Abstract
CRISPR/Cas9 possesses the most promising prospects as a gene-editing tool in post-genomic researches. It becomes an epoch-marking technique for the features of speed and convenience of genomic modification. However, it is still unclear whether CRISPR/Cas9 gene editing can cause irreversible damage to the genome. In this study, we successfully knocked out the WHITE gene in Drosophila, which governs eye color, utilizing CRISPR/Cas9 technology. Subsequently, we conducted high-throughput sequencing to assess the impact of this editing process on the stability of the entire genomic profile. The results revealed the presence of numerous unexpected mutations in the Drosophila genome, including 630 SNVs (Single Nucleotide Variants), 525 Indels (Insertion and Deletion) and 425 MSIs (microsatellite instability). Although the KO (knockout) specifically occurred on chromosome X, the majority of mutations were observed on chromosome 3, indicating that this effect is genome-wide and associated with the spatial structure between chromosomes, rather than being solely limited to the location of the KO gene. It is worth noting that most of the mutations occurred in the intergenic and intron regions, without exerting any significant on the function or healthy of the animal. In addition, the mutations downstream of the knockout gene well beyond the upstream. This study has found that gene editing can lead to unexpected mutations in the genome, but most of these mutations are harmless. This research has deepened our understanding of CRISPR/Cas9 and broadened its application prospects.
Keywords: Drosophila, CRISPR/Cas9, Mutations, SNV, Indel, MSI
Abbreviations
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- SNV
single nucleotide variants
- Indel
Insertion and Deletion
- MSI
microsatellite instability
- KO
Knockout
- sgRNA
small-guide RNA
- CDS
coding sequences
- UTR
untranslated Regions
1. Introduction
In the past decades, genome manipulation technology has experienced rapid advancements across the globe [1]. Currently, the most favored genome editing tool is CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/associated nuclease 9) systems [2,3], which has revolutionized the field of genome engineering and regulation since its emergence. CRISPR/Cas9 system serves as an RNA-guided nuclease capable of simultaneously editing multiple sites in the genome by encoding several guide sequences. Due to the characteristics of simple composition and high editing efficiency [[4], [5], [6]], it has rapidly gained popularity and become as a hallmark in biomedicine field. CRISPR/Cas9 can be universally applied to a wide range of biological studies, including nonhuman primates [7], mice [8], rat [9], zebra fish [10], Drosophila [11], helminth [12], as well as a myriad of common plants [13] and microbes [14]. CRISPR/Cas9 gene editing has found widespread application, however, there remains no definitive conclusion regarding whether the process of gene editing and repair can potentially induce harmful effects on the genome.
The CRISPR/Cas9 system is composed of Cas9 protein and sgRNA [15]. In the progression of gene editing, Cas9 protein initially recognizes a specific Protospacer adjacent motif (PAM). Subsequently, the sgRNA binds to the target gene sequence, followed by the Cas9 protein cleaving the double-stranded DNA, thus producing double-stranded DNA breaks (DSB). Eventually the damaged DNA could be repaired by non-homologous end joining (NHEJ) or homologous recombination (HR) [[16], [17], [18]]. Compared to HDR, NHEJ is more common in the process of genome repair, as it does not require a template, thus resulting in a faster repair rate. However, this repair is less precise and often leads to base mismatches, insertions, or deletions, ultimately triggering mutations [19]. While the CRISPR/Cas9 gene editing system exhibits remarkable efficiency in cutting target genes, the off-target effects and unexpectable mutations beyond target sites seem inevitable due to the activities of DNA damage and repair. Comprehensively understanding the “side-effect” of CRISPR/Cas9 remains a challenge in the development of CRISPR/Cas9 gene editing system [20]. Mutations caused by Cas9 can lead to changes in gene expression patterns [21], chromosomal rearrangements [22,23], and even promote the development of certain diseases such as cancer [24,25]. The impact of mutations on the function of genes and proteins depends on the location of the mutation within the gene. Mutations occurring in promoters may inhibit DNA transcription and translation [26,27], while missense mutations in exons can alter the amino acid sequence of a protein, potentially disrupting its structure and function [28]. Regarding that CRISPR/Cas has been applied in treatment of human inherited diseases, exploring the safety of CRISPR/Cas9 gene editing is crucial for the health and well-being of human life. Investigation of the safety of CRISPR/Cas9 gene editing and its impact on genomic stability has been carried out in various organisms, such as mouse [22], livestock [29], zebrafish [30], and human cells [31,32]. However, the influence of CRISPR/Cas9 on the whole genome stability of lower animals, such as Drosophila, may have been underestimated and remains unexplored.
Drosophila, belonging to the phylum Arthropoda, class Insecta, and order Diptera, has the features of easy feeding, short reproductive cycle, and rich morphology, making it a widely used model animal [33]. The eye color of Drosophila is controlled by the WHITE gene, which is located on the chromosome X of Drosophila. The silencing of the WHITE gene will turn the eyes of Drosophila from white to red [34,35]. This phenotypic change serves as a convenient indicator for determining the success of gene knockdown experiments, particularly when utilizing CRISPR/Cas9. By monitoring this visible phenotype, researchers can efficiently assess whether the target gene has been effectively silenced. Recognizing the benefits of studying WHITE gene, we employed the CRISPR/Cas9 gene editing system to generate Drosophila with WHITE knockout. This allowed us to assess the potential risks and limitations of CRISPR/Cas9 for genetic modifications.
Our final results indicate that CRISPR/Cas9 gene editing can lead to the occurrence of unexpected mutations. The location of these mutations may be related to the distance and spatial structure from the Cas9 target gene. Fortunately, most mutations occur in non-coding regions and do not affect the survival and phenotype of Drosophila.
2. Materials and methods
2.1. Drosophila sample source
The Drosophila samples were donated by Professor Ni Jianquan, Dr. Jin Sun from Tsinghua University. The genotype of Drosophila is y[1]sc[*]v[1]sev[21]; P{y[+t7.7]v[+t1.8] = nos-Cas9.R}attP40 [36].
2.2. Knockout of WHITE gene via CRISPR/Cas9 system
The Cas protein derived by nos promotor was transfected into the Drosophila fertilized eggs (nos > cas9). The sgRNAs were obtained from our previous study. The sequence of sgRNAs: w1: CAGGAGCTATTAATTCGCGGAGG, w2: TAGTTGGCCGCTCCCTGAACCGG [36]. The sgRNAs were injected into the nos > cas9 fertilized eggs (G0). The hatched G0 were hybrid with nos > cas9 to obtain the G1. The female G1 were hybrid with nos > cas9 to obtain the G2. The white-eye male Drosophila were chosen as the positive group, and the red-eye male Drosophila as negative group to perform the whole genome sequencing. Each group was composed more than 50 flies.
2.3. Whole genome sequencing of Drosophila
The whole genomic DNA (gDNA) of Drosophila were extracted by Invitrogen DNA kit (Thermofisher), according to the manufacturer's instructions. The WGS DNA library was prepared to perform paired-end sequencing by MGISEQ-2000 to obtain raw sequencing data with sequencing read length of 100 bp. The sequencing depth of control group was 175 × , of WHITE-KO group was 154 × . On average 20 Gb raw data were obtained for each sample. The raw data was purified via Cutadapt (version 1.12) to obtain the clean data. The clean data was mapped to the Drosophila genome (Dmel, release = r6.30) by BWA Aln(Version: 0.7.12-r1039), then the mapped results (.Sam, The Sequence Alignment/Map format) were analyzed by Samtools (version: 1.3.1) to remove the duplication. The mutations of clean data were compared with the Drosophila reference genome to exclude the SNPs. Subsequently, mutations with a mutation rate of more than 10% were retained.
2.4. Analysis of mutations on the drosophila chromosomes
The mutations acquired from the sequencing were mapped to the Genome Data Viewer of NCBI (ncbi.nlm.nih.gov/genome/gdv/) to obtain the specific information of mutations. SNV and indel site filtering and genotype filtering were performed to obtain high-confidence variants.
