in cytoplasmic processing. As mentioned above, increased cytoplasmic processing could explain the decrease in the p50 bands detected by the anti-HIV-1 immunoglobulin (Fig. 6C, lane 2). In comparing the immunoreactive proteins from the same CH-1 cell lysates, it was apparent that RG501 had little effect on the levels of Gag p55 (Fig. 6C, lanes 1 and 2) yet there was an increase in the amount of Gag-Pol p160 (Fig. 6D, lanes 1 and 2). Thus, the inhibition of viral particle assembly and therefore viral replication by RG501 appears to be a direct result of changing the Gag/Gag-Pol ratio through stimulating frameshifting.
There are two conclusions to be drawn from the data presented in this report. First, we have shown that there is a direct link between the Gag/Gag-Pol ratio and HIV-1 particle assembly. Stimulating frameshifting resulted in only a modest effect on overall viral gene expression (a two- to threefold change in the Gag/Gag-Pol ratio) yet viral particle formation was inhibited. Two possible explanations for why increasing the levels of Gag-Pol would be detrimental to particle formation are as follows. (i) During particle formation, Gag-Pol represents the nucleation point for condensation of Gag around the viral genomic RNA, and when the amount of Gag-Pol increases, more particles start to form so Gag becomes limiting. (ii) The condensation of Gag and Gag-Pol around the viral genomic RNA is more random, and increased levels of Gag-Pol may inhibit particle formation because too many Gag-Pol molecules become associated with the viral RNA to be incorporated into the constrained structure of the HIV-1 particle. In each case, it would be expected that incomplete particles are formed; we are currently initiating studies to investigate this phenomenon. The second conclusion from these experiments is that frameshifting is a suitable target for HIV-1 drug discovery. Since frameshifting controls the precise ratio of Gag to Gag-Pol which is critical for viral particle assembly, a drug that affects frameshifting would effectively block viral replication. Although RG501 itself is not a suitable candidate for use as a therapeutic agent, if such an agent that acts against frameshifting is identified, it should complement the current portfolio of drugs available for combination therapy treatment of HIV-1-infected individuals.
ACKNOWLEDGMENTS
We are indebted to D. Boykin for kindly providing RG501 (DB213). We thank C. Craik and L. Babé for the generous gift of CH-1 and 12A2 cell lines and K. Li and W. D. Wilson for sharing results prior to publication. We also thank M. B. Mathews, C. M. Moehle, J. Harford, J. C. Watson, and G. W. Witherell for helpful discussions and criticisms of the manuscript. In addition, we thank K. Steimer and A. Prince for making antibodies available through the AIDS Research and Reference Reagent Program, Division of AIDS Program, NIAID, NIH.
This work was supported in part by a Small Business Innovative Research grant (AI36728) to S.R.G. from the National Institutes of Health.
REFERENCES
- 1.Atkins J F, Gesteland R F. Regulatory recoding. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 653–684. [Google Scholar]
- 2.Babé L M, Craik C S. The production of nonenveloped human immunodeficiency virus type 1 particles by a mammalian cell line and effects of a protease inhibitor on particle maturation. Antimicrob Agents Chemother. 1994;38:2430–2439. doi: 10.1128/aac.38.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babé L M, Rosé J, Craik C S. Trans-dominant inhibitory human immunodeficiency virus type 1 protease monomers prevent protease activation and virion maturation. Proc Natl Acad Sci USA. 1995;92:10069–10073. doi: 10.1073/pnas.92.22.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J M. Retroviridae and their replication. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. New York, N.Y: Raven Press Ltd.; 1990. pp. 1437–1500. [Google Scholar]
- 6.Dinman J D, Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Peptidyl-transferase inhibitors have antiviral properties by altered programmed-1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franchini G, Gurgo C, Guo H G, Gallo R C, Collalti E, Fargnoli K A, Hall L F, Wong-Staal F, Reitz M S. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 10.Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987;326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield D, Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue J-I, Watanabe T, Sato M, Oda A, Toyoshima K, Yoshida M, Seiki M. Nucleotide sequence of the protease-coding region in an infectious DNA of simian retrovirus (STLV) of the HTLV-1 family. Virology. 1986;150:187–195. doi: 10.1016/0042-6822(86)90278-3. [DOI] [PubMed] [Google Scholar]
- 13.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 14.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 15.Kang H, Hines J V, Tinoco I. Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J Mol Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- 16.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami K, Pande S, Faiola B, Moore D P, Boeke J D, Farabaugh P J, Strathern J N, Nakamura Y, Garfinkel D J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kollmus H, Hentze M W, Hauser H. Regulated ribosomal frameshifting by an RNA-protein interaction. RNA. 1996;2:316–323. [PMC free article] [PubMed] [Google Scholar]
- 19.Larder B A, Kellam P, Kemp S D. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature. 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 20.Li, K., et al. 1998. Unpublished data.
- 21.Millan J L. Molecular cloning and sequence analysis of human placental alkaline phosphatase. J Biol Chem. 1986;261:3112–3115. [PubMed] [Google Scholar]
- 22.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Morrow C D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter D C, Ansardi D C, Choi W S, Morrow C D. Encapsidation of genetically engineered poliovirus minireplicons which express human immunodeficiency virus type 1 Gag and Pol proteins upon infection. J Virol. 1993;67:3712–3719. doi: 10.1128/jvi.67.7.3712-3719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prince A M, Horowitz B, Baker L, Shulman R W, Ralph H, Valinsky J, Cundell A, Brotman B, Boehle W, Rey F, Piet M, Reesink H, Lelie N, Tersmette M, Miedema F, Barbosa L, Nemo G, Nastala C L, Allan J S, Lee D R, Eichberg J W. Failure of human immunodeficiency virus (HIV) immune globulin to protect chimpanzees against experimental challenge with HIV. Proc Natl Acad Sci USA. 1988;85:6944–6948. doi: 10.1073/pnas.85.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prince A M, Reesink H, Pascual D, Horowitz B, Hewlett I, Murthy K K, Cobb K K, Eichberg J W. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res Hum Retroviruses. 1991;7:971–973. doi: 10.1089/aid.1991.7.971. [DOI] [PubMed] [Google Scholar]
- 27.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 28.Somogyi P, Jenner J A, Brierley I, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P, Barr P J, Hallewell R A. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p24 gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 30.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 31.Weislow O S, Kiser R, Fine D L, Bader J, Shoemaker R H, Boyd M R. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J Natl Cancer Inst. 1989;81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]
- 32.Weiss R, Teich N, Varmus H, Coffin J. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. [Google Scholar]
- 33.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Xu H, Boeke J D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci USA. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]