Abstract
To ensure biological validity in metabolic phenotyping, findings must be replicated in independent sample sets. Targeted workflows have long been heralded as ideal platforms for such validation due to their robust quantitative capability. We evaluated the capability of liquid chromatography-mass spectrometry (LC-MS) assays targeting organic acids and bile acids to validate metabolic phenotypes of SARS-CoV-2 infection. Two independent sample sets were collected: (1) Australia: plasma, SARS-CoV-2 positive (n = 20), noninfected healthy controls (n = 22) and COVID-19 disease-like symptoms but negative for SARS-CoV-2 infection (n = 22). (2) Spain: serum, SARS-CoV-2 positive (n = 33) and noninfected healthy controls (n = 39). Multivariate modeling using orthogonal projections to latent structures discriminant analyses (OPLS-DA) classified healthy controls from SARS-CoV-2 positive (Australia; R2 = 0.17, ROC-AUC = 1; Spain R2 = 0.20, ROC-AUC = 1). Univariate analyses revealed 23 significantly different (p < 0.05) metabolites between healthy controls and SARS-CoV-2 positive individuals across both cohorts. Significant metabolites revealed consistent perturbations in cellular energy metabolism (pyruvic acid, and 2-oxoglutaric acid), oxidative stress (lactic acid, 2-hydroxybutyric acid), hypoxia (2-hydroxyglutaric acid, 5-aminolevulinic acid), liver activity (primary bile acids), and host-gut microbial cometabolism (hippuric acid, phenylpropionic acid, indole-3-propionic acid). These data support targeted LC-MS metabolic phenotyping workflows for biological validation in independent sample sets.
Keywords: metabolic phenotyping array, metabolic phenotyping, validation, LC-MS, SARS-CoV-2, COVID-19, TCA cycle, hypoxia, organic acids, oxidative stress, bile acids
Introduction
Metabolic phenotyping is a powerful tool that enables for the comprehensive analysis of the molecular content of biological fluids or tissue.1 The technique allows for the identification of discriminatory markers and multimarker signatures capable of both stratification and classification at the individual level as well as identification of metabolic risk markers of disease at the population level.1 Advances in mass spectrometry protocols that target known metabolites enable the quantification of large panels of metabolites to capture precise detail on known biochemical pathways within a system.2
To ensure validity of results, once a discovery sample set has revealed putative metabolic phenotypes, it is widely accepted that validation must be performed in independent sample sets.3 Targeted workflows have long been heralded as ideal platforms for such biomarker validation due to their robust quantitative capability2 derived from substantial bioanalytical validation.4 However, the performance of targeted assays in biomarker validation when faced with intercohort sampling variation that can be experienced in real-world scenarios is rarely reported.
In ideal scenarios, validation is performed in independent sample sets collected under matched protocols, designed from the ground up. However, this can often be challenging to achieve, particularly in cases where sample availability is suboptimal or in cases where validation assays are performed on previously collected samples, where collection protocols cannot be retrospectively matched. A pertinent example of this is during the 2019 SARS-CoV-2 outbreak that caused unprecedented demand on international health care systems, with over 750 million confirmed cases to the world health organization (WHO) and over 7 million individuals reported to have died as a result of infection.5 The immense pressure that the initial phase of the pandemic caused on clinical healthcare providers often meant research was reactive rather than proactive, resulting in sample sets that were collected using unmatched protocols. Therefore, we sought to evaluate the feasibility of using targeted metabolic phenotyping workflows to perform biomarker validation in such a real-world scenario, where sample sets were not optimally collected using standardized protocols.
SARS-CoV-2 was identified as an ideal exemplar for this evaluation. Metabolic phenotyping has been widely employed throughout the pandemic to document the metabolic implications of infection, with reported perturbations in a range of metabolic pathways including amino acids,6−10 biogenic amines,6,8,10,11 and the lipidome.12 Indeed, previous work from our group has demonstrated that a metabolic phenoconversion exists indicative of a multiorgan systemic impact of infection,10 resulting in a phenotype that is predictive of severity13 and prognosis,14 undergoing phenoreversion throughout the recovery phase of the disease.15 However, due to the aforementioned challenges with the pandemic, comprehensive sample sets collected from independent populations were not always available. In this study, we evaluated whether targeted metabolic phenotyping platforms applied to different blood sample types (EDTA plasma vs serum) collected at the height of the pandemic and therefore corresponding to similar strains of the virus would reveal complementary results regarding the biochemical response associated with acute infection. To achieve this we employed two ultrahigh performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS) metabolite arrays, the first designed to target organic acids, capturing metabolites that are key in mitochondrial energy metabolism and the tricarboxylic acid (TCA) cycle, as well as metabolites that are indicative of oxidative stress,16 providing a snapshot of host cellular energy generation during infection. Furthermore, the assay was adept at capturing gut-microbial/host cometabolites, considered as an important link in the synergistic microbiome–host relationship increasingly thought to influence host immune function17,18 and linked to severity of COVID-19 disease outcomes.19 The second assay targeted host bile acid metabolism, a poorly studied area of SARS-CoV-2 infection. Bile acids are a key class of bioactive metabolites that result from the catabolism of cholesterol. They have multiple physiological roles, including modulation of bile flow and lipid secretion and metabolism, and have also been implicated in the regulation of cholesterol homeostasis. Bile acids are also modulated by the gut-microbiome and can influence receptors and cells of the host immune system.20 These complementary assays were applied to blood serum and plasma samples collected from two independent population cohorts and include noninfected healthy controls, individuals who tested positive for SARS-CoV-2 infection and individuals who reported with COVID-19 disease like symptoms but tested negative for the SARS-CoV-2 virus.
Materials and Methods
Study Samples
The study consisted of two independent cohorts: a cohort of plasma samples collected from Australia and serum samples collected from Spain. Quality control samples consisted of a commercial pooled sample matched to the biofluid for each cohort (plasma/serum).
Australian Cohort
The Australian cohort consisted of plasma samples collected from Perth, WA, Australia. The study was initiated at Fiona Stanley Hospital by the COVID Research Response Collaboration (https://research-au.net/covid-research-response/, accessed on December 12, 2022) as part of the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC)/World Health Organization (WHO) pandemic trial framework (SMHS Research Governance Office PRN:3976 and Murdoch University Ethics no. 2020/052, and no. 2020/053).
Plasma samples were collected from 64 individuals who were either (i) SARS-CoV-2 positive (n = 20), (ii) healthy control individuals who displayed no signs or symptoms of infection (n = 22), and (iii) SARS-CoV-2-negative (n = 22) individuals, where the individuals reported to COVID-19 clinics with signs and symptoms but tested negative for SARS-CoV-2 infection. Cohort demographics and descriptions are given in Table 1.
Table 1. Patient Samples in the Australia and Spain Sample Sets.
| healthy control | SARS-CoV-2 positive | SARS-CoV-2 negative | |
|---|---|---|---|
| Australia | |||
| n | 22 | 20 | 22 |
| age (mean ± [sd]) | 54.35 [18.05] | 67.32 [12.76] | 47.64 [15.05] |
| biological sex (M/F) | 14/8 | 6/13a | 9/13 |
| Spain | |||
| n | 39 | 33 | NA |
| age (mean ± [sd]) | 42.15 [14.57] | 72.09 [20.32] | NA |
| biological sex (M/F) | 22/17 | 10/22a | NA |
Missing some clinical annotation data.
Spanish Cohort
A cohort of serum samples were obtained from collaborators at the Centro de Investigación Cooperativa en Biociencias—CIC bioGUNE, Derio, Bizkaia, Spain. Patients were recruited at the Basurto University Hospital and Cruces University Hospital in the Basque Country (Spain), with 33 samples collected from individuals who tested positive for SARS-CoV-2 and 39 samples collected from healthy controls. The Spanish cohort did not contain SARS-CoV-2 negative symptomatic participants. A demographic description of the cohort is provided in Table 1. The project was evaluated and approved by the Comité de Ética de Investigación con Medicamentos de Euskadi (CEIm-E, PI + CES-BIOEF 2020–04 and PI219130). All samples were supplied by the Basque Biobank for Research (BIOEF). Samples were imported under Import Permit 0004275122 issued by the Australian Government’s Department of Agriculture, Water, and the Environment.
Metabolite Phenotyping Array
Targeted metabolite phenotyping analysis was performed using a commercial analytical array (Q300 metabolite array kit, Metabo-Profile, Human Metabolome Institute, Shenzhen, Guangdong, China) previously published,16 which contained all reagents, standards, and internal standards required for analysis. Analysis was performed as per the standard operating protocol supplied by the manufacturer and produced quantitative data for 115 target metabolites (Table S1).
In detail, 20 μL aliquots of plasma (Australia) or serum (Spain) were transferred to 96 well plates. To each well 120 μL of methanol containing stable isotope labeled internal standards (Table S1) was added, followed by 20 min on a plate shaker and 20 min of centrifugation at 4000g. Following centrifugation, 30 μL of the supernatant was removed and transferred to a fresh 96-well plate.
Resultant extracts then underwent a derivatization procedure with the addition of 20 μL of methanol containing 200 mM 3-nitrophenylhydrazine (3-NPH) and 20 μL of methanol containing 6% pyridine (v/v) and 96 mM 1-ethyl-3(3-(dimethylamino)propyl)carbodiimide (EDC). Derivatization of target metabolites was achieved following incubation at 30 °C for 60 min on a plate mixer. Extract preparations were diluted with 330 μL of 1:1 methanol:water (v/v) and mixed for 5 min on a plate shaker. Samples underwent centrifugation for 20 min at 4000g. Finally 140 μL of supernatant were transferred to a fresh 96 well plate, foil sealed and transferred to a UPLC-MS/MS system consisting of a Waters ACQUITY UPLC (Waters Corp., Milford, MA, USA) coupled to a Waters Xevo TQ-XS MS (Waters Corp., Wilmslow, UK) for analysis. A 5 μL injection volume was performed.
Chromatographic separations were performed on an ACQUITY BEH C18 analytical column (1.7 μm, 100 mm × 2.1 mm) (Waters, Milford, MA) with an additional ACQUITY BEH C18 VanGuard precolumn (1.7 μm, 5 mm × 2.1 mm) (Waters, Milford, MA). The solvent system consisted of (A) water (0.1% formic acid) and (B) acetonitrile–2-propanol (70:30 v/v) (0.1% formic acid). Solvent was delivered at 0.4 mL/min using the following linear gradient: 0–1 min (hold at 5% B), 1–5 min (5–30% B), 5–9 min (30–50% B), 9–12 min (50–78% B), 12–15 min (78–95% B), 15–16 min (95–100%B), 16–18 min (hold at 100%B), 18–18.1 min (100–5% B), 18.1–20 min (hold at 5% B).
The TQ-XS MS instrument was operated in both positive (1.5 kV) and negative (2 kV) ion modes, with a cone voltage of 60 V, source temperature of 150 °C, desolvation gas (nitrogen) temperature of 550 °C and flow of 1000 L/h, cone gas (nitrogen), 150 L/h; nebulizer gas (nitrogen), 7 bar. Multiple reaction monitoring (MRM) transition and retention time data is reported in Table S2.