2.5. Analysis of MSI on the Drosophila chromosomes
MSIsensor (version: v0.6) was performed to assess the MSI of WHITE-KO Drosophila compared to the control group using Pearson's Chi-Squared Test. The statistical results of mutations were analyzed and visualized by Graphpad Prism 8.0.
3. Results
3.1. Global mutation on Drosophila chromosomes
To investigate the impact of Cas9 gene editing on the Drosophila genome, we designed two sgRNAs targeting the WHITE gene. These sgRNAs were derived from our previous work, ensuring no potential off-target effects [36,37]. sgRNA targeting the WHITE gene was injected into Drosophila embryos, and the flies were subsequently grouped into positive knockout (red-eyed) and negative knockout (white-eyed) cohorts based on their eye color phenotype. Globally, we found 630 SNVs (single nucleotide variants), 525 indels (insertion and deletion) and 425 MSIs (microsatellite instability), distributed on all chromosomes. To assess whether the mutations introduced by Cas9 editing contained off-target sites, we compared the identified mutations with the 1771 potential off-target sites predicted using the Cas-OFFinder tool (www.rgenome.net/cas-offinder/). Upon comparison, no match was found between the mutations and the potential off-target sites predicted by Cas-OFFinder. This result indicates that the sgRNA designed to target the WHITE gene was effective and specific, and the mutations we identified are not related to off-targets effect (Supplementary material).
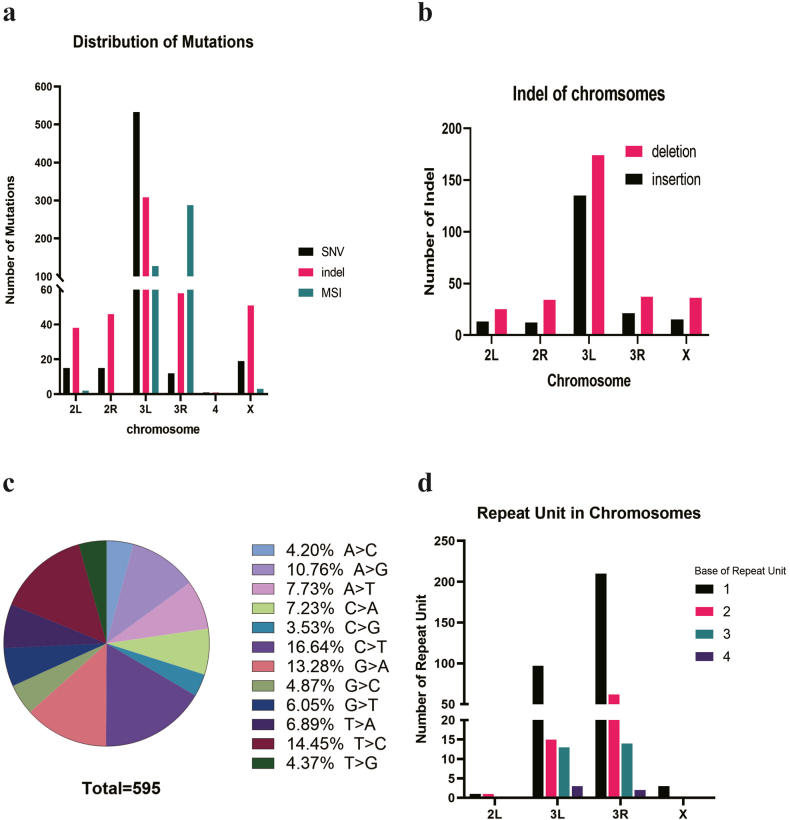
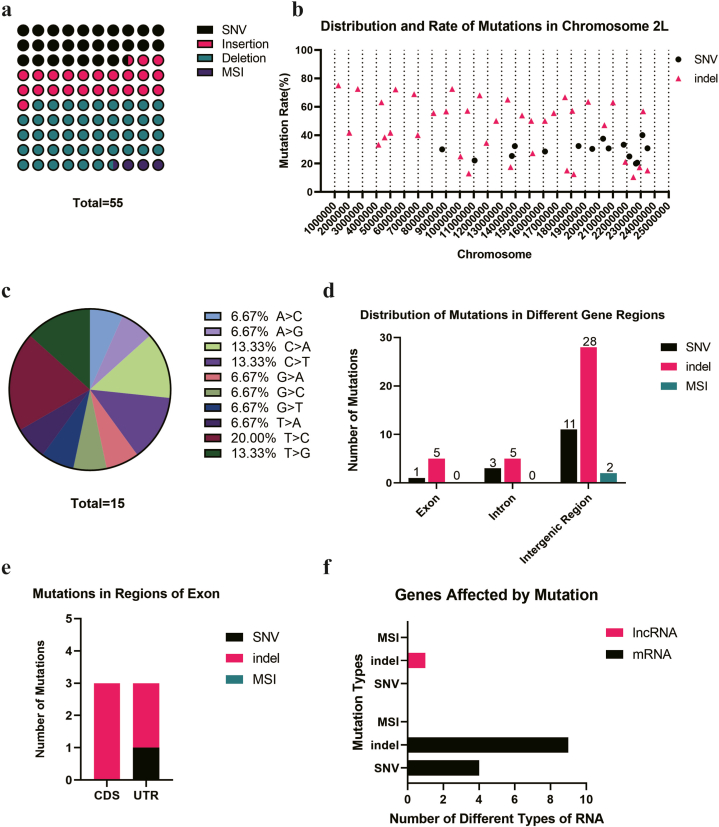
Although the target gene WHITE is located on the chr X: 2790599–2796466, the X chromosome does not harbor the highest number of mutations. Unexpectedly, the majority of single nucleotide variants (SNVs) and insertions/deletions (indels) are situated on chromosomes 3L and 3R, regions that also exhibited massive MSIs (Fig. 1a). The number of deletions on each chromosome surpasses the count of insertions, aligning with observations made in mice embryos [38] (Fig. 1b). In deeper exploration of SNV types, we discovered that cytosine to thymine (C > T) transitions account for the highest proportion, while cytosine to guanine (C > G) transitions constitute the lowest proportion (Fig. 1c). MSIs are mainly concentrated on the 3L and 3R chromosomes, with the core repeat units primarily composed of mononucleotide repetitions (Fig. 1d). No mutation has been found on chromosome 4.
Fig. 1.
The distribution of mutations on the genome of CRISPR/Cas9 edited Drosophila. a. There were 1519 mutations, including 116 mutations on the chr 2, 1328 mutations on the chr 3, 2 mutations on the chr 4, and 73 mutations on the chr X; b. There were more deletions than insertions on every chromosome of Drosophila; c. There were 595 SNVs, including A > C(25/595), A > G(64/595), A > T(46/595), C > A(43/595), C > G(21/595), C > T(99/595), G > A(79/595), G > C(29/595), G > T(36/595), T > A(41/595), T > C(86/595), T > G(26/595). d. 128 MSIs were distributed on the chromosome 3L, single base repeats accounting for 75.58% (97/128), and 288 MSIs were distributed on the 3R, single base repeats accounting for 72.92% (210/288).
