Abstract
Purpose
IKAROS, encoded by IKZF1, is a member of the IKAROS family of zinc-finger transcription factors playing critical roles in lymphocyte development, differentiation, and tumor suppression. Several studies demonstrated that IKZF1 mutations affecting DNA binding or homo-/hetero-dimerization are mostly associated with common variable immunodeficiency, combined immunodeficiency, or hematologic manifestations. Herein we report a likely de novo, nonsense IKZF1 mutation (p.C182*) in a baby with low T cell receptor excision circles (TREC) identified by newborn screening testing for severe combined immunodeficiency. The patient also presented a profound B cell deficiency at birth.
Methods
Genetic, functional, immunologic, and clinical outcome data associated with this patient and her mutation were evaluated.
Results
Mutant p.C182* was detected in the cytoplasm of the patient’s primary cells, in contrast to wild type (WT) IKAROS protein, only detected in the nucleus. Functional in vitro assessments revealed that p.C182* was less stable than WT IKAROS protein and failed to bind to its target DNA binding sequence and dimerize with WT IKAROS protein, resulting in impaired pericentromeric targeting and transcriptional repression by means of haploinsufficiency. During follow-up, while a spontaneous recovery of TREC and T cells was observed, B cells improved but not to sustained normal ranges.
Conclusions
Patients with IKAROS-associated diseases can present with SCID-like TREC values through newborn screening testing. IKZF1 mutations should be added to the low TREC differential, although spontaneous recovery has to be considered.
Keywords: TREC, T cells, B cells, lymphopenia, CVID, CID, NBS
Introduction
Much is understood about the complex biology of the transcription factor IKAROS/IKZF1, but its story continues to unfold. IKAROS is part of a family of zinc-finger proteins (IKZF1–IKZF5) that bind DNA through four N-terminal zinc-finger domains and homo- or heterodimerize through two C-terminal zinc-fingers [1]. IKAROS can act both as an activator and as a repressor of transcription, exerting its function by targeting DNA sequences at pericentromeric heterochromatin (PC-HC) regions of particular genes and regulating the nucleosome remodeling and histone deacetylase complex [2]. In humans, both somatic and germline IKZF1 mutations have been implicated in distinctive, although related disease presentations: Somatic changes are associated with B cell precursor acute lymphoblastic leukemia (B-ALL) in children and adults [3]; germline heterozygous mutations contribute mostly to common variable immune deficiency (CVID) when acting by haploinsufficiency [4], to a severe form of combined immune deficiency (CID) with susceptibility to pneumocystis pneumonia by a dominant negative effect [5], and mostly to hematologic cytopenia/Evans syndrome and malignancies through dimerization defects [6]. Here, we describe and functionally characterize a novel IKAROS mutant identified in a baby presenting with low T cell receptor excision circles (TREC) through severe combined immunodeficiency (SCID) newborn screening.
Methods
Patients and Samples
All patients or their guardians provided informed consent in accordance with the Declaration of Helsinki under institutional review board–approved protocols of the National Institute of Allergy and Infectious Diseases, NIH, and Western IRB (Pr #:20171726); ClinicalTrials.gov Identifier: NCT03385876. Blood from healthy donors and patients were obtained under approved protocols. All procedures were based on standard of care and established clinical guidelines were followed.
Whole Genome Sequencing (WGS) and Sanger Validation
Consent for rapid WGS was obtained for the proband and her parents. Following DNA extraction from whole blood, samples were processed and analyzed according to the ACMG/AMP guidelines for the interpretation of sequence variants as described elsewhere [7, 8]. IKZF1 mutations were Sanger validated from gDNA using specific primers and standard techniques (primers sequence available upon request).
cDNA Sequencing
Total RNA was isolated from total PBMCs using the Qiagen RNeasy Plus mini kit and reverse transcribed using the QuantiTech Reverse Transcription Kit (Qiagen). cDNA was PCR-amplified and sequenced using IKZF1 primers, forward: 5′-CAAATCCACATAACCTGAGGAC-3′; forward: 5′-TTCCAGTGCAATCAGTGCGGGGCCT-3′; and reverse: 5′-CTGGGTACAGTGTGC-3′. To quantify the ratio of IKZF1 WT and mutant transcripts, the cDNA was first PCR-amplified for 30 cycles using primers (forward: 5′-CAAA TCCACATAACCTGAGGAC-3′, reverse: 5′-CTGG GTACAGTGTGC-3′), and an aliquot of this reaction was further amplified for 30 cycles using primers (forward: 5′-TTCCAGTGCAATCAGTGCGGGGCCT-3′; reverse: 5′-CTGGGTACAGTGTGC-3′). PCR products were cloned into pGEM-TA Easy Vector by TA cloning (Promega, A1360) according to the manufacturer’s instruction. Following bacterial transformation, 27 colonies were sequenced for IKZF1 using primers spanning the mutated area.
