Abstract
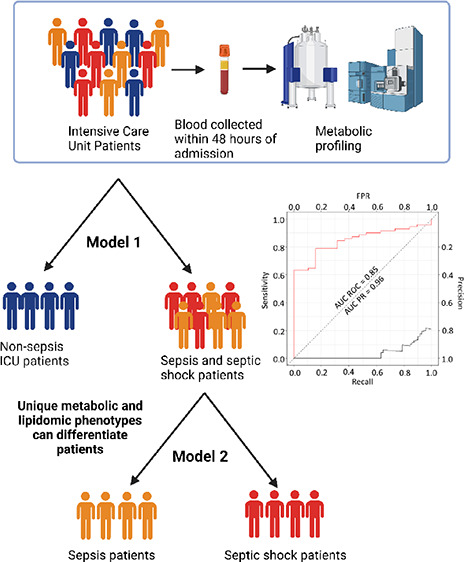
Delayed diagnosis of patients with sepsis or septic shock is associated with increased mortality and morbidity. UPLC-MS and NMR spectroscopy were used to measure panels of lipoproteins, lipids, biogenic amines, amino acids, and tryptophan pathway metabolites in blood plasma samples collected from 152 patients within 48 h of admission into the Intensive Care Unit (ICU) where 62 patients had no sepsis, 71 patients had sepsis, and 19 patients had septic shock. Patients with sepsis or septic shock had higher concentrations of neopterin and lower levels of HDL cholesterol and phospholipid particles in comparison to nonsepsis patients. Septic shock could be differentiated from sepsis patients based on different concentrations of 10 lipids, including significantly lower concentrations of five phosphatidylcholine species, three cholesterol esters, one dihydroceramide, and one phosphatidylethanolamine. The Supramolecular Phospholipid Composite (SPC) was reduced in all ICU patients, while the composite markers of acute phase glycoproteins were increased in the sepsis and septic shock patients within 48 h admission into ICU. We show that the plasma metabolic phenotype obtained within 48 h of ICU admission is diagnostic for the presence of sepsis and that septic shock can be differentiated from sepsis based on the lipid profile.
Keywords: ICU, sepsis, septic shock, NMR spectroscopy, mass spectrometry, pharmaco-metabonomics, plasma IVDr, metabolic phenotyping, diagnostic modeling, lipoproteins, lipids, SPC, APACHE
Introduction
Despite notable improvements in the delivery of intensive care in recent years, severe sepsis and septic shock remain a significant clinical problem with a substantial morbidity, mortality, and economic burden.1,2 Globally, almost 50 million sepsis cases were reported in 2017 with mortality rates at 20–50% depending on age and regional disparities.3 Sepsis is defined as a life threatening organ dysfunction condition caused by the dysregulated host response to infection4 but due to the heterogeneity of the timing and presentation of symptoms, diagnosis and prognostication of patients with sepsis is challenging.5 The molecular mechanisms underpinning sepsis pathobiology are incompletely understood but host oxidative stress,6 endothelial disruption,7 and mitochondrial dysfunction8 contribute to the dysregulated host response in sepsis and may result in significant systemic metabolic changes.9 Metabolic phenotyping methodologies may therefore be useful for understanding sepsis pathogenesis.10 For example, Mao et al. and others have found disrupted amino acid and carbohydrate metabolism in patients with systemic inflammatory response syndrome, which shifted to disrupted fat metabolism in multiple organ dysfunction syndrome.11−13
The diagnosis of sepsis relies on a clinical assessment of the likelihood of infection and associated systemic features. Gold standard diagnosis is subsequently dependent on the positive identification of a pathogenic organism using standard clinical pathogen culturing,14 which occurs in only a minority of patients with sepsis and may take days to confirm. Around 20% of patients in the intensive care unit with sepsis will progress to septic shock, which is associated with multi organ failure and 9.8% mortality.15−17 Clinical scoring systems such as the Acute Physiology and Chronic Health Evaluation (APACHE), APACHE II, APACHE III, Simplified Acute Physiology Score, Sequential Organ Failure Assessment, and Mortality Probability Model II can be used to assess the illness severity level but these scoring systems have low specificity and sensitivity, particularly with respect to predicting clinical outcomes.18 Although improved patient outcomes have been achieved over the last three decades through the implementation of these predictive severity scores, together with evidence-based management strategies,19 mortality remains high. Effective treatment of sepsis relies on rapid diagnosis and subsequent, goal directed therapy since the mortality rate of patients with septic shock can increase 8% for every hour delay in antimicrobial therapy.20 Thus, early identification of sepsis and progression to septic shock present a critical unmet clinical need.
We have previously shown that preinterventional metabolic phenotyping can enable prediction of interventional outcomes such as drug toxicity,21 drug metabolism,22 and anticancer drug efficacy in animals and humans.23 The same principle using a training-test set model can be applied to predict disease trajectory outcomes including recovery or progression to long-COVID in SARS CoV-2 infection24 or even COVID-19 induced death in intensive care unit (ICU) patients.25 Since the biomarkers that are currently used to identify sepsis are inadequate for discriminating between sepsis and septic shock patients in ICU, we applied a multiplatform metabolic profiling approach to ascertain whether plasma profiles could be used to differentiate patients with no sepsis, sepsis, and septic shock at admission into ICU regardless of admission route or type.
