Abstract
Macrophage cholesterol homeostasis is crucial for health and disease and has been linked to the lipid-peroxidizing enzyme arachidonate 15-lipoxygenase type B (ALOX15B), albeit molecular mechanisms remain obscure. We performed global transcriptome and immunofluorescence analysis in ALOX15B-silenced primary human macrophages and observed a reduction of nuclear sterol regulatory element-binding protein (SREBP) 2, the master transcription factor of cellular cholesterol biosynthesis. Consequently, SREBP2-target gene expression was reduced as were the sterol biosynthetic intermediates desmosterol and lathosterol as well as 25- and 27-hydroxycholesterol. Mechanistically, suppression of ALOX15B reduced lipid peroxidation in primary human macrophages and thereby attenuated activation of mitogen-activated protein kinase ERK1/2, which lowered SREBP2 abundance and activity. Low nuclear SREBP2 rendered both, ALOX15B-silenced and ERK1/2-inhibited macrophages refractory to SREBP2 activation upon blocking the NPC intracellular cholesterol transporter 1. These studies suggest a regulatory mechanism controlling macrophage cholesterol homeostasis based on ALOX15B-mediated lipid peroxidation and concomitant ERK1/2 activation.
Keywords: Arachidonate 15-lipoxygenase type B, 15-LO2, Sterol regulatory element-binding protein 2, Lipid peroxidation, Reactive oxygen species, MAPK
Graphical abstract
Highlights
-
•
Suppressing macrophage ALOX15B extensively reduced SREBP2-target genes.
-
•
Silencing ALOX15B decreased sterol intermediates and oxysterols in macrophages.
-
•
ALOX15B-suppression lowered macrophage lipid peroxidation (LPO) and oxylipins.
-
•
LPO controls macrophage ERK1/2 activation and SREBP2-target gene expression.
-
•
Macrophage cholesterol homeostasis is regulated by an ALOX15B–LPO–ERK1/2–SREBP2 axis.
Abbreviations:
- ALOX15
arachidonate 15-lipoxygenase type A
- ALOX15B
arachidonate 15-lipoxygenase type B
- CYP51A1
cytochrome P450 family 51 subfamily A member 1
- DHCR24
24-dehydrocholesterol reductase
- DMNQ
2,3-dimethoxy-1,4-naphthoquinone
- ERK1
MAPK3
- ERK2
MAPK1
- HMGCS1
3-hydroxy-3-methylglutaryl-CoA synthase 1
- JNK
Jun N-terminal kinase
- KD
knockdown
- LDLR
low density lipoprotein receptor
- LOX
lipoxygenase
- LXR
liver X receptor
- MAPK
mitogen-activated protein (MAP) kinase
- MSMO1
methylsterol monooxygenase 1
- MVK
mevalonate kinase
- NPC1
NPC intracellular cholesterol transporter 1
- PUFA
polyunsaturated fatty acid
- qPCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
- RSL3
RAS-selective lethal 3
- SCAP
SREBP-cleavage activating protein
- SREBP1
sterol regulatory element-binding protein 1
- SREBP2
sterol regulatory element-binding protein 2
1. Introduction
Peroxidation of polyunsaturated fatty acids (PUFAs) is initiated either non-enzymatically by free radicals [1] or enzymatically by lipoxygenases (LOXs) [2]. LOXs are a heterogeneous family of non-heme iron containing dioxygenases, which catalyze the stereo-specific peroxidation of PUFAs to their corresponding hydroperoxyl derivatives through insertion of molecular oxygen at the (1Z, 4Z) pentadiene moiety [3]. The human genome encodes six functional LOX genes, including two 15-LOXs that oxygenate arachidonic acid at carbon atom 15, classified as arachidonate 15-lipoxygenase (ALOX15) and arachidonate 15-lipoxygenase type B (ALOX15B) [4]. Whereas ALOX15 is absent in unstimulated primary human macrophages and its induction is strictly dependent on T helper cell type 2 cytokines interleukin 4 and/or interleukin 13, ALOX15B is constitutively expressed at both messenger RNA and protein levels [5]. Although the biological function of ALOX15B has not been fully characterized, accumulating data indicate a role for ALOX15B in cellular lipid homeostasis. In primary human macrophages, ALOX15B-depletion reduced lipid accumulation and inflammation in atherosclerosis [6]. Furthermore, suppressing macrophage ALOX15B limited signaling of sterol regulatory element-binding protein (SREBP) 2, the master transcription factor of endogenous cholesterol biosynthesis [5].
Cholesterol is an essential component of mammalian cell membranes [7] and serves as a precursor for oxysterols, bile acids, and steroid hormones [8]. Cellular cholesterol homeostasis is tightly controlled through a combination of synthesis, uptake from lipoproteins, storage, and efflux pathways [9]. SREBP2 is synthesized as an endoplasmic reticulum (ER)-anchored precursor. To become active, SREBP2 must translocate to the Golgi apparatus in a process regulated by ER membrane cholesterol content and SREBP-cleavage activating protein (SCAP). When ER membrane cholesterol is depleted, SCAP escorts SREBP2 in coat protein complex II (COPII) vesicles to the Golgi for proteolytic activation [10,11]. Conversely, when ER cholesterol levels rise, excess cholesterol binds to SCAP to prevent the recruitment of SCAP-SREBP2 complexes into COPII vesicles. However, not only cholesterol but also biosynthetic intermediates, including desmosterol and cholesterol-derived oxysterols such as 25-hydroxycholesterol, control translocation of the SCAP-SREBP2 complex by binding to SCAP [12], or the SCAP interaction partner insulin-induced gene (INSIG) 1 [13,14].
Processed SREBP2 enters the nucleus where it binds sterol regulatory element (SRE) sequences in the promotors of target genes to upregulate their transcription [15,16]. Preventing SREBP2 cleavage stalls transcriptional activation of cholesterol related genes, including those facilitating cholesterol uptake, to maintain homeostasis [17]. SREBP2-dependent cholesterol biosynthesis is controlled by feedback regulation via sterols, and post-transcriptional regulation of SREBP2, including phosphorylation [18,19], sumoylation [20] as well as acetylation [21].
We provide evidence that suppression of ALOX15B in primary human macrophages reduces SREBP2-dependent gene expression and lowers the amount of sterol intermediates and oxysterols by limiting mitogen-activated protein kinase (MAPK) ERK1 and ERK2 (MAPK3 and MAPK1) activation due to reduced lipid peroxidation. This highlights a so far unappreciated role of lipid peroxidation, ERK1/2 activation and SREBP2 signaling in primary human macrophages and links this signaling axis to ALOX15B activity.
2. Results
2.1. Silencing ALOX15B in macrophages lowered SREBP2-dependent gene expression
Previous studies indicated that ALOX15B suppression disrupts SREBP2-mediated cholesterol homeostasis in macrophages [5]. To explore underlying molecular mechanisms how ALOX15B regulates SREBP2-dependent cholesterol biosynthesis we performed RNA sequencing (RNA-seq) of primary human macrophages, transiently transfected with control or ALOX15B-targeted small-interfering RNA (siRNA).
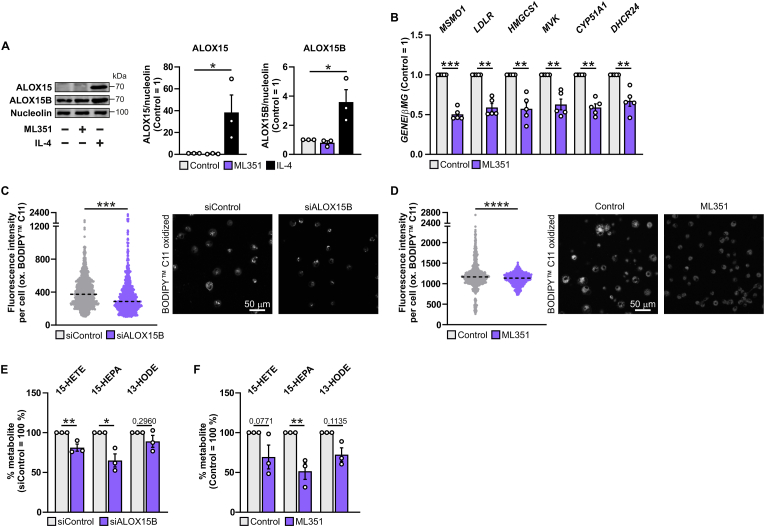
Knockdown (KD) of ALOX15B markedly reduced ALOX15B gene and protein expression as determined by quantitative polymerase-chain reaction (qPCR) and Western analysis (SI Fig. 1A and B). Silencing ALOX15B followed by global RNA-seq analysis revealed 711 downregulated genes defined by a log2 fold change ≤ −0.58 and 416 upregulated genes defined by a log2 fold change ≥0.58 (Fig. 1A). Gene set enrichment analysis disclosed that ALOX15B KD macrophages exhibit transcriptional changes related to several metabolic and homeostatic pathways including cholesterol homeostasis and steroid biosynthesis (Fig. 1B–SI Fig. 1C). Analysis of differentially expressed genes indicated that SREBF2, the gene encoding SREBP2, was among the most highly downregulated in ALOX15B KD macrophages (Fig. 1C). In line with its function as a master transcriptional regulator of cholesterol biosynthesis, several SREBP2-dependent genes [15] were also significantly downregulated in ALOX15B KD cells (Fig. 1C), which was confirmed using qPCR and Western analysis (Fig. 1D–SI Fig. 1D). Highlighted in blue, Fig. 1E illustrates the abundance of genes involved in cholesterol biosynthesis that are significantly downregulated [adjusted p-value (padj) ≤ 0.05] in ALOX15B KD macrophages.
Fig. 1.