3.2. Mutation on chromosome X
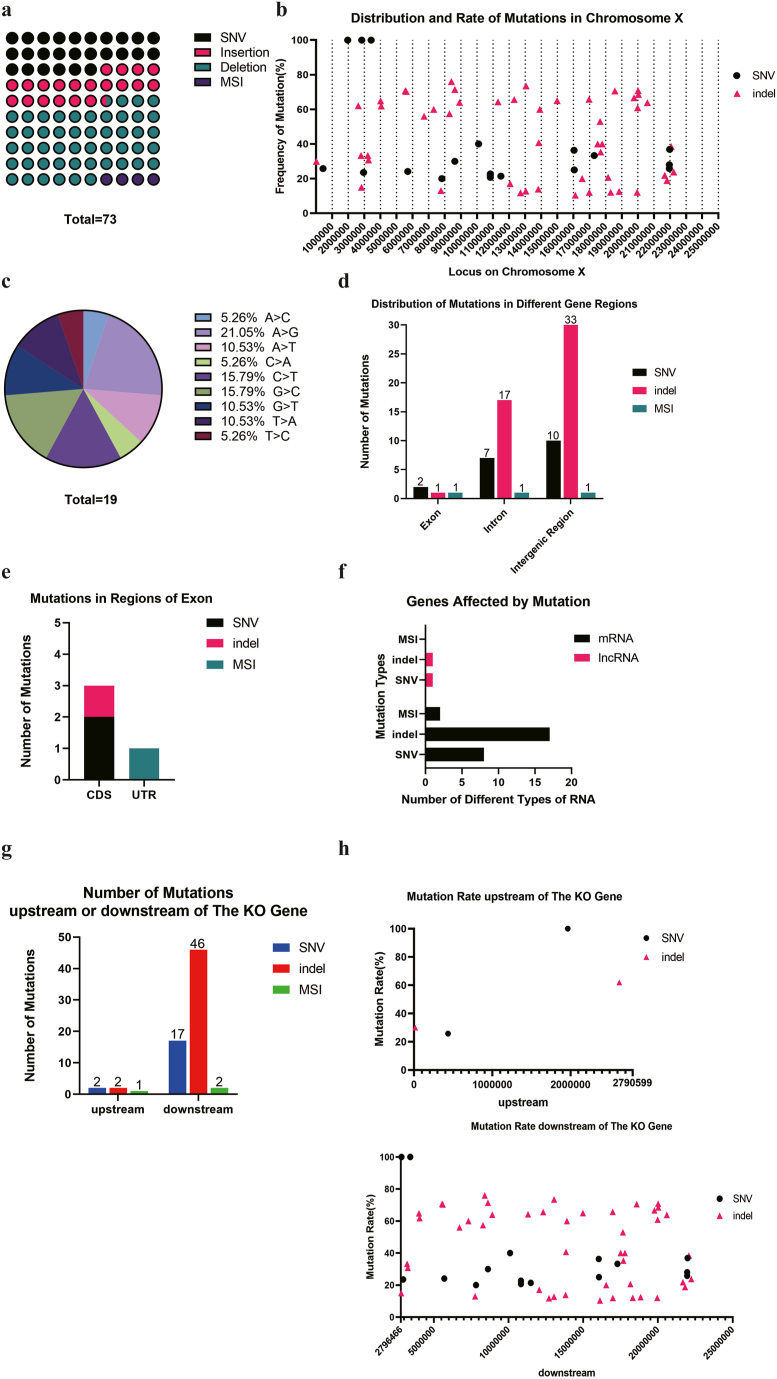
Since WHITE gene knocked-out by Cas9 is located on the chr X: 2790599–2796466, we prioritized the analysis of mutations on this chromosome. Sequencing data revealed that indels constitute the majority of mutations on chromosome X (Fig. 2a). No clustering of mutations was observed on chromosome X, but the count and average mutation rate of indels were notably higher than those of SNVs (Fig. 2b). Remarkably, three specific SNV sites exhibited a mutation rate of 100% (Table 1). Among all the SNVs, the A > G point mutation constituted 21.05% of the total (Fig. 2c). On chromosome X, the majority of the mutations, particularly indels, occurred in intron and intergenic regions (Fig. 2d and e). After examining the gene types impacted by mutations, we found that mRNA was the gene type most severely affected by the mutations detected on chromosome X, which is consistent with observations made on other chromosomes (Fig. 2f). Based on the sequencing data, we identified 2 SNVs, 2 indels and 1 MSIs located upstream of WHITE, while the majority of mutations were situated downstream of WHITE (Fig. 2g). Notably, while the number of mutations downstream of WHITE increased substantially, no correlation was observed between the mutation rate and the distance from the WHITE gene (Fig. 2h). In addition, we discovered 3 mononucleotide microsatellites on chromosome 3 that exhibited more than 10 repeats (Table 2).
Fig. 2.
The mutations on the chromosome X of CRISPR/Cas9 edited Drosophila. a. There were 19 SNVs, 15 insertions, 36 deletions and 3 MSIs on the chromosome X; b. The distribution and rate of mutations on the chromosome X; c. There were 19 SNVs, including A > C(1/19), A > G(4/19), A > T(2/19), C > A(1/19), C > T(3/19), G > C(3/19), G > T(2/19), T > A(2/19), T > C(1/19) on the chromosome X. d. There were 2 SNVs, 1 indel, and 1 MSI located on the exon, 7 SNVs, 17 indels, 1 MSI located on the intron, 10 SNVS, 33 indels and 1 MSIs located on the intergenic region of the chromosome X; e. The mutations on the intergenic region were located on the CDS and UTR region; f. The gene type that affected by mutations included 2 lncRNAs and 25 mRNAs; g. There were 5 mutations located on the upstream of WHITE gene and 65 mutations located on the downstream of WHITE gene; h. The mutation rate and number showed no relationship with distance to the KO gene.
Table 1.
The mutation frequency of 3 SNV is 100%.
| Chromosome | SNV site | Mutation Frequency | FDR | Gene | Gene Type | Location |
|---|---|---|---|---|---|---|
| X | 1963065 | 100.00% | 1.03E-28 | Hr4 | mRNA | intron |
| X | 2836289 | 100.00% | 7.58E-29 | kirre | mRNA | intron |
| X | 3425585 | 100.00% | 5.03E-18 | CR44999 | lncRNA | intron |
Table 2.
The repeat of 3 MSI located on the Chr X.