Peak integration, calibration, and quantification for each metabolite from raw data was performed using automated Q300 processing with the Targeted Metabolome Batch Quantification (TMBQ) software (v1.0, HMI, Shenzhen, Guangdong, China).16
Bile Acids Assay
Bile acid data were collected using an in-house protocol developed and validated following fit-for-purpose recommendations for bioanalytical workflows from the European Bioanalysis Forum adapted from the US Food and Drug administration.21,22 Bile acid standards: chenodeoxycholic acid, cholic acid, glycochenodeoxycholic acid, glycocholic acid, glycodeoxycholic acid, glycoursodeoxycholic acid, taurocholic acid, taurochenodeoxycholic acid, taurodeoxycholic acid, ursodeoxycholic acid, deoxycholic acid, glycolithocholic acid, lithocholic acid, taurohyodeoxycholic acid, and taurolithocholic acid were purchased from Steraloids (Newport, RI, USA). Stable isotope labeled internal standard mixtures of unconjugated and conjugated bile acids were purchased from Cambridge Isotope Laboratories (Tewksbury, MA, USA) (Table S1). Optima grade solvents, acetonitrile, methanol, propan-2-ol, dimethyl sulfoxide (DMSO), and Lichropur grade ammonium acetate were purchased from Thermo Fisher Scientific (Malaga, WA, Australia). Water was obtained from a Milli-Q IQ-7000 system (Merck, Bayswater, VIC, Australia).
Stock solutions of bile acid standards were prepared at 1000 μg/mL in methanol (except for GCA that was dissolved in DMSO). Individual stock solutions were combined to generate an equimolar working stock solution at 1000 ng/mL in 50/50 water/methanol (v/v). Calibrators and quality controls were prepared from the working stock solution and further diluted with 50/50 water/methanol (v/v) to 1000, 600, 300, 150, 75, 25, 15, and 5 ng/mL for calibrators and 800, 320, 32, and 8 ng/mL for quality controls. The stable isotope internal standard solution mixture (ISTD, 200 ng/mL in methanol) was prepared by diluting stock solutions at 100 μg/mL in methanol.
Plasma samples were thawed at 4 °C, vortex mixed, and centrifuged, and 20 μL was transferred to a 500 μL 96-well plate (Eppendorf). 20 μL of each calibrator and quality control were transferred to the sample plate. 40 μL of ISTD solution (200 ng/mL in methanol) was added to each well followed by 200 μL methanol for protein precipitation. The 96-well plate was heat sealed and mixed at 1600 rpm for 10 min at 6 °C. Following centrifugation for 30 min at 6 °C, 180 μL of supernatant was transferred to a 350 μL 96-well plate (Eppendorf) and dried by using a SpeedVac vacuum concentrator (Thermo Fisher Scientific, MA, USA). Dried extracts were reconstituted in 50 μL of 50/50 water/methanol (v/v) containing 5 mM ammonium acetate prior to UPLC-MS/MS analysis.
UPLC-MS/MS analysis was performed using a Waters ACQUITY UPLC (Waters Corp., Milford, MA, USA) coupled to a Waters Xevo TQ-XS MS (Waters Corp., Wilmslow, UK). Chromatographic separation was performed on an ACQUITY HSS T3 2.1 × 150 mm, 1.8 μm column (Waters Corp., Milford, MA, USA) maintained at 40 °C. Linear gradient elution was performed at 0.4 mL/min. The mobile phase was composed of 5 mM ammonium acetate in water (A) and 5 mM ammonium acetate in 50/50 methanol/acetonitrile (v/v) (B). The gradient started at 20% B, increasing to 40% B over 1 min, 95% B at 6 min, 98% B at 7 min held until 9 min before returning to 20% B at 9.2 min for re-equilibration until 10.5 min. The weak and the strong washes were 90:10 water/propanol-2-ol (v/v) and 100% propanol-2-ol, respectively. A 5 μL injection volume was performed.
Mass spectrometry detection was performed using negative electrospray ionization (ESI) operated in multiple reaction monitoring (MRM) mode. The ion source settings were: capillary voltage, 2 kV; cone voltage, 60 V; desolvation temperature, 650 °C; desolvation gas flow (nitrogen), 900 L/h; cone gas (nitrogen), 150 L/h; nebulizer gas (nitrogen), 7 bar. MRM transition and retention time data is reported in Table S3. Peak integration, calibration, and quantification for each metabolite from raw data was performed using Waters TargetLynx package within MassLynx software (v4.2, Waters Corp., Wilmslow, UK).
Data Preprocessing and Statistical Analysis
Metabolites with >50% missing values were excluded from further statistical analysis. Remaining missing values were assumed to be missing as a result of being below the limit of detection and were therefore imputed using the half minimum concentration method. Metabolites with a concentration relative standard deviation of >30% across replicate preparations of a quality control reference sample were excluded. Metabolites were only included in the statistical analysis if they passed the above criteria in both the Australian and Spanish cohorts. Data describing the QC performance of each metabolite can be found in Table S1.
Multivariate and univariate statistical analysis was performed separately on each cohort due to the potential metabolite concentration differences between serum and plasma biofluids. For multivariate analysis, both principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA) were implemented from the metabom8 package (v1.0.0, https://github.com/tkimhofer/metabom8) in R (v4.2.2).
Univariate analysis was performed as follows: For the Australian cohort, which consisted of three participant classes (healthy control, SARS-CoV-2 negative, and SARS-CoV-2 positive), nonparametric Kruskal–Wallis and Dunn’s tests for pairwise multiple comparisons were performed. For the Spain cohort, which consisted of two participant classes (healthy control and SARS-CoV-2 positive), nonparametric Mann–Whitney U-tests were performed. To control the false discovery rate the method proposed by Benjamini–Hochburg was implemented to generate q-values.23 Metabolites were deemed significant if they returned a BH q-value of <0.1.
Results
A panel of 106 (Q300 metabolite array = 96; bile acid method = 10) metabolites passed quality control (QC) data filtering (RSD% < 30%; missing values <50% in replicate pooled QC analysis) and were matched across both the sample sets (Australia and Spain) (Table S1). Principal component analysis (PCA) was performed on the biological samples and replicate QC samples and demonstrated clustering of the QC samples and no biological sample outliers, indicating that the analysis had a technical variation lower than biological variation. The replicate QC samples clustered away from the biological samples in the Spanish cohort, since the QC was an external pooled reference serum (Figure S1A, S1B).
PCA using only study samples resulted in an unsupervised separation of healthy controls and SARS-CoV-2 positive samples in both cohorts, indicating a strong metabolic pattern that is indicative of SARS-CoV-2 infection. In the Australian cohort, the symptomatic but SARS-CoV-2 negative group clustered closely with the healthy controls, indicating a stronger metabolic signature associated with the SARS-CoV-2 positive group in contrast to the symptomatic (SARS-CoV-2 negative) group when comparing to healthy controls (Figure S1C, S1D).
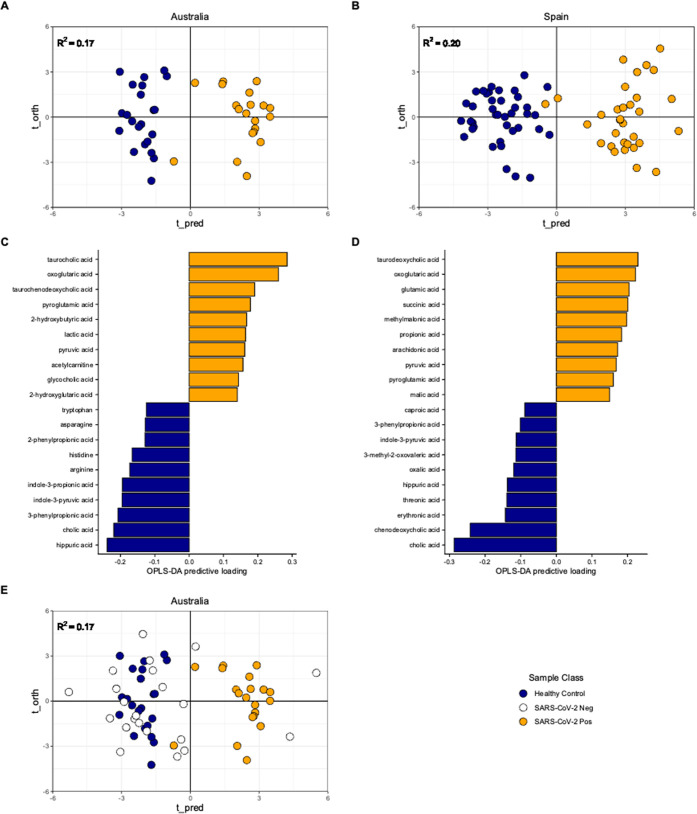
Supervised orthogonal projections to latent structures discriminant analysis (OPLS-DA) was performed independently on each cohort and revealed distinct metabolic signatures when comparing between the healthy control and SARS-CoV-2 positive groups. Model scores were: Australia, R2 = 0.17, ROC-AUC = 1.0; Spain R2 = 0.20, ROC-AUC = 1.0 (Figure 1, A and B). OPLS-DA loadings plots indicated a panel of common metabolites across the two cohorts that influenced separation consisting of lower concentrations of tryptophan, hippuric acid, indole-3-pyruvic acid, hydrocinnamic acid, and higher concentrations of glutamic acid, pyruvic acid, and oxoglutaric acid associated with SARS-CoV-2 infection (Figure 1C, 1D).
Figure 1.
Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) models of the study samples. Panels A and B present OPLS-DA score plots indicating supervised separation of healthy control and SARS-CoV-2 positive groups in both studies and biofluid types. Panels C and D present the metabolite feature loadings that have the greatest influence and drive the OPLS-DA model separation. Panel E presents SARS-CoV-2 negative data from the Australian cohort projected onto the Healthy Control vs SARS-CoV-2 positive Australian model in panel A.
Data acquired from individuals in the Australian plasma cohort who reported COVID-19 like symptoms but tested negative for SARS-CoV-2 infection (SARS-CoV-2 negative group) were projected into the Australian OPLS-DA healthy control vs SARS-CoV-2 positive model. The resultant scores plot indicated that only 3 of 22 were projected to cluster with the SARS-CoV-2 positive samples, with the majority (19 of 22), clustering on the healthy control side of the model (Figure 1E).
To validate and report the metabolites that were consistently perturbed across both cohorts and biofluids, univariate analysis using nonparametric Kruskal–Wallis (Australian cohort, three groups) and Mann–Whitney (Spain, two groups) were then performed on the 106 metabolites. The false discovery rate was controlled using the method described by Benjamini and Hochburg, resulting in the generation of q-values.21 A panel of 24 metabolites returned a matched q-value of <0.1 across both the Australian and Spanish cohorts (Table 2, Figure S2). Full univariate outputs are reported in Table S4.