Silencing ALOX15B in primary human macrophages reduces SREBP2-dependent gene expression and nuclear SREBP2 protein
(A) Global heatmap showing all downregulated genes in ALOX15B KD macrophages with a log2FC ≤ −0.58 and all upregulated genes with a log2FC ≥ 0.58. (B) Enrichment analysis of RNA-seq data comparing control to ALOX15B KD macrophages using Kyoto Encyclopedia of Genes and Genomes (KEGG) and REACTOME gene sets. (C) Heat map displaying differentially expressed SREBP2-target genes in control and ALOX15B KD cells as count-per-million (CPM) with annotated log2FC and p-adjusted value. (D) Validation of MSMO1, LDLR, HMGCS1, MVK, CYP51A1, and DHCR24 gene expression by real-time qPCR of control and ALOX15B KD macrophages. (E) Schematic representation of the Bloch and Kandutsch-Russell cholesterol biosynthesis pathways. Broken arrows indicate potential crossover sites. Significantly downregulated genes in ALOX15B-silenced macrophages are shown in blue [log2FC ≤ −0.58]. (F) SREBP2 immunofluorescence microscopy in control and ALOX15B KD macrophages. Densitometry of SREBP2 fluorescence signal in whole cell, nucleus and cytoplasm. Data are presented as mean ± SE from at least five independent experiments. Statistical analysis was performed using one sample t-test for D and F (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 vs siControl).
Considering that nuclear translocation of proteolytically processed SREBP2 is a prerequisite for the transcription of genes involved in cholesterol biosynthesis [16], we next determined cellular localization of SREBP2 using immunofluorescence microscopy. Consistent with reduced SREBF2 expression, we identified decreased nuclear SREBP2 protein in ALOX15B KD cells (Fig. 1F).
Although reduced SREBP2-dependent cholesterol biosynthesis is often associated with reciprocal regulation of the liver-X-receptor (LXR) transcriptional program [22], silencing of ALOX15B did not affect LXR-dependent gene expression [23] (SI Fig. 1E). Additionally, SREBF1, the gene encoding SREBP1, whose activity is regulated in the ER in a SREBP2-like manner [24], and moreover is an LXR target [25,26], was not differentially expressed nor were SREBP1 target genes [15] in ALOX15B KD macrophages (SI Fig. 1F). Conclusively, silencing ALOX15B specifically inhibits SREBP2 target gene expression without affecting the expression of LXR-dependent pathways or SREBP1 target genes.
2.2. In ALOX15B-silenced macrophages sterol intermediates and oxysterols are reduced
To determine whether altered SREBP2-dependent gene expression in ALOX15B KD macrophages evokes changes in cellular sterol content, we measured total cholesterol using gas chromatography-flame ionization detection (GC-FID) and non-cholesterol sterols by gas chromatography-mass spectrometry in selected ion monitoring mode (GC-MS-SIM). While sterol intermediates lanosterol and dihydrolanosterol remained unchanged, ALOX15B KD cells contained reduced levels of the Bloch pathway intermediate desmosterol [27] and the Kandutsch-Russell intermediate lathosterol [28,29] (Fig. 2A), both markers of active cholesterol biosynthesis [30]. ALOX15B-silencing also significantly lowered cholesterol-derived 25- and 27-hydroxycholesterol (Fig. 2B). Analysis of total or unesterified cholesterol in ALOX15B KD macrophages revealed only minor changes (Fig. 2C and D).
Fig. 2.
ALOX15B-silenced macrophages contain reduced levels of sterol intermediates and oxysterols but not cholesterol
(A) Sterol intermediates, (B) cholesterol-derived oxysterols, and (C) total cholesterol in control and ALOX15B KD macrophages measured using GC-MS-SIM for A and B or GC-FID for C, respectively. (D) Free and esterified cholesterol and (E) phytosterols in control and ALOX15B KD macrophages. (F) Flow cytometry determined median fluorescence intensity of uptaken green-fluorescent LDL (10 μg/ml) in control and ALOX15B KD macrophages. (G) Real-time qPCR analysis of MSMO1, HMGCS1, CYP51A1, and DHCR24 gene expression in control and ALOX15B KD cells with or without addition of NPC1 inhibitor U18666A (5 μM) for 24 h. Data are presented as mean ± SE from at least four independent experiments. Statistical analysis was performed using one sample t-test for A-B (*P < 0.05, **P < 0.01, and ****P < 0.0001 vs siControl. ns = not significant), two-tailed student's t-test for C–F (p-values are indicated. ns = not significant), and one-way ANOVA with Sidak's multiple comparisons test for G (**P < 0.01 and ****P < 0.0001 vs siControl).
Since ALOX15B-silenced macrophages contained lower levels of desmosterol and lathosterol as well as cholesterol-derived oxysterols we questioned whether cholesterol levels in ALOX15B KD macrophages reflect altered uptake of extracellular sterols through either receptor- or non-receptor-mediated processes [31]. To follow sterol uptake, we measured cellular accumulation of exogenous phytosterols, ubiquitously found in cell culture media [32] as well as fluorescently labeled low density lipoprotein (LDL). Analysis of intracellular campesterol, sitosterol, brassicasterol, and stigmasterol levels measured by GC-MS-SIM revealed no difference in phytosterol accumulation between control and ALOX15B KD macrophages (Fig. 2E). In line, ALOX15B-silenced macrophages did not accumulate LDL compared to control macrophages. They rather exhibited a trend towards reduced LDL uptake (Fig. 2F).
We next examined how ALOX15B-silenced macrophages respond to disruption of intracellular cholesterol transport since nuclear translocation of SREBP2 is tightly regulated by the ER cholesterol amount [17]. We used the cationic amphiphile U18666A to block cholesterol transport from lysosomes by inhibiting the NPC intracellular cholesterol transporter 1 (NPC1) [33]. Comparing SREBP2-dependent gene expression in control and ALOX15B-silenced macrophages revealed that U18666A increased expression of SREBP2-dependent genes methylsterol monooxygenase 1 (MSMO1), 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), cytochrome P450 family 51 subfamily A member 1 (CYP51A1), and 24-dehydrocholesterol reductase (DHCR24) in control cells. However, U18666A failed to increase expression of these genes in ALOX15B KD cells (Fig. 2G).
It appears that ALOX15B KD macrophages exhibited significantly reduced SREBP2-dependent gene expression and sterol intermediates, while total cellular cholesterol levels remained unchanged. These data suggest a sterol-independent impairment of SREBP2 activation in ALOX15B-silenced cells, as these cells were unable to induce SREBP2-target gene expression after blocking NPC1.
2.3. Suppressing ALOX15B reduced lipid peroxidation and 15-LOX-derived oxylipins
To investigate whether inhibition of 15-LOX using a small molecule inhibitor recapitulates altered SREBP2-dependent gene expression seen in ALOX15B-silenced macrophages, we used the ALOX15 inhibitor ML35148 (ML351) [34]. To verify the presence of ALOX15B, we performed Western analysis of ALOX15 and ALOX15B and confirmed constitutive expression of ALOX15B but the absence of ALOX15 in unstimulated and inhibitor-treated macrophages. Moreover, we corroborated induction of ALOX15 following 48 h stimulation with interleukin 4 [5] (Fig. 3A). Therefore, in resting macrophages ML351 selectively targets ALOX15B due to the absence of ALOX15.
Fig. 3.
ALOX15B KD or inhibition by ML351 reduce cellular lipid peroxidation, oxylipins and SREBP2-target gene expression
(A) Representative Western analysis and densitometry of ALOX15 and ALOX15B in macrophages treated with ML351 (10 μM) for 24 h or interleukin 4 (IL-4) (20 ng/ml) for 48 h. (B) Validation of MSMO1, LDLR, HMGCS1, MVK, CYP51A1, and DHCR24 gene expression by real-time qPCR of macrophages treated with DMSO and ML351 (10 μM) for 24 h. Confocal microscopy of BODIPY™ 581/591 C11 lipid peroxidation sensor in (C) control and ALOX15B KD macrophages as well as (D) macrophages treated for 30 min with DMSO or ML351 (10 μM). Graphs depict oxidized BODIPY C11 fluorescence intensity per cell including median. Formation of esterified 15-LOX-specific hydroxy-fatty acids in macrophages treated with (E) control and ALOX15B siRNA as well as (F) DMSO and ML351 (10 μM) for 24 h. Data are presented as mean ± SE from at least three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett's multiple comparisons test for A (*P < 0.05 vs untreated control), one sample t-test for B (**P < 0.01 and ***P < 0.001 vs DMSO control), two-tailed student's t-test for C-D (***P < 0.001 and ****P < 0.0001 vs siControl or DMSO control), and one-way ANOVA with Sidak's multiple comparisons test for E and F (*P < 0.05 and **P < 0.01 vs siControl or DMSO control).
ML351 reduced the expression of SREBP2-dependent genes MSMO1, low density lipoprotein receptor (LDLR), HMGCS1, mevalonate kinase (MVK), CYP51A1, and DHCR24 (Fig. 3B) and thus, recapitulates the situation seen in ALOX15B KD macrophages.
To obtain data on the enzymatic activity of ALOX15B in ALOX15B-silenced and ML351-treated macrophages we measured lipid peroxidation using confocal microscopy. In ALOX15B-silenced (Fig. 3C) and ML351-treated macrophages (Fig. 3D) lipid peroxidation, determined using BODIPY 581/591C11, was significantly decreased. In addition, lipid hydroxide analysis using liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) revealed reduced levels of 15-LOX specific oxylipins [35] 15-hydroxyeicosatetraenoic acid (HETE) and 15-hydroxyeicosapentaenoic acid (HEPA) and a modest reduction of 13-hydroxyoctadecadienoic acid (HODE) in ALOX15B-silenced (Fig. 3E) and ML351-treated macrophages (Fig. 3F). Obviously, inhibition of ALOX15B by ML351 reflects results seen in ALOX15B-silenced macrophages.
2.4. Lipid peroxidation increased SREBP2-target gene expression in human macrophages
ALOX15B-silenced macrophages exhibit reduced SREBP2-target gene expression and lower lipid peroxidation. To investigate the relationship between SREBP2-target genes and lipid peroxidation we used RAS-selective lethal (RSL) 3 to inhibit glutathione peroxidase 4 (GPX4) [36], which prevents accumulation of lipid peroxides by reducing them to the respective alcohols [36,37].