Plasma samples collected from patients within 48 h of admission into ICU were analyzed by ultra-high performance liquid chromatography mass spectrometry (UPLC-MS) to obtain 36 amino acid and tryptophan pathway intermediates and 975 lipids. In addition, nuclear magnetic resonance (NMR) spectroscopy was used to quantify 117 lipoprotein parameters, glycoproteins and a Supramoleculer Phosphoplipid Composite (SPC) known to be associated with inflammatory processes.26,27 For this Western Australian cohort, we demonstrate that a subset of 15 metabolites can successfully stratify patients into the correct sepsis clinical outcome upon admission into ICU.
Materials and Methods
Participant Enrolment and Sample Collection
Blood plasma samples were collected from 152 patients within 48 h of admission into ICU where 62 patients did not have sepsis, 71 patients had sepsis without shock, and 19 had septic shock. A second blood collection was completed 48 h after the first blood collection (sample n = 104) and a third completed at 7 days (sample n = 46) post admission. All participants provided informed consent to clinical investigations, according to the Declaration of Helsinki, and the data were anonymized to protect their confidentiality. Samples were approved for analysis as part of the ROCIT (Restoration of the microbiome in critical illness) study,28 which was conducted in the ICU’s of five hospitals in Perth, Western Australia (ANZCTR 12617000783325). Research ethics committee approvals were completed (South Metropolitan Health Service Human Research Ethics Committee: RGS000004 and Murdoch University Ethics: 2019/037). Plasma samples were stored at −80 °C until analysis.
For the purpose of comparison of healthy individuals with the ICU patients, control samples (n = 50) were collected by the Basque Biobank for research (BIOEF) prior to the COVID-19 pandemic from an apparently healthy population. All participants provided informed consent to the clinical investigation, according to the Declaration of Helsinki, and the data were anonymized to protect their confidentiality. The sample handling protocol was evaluated and approved by the Comité de Ética de Investigación con medicamentos de Euskadi (CEIm-E, PI+CES-BIOEF 2020–04 and PI219130). Shipment of human samples to the ANPC had the approval of the Ministry of Health of the Spanish Government. Samples were stored at −80 °C. Samples were approved for analysis as part of the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC)/World Health Organisation (WHO) pandemic trial framework (SMHS Research governance office PRN:3976 and Murdoch University Ethics no. 2020/052, and no. 2020/053).
1H NMR Sample Preparation
Samples were defrosted at room temperature for 1 h. NMR samples were prepared in a SamplePro Tube (Bruker Biospin) robot system for liquid handling. Every sample was automatically prepared as a mixture of phosphate buffer (75 mM Na2HPO4, 2 mM NaN3, 4.6 mM sodium trimethylsilyl propionate-[2,2,3,3–2H4] (TSP) in H2O/D2O 4:1, pH 7.4 ± 0.1) and plasma at a 1:1 ratio for a final volume of 600 μL into 5 mm SampleJet NMR tubes.
1H NMR Spectroscopy Data Acquisition and Processing Parameters
NMR spectroscopic analyses were performed on a 600 MHz Bruker Avance III HD spectrometer, equipped with a 5 mm BBI probe and fitted with the Bruker SampleJet robot cooling system set to 5 °C. A full quantitative calibration was completed prior to the sample analysis using a protocol described elsewhere.29 All experiments were acquired using Bruker In Vitro Diagnostics research (IVDr) methods. For each sample prepared, two experiments were run with a total analysis time of 8.5 min: (i) a standard 1D experiment with solvent presaturation (32 scans, 98K data points, spectral width of 30 ppm) and (ii) a DIRE experiment (64 scans, 98K data points, spectral width of 30 ppm).30
From the standard 1D experiment, a total of 112 lipoprotein parameters for each sample were generated using the Bruker IVDr Lipoprotein Subclass Analysis (B.I.-LISA) method whereby the −(CH2)n at δ = 1.25 and −CH3 at δ = 0.80 peaks of the 1D spectrum after normalization to the Bruker QuantRef TM manager within Topspin were quantified using a PLS-2 regression model.31 B.I.LISA data consist of total plasma lipid analytes cholesterol, free cholesterol, phospholipids, triglycerides, Apolipoproteins A1/A2/B100 and the B100/A1 ratio, and analyte distributions in different density classes of plasma-lipoproteins: high-density lipoprotein (HDL, density 1.063–1.210 kg/L), intermediate-density lipoprotein (IDL, density 1.006–1.019 kg/L), low-density lipoprotein (LDL, density 1.019–1.063 kg/L), and very low-density lipoprotein (VLDL, 0.950–1.006 kg/L). The main lipoprotein classes HDL, LDL, and VLDL are subdivided into different density subclasses. LDL subdivisions included LDL1: 1.019–1.031 kg/L, LDL2: 1.031–1.034 kg/L, LDL3: 1.034–1.037 kg/L, LDL4: 1.037–1.040 kg/L, LDL5: 1.040–1.044 kg/L, LDL6: 1.044–1.063 kg/L). HDL subfractions were also assigned to 4 density classes: HDL1 1.063–1.100 kg/L, HDL2 1.100–1.112 kg/L, HDL3 1.112–1.125 kg/L, and HDL4 1.125–1.210 kg/L, and the VLDL subfractions were divided into 5 density classes. A list of all the 112 lipoprotein subfractions and parameter annotations are shown in Table S1. DIRE spectra preprocessing included baseline correction using an asymmetric least-squares routine and the spectra were normalized to the eretic signal using the R package metabom8 (version 1.0.0, github.com/tkimhofer/metabom8). To estimate the signal intensities of the glycoprotein peaks, GlycA and GlycB spectral regions were integrated (δ2.03 and δ2.07 ppm, respectively). The GlycA signal (δ2.03) is a composite of N-acetyl signals from five proteins: α-1-acid glycoprotein, α-1-antitrypsin, α-1-antichymotrypsin, haptoglobin, and transferrin. The GlycB acetyl signal (δ2.07) arises from glycoprotein N-acetylneuraminidino groups.27 The region containing the Supramolecular Phospholipid Composite (SPC) peak was integrated to determine SPC1 (δ3.200–3.236) corresponding predominantly to small HDL (HDL4) phospholipids, SPC2 (δ3.236–3.252) corresponding to larger HDL phospholipid particles (HDL1-HDL3) and SPC3 (δ3.252–3.300) corresponding to LDL phospholipids (26).