Table 2. Univariate Analysis Resultsa.
| Australia
(Kruskal–Wallis (KW)
with post hoc Dunn’s test (DT), three groups: healthy; SARS-CoV-2
neg; SARS-CoV-2 pos) |
Spain
(Mann–Whitney (MW), two
groups: healthy; SARS-CoV-2 pos) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| metabolite | direction of change | KW p | BH q | healthy control mean conc. (sd) [μM] | SARS-CoV-2 pos mean conc. (sd) [μM] | SARS-CoV-2 neg mean conc. (sd) [μM] | QC mean conc. (sd) [μM] | healthy control – SARS-CoV-2 neg | healthy control – SARS-CoV-2 pos | SARS-CoV-2 neg – SARS-CoV-2 pos | MW p | BH q | healthy control mean conc. (sd) [μM] | SARS-CoV-2 pos mean conc. (sd) [μM] | QC mean conc. (sd) [μM] |
| 2-hydroxy-butyric acid | ↑ | 4.20 × 10–04 | 2.34 × 10–03 | 68.97 (27.87) | 126.98 (39.67) | 70.58 (69.3) | 48.78 (2.94) | 3.52 × 10–01 | 1.52 × 10–04 | 5.97 × 10–04 | 6.08 × 10–04 | 2.21 × 10–03 | 62.02 (43.92) | 95.12 (78.59) | 74.52 (17.2) |
| 2-hydroxy-glutaric acid | ↑ | 5.31 × 10–03 | 1.76 × 10–02 | 3.81 (1.29) | 4.74 (1.88) | 3.85 (2.02) | 1.86 (0.3) | 1.89 × 10–01 | 8.19 × 10–04 | 1.11 × 10–02 | 2.39 × 10–03 | 6.18 × 10–03 | 3.34 (2.31) | 5.44 (4.86) | 7.89 (1.37) |
| 2-phenyl-propionic acid | ↓ | 6.07 × 10–06 | 9.18 × 10–05 | 0.32 (0.24) | 0.01 (0.16) | 0.2 (0.22) | 0.16 (0.01) | 3.18 × 10–02 | 5.71 × 10–07 | 1.12 × 10–03 | 5.93 × 10–04 | 2.21 × 10–03 | 0.48 (0.38) | 0.14 (0.24) | 0.27 (0.03) |
| 3-phenyl-propionic acid | ↓ | 4.02 × 10–06 | 8.52 × 10–05 | 1.14 (0.86) | 0.06 (0.51) | 0.6 (0.74) | 0.65 (0.15) | 1.53 × 10–02 | 3.25 × 10–07 | 2.08 × 10–03 | 6.74 × 10–04 | 2.23 × 10–03 | 0.98 (0.84) | 0.32 (0.52) | 0.57 (0.1) |
| 5-amino-levulinic acid | ↑ | 3.26 × 10–04 | 1.96 × 10–03 | 3.17 (0.63) | 4.06 (0.97) | 3.03 (0.92) | 4.09 (0.58) | 4.47 × 10–01 | 1.85 × 10–04 | 3.01 × 10–04 | 1.01 × 10–04 | 4.88 × 10–04 | 2.82 (0.74) | 3.58 (1.5) | 6.27 (0.99) |
| aspartic acid | ↑ | 5.60 × 10–05 | 5.40 × 10–04 | 5.8 (2.13) | 9.43 (3.38) | 7.3 (5.06) | 14.11 (1.21) | 7.88 × 10–02 | 6.82 × 10–06 | 1.49 × 10–03 | 5.47 × 10–05 | 3.04 × 10–04 | 21.89 (10.52) | 37.41 (17.81) | 72.87 (16.88) |
| glutamic acid | ↑ | 1.41 × 10–03 | 6.49 × 10–03 | 9.76 (7.99) | 19.02 (8.93) | 12.98 (8.17) | 33.56 (3.15) | 1.32 × 10–01 | 1.90 × 10–04 | 6.87 × 10–03 | 5.33 × 10–11 | 3.82 × 10–09 | 11.46 (6.31) | 26.85 (13.3) | 74.13 (9.63) |
| glutamine | ↓ | 2.32 × 10–03 | 9.47 × 10–03 | 367.41 (89.23) | 311.36 (50.55) | 316.07 (55.08) | 216.2 (9.77) | 2.24 × 10–03 | 8.33 × 10–04 | 3.55 × 10–01 | 2.27 × 10–02 | 4.23 × 10–02 | 109752.03 (14792.38) | 98053.08 (23220.43) | 46951.44 (7198.67) |
| hippuric acid | ↓ | 1.60 × 10–04 | 1.21 × 10–03 | 7.11 (8.48) | 2.2 (4.79) | 3.85 (3.91) | 4 (0.15) | 1.01 × 10–02 | 1.53 × 10–05 | 2.87 × 10–02 | 7.63 × 10–04 | 2.45 × 10–03 | 9.55 (9.68) | 4.12 (7.41) | 5.57 (0.91) |
| indole-3-propionic acid | ↓ | 1.09 × 10–05 | 1.45 × 10–04 | 1.84 (1.38) | 0.23 (17.33) | 1.81 (1.68) | 1.14 (0.14) | 4.94 × 10–01 | 1.46 × 10–05 | 1.56 × 10–05 | 1.96 × 10–02 | 3.77 × 10–02 | 1.67 (1.21) | 0.98 (1.81) | 1.44 (0.18) |
| indole-3-pyruvic acid | ↓ | 1.66 × 10–06 | 4.41 × 10–05 | 83.33 (22.64) | 40.1 (14.28) | 73.88 (23.17) | 20.43 (1.78) | 2.85 × 10–01 | 1.01 × 10–06 | 1.34 × 10–05 | 8.99 × 10–04 | 2.72 × 10–03 | 146.58 (56.97) | 101.7 (60.15) | 2.94 (0.66) |
| lactic acid | ↓ | 2.75 × 10–05 | 2.92 × 10–04 | 614.23 (136.68) | 960.34 (422.92) | 550.57 (166.95) | 960.68 (100.36) | 2.23 × 10–01 | 1.76 × 10–04 | 7.95 × 10–06 | 1.20 × 10–08 | 1.41 × 10–07 | 817.58 (165.75) | 1319.36 (504.51) | 886.62 (105.83) |
| malic acid | ↓ | 5.23 × 10–03 | 1.76 × 10–02 | 4.18 (1.52) | 6.32 (3.74) | 3.64 (2.5) | 4.71 (0.23) | 9.41 × 10–02 | 2.60 × 10–02 | 6.24 × 10–04 | 8.83 × 10–08 | 7.80 × 10–07 | 3.22 (1.13) | 5.75 (3.64) | 4.3 (0.47) |
| n-acetyl-neuraminic acid | ↓ | 1.24 × 10–06 | 4.37 × 10–05 | 1.1 (0.17) | 1.58 (0.86) | 1.1 (0.43) | 1.05 (0.09) | 4.97 × 10–01 | 2.60 × 10–06 | 2.70 × 10–06 | 4.54 × 10–10 | 9.63 × 10–09 | 0.84 (0.22) | 1.9 (0.91) | 0.85 (0.13) |
| o-adipoyl-carnitine | ↑ | 1.10 × 10–06 | 4.37 × 10–05 | 0.01 (0) | 0.04 (0.05) | 0.01 (0.01) | 0.01 (0) | 9.83 × 10–02 | 2.08 × 10–07 | 7.20 × 10–05 | 9.45 × 10–03 | 1.96 × 10–02 | 0.02 (0.02) | 0.03 (0.11) | 0.01 (0) |
| oleylcarnitine | ↑ | 1.21 × 10–02 | 3.68 × 10–02 | 0.21 (0.05) | 0.27 (0.15) | 0.23 (0.08) | 0.13 (0.01) | 2.40 × 10–01 | 2.10 × 10–03 | 1.48 × 10–02 | 1.08 × 10–04 | 4.96 × 10–04 | 0.22 (0.11) | 0.34 (0.18) | 0.12 (0.03) |
| oxoglutaric acid | ↑ | 8.26 × 10–07 | 4.37 × 10–05 | 5.15 (2.38) | 15.32 (4.85) | 5.02 (9.7) | 52.44 (4.09) | 4.01 × 10–01 | 1.08 × 10–06 | 3.53 × 10–06 | 5.69 × 10–10 | 1.00 × 10–08 | 22.61 (14.55) | 52.38 (62.12) | 18.88 (4.08) |
| palmitoyl-carnitine | ↑ | 3.12 × 10–02 | 8.49 × 10–02 | 0.14 (0.03) | 0.16 (0.06) | 0.12 (0.04) | 0.11 (0.01) | 4.87 × 10–01 | 1.03 × 10–02 | 1.12 × 10–02 | 1.40 × 10–03 | 4.01 × 10–03 | 0.08 (0.03) | 0.12 (0.04) | 0.07 (0.01) |
| pyroglutamic acid | ↑ | 1.87 × 10–05 | 2.20 × 10–04 | 16.87 (8.23) | 30.82 (11.19) | 19.88 (8.6) | 38.48 (3.81) | 9.35 × 10–02 | 2.67 × 10–06 | 5.52 × 10–04 | 4.11 × 10–10 | 9.63 × 10–09 | 23.62 (7.7) | 40.47 (15) | 98.7 (15.4) |
| pyruvic acid | ↑ | 9.70 × 10–05 | 8.57 × 10–04 | 112.94 (46.59) | 221.69 (93.44) | 105.9 (42.89) | 141.34 (8.33) | 1.64 × 10–01 | 7.47 × 10–04 | 1.80 × 10–05 | 2.43 × 10–06 | 1.84 × 10–05 | 51.91 (31.8) | 118.32 (69.7) | 9.34 (1.29) |
| tryptophan | ↓ | 2.35 × 10–04 | 1.56 × 10–03 | 151.27 (35.11) | 107.71 (24.65) | 143.68 (33.57) | 106.68 (10.1) | 4.36 × 10–01 | 2.45 × 10–04 | 1.34 × 10–04 | 6.47 × 10–04 | 2.21 × 10–03 | 162.99 (26.31) | 128.91 (43.65) | 181.81 (26.88) |
| glycocholic acid | ↑ | 3.65 × 10–02 | 9.43 × 10–02 | 63.12 (81.07) | 197.33 (209.57) | 147.96 (135.02) | 181.37 (2.09) | 5.76 × 10–02 | 5.47 × 10–03 | 1.57 × 10–01 | 6.31 × 10–03 | 1.36 × 10–02 | 0.23 (0.36) | 0.57 (0.68) | 1.67 (0.05) |
| taurocholic acid | ↑ | 8.58 × 10–04 | 4.33 × 10–03 | 7.82 (29.01) | 50.48 (113) | 25.77 (54.24) | 49.95 (1.72) | 2.32 × 10–02 | 8.73 × 10–05 | 3.52 × 10–02 | 1.90 × 10–03 | 5.17 × 10–03 | 7.96 (26.19) | 26.75 (115.29) | 121.47 (3.33) |
An additional 52 metabolites (17 Australia, 35 Spain) were reported as significant in only a single cohort. For Australia, this included bile acids (taurochenodeoxycholic acid), carnitines (acetylcarnitine, butyrylcarnitine, carnitine, propionylcarnitine), carbohydrates (glucose, glyceric acid), amino acids (alanine, alpha-aminobutyric acid, arginine, histidine, phenylalanine, proline, sarcosine, threonine), and tryptophan metabolites (kynurenine, indole-3-acetic acid). In comparison, metabolites only significant in the Spain cohort included, bile acids (chenodeoxycholic acid, cholic acid, glycoursodeoxycholic acid, taurodeoxycholic acid), amino acids (beta-alanine, creatine, glycine, n-acetyaspartic acid, ornithine, phenylacetylglutamine, serine), carbohydrates (phenylacetic acid, erythronic acid), carnitines (linoleyl carnitine, stearoylcarnitine), short and medium chain fatty acids (10z-nonadecenoic acid, alpha-linolenic acid, arachidonic acid, caproic acid, docosahexaenoic acid, isovaleric acid, linoleic acid, myristoleic acid, propionic acid, sebacic acid, valeric acid), organic carboxylic acids (3-hydroxybutyric acid, 3-methyl-2-oxovaleric acid, glutaric acid, ketoleucine, methylmalonic acid, oxalic acid, succinic acid, threonic acid) (Table S4).