In naïve macrophages, lipid peroxides accumulated following treatment with RSL3 for 6 h (SI Fig. 2A and B) and less pronounced after 24 h (SI Fig. 2C). RSL3-treated macrophages increased SREBP2-target genes (Fig. 4A) and corresponding proteins (Fig. 4B) after 24 h, an effect not seen at early time points, i.e. 6 h (SI Fig. 2D). In addition, 15-hydroperoxyeicosatetraenoic acid (HpETE), the ALOX15B-catalyzed AA oxygenation product, increased upon treatment with RSL3 and decreased with the ALOX15B inhibitor ML351 (SI Fig. 2E). When suppressing lipid peroxide accumulation with the radical-trapping antioxidant liproxstatin-1 (SI Fig. 2F) [37,38] SREBP2-dependent genes were significantly lowered (Fig. 4C). Also, the lipophilic antioxidant α-tocopherol [39] attenuated lipid peroxidation (SI Fig. 3A) as well as SREBP2-target gene expression to a similar degree as the 15-LOX inhibitor ML351 (SI Fig. 3B). The redox cycler 2,3-dimethoxy-1,4-naphthoquinone (DMNQ) [40], which elevated reactive oxygen species (ROS) in naïve macrophages (SI Fig. 3C), increased expression of SREBP2-dependent genes, while DMNQ (SI Fig. 3D), as well as RSL3 (Fig. 4D), partially reversed SREBP2-target genes attenuated by ML351. These studies imply a link between lipid peroxidation and the expression of SREBP2-dependent genes.
Fig. 4.
Modulating lipid peroxidation by RSL3 and liproxstatin-1 affects SREBP2-dependent gene expression
(A) Gene expression of MSMO1, MVK, LDLR, CYP51A1, and DHCR24 via real-time qPCR of macrophages treated with RSL3 (10 μM) for 24 h. (B) Western analysis and corresponding densitometry of DHCR24 and CYP51A1 of macrophages treated with DMSO or RSL3 (10 μM) for 24 h. (C) Gene expression analysis of MSMO1, MVK, LDLR, CYP51A1, and DHCR24 via real-time qPCR of macrophages treated with DMSO or liproxstatin-1 (1 μM) for 6 h. (D) Analysis of SREBP2-target genes in macrophages treated with RSL3 (10 μM) and ML351 (10 μM) for 24 h. For combined treatments, macrophages were exposed to RSL3 for 24 h, while ML351 was added during the last 6 h. Data are presented as mean ± SE from at least ten independent experiments. Statistical analysis was performed using one sample t-test for A, B and C (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 vs DMSO Control) and one-way ANOVA with Tukey's multiple comparisons test for D (***P < 0.001 and ****P < 0.0001 vs RSL3).
2.5. Lipid peroxidation activated expression of SREBP2 target genes via ERK1/2
Cellular cholesterol biosynthesis is governed by a multitude of factors, including negative feedback regulation via sterol intermediates [41] or oxysterols [42], a restricted SREBP2 processing by high ER cholesterol [17] as well as post-transcriptional regulation of SREBP2 [18,20,43]. Therefore, we searched for mechanisms of how lipid peroxidation modulates SREBP2 (Fig. 5A). Gene set enrichment analysis of our RNA-seq data using REACTOME gene sets showed downregulation of genes involved in phagocyte ROS production, nuclear events facilitated by MAPK signaling, in addition to ERK1/2 targets in ALOX15B KD macrophages (Fig. 5B and SI Fig. 4A).
Fig. 5.
ALOX15B-suppressed macrophages exhibit reduced ERK1/2 activity
(A) Schematic illustration of ERK1/2 and AKT as potential signaling hubs that reduce SREBP2 activation in ALOX15B-suppressed macrophages. (B) Gene Set Enrichment Analysis blots of downregulated REACTOME pathways in ALOX15B KD macrophages, including ROS production, signaling by MAPKs as well as ERK1/2. (C) Real-time qPCR of SREBP2-target genes in macrophages treated with ML351 (15-LOX inhibitor, 10 μM), PD98059 (ERK1/2 inhibitor, 10 μM), AKTVIII (AKT inhibitor, 1 μM), and PF4708671 (ribosomal protein S6 kinase B1 inhibitor, 10 μM) for 6 h. Western analysis of total and phosphorylated ERK1 (p44 MAPK) and ERK2 (p42 MAPK) in (D) control and ALOX15B-silenced macrophages as well as (E) DMSO- and ML351-treated macrophages. (F) Western analysis of total and phosphorylated ERK1 (p44 MAPK) and ERK2 (p42 MAPK) of macrophages treated with DMSO, RSL3 (10 μM) or liproxstatin-1 (1 μM) for 24 h. Data are presented as mean ± SE from at least four independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett's multiple comparisons test for C (**P < 0.01 and ****P < 0.0001 vs DMSO control) and one sample t-test for D, E and F (*P < 0.05 and **P < 0.01 vs siControl or DMSO control).
As both, signaling of AKT serine/threonine kinase (AKT) and ERK1/2 activation are known to be redox sensitive [[44], [45], [46], [47], [48]], as well as being implicated in SREBP2 activation [[18], [19], [20],[49], [50], [51]], we analyzed their involvement in SREBP2-dependent gene expression in primary human macrophages (Fig. 5A). In addition to the mechanistic target of rapamycin (mTOR) upstream kinase AKT, also the mTOR downstream target ribosomal protein S6 kinase B1 regulates SREBP2 [43,52]. Hence, we analyzed SREBP2-dependent gene expression under conditions of blocking ERK1/2, AKT, and ribosomal protein S6 kinase B1. Inhibition of ERK1/2 by PD98059 [53] decreased SREBP2 target gene expression to a similar extent as the 15-LOX inhibitor ML351. Blocking either AKT via AKTVIII [54] or ribosomal protein S6 kinase B1 via PF4708671 [55] left SREBP2-dependent gene expression unaltered (Fig. 5C). Moreover, Western analysis showed reduced phosphorylation of ERK1/2 in ALOX15B-silenced (Fig. 5D) and 15-LOX inhibited macrophages (Fig. 5E). Although AKT phosphorylation at Ser473 and Thr308 was attenuated in ALOX15B-silenced macrophages (SI Fig. 4B), only AKT phosphorylation at Ser473 was reduced in cells incubated with ML351 (SI Fig. 4C). Considering redundancies in MAPK signaling, we additionally performed Western analysis of p38 MAPK and Jun N-terminal kinase (JNK). Whereas no change in phosphorylation of p38 MAPK was detectable with our experimental setup, JNK phosphorylation modestly increased with silencing of ALOX15B or following liproxstatin-1 treatment (SI Fig. 4D). However, JNK inhibition with SP600125 did not increase SREBP2-target gene expression (SI Fig. 4E).
To determine whether a decrease in ERK1/2 phosphorylation in ALOX15B-suppressed macrophages results from reduced lipid peroxidation, we monitored ERK1/2 phosphorylation in RSL3-and liproxstatin-1-treated cells. While liproxstatin-1 decreased ERK1 phosphorylation, RSL3 increased ERK1/2 phosphorylation in primary human macrophages (Fig. 5F), implying that ERK activity is sensitive to the cellular amount of lipid peroxides. It appears that reduced lipid peroxidation, resulting from ALOX15B suppression, decreased ERK1/2 activation, which lowered SREBP2-dependent gene expression.
2.6. Inhibiting ERK1/2 reduced nuclear SREBP2 and prevented the induction of cholesterol biosynthesis genes in response to blocking NPC1
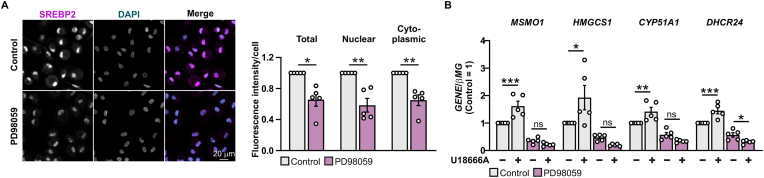
Our data suggested regulation of SREBP2-target gene expression via ERK1/2 in ALOX15B-suppressed macrophages. Thus, we analyzed cellular localization of SREBP2 protein in ERK1/2-inhibited macrophages. Inhibition of ERK1/2 lowered nuclear and cytoplasmic SREBP2 protein (Fig. 6A), which corroborates findings in ALOX15B-silenced cells (Fig. 1F). Analysis of SREBP2 target gene expression in ERK1/2-inhibited cells after disrupting intracellular cholesterol transport via NPC1 inhibition using U18666A (Fig. 6B) recapitulated results from ALOX15B-suppressed macrophages (Fig. 2G). With inhibited ERK1/2, macrophages exposed to the NPC1 inhibitor were unable to induce transcription of the SREBP2 target genes MSMO1, HMGCS1, CYP51A1, and DHCR24 in comparison to control conditions (Fig. 6B).
Fig. 6.
ERK1/2 inhibition reduces nuclear SREBP2 and prevents SREBP2-target gene expression upon NPC1 blockade
(A) SREBP2 immunofluorescence microscopy in macrophages treated with DMSO or the ERK1/2 inhibitor PD98059 (10 μM) for 3 h. Mean pixel intensity of SREBP2 fluorescence signal in whole cell, nucleus and cytoplasm. (B) Real-time qPCR analysis of MSMO1, HMGCS1, CYP51A1, and DHCR24 gene expression in macrophages treated with DMSO and the ERK1/2 inhibitor PD98059 (10 μM) in combination with or without the NPC1 inhibitor U18666A (5 μM) for 24 h. Data are presented as mean ± SE from at least four independent experiments. Statistical analysis was performed using one sample t-test for A (*P < 0.05 and **P < 0.01 vs DMSO control) and one-way ANOVA with Sidak's multiple comparisons test for B (*P < 0.05, **P < 0.01, ***P < 0.001 vs DMSO control).