| Chromosome | Site | Left Flank | Repeated Base | Number of replicates | Right Flank | P_value | FDR | Gene | Gene Type | Location |
|---|---|---|---|---|---|---|---|---|---|---|
| X | 4227835 | TGAGC | A | 12 | TTGTA | 1.69E-05 | 0.00705 | intergenetic region | ||
| X | 1660613 | AACGG | T | 11 | AATAG | 9.63E-05 | 0.019098 | br | mRNA | exon |
| X | 4847161 | GCCAT | A | 11 | GCACA | 0.000119 | 0.022685 | CG42594 | mRNA | intron |
3.3. Mutation on chromosome 3
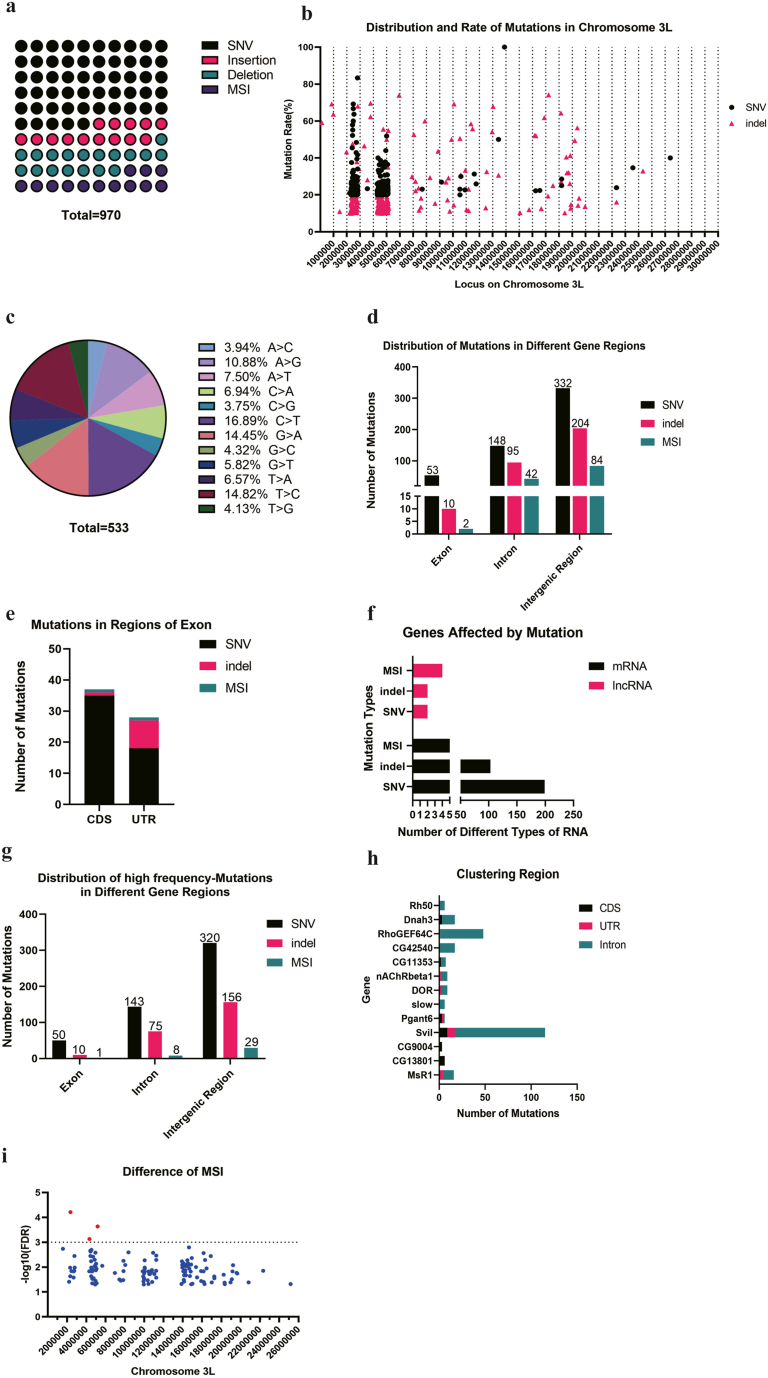
Chromosome 3 is the largest chromosome in Drosophila, and consequently, we prioritized the analysis of its mutation characteristics. On chromosome 3L, we detected 533 SNVs, representing 54.89% of all mutations, and 309 indels, of which 135 were insertions and 174 were deletions (Fig. 3a). The distribution of SNVs and indels on chromosome 3L revealed a pattern of clustering, with the highest concentration of SNVs observed in the chromosomal regions of chr: 3L 2000000–3000000 and 4000000–5000000 bp. Similarly, the majority of indels were clustered in the regions of chr: 3L 2000000–3000000 and 4000000–5500000 bp. Intriguingly, the high-frequency mutation of SNV and indel exhibited a significant regional coincidence (Fig. 3b). Regarding the SNVs, the C > T mutation accounted for 16.89%, while the C > G comprised 3.75% of the total (Fig. 3c). The majority of mutations were detected in intron and intergenic regions (Fig. 3d). When mutations occurred in exons, they exhibited a higher frequency in the CDS region compared to the UTR (Fig. 3e). When analyzing the type of genes harboring mutations, we discovered that mRNA was more susceptible to mutations compared to lncRNA, regardless of MSI, indel or SNV (Fig. 3f). In the regions of the high-frequency, the majority of mutations still occurred in the intergenic region (Fig. 3g). Further investigation about the mutational position displayed that mutations were concentrated in the Svil gene, including the introns, CDS and UTR (Fig. 3h). Considering most of MSIs were located on the chr 3, we screened MSI sites based on FDR values and found out 3 MSI sites with significant difference (Fig. 3i and Table 3).
Fig. 3.
The mutations on the chromosome 3L of CRISPR/Cas9 edited Drosophila. a. There were 533 SNVs, 135 insertions, 174 deletions and 128 MSIs on the chromosome 3L; b. The distribution and rate of mutations on the chromosome 3L; c. There were 533 SNVs, including A > C(21/533), A > G(58/533), A > T(40/533), C > A(37/533), C > G(20/533), C > T(90/533), G > A(77/533), G > C(23/533), G > T(31/533), T > A(35/533), T > C(79/533), T > G(22/533) on the chromosome 3L; d. There were 53 SNVS, 10 indels and 2 MSIs located on the exon, 148 SNVS, 95 indels and 42 MSIs located on the intron, 332 SNVS, 204 indels and 84 MSIs located on the intergenic region of chromosome 3L; e. The mutations on the exon region were located on the CDS and UTR region; f. The gene type that affected by mutations included 8 lncRNAs and 342 mRNAs; h. There were 58 single base insertions and 61 single base deletions of all the indels on the chromosome 3L; i-j. There were 81 short insertions and 89 short deletions that shorter than 10 bp, and 11 long insertions and 29 long deletions; g. In the high-frequency mutation region, there were 50 SNVS, 10 indels and 1 MSI located on the exon, 141 SNVS, 75 indels and 8 MSIs located on the intron, 320 SNVS, 156 indels and 29 MSIs located on the intergenic region; h. There were 9 mutations located on the CDS of Svil, 8 on the UTR and 98 on the intron;
i. There were 3 significantly different MSI sites that -log10(FDR) > 3.
Table 3.
3 MSI with significant difference located on the Chr 3L.
| Chromosome | Site | Left Flank | Repeated Base | Number of replicates | Right Flank | P_value | FDR | -LOG10(FDR) | Location |
|---|---|---|---|---|---|---|---|---|---|
| 3L | 2351768 | AATAG | A | 10 | GAGCA | 9.51E-09 | 6.18E-05 | 4.208759 | intergenetic region |
| 3L | 5169364 | TGGGT | G | 10 | CGGCA | 4.28E-08 | 0.000232 | 3.634624 | intergenetic region |
| 3L | 4324818 | GAAAG | AT | 7 | AAACA | 2.27E-07 | 0.000739 | 3.131403 | intergenetic region |
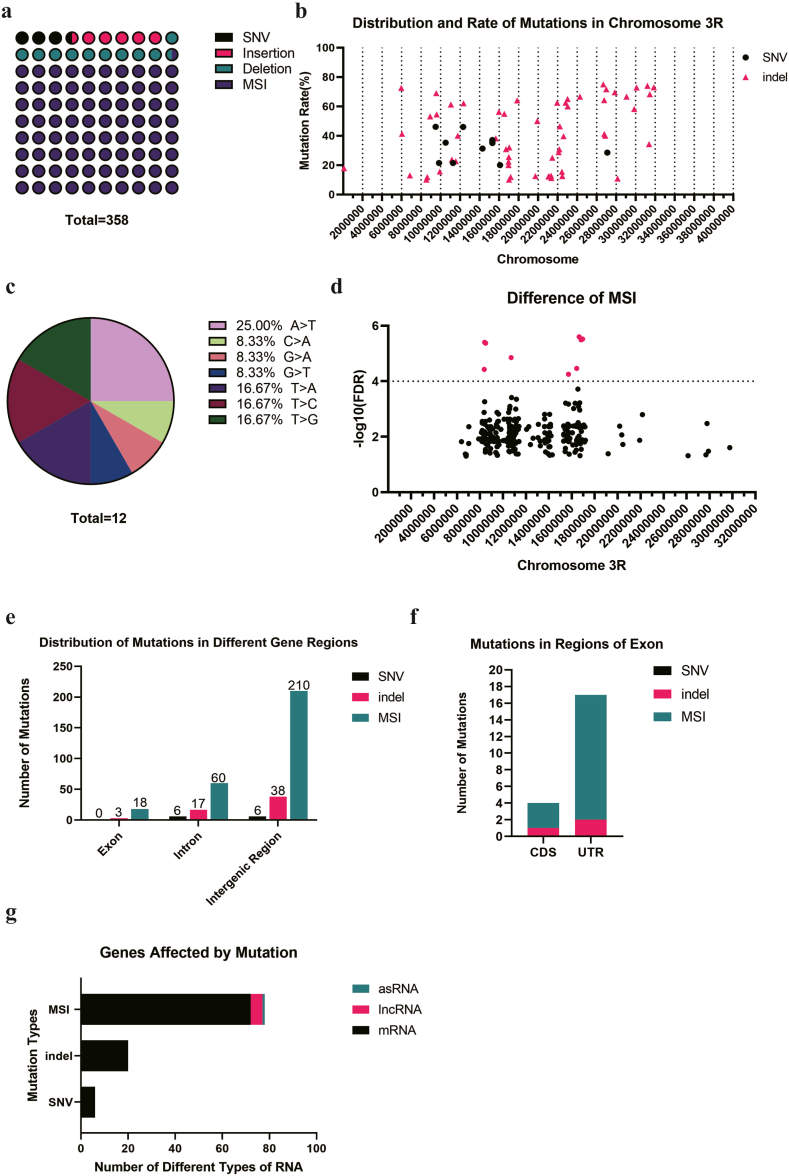
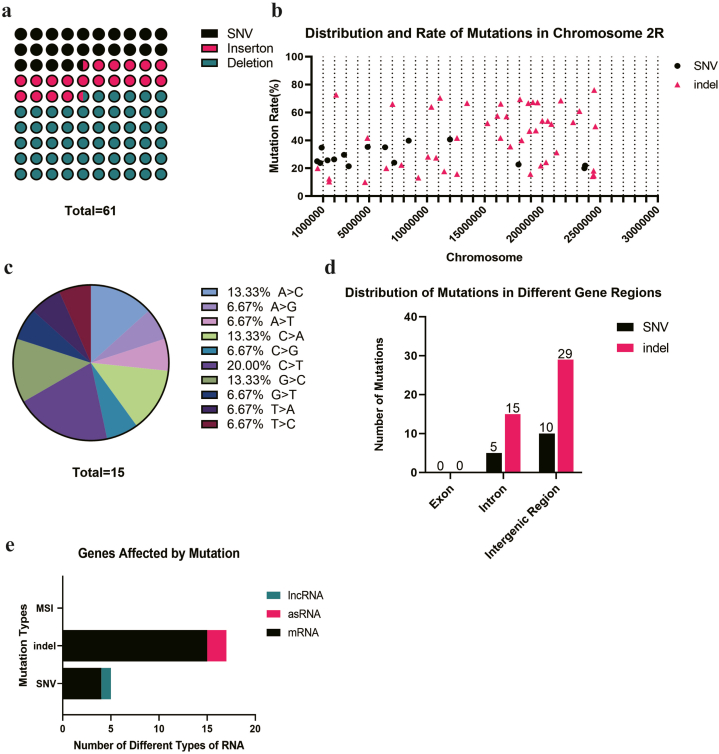