Discussion
To establish the capability of targeted mass spectrometry to deliver robust and consistent metabolic phenotyping, we evaluated the performance of two methods when applied across independent sample sets collected from two different geographical locations consisting of different biofluids (Australia = plasma; Spain = serum).
Following data preprocessing and subsequent statistical analysis, independent OPLS-DA models were able to classify SARS-CoV-2 patients from healthy controls in both the Australian and Spanish cohorts, indicating a systemic metabolic shift following infection that is observed in both plasma and serum. Using the loadings of the OPLS-DA models, common metabolites that were discriminant in both cohorts included oxoglutaric acid, pyruvic acid, lactic acid, and glutamic acid, providing robust evidence that perturbations occur in the TCA cycle in response to viral infection. Further univariate analysis resulted in the generation of a panel of 23 metabolites that were present at significantly different concentrations in the healthy controls and SARS-CoV-2 positive participant groups in independent analyses of both the Australian plasma and Spanish serum sample sets (Table 2). This finding gives encouragement that targeted mass spectrometry metabolic phenotyping assays can be used to validate biological phenotypes across independent sample sets, even in cases in which collection protocols are unmatched.
Despite the consistency within the multivariate and univariate testing across the two sample sets, there were some discrepancies, with 17 metabolites statistically significant in univariate analysis only in the Australian plasma cohort and 35 statistically significant only in the Spanish serum cohort (Table S2). The reasons for this are unclear, however may be due to the sampling workflows and biofluid types used by each study location, with known reports of small molecule compositional difference between plasma and serum, with the protocols for each known to impact the integrity and stability of certain metabolites.24
Metabolite concentrations across the two cohorts were largely comparable (Table S4), with 62/82 (75%) metabolites reporting median concentrations within a factor of 2 when comparing the healthy control groups across the two cohorts. Interestingly, metabolites from the panel of 23 biomarkers had some of the highest variation in concentration between the two data sets, including oxoglutaric acid, aspartic acid, and glutamine that reported 4.5, 3.8, and 299.1× greater median concentrations in the Spanish serum cohort respectively (Figure S2, Table S4). Some metabolites, however, shared consistent concentrations, yet had differing univariate statistical outcomes. Specifically, succinic acid had a factor difference of 1.072 between cohorts, yet returned p = 4.48 × 10–11 in the Spanish cohort and p = 0.90 in the analysis of the Australian cohort. The reasons behind this are unclear, especially with much of the TCA cycle reporting differences between the healthy controls and the SARS-CoV-2 groups. However, as discussed previously, this may be due to the discrepancy in collection protocols and biofluid types between the study locations with differences in the performance of plasma and serum in small molecule bioanalysis previously reported in the literature.24
Despite this, the general distribution and direction of change between healthy controls and SARS-CoV-2 positive groups were consistent across both data sets, supporting the concept of an overall biomarker signature that can be achieved across independent sample sets regardless of sampling protocol, biofluid type, and concentration range encountered. From the analyses of both cohorts, a distinct phenotypic signature was identified that indicated perturbations in systemic metabolism in response to infection from the SARS-CoV-2 virus; these are discussed in the following sections.
Mitochondrial Central Cellular Energy Metabolism and the TCA Cycle
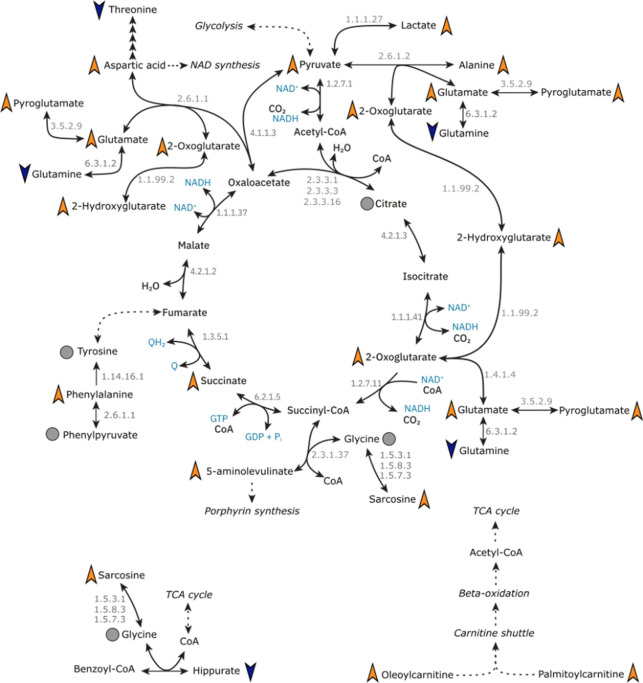
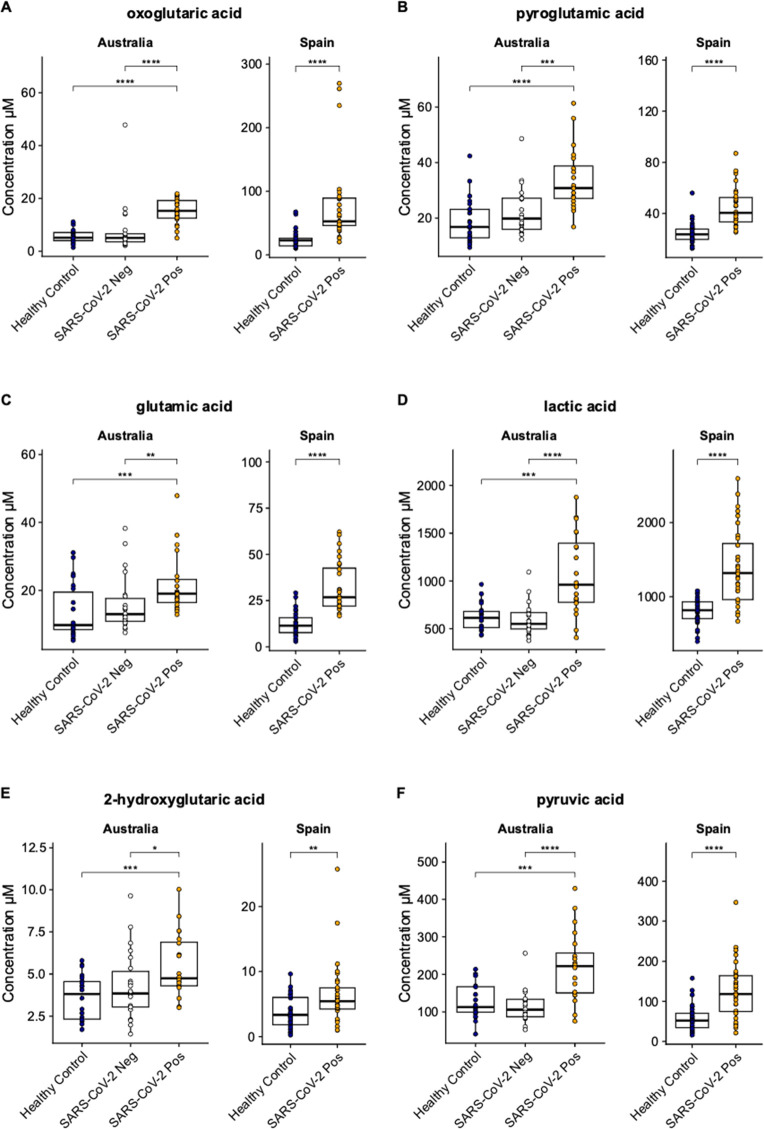
The TCA cycle is integral to mammalian cellular energy generation. The cycle consists of a series of organic acids that are also precursors of amino acids and starting products of key biochemical pathways. The derivatization reagent, 3-NPH, used in the Q300 metabolite array in this study targets carboxyl, carbonyl and phosphoryl groups, and therefore was especially adept at targeting the organic acids and amino acids that are either directly in the pathway, are peripheral to the cycles function or are downstream products of the cycle metabolites (Figure 2). Of these, pyruvic acid, 2-oxoglutaric acid, lactic acid, glutamic acid, pyroglutamic acid, 2-hydroxyglutaric acid, aspartic acid, alanine, and 5-aminolevulinic acid were all present at significantly higher concentrations in both the plasma and serum of the SARS-CoV-2 positive groups in both cohorts, when compared to the controls (Figure 3). Succinic acid, however, was only reported at a significantly higher concentration in the SARS-CoV-2 positive group in the Spain cohort, and not in the Australian cohort.
Figure 2.
Mapping systemic tricarboxylic acid (TCA) cycle and associated metabolite changes in SARS-CoV-2. The figure presents metabolites of the TCA cycle and peripheral metabolites that are perturbed following SARS-CoV-2 infection in our analyses in both plasma and serum biofluids. Orange triangles indicated higher concentration in SARS-CoV-2 positive groups compared with healthy controls, while blue triangles indicate a lower concentration in SARS-CoV-2 positive groups compared with healthy controls. Gray circle dots indicate the metabolite was measured, but there was no difference between SARS-CoV-2 positive and healthy control groups. Metabolites without annotation were not measured by the metabolite array.
Figure 3.
Population variations in individual metabolite levels. Boxplot visualizations of key metabolites from the TCA cycle that are present at significantly higher concentrations in SARS-CoV-2 infection compared with healthy controls in both the Australian plasma and Spanish serum samples. Key metabolites were (A) oxoglutaric acid; (B) pyroglutamic acid; (C) glutamic acid; (D) lactic acid; (E) 2-hydroxyglutaric acid; and (F) pyruvic acid. Boxplot annotations represent: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. A schematic of the pathway is presented in Figure 2. Additional boxplots of the remaining significant metabolites from the study are presented in Figure S2.
The presence of multiple TCA cycle metabolites indicates that the SARS-CoV-2 infection results in the occurrence of cellular hypermetabolism. This is supported further by the data with significant increases in concentration in two carnitine species, oleoylcarnitine and palmitoylcarnitine, members of the mitochondrial carnitine shuttle, the process adjacent to the TCA cycle, whereby free fatty acids are transferred to the mitochondria for beta-oxidation and generation of acetyl-CoA for use in the TCA cycle. This was observed in both plasma and serum, providing further indications of altered mitochondrial metabolism and TCA cycle activity after infection with the virus.
Previous literature has reported perturbations in the TCA cycle in SARS-CoV-2, consistent with our observations with reports of increases in the concentration of 2-oxoglutaric acid,9 glutamic acid,9,10,25 and lactic acid,9,25 as well as decreases in glutamic acid9,10,25 and no significant change in citric acid concentrations.9 However, inconsistencies in the literature exist for pyruvic acid, the initiating metabolite of the cycle, with studies reporting both an increase25 and decrease9 following SARS-CoV-2 infection. Our analyses presented here report a significant increase in pyruvic acid in both cohorts, further reinforcing the value in validating biological findings in multiple independent cohorts. However, our data reported different intercohort/sample type results for succinate, with a significant increase in concentration in SARS-CoV-2 positive individuals from Spain, but no change reported in the Australian group. The reasons for this are not immediately clear, with previous literature having also reported no significant change in succinic acid with SARS-CoV-2 infection,9 in agreement with our Australian plasma cohort data, and higher concentrations associated with infection as seen in the Spanish serum cohort.26
Mechanistically, previous in vitro work has observed increases in TCA cycle activity following SARS-CoV-2 infection, and was reported to be a result of increased glucose carbon entry into the cycle, due to increased expression of the enzyme pyruvate carboxylase.27 Our data across both biofluids would appear to support this hypermetabolism of the cycle, with increases in concentration of key TCA cycle metabolites including pyruvic acid and 2-oxoglutaric acid in the SARS-CoV-2 positive groups; however, there was no significant difference in the concentration of citric acid in either biofluid.