Liquid Chromatography Mass Spectrometry (LC-MS)
Biogenic amines, amino acids, and tryptophan metabolites were measured using two LC-MS quantification methods following previously reported methods for tryptophan and associated catabolites32 and amino acids.33 In brief, samples were thawed at 4 °C and prepared for analysis. For the quantification of the biogenic amines and amino acid metabolites, a Bruker Impact II QToF mass spectrometer (Bruker, Daltonics, Billerica, MA) coupled to a Waters Acquity I-class UPLC system (Waters Corp., Milford, MA) was used. Full scan mass spectrometry data in high resolution were acquired using electrospray ionization positive in a mass range of m/z 30–1000, operated in broadband collision-induced dissociation (bbCID) function to generate fragmentation data. Resulting data files were processed for peak integration and quantification using the Target Analysis for Screening Quantification (TASQ; v2.2) software (Bruker Daltonics, Bremen, Germany), where calibration curves were linearly fitted with a weighting factor of 1/x. For the measurement of tryptophan and associate catabolites, a Waters TQ-XS triple quadrupole (QQQ) coupled to a Waters Acquity I-class UPLC system (Waters, Wilmslow, UK) was used operating in positive electrospray ionization using multiple reaction monitoring (MRM). Obtained raw files were processed for peak integration and metabolite quantifications using the TargetLynx package within MassLynx v4.2 (Waters Corp., Milford, MA) where calibration curves were linearly fitted using a weighting factor of 1/x. Resulting data matrices were combined, and quality control was checked prior to statistical analysis.
LC-MS Lipid Analysis
Plasma lipid analysis was performed by ultrahigh-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) using a Exion UHPLC system coupled to a QTRAP 6500+ mass analyzer (Sciex Concord, CA) from a previously published method.34 In brief, plasma samples (10 μL) were thawed at 4 °C and combined with 90 μL of stable isotopically labeled internal standards diluted in isopropyl alcohol for extraction, utilizing a Biomek i5 liquid handling system for the preparation (Beckman Coulter, Mount Waverley, Victoria 3149, Australia). Samples were chilled and centrifuged for 15 min at 14000g before 50 μL of supernatant was transferred into a 350 μL 96-well plate for analysis (Eppendorf, Macquarie Park, NSW, Australia). For quality control (QC) an independent plasma pool was prepared and injected following each block of 9 experimental samples throughout the analytical sequence, which were used for the assessment of analytical precision. The raw files generated were preprocessed using SkylineMS,35 and quality control random forest signal correction (QC-RFSC) from the statTarget package was used to correct for analytical drift.36 Feature filtering, RSD QC > 30%, and feature intensity threshold filtering < 5000 in > 50% of the QCs were applied and metabolites were removed from further statistical analysis based on their failure to meet acceptable analytical precision. The comprehensive analysis covered 20 subclasses of lipids including cholesterol esters (CEs), ceramides (CERs), diacylglycerides (DAGs), dihydroceramides (DCERs), free fatty acids (FFAs), hexosylceramides (HCERs), lactosylceramides (LCERs), lysophosphatidylphocholines (LPCs), lysophosphatidylphoethanolamines (LPEs), lysophosphatidylglycerides (LPGs), and lysophosphatidylinositols (LPIs), lysophosphatidylserines (LPSs), monoacylglycerols (MAGs), phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), phosphatidylglycerides (PGs), phosphatidylinositols (PIs), phosphatidylserines (PSs), sphingomyelins (SMs), and triacylglycerides (TAGs).