3. Discussion
Previous evidence suggested that 15-lipoxygenases, i.e. ALOX15 and/or ALOX15B regulate SREBP2-dependent cholesterol homeostasis in primary human macrophages, albeit molecular mechanisms remained obscure [5]. Working in the absence of Th2 cytokines, we proved that in naïve macrophages the constitutively expressed ALOX15B accounts for these effects, because ALOX15 expression, depending on interleukin-4/interleukin-13, was absent. To explore molecular mechanisms, how ALOX15B affects SREBP2-signaling, we transfected primary human macrophages with ALOX15B-targeted siRNA and/or employed the 15-LOX inhibitor ML351. Global transcriptome analysis in ALOX15B-silenced cells revealed broad transcriptional changes with an obvious focus on SREBP2-target genes, without affecting LXR- or SREBP1-transcriptional programs. In ALOX15B-suppressed cells, nuclear SREBP2 was decreased, likewise sterol intermediates and cholesterol-derived oxysterols. Attenuating ALOX15B activity reduced lipid peroxidation, along with decreasing 15-LOX-specific oxylipins. Modulating the extent of lipid peroxidation by using RSL3 and liproxstatin-1 altered SREBP2-dependent gene expression in an ERK1/2-dependent manner.
While indicators of an active cholesterol biosynthesis [30], desmosterol and lathosterol, were lower in ALOX15B KD macrophages, reduction of total cellular cholesterol was modest. Considering that cellular cholesterol concentrations are three orders of magnitude higher than concentrations of sterol intermediates, a small, non-significant reduction in cellular cholesterol levels can be anticipated in ALOX15B-silenced macrophages. This is consistent with reports from SCAP-deficient macrophages, which, despite markedly reduced SREBP2-dependent gene expression, exhibit normal levels of cellular cholesterol [56].
Suppressing ALOX15B in human macrophages reduced cellular lipid peroxidation and 15-LOX-derived lipid hydro(pero)xides. As ALOX15B peroxidizes both free [35] and esterified PUFAs [57,58], we assume a lower amount of peroxyl radicals in ALOX15B-suppressed cells. Consistently, attenuating lipid peroxidation via liproxstatin-1 and radical trapping via α-tocopherol hampered SREBP2 target gene expression. In contrast, RSL3 as well as the redox cycler DMNQ enhanced SREBP2-dependent gene expression and partially reversed the attenuated cholesterol biosynthetic gene expression during ALOX15B inhibition. Indeed, ROS are known to affect cholesterol homeostasis. The antioxidant N-acetyl-cysteine reduced SREBP2 gene expression in human hepatoma Huh7 cells [59], while lipid peroxidation induced by iron and ascorbate [60] as well as the linoleic acid oxygenation product 13-hydroxyoctadecadienoic acid (HODE) [61] promoted cholesterol efflux from human THP-1-derived and murine macrophages, respectively. Therefore, a role of lipid peroxidation in affecting SREBP2-dependent cholesterol homeostasis in macrophages is apparent.
Besides enzymatically driven lipid peroxidation, ROS initiate oxidative degradation of lipids by radical induced hydrogen abstraction [62]. Free radicals formed during Fenton chemistry contribute to lipid peroxidation via secondary formation of peroxyl radicals (ROO•) [63], which in addition to the classical ROS species, i.e. superoxide (O2−•), hydrogen peroxide (H2O2), and the hydroxyl radical (HO•), contribute to degrade lipids, proteins, and carbohydrates and crosslink nucleic acids [64,65]. Due to their numerous bis-allylic methylene groups, PUFAs, which are essential cell membrane components, are particularly susceptible to free radical damage [62,66]. In this context, we noticed that both RSL3, a GPX4 inhibitor with the ability to elevate cellular lipid peroxides, as well as the redox cycler DMNQ increased SREBP2-dependent gene expression in primary human macrophages. Therefore, we assume that both enzymatic as well as non-enzymatic lipid peroxidation elevate SREBP2-target gene transcription. Reduced lipid peroxidation and decreased formation of 15-LOX-specific lipid hydro(pero)xides in ALOX15B-depleted cells may suggest that the reduced SREBP2 activity is initially facilitated by decreased lipid peroxide signaling and presumably not by decreased ROS formation.
A possible explanation for the increase in cholesterol biosynthesis upon accumulation of lipid peroxides could be a reorganization of membrane cholesterol into domains [67,68], that might alter membrane fluidity [69]. Indeed, lipid peroxidation has been associated with changes in membrane fluidity [70,71]. In cancer cells, accumulation of cholesterol limited the vicious cycle of lipid peroxidation through formation of cholesterol-rich lipid rafts and the concomitant decrease in membrane fluidity. In mice, depletion of cholesterol in the microenvironment of malignant melanoma potentiated lipid peroxidation and the antitumor activity of a GPX4 inhibitor [72]. Conclusively, propagating lipid peroxidation demands diffusion of PUFA-phospholipids, while accumulating cholesterol may increase membrane stiffness [73]. Therefore, it is reasonable that increased cholesterol biosynthesis may slow down lipid peroxidation [72]. It is further hypothesized that cholesterol-mediated stabilization of oxidized phospholipids and hydroperoxides may enhance the stability of membrane bilayers [72,74]. Also, accumulation of the cholesterol precursor 7-dehydrocholesterol has been reported to protect cellular membranes from (phospho)lipid peroxidation and lipid peroxidation-associated cell death [75]. Moreover, squalene accumulation prevented oxidative cell death in cholesterol auxotrophic lymphomas [76]. In addition to sterol-dependent protection from lipid peroxide accumulation, an increased flux through the cholesterol biosynthetic pathway generates coenzyme Q10 (ubiquinone-10) and ubiquinol, which acts as a lipophilic radical scavenger, known to reduce lipid peroxidation [77].
The cellular redox status is linked to various cell signaling pathways including ERK1/2 [44,78] and AKT/mTOR [46,47]. We noticed diminished ERK1/2 signaling in ALOX15B-silenced macrophages based on not only REACTOME gene set enrichment analysis but also impaired ERK1/2 phosphorylation and reduced SREBP2-dependent gene expression upon ERK1/2 inhibition. Regarding cholesterol biosynthetic gene expression, no implication of related MAPKs, in particular p38 MAPK or JNK, was apparent in ALOX15B-attenuated cells. Although not appearing in the top 25 hits of differentially regulated REACTOME pathways (SI Fig. 4A), we also observed reduced AKT phosphorylation at Ser473 and Thr308 in ALOX15B-silenced cells (SI Fig. 4B). However, this effect was neither seen in 15-LOX-inhibited cells (SI Fig. 4C), nor did inhibition of the mTOR upstream signaling hub AKT or the mTOR downstream target ribosomal protein S6 kinase B1 reduce SREBP2 target gene expression. Nevertheless, the link between SREBP2 and mTOR is controversial. Some reports claim SREBP2 as an mTOR signaling target [43,51,52], others show unaltered SREBP2 signaling by mTOR silencing [51], while also SREBP1 is highlighted to be regulated by mTOR [51,79]. However, it is undisputed that both SREBP2 [18,19,50,80] as well as SREBP1 [19,81] are recognized as ERK1/2-regulated targets.
ALOX15B-silenced macrophages show reduced lipid peroxidation and attenuated ERK1/2 phosphorylation. ERK1/2 phosphorylation also decreased upon treatment with liproxstatin-1 but increased by adding RSL3. This corroborates previous reports describing ERK1/2 as a redox target in macrophages [44,78] as well as a target of lipid peroxidation in pulmonary epithelial cells [78] or mouse carotid artery [44]. Upon inhibition of ERK1/2, we observed lower amounts of cytoplasmic as well as nuclear SREBP2 protein, similar to what happened in ALOX15B-silenced macrophages. In some analogy, inhibition of ERK1/2 in HepG2 cells decreased both the precursor and the processed form of SREBP2 [50]. A lower amount of active SREBP2 along with its potential self-regulatory decrease [82] rendered both, ALOX15B-silenced as well as ERK1/2-inhibited macrophages, refractory to SREBP2-target gene induction when blocking the intracellular cholesterol transporter NPC1. Although SREBP2 phosphorylation can be anticipated from literature [18,19,80] we failed to proove this, most likely due to the lack of appropriate, commercially available antibodies.
Various pathologies are associated with both lipid peroxidation and altered cholesterol metabolism [[83], [84], [85]]. Although this offers a therapeutic potential, underlying pathways are not yet fully understood. With this study in primary human macrophages, we describe an under-appreciated ALOX15B – lipid peroxidation – ERK1/2-signaling axis in controlling SREBP2-dependent cholesterol homeostasis. The role of ALOX15B in regulating and connecting lipid homeostasis and lipid peroxidation under pathophysiological conditions should be the topic of further studies.
4. Conclusion
Suppressing ALOX15B in primary human macrophages impairs SREBP2-dependent cholesterol biosynthesis by reducing cellular lipid peroxidation, ERK1/2 signaling and SREBP2 target gene expression. This reduces the amount of cholesterol precursors as well as cholesterol-derived oxysterols. Future studies will explore the relevance of ALOX15B in affecting cholesterol homeostasis for human macrophage biology.
5. Materials and methods
5.1. Reagents
ML351 (cat. no. 6448) and PF4708671 (cat. no. 4032) were purchased from Tocris (Wiesbaden-Nordenstadt, Germany), TRIzol™ Reagent (cat. no. L34355) and CellROX™ Deep Red Reagent (cat. no. C10422) from Thermo Fisher Scientific (Dreieich, Germany), RSL3 (cat. no. SML 2239), α-tocopherol (cat. no. 258024) and AKTVIII (cat. no. 124018) from Sigma-Aldrich (Munich, Germany), RNAPure™ peqGOLD from PeqLab Biotechnology (Erlangen, Germany), human interleukin 4 from PeproTech (Hamburg, Germany), U18666A (cat. no. BML-S200) and DMNQ (cat. no. ALX-420-027-M005) from Enzo Life Sciences (Lausen, Switzerland), PD98059 (cat. no. 9900L) and SP600125 (cat. no. 8177S) from Cell Signaling Technology (Danvers, MA, USA), liproxstatin-1 (cat. no. 17730) from Cayman Chemical (Ann Arbor, MI, USA). Human gene primers were purchased from Biomers GmbH (Ulm, Germany). All chemicals were of the highest grade of purity and commercially available and were dissolved according to the manufacturer's instructions.