A further mechanistic source of increased TCA cycle activity may be a result of dysregulated glucose metabolism and glycolysis as an entry carbon source for the cycle. Plasma glucose concentrations have previously been reported at elevated concentrations following SARS-CoV-2 infection when compared with healthy controls10,25 in cross-sectional analyses. In longitudinal analyses, glucose has been observed to elevate in the active phase of the disease, but return to healthy levels following recovery from infection,15 and glucose dysregulation has been reported to adversely affect mortality and length of hospital stay in the disease.28 Furthermore, mechanistic in vitro work has indicated that increased rates of glycolysis are essential for SARS-CoV-2 viral replication.25 In the present study, we observed glucose levels to be increased for the Australian cohort and no change observed in the Spanish cohort. One possible explanation for the discrepancy in glucose findings is the timing of the sample collection in patients and what phase they were in at the time of collection, which may influence the metabolite concentrations.
Influence of Hypoxia in Acute Patients
Dysregulation of the TCA cycle occurs following systemic hypoxia (defined as low oxygen saturation levels SpO2 < 90–95%) that have been reported following infection with the virus.29 Previous literature indicates that hypoxia leads to the upregulation of pyruvate dehydrogenase kinase1 (PDK1) expression and inhibits glycolysis. PDK1 acts on the enzyme pyruvate dehydrogenase, phosphorylating it, and inhibiting its action on pyruvic acid, in turn disrupting the TCA cycle.30 This results in an excess of pyruvate and, subsequently, the conversion to lactic acid, a known metabolic product of glucose. In times of tissue hypoxia, the accumulation of lactic acid during reduced levels of oxygen saturation likely reflects a mitochondrial limitation which coincides with greater lactate accumulation.31 The response is rapid and as such has been proposed as a potential clinical biomarker for the monitoring of critically ill patients in intensive care units.31
Our data indicated the presence of hypoxia including observations of elevations of 2-hydroxyglutaric acid in both biofluid sample sets. 2-Hydroxyglutaric acid is an intermediary metabolite resulting from the reduction of the ketone group 2-oxoglutaric acid to a hydroxy group.32 2-Hydroxyglutaric acid has mostly been studied due to its propensity as a tumorigenic metabolite that is produced by cancerous cells which exhibit gain-of-function isocitrate dehydrogenase 1/2 mutations.32,33 When considering COVID-19, to the best of the author’s knowledge, changes in 2-hydroxyglutaric acid are yet to be reported, yet 2-hydroxyglutaric acid is known to be produced at significantly increased concentrations during times of tissue hypoxia.33,34 The mechanism of this hypoxic response has been demonstrated in mice, and reported to be a result of a hypoxia-inducible factor 1-alpha (HIF-1α)-dependent mechanism.33 It is possible that increases in 2-hydroxyglutaric acid may influence the adaptive immune response to SARS-CoV-2, with reports of accumulation, activation and differentiation of CD8+ T-cells,33 reported to play a role in the immune response to the disease35 and promote proinflammatory cytokine IL-1β and create an inflammatory glycolytic state.
Metabolic phenotyping during acute SARS-CoV-2 infection revealed higher concentrations of 5-aminolevulinic acid (5-ALA), a metabolic byproduct of the TCA cycle involved in the early phase of biosynthesis of porphyrins, in SARS-2-CoV positive individuals in both the Australian plasma and Spanish serum sample sets. Porphyrins are essential for the production of heme and oxygen carriage throughout the host system,36 hence elevations of in 5-ALA may result from a compensatory mechanism of the host attempting to increase heme production and subsequent oxygen carriage around the system. Interestingly, 5-ALA has been reported to exhibit in vitro antipathogenic effects against a range of pathogens including the SARS-CoV-2 virus.37,38
Oxidative Stress
The metabolic signatures observed in both cohorts of the study indicate increased systemic oxidative stress occurring postinfection. For example, 2-hydroxybutyric acid was present at significantly higher concentrations in the SARS-CoV-2 positive groups in both the Australian plasma and Spanish serum sample sets. 2-hydroxybutyric acid is formed in hepatic tissue as a byproduct of glutathione synthesis from cysteine. When the hepatic system undergoes states of oxidative stress, for example, in the detoxification of xenobiotics, the rate of hepatic glutathione synthesis increases, thereby increasing the concentration of 2-hydroxybutyric acid. The findings in our data support previous reports of elevated 2-hydroxybutyric acid concentrations in SARS-CoV-2 infection.26,39 As it is a marker of oxidative stress in the hepatic system, findings of increased concentrations re-enforce our earlier work of a multiorgan systemic signature of SARS-CoV-2 infection.10 Furthermore, as SARS-CoV-2 infection can lead to hypoglycemia and insulin resistance,40 an association of note is that 2-hydroxybutyric acid has also been reported to be useful as an early indicator of insulin resistance in nondiabetic subjects,41 while elevated serum 2-hydroxybutyric acid has been shown to predict glucose intolerance,42 and therefore it may be an overlap of underlying mechanism that leads to the elevations of the metabolite in SARS-CoV-2 infection.
Host–Microbiome Cometabolites
The host microbiome is known to influence on host immune function, with particular attention on the microbiome of the host gut18 and the production of host–microbiome cometabolites that play roles in both human and microbial metabolism. Interestingly the makeup of the host gut microbiome has been associated with outcomes in SARS-CoV-2 infection,19 and a panel of metabolites known to be host–microbiome cometabolites were reported in our data to alter in response to infection from the virus.
Hippuric acid is one such metabolite that is known to be a host-microbial cometabolite. Hippuric acid is omnipresent in human biology as a result of the mitochondrial catabolism and subsequent glycine conjugation of phenolic and benzoic acid containing compounds, including 2- and 3- phenylpropionic acid and phenylalanine, however it is also formed from microbially derived benzoic acid in the gut43 and known to be influenced by the composition of the gut microbiome.43 Indeed, germ-free in vivo models have been shown to renally excrete hippuric acid at significantly reduced concentrations compared with wild-type.44 Furthermore, hippuric acid has also been associated with increased gut-microbial diversity.43 Our data indicated hippuric acid, as well its precursors 2- and 3-phenylpropionate and phenylalanine were present at significantly lower concentrations in the plasma and serum of individuals infected with the SARS-CoV-2 virus. This was in agreement with previous observations in the literature,26,45,46 with hippuric acid also reported to associate with disease severity.19
Phenylpropionic acid precursors to hippuric acid are also associated with gut microbiome diversity. While the detailed biological role of 2-phenylpropionic acid has not been extensively described in the literature to date, however, isomeric phenylpropionate metabolites including 3-phenylpropionic acid, are reported to be derived from the amino acids phenylalanine and tyrosine by anaerobic gut microbial species including Clostridium,47 and are reported to be indicators of high gut microbial alpha diversity.48,49 While our analyses are the first time 2-phenylpropionic acid has been associated with SARS-CoV-2 infection, lower concentrations of 3-phenylpropionic acid have been previously reported in those infected with the virus and is supported by our findings in both biofluid types.50
The metabolic fate of the essential amino acids phenylalanine and tryptophan have also been hypothesized to be influenced by the gut microbiome and act as host–microbiome cometabolites51,52 and were found to be significantly different in the plasma and serum of the two SARS-CoV-2 positive groups when compared to the corresponding controls in each cohort. In the case of phenylalanine, a precursor for the aforementioned hippuric acid and phenylpropionic acid isoforms, we observed significantly higher concentrations in the SARS-CoV-2 group compared with the control groups in both the Australian plasma and Spanish serum sample sets, supporting previous observations in the literature,6,10,53 which also report that phenylalanine is associated with disease severity.54
Tryptophan, on the other hand, was observed to have a significantly lower concentration in plasma and serum of SARS-CoV-2 positive groups compared with the healthy controls, again supporting previous literature.6,10,11 The observation of tryptophan perturbation is curious and likely due to a combination of both host and microbial metabolism. For the host, elevations in the activity of indoleamine-2,3-dioxygenase (IDO) as a result of upregulated macrophage activity in response to infection metabolizes tryptophan via the kynurenine pathway, resulting in reduced concentrations of tryptophan and elevations in kynurenine and the subsequent kynurenine/tryptophan ratio.6,10,11 However, the observation of significant decreases in indole-3-pyruvic acid in our analyses indicate a parallel metabolic influence of the gut microbiome, as indole-3 pyruvic acid is thought to be microbially derived from gut microbial species including Clostridium sporogenes(55) and Trypanosoma brucei.56
Despite these observations, it should be noted that diet and medication, especially antibiotics, can impact the composition of the microbiome57,58 and subsequently impact host-microbial cometabolites such as hippuric acid and tryptophan. To confirm causative links between SARS-CoV-2 viral infection, the microbiome, and metabolic phenotypes, extensive additional work would be required that captures microbial compositional data, dietary, and medication intake.
Bile Acids
Bile acids are a key class of bioactive metabolites that result from the catabolism of cholesterol. They have multiple physiological roles including modulation of bile flow and lipid secretion and metabolism and have also been implicated in the regulation of cholesterol homeostasis. Results indicated both agreement and discrepancies between biofluid types, with cholic acid reported at lower concentrations in the SARS-CoV-2 positive groups in both types of biofluids, whereas taurocholic acid, taurochenodeoxycholic acid, and glycocholic acid were only significant in the Australia plasma sample set and taurodeoxycholic acid and glycoursodeoxycholic acid were only significant in the Spain serum sample set. It is unclear why there are discrepancies between the two cohorts and could be as a result of many factors, including the time of collection in relation to the patient journey, lifestyle and dietary factors, or true differences in biofluid type.
Cholic acid, taurocholic acid, and taurodeoxycholic acid are major primary bile acids and are synthesized by hepatocytes in the liver, while glycoursodeoxycholic acid and glycocholic acid are secondary bile acids produced by the colonic gut microbiome. Results from the analysis indicate that individuals infected with SARS-CoV-2 infection have increased metabolism of cholic acid to both its primary and secondary conjugates. Previously in the literature there have been minimal investigations into bile acid metabolism in SARS-CoV-2, however it has been reported that taurocholic acid, taurodeoxycholic acid, glycodeoxycholic acid, glycocholic, and glycoursodeoxycholic acid increase following infection with the virus46 which is in agreement with our findings. It is unclear as to the mechanistic relevance of increased bile acid conjugation from cholic acid to its derivatives, however adds further evidence to our previous reports that SARS-CoV-2 has a multiorgan signature indicating systemic metabolic impact reaching beyond the respiratory system.10 Unfortunately, clinical measures indicative of liver function, for example, measures of alanine transaminase (ALT) and/or total bile acids, were unavailable for the samples; however, future work that captures this information may be valuable in determining the systemic relevance of the observation in SARS-CoV-2 infection.
Interestingly, bile acids have been investigated as a therapeutic agent for SARS-CoV-2, in particular ursodeoxycholic acid, to inhibit farnesoid X receptor (FXR), reducing expression of angiotensin-converting enzyme 2 (ACE2), a key enzyme in the infection cycle of the virus,59 indicating that there may be benefits of bile acid conjugation in the systemic response to the disease.