The situation on the chromosome 3R is quite different from that on 3L. MSIs comprised the largest proportion of mutations at 80.45%, followed by deletions at 10.34%. The number of deletions exceeded insertions, and SNVs accounted for the smallest proportion at 3.35% (Fig. 4a). Unlike the chromosome 3L, the SNVs and indels on chromosome 3R did not demonstrate a tendency of aggregation (Fig. 4b). Of all the SNVs, the A > T point mutation constituted the largest proportion at 25.00%, followed by T > A, T > C, and T > G, each accounting for 16.67% respectively (Fig. 4c). Although rarely observed in other chromosomes, a significant number of MSIs (288) were present on the chromosome 3R. Most of MSIs were located within the region chr: 10000000–18000000 on the chromosome 3R (Fig. 4d). After sequencing, we identified the most significantly different MSI sites based on the FDR values (Table 4). Similarly to other chromosomes, the majority of these mutations were located in the intergenic region and intron (Fig. 4e). Within the exon mutations, we observed 3 indels and 18 MSIs. The majority of these mutations were situated in the UTR sequence, while the remaining mutations resided in the CDS of 3 genes detailed in Table 5 (Fig. 4f). The mutated genes primarily consisted of mRNA, along with lncRNA and antisense-RNA (asRNA), with mRNA being the most commonly affected (Fig. 4g).
Fig. 4.
The mutations on the chromosome 3R of CRISPR/Cas9 edited Drosophila.a. There were 12 SNVs, 21 insertions, 37 deletions and 288 MSIs on the chromosome 3R; b. The distribution and mutation rate of mutations on the chromosome 3R; c. There were 533 SNVs, including A > T(3/12), C > A(1/12), G > A(1/12), G > T(1/12), T > A(2/12), T > C(2/12), T > G(2/12) on the chromosome 3R; d. There were 9 significantly different MSI sites which are with -log10(FDR) > 3; e. There were 3 Indels and 18 MSIs located on the exon, 6 SNVS, 17 Indels and 60 MSIs located on the intron, 6 SNVS, 38 Indels and 210 MSIs located on the intergenic region of the chromosome 3R; f. The mutations on the exon region were located on the CDS and UTR region; g. The gene type that affected by mutations included 5 lncRNAs, 98 mRNAs and 1 asRNA.
Table 4.
9 MSI with significant difference located on the Chr 3R.
| Chromosome | Site | Left Flank | Repeated Base | Number of replicates | Right Flank | P_value | FDR | -LOG10(FDR) | Location |
|---|---|---|---|---|---|---|---|---|---|
| 3R | 16644912 | TACAG | A | 10 | CTCGG | 7.68E-11 | 2.50E-06 | 5.602616 | exon |
| 3R | 16942403 | CCCGG | T | 10 | CATTT | 1.39E-10 | 3.01E-06 | 5.521737 | intergenetic region |
| 3R | 16813319 | AGCTG | A | 10 | CAGCG | 4.96E-11 | 3.23E-06 | 5.491228 | intergenetic region |
| 3R | 8402572 | GAGCC | TG | 6 | TATGT | 3.04E-10 | 3.96E-06 | 5.402788 | intergenetic region |
| 3R | 8493033 | GCAGT | TGC | 6 | AGTTG | 2.62E-10 | 4.25E-06 | 5.371427 | intergenetic region |
| 3R | 10721790 | AACTA | T | 10 | CAGTG | 1.30E-09 | 1.41E-05 | 4.852046 | intergenetic region |
| 3R | 16427824 | TGTGA | GT | 8 | CTGGC | 4.24E-09 | 3.45E-05 | 4.462068 | intergenetic region |
| 3R | 8366486 | GTCGA | TGT | 6 | TGATG | 4.02E-09 | 3.73E-05 | 4.427884 | intergenetic region |
| 3R | 15712602 | TTTGA | T | 10 | ACCAC | 7.77E-09 | 5.61E-05 | 4.250867 | intergenetic region |
Table 5.
MSI located in the CDS of 3 genes.
| Chromosome | Site | Left Flank | Repeated Base | Number of replicates | Right Flank | P_value | FDR | -LOG10(FDR) | Gene | Gene Type | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3R | 11047920 | ATGAT | GCA | 7 | ACAGC | 1.04E-05 | 0.005538 | 2.256624 | side-Ⅵ | mRNA | exon-CDS |
| 3R | 9437033 | AGCAC | CAG | 7 | GATCC | 0.000244 | 0.040207 | 1.395698 | ps | mRNA | exon-CDS |
| 3R | 14081682 | TGCAA | CAG | 7 | GCGCA | 0.000295 | 0.046597 | 1.331642 | foxo | mRNA | exon-CDS |
3.4. Mutation on chromosome 2
Lastly, we analyzed the mutational profile of chromosome 2, finding a total of 116 mutations, which were fewer than those on other chromosomes. As to chromosome 2L, 69.09% of the mutations were indels, of which deletion was predominant, accounting for 45.45% of all mutations (Fig. 5a). On chromosome 2L, the mutations did not exhibit significant clustering, and the mutation rate of each indel was generally higher than that of SNV, typically exceeding 40%. In contrast, the mutation rate for SNVs was below 40% (Fig. 5b). Among all SNVs on chromosome 2L, the T > C point mutation comprised 20.00% of the total (Fig. 5c). Mutations were observed predominantly in intergenic regions, totaling 41 mutations, while exons and introns exhibited fewer mutations, with 6 and 8 mutations, respectively (Fig. 5d). Besides, there were 3 indels located in CDS region (Fig. 5e). To identify the specific types of genes affected by these mutations, we mapped the precise location of these mutations and found that most mutations were situated within mRNA region (Fig. 5f).