5.2. Primary human macrophage generation
Human peripheral blood mononuclear cells were isolated from commercially obtained buffy coats of anonymous donors (DRK-Blutspendedienst Baden-Württemberg-Hessen, Institut für Transfusionsmedizin und Immunhämatologie, Frankfurt, Germany) using Ficoll density centrifugation. Peripheral blood mononuclear cells were washed twice with PBS containing 2 mM EDTA and subsequently incubated for 1.5 h under growth conditions in RPMI 1640 medium supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) to allow their adherence to culture dishes (Sarstedt, Nümbrecht, Germany). Non-adherent cells were removed. Monocytes were then differentiated into naïve macrophages with RPMI 1640 medium (Gibco, Dreieich, Germany) containing 3% AB-positive human serum (DRK-Blutspendedienst Baden-Württemberg-Hessen, Frankfurt, Germany) (complete media) for at least 7 days.
5.3. siRNA transfections
A final concentration of 50 nM of either non-targeting control siRNA (ON-TARGETplus Non-targeting Control Pool, SMARTpool, cat. no. D-001810-10, Horizon Discovery Biosciences (Cambridge, United Kingdom)) or siRNA targeting human ALOX15B (ON-TARGETplus Human ALOX15B siRNA, SMARTpool, cat. no. L-009026-00-0005, Horizon Discovery Biosciences) were transfected into primary human macrophages using HiPerFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's recommendations for a total duration of 5 days. Each knockdown was routinely confirmed by quantitative polymerase chain reaction for each experiment.
5.4. RNA extraction and quantitative real-time PCR
Total RNA from macrophages was isolated using PeqGOLD RNAPure reagent or TRIzol reagent following the manufacturer's protocol and quantified using the NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, USA). Total RNA was transcribed into cDNA using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, cat. no. K1641). Quantitative real-time PCR was performed using Power up SYBR green mastermix (Applied Biosystems, Darmstadt, Germany) and the QuantStudio™ 3 and 5 Real-Time PCR System. β2 microglobulin (βMG) was used as an endogenous control for human macrophages. Primer sequences were used as follows: ALOX15B (Fwd: 5′-aag ggc ttc cta aac cag ga-3′, Rev: 5′-tga cat cac atg tgg cat tg-3′), βMG (Fwd: 5′-act cac gtc atc cag cag ag-3′, Rev: 5′-tca aac ctc cat gat gct gct-3′), CYP51A1 (Fwd: 5′-gaa acg cag aca gtc tca aga-3′, Rev: 5′-acg ccc atc ctt gta tgt agc-3′), DHCR24 (5′-cac tgt ctc act acg tgt cgg-3′, Rev: 5′-cca gcc aat gga ggt cag c-3′), HMGCS1 (Fwd: 5′-cat tag acc gct gct att ctg tc-3′, Rev: 5′-ttc agc aac atc cga gct aga-3′), LDLR (Fwd: 5′-tac ccc tcg aga cag atg gt-3′, Rev: 5′-act gtc cga agc ctg ttc tg-3′), MSMO1 (Fwd: 5′-tat gct ggt tct cgg cat cat-3′, Rev: 5′-cca aaa att cga tcc cac cat gt-3′), MVK (Fwd: 5′-cat ggc aag gta gca ctg g-3′, Rev: 5′-gat acc aat gtt ggg taa gct ga-3′).
5.5. RNA-sequencing library preparation
Total RNA of control and ALOX15B-silenced macrophages was isolated using RNeasy Mini Kit (Qiagen, cat. no. 74104) and purified by precipitation. RNA quality was assessed using an Agilent High Sensitivity RNA ScreenTape (cat. no. 5067-5579) with an Agilent TapeStation 4150 (Agilent Technologies, Waldbronn, Germany). RNA was quantified using a Qubit HS RNA Assay Kit (cat. no. Q32852) and Qubit 3 fluorometer (Thermo Fisher Scientific). Subsequently, library preparation of 500 ng RNA was performed using the Lexogen QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen GmbH, Vienna, Austria, cat. no. 015.24). Library quality and quantity were controlled by Qubit 3 fluorometer and Agilent D1000 DNA ScreenTape (cat. no. 5067-5582). The average size of the libraries was calculated within 150–1000 bp and a 1 nM equimolar pool of all libraries was generated. Single-end sequencing was performed with 75 cycles using the High Output Kit v2.5 (cat. no. 20024906) on an Illumina NextSeq 500 sequencer (Illumina, San Diego, CA, USA). Sequencing data have been deposited under the GEO accession number GSE208160.
5.6. RNA-sequencing data analysis
FASTQ data processing was performed within the Lexogen QuantSeq 3′ mRNA-Seq integrated data analysis pipeline on the BlueBee on-line platform (BlueBee Holding BV, Rijswijk, The Netherlands). FASTQ reads were trimmed (bbduk v35.92), mapped to the hg38 human genome assembly (STAR Aligner v2.5.2a with modified ENCODE settings), and gene-specific read counts were determined (HTSeq-count v0.6.0). Differential gene expression analysis of read counts was conducted by DeSeq2 and gained data were filtered by log2 fold change and p-adjusted (FDR) value. Gene Set Enrichment Analysis and pathway analysis was performed using GenePattern [86] and heat maps were generated using MORPHEUS (https://software.broadinstitute.org/morpheus) or R version 4.2.1 (2022-06-23 ucrt) (https://www.R-project.org).
5.7. Immunoblots
Macrophage lysates were resolved on polyacrylamide gels followed by semi-dry transfer onto nitrocellulose membranes. Membranes were blocked with 5% milk or 5% bovine serum albumin (BSA)/100 mM Tris–HCl, 150 mM sodium chloride, 0.01% (v/v) Tween 20 (TTBS) followed by incubation with Revert 700™ Total Protein Stain (LI-COR Biosciences, Bad Homburg, Germany, cat. no. 926-11021) and Revert™ Wash Solution (LI-COR Biosciences, cat. no. 926-11022) and with antibodies against ALOX15B (cat. no. 10004454, Cayman Chemicals), ALOX15 (cat. no. ab119774, Abcam (Cambridge, UK)), DHCR24 (cat. no. 10471-1-AP) and LSS (cat. no. 13715-1-AP) from Proteintech (St. Leon-Rot, Germany), CYP51A1 (cat. no. H00001595-A01, Abnova (Tapei City, Taiwan)), phospho-p44/42 MAPK (phospho-ERK1/2, Thr202/Tyr204, cat. no. 9101), p44/42 MAPK (ERK1/2, cat. no. 4695), phospho-AKT (Thr308, cat. no. 5106), phospho-AKT (Ser473, cat. no. 4051), AKT (cat. no. 9272), SAPK/JNK (cat. no. 9252), phospho-SAPK/JNK (Thr183/Tyr185, cat. no. 9255), p38 MAPK (cat. no. 9212), and phospho-p38 MAPK (Thr180/Tyr182, cat. no. 9211) from Cell Signaling Technology (Danvers, MA, USA), or nucleolin (cat. no. sc-55486) from Santa Cruz Biotechnology (Heidelberg, Germany). For protein detection, the membrane was incubated with IRDye secondary antibodies (LI-COR Biosciences, Bad Homburg, Germany) in 5% BSA/TTBS. Membranes were visualized with the Odyssey infrared imaging system (LI-COR Biosciences) and quantified with Image Studio Digits 5.0 (LI-COR Biosciences). The signal of individual primary antibodies was normalized either to nucleolin or, in the case of phosphorylated proteins, the signal of the primary antibody against the phosphorylated protein was normalized to the respective total protein.
5.8. Immunocytochemistry and lipid peroxidation
Peripheral blood mononuclear cells were seeded on Poly-l-lysine coated glass bottomed 6 well plates (IBL, Gerasdorf, Austria, cat. no. 220200042) and differentiated into macrophages for a minimum of 7 days prior to siRNA transfection. For SREBP2 staining, cells were washed 1x with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. Subsequently, cells were washed 3x with PBS and then permeabilized with 0.1% Triton-X 100 in PBS for 10 min, then the cells were washed 3x with PBS and blocked with 10% normal goat serum (NGS) containing 100 mM glycine. SREBP2 primary antibody (Cayman chemicals, cat. no. 10007663) was diluted 1:500 in 2% normal goat serum with 10 mM glycine and incubated overnight at 4 °C. Samples were washed 3x with PBS and incubated with secondary antibodies F(ab')2-Goat anti-Rabbit IgG Alexa Fluor Plus 555 (Thermo Fisher Scientific, cat. no. A48283) at 1:500 dilution in 2% normal goat serum with 10 mM glycine for 45 min at room temperature. Thereafter, cells were washed 3x with PBS then incubated with 1 μg/ml DAPI (spectral 4′,6-diamidino-2-phenylindole, PerkinElmer, Rodgau, Germany) in PBS containing 0.1% Tween 20 for 1 min before further 3x washes with PBS. For BODIPY™ 581/591 C11 lipid peroxide staining, cells were incubated with DMSO or ML351 (10 μM) for 30 min. Then, KD macrophages or ML351 treated cells were incubated with BODIPY™ 581/591 C11 (5 μM) for 30 min in complete media under growth conditions. Afterwards, cells were washed with warm PBS and complete media was added. Cells were imaged immediately at 37 °C.
5.9. Microscopy
Confocal microscopy was performed using a Zeiss laser scanning microscope 800 Axio-Observer using Zen Blue v2.3 software for acquisition (Carl Zeiss, Oberkochen, Germany). All images were acquired using GaAsP detector with a Plan-Apochromat 40x/1.4 Oil DIC M27 objective. 405 nm, 488 nm and 561 nm diode lasers were used at 0.4/0.5%, detector gain setting varied from 600 to 800 V. Images were acquired at 16 bit with a scaling of 0.085 × 0.085 μm per pixel. For BODIPY™ 581/591C11, confocal images were acquired via simultaneous excitation of the 488 nm and the 568 nm lasers with emission detection acquired between 400 and 590 nm for oxidized and 600–700 nm for non-oxidized, respectively.
5.10. Image analysis
Intensity analysis for mean pixel intensity was performed on segmented images for whole cell or nucleus via thresholding following Gaussian blurring. Cytoplasmic measurements were generated via subtracting nuclear region from whole cell region using the image calculator. Macros are available upon request.