Data Analysis
All computation and data visualization were performed using R and RStudio IDE with the open-source R package metabom8 (version 1.0.0), available from GitHub (github.com/tkimhofer/metabom8). Mann–Whitney U tests were performed to compare metabolite concentrations within different groups. The Cliff’s delta statistic was calculated for all parameters to assess the overall effect size for the intergroup differences.37 Absolute Cliff’s delta scores were interpreted as 1 indicating maximum difference (regardless of sign) and 0 indicating no difference. Orthogonal projection to latent structures-discriminant analysis (O-PLS-DA)38 was used to model variance in the data between different sepsis groups and to extract discriminating features between the groups. The optimal number of orthogonal components for each model was determined using the area under the receiver operator characteristic curve (AUROC) calculated from predictive component scores, generated using a standard 7-fold cross-validation (CV) procedure. Adjusted p-values were used to assess the significance of metabolites, lipoproteins and lipids.
Results and Discussion
Demographics of the Patient Cohort
Admission into ICU came via one of four routes, the emergency department, ward, and operating theater either elective or emergency (Table 1) and were classified into four different types of admissions, cardiothoracic, medical, surgery, or trauma. On admission into the ICU, the Apache II (Acute Physiology and Chronic Health Evaluation II) score was significantly different between the no sepsis and sepsis groups (p = 3.5 × 10–4) and the no sepsis versus septic shock groups (p = 2.5 × 10–3), while it was not significantly different between the sepsis and septic shock patients. C-Reactive protein was significantly higher in the sepsis group compared to the no sepsis group (p = 9.3 × 10–4) and higher in the septic shock group compared to the no sepsis group (p = 0.01). However, there was no significant difference between the sepsis and septic shock groups. Overall, survival at 60 days post-admission into ICU was 94%.
Table 1. Full Cohort Demographicsa.
| ICU patients no sepsis(n = 62) | ICU patients sepsis(n = 71) | ICU patients septic shock (n= 19) | healthy(n = 50) | |
|---|---|---|---|---|
| sex, male | 38 (61.30%) | 42 (59.15%) | 8 (42.10%) | 35 (70.00%) |
| age, years [SD] | 64.00 [±17.34] | 67.00 [±12.98] | 64.00 [±14.80] | 62.55 [±1.48] |
| BMI, kg/m2 [SD] | 25.47 [±2.37] | |||
| total days in hospital | 8 [±8.40] | 11 [±10.38] | 11 [±13.73] | |
| total days in ICU | 2 [±3.71] | 4 [±3.93] | 4 [±6.23] | |
| source of admission to ICU | ||||
| ED | 19 (30.65%) | 37 (52.11%) | 8 (42.11%) | |
| OT elective | 22 (35.48%) | 13 (18.31%) | 1 (5.26%) | |
| OT emergency | 7 (11.29%) | 6 (8.45%) | 4 (21.05%) | |
| ward | 14 (22.59%) | 15 (21.13%) | 6 (31.58%) | |
| admission type to ICU | ||||
| cardiothoracic | 18 (29.03%) | 8 (11.27%) | 0 (0.00%) | |
| medical | 26 (41.94%) | 43 (60.56%) | 11 (57.89%) | |
| surgery | 13 (20.97%) | 19 (26.76%) | 8 (42.11%) | |
| trauma | 5 (8.06%) | 1 (1.41%) | 0 (0.00%) | |
| chronic respiratory disease | 6 (9.68%) | 5 (7.04%) | 0 | 0 |
| chronic cardiovascular disease | 6 (9.68%) | 13 (18.31%) | 1 (5.26%) | 0 |
| chronic liver disease | 1 (1.62%) | 1 (1.41%) | 0 | 0 |
| chronic renal disease | 4 (6.45%) | 4 (5.63%) | 2 (10.53%) | 0 |
| chronic immune disease | 1 (1.62%) | 0 | 0 | 0 |
| Apache II score | 11.5 [±6.15] | 15.0 [±6.08] | 15.0 [±7.32] | |
| CRP at enrolment (mg/L) | 66.2 [±121.0] | 220.0 [±127.8] | 210.0 [±134.7] | |
| patients who did not survive past 60 days | 2 | 3 | 1 |
Definition of abbreviations: APACHE II = Acute Physiology and Chronic Health Evaluation II; BMI = body mass index; CRP = C-Reactive protein; ED = emergency department; ICU = intensive care unit; OT = operating theater.
Here, we aimed to stratify patients based on their plasma metabolic profiles obtained within 48 h admission into ICU according to their clinical diagnosis of (i) no sepsis, (ii) sepsis, or (iii) septic shock. In the first instance, an O-PLS-DA model was built to establish whether the samples obtained at ICU admission could differentiate patients who went on to develop sepsis or septic shock from those who did not develop sepsis. Subsequently, a second model was built to establish whether there was a difference in the plasma profiles of patients who developed sepsis versus septic shock at the baseline. The variables were selected using a Cliff’s delta value of above 0.5 or below −0.5 obtained by univariate analysis. This resulted in four variables included in model 1 and 10 variables included in model 2.
Stratification of Nonseptic Patients versus Septic and Septic Shock Patients within 48 h of Admission to ICU (Model 1)
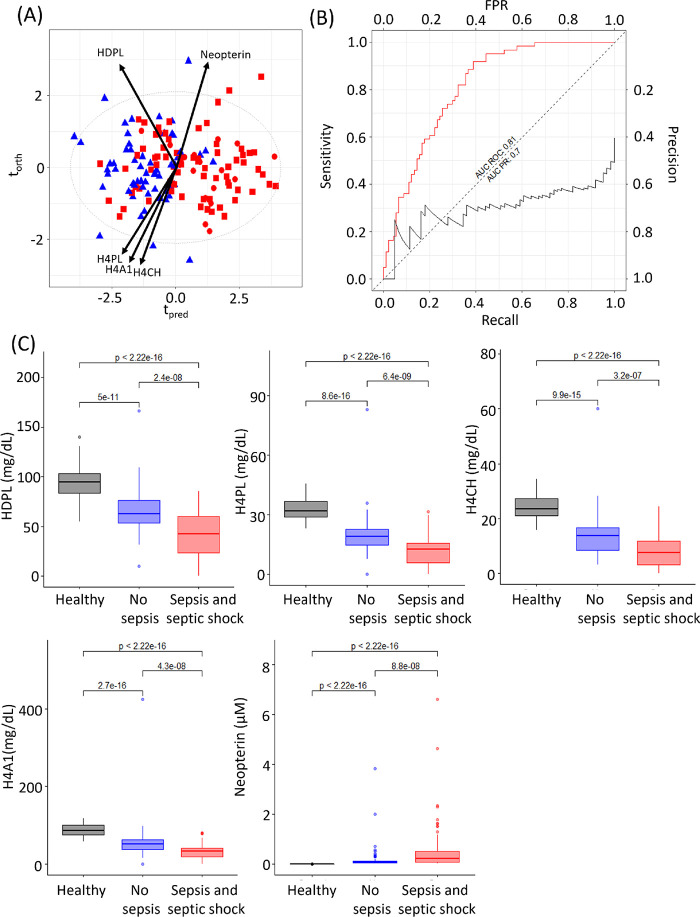
Higher concentrations of neopterin and lower concentrations of four lipoproteins, high density phospholipid (HDPL), high density cholesterol subfraction 4 (H4CH), high density phospholipid subfraction 4 (H4PL), and high density lipoprotein Apolipoprotein A1 subfraction 4 (H4A1) were selected as being characteristic of the sepsis/septic shock group. An O-PLS-DA model was constructed using these 5 parameters, which resulted in a model with a CV-AUROC 0.80 (Figure 1A). The AUC ROC of model 1 was 0.81, and the area under the precision recall (AUC PR) curve was 0.70. The use of this metabolite panel proved superior to CRP for differentiating non sepsis patients from those that had sepsis and septic shock at admission into ICU, where the AUC ROC of C-reactive protein was found to be 0.76 with an AUC PR of 0.66 (Figure S1a).
Figure 1.
O-PLS-DA of nonsepsis (blue triangle) vs sepsis (red square) and septic shock (red circle) within 48 h of admission into ICU (R2X = 0.72, CV-AUROC = 0.80), arrows indicate loadings of neopterin, HDPL, H4PL, H4A1, and H4CH. (B) Area under the ROC and area under the precision recall curve for this model (model 1 AUC ROC = 0.81 (red line), model 1 AUC PR = 0.7 (black line). (C) Box plots of HDPL, H4PL, H4CH, H4A1 and neopterin for healthy participants, no sepsis ICU patients, and sepsis and septic shock ICU patients. Significance levels of the Mann–Whitney tests are shown for comparison of the groups.
The data sets for the assays were also modeled independently, stratified by no sepsis versus the sepsis and septic shock group within 48 h of admission into ICU using O-PLS-DA. In each case, models were built using a set of combined variables from the individual analyte sets: lipids (n = 975), lipoproteins (n = 112), amino acid and tryptophan pathway intermediates (n = 36), and the inflammatory markers, the glycoproteins, and SPC (n = 5). When all participants were included in the analysis of the lipid data set, a significant model could not be generated with the resulting CV-AUROC = 0.59. For the other matrices, the baseline sample taken on admission to ICU was able to differentiate those patients who went on to develop sepsis and/or septic shock from those who did not, yielding CV-AUROCs of 0.66, 0.78, and 0.68, respectively, for the MS-derived amino acid and tryptophan pathway (AATr) intermediates (Figure S2), the NMR derived lipoproteins (Figure S3), and the NMR derived glycoproteins and SPC (Figure S4). OPLS loadings, Cliff’s delta, and adjusted p-values can be found for the independent assays in Tables S2–S4.
While the four lipoproteins were found at higher concentrations in no sepsis versus the sepsis and septic shock group, neopterin was higher in the sepsis and septic shock group. All these parameters are significantly different between the sepsis and nonsepsis groups, p-values between 6.4 × 10–9 and 3.2 × 10–7 (Figure 1c). Values for a healthy control group are included for comparison, and it can be seen that the selected parameters all strongly differentiate both ICU groups from the control group (Figure 1). Of the five parameters that were able to predict development of sepsis or septic shock versus no sepsis, only neopterin (p-value = 0.04) was able to discriminate between sepsis and septic shock (Figure S5). In addition, comparison of the parameters for each of the three groups versus the healthy controls yielded significantly different p-values, reflecting the systemic perturbation following inflammation, infection, and sepsis.
High density lipoprotein has been previously shown to be reduced in sepsis patients.39−41 HDL contains apoprotein A1 (ApoA1) and is primarily involved in transporting cholesterol from peripheral tissues back to the liver and as such has a cardioprotective function, in addition to having antiapoptotic, antithrombotic, and anti-inflammatory properties.42 Upon infection, HDL binds and neutralizes the bacterial lipopolysaccharide and lipoteichoic acid.43,44 The apolipoprotein A2 in HDL has been shown to suppress the inhibitory effect of high concentrations of lipopolysaccharide binding protein (LPS) prior to HDL binding to LPS, which also serves to augment monocyte activity and control the response to sepsis.45 Although H4A2 was not selected based on the Cliff’s delta (cd = −0.46) cut off, it was nevertheless significant (p-value = 2.32 × 10–4) and is consistent with prior literature. It has been hypothesized that this renders the toxins inaccessible to their receptor CD14 on macrophages, preventing the release of cytokines, such as IL-1, IL-6, and TNF-α, thus protecting against progression to septic shock.46,47 In addition, it has been shown that physiological concentrations of HDL can exert an anti-inflammatory effect by inhibiting the activation of adhesion molecules on endothelial cells.48 Previously, studies have reported that HDL levels at time of admission to ICU were more predictive of 28 day mortality than any other measured parameter.49 Here, we show that the HDL4 concentrations were reduced in all ICU patients at admission into the ICU, but notably, patients who were diagnosed with sepsis and septic shock had particularly low HDL4 levels in comparison with those who did not. Not only have decreased levels of HDL been reported with sepsis, but they have also been noted with a plethora of other inflammatory diseases for example coronary artery disease;50 diabetes51,52 and SARS-CoV-2 infection, where the reduced levels of HDL4 are predictive of disease severity.25,53−55 HDL4-apolipoprotein A1, HDL4-apolipoprotein A2, and HDL4 phosphospholipids were found to be associated with survival in a study on pulmonary arterial hypertension and HDL4 particles were shown to act as carriers for proteins prekallikrein (a precursor of kallikrein) and neurolipin-1, which act as modulators of inflammatory cascades.56
In contrast to the lower levels of HDL4 in patients with sepsis and septic shock, plasma neopterin levels were elevated. Neopterin is a marker of oxidative stress and inflammation.57 It is produced as the oxidation product of 7,8-dihydroneopterin, which is generated by IFN-γ (produced by T lymphocytes)-activated macrophages.58 Levels of neopterin can indicate infection, trauma, cancer, and cardiovascular disease59−61 and neopterin levels have been shown to be increased in sepsis patients,62 which is consistent with the findings in the current study. Some studies have shown that neopterin correlates with mortality in patients with sepsis63 and in one study in liver transplant patients, early post-transplant neopterin plasma levels were identified as a sensitive predictive marker of bacteremia and mortality 1 year post-transplant.64 Although neopterin concentrations were different between the no sepsis versus sepsis/septic shock in the current study, they did not differentiate survivors from nonsurvivors, most likely due to the fact that the mortality in the ROCIT study was only 6% and therefore survival analysis was underpowered. Another study monitored the dynamic profile of neopterin and found the maximum elevation in neopterin levels was reached 12 h after admission to ICU,65 suggesting that the time window for analyzing circulatory fluids is important.
Stratification of Septic versus Septic Shock Patients within 48 h of Admission into ICU (Model 2)
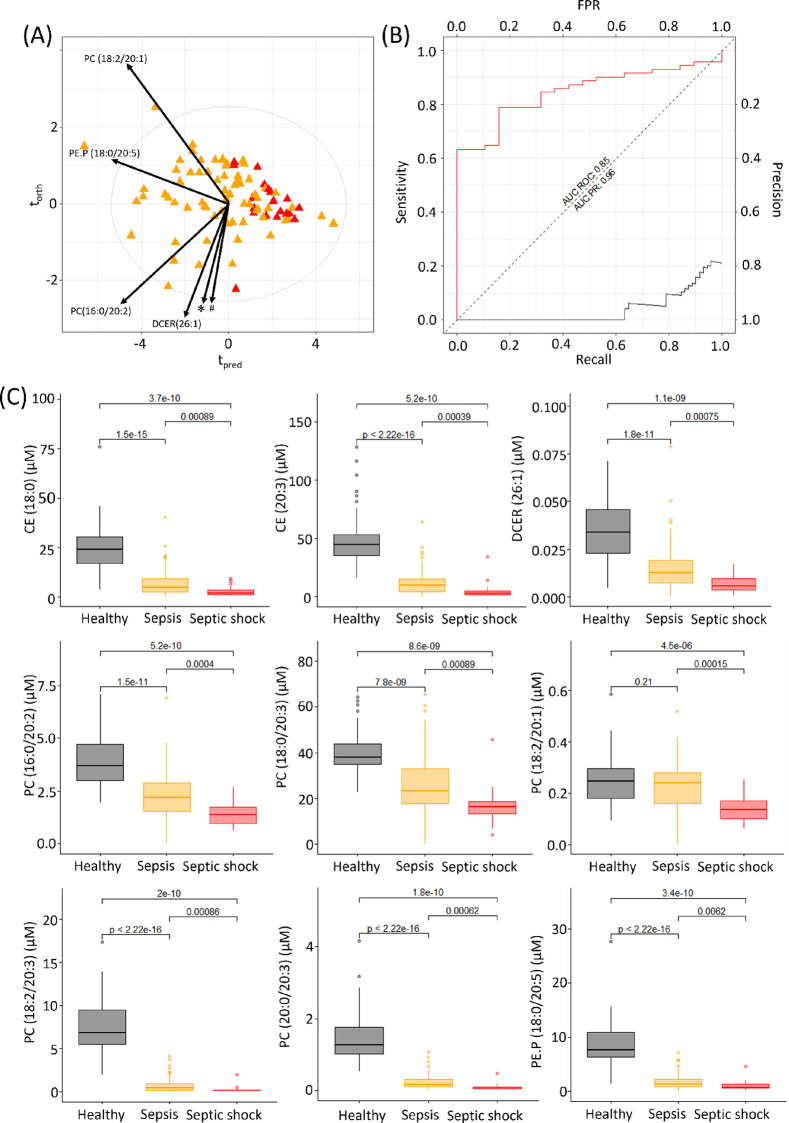
As before, metabolites that had a Cliff’s delta above 0.5 or below −0.5 were selected, yielding a final panel of ten differential lipids including three cholesterol esters (CE(18:0), CE(18:2), and CE (20:3)), one dihydroceramide (DCER(26;1)), one phosphatidylethanolamine (PE.P(18:0/20:5)), and five phosphatidylcholines (PC(16:0/20:2), PC(18:0/20:3), PC(18:2/20:1), PC(18:2/20:3), and PC(20:0/20:3)). An O-PLS-DA model was constructed using these 10 lipids, which resulted in a model with a CV-AUROC 0.81 (Figure 2A), indicating a predictive model for differentiating sepsis from septic shock. The AUC ROC of model 2 was 0.85 and AUC PR was 0.96. The AUC ROC built from the metabolite panel outperformed that of C-reactive protein, which was found to be 0.51 with an AUC PR to be 0.78 for this cohort (Figure S1b). All 10 lipids were significantly reduced in septic shock patients in comparison to sepsis patients (p-values 1.5 × 10–4–8.9 × 10–4) (Figure 2c). In comparison to the healthy controls, all the lipids were highly significantly different except for PC(18:2/20:1).
Figure 2.
O-PLS-DA of septic (orange) versus septic shock (red) patients within 48 h of admission into ICU (R2X = 0.64, CV-AUROC = 0.81), arrows indicate loadings of the lipids, * = CE(18:2), PC(18:0/20:3); # = PC(20:0/20:3), CE(18:0), PC(18:2/20:3), and CE(20:3). (B) Area under the ROC and area under the precision recall curve for this model (AUC ROC = 0.85 (red line), model 2 AUC PR = 0.96 (black line). (C) Box plots of the lipids included in the model 2 analysis for healthy participants, sepsis, and septic shock patients. Significance levels of the Mann–Whitney tests are shown for the comparison of the 3 groups. CE(18:2) is not shown here as a box plot comparison as it did not pass quality control requirements in the healthy control cohort (variability was above 30% in the long term reference QC samples).
O-PLS-DA models were built to compare the sepsis versus septic shock patients within 48 h of admission into ICU for each of the independent assays: the NMR-derived lipoproteins (CV-AUROC = 0.60, Figure S6); the MS derived lipids (CV-AUROC = 0.68, Figure S7); and the NMR derived inflammatory markers (CV-AUROC = 0.63, Figure S8). A model could not be generated from the MS derived amino acid and tryptophan pathway intermediates (CV-AUROC = 0.55), indicating that these parameters were not significantly different between the two groups. OPLS loadings, Cliff’s delta, and adjusted p-values can be found for the independent assays in Tables S5–S7.
It is known that sepsis and septic shock cause extensive lipid dysregulation with the severity of dysregulation mirroring the progression from sepsis to septic shock.66,67 Four phosphatidylcholine species were found in this study to be markedly reduced in septic shock patients in comparison to sepsis patients on admission into ICU (p-values 4.0 × 10–4–8.9 × 10–4). Previous studies have shown total phosphatidylcholine species to be reduced in septic shock in comparison to the baseline measurements.68,69 The liver is the major source of phosphatidylcholines,70 and it has been hypothesized that reduction in plasma phosphatidylcholine levels is caused by pro-inflammatory cytokines such as TNF-α, where the cytokines influence the expression of hepatic lipid-modifying enzymes.71
Two cholesterol esters were significantly different between the sepsis and septic shock patients at admission into ICU (CE (18:0), p-value = 8.9 × 10–4 and CE (20:3), p-value = 3.9 × 10–4). Cholesterol esters are produced by lecithin-cholesterol transferase (LCET) enzyme from cholesterol and phosphatidylcholine. This is consistent with the fact that during sepsis and septic shock LCET activity is decreased.72 In addition, we have demonstrated that the precursor of both CE (18:0) and CE (20:3), PE.P (18:0/20:5), is decreased in sepsis and septic shock patients. Depleted plasma concentrations of CE (18:0) together with a second cholesterol ester CE (16:0) have been reported as predictors of sepsis following cardiac surgery.73 Cholesteryl ester transfer protein (CETP) is another plasma protein that regulates HDL composition by facilitating the transfer of cholesteryl esters from HDL to LDL and VLDL lipoproteins. Infection and inflammation have been shown to cause a decrease in CETP, which is bound to HDL in blood,74,75 which may impact levels of free cholesteryl esters in blood, although their concentrations are determined by a number of factors and the relationship is not simple. DCER (26:1) and PE.P (18:0/20:5) are also significantly reduced in comparison to the healthy controls, indicating an impairment of the antioxidant defense system and an increase in oxidative stress.
Glycoproteins and SPC as Inflammatory Markers
The glycoprotein NMR signal contains contributions from α-1-acid glycoprotein, α-1-antichymotrypsin, α-1-antitrypsin, haptoglobin, and transferrin30 and are known to be markers of inflammation, and studies have shown that these markers (GlycA and GlycB) are more reliable than high sensitivity CRP in reflecting inflammation profiles.76 SPC signals arise from the trimethylammonium headgroups of phospholipids in lipoprotein subcompartments, where SPC1 represents the phospholipid content of HDL4, SPC2 is the phospholipid of HDL1–3, and SPC3 is the phospholipid from LDL.26 We have previously shown with SARS-CoV-2 infection that the relative levels of glycoproteins and SPC gives insight into the inflammatory status of a patient where the glycoproteins are increased and SPC decreased in comparison to healthy control samples.25,28 We have also shown that the SPC regions change differently in nonsevere burns patients and remain perturbed at 6 weeks post-injury.77
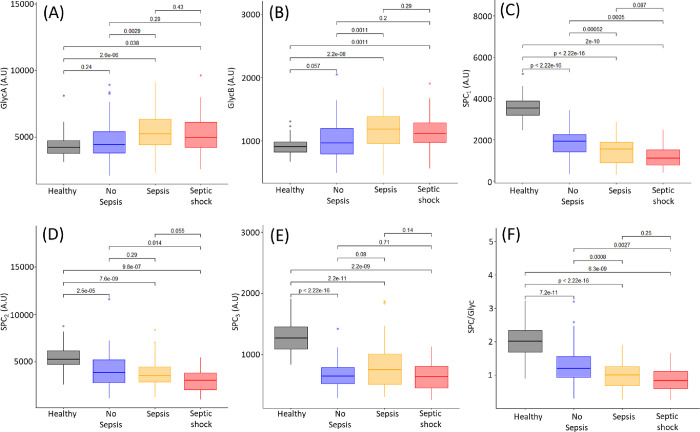
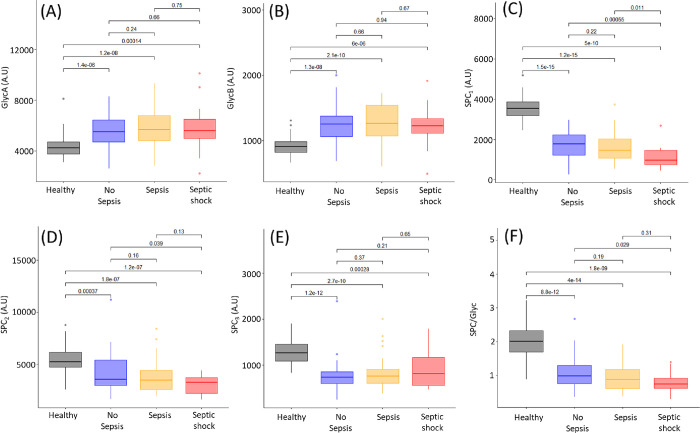
At admission into ICU, GlycA and GlycB values in sepsis and septic shock patients were significantly different from those of the healthy controls but were not elevated in the ICU patients who did not develop sepsis. However, the sepsis patients were significantly different from the ICU patients who did not develop sepsis (Figure 3A,B). There was no significant difference between the sepsis and septic shock patients. At the second blood collection, 48 h after the initial collection, the no sepsis, sepsis, and septic shock patients were all significantly different from the healthy controls for GlycA and GlycB, while there was no significant difference between the three sepsis groups (Figure 4A,B). Interestingly, 7 days post-admission, the healthy controls are significantly different for GlycA from the no sepsis (p-value = 2.5 × 10–4), sepsis (p-value = 2.7 × 10–6), and septic shock (p-value = 1.9 × 10–4) groups (Figure S9). Similarly, the healthy controls exhibited significantly lower concentrations of GlycB from the ICU no sepsis (p-value = 1.0 × 10–5), sepsis (p-value = 6.9 × 10–6), and septic shock groups (p-value = 2.4 × 10–5). There was no significant difference between the no sepsis, sepsis, and septic shock groups for GlycA and GlycB, indicating that 48 h after entry into ICU, all patients had elevated GlycA and GlycB regardless of cause of entry to ICU and whether they had sepsis or not. Multiple studies have reported a correlation between GlycA and cytokine concentrations, particularly SCF, IL6, HGF, IL-