Fig. 5.
The mutations on the chromosome 2L of CRISPR/Cas9 edited Drosophila.a. There were 15 SNVs, 13 insertions, 25 deletions and 2 MSIs on the chromosome 2L; b. The distribution and rate of mutations on the chromosome 2L; c. There were 15 SNVs including A > C(1/15), A > G(1/15), C > A(2/15), C > T(2/15), G > A(1/15), G > C(1/15), G > T(1/15), T > A(1/15), T > C(3/15), T > G(2/15) on the chromosome 2L; d. There were 1 SNV, 5 Indels located on the exon, 3 SNVS, 5 Indels located on the intron, 11 SNVS, 28 Indels and 2 MSIs located on the intergenic region of the chromosome 2L; e. The mutations on the exon region were located on the CDS and UTR region;
f. The gene type that affected by mutations included 1 lncRNA and 13 mRNAs.
Mutations detected on chromosome 2R were exclusively SNVs and indels, with MSI mutations absent. Specifically, SNVs made up 24.59% of the total mutations, while deletions were the most prevalent type of indel, accounting for 55.74% of all mutations (Fig. 6a). On chromosome 2R, mutations did not exhibit significant clustering, but were instead concentrated primarily within the first 250,000 bp of the chromosome. The majority of SNVs had mutation rates below 40%, while the average mutation rate of indels was significantly elevated compared to SNV mutation rates (Fig. 6b). Of all the SNVs, the C > T point mutation accounted for 20.00% (Fig. 6c). Similar to chromosome 2L, mutations in intergenic regions were more prevalent on chromosome 2R compared to intronic regions, with no mutations observed in exon regions (Fig. 6d). Consistently, the majority of mutations were located on the mRNA-coding sequence (Fig. 6e), rather than within exons.
Fig. 6.
The mutations on the chromosome 2R of CRISPR/Cas9 edited Drosophila. a. There were 15 SNVs,12 insertions and 34 deletions on the chromosome 2R; b. The distribution and mutation rate of mutations on the chromosome 2R; c. There were 15 SNVs including A > C(2/15), A > G(1/15), A > T(1/15), C > A(2/15), C > G(1/15), C > T(3/15), G > C(2/15), G > T(1/15), T > A(1/15), T > C(1/15) on the chromosome 2R; d. There were no mutations located on the exon, 5 SNVS and 10 indels located on the intron, 10 SNVS and 29 indels located on the intergenic region of the chromosome 2R. e. The gene type that affected by mutations included 1 lncRNA, 2 asRNA and 19 mRNAs.
4. Discussion
The widespread application of CRISPR/Cas9 gene editing systems across various fields, including agriculture, biology, and particularly clinical gene therapy, has led to a significant increase in the scrutiny of associated biosafety and side effects [4,39,40]. Currently, there are successful cases of CRISPR/Cas9 gene editing being applied in clinical therapy to treat diseases. In 2016, Professor Lu from the West China Hospital in China completed the world's first human injection of gene-edited cells. Lu and his team extracted T-cells from the blood of NSCLC patients, knocked out the PD-1 gene using CRISPR/Cas9 technology, and then reinfused the edited T-cells into the patient's bloodstream. After four years of evaluation, it was found that CRISPR/Cas9 had improved the patient's survival period, confirming the safety and effectiveness of CAS9 in human applications [41]. In another study, Corbacioglu et al. targeted and knocked out the BCL11A erythroid-specific enhancer using Cas9, which is a transcriptional factor that suppresses the expression of erythroid γ-globin and fetal hemoglobin. Two patients, one with transfusion-dependent β-thalassemia (TDT) and the other with sickle cell disease (SCD), were effectively cured after receiving treatment for one year [42].
However, as CRISPR/Cas9 has become widely used in clinical treatments and scientific research, there have also been concerns raised about its safety. Bao et al. revealed that CRISPR-Cas9 genome editing can induce large gene modifications, such as deletions, insertions, and complex local rearrangements in different primary cells and cell lines [43]. Previous studies have established the influence of CRISPR/Cas9 gene editing in higher animals, including humans [5], orangutans [44], and mice [22]. However, few studies have delved into whether CRISPR/Cas9 has any adverse effects on lower animals. Herein, we present the first systematic analysis of the CRISPR/Cas9 gene editing impact on the Drosophila genome. By performing whole genome sequencing after successfully knocking out WHTIE gene in Drosophila genome via CRISPR/Cas9, we identified a significant number of mutations induced during the process. These mutations included 630 SNVs, 525 indels and 425 MSIs. Given the relatively small size of the Drosophila genome, totaling approximately 180 Mb, compared to the much larger genomes of humans or mice, the identification of unexpected mutations within the Drosophila genome was unexpected.
According to the data of previous studies, the spontaneous mutation rate in Drosophila genome is approximately 3 × 10−9 [[45], [46]]. In contrast, the spontaneous mutation rate in human is about 1.61 ± 0.13 × 10−8 [47], of mice is about 5.4 × 10−9 [48]. Therefore, we firmly believe that spontaneous mutations within the genome will not influence our research. In addition, we have meticulously compared the sequence data with the reference genome of Drosophila, thus ruling out any interference from known SNPs. Furthermore, it is imperative to eliminate any potential interference from off-target sites of the sgRNA in our mutational analysis. Fortunately, the sgRNA we have utilized to target the WHITE gene has undergone rigorous validation in numerous previous studies, and no off-target sites have been identified [37,49]. Moreover, our analysis revealed that none of the 1771 predicted off-target sites were present among the genome-wide mutations we identified. Taken together, we confidently assert that all the mutations we detected are independent of SNP, off-target or spontaneous mutations.
Previous studies have shown that the CRISPR/Cas9 gene editing system could have adverse effects on the stability of genome. For example, sgRNA transfected in mice could lead to significant deletions and intricate genomic rearrangements proximal to the target site [31]. It has been found that about half of DNA breaks persist as unrepaired after treatment, leading to the loss of several chromosome arms [50]. This indicates that the repair mechanisms for DNA breaks are not always effective, resulting in genomic instability. In addition, it has been revealed that Cas9 can directly induce DNA double-strand breaks and genomic instability in human cells lines [51]. These breaks can lead to structural defects within the nucleus, including the formation of micronuclei and chromosome bridges. These structural defects, in turn, can initiate a mutational process known as chromothripsis [52]. There are two main factors that influence CRISPR/Cas9 gene editing, leading to off-target effects and mutations [53,54]. The first is the misrecognition of PAM sequences by the Cas9 protein. PAM sequences, which come in multiple patterns such as "NAG" and "NGA", can be erroneously recognized by the CRISPR/Cas9 system, leading to varying cleavage efficiencies depending on the PAM pattern [55]. The second factor is a mismatch between the sgRNA and the target gene sequence. Studies have shown that sgRNAs exhibit a degree of fault tolerance, accommodating up to 1–5 base mismatches. Excessive mismatches could be responsible for mutations such as insertions or deletions [56]. However, recent studies have demonstrated that the mutation rate of SNV caused by Cas9 is statistically indistinguishable from the probability of SNVs arising from other reasons [57]. We have also discovered that almost all observed SNVs occurred in intronic and intergenic regions of genes, which exert far less effect on gene expression and function. Additionally, several studies have demonstrated that Cas9 gene editing in human embryos and mice could lead to large chromosomal deletions [22,31]. However, our analysis did not reveal any abnormalities at the chromosomal level within the Drosophila genome. Similarly, we found that indel mutations mostly occurred in introns and intergenic regions, which would have minimal effects on gene function as well.