5.11. Uptake of pHrodo LDL particles
Transfected macrophages were serum starved at day 4 for 24 h with serum-free RPMI 1640 supplemented with 0.22 μm sterile-filtered 0.3% bovine serum albumin. Afterwards, cells were washed and incubated with pHrodo™ Green-LDL (10 μg/ml) for 3 h at growth conditions. Macrophages were then extensively washed and analyzed by a LSR II/Fortessa flow cytometer (BD Life Sciences). Data were analyzed using FlowJo V10 (TreeStar, Ashland, OR, USA).
5.12. Flow cytometric detection of lipid peroxidation
Differentiated macrophages were treated with RSL3 (10 μM) for 6 h in complete RPMI. Cells were scraped, washed, and incubated with BODIPY™ 581/591C11 (5 μM) for 30 min at 37 °C. After two washes, fluorescence in BB515 channel was recorded by a LSR II/Fortessa flow cytometer (BD Life Sciences). Data were analyzed using FlowJo V10 (TreeStar, Ashland, OR, USA).
5.13. Flow cytometric detection of cellular ROS
Differentiated macrophages were cultured with complete RPMI, supplemented with DMNQ (10 μM) and CellROX™ (5 μM) for 3 h under growth conditions. Cells were washed, scraped and directly analyzed by a LSR II/Fortessa flow cytometer (BD Life Sciences). Data analysis was performed with FlowJo V10 (TreeStar, Ashland, OR, USA).
5.14. Measurement of cellular cholesterol, non-cholesterol sterol, and oxysterol content
The cellular content of cholesterol was measured by gas chromatography-flame ionization detection (GC-FID) and of non-cholesterol sterols and oxysterols by gas chromatography-mass spectrometry in selected ion monitoring mode (GC-MS-SIM). Cell pellets were spun in a speedvac concentrator (12 mbar; Savant AES 1000) and weighed. Cholesterol, non-cholesterol sterols, and oxysterols were extracted using chloroform. After alkaline hydrolysis, the concentrations of cholesterol precursors were measured with GC-MS-SIM as previously described [5,87]. The trimethylsilyethers of the (oxy)sterols were separated on a DB-XLB (30 m length × 0.25 mm internal diameter, 0.25 μm film) column (Agilent Technologies) using the 6890 N Network GC system (Agilent Technologies). Epicoprostanol (Steraloids, Newport, RI, USA) was used as an internal standard, to quantify the non-cholesterol sterols (Medical Isotopes, Pelham, NH, USA) on a 5973 Network MSD (Agilent Technologies). Total cholesterol was measured by GC-FID on an HP 6890 GC system (Hewlett Packard, Waldbronn, Germany), equipped with a DB-XLB (30 m length × 0.25 mm internal diameter, 0.25 μm film) column (Agilent Technologies) using 5α-cholestane (Steraloids) as internal standard [32,88].
5.15. Oxylipin analysis
In order to analyze esterified oxylipins, total (esterified and non-esterified) oxylipins and non-esterified oxylipins were analyzed by LC-MS/MS as described [91,92].
Cell pellets were resuspended and sonicated in PBS containing inhibitor mix and antioxidants solution and protein content was determined via bicinchoninic acid assay [91,92]. For free oxylipin determination, one part of the cell lysate was mixed with an internal standard solution and subsequently processed by solid phase extraction following protein precipitation with methanol. For total oxylipin determination, another part of the cell lysate was mixed with internal standards solution and proteins were precipitated with isopropanol and the supernatant was hydrolyzed using potassium hydroxide solution. Following hydrolysis, samples were neutralized and extracted by solid phase extraction [91,92]. The extracts were analyzed by means of liquid chromatography-mass spectrometry. Oxylipins were separated using a Zorbax Eclipse Plus C18 reversed-phase column (2.1 × 150 mm, particle size 1.8 μm, pore size 9.5 nm, Agilent, Waldbronn, Germany). Detection was carried out following electrospray ionization in negative mode by a triple quadrupole mass spectrometer (5500 QTRAP, SCIEX, Darmstadt Germany). Quantification was based on an external calibration with characterized standards [94] using internal standards. All methods parameters including the transitions are reported in Refs. [89,90,92]. The esterified oxylipin concentration was calculated by the difference of the determined total and free oxylipin concentration in each sample.
In order to determine the amount of hydroperoxy fatty acid, the sonicated cell-suspension was split for two analyses with and without reduction. For the reduction, 30 μl 10 mg/mL tin (II) chloride/methanol was added for converting hydroperoxy fatty acids such as 15-HpETE to hydroxy fatty acids such as 15-HETE after neutralization prior to solid-phase-extraction [93,95]. 15-HpETE was calculated based on the 15-HETE concentrations determined in the cells after reduction subtracting the amount in cells without the reduction.
5.16. Statistical analysis
Graphical data are presented as mean ± SE of at least three independent experiments using human monocyte-derived macrophages from different individual donors, unless noted otherwise. Differences were considered significant when P < 0.05 (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns = not significant). Statistical significance was calculated using one-way analysis of variance (ANOVA) as stated with either Tukey's, Sidak's or Dunnett's post hoc test, two-tailed Student's t-test with significance level set at 0.05, or one sample t-test with significance level set at 0.05 using Prism 9 and 10 software (GraphPad, La Jolla, CA, USA). Final assembly and preparation of all figures was done using CorelDRAW 2021 (Corel Corporation, Ottawa, Canada).
Data availability
All relevant data are within the paper and its supplementary information files. Sequencing data have been deposited under the GEO accession number GSE208160.
CRediT authorship contribution statement
Yvonne Benatzy: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. Megan A. Palmer: Data curation, Investigation, Methodology, Writing – original draft. Dieter Lütjohann: Data curation, Methodology. Rei-Ichi Ohno: Data curation, Methodology. Nadja Kampschulte: Data curation, Methodology. Nils Helge Schebb: Data curation, Methodology. Dominik C. Fuhrmann: Data curation, Investigation, Methodology, Writing – original draft. Ryan G. Snodgrass: Conceptualization, Data curation, Investigation, Writing – review & editing. Bernhard Brüne: Conceptualization, Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by Deutsche Forschungsgemeinschaft SFB1039 TP B04 and Br 999/26-1. We thank Gudrun Beyer, Andrea Duchêne, Tanja Keppler and Anja Kerksiek for their excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103149.
Contributor Information
Dominik C. Fuhrmann, Email: fuhrmann@biochem.uni-frankfurt.de.
Ryan G. Snodgrass, Email: ryan.snodgrass@usda.gov.
Bernhard Brüne, Email: b.bruene@biochem.uni-frankfurt.de.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Yin H., Xu L., Porter N.A. Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 2.Shah R., Shchepinov M.S., Pratt D.A. Resolving the role of lipoxygenases in the initiation and execution of ferroptosis. ACS Cent. Sci. 2018;4:387–396. doi: 10.1021/acscentsci.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn H., Humeniuk L., Kozlov N., Roigas S., Adel S., Heydeck D. The evolutionary hypothesis of reaction specificity of mammalian ALOX15 orthologs. Prog. Lipid Res. 2018;72:55–74. doi: 10.1016/j.plipres.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn H., Banthiya S., van Leyen K. Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta. 2015;1851:308–330. doi: 10.1016/j.bbalip.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snodgrass R.G., Zezina E., Namgaladze D., Gupta S., Angioni C., Geisslinger G., Lütjohann D., Brüne B. A novel function for 15-lipoxygenases in cholesterol homeostasis and CCL17 production in human macrophages. Front. Immunol. 2018;9:1906. doi: 10.3389/fimmu.2018.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magnusson L.U., Lundqvist A., Karlsson M.N., Skålén K., Levin M., Wiklund O., Borén J., Hultén L.M. Arachidonate 15-lipoxygenase type B knockdown leads to reduced lipid accumulation and inflammation in atherosclerosis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange Y., Swaisgood M.H., Ramos B.V., Steck T.L. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- 8.Midzak A., Papadopoulos V. Binding domain-driven intracellular trafficking of sterols for synthesis of steroid hormones, bile acids and oxysterols. Traffic. 2014;15:895–914. doi: 10.1111/tra.12177. [DOI] [PubMed] [Google Scholar]
- 9.Luo J., Yang H., Song B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 10.Hua X., Nohturfft A., Goldstein J.L., Brown M.S. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage–activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 11.Rawson R.B., Zelenski N.G., Nijhawan D., Ye J., Sakai J., Hasan M.T., Chang T.Y., Brown M.S., Goldstein J.L. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 12.Spann N.J., Garmire L.X., McDonald J.G., Myers D.S., Milne S.B., Shibata N., Reichart D., Fox J.N., Shaked I., Heudobler D., Raetz C.R.H., Wang E.W., Kelly S.L., Sullards M.C., Murphy R.C., Merrill A.H., Brown H.A., Dennis E.A., Li A.C., Ley K., Tsimikas S., Fahy E., Subramaniam S., Quehenberger O., Russell D.W., Glass C.K. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radhakrishnan A., Ikeda Y., Kwon H.J., Brown M.S., Goldstein J.L. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang T., Espenshade P.J., Wright M.E., Yabe D., Gong Y., Aebersold R., Goldstein J.L., Brown M.S. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 15.Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., Goldstein J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 17.Radhakrishnan A., Goldstein J.L., McDonald J.G., Brown M.S. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metabol. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotzka J., Lehr S., Roth G., Avci H., Knebel B., Muller-Wieland D. Insulin-activated Erk-mitogen-activated protein kinases phosphorylate sterol regulatory element-binding Protein-2 at serine residues 432 and 455 in vivo. J. Biol. Chem. 2004;279:22404–22411. doi: 10.1074/jbc.M401198200. [DOI] [PubMed] [Google Scholar]