Western Blotting and Immunoprecipitation
To generate T cell blasts, total PBMCs were stimulated with anti-CD3 and anti-CD28 (1 μg/ml) in the presence of IL-2 (10 ng/ml) for 7 days. IL-2 was added every 2–3 days. Nuclear and cytoplasmic extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). For the immunoprecipitation, 293 T cells were transfected with HA tagged IKAROS WT or mutant together with flag-tagged IKAROS WT. Twenty-four hours after transfection, cell lysates were prepared in IP buffer (50-mM Tris pH 7.4, 150-mM NaCl, 2-mM EDTA, 0.5% Triton X-100 protease, and phosphatase inhibitor cocktail [Sigma]), and protein lysates were incubated with a rabbit anti-FLAG antibody and 40 μl protein A/G agarose beads (Pierce) for 4 h at 4 °C. Beads were washed three times with IP buffer; samples were prepared and separated on NuPAGE® Novex® 4–12% Bis-Tris Protein Gels (Life Technology). The following antibodies were used in the study: anti-IKAROS (Abcam; 229275); anti-HA antibody (Biolegend; 901501); anti-flag antibody (cell signaling; 14793); Lamin A/C (cell signaling; 4777); and GAPDH (cell signaling; 2118).
Fluorescence Microscopy
Immunofluorescence was performed essentially as described previously [4–6]. Briefly, NIH3T3 cells were transfected with pcDNA3-HA WT or mutant IKAROS with or without pCMV6-AC- MYC-DDK-WT IKAROS using Effectene (Qiagen). The cells were fixed, permeabilized, and stained with anti-HA antibody (Biolegend; 901501) and/or anti-flag antibody (cell signaling; 14793) for 2 h. The cells were then washed and stained with Alexa Fluor 488 or Alexa Fluor 568 - conjugated secondary antibodies for an hour. Images were obtained with an EVOS M5000 fluorescence microscope (× 40 objective, Thermo Fisher scientific).
LightShift Chemiluminescent EMSA
HEK293T cells were transfected with pCMV6-AC-MYC-DDK-WT or mutant C182* IKAROS. Forty-eight hours after transfection, nuclear extracts were prepared, and gel mobility shift assays were performed using LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) as described previously [5, 6]. The mutant protein was detected in the nuclear extracts, and the same amount of nuclear extracts were allowed to bind to IKAROS probes. Sequences for the probes were as follows: γ satellite from human chromosome 8 (γ-Sat8) (5′-BIOTIN-GCGAGACCGCAGGG AATGCTGGGAGCCTCCC) and IKAROS consensus-binding sequence 1 (IKBS1) (5′-BIOTIN- TCAGCTTT TGGGAATACCCTGTCA). The EMSA images were acquired with C-Digit Scanner using Image Studio Software (Li-Cor).
Dual Luciferase Reporter Assay
The IKBS1 reporter plasmid (pGL4.11-Luc2P, Promega) containing four repeats of the high affinity IKAROS binding site (IKBS1, TCAGCTTTTGGGAATACCCTGTCA) was described previously [6]. HEK293T cells were transiently transfected with pcDNA3-HA-IKAROS WT and/or mutant C182*, pGL4.11-IKBS1, and pRL-TK (Renilla luciferase) using Effectine (Qiagen). Cells were harvested after 20–24 h and analyzed for luciferase activity using a Dual-Luciferase reporter assay system (Promega) on a GloMax microplate reader (Promega) according to manufacturer’s instructions. The luciferase activity was calculated by dividing the luciferase activity of IKBS1 by the internal Renilla luciferase activity.
Protein Stability Test
HEK293T cells were transfected with pcDNA3-HA-WT or C182* IKAROS. After 20–24 h, cells were treated/untreated with cycloheximide (CHX, 20 μg/ml) for additional 24 h. Whole cell lysates were prepared and analyzed by immunoblot analysis using an anti-HA antibody to test for IKAROS expression. Anti-vinculin (Santa Cruz, sc-73614) was used as a loading control. The relative IKAROS protein stability was calculated by dividing CHX treated sample by untreated sample (× 100) after normalization with vinculin to show the remaining protein amount after CHX treatment.
T and B Cell Proliferation
Total PBMCs were incubated with CellTrace Violet Cell Proliferation Kit (Invitrogen, following manufacturer’s instructions). Total of 1 × 105 cells were seeded into 96-well plates, stimulated with anti-CD3 and anti-CD28 antibodies (1 μg/ml each, clones OKT3 and CD28.2, respectively, eBioscience), CD40L (Enzo 100 ng/ml), IL-21 (Peprotech, 50 ng/ml), CpG (Enzo 1 μM), and anti-IgM (Jackson Lab, 10 μg/ml) as indicated in the figure. After 3 days (for T cells) or 4 days (for B cells), cells were stained with fluorochrome-conjugated CD4, CD8, or CD19 antibodies (BD Biosciences). Cells were washed two times with PBS and acquired and analyzed by flow cytometry (Becton Dickinson FACSCanto II) and FlowJo software (TreeStar).
Results
A full-term Hispanic girl born at 40 weeks gestational age was found to have a confirmed abnormal newborn screen for SCID, with low TREC at 17 copies/μL (reference range > 18). Call back for liquid blood testing with a complete blood count and peripheral blood flow cytometry as requested by the State of California [9] was normal for age at 10 days of life (Table 1). Flow cytometry demonstrated normal CD3+, CD4+, CD8+ T, and NK cells numbers but low B cells. Most CD4+ and CD8+ T cells expressed CD45RA indicating normal thymic generation of naïve T cells (Table 1). Mitogen testing showed normal proliferation to PHA and PWM for CD3+ and CD45+ cells but was unable to be done on B cells due to low numbers. Repeat analysis at 8 weeks of life showed persistently low but improving B cell counts from 30/μL (day 10) to 368/μL (8 weeks) (Table 1). Recent thymic emigrants (CD3 + CD4 + CD45RA + CD31+; 79% in CD4+ T cells) were normal at age 21 weeks.
Table 1.
Laboratory findings
| Day of life 1 | Day of life 10 | Day of life 18 | 8 Weeks | 14 Weeks | 17 Months | |
|---|---|---|---|---|---|---|
|
| ||||||
| TREC (reference range) | 17 (L)* (> 18) | 27,040# (> 6794) | ||||
| Lymphocyte subsets (absolute cells/μL and %) | ||||||
| Lymphocytes absolute (reference range) | 4300 (2000–17,000) | 4070 (1930–7460) | 4908 (3300–15,000) | 7948 (4000–13,500) | 4815 (4000–10,500) | |
| CD3 absolute (reference range) | 4048 (2500–5500) | 3768 (1484–5327) | 4092 (2160–5540) | 5777 (H) (2160–5540) | 3804 (2030–5150) | |
| CD3% (reference range) | 94.1% (H) (53–84) | 93% (H) (54–82) | 83% (H) (54–76) | 73% (54–76) | 79% (H)1 (49–75) | |
| CD3−CD16+CD56+ (reference range) | 203 (170–1100) | 245 (43–526) | 363 (160–930) | 1037 (H) (160–930) | 266 (95–620) | |
| CD3−CD16+CD56+ (%) (reference range) | 4.7% (4–18) | 6% (2–10) | 7% (3–14) | 14% (3–14) | 5% (2–16) | |
| CD4 absolute (reference range) | 3357 (1600–4000) | 3037 (733–3181) | 3229 (1390–4080) | 4610 (H) (1390–4080) | 2576 (1220–3550) | |
| CD4% (reference range) | 78.1% (H) (35–64) | 75% (H) (34–62) | 66% (H) (36–55) | 58% (H) (36–55) | 56% (30–60) | |
| CD4:CD8 ratio (reference range) | 5.0 | 4.3 (≥ 1.0) | 3.7 (1.7–3.9) | 4.1 (H) (1.7–3.9) | 2.8 (1.4–3.9) | |
| CD8 absolute (reference range) | 673 (560–1700) | 710 (370–2555) | 868 (600–1490) | 1159 (600–1490) | 912 (530–1590) | |
| CD8% (reference range) | 15.7% (12–28) | 17% (15–36) | 18% (12–24) | 14% (12–24) | 20% (13–28) | |
| CD3+CD4+CD45RA+ absolute (reference range) | 2746 (1200–3700) | |||||
| CD45RA+ (%) of CD4 | 81.8% | |||||
| CD3+CD8+CD45RA+ absolute (reference range) | 572 (450–1500) | |||||
| CD45RA+ (%) of CD8 | 85.0% | |||||
| CD19 absolute (reference range) | 30 (L) (300–2000) | 36 (L) (370–2306) | 368 (L) (740–2560) | 913 (740–2560) | 651 (L) (830–1880) | |
| CD19% (reference range) | 0.7% (L) (6–32) | 1% (L) (13–39) | 7% (L) (17–36) | 12% (L) (17–36) | 13% (L) (20–38) | |
| CD27+IgM−IgD−(%) of CD19 (reference range) | 0.9% (L) (1.9–30.4) | |||||
| CD38+IgM+(%) of CD19 (reference range) | 69.5% (H) (7.6–48.6) | |||||
| Immunoglobulins | ||||||
| Total IgG (reference range) | 691 (162–872) | 943 (311–664) | 382 (L) (451–1202) | |||
| IgM (reference range) | 31 (1–57) | 25 (0–127) | 51 (35–184) | |||
| IgA (reference range) | 8 (0–10) | 85 (H) (0–42) | 18(15–111) | |||
| IgE (reference range) | < 2.00 (0.00–8.00) | |||||
| Vaccine titers (IU/ml) | Tetanus: 0.242 Diphtheria: 1.643 HBs: reactive | |||||
| Mitogens | ||||||
| Viab. of lymphs at day 0 | 89.7% | |||||
| Max prolif of PWM as %CD45 | 56.1% | |||||
| Max prolif of PWM as %CD3 | 63% | |||||
| Max prolif of PHA as %CD45 | 92.5% | |||||
| Max prolif of PHA as %CD3 | 95.2% | |||||
H and L indicate values above/below the age matched nomral range (respectively) for inhouse or CLIA-certified commercially available tests. HBs hepatitis B surface.
EnLite (PerkinElmer) kit was used to measure TREC copies per μL of peripheral blood as previously described [9].
Copies per 106 CD3+ T cells.
A CD3+/CD4−/CD8− double negative T cell population at 316 cells/μL; 6.6% of lymphocytes was detected; no αβ/γδ markers were included in this test.
Protective titers, > 0.15 IU/ml.
Protective titers, > 0.1 IU/ml
A commercially available 207 primary immunodeficiency gene panel showed heterozygous variants of unknown significance in CD8A, LYST, SEMA3E, STAT5B, and TTC7A in the patient that did not correlate or fully explained the patient’s phenotype (Table 2). Due to low TREC and persistently low B cells at 8 weeks of life, parents were consented to an institutional review board–approved study for rapid WGS for the trio. A pathogenic heterozygous mutation was identified in IKZF1 c.546C > A (p.C182*) in the patient, but not in the parents (likely de novo; parental gonadal mosaicism was not ruled out); the mutation was confirmed by Sanger sequencing. This nonsense variant, mapping to exon 5 (of 8) of the gene, has not been previously reported. Mutation p.C182* is predicted to encode for an ~ 20 kDa truncated protein lacking part of the DNA binding domain (partial ZF3 and complete ZF4-deletion) and the whole C-terminus dimerization domain (ZF5 and ZF6), likely severely impacting protein function. Of note, IKZF1 was not included in the 207 gene panel tested in the patient or the updated 452 gene panel from this vendor.
Table 2.
Variants of uncertain significance
| Gene | Transcript | Variant | Zygosity | GnomAD allele frequency | dbSNP ID |
|---|---|---|---|---|---|
|
| |||||
| CD8A | NM_001768.6 | c.63_64delinsTG(p.Ser22Gly) | Heterozygous | 0.000039 | rs776230766, rs766042221 |
| LYST | NM_000081.3 | c.8806G>A (p.Val2936Ile) | Heterozygous | 0.000580 | rs2753327 |
| SEMA3E | NM_012431.2 | c.475G>C (p.Glu159Gln) | Heterozygous | 0.000012 | rs1241693530 |
| STAT5B | NM_012448.3 | c.650G>A (p.Arg217His) | Heterozygous | 0.000021 | rs200900246 |
| TTC7A | NM_020458.3 | c.154C>A (p.Pro52Thr) | Heterozygous | 0.000014 | rs906523542 |
At 14 weeks of life, B lymphocytes increased to 913 cells/uL. B cell subsets were essentially normal except for increased transitional B cells (CD19+/CD38+/IgM+) and decreased class-switched memory B cells (CD19+/CD27+/IgM−/IgD−). TREC analysis was also tested and was normal at 27,040 copies per 106 CD3+ T cells (Table 1). Repeated T and B cell proliferation upon TCR and BCR stimulation (respectively) showed a normal-to-slightly-diminished response (Fig. 1 a and b). A revertant mutation in T- and monocyte-enriched cells from a 21-week sample was ruled out (Fig. 1c; limited sample volume did not allow for B cell enrichment and testing).
Fig. 1.
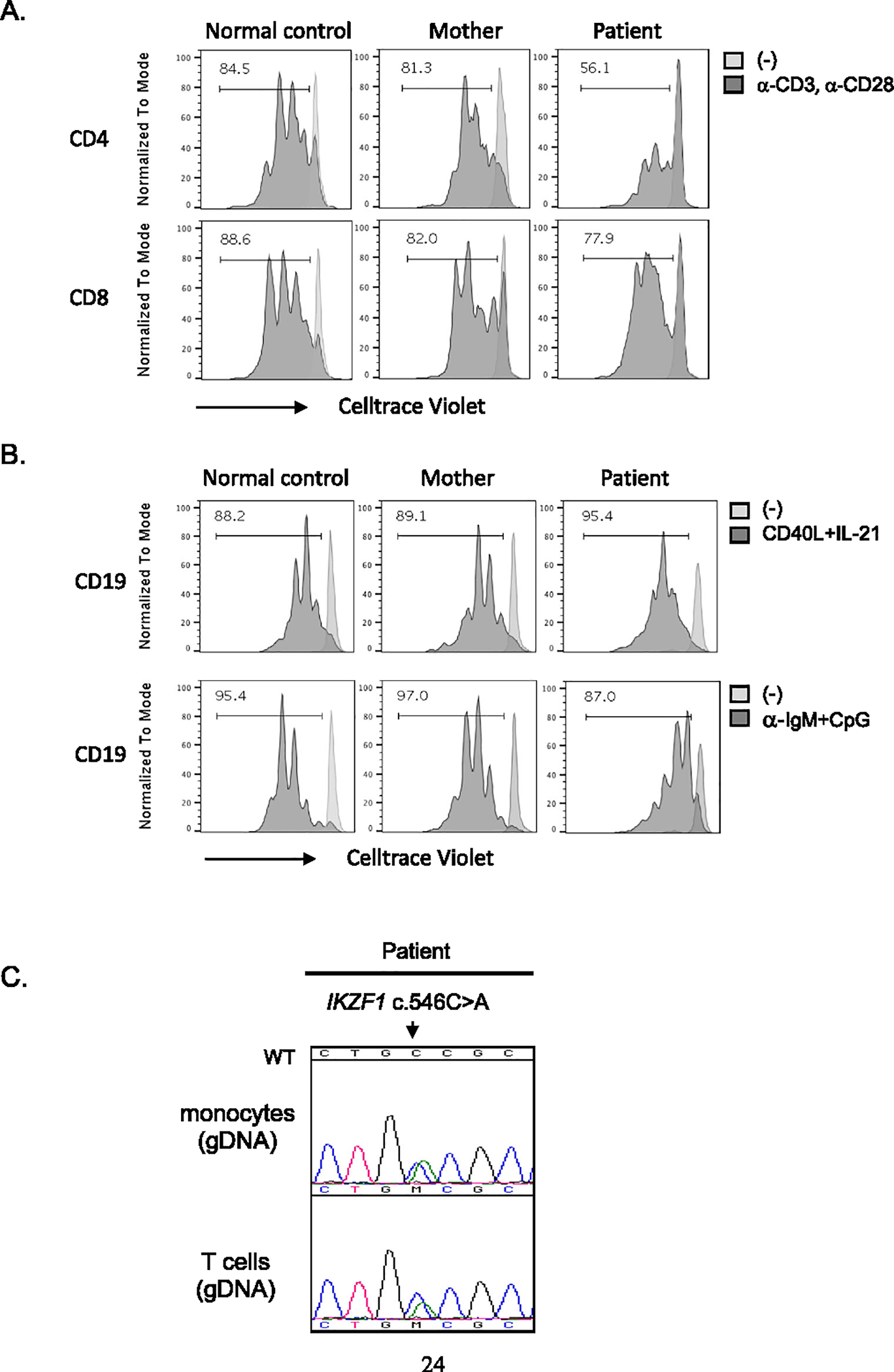
T and B cell proliferation. a Total PBMCs (21-week sample from the patient) stained with CellTrace Violet reagent and stimulated with anti-CD3 and anti-CD28 antibodies for 3 days. b For B cell proliferation, cells were stimulated with CD40L and IL-21, or CpG and a-IgM as indicated in the figure. After 3 (for T cells) or 4 days (for B cells), cells were acquired and analyzed by flow cytometry and FlowJo software, respectively. Numbers in the box indicate the percentage of cells having undergone at least one cellular division assessed by CellTrace Violet dye dilution. c CD3 T cells and monocytes were enriched from the patient’s PBMCs (21-week sample). Genomic DNA was prepared from the cells, and Sanger sequencing was performed. WT indicates reference sequence for wild type IKZF1
When IKZF1 mRNA/cDNA was evaluated, both WT and mutant transcripts were detected at similar levels in the patient’s PBMCs after subcloning and sequencing (Fig. 2a). In terms of IKAROS protein, the patient showed reduced levels of WT IKAROS expression in the nucleus compared to the normal controls, while the mutant protein was readily found in the cytosol (Fig. 2b). To further assess the function of the variant protein, a mutant construct was tested for IKAROS PC-HC targeting, DNA binding, transcription activity, and protein stability. While WT IKAROS exhibited the characteristic punctate staining pattern of PC-HC localization, the mutant lost its ability to target this region and was abundantly detected in the cytoplasm (Fig. 2c). When co-expressed with the WT vector, IKAROS p.C182* did not exert any dominant effect on pericentromeric targeting, and its mechanism of action was determined as haploinsufficiency for this function (Fig. 2d). The mutant also failed to dimerize with WT IKAROS due to the lack of ZF5 and ZF6 (Fig. 2e, and as suggested in Fig. 2d), and to bind to specific DNA targets (Fig. 2f). While WT IKAROS repressed basal IKBS1 transcription activity by ~ 60%, mutant C182* completely failed to do so. In WT/C182* co-transfection experiments at three different ratios (WT/C182* 25/75; 50/50 and 75/25), WT expression repressed the transcription activity in a progressive and dose-dependent manner (WT/C182*: 25/75, ~ 37%, 50/50, ~ 40%, 75/25, ~ 47%), arguing against a dominant negative effect but in favor of a haploinsufficiency mechanism by C182* for this function too (Fig. 2g). Mutant C182* protein stability was also reduced when compared to WT IKAROS (Fig. 2h), being this feature common in dimerization defective, but not in classical haploinsufficient variants [6]. This reduced protein stability may as well contribute to explain the apparent discrepancy between C182* nuclear localization in primary cells (absent; Fig. 2b) vs. transfected and overexpressed in HEK293T cells (present, although failing to PC-HC localization, targeted DNA binding and transcription activity; Fig. 2 c, d, f, and g). These results suggest that mutant p.C182* is retained in the cytosol, is less stable than WT IKAROS, fails to efficiently localize to the nucleus or to dimerize with the WT protein, and is also unable to bind target DNA sequences, without directly affecting the WT protein. As shown, this new IKZF1 mutation displays distinctive mechanistic and clinical features when compared with previously reported allelic variants [4–6, 10].
Fig. 2.
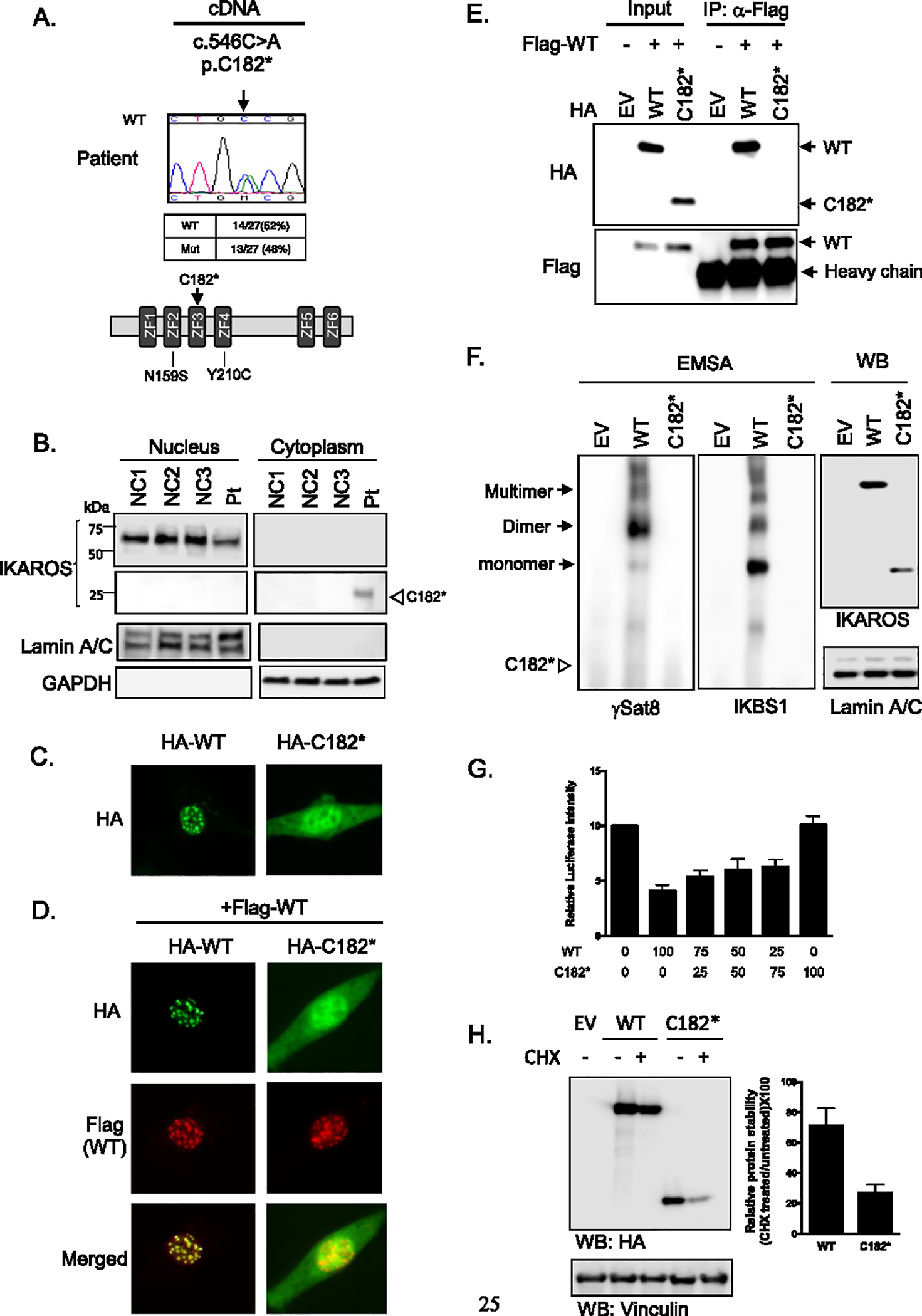
Mutant IKAROS p.C182* expression and its functional tests. a Sequence chromatograms from cDNA from PBMCs in the patient. To quantify the wild type and mutant ratio, PCR products were cloned into pGEM-TA Easy Vectors, and 27 colonies were sequenced. Schematic representation of the IKAROS protein. ZF indicates zinc figure. Previously reported IKZF1 mutations associated with transient SCID presentation are indicated below the ZF2 and ZF4. b IKAROS expression in T cell blasts from the patient and normal controls (NC). Immunoblotting of Lamin A/C and GAPDH was used as a marker for the nuclear and cytoplasmic fraction, respectively. c,d Immunofluorescence staining of WT and mutant proteins transfected into NIH3T3 cells. Cells were visualized by using an EVOS fluorescence microscope (× 40 objective). e HEK 293 T cells were co-transfected with flag-tagged wild-type IKAROS WT and HA-IKAROS WT or mutant. EV indicates empty vector control. Immunoprecipitations and immunoblotting were performed using indicated antibodies. f HEK 293 T cells were transfected with WT and mutant IKAROS DNA, and nuclear extracts were used for the EMSA assay. The IKAROS WT and C182* expression was tested from the nuclear extracts. Lamin A/C was used as a marker for the nuclear fraction. Triangle indicates the expected mutant protein binding site. g HEK293T cells were co-transfected with expression plasmids for IKAROS WT and/or mutant and pGL4.11-IKBS1 and pRL-TK (Renilla). Numbers indicate the percentage of WT and mutant IKAROS DNA used for the co-transfection. The firefly luciferase activity was normalized to the Renilla luciferase activity, and then the results were normalized to the empty vector control. Data are means ± SEM from four independent experiments. h HEK293T cells transfected with WT or mutant IKAROS were treated with cycloheximide (CHX) for additional 24 h. IKAROS expression after CHX treatment was analyzed by immunoblotting. Vinculin was used as a loading control. The relative IKAROS protein stability was shown in the graph. Data are mean value + SD of three independent experiments. Data shown are representative of 2 (Fig. 1e) or 3 (Fig. 1 c, d, f, and h) independent experiments
Discussion
Clinically, our patient presented with two main immune characteristics: very low B cells and abnormally low TREC at newborn screen testing. Low B cells, either since very early in life or progressively declining, have been previously described to be associated with all IKZF1 allelic variants [4–6, 10]. During the first months of follow-up in our patient, B cells progressively increased although not completely normalized for age. In association with the low TREC, low T cell numbers, maturation, or functional T cell defects were not detected when evaluated at 10 days (or later timepoints) after the SCID newborn screen test was collected. While the chance of a “false positive” SCID newborn screen test remains possible, the sequential rather than simultaneous testing does not allow us to definitively confirm or rule out this hypothesis based exclusively in those tests. On the other hand, the experience accumulated from SCID newborn screening demonstrated that primary immunodeficiencies as DiGeorge syndrome, Ataxia-Telangiectasia, or Wiskott-Aldrich syndrome (as well as secondary forms of lymphocyte loss) can present with low TREC and lymphopenia at birth with subsequent values normalization [9, 11, 12]. More closely to our case, the fact that two previously reported IKZF1-mutated infants presented with SCID-like manifestations followed by spontaneous immunologic recovery raises the possibility of “transient SCID” being part of the IKAROS-associated diseases spectrum (Fig. 2a). The patient herein described is the third IKZF1-mutated infant to present with such features in a disease with less than 100 cases described in the literature [13]. In 2016, Hoshino et al. reported a newborn male patient diagnosed with pancytopenia and virtual alymphocytosis at birth (absolute lymphocyte count (ALC), 150/μL) in whom bone marrow transplantation was recommended [10]. However, after 1 month, the patient showed spontaneous recovery of all hematologic parameters including lymphocyte counts (ALC, 6030/μL). This patient carried a germline heterozygous IKZF1 p.Y210C mutation believed to be acting by DNA binding haploinsufficiency. In 2017, Yoshida et al. reported a 9-month-old female patient presenting with a severe pulmonary infection/acute respiratory distress and a T-/B-SCID immunophenotype (260/μL lymphocytes, 192/μL T cells; 15/μL B cells; agammaglobulinemia; and markedly reduced T cell proliferation) [14]. While the patient underwent a purified CD34-positive bone marrow transplant from her haploidentical mother without preconditioning at 10 months of age, the graft was rejected and followed with complete autologous recovery. At the age of 4 years, the patient presented an ALC of 2054/μL; 1970/μL T cells; 26/μL B cells; and normal-low proliferative responses. This patient carried a germline heterozygous IKZF1 mutation p.N159S, later reported to act in a dominant negative manner [5].
Interestingly, in our patient, the number of B and T cells (assuming a direct correlation between low TREC and low thymic output/T cell lymphopenia) increased over her first weeks/months of life. These results may suggest that having one WT and one mutant IKAROS copy (i.e., pY210C, p.N159S, or p.C182*) during the prenatal period and under certain circumstances might not be sufficient to achieve normal B- or T- cell production at birth but might be enough to partially or completely restore lymphocyte development postnatally. On the other hand, as IKAROS-associated disease manifestations could be progressive and appear later in life, long-term follow-up for B- and T cell-related defects (e.g., B cell lymphopenia, hypogammaglobulinemia, B or T cell malignancy) is warranted in this patient, as evidenced by decreasing B cells and IgG levels at 17 months evaluation [4, 6, 13]. Although the patient remains asymptomatic, her clinical and immune status is regularly monitored in case any medical intervention is needed (e.g., IgG replacement therapy). Besides, IKZF1 mutations should be added to the list of genes associated with abnormal SCID newborn screens although spontaneous recovery should be considered.
Acknowledgements
We thank Julie Niemela (NIH) for her contribution performing Sanger sequencing. The content of this article does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. We would like to thank the patient and her family for participating in this research.
Funding
These studies were supported by grant U19HD077693 from NICHD and NHGRI and gifts from the Liguori Family, John Motter and Effie Simanikas, Ernest and Evelyn Rady, and Rady Children’s Hospital, San Diego; and the Intramural Research Program, NIH Clinical Center, US National Institutes of Health (NIH).
Abbreviations
- CVID
Common variable immunodeficiency
- CID
Combined immunodeficiency
- TREC
T cell receptor excision circles
- PC-HC
Pericentromeric heterochromatin
- SCID
Severe combined immunodeficiency
Footnotes
Conflict of Interest The authors declare no competing interests.
Ethics Approval Yes.
Consent to Participate All patients or their guardians provided informed consent in accordance with the Declaration of Helsinki under institutional review board–approved protocols of the National Institute of Allergy and Infectious Diseases, NIH, and Western IRB (Pr #:20171726); ClinicalTrials.gov Identifier: NCT03385876.
Consent for Publication Yes.
References
- 1.Fan Y, Lu D. The Ikaros family of zinc-finger proteins. Acta Pharm Sin B. 2016;6(6):513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14(17):2146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vairy S, Tran TH. IKZF1 alterations in acute lymphoblastic leukemia: the good, the bad and the ugly. Blood Rev. 2020;44:100677. [DOI] [PubMed] [Google Scholar]
- 4.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374(11):1032–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest. 2018;128(7):3071–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehn HS, Niemela JE, Stoddard J, Ciullini Mannurita S, Shahin T, Goel S, et al. Germline IKAROS dimerization haploinsufficiency causes hematologic cytopenias and malignancies. Blood. 2020;137(3):349–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingsmore SF, Cakici JA, Clark MM, Gaughran M, Feddock M, Batalov S, et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105(4):719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amatuni GS, Currier RJ, Church JA, Bishop T, Grimbacher E, Nguyen AA, et al. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia in California, 2010–2017. Pediatrics. 2019;143(2):e20182300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol. 2017;140(1):223–31. [DOI] [PubMed] [Google Scholar]
- 11.Mallott J, Kwan A, Church J, Gonzalez-Espinosa D, Lorey F, Tang LF, et al. Newborn screening for SCID identifies patients with ataxia telangiectasia. J Clin Immunol. 2013;33(3):540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borte S, Fasth A, von Dobeln U, Winiarski J, Hammarstrom L. Newborn screening for severe T and B cell lymphopenia identifies a fraction of patients with Wiskott-Aldrich syndrome. Clin Immunol. 2014;155(1):74–8. [DOI] [PubMed] [Google Scholar]
- 13.Kuehn HS, Nunes-Santos CJ, Rosenzweig SD, IKAROS-Associated Diseases in 2020. Genotypes, phenotypes, and outcomes in primary immune deficiency/inborn errors of immunity. J Clin Immunol. 2021;41(10):1–10. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida N, Sakaguchi H, Muramatsu H, Okuno Y, Song C, Dovat S, et al. Germline IKAROS mutation associated with primary immunodeficiency that progressed to T-cell acute lymphoblastic leukemia. Leukemia. 2017;31(5):1221–3. [DOI] [PubMed] [Google Scholar]