Microsatellite instability is a key indicator of genomic stability and chromatin mismatch repair capacity. Our previous studies have discovered a high occurrence of MSI in CRISPR/Cas9 gene-edited mice when detected by hotspot MSIs loci or KO gene-linked loci [58,59]. Meanwhile, we compared CRISPR/Cas9 gene editing with traditional gene targeting techniques and found that mice edited with CRISPR/Cas9 exhibited a significantly higher incidence of MSI [59]. In the present study, the MSIs identified in Drosophila are primarily concentrated on the 3R chromosome, in contrast to the randomized distribution observed in mice. Furthermore, our recent findings indicate that there is rarely any unexpected mutation in CRISPR/Cas9 edited pigs, whether it be SNV, Indel, or MSI (data not shown). Therefore, we speculate that animals with a higher level of evolution would exhibit greater stability in their genomes during the process of DNA damage repair.
5. Conclusion
Collectively, our study described the effect of the CRISPR/Cas9 gene editing system on the stability of Drosophila genome for the first time. Through a thorough analysis of mutations including SNV, indel, and MSI, our study revealed numerous mutations in the genome of CRISPR/Cas9 edited Drosophila. Fortunately, the majority of mutations occurred in the non-coding region, ensuring that the structure and stability of the chromatin remained uncompromised. This study provides a novel insight into the application of the CRISPR/Cas9 gene editing system in lower animals.
Funding
This work was supported by National Natural Science Foundation of China (No. 31970512).
Ethical Statement
Animal experiments has been accepted by the animal welfare ethics revies of Capital Medical University (No. AEEI-2020-001).
Data availability
The raw data of Drosophila genome sequence has already been uploaded to the SRA database (SRA: PRJNA1083262).
CRediT authorship contribution statement
Zhu Xiao: Writing – original draft, Visualization. Wu Ying: Validation. Zhang Xing: Software, Methodology. Li Zhihui: Software, Methodology. Zhang Qiuyu: Visualization. Hu Caijiao: Validation. Li Changlong: Methodology, Conceptualization. Hanping Shi: Writing – original draft. Li Deng: Writing – review & editing. Chen Zhenwen: Validation, Methodology, Conceptualization. Ni Jianquan: Resources, Conceptualization. Huo Xueyun: Validation, Methodology, Conceptualization. Du Xiaoyan: Writing – review & editing, Writing – original draft.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Xiaoyan Du reports financial support was provided by National Natural Science Foundation of China. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank native English speakers for language editing and review services.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29061.
Contributor Information
Huo Xueyun, Email: huoxueyun@126.com.
Du Xiaoyan, Email: duduyan@ccmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Khalil A.M. The genome editing revolution: review. J. Genet. Eng. Biotechnol. 2020;18:68. doi: 10.1186/s43141-020-00078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., E P.R.I., Lin S., Kiani S., Guzman C.D., Wiegand D.J., Ter-Ovanesyan D., Braff J.L., Davidsohn N., Housden B.E., Perrimon N., Weiss R., Aach J., Collins J.J., Church G.M. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong L., Huang Y., He J., Yang N., Xu B., Ma Y., Liu J., Tang C., Luo C., Wu P., Lai Z., Huo Y., Lu T., Huang D., Gong W., Gan L., Luo Y., Zhang Z., Liu X., Zhao Y. Generation of in situ CRISPR-mediated primary and metastatic cancer from monkey liver. Signal Transduct Target Ther. 2021;6:411. doi: 10.1038/s41392-021-00799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remmel A. CRISPR-based gene therapy dampens pain in mice. Nature. 2021;591:359. doi: 10.1038/d41586-021-00644-5. [DOI] [PubMed] [Google Scholar]
- 9.Krohn P., Rega L.R., Harvent M., Festa B.P., Taranta A., Luciani A., Dewulf J., Cremonesi A., Camassei F.D., Hanson J.V.M., Gerth-Kahlert C., Emma F., Berquez M., Devuyst O. Multisystem involvement, defective lysosomes and impaired autophagy in a novel rat model of nephropathic cystinosis. Hum. Mol. Genet. 2022;31:2262–2278. doi: 10.1093/hmg/ddac033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anton-Galindo E., Dalla Vecchia E., Orlandi J.G., Castro G., Gualda E.J., Young A.M.J., Guasch-Piqueras M., Arenas C., Herrera-Ubeda C., Garcia-Fernandez J., Aguado F., Loza-Alvarez P., Cormand B., Norton W.H.J., Fernandez-Castillo N. Deficiency of the ywhaz gene, involved in neurodevelopmental disorders, alters brain activity and behaviour in zebrafish. Mol Psychiatry. 2022;27:3739–3748. doi: 10.1038/s41380-022-01577-9. [DOI] [PubMed] [Google Scholar]
- 11.Ullastres A., Merenciano M., Gonzalez J. Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol. 2021;22:265. doi: 10.1186/s13059-021-02471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei S., Chen H., Dzakah E.E., Yu B., Wang X., Fu T., Li J., Liu L., Fang S., Liu W., Shan G. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol. 2019;20:7. doi: 10.1186/s13059-018-1619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant Breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 14.Jakociunas T., Jensen M.K., Keasling J.D. CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 2016;34:44–59. doi: 10.1016/j.ymben.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Wright A.V., Nunez J.K., Doudna J.A. Biology and applications of CRISPR systems: Harnessing Nature's Toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Read.) 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 17.Mojica F.J.M., Diez-Villasenor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Read.) 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Javed M.R., Sadaf M., Ahmed T., Jamil A., Nawaz M., Abbas H., Ijaz A. CRISPR-cas system: History and prospects as a genome editing tool in Microorganisms. Curr. Microbiol. 2018;75:1675–1683. doi: 10.1007/s00284-018-1547-4. [DOI] [PubMed] [Google Scholar]
- 19.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolukbasi M.F., Gupta A., Wolfe S.A. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat. Methods. 2016;13:41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- 21.Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C., Randhawa R., Kulkarni T., Yang Z., McAllister G., Russ C., Reece-Hoyes J., Forrester W., Hoffman G.R., Dolmetsch R., Kaykas A. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., Kiezun A., Hammerman P.S., McKenna A., Drier Y., Zou L., Ramos A.H., Pugh T.J., Stransky N., Helman E., Kim J., Sougnez C., Ambrogio L., Nickerson E., Shefler E., Cortes M.L., Auclair D., Saksena G., Voet D., Noble M., DiCara D., Lin P., Lichtenstein L., Heiman D.I., Fennell T., Imielinski M., Hernandez B., Hodis E., Baca S., Dulak A.M., Lohr J., Landau D.A., Wu C.J., Melendez-Zajgla J., Hidalgo-Miranda A., Koren A., McCarroll S.A., Mora J., Crompton B., Onofrio R., Parkin M., Winckler W., Ardlie K., Gabriel S.B., Roberts C.W.M., Biegel J.A., Stegmaier K., Bass A.J., Garraway L.A., Meyerson M., Golub T.R., Gordenin D.A., Sunyaev S., Lander E.S., Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R., Desmedt C., Eils R., Eyfjord J.E., Foekens J.A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jager N., Jones D.T., Jones D., Knappskog S., Kool M., Lakhani S.R., Lopez-Otin C., Martin S., Munshi N.C., Nakamura H., Northcott P.A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J.V., Puente X.S., Raine K., Ramakrishna M., Richardson A.L., Richter J., Rosenstiel P., Schlesner M., Schumacher T.N., Span P.N., Teague J.W., Totoki Y., Tutt A.N., Valdes-Mas R., van Buuren M.M., van 't Veer L., Vincent-Salomon A., Waddell N., Yates L.R., Consortium I.B.C., Consortium I.M.-S., PedBrain I., Zucman-Rossi J., Futreal P.A., McDermott U., Lichter P., Meyerson M., Grimmond S.M., Siebert R., Campo E., Shibata T., Pfister S.M., Campbell P.J., Stratton M.R. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q., Qi Y., Wang S., Liu J., Geng P., Zhou Q., Zhang W., Cai J., Hu B., Dai D., Li H. vol. 13. Thorac Cancer; 2022. pp. 853–857. (Polymorphic Mutations in the Polb Gene Promoter and Their Impact on Transcriptional Activity). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell R.J., Rube H.T., Xavier-Magalhaes A., Costa B.M., Mancini A., Song J.S., Costello J.F. Understanding TERT promoter mutations: a common path to immortality. Mol. Cancer Res. 2016;14:315–323. doi: 10.1158/1541-7786.MCR-16-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robichaux J.P., Le X., Vijayan R.S.K., Hicks J.K., Heeke S., Elamin Y.Y., Lin H.Y., Udagawa H., Skoulidis F., Tran H., Varghese S., He J., Zhang F., Nilsson M.B., Hu L., Poteete A., Rinsurongkawong W., Zhang X., Ren C., Liu X., Hong L., Zhang J., Diao L., Madison R., Schrock A.B., Saam J., Raymond V., Fang B., Wang J., Ha M.J., Cross J.B., Gray J.E., Heymach J.V. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature. 2021;597:732–737. doi: 10.1038/s41586-021-03898-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennig S.L., Owen J.R., Lin J.C., Young A.E., Ross P.J., Van Eenennaam A.L., Murray J.D. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoijer I., Emmanouilidou A., Ostlund R., van Schendel R., Bozorgpana S., Tijsterman M., Feuk L., Gyllensten U., den Hoed M., Ameur A. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 2022;13:627. doi: 10.1038/s41467-022-28244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V., Bibeyran A., Lalanne M., Prouzet-Mauleon V., Turcq B., Ged C., Blouin J.M., Richard E., Dabernat S., Moreau-Gaudry F., Bedel A. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019;10:1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha S., Barbosa K., Cheng K., Leiserson M.D.M., Jain P., Deshpande A., Wilson D.M., 3rd, Ryan B.M., Luo J., Ronai Z.A., Lee J.S., Deshpande A.J., Ruppin E. A systematic genome-wide mapping of oncogenic mutation selection during CRISPR-Cas9 genome editing. Nat. Commun. 2021;12:6512. doi: 10.1038/s41467-021-26788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celniker S.E., Rubin G.M. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 34.Yan Y., Ziemek J., Schetelig M.F. CRISPR/Cas9 mediated disruption of the white gene leads to pigmentation deficiency and copulation failure in Drosophila suzukii. J. Insect Physiol. 2020;126 doi: 10.1016/j.jinsphys.2020.104091. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki A., Nishimura T., Takano T., Naito S., Yoo S.K. White regulates proliferative homeostasis of intestinal stem cells during ageing in Drosophila. Nat. Metab. 2021;3:546–557. doi: 10.1038/s42255-021-00375-x. [DOI] [PubMed] [Google Scholar]
- 36.Ren X., Sun J., Housden B.E., Hu Y., Roesel C., Lin S., Liu L.P., Yang Z., Mao D., Sun L., Wu Q., Ji J.Y., Xi J., Mohr S.E., Xu J., Perrimon N., Ni J.Q. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q., Deng P., Liu L.P., Ji J.Y., Li J.B., Ni J.Q. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H.Y., Wang C., Lee H.K., Yoo K.H., Zeng X., Kuhns T., Yang C.M., Mohr T., Liu C., Hennighausen L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017;8 doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai S.Q., Joung J.K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao A., Burritt D.J., Chen H., Zhou X., Cao D., Tran L.P. The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 2019;39:321–336. doi: 10.1080/07388551.2018.1554621. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., Shen H., Tong R., Wang W., Li L., Song J., Li J., Su X., Ding Z., Gong Y., Zhu J., Wang Y., Zou B., Zhang Y., Li Y., Zhou L., Liu Y., Yu M., Wang Y., Zhang X., Yin L., Xia X., Zeng Y., Zhou Q., Ying B., Chen C., Wei Y., Li W., Mok T. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 42.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., Ho T.W., Kattamis A., Kernytsky A., Lekstrom-Himes J., Li A.M., Locatelli F., Mapara M.Y., de Montalembert M., Rondelli D., Sharma A., Sheth S., Soni S., Steinberg M.H., Wall D., Yen A., Corbacioglu S. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 43.Park S.H., Cao M., Pan Y., Davis T.H., Saxena L., Deshmukh H., Fu Y., Treangen T., Sheehan V.A., Bao G. Comprehensive analysis and accurate quantification of unintended large gene modifications induced by CRISPR-Cas9 gene editing. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W., Li S., Li X.J. A CRISPR monkey model unravels a unique function of PINK1 in primate brains. Mol. Neurodegener. 2019;14:17. doi: 10.1186/s13024-019-0321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keightley P.D., Trivedi U., Thomson M., Oliver F., Kumar S., Blaxter M.L. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keightley P.D., Ness R.W., Halligan D.L., Haddrill P.R. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics. 2014;196:313–320. doi: 10.1534/genetics.113.158758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipson M., Loh P.R., Sankararaman S., Patterson N., Berger B., Reich D. Calibrating the human mutation rate via ancestral recombination density in diploid genomes. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohno M. Spontaneous de novo germline mutations in humans and mice: rates, spectra, causes and consequences. Genes Genet. Syst. 2019;94:13–22. doi: 10.1266/ggs.18-00015. [DOI] [PubMed] [Google Scholar]
- 49.Xu R.G., Wang X., Shen D., Sun J., Qiao H.H., Wang F., Liu L.P., Ni J.Q. Perspectives on gene expression regulation techniques in Drosophila. J Genet Genomics. 2019;46:213–220. doi: 10.1016/j.jgg.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Zuccaro M.V., Xu J., Mitchell C., Marin D., Zimmerman R., Rana B., Weinstein E., King R.T., Palmerola K.L., Smith M.E., Tsang S.H., Goland R., Jasin M., Lobo R., Treff N., Egli D. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell. 2020;183:1650–1664 e15. doi: 10.1016/j.cell.2020.10.025. [DOI] [PubMed] [Google Scholar]
- 51.Xu S., Kim J., Tang Q., Chen Q., Liu J., Xu Y., Fu X. CAS9 is a genome mutator by directly disrupting DNA-PK dependent DNA repair pathway. Protein Cell. 2020;11:352–365. doi: 10.1007/s13238-020-00699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leibowitz M.L., Papathanasiou S., Doerfler P.A., Blaine L.J., Sun L., Yao Y., Zhang C.Z., Weiss M.J., Pellman D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021;53:895–905. doi: 10.1038/s41588-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H.W., Listgarten J., Root D.E. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iyer V., Boroviak K., Thomas M., Doe B., Riva L., Ryder E., Adams D.J. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huo X., Du Y., Lu J., Guo M., Li Z., Zhang S., Li X., Chen Z., Du X. Analysis of microsatellite instability in CRISPR/Cas9 editing mice. Mutat. Res. 2017;797–799:1–6. doi: 10.1016/j.mrfmmm.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 59.Huo X., Zhang X., Liu Y., Sun Y., Ren Y., Li C., Du X., Chen Z. Instability of microsatellites linked to targeted genes in CRISPR/Cas9-edited and traditional gene knockout mouse strains. J Genet Genomics. 2018;45:553–556. doi: 10.1016/j.jgg.2018.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of Drosophila genome sequence has already been uploaded to the SRA database (SRA: PRJNA1083262).