- 19.Kotzka J., Müller-Wieland D., Roth G., Kremer L., Munck M., Schürmann S., Knebel B., Krone W. Sterol regulatory element binding proteins (SREBP)-1a and SREBP-2 are linked to the MAP-kinase cascade. J. Lipid Res. 2000;41:99–108. doi: 10.1016/S0022-2275(20)32079-4. [DOI] [PubMed] [Google Scholar]
- 20.Arito M., Horiba T., Hachimura S., Inoue J., Sato R. Growth factor-induced phosphorylation of sterol regulatory element-binding proteins inhibits sumoylation, thereby stimulating the expression of their target genes, low density lipoprotein uptake, and lipid synthesis. J. Biol. Chem. 2008;283:15224–15231. doi: 10.1074/jbc.M800910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giandomenico V., Simonsson M., Grönroos E., Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muse E.D., Yu S., Edillor C.R., Tao J., Spann N.J., Troutman T.D., Seidman J.S., Henke A., Roland J.T., Ozeki K.A., Thompson B.M., McDonald J.G., Bahadorani J., Tsimikas S., Grossman T.R., Tremblay M.S., Glass C.K. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc. Natl. Acad. Sci. USA. 2018;115:E4680–E4689. doi: 10.1073/pnas.1714518115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tontonoz P., Mangelsdorf D.J. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 24.Smulan L.J., Ding W., Freinkman E., Gujja S., Edwards Y.J.K., Walker A.K. Cholesterol-independent SREBP-1 maturation is linked to ARF1 inactivation. Cell Rep. 2016;16:9–18. doi: 10.1016/j.celrep.2016.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rong S., Cortés V.A., Rashid S., Anderson N.N., McDonald J.G., Liang G., Moon Y.-A., Hammer R.E., Horton J.D. Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXR in livers of mice. Elife. 2017;6 doi: 10.7554/eLife.25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz J.R., Tu H., Luk A., Repa J.J., Medina J.C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D.J., Lustig K.D., Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloch K. The biological synthesis of cholesterol. Science. 1965;150:19–28. doi: 10.1126/science.150.3692.19. [DOI] [PubMed] [Google Scholar]
- 28.Kandutsch A.A., Russell A.E. Preputial gland tumor sterols. 2. The identification of 4 alpha-methyl-Delta 8-cholesten-3 beta-ol. J. Biol. Chem. 1960;235:2253–2255. [PubMed] [Google Scholar]
- 29.Kandutsch A.A., Russell A.E. Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J. Biol. Chem. 1960;235:2256–2261. [PubMed] [Google Scholar]
- 30.Miettinen T.A., Gylling H., Nissinen M.J. The role of serum non-cholesterol sterols as surrogate markers of absolute cholesterol synthesis and absorption. Nutr. Metabol. Cardiovasc. Dis. 2011;21:765–769. doi: 10.1016/j.numecd.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Kruth H.S. Fluid-phase pinocytosis of LDL by macrophages: a novel target to reduce macrophage cholesterol accumulation in atherosclerotic lesions. Curr. Pharmaceut. Des. 2013;19:5865–5872. doi: 10.2174/1381612811319330005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay D.S., Jones P.J.H., Myrie S.B., Plat J., Lütjohann D. Methodological considerations for the harmonization of non-cholesterol sterol bio-analysis. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2014;957:116–122. doi: 10.1016/j.jchromb.2014.02.052. [DOI] [PubMed] [Google Scholar]
- 33.Lu F., Liang Q., Abi-Mosleh L., Das A., de Brabander J.K., Goldstein J.L., Brown M.S. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife. 2015;4 doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai G., Joshi N., Jung J.E., Liu Y., Schultz L., Yasgar A., Perry S., Diaz G., Zhang Q., Kenyon V., Jadhav A., Simeonov A., Lo E.H., van Leyen K., Maloney D.J., Holman T.R. Potent and selective inhibitors of human reticulocyte 12/15-lipoxygenase as anti-stroke therapies. J. Med. Chem. 2014;57:4035–4048. doi: 10.1021/jm401915r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kutzner L., Goloshchapova K., Heydeck D., Stehling S., Kuhn H., Schebb N.H. Mammalian ALOX15 orthologs exhibit pronounced dual positional specificity with docosahexaenoic acid. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:666–675. doi: 10.1016/j.bbalip.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Rådmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Förster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O'Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zilka O., Shah R., Li B., Friedmann Angeli J.P., Griesser M., Conrad M., Pratt D.A. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 2017;3:232–243. doi: 10.1021/acscentsci.7b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burton G.W., Ingold K.U. Vitamin E: application of the principles of physical organic chemistry to the exploration of its structure and function. Acc. Chem. Res. 1986;19:194–201. doi: 10.1021/ar00127a001. [DOI] [Google Scholar]
- 40.Watanabe N., Forman H.J. Autoxidation of extracellular hydroquinones is a causative event for the cytotoxicity of menadione and DMNQ in A549-S cells. Arch. Biochem. Biophys. 2003;411:145–157. doi: 10.1016/s0003-9861(02)00716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Ma M.-Y., Sun M., Jiang L.-Y., Zhao X.-T., Fang X.-X., Man Lam S., Shui G.-H., Luo J., Shi X.-J., Song B.-L. Endogenous sterol intermediates of the mevalonate pathway regulate HMGCR degradation and SREBP-2 processing. J. Lipid Res. 2019;60:1765–1775. doi: 10.1194/jlr.RA119000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams C.M., Reitz J., de Brabander J.K., Feramisco J.D., Li L., Brown M.S., Goldstein J.L. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 43.Düvel K., Yecies J.L., Menon S., Raman P., Lipovsky A.I., Souza A.L., Triantafellow E., Ma Q., Gorski R., Cleaver S., Vander Heiden M.G., MacKeigan J.P., Finan P.M., Clish C.B., Murphy L.O., Manning B.D. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Squadrito F., Minutoli L., Esposito M., Bitto A., Marini H., Seminara P., Crisafulli A., Passaniti M., Adamo E.B., Marini R., Guarini S., Altavilla D. Lipid peroxidation triggers both c-Jun N-terminal kinase (JNK) and extracellular-regulated kinase (ERK) activation and neointimal hyperplasia induced by cessation of blood flow in the mouse carotid artery. Atherosclerosis. 2005;178:295–302. doi: 10.1016/j.atherosclerosis.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Traore K., Sharma R., Thimmulappa R.K., Watson W.H., Biswal S., Trush M.A. Redox-regulation of Erk1/2-directed phosphatase by reactive oxygen species: role in signaling TPA-induced growth arrest in ML-1 cells. J. Cell. Physiol. 2008;216:276–285. doi: 10.1002/jcp.21403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calamaras T.D., Lee C., Lan F., Ido Y., Siwik D.A., Colucci W.S. The lipid peroxidation product 4-hydroxy-trans-2-nonenal causes protein synthesis in cardiac myocytes via activated mTORC1-p70S6K-RPS6 signaling. Free Radic. Biol. Med. 2015;82:137–146. doi: 10.1016/j.freeradbiomed.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yalcin S., Marinkovic D., Mungamuri S.K., Zhang X., Tong W., Sellers R., Ghaffari S. ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3(-/-) mice. EMBO J. 2010;29:4118–4131. doi: 10.1038/emboj.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keyes J.D., Postiglione A., Patwardhan S., Smith-Pearson P., Parsonage D., Newman R.H., Poole L.B. Impact of redox modifications on ERK2 substrate phosphorylation. Free Radic. Biol. Med. 2017;112:193–194. doi: 10.1016/j.freeradbiomed.2017.10.305. [DOI] [Google Scholar]
- 49.Luu W., Sharpe L.J., Stevenson J., Brown A.J. Akt acutely activates the cholesterogenic transcription factor SREBP-2. Biochim. Biophys. Acta. 2012;1823:458–464. doi: 10.1016/j.bbamcr.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 50.Abidi P., Zhang F., Li C., Liu J. Blockage of the ERK signaling pathway abrogates the SCAP ligand-induced transcriptional activation of the LDL receptor gene in HepG2 cells. Int. J. Mol. Med. 2005;16:779–785. [PubMed] [Google Scholar]
- 51.Porstmann T., Santos C.R., Griffiths B., Cully M., Wu M., Leevers S., Griffiths J.R., Chung Y.-L., Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metabol. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B.T., Ducker G.S., Barczak A.J., Barbeau R., Erle D.J., Shokat K.M. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc. Natl. Acad. Sci. USA. 2011;108:15201–15206. doi: 10.1073/pnas.1103746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alessi D.R., Cuenda A., Cohen P., Dudley D.T., Saltiel A.R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 54.Lindsley C.W., Zhao Z., Leister W.H., Robinson R.G., Barnett S.F., Defeo-Jones D., Jones R.E., Hartman G.D., Huff J.R., Huber H.E., Duggan M.E. Allosteric Akt (PKB) inhibitors: discovery and SAR of isozyme selective inhibitors. Bioorg. Med. Chem. Lett. 2005;15:761–764. doi: 10.1016/j.bmcl.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Pearce L.R., Alton G.R., Richter D.T., Kath J.C., Lingardo L., Chapman J., Hwang C., Alessi D.R. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1) Biochem. J. 2010;431:245–255. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- 56.York A.G., Williams K.J., Argus J.P., Zhou Q.D., Brar G., Vergnes L., Gray E.E., Zhen A., Wu N.C., Yamada D.H., Cunningham C.R., Tarling E.J., Wilks M.Q., Casero D., Gray D.H., Yu A.K., Wang E.S., Brooks D.G., Sun R., Kitchen S.G., Wu T.-T., Reue K., Stetson D.B., Bensinger S.J. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bender G., Schexnaydre E.E., Murphy R.C., Uhlson C., Newcomer M.E. Membrane-dependent activities of human 15-LOX-2 and its murine counterpart: implications for murine models of atherosclerosis. J. Biol. Chem. 2016;291:19413–19424. doi: 10.1074/jbc.M116.741454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coffa G., Brash A.R. A single active site residue directs oxygenation stereospecificity in lipoxygenases: stereocontrol is linked to the position of oxygenation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15579–15584. doi: 10.1073/pnas.0406727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seo K., Shin S.M. Induction of Lipin1 by ROS-dependent SREBP-2 activation. Toxicol. Res. 2017;33:219–224. doi: 10.5487/TR.2017.33.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcil V., Delvin E., Sané A.T., Tremblay A., Levy E. Oxidative stress influences cholesterol efflux in THP-1 macrophages: role of ATP-binding cassette A1 and nuclear factors. Cardiovasc. Res. 2006;72:473–482. doi: 10.1016/j.cardiores.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 61.Kämmerer I., Ringseis R., Biemann R., Wen G., Eder K. 13-hydroxy linoleic acid increases expression of the cholesterol transporters ABCA1, ABCG1 and SR-BI and stimulates apoA-I-dependent cholesterol efflux in RAW264.7 macrophages. Lipids Health Dis. 2011;10:222. doi: 10.1186/1476-511X-10-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gardner H.W. Oxygen radical chemistry of polyunsaturated fatty acids. Free Radic. Biol. Med. 1989;7:65–86. doi: 10.1016/0891-5849(89)90102-0. [DOI] [PubMed] [Google Scholar]
