Abstract
We have studied the phenotypic impact of adaptative Gag cleavage site mutations in patient-derived human immunodeficiency virus type 1 (HIV-1) variants having developed resistance to the protease inhibitor ritonavir or saquinavir. We found that Gag mutations occurred in a minority of resistant viruses, regardless of the duration of the treatment and of the protease mutation profile. Gag mutations exerted only a partial corrective effect on resistance-associated loss of viral fitness. Reconstructed viruses with resistant proteases displayed multiple Gag cleavage defects, and in spite of Gag adaptation, several of these defects remained, explaining the limited corrective effect of cleavage site mutations on fitness. Our data provide clear evidence of the interplay between resistance and fitness in HIV-1 evolution in patients treated with protease inhibitors.
Infectivity of newly assembled retroviral particles is dependent upon a late maturation event associated with processing of the Gag and Gag-Pol polyprotein precursors by a Gag-Pol-encoded aspartic protease (PR) (7, 19, 22, 24, 37). As a consequence, occupation of the catalytic site of the PR by synthetic peptidomimetic molecules can profoundly inhibit viral replication (2, 11, 27). Such inhibitors are now widely used in the treatment of patients infected with human immunodeficiency virus type 1 (HIV-1), leading to durable near extinction of virus replication and significant improvement of the prognosis of the infection, especially when administered in association with other antiretroviral drugs (15, 25, 26). However, viral resistance to PR inhibitors can emerge in the course of the treatment, leading to a gradual viral escape replication (8, 9, 13, 29, 34). HIV-1 resistance to PR inhibitors is the consequence of stepwise accumulation of mutations in the viral PR (8, 29). Many mutations associated with HIV-1 resistance to PR inhibitors have now been identified (28). Although they can be sporadically witnessed in viral quasispecies, most of these mutations are not found in PR inhibitor-naive HIV-1 infected patients, suggesting that they confer a selective disadvantage to the virus (3, 20, 30, 38). Indeed, some resistance mutations, alone or in combination, can significantly reduce the catalytic activity of the protease and consequently affect the replicative capacity of the virus (4, 16–18, 31, 35). This phenomenon, which can be observed both in viruses selected for resistance in tissue culture (4, 10, 17, 18) and in treated patients (39, 40), may have crucial clinical implications. It was recently observed that in some resistant viruses, obtained by in vitro selection or from treated patients, resistance-associated loss of viral fitness can be significantly corrected by the emergence of compensatory mutations in two C-terminal Gag cleavage sites (12, 40).
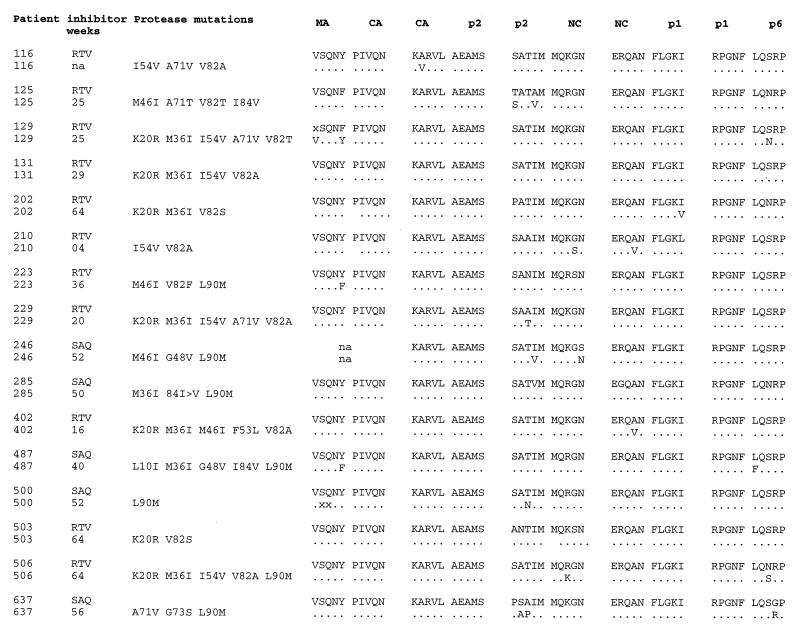
In the present study we wished to further investigate the conditions that favor the emergence of Gag cleavage site mutations and to evaluate the phenotypic impact of these mutations on HIV-1 fitness and resistance to PR inhibitors. We studied gag sequences from 16 patients treated with PR inhibitors, presenting different profiles of mutations in the PR (Fig. 1). The selected samples were obtained from patients treated for various periods with ritonavir (RTV) or saquinavir (SQV) (11 and 5 patients, respectively) (Fig. 1). Patients 116, 125, 129, 131, 210, 223, and 229 were treated with RTV monotherapy; patients 246, 285, 487, 500, and 637 were treated with SQV monotherapy or SQV in combination with dideoxycytosine; and patients 202, 402, 503, and 506 received a combination therapy regimen that included RTV, zidovudine, and dideoxycytosine. The PR sequences of patients 202, 246, 402, 503, and 506 were determined from bulk PCR products amplified from plasma virus as previously described (39). We compared the samples obtained shortly before the initiation of PR inhibitor therapy with those obtained after the development of resistance.