N-Acetylneuraminic Acid
Finally, N-acetylneuraminic acid, the most abundant sialic acid in mammals, was also found at elevated concentrations in the plasma and serum of both cohorts. This observation is of particular note as N-acetylneuraminic acid containing glycolipids have been reported to be a critical component in the infection process of respiratory viruses, including SARS-CoV-2,60 MERS-CoV61 and influenza viruses.62 Furthermore, N-acetylneuraminic acid has been reported to bind in vitro to the SARS-CoV-2 spike protein.63 The mechanistic reasons behind elevations in free plasma/serum N-acetylneuraminic acid following infection with the virus are unclear and associations have not been previously reported in the literature; however, the association with spike protein binding and the host cell infection process are potentially worthy of future investigation.
Phenotypes of Non-SARS-CoV-2 Viral Induced Respiratory Infections
It should also be noted that many of the observations within this work are not unique to SARS-CoV-2 infection, with other respiratory infections reporting phenotypic similarities. Examples include influenzas (TCA cycle64), respiratory syncytial viruses (RSV) (TCA cycle65), human adenoviral infections (TCA cycle,66 bile acids67), and Mycoplasma pneumoniae bacterial infection (bile acids,68 amino acids68), indicating that such phenotypes are not unique and are not specific to act as diagnostic biomarkers in SARS-CoV-2 infection. However, despite this, understanding the metabolic consequence of viral infection can offer value beyond diagnostic capability, where the gold standard in SARS-CoV-2 remains RT-PCR.69 Indeed, insights into the systemic response to infection, regardless of viral cause, may help determine severity of infection,13,25 predict patient prognosis,13 and identify those who have prolonged phenotypic perturbations15 and may therefore require personalized intervention strategies.
When considering the third group of participants in the Australia cohort, who reported COVID-19 disease like symptoms but tested negative for SARS-CoV-2 infection, only 3/22 clustered in OPLS-DA models with the SARS-CoV-2 positive group. While the source of their respiratory symptoms was unconfirmed in this study, it indicated that the phenotypic response to SARS-CoV-2 infection may have some differences to other infectious pathogens. Unfortunately, there was not a corresponding SARS-CoV-2 negative group in the Spanish sample set, reinforcing the need to biologically validate findings across independent sample sets. To advance findings in future studies, the inclusion of multiple infection or insult types with orthogonal confirmation of the responsible infection pathogen would be of significant value.
Conclusions
Biological validation of the results in metabolic phenotyping studies is critical for clinical translation. We therefore evaluated the use of targeted liquid chromatography-mass spectrometry to generate and then biologically validate systemic metabolic phenotypes in the acute infection phase of SARS-CoV-2 infection. Signatures were compared in independent cohorts collected using unmatched protocols (plasma vs serum) at the height of the SARS-CoV-2 pandemic. Data resulted in agreement of 23 metabolites that were significantly perturbed following infection in both biofluids, providing validation confidence in the metabolite changes observed in response to infection. Our analyses support the concept of metabolic phenoconversion introduced by our group previously,10 with evidence of disruption to mitochondrial energy metabolism, oxidative stress, and dysbiosis of the host microbiome. These SARS-CoV-2 mediated disturbances contribute further insights into the systemic responses to infection and present potential markers for diagnostic and prognostic testing and targets for therapeutic action.
Furthermore, our findings reinforce the consensus that targeted metabolic phenotyping is robust when used for the biological validation of metabolic phenotypes in health and disease. In cases of optimal study design, samples are collected according to standardized protocols to minimize potential sources of external variation that might compromise the integrity of findings. However, we provide evidence that using such an approach can provide value in the biological validation of phenotypes where matched biofluids are not available, indicating that biofluids independently collected using nonmatched protocols (plasma vs serum) may still serve as a valuable validation set to corroborate phenotypic trends and signatures. This builds upon our previous work where we have demonstrated that targeted phenotyping workflows can be robust to different sampling conditions, including blood collection type and degrees of hemolysis when using targeted lipidomic70 and lipoprotein phenotyping.71 These findings justify that there remains value in validating biomarker signatures in cohorts in which samples may have been collected or treated differently, withstanding that the methodology is robust to external influences.
Acknowledgments
We would like to acknowledge the Western Australian COVID Research Response team (https://research-au.net/covid-research-response/ (accessed on October 24, 2023)).
Data Availability Statement
Raw mass spectrometry data for the project is freely available on the MassIVE server (project identifier: MSV000094022).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00797.
Table S1. Metabolite feature quality control performance metrics; Table S2. Mass spectrometry multiple reaction monitoring (MRM) transition and chromatographic retention time details for the Q300 metabolite array; Table S3. Mass spectrometry multiple reaction monitoring (MRM) transition and chromatographic retention time details for the quantitative bile acid method; Table S4. Results from univariate statistical analysis; Figure S1. Principal Component Analysis (PCA); Figure S2. Boxplot visualizations of the 23 metabolites that returned a Benjamini-Hochberg FDR q < 0.01 following univariate testing across both Australian and Spain cohorts (PDF)
We thank the Spinnaker Health Research Foundation (Fiona Stanley Hospital, 11 Robin Warren Drive, WA 6150, Australia), the McCusker Foundation, (186 Hampden Road, Nedlands, WA 6009, Australia), the Western Australian State Government, and the National Health and Medical Research Council Medical Research Future Fund (grant number 2014349) for funding the Australian National Phenome Centre for this and related work, and the Basque Country (Elkartek BG2019) and the Agencia Estatal de Investigación (Spain) for grant RTI2018-101269-B-I00. We thank the Department of Jobs, Tourism, Science, and Innovation, Government of Western Australian Premier’s Fellowship for funding EH, and ARC Laureate Fellowship funding for EH. JW thanks Ministerio de Ciencia, Tecnología e Innovación (Minciencias), Ministerio de Educación Nacional, Ministerio de Industria, Comercio y Turismo e ICETEX (792-2017) 2a Convocatoria Ecosistema Científico-Colombia Científica para la Financiación de Proyectos de I + D + (i), World Bank and Vicerrectoría de Investigaciones, Pontificia Universidad Javeriana, Bogotá, Colombia (contract no. FP44842-221-2018).
The authors declare no competing financial interest.
Supplementary Material
References
- Nicholson J. K.; Holmes E.; Kinross J. M.; Darzi A. W.; Takats Z.; Lindon J. C. Metabolic Phenotyping in Clinical and Surgical Environments. Nature 2012, 491 (7424), 384–392. 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- Roberts L. D.; Souza A. L.; Gerszten R. E.; Clish C. B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 10.1002/0471142727.mb3002s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. R.; Cristea I. M.; Dong M.-Q.; Eyers C. E.; LaBaer J.; Li J. V.; Nicholson J. K.; Overall C. M.; Palmblad M.; Slavov N. Want to Publish in JPR? This Is What You Need to Know!. J. Proteome Res. 2022, 21 (12), 2837–2839. 10.1021/acs.jproteome.2c00704. [DOI] [PubMed] [Google Scholar]
- Sarmad S.; Viant M. R.; Dunn W. B.; Goodacre R.; Wilson I. D.; Chappell K. E.; Griffin J. L.; O’Donnell V. B.; Naicker B.; Lewis M. R.; Suzuki T. A Proposed Framework to Evaluate the Quality and Reliability of Targeted Metabolomics Assays from the UK Consortium on Metabolic Phenotyping (MAP/UK). Nat. Protoc. 2023, 18 (4), 1017–1027. 10.1038/s41596-022-00801-8. [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int (accessed on February 5, 2024).
- Lawler N. G.; Gray N.; Kimhofer T.; Boughton B.; Gay M.; Yang R.; Morillon A.-C.; Chin S.-T.; Ryan M.; Begum S.; Bong S. H.; Coudert J. D.; Edgar D.; Raby E.; Pettersson S.; Richards T.; Holmes E.; Whiley L.; Nicholson J. K. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J. Proteome Res. 2021, 20 (5), 2796–2811. 10.1021/acs.jproteome.1c00052. [DOI] [PubMed] [Google Scholar]
- Gray N.; Lawler N. G.; Yang R.; Morillon A.-C.; Gay M. C. L.; Bong S.-H.; Holmes E.; Nicholson J. K.; Whiley L. A Simultaneous Exploratory and Quantitative Amino Acid and Biogenic Amine Metabolic Profiling Platform for Rapid Disease Phenotyping via UPLC-QToF-MS. Talanta 2021, 223, 121872. 10.1016/j.talanta.2020.121872. [DOI] [PubMed] [Google Scholar]
- Shen B.; Yi X.; Sun Y.; Bi X.; Du J.; Zhang C.; Quan S.; Zhang F.; Sun R.; Qian L.; Ge W.; Liu W.; Liang S.; Chen H.; Zhang Y.; Li J.; Xu J.; He Z.; Chen B.; Wang J.; Yan H.; Zheng Y.; Wang D.; Zhu J.; Kong Z.; Kang Z.; Liang X.; Ding X.; Ruan G.; Xiang N.; Cai X.; Gao H.; Li L.; Li S.; Xiao Q.; Lu T.; Zhu Y.; Liu H.; Chen H.; Guo T. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182 (1), 59–72. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.; Liu C.; Li D.; Huang Q.; Liu D.; Zhang Y.; Ye C.; Zhou D.; Wang Y.; Tan Y.; Li K.; Lin F.; Zhang H.; Lin J.; Xu Y.; Liu J.; Zeng Q.; Hong J.; Chen G.; Zhang H.; Zheng L.; Deng X.; Ke C.; Gao Y.; Fan J.; Di B.; Liang H. Metabolomic Analyses Reveal New Stage-Specific Features of COVID-19. Eur. Respir. J. 2022, 59 (2), 2100284. 10.1183/13993003.00284-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimhofer T.; Lodge S.; Whiley L.; Gray N.; Loo R. L.; Lawler N. G.; Nitschke P.; Bong S.-H.; Morrison D. L.; Begum S.; Richards T.; Yeap B. B.; Smith C.; Smith K. G. C.; Holmes E.; Nicholson J. K. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19 (11), 4442–4454. 10.1021/acs.jproteome.0c00519. [DOI] [PubMed] [Google Scholar]