- 63.Feng H., Stockwell B.R. Unsolved mysteries: how does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blokhina O., Virolainen E., Fagerstedt K.V. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 2003;91:179–194. doi: 10.1093/aob/mcf118. Spec No. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaschler M.M., Stockwell B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017;482:419–425. doi: 10.1016/j.bbrc.2016.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagner B.A., Buettner G.R., Burns C.P. Free radical-mediated lipid peroxidation in cells: oxidizability is a function of cell lipid bis-allylic hydrogen content. Biochemistry. 1994;33:4449–4453. doi: 10.1021/BI00181A003. [DOI] [PubMed] [Google Scholar]
- 67.Jacob R.F., Aleo M.D., Self-Medlin Y., Doshna C.M., Mason R.P. 1,2-naphthoquinone stimulates lipid peroxidation and cholesterol domain formation in model membranes. Invest. Ophthalmol. Vis. Sci. 2013;54:7189–7197. doi: 10.1167/iovs.13-12793. [DOI] [PubMed] [Google Scholar]
- 68.Jacob R.F., Mason R.P. Lipid peroxidation induces cholesterol domain formation in model membranes. J. Biol. Chem. 2005;280:39380–39387. doi: 10.1074/jbc.M507587200. [DOI] [PubMed] [Google Scholar]
- 69.Horváth Á., Erostyák J., Szőke É. Effect of lipid raft disruptors on cell membrane fluidity studied by fluorescence spectroscopy. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232213729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borst J.W., Visser N.V., Kouptsova O., Visser A.J. Oxidation of unsaturated phospholipids in membrane bilayer mixtures is accompanied by membrane fluidity changes. Biochim. Biophys. Acta. 2000;1487:61–73. doi: 10.1016/s1388-1981(00)00084-6. [DOI] [PubMed] [Google Scholar]
- 71.Wong-Ekkabut J., Xu Z., Triampo W., Tang I.-M., Tieleman D.P., Monticelli L. Effect of lipid peroxidation on the properties of lipid bilayers: a molecular dynamics study. Biophys. J. 2007;93:4225–4236. doi: 10.1529/biophysj.107.112565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao X., Lian X., Xie J., Liu G. Accumulated cholesterol protects tumours from elevated lipid peroxidation in the microenvironment. Redox Biol. 2023;62 doi: 10.1016/j.redox.2023.102678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakraborty S., Doktorova M., Molugu T.R., Heberle F.A., Scott H.L., Dzikovski B., Nagao M., Stingaciu L.-R., Standaert R.F., Barrera F.N., Katsaras J., Khelashvili G., Brown M.F., Ashkar R. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. USA. 2020;117:21896–21905. doi: 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khandelia H., Loubet B., Olzyńska A., Jurkiewicz P., Hof M. Pairing of cholesterol with oxidized phospholipid species in lipid bilayers. Soft Matter. 2014;10:639–647. doi: 10.1039/c3sm52310a. [DOI] [PubMed] [Google Scholar]
- 75.Angeli J.P.F., Shah R., Pratt D.A., Conrad M. Ferroptosis inhibition: mechanisms and opportunities. Trends Pharmacol. Sci. 2017;38:489–498. doi: 10.1016/j.tips.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Garcia-Bermudez J., Baudrier L., Bayraktar E.C., Shen Y., La K., Guarecuco R., Yucel B., Fiore D., Tavora B., Freinkman E., Chan S.H., Lewis C., Min W., Inghirami G., Sabatini D.M., Birsoy K. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature. 2019;567:118–122. doi: 10.1038/s41586-019-0945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., Roberts M.A., Tong B., Maimone T.J., Zoncu R., Bassik M.C., Nomura D.K., Dixon S.J., Olzmann J.A. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iles K.E., Dickinson D.A., Wigley A.F., Welty N.E., Blank V., Forman H.J. HNE increases HO-1 through activation of the ERK pathway in pulmonary epithelial cells. Free Radic. Biol. Med. 2005;39:355–364. doi: 10.1016/j.freeradbiomed.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yecies J.L., Zhang H.H., Menon S., Liu S., Yecies D., Lipovsky A.I., Gorgun C., Kwiatkowski D.J., Hotamisligil G.S., Lee C.-H., Manning B.D. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metabol. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sundqvist A., Bengoechea-Alonso M.T., Ye X., Lukiyanchuk V., Jin J., Harper J.W., Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metabol. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 81.Roth G., Kotzka J., Kremer L., Lehr S., Lohaus C., Meyer H.E., Krone W., Müller-Wieland D. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J. Biol. Chem. 2000;275:33302–33307. doi: 10.1074/jbc.M005425200. [DOI] [PubMed] [Google Scholar]
- 82.Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 83.Chistiakov D.A., Melnichenko A.A., Myasoedova V.A., Grechko A.V., Orekhov A.N. Mechanisms of foam cell formation in atherosclerosis. J. Mol. Med. (Berl.) 2017;95:1153–1165. doi: 10.1007/s00109-017-1575-8. [DOI] [PubMed] [Google Scholar]
- 84.Guo C., Chi Z., Jiang D., Xu T., Yu W., Wang Z., Chen S., Zhang L., Liu Q., Guo X., Zhang X., Li W., Lu L., Wu Y., Song B.-L. Di Wang, cholesterol homeostatic regulator SCAP-SREBP2 integrates NLRP3 inflammasome activation and cholesterol biosynthetic signaling in macrophages. Immunity. 2018;49:842–856. doi: 10.1016/j.immuni.2018.08.021. e7. [DOI] [PubMed] [Google Scholar]
- 85.Liu W., Chakraborty B., Safi R., Kazmin D., Chang C.-Y., McDonnell D.P. Dysregulated cholesterol homeostasis results in resistance to ferroptosis increasing tumorigenicity and metastasis in cancer. Nat. Commun. 2021;12:5103. doi: 10.1038/s41467-021-25354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 87.Snodgrass R.G., Benatzy Y., Schmid T., Namgaladze D., Mainka M., Schebb N.H., Lütjohann D., Brüne B. Efferocytosis potentiates the expression of arachidonate 15-lipoxygenase (ALOX15) in alternatively activated human macrophages through LXR activation. Cell Death Differ. 2021;28:1301–1316. doi: 10.1038/s41418-020-00652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Šošić-Jurjević B., Lütjohann D., Renko K., Filipović B., Radulović N., Ajdžanović V., Trifunović S., Nestorović N., Živanović J., Manojlović Stojanoski M., Kӧhrle J., Milošević V. The isoflavones genistein and daidzein increase hepatic concentration of thyroid hormones and affect cholesterol metabolism in middle-aged male rats. J. Steroid Biochem. Mol. Biol. 2019;190:1–10. doi: 10.1016/j.jsbmb.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 89.Rund K.M., Ostermann A.I., Kutzner L., Galano J.-M., Oger C., Vigor C., Wecklein S., Seiwert N., Durand T., Schebb N.H. Development of an LC-ESI(-)-MS/MS method for the simultaneous quantification of 35 isoprostanes and isofurans derived from the major n3- and n6-PUFAs. Anal. Chim. Acta. 2018;1037:63–74. doi: 10.1016/j.aca.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 90.Kutzner L., Rund K.M., Ostermann A.I., Hartung N.M., Galano J.-M., Balas L., Durand T., Balzer M.S., David S., Schebb N.H. Development of an optimized LC-MS method for the detection of specialized pro-resolving mediators in biological samples. Front. Pharmacol. 2019;10:169. doi: 10.3389/fphar.2019.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ostermann A.I., Koch E., Rund K.M., Kutzner L., Mainka M., Schebb N.H. Targeting esterified oxylipins by LC-MS - effect of sample preparation on oxylipin pattern. Prostag. Other Lipid Mediat. 2020;146 doi: 10.1016/j.prostaglandins.2019.106384. [DOI] [PubMed] [Google Scholar]
- 92.Hartung N.M., Mainka M., Pfaff R., Kuhn M., Biernacki S., Zinnert L., Schebb N.H. Development of a quantitative proteomics approach for cyclooxygenases and lipoxygenases in parallel to quantitative oxylipin analysis allowing the comprehensive investigation of the arachidonic acid cascade. Anal. Bioanal. Chem. 2023;415:913–933. doi: 10.1007/s00216-022-04489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koch E., Wiebel M., Löwen A., Willenberg I., Schebb N.H. Characterization of the oxylipin pattern and other fatty acid oxidation products in freshly pressed and stored plant oils. J. Agric. Food Chem. 2022;70:12935–12945. doi: 10.1021/acs.jafc.2c04987. [DOI] [PubMed] [Google Scholar]
- 94.Hartung N.M., Mainka M., Kampschulte N., Ostermann A.I., Schebb N.H. A strategy for validating concentrations of oxylipin standards for external calibration. Prostag. Other Lipid Mediat. 2019;141:22–24. doi: 10.1016/j.prostaglandins.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 95.Koch E., Löwen A., Nikolay S., Willenberg I., Schebb N.H. Trans-hydroxy, trans-epoxy, and erythro-dihydroxy fatty acids increase during deep-frying. J. Agric. Food Chem. 2023;71:7508–7513. doi: 10.1021/acs.jafc.3c00964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its supplementary information files. Sequencing data have been deposited under the GEO accession number GSE208160.