FIG. 1.
Sequences of Gag cleavage sites and PRs from studied patients. RTV- and SQV (SAQ)-treated patients were selected to cover a variety of PR mutation profiles and durations of therapy. The amino acid substitutions involved in PR inhibitor resistance that were found between the pretherapy and resistant virus isolates are shown. Gag cleavage site sequences for the pretherapy viruses and amino acid substitutions found in postresistance isolates are reported. A dot indicates that the resistant isolate carried the same residue as the pretherapy one; an x indicates a position at which the nature of the residue is not defined, possibly because multiple isolates were amplified by PCR. na, not available.
To determine the Gag cleavage site sequences of all the patients’ samples, viral RNA was purified from plasma or supernatant of a single-passage peripheral blood lymphocyte infection (patients 116, 125, 129, 131, 210, 223, and 229), reverse transcribed, and amplified by nested PCR, and bulk PCR products were sequenced. The primer pairs GagB+ (5′CTTGCTGAAGCGCGCACGGCAAGAGG3′) and GagCS (5′TCTTGTGGGGTGGCTCCTTCTG3′) and the primer pair 17/24+ (5′AACATATAGTATGGGCAAGC3′) and 17/24− (5′TACCCATGCATTTAAAGTTC3′) were used for the matrix/capsid (MA/CA) cleavage site. For all other Gag cleavage sites we used the following primers: Cliv1 (5′GACAGAAACCTTGTTGGTCC3′), Cliv2 (5′CGCTGCCAAAGAGTGATCT3′), ClivN1 (5′TGGTCCAAAATGCGAACC3′), and ClivN2 (5′AAAGAGTGATCTGAGGGAAG3′). As expected the p2/nucleocapsid (p2/NC) cleavage site displayed the highest level of intrapatient variability (Fig. 1), but the observed changes involved residues which are variable also in PR inhibitor-naive patients (3, 20, 23, 30). Two RTV-resistant viruses, viruses 210 and 402 (patient numbers are also used as virus numbers in this work), presented an A-to-V mutation at position P2 of the NC/p1 cleavage site. SQV-resistant virus 487 displayed a cleavage site mutation located at the P1′ position of the p1/p6 cleavage site (L to F) in addition to the MA/CA substitution. Both of these Gag cleavage site mutations were reported in viruses that developed resistance to PR inhibitors in vitro or in vivo (12, 40). One study suggested that the development of Gag cleavage site mutations is associated with heavily mutated PRs (“dead end”) for which the concomitant evolution of additional mutations in the PR and in the Gag substrate may be the only way for the virus to survive in an increasingly selective environment (12). More recently, the analysis of resistant viral isolates from indinavir-treated patients indicated Gag adaptation as a common evolutionary pathway (six out six patients), taking place as early as 6 weeks after the start of therapy and in the presence of as few as two PR mutations (40). While some of the mutations that we observed in the present study are identical to the previously described ones, we did not find common correlates for the emergence of Gag cleavage site mutations in terms of their association with particular PR mutations or duration of treatment (Fig. 1). Interestingly, we observed for the first time substitutions in the MA/CA (patients 223, 487, and 129) and CA/p2 cleavage sites (patient 116) and a K-to-N substitution at position 38 of the NC protein in two resistant viruses (from patients 223 and 503 [data not shown]), suggesting that there are additional possibilities for Gag adaptation besides the previously described substitutions in the cleavage sites surrounding the p1 peptide (12, 40).
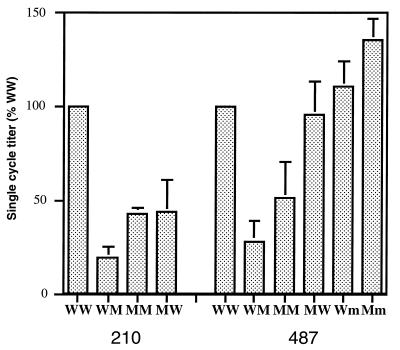
We have recently reported that reconstructed HIV-1 molecular clones carrying inhibitor-resistant proteases displayed a reduction in replicative capacity with respect to clones carrying the corresponding parental pretherapy PRs (39). To study the effect of the observed Gag cleavage site mutations on viral infectivity, we constructed viral clones with the four possible combinations of pretherapy and postresistance gag and PR sequences from one RTV- and one SQV-treated patient (patients 210 and 487, respectively), using an infectious molecular clone of HIV-1 (1, 32). To this end, the gag gene was reverse transcribed and PCR amplified with the primer pair GagA+ (5′CCAGAGGAGATCTCTCGACGC3′) and ClivN2 (see above) and the primer pair GagB+ (see above) and GagB− (5′TTCCTTGTCTAGAGGCTCCTGCTTC3′). In this set- ting, the pretherapy Gag precursor molecule was associated with the pretherapy Protease (wild-type clones [WW]) and, independently, with the mutated PR allele (clones WM). Similarly, the mutated Gag precursor molecule was associated with the PRs obtained before (clones MW) and after (clones MM) the development of resistance. The entire gag genes from the patients isolates were cloned to take into account the influence of distal residues on the overall conformation of Gag precursor. Infectious supernatants obtained from transfected HeLa cells were normalized by measurement of HIV-1 p24 antigen and used to infect P4 indicator cells as reported previously (5, 14, 39). The infectivity of each Gag-PR combination was expressed as a percentage of the corresponding pretherapy (WW) clone (Fig. 2). For the Gag-PR combinations from patient 210 (RTV treated), the association of the resistant PR and the pretherapy Gag (clone 210WM) resulted in a fivefold reduction in infectivity with respect to the pretherapy combination (Fig. 2, compare 210WM to 210WW). A significant but partial rescue was observed upon expression of the adapted Gag precursor with the resistant PR (clone 210MM) (Fig. 2); in this case the reduction in infectivity was only 2.5-fold. A similar trend was observed for the 487-derived virus (from an SQV-treated patient), for which the reduction in infectivity due to the resistant PR was about fourfold (Fig. 2, compare 487WW and 487WM), and the partial rescue attributable to Gag adaptation reduced the defect to twofold (487MM). Unexpectedly, the expression of the Gag cleavage site mutation of virus 210 in the presence of the pretherapy protease (210MW) resulted in a marked reduction of infectivity (Fig. 2). The same Gag-PR combination for virus 487 was as infectious as its pretherapy counterpart (compare 487MW and 487WW).
FIG. 2.
Single-cycle infectivity of reconstructed viruses. The single-cycle titer of each reconstructed virus was measured by titration on P4 (HeLa-CD4 LTR-LacZ) cells and expressed as a percentage of the titer of the pretherapy virus (WW) from the same patient. The results presented are the means and the standard deviations (error bars) of at least three independent transfection and infection experiments.
As shown in Fig. 1, the resistant virus from patient 487 acquired the typical SQV resistance mutation G to V at position 48 (G48V) in combination with other substitutions. The passage of this virus in peripheral blood lymphocyte cultures resulted in the rapid reversion of this substitution (33a), suggesting that it confers a strong disadvantage to the virus in the absence of SQV. To evaluate the extent of this phenomenon, we measured the infectivities of two reconstructed viral clones carrying pretherapy and postresistance gag alleles upstream of the mutated PR lacking the G48V mutation. These two clones (487Wm, and 487Mm) are identical to the clones 487WM and 487MM, respectively, except for the nature of the residue in position 48. Strikingly, their infectivities were not reduced with respect to their pretherapy (487WW) counterparts (Fig. 2). The mutation G48V is thus responsible for the dramatic decrease in infectivity observed for the 487WM virus.
To evaluate the selective advantage in terms of resistance that determined the evolution of such a detrimental mutation, we calculated the 50% inhibitory concentration (IC50) and IC90 of SQV (concentrations that inhibit 50 and 90 percent of infectious events, respectively) for the reconstructed 487 viral clones (6, 39). We found typical PR inhibitor-naive levels of resistance for the clones expressing the pretherapy PR alleles 487WW and 487MW (Table 1). The accumulation of five mutations in the PR (clone 487WM) caused a remarkable increase in the level of resistance (1.5 and more than 3 μM for IC50 and IC90, respectively), which was not significantly altered by the association of the mutated PR with the adapted Gag in clone 487MM (Table 1). Interestingly, the 487Wm and 487Mm clones, expressing the mutated PR lacking the G48V mutation, displayed significantly lower levels of resistance (Table 1). It is likely that a strong selective pressure due to SQV treatment in patient 487 favored the fixation of the G48V mutation for its effect on SQV resistance, despite the marked reduction of viral replication associated with such substitution.
TABLE 1.
Inhibitor resistance of 487-derived virusesa
| Virus clone | IC50 (SD) (nM) | IC90 (SD) (nM) |
|---|---|---|
| 487WW | 2.4 (1.5) | 9 (6.6) |
| 487WM | 1,502 (487) | >3,125 |
| 487MM | 1,933 (322) | >3,125 |
| 487MW | 6.7 (2.4) | 23 (12) |
| 487Wm | 99 (59) | 448 (144) |
| 487Mm | 302 (195) | 1,041 (359) |
SQV IC50s and IC90s were measured in a single-cycle resistance assay on P4 cells.
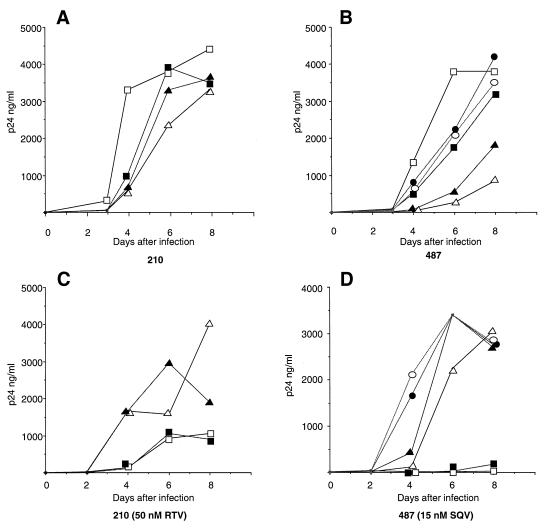
To validate the effect of Gag substitutions in a virus propagation assay, T-lymphoblastoid MT4 cells (106 cells in 5 ml) were infected with infectious supernatant from transfected HeLa cells (corresponding to 30 ng of HIV p24). The cultures were maintained for 8 days, aliquots of supernatant (50 μl) were collected every 2 days, and their p24 content was analyzed. For both 210- and 487-derived viruses (Fig. 3A and B, respectively), the pretherapy (WW) virus was characterized by the fastest growth kinetics, indicating that in the absence of treatment this combination results in the fittest virus. Interestingly, for both viral series the coexpression of the mutated PR with pretherapy Gag (clones WM) resulted in the slowest-growing virus, in complete agreement with the single-cycle infectivity data. The rescue of viral infectivity due to Gag adaptation (clone MM) was reproducible for both viruses but never complete, confirming the effect observed in the single-cycle assay. The Gag-adapted but pretherapy protease (MW) viruses displayed growth kinetics slower than those of the respective pretherapy viruses (WW) and faster than those of the WM viruses. However, for the isolates from patient 487 the MW virus grew reproducibly faster than the MM virus (Fig. 3B), while for isolates from patient 210 the viruses MW and MM had similar kinetics (Fig. 3A), reflecting the reduction in infectivity observed in the single-cycle assay for virus 210MW. The 487Wm and 487Mm clones, lacking the G48V mutations, grew with kinetics reproducibly slower than those of the pretherapy virus 487WW but clearly faster than those of the PR-mutated viruses (487WM and 487MM). This observation confirms the replicative impairment due to the G48V mutation. The reduced growth kinetics reported in Fig. 3 contrast with the absence of titer difference observed in the single-cycle assay and may relate to differences in assay sensitivities to replicative defects involving late stages of the viral life cycle.
FIG. 3.
Growth kinetics of the reconstructed viruses in lymphoid cells. MT4 cells were infected with p24-normalized amounts of particles from the reconstructed 210- and 487-derived viruses in the absence (A and B) and in the presence (C and D) of PR inhibitor. Virus production was monitored by measuring HIV-1 p24 antigen concentration in the culture supernatant. For all panels, open squares correspond to pretherapy WW viruses, open triangles correspond to PR-mutated WM viruses, filled triangles correspond to PR-mutated and Gag-adapted MM viruses, and filled squares correspond to Gag-adapted but wild-type PR MW viruses. (B and D) Circles correspond to the reconstructed viruses lacking the G48V protease mutation; open circles correspond to the 487Wm clone, and filled circles correspond to the 487Mm clone. The data presented are from one of two independent experiments, in which comparable results were obtained.
The development of mutated PR alleles and the coevolution of Gag cleavage sites took place under the selective pressure exerted by PR inhibitors in the treated patients. Therefore, we could expect that the selective advantage conferred by PR mutations and by Gag adaptation would be revealed best in the presence of the specific PR inhibitor. In the presence of 50 nM RTV, the 210-derived viruses carrying the naive PR (210WW and 210MW) displayed a reduced viral production and delayed growth kinetics compared to the resistant viruses (210WM and 210MM) (Fig. 3C). Under these conditions the 210MM virus displayed a reproducible replicative advantage toward 210WM, indicating that the specific Gag adaptation, which plays a significant role in improving viral fitness in the absence of drug (see above) confers a sensible advantage also in terms of viral growth in the presence of RTV. A conceptually identical situation was observed when the 487-derived clones were mutually compared. In this case, however, in the presence of 15 nM SQV, the growth of inhibitor-naive viruses (WW and MW) was hardly detectable, while accumulation of p24 antigen in the culture supernatant was readily detected for all viruses with a mutated PR (Fig. 3D). Again, the Gag-adapted virus 487MM had a clear selective advantage with respect to 487WM. Remarkably, the viruses lacking the G48V mutation (487Wm and 487Mm) grew reproducibly faster than any other 487-derived virus (Fig. 3D), indicating that under these conditions the positive effect on replication due to the absence of the G48V substitution is more important than the impact of this mutation on resistance. In the presence of such a modest concentration of SQV, the increase in resistance associated with the G48V mutation does not confer a selective advantage to the virus.
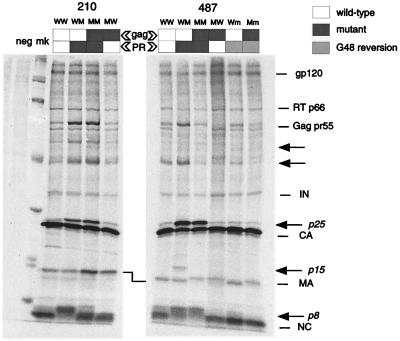
To characterize from a molecular perspective the replicative defect associated with the resistant PRs and the partial rescue consequent to Gag adaptation, we analyzed the particle protein content of the reconstructed viruses. HeLa cells were transfected with the 210- and 487-derived clones; metabolically labelled viral particles released in the culture supernatant were partially purified by ultracentrifugation on a sucrose cushion, lysed, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as previously described (39). Particle-associated material from three independent experiments were compared, with a representative experiment being shown in Fig. 4. Even in the absence of immunoprecipitation, the major Gag cleavage products, as well as some of the Pol products and the mature gp120, were readily detected (Fig. 4). Remarkably, the expression of mutated PRs (lanes WM and MM) was always associated with the presence of several cleavage intermediates, absent in the corresponding pretherapy PRs (lanes WW). Among the incompletely processed precursors, we could identify the full-length Gag precursor (Gag pr55); a p25 protein, which corresponds to the CA p24 protein still bound to the p2 spacer peptide and could be immunoprecipitated by a p24 monoclonal antibody (data not shown); the p8 intermediate, which represents the NC p7 protein plus the p1 peptide; and a previously described p15 band, corresponding to the NC/p1/p6 cleavage intermediate (12, 33, 36). Interestingly, we observed that Gag adaptation determined a clear correction of some of the incomplete cleavage events. As shown in Fig. 4, the presence of the 210 mutated PR caused a complete shift of the mature NC protein band from 7 to 8 kDa (compare lanes 210WW and 210WM). In the presence of the NC/p1 cleavage site adaptation, the NC protein band regained the pretherapy localization (compare lanes 210MM and 210WW), while only minor amounts of intermediate p8 band remained visible. The detection of several other intermediate precursor bands, in particular p25 and the full-length Gag pr55, consequent to impaired cleavage by the mutated protease (lane 210WM) was not altered by Gag adaptation (lane 210MM), indicating that the impact of Gag adaptation was limited to processing of NC. Correspondingly, resistance-associated loss of viral fitness in virus 210 was only partially corrected by Gag adaptation.
FIG. 4.
Protein maturation profile of reconstructed viruses. HeLa cells were transfected with reconstructed viral clones carrying the indicated Gag-PR combinations. Metabolically labelled viral particles released in the supernatant were purified on a sucrose cushion, lysed, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Mature Gag proteins—MA, capsid, and NC—as well as mature Pol proteins—integrase (IN) and reverse transcriptase (RT)—are indicated, together with the full-length Gag precursor (Gag pr55) and the mature surface glycoprotein gp120. Identified cleavage intermediates are indicated in italics. Arrows point to additional partially cleaved precursors associated with mutated PR processing. Lane neg corresponds to mock transfected cells; lane mk contains the molecular weight marker proteins (with molecular masses of 220, 97, 66, 46, 30, 21, and 14 kDa).
In the 487 series the cleavage site adaptations involved the MA/CA and p1/p6 sites. The mature protein p6 does not contain methionine or cysteine residues, but it could be seen as part of the previously described cleavage intermediate p15 when the pretherapy Gag precursor was inefficiently cleaved by the resistant PR (lane 487WM). As a consequence of Gag adaptation, an improved proteolysis at the p6/p1 site determined the disappearance of the cleavage intermediate p15 (compare lanes 487WM and 487MM). In contrast with what we reported for the 210-derived virus, Gag adaptation in 487-derived virus corrected also the accumulation of the full-length Gag precursor (compare lanes 487WM and 487MM). Both changes in the p1/p6 and the MA/CA cleavage sites of virus 487MM could be responsible for this phenomenon. Another clear processing defect associated with resistant PR in virus 487WM concerns the CA/p2 site, translating into a high amount of particle-associated cleavage intermediate p25. Since no change in the CA/p2 cleavage site sequence emerged along with resistance, no cleavage improvement at this site was produced (compare lanes 487WM and 487MM). Similar to virus 210WM, cleavage of pretherapy Gag by the mutated PR in virus 487WM resulted in the appearance of the p8 intermediate band among others, but in this case a discrete amount of completely processed NC protein could still be distinguished below it. Thus, the relative impairment at the NC/p1 site seems less dramatic for virus 487 than for virus 210. As expected, since the NC/p1 site is not involved in Gag adaptation in the 487-derived virus, no improvement of NC protein maturation was observed as a consequence of Gag cleavage site mutation (compare lanes 487WM and 487MM). Again, incomplete correction of Gag processing impairment must account for the merely partial improvement in viral fitness conferred by Gag adaptation also in the 487MM virus.
The probability for selection of adaptative substitutions in HIV-1 Gag cleavage sites as a response to a functionally impaired PR should be a function of several parameters. First, it should be affected by the extent of the cleavage defect at each of the affected sites. This is illustrated by the occurrence of cleavage site adaptation at the NC/p1 site in virus 210, where the cleavage was almost completely abolished. In this respect, it is interesting that the most extensive cleavage defects observed here and the most frequently described Gag corrections involve the p1 spacer peptide. Second, the selection of adaptative Gag mutations should be conditioned by the potential impact of individual cleavage defects on virus infectivity. Indeed, we surmise that the impairment of cleavage at some particular sites may have a more radical effect than at other sites. Third, if cleavage is impaired at a single site, selection of a corresponding adaptive mutation will be more likely than when, as shown here, PR resistance results in a marked reduction of Gag cleavage at several sites. In this case adaptation should still occur at those sites for which cleavage impairment is more rate limiting. In addition, while substitutions involving a limited number of cleavage sites with slow cleavage kinetics may be tolerable, this may not be the case for corrections involving multiple sites. In particular, there appears to be a delicate balance in the efficiency of cleavage by the PR at the different sites in HIV-1 Gag and Gag-Pol, resulting in a timely coordination of processing and assembly of the mature virion proteins (21, 33). Therefore, it is possible that too many constraints are opposed to changes at multiple Gag sites for full rescue of resistance-associated loss of viral fitness. This question, which may have a considerable impact on the clinical outcome of resistance to PR inhibitors in treated patients, will require careful long-term genotypic and phenotypic analysis of HIV-1 evolution in the course of antiretroviral treatments.
Acknowledgments
We thank Esther Race and Ian Duncan (Roche, Welwyn Garden City, United Kingdom), Akhter Molla and John Leonard (Abbott Laboratories, Chicago, Ill.), Dominique Mathez and Jacques Leibowitch (Unité d’Immuno-virologie, Hôpital Raymond Poincaré, Garches, France), and Françoise Brun-Vézinet and Diane Descamps (Laboratoire de Virologie, Hôpital Bichat-Claude Bernard, Paris, France) for virus samples and for some of the PR sequences; Sylvie Paulous and Laurent Guillemot for technical assistance; and Luc Montagnier for his support.
This work was supported in part by the Agence Française de Recherches sur le Sida (ANRS). F.M. is the recipient of a fellowship from ANRS.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashorn P, McQuade T J, Thaisrivongs S, Tomasselli A G, Tarpley W G, Moss B. An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc Natl Acad Sci USA. 1990;87:7472–7476. doi: 10.1073/pnas.87.19.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within Gag/Pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 4.Borman A, Paulous S, Clavel F. Resistance of HIV-1 to protease inhibitors: selection of resistance mutations in the presence and in the absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 5.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription: a termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 6.Chou T C, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M. Retroviridae and their replication. In: Fields B, et al., editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1991. pp. 645–708. [Google Scholar]
- 8.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Hobbins H L, Roth E, Shivaprakash M, Titus D L, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 10.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retroviruses. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 12.Doyon L, Poulin F, Pilote L, Clouette C, Thibeault D, Croteau G, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson J W. The not-so-great escape. Nat Struct Biol. 1995;2:523–529. doi: 10.1038/nsb0795-523. [DOI] [PubMed] [Google Scholar]
- 14.Eustice D, Feldman P, Colberg-Poley A, Buckery R, Neubauer R. A sensitive method for the detection of β-galactosidase in transfected mammalian cells. BioTechniques. 1991;11:739–743. [PubMed] [Google Scholar]
- 15.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 16.Gulnick S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 17.Ho D, Toyoshima T, Mo H, Kempf D, Norbeck D, Chen C, Wideburg N, Burt S, Erickson J, Singh M. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan A, Michael S, Wehbie R, Knigge M, Paul D, Everitt L, Kempf D, Norbeck D, Erickson J, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz R, Skalka A. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 20.Kozal M, Shah N, Shen N, Yang R, Fucini R, Merigan T, Richman D, Morris D, Hubbell E, Chee M, Gingeras T. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 21.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus type 1 capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kräusslich H-G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 23.Lech W, Wang G, Yang Y, Chee Y, Dorman K, McCrae D, Lazzeroni L, Erickson J, Sinsheimer J, Kaplan A. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb D D, Hutchison III C A, Edgell M H, Farmerie W G, Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J Virol. 1989;63:111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz M, Cao Y, Hurley A, et al. Abstracts of the XI International Conference on AIDS, Vancouver, British Columbia, Canada. 1996. Triple therapy with AZT, 3TC and Ritonavir in 12 subjects newly infected with HIV-1, abstr. Th. B. 933. [Google Scholar]
- 26.Mathez D, de Truchis P, Gorin I, Katlama C, Pialoux G, Saimot A G, Tubiana R, Chauvin J P, Bagnarelli P, Clementi M, Leibowitch J. Third Conference on Retroviruses and Opportunistic Infections, Washington D.C. 1996. Ritonavir, AZT, ddC, as a triple combination in AIDS patients, abstr. 285. [Google Scholar]
- 27.McQuade T J, Tomasselli A G, Liu L, Karacostas V, Moss B, Sawyer T K, Heinrikson R L, Tarpley W G. A synthetic HIV-1 protease inhibitor with antiviral activity arrests HIV-like particle maturation. Science. 1990;247:454–456. doi: 10.1126/science.2405486. [DOI] [PubMed] [Google Scholar]
- 28.Mellors J W, Larder B A, Schinazi R F. Mutations in HIV-1 reverse transcriptase and protease associated with drug resistance. Int Antiviral News. 1995;3:8–13. [Google Scholar]
- 29.Molla A, Kempf D, Korneyeva M, Gao Q, Shipper P, Mo H, Markowitz M, Vasavanonda S, Chernyavskyi T, Niu P, Lyons N, Hsu A, Granneman G, Ho D, Boucher C, Leonard J, Norbeck D. Ordered accumulation of mutations in HIV protease confers resistance to Ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 30.Myers G, Wain-Hobson S, Henderson L, Korber B, Jeang K, Pavlakis G. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 31.Nijhuis M, Schuurman R, Schipper P, de Jong D, van Bommel T, de Groot T, Molla A, Borleffs J, Danner S, Boucher C. Abstracts of the International Discussion Meeting on HIV Population Dynamics, Variation and Drug Resistance, Edinburgh, United Kingdom. 1997. Reduced replication potential of HIV-1 variants initially selected under Ritonavir therapy is restored upon selection of additional substitutions. [Google Scholar]
- 32.Patick A, Rose R, Greytok J, Bechtold C, Hermsmeier M, Chen P, Barrish J, Zahler R, Colonno R, Lin P. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettit S, Moody M, Wehbie R, Kaplan A, Nantermet P, Klein C, Swanstrom R. The p2 domain of HIV-1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Race, E. Personal communication.
- 34.Roberts N A. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS. 1995;9:S27–S32. [PubMed] [Google Scholar]
- 35.Rose R, Gong Y, Greytok J, Bechtold C, Terry B, Robinson B, Alam M, Colonno R, Lin P. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trich R, Cheng Y S E, Yin F H, Erickson-Vitannen S. Mutagenesis of protease cleavage sites in HIV-1 gag polyprotein. J Virol. 1991;65:922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogt V. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 38.Winslow D L, Stack S, King R, Scarnati H, Bincsik A, Otto M J. Limited sequence diversity in the HIV type 1 protease gene from clinical isolates and in vivo susceptibility to HIV protease inhibitors. AIDS Res Hum Retroviruses. 1995;11:107–113. doi: 10.1089/aid.1995.11.107. [DOI] [PubMed] [Google Scholar]
- 39.Zennou, V., F. Mammano, S. Paulous, D. Mathez and F. Clavel. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300–3306. [DOI] [PMC free article] [PubMed]
- 40.Zhang Y, Imamichi H, Imamichi T, Lane H, Falloon J, Vasudevachari M, Salzman N. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]