- Almulla A. F.; Supasitthumrong T.; Tunvirachaisakul C.; Algon A. A. A.; Al-Hakeim H. K.; Maes M. The Tryptophan Catabolite or Kynurenine Pathway in COVID-19 and Critical COVID-19: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2022, 22 (1), 615. 10.1186/s12879-022-07582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.; Lawler N. G.; Zeng A. X.; Ryan M.; Bong S. H.; Boughton B. A.; Bizkarguenaga M.; Bruzzone C.; Embade N.; Wist J.; Holmes E.; Millet O.; Nicholson J. K.; Whiley L. Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection. Metabolites 2021, 11 (7), 467. 10.3390/metabo11070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge S.; Lawler N. G.; Gray N.; Masuda R.; Nitschke P.; Whiley L.; Bong S.-H.; Yeap B. B.; Dwivedi G.; Spraul M.; Schaefer H.; Gil-Redondo R.; Embade N.; Millet O.; Holmes E.; Wist J.; Nicholson J. K. Integrative Plasma Metabolic and Lipidomic Modelling of SARS-CoV-2 Infection in Relation to Clinical Severity and Early Mortality Prediction. Int. J. Mol. Sci. 2023, 24 (14), 11614. 10.3390/ijms241411614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffieux H.; Hanson A. L.; Lodge S.; Lawler N. G.; Whiley L.; Gray N.; Nolan T. H.; Bergamaschi L.; Mescia F.; Turner L.; de Sa A.; Pelly V. S.; Kotagiri P.; Kingston N.; Bradley J. R.; Holmes E.; Wist J.; Nicholson J. K.; Lyons P. A.; Smith K. G. C.; Richardson S.; Bantug G. R.; Hess C. A Patient-Centric Modeling Framework Captures Recovery from SARS-CoV-2 Infection. Nat. Immunol. 2023, 24, 1–10. 10.1038/s41590-022-01380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.; Wist J.; Masuda R.; Lodge S.; Nitschke P.; Kimhofer T.; Loo R. L.; Begum S.; Boughton B.; Yang R.; Morillon A.-C.; Chin S.-T.; Hall D.; Ryan M.; Bong S.-H.; Gay M.; Edgar D. W.; Lindon J. C.; Richards T.; Yeap B. B.; Pettersson S.; Spraul M.; Schaefer H.; Lawler N. G.; Gray N.; Whiley L.; Nicholson J. K. Incomplete Systemic Recovery and Metabolic Phenoreversion in Post-Acute-Phase Nonhospitalized COVID-19 Patients: Implications for Assessment of Post-Acute COVID-19 Syndrome. J. Proteome Res. 2021, 20 (6), 3315–3329. 10.1021/acs.jproteome.1c00224. [DOI] [PubMed] [Google Scholar]
- Xie G.; Wang L.; Chen T.; Zhou K.; Zhang Z.; Li J.; Sun B.; Guo Y.; Wang X.; Wang Y.; Zhang H.; Liu P.; Nicholson J. K.; Ge W.; Jia W. A Metabolite Array Technology for Precision Medicine. Anal. Chem. 2021, 93 (14), 5709–5717. 10.1021/acs.analchem.0c04686. [DOI] [PubMed] [Google Scholar]
- Kinross J. M.; Darzi A. W.; Nicholson J. K. Gut Microbiome-Host Interactions in Health and Disease. Genome Med. 2011, 3 (3), 14. 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D.; Liwinski T.; Elinav E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30 (6), 492–506. 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y. K.; Zuo T.; Lui G. C.-Y.; Zhang F.; Liu Q.; Li A. Y.; Chung A. C.; Cheung C. P.; Tso E. Y.; Fung K. S.; Chan V.; Ling L.; Joynt G.; Hui D. S.-C.; Chow K. M.; Ng S. S. S.; Li T. C.-M.; Ng R. W.; Yip T. C.; Wong G. L.-H.; Chan F. K.; Wong C. K.; Chan P. K.; Ng S. C. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients with COVID-19. Gut 2021, 70 (4), 698–706. 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik D.; Yao L.; Zhang Y.; Bae S.; D’Agostino G. D.; Zhang M.; Kim E.; Franzosa E. A.; Avila-Pacheco J.; Bisanz J. E.; Rakowski C. K.; Vlamakis H.; Xavier R. J.; Turnbaugh P. J.; Longman R. S.; Krout M. R.; Clish C. B.; Rastinejad F.; Huttenhower C.; Huh J. R.; Devlin A. S. Human Gut Bacteria Produce ΤΗ17-Modulating Bile Acid Metabolites. Nature 2022, 603 (7903), 907–912. 10.1038/s41586-022-04480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . Bioanalytical Method Validation Guidance for Industry; FDA. https://www.fda.gov/media/70858/download (accessed on February 5, 2024). [Google Scholar]
- Timmerman P.; White S.; McDougall S.; Kall M. A.; Smeraglia J.; Fjording M. S.; Knutsson M. Tiered Approach into Practice: Scientific Validation for Chromatography-Based Assays in Early Development - a Recommendation from the European Bioanalysis Forum. Bioanalysis 2015, 7 (18), 2387–2398. 10.4155/bio.15.168. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Liu X.; Hoene M.; Wang X.; Yin P.; Häring H.-U.; Xu G.; Lehmann R. Serum or Plasma, What Is the Difference? Investigations to Facilitate the Sample Material Selection Decision Making Process for Metabolomics Studies and Beyond. Anal. Chim. Acta 2018, 1037, 293–300. 10.1016/j.aca.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Krishnan S.; Nordqvist H.; Ambikan A. T.; Gupta S.; Sperk M.; Svensson-Akusjärvi S.; Mikaeloff F.; Benfeitas R.; Saccon E.; Ponnan S. M.; Rodriguez J. E.; Nikouyan N.; Odeh A.; Ahlén G.; Asghar M.; Sällberg M.; Vesterbacka J.; Nowak P.; Végvári A.; Sönnerborg A.; Treutiger C. J.; Neogi U. Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication. Mol. Cell. Proteomics 2021, 20, 100159. 10.1016/j.mcpro.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.; Yan R.; Lv L.; Jiang H.; Lu Y.; Sheng J.; Xie J.; Wu W.; Xia J.; Xu K.; Gu S.; Chen Y.; Huang C.; Guo J.; Du Y.; Li L. The Serum Metabolome of COVID-19 Patients Is Distinctive and Predictive. Metabolism 2021, 118, 154739. 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen P. J.; Garcia G.; Purkayastha A.; Matulionis N.; Schmid E. W.; Momcilovic M.; Sen C.; Langerman J.; Ramaiah A.; Shackelford D. B.; Damoiseaux R.; French S. W.; Plath K.; Gomperts B. N.; Arumugaswami V.; Christofk H. R. SARS-CoV-2 Infection Rewires Host Cell Metabolism and Is Potentially Susceptible to mTORC1 Inhibition. Nat. Commun. 2021, 12 (1), 1876. 10.1038/s41467-021-22166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabella S.; Gomez-Paz S.; Lam E.; Gonzalez-Mosquera L.; Fogel J.; Rubinstein S. Glucose Dysregulation and Its Association with COVID-19 Mortality and Hospital Length of Stay. Diabetes Metab. Syndr. 2022, 16 (3), 102439. 10.1016/j.dsx.2022.102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A.; Tabassum T.; Araf Y.; Al Nahid A.; Ullah Md. A.; Hosen M. J. Silent Hypoxia in COVID-19: Pathomechanism and Possible Management Strategy. Mol. Biol. Rep. 2021, 48 (4), 3863–3869. 10.1007/s11033-021-06358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I.; Cairns R. A.; Fontana L.; Lim A. L.; Denko N. C. HIF-1 Mediates Adaptation to Hypoxia by Actively Downregulating Mitochondrial Oxygen Consumption. Cell Metab. 2006, 3 (3), 187–197. 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Bakker J.; Nijsten M. W.; Jansen T. C. Clinical Use of Lactate Monitoring in Critically Ill Patients. Ann. Intensive Care 2013, 3 (1), 12. 10.1186/2110-5820-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X.; Hu H. The Roles of 2-Hydroxyglutarate. Front. Cell Dev. Biol. 2021, 10.3389/fcell.2021.651317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrakis P. A.; Palazon A.; Macias D.; Lee K. L.; Phan A. T.; Veliça P.; You J.; Chia G. S.; Sim J.; Doedens A.; Abelanet A.; Evans C. E.; Griffiths J. R.; Poellinger L.; Goldrath A. W.; Johnson R. S. S-2-Hydroxyglutarate Regulates CD8+ T-Lymphocyte Fate. Nature 2016, 540 (7632), 236–241. 10.1038/nature20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. L. A New Hydroxy Metabolite of 2-Oxoglutarate Regulates Metabolism in Hypoxia. Cell Metab. 2015, 22 (2), 198–200. 10.1016/j.cmet.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Rha M.-S.; Shin E.-C. Activation or Exhaustion of CD8+ T Cells in Patients with COVID-19. Cell. Mol. Immunol. 2021, 18 (10), 2325–2333. 10.1038/s41423-021-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner L. C.; Cox T. M. Biosynthesis of Heme in Immature Erythroid Cells. The Regulatory Step for Heme Formation in the Human Erythron. J. Biol. Chem. 1988, 263 (14), 6676–6682. 10.1016/S0021-9258(18)68695-8. [DOI] [PubMed] [Google Scholar]
- Sakurai Y.; Ngwe Tun M. M.; Kurosaki Y.; Sakura T.; Inaoka D. K.; Fujine K.; Kita K.; Morita K.; Yasuda J. 5-Amino Levulinic Acid Inhibits SARS-CoV-2 Infection in Vitro. Biochem. Biophys. Res. Commun. 2021, 545, 203–207. 10.1016/j.bbrc.2021.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwe Tun M. M.; Sakura T.; Sakurai Y.; Kurosaki Y.; Inaoka D. K.; Shioda N.; Yasuda J.; Kita K.; Morita K. Antiviral Activity of 5-Aminolevulinic Acid against Variants of Severe Acute Respiratory Syndrome Coronavirus 2. Trop. Med. Health 2022, 50 (1), 6. 10.1186/s41182-021-00397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone C.; Bizkarguenaga M.; Gil-Redondo R.; Diercks T.; Arana E.; García de Vicuña A.; Seco M.; Bosch A.; Palazón A.; San Juan I.; Laín A.; Gil-Martínez J.; Bernardo-Seisdedos G.; Fernández-Ramos D.; Lopitz-Otsoa F.; Embade N.; Lu S.; Mato J. M.; Millet O. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23 (10), 101645. 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govender N.; Khaliq O. P.; Moodley J.; Naicker T. Insulin Resistance in COVID-19 and Diabetes. Prim. Care Diabetes 2021, 15 (4), 629–634. 10.1016/j.pcd.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall W. E.; Beebe K.; Lawton K. A.; Adam K.-P.; Mitchell M. W.; Nakhle P. J.; Ryals J. A.; Milburn M. V.; Nannipieri M.; Camastra S.; Natali A.; Ferrannini E. α-Hydroxybutyrate Is an Early Biomarker of Insulin Resistance and Glucose Intolerance in a Nondiabetic Population. PLoS One 2010, 5 (5), e10883 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrannini E.; Natali A.; Camastra S.; Nannipieri M.; Mari A.; Adam K.-P.; Milburn M. V.; Kastenmüller G.; Adamski J.; Tuomi T.; Lyssenko V.; Groop L.; Gall W. E. Early Metabolic Markers of the Development of Dysglycemia and Type 2 Diabetes and Their Physiological Significance. Diabetes 2013, 62 (5), 1730–1737. 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brial F.; Chilloux J.; Nielsen T.; Vieira-Silva S.; Falony G.; Andrikopoulos P.; Olanipekun M.; Hoyles L.; Djouadi F.; Neves A. L.; Rodriguez-Martinez A.; Mouawad G. I.; Pons N.; Forslund S.; Le-chatelier E.; Lay A. L.; Nicholson J.; Hansen T.; Hyötyläinen T.; Clément K.; Oresic M.; Bork P.; Ehrlich S. D.; Raes J.; Pedersen O. B.; Gauguier D.; Dumas M.-E. Human and Preclinical Studies of the Host-Gut Microbiome Co-Metabolite Hippurate as a Marker and Mediator of Metabolic Health. Gut 2021, 70 (11), 2105–2114. 10.1136/gutjnl-2020-323314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S. P.; Tsang T. M.; Wang Y.; Cloarec O.; Skordi E.; Martin F.-P.; Rezzi S.; Ross A.; Kochhar S.; Holmes E.; Nicholson J. K. Systemic Multicompartmental Effects of the Gut Microbiome on Mouse Metabolic Phenotypes. Mol. Syst. Biol. 2008, 4, 219. 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley E. L. Metabolites of Shikimate and Tryptophan Pathways in Coronavirus Disease (COVID-19). Microb. Metab. Dis. 2021, 87–104. 10.1016/B978-0-323-88445-7.00009-9. [DOI] [Google Scholar]
- Valdés A.; Moreno L. O.; Rello S. R.; Orduña A.; Bernardo D.; Cifuentes A. Metabolomics Study of COVID-19 Patients in Four Different Clinical Stages. Sci. Rep. 2022, 12, 1650. 10.1038/s41598-022-05667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloborodov N. V.; Khodakova A. S.; Bairamov I. T.; Olenin A. Yu. Microbial Origin of Phenylcarboxylic Acids in the Human Body. Biochem. Mosc. 2009, 74 (12), 1350–1355. 10.1134/S0006297909120086. [DOI] [PubMed] [Google Scholar]
- Menni C.; Zhu J.; Le Roy C. I.; Mompeo O.; Young K.; Rebholz C. M.; Selvin E.; North K. E.; Mohney R. P.; Bell J. T.; Boerwinkle E.; Spector T. D.; Mangino M.; Yu B.; Valdes A. M. Serum Metabolites Reflecting Gut Microbiome Alpha Diversity Predict Type 2 Diabetes. Gut Microbes 2020, 11 (6), 1632–1642. 10.1080/19490976.2020.1778261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister T.; Jackson M. A.; Martin T. C.; Zierer J.; Jennings A.; Mohney R. P.; MacGregor A.; Steves C. J.; Cassidy A.; Spector T. D.; Menni C. Hippurate as a Metabolomic Marker of Gut Microbiome Diversity: Modulation by Diet and Relationship to Metabolic Syndrome. Sci. Rep. 2017, 7, 13670. 10.1038/s41598-017-13722-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgin M.; Derosa L.; Silva C. A. C.; Goubet A.-G.; Dubuisson A.; Danlos F.-X.; Grajeda-Iglesias C.; Cerbone L.; Geraud A.; Laparra A.; Aprahamian F.; Nirmalathasan N.; Madeo F.; Zitvogel L.; Kroemer G.; Durand S. Circulating Acetylated Polyamines Correlate with Covid-19 Severity in Cancer Patients. Aging 2021, 13 (17), 20860–20885. 10.18632/aging.203525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton T. A. Metabolic Differences Underlying Two Distinct Rat Urinary Phenotypes, a Suggested Role for Gut Microbial Metabolism of Phenylalanine and a Possible Connection to Autism. FEBS Lett. 2012, 586 (7), 956–961. 10.1016/j.febslet.2012.01.049. [DOI] [PubMed] [Google Scholar]
- Kaur H.; Bose C.; Mande S. S. Tryptophan Metabolism by Gut Microbiome and Gut-Brain-Axis: An in Silico Analysis. Front. Neurosci. 2019, 10.3389/fnins.2019.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atila A.; Alay H.; Yaman M. E.; Akman T. C.; Cadirci E.; Bayrak B.; Celik S.; Atila N. E.; Yaganoglu A. M.; Kadioglu Y.; Halıcı Z.; Parlak E.; Bayraktutan Z. The Serum Amino Acid Profile in COVID-19. Amino Acids 2021, 53 (10), 1569–1588. 10.1007/s00726-021-03081-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luís Luporini R.; Pott-Junior H.; Carolina B.; Di Medeiros Leal M.; Castro A.; Gilberto Ferreira A.; Regina Cominetti M.; de Freitas Anibal F. Phenylalanine and COVID-19: Tracking Disease Severity Markers. Int. Immunopharmacol. 2021, 101, 108313. 10.1016/j.intimp.2021.108313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D.; Spitzer M. H.; Van Treuren W.; Merrill B. D.; Hryckowian A. J.; Higginbottom S. K.; Le A.; Cowan T. M.; Nolan G. P.; Fischbach M. A.; Sonnenburg J. L. A Gut Bacterial Pathway Metabolizes Aromatic Amino Acids into Nine Circulating Metabolites. Nature 2017, 551 (7682), 648–652. 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettrick A. F.; Corcoran S. E.; Barry P. J. G.; McFarland J.; Crès C.; Curtis A. M.; Franklin E.; Corr S. C.; Mok K. H.; Cummins E. P.; Taylor C. T.; O’Neill L. A. J.; Nolan D. P. Trypanosoma Brucei Metabolite Indolepyruvate Decreases HIF-1α and Glycolysis in Macrophages as a Mechanism of Innate Immune Evasion. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (48), E7778-E7787 10.1073/pnas.1608221113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A.; Maurice C. F.; Carmody R. N.; Gootenberg D. B.; Button J. E.; Wolfe B. E.; Ling A. V.; Devlin A. S.; Varma Y.; Fischbach M. A.; Biddinger S. B.; Dutton R. J.; Turnbaugh P. J. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505 (7484), 559–563. 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein S. R. S.; Mahmud B.; Dantas G. Antibiotic Perturbations to the Gut Microbiome. Nat. Rev. Microbiol. 2023, 21 (12), 772–788. 10.1038/s41579-023-00933-y. [DOI] [PubMed] [Google Scholar]
- Brevini T.; Maes M.; Webb G. J.; John B. V.; Fuchs C. D.; Buescher G.; Wang L.; Griffiths C.; Brown M. L.; Scott W. E.; Pereyra-Gerber P.; Gelson W. T. H.; Brown S.; Dillon S.; Muraro D.; Sharp J.; Neary M.; Box H.; Tatham L.; Stewart J.; Curley P.; Pertinez H.; Forrest S.; Mlcochova P.; Varankar S. S.; Darvish-Damavandi M.; Mulcahy V. L.; Kuc R. E.; Williams T. L.; Heslop J. A.; Rossetti D.; Tysoe O. C.; Galanakis V.; Vila-Gonzalez M.; Crozier T. W. M.; Bargehr J.; Sinha S.; Upponi S. S.; Fear C.; Swift L.; Saeb-Parsy K.; Davies S. E.; Wester A.; Hagström H.; Melum E.; Clements D.; Humphreys P.; Herriott J.; Kijak E.; Cox H.; Bramwell C.; Valentijn A.; Illingworth C. J. R.; Dahman B.; Bastaich D. R.; Ferreira R. D.; Marjot T.; Barnes E.; Moon A. M.; Barritt A. S.; Gupta R. K.; Baker S.; Davenport A. P.; Corbett G.; Gorgoulis V. G.; Buczacki S. J. A.; Lee J.-H.; Matheson N. J.; Trauner M.; Fisher A. J.; Gibbs P.; Butler A. J.; Watson C. J. E.; Mells G. F.; Dougan G.; Owen A.; Lohse A. W.; Vallier L.; Sampaziotis F. FXR Inhibition May Protect from SARS-CoV-2 Infection by Reducing ACE2. Nature 2023, 615 (7950), 134–142. 10.1038/s41586-022-05594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.; McCord K. A.; Bui D. T.; Bouwman K. M.; Kitova E. N.; Elaish M.; Kumawat D.; Daskhan G. C.; Tomris I.; Han L.; Chopra P.; Yang T.-J.; Willows S. D.; Mason A. L.; Mahal L. K.; Lowary T. L.; West L. J.; Hsu S.-T. D.; Hobman T.; Tompkins S. M.; Boons G.-J.; de Vries R. P.; Macauley M. S.; Klassen J. S. Sialic Acid-Containing Glycolipids Mediate Binding and Viral Entry of SARS-CoV-2. Nat. Chem. Biol. 2022, 18 (1), 81–90. 10.1038/s41589-021-00924-1. [DOI] [PubMed] [Google Scholar]
- Li W.; Hulswit R. J. G.; Widjaja I.; Raj V. S.; McBride R.; Peng W.; Widagdo W.; Tortorici M. A.; van Dieren B.; Lang Y.; van Lent J. W. M.; Paulson J. C.; de Haan C. A. M.; de Groot R. J.; van Kuppeveld F. J. M.; Haagmans B. L.; Bosch B.-J. Identification of Sialic Acid-Binding Function for the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (40), E8508-E8517 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M. The War against Influenza: Discovery and Development of Sialidase Inhibitors. Nat. Rev. Drug Discovery 2007, 6 (12), 967–974. 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- Baker A. N.; Richards S.-J.; Guy C. S.; Congdon T. R.; Hasan M.; Zwetsloot A. J.; Gallo A.; Lewandowski J. R.; Stansfeld P. J.; Straube A.; Walker M.; Chessa S.; Pergolizzi G.; Dedola S.; Field R. A.; Gibson M. I. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6 (11), 2046–2052. 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoei M. M.; Vogel H. J.; Weljie A. M.; Kumar A.; Yende S.; Angus D. C.; Winston B. W. the Canadian Critical Care Translational Biology Group (CCCTBG). Plasma Metabolomics for the Diagnosis and Prognosis of H1N1 Influenza Pneumonia. Crit. Care 2017, 21 (1), 97. 10.1186/s13054-017-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Vicente M.; González-Riaño C.; Barbas C.; Jiménez-Sousa M. Á.; Brochado-Kith O.; Resino S.; Martínez I. Metabolic Changes during Respiratory Syncytial Virus Infection of Epithelial Cells. PLoS One 2020, 15 (3), e0230844 10.1371/journal.pone.0230844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-García F. J.; Pérez-Hernández C. A.; Rodríguez-Murillo M.; Moreno-Altamirano M. M. B. The Role of Tricarboxylic Acid Cycle Metabolites in Viral Infections. Front. Cell. Infect. Microbiol. 2021, 11, 725043. 10.3389/fcimb.2021.725043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Du J.; Wei T.-T.; Chen L.-Y.; Yang X.-X.; Bo T.; Liu H.-Y.; Xie M.-Z.; Zhao T.-S.; Yang J.-L.; Cui F.; Chen W.-W.; Lu Q.-B. Alterations in Bile Acids as Metabolic Signatures in the Patients with Human Adenovirus Type 7 Infection. Front. Med. 2022, 10.3389/fmed.2022.896409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Luu L. D. W.; Wang X.; Cui X.; Huang X.; Fu J.; Zhu X.; Li Z.; Wang Y.; Tai J. Metabolomic Analysis Reveals Potential Biomarkers and the Underlying Pathogenesis Involved in Mycoplasma Pneumoniae Pneumonia. Emerg. Microbes Infect. 2022, 11 (1), 593–605. 10.1080/22221751.2022.2036582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D.; Naiyer S.; Mansuri S.; Soni N.; Singh V.; Bhat K. H.; Singh N.; Arora G.; Mansuri M. S. COVID-19 Diagnosis: A Comprehensive Review of the RT-qPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12 (6), 1503. 10.3390/diagnostics12061503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. J.; Grant-St James A.; Lawler N. G.; Fear M. W.; Raby E.; Wood F. M.; Maker G. L.; Wist J.; Holmes E.; Nicholson J. K.; Whiley L.; Gray N. Comprehensive Lipidomic Workflow for Multicohort Population Phenotyping Using Stable Isotope Dilution Targeted Liquid Chromatography-Mass Spectrometry. J. Proteome Res. 2023, 22, 1419. 10.1021/acs.jproteome.2c00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. L.; Whiley L.; Gray N.; Gay M.; Nitschke P.; Masuda R.; Holmes E.; Nicholson J. K.; Wist J.; Lawler N. Rapid and Self-Administrable Capillary Blood Microsampling Demonstrates Statistical Equivalence with Standard Venous Collections in NMR-Based Lipoprotein Analysis. Anal. Chem. 2024, 10.1021/acs.analchem.3c05152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data for the project is freely available on the MassIVE server (project identifier: MSV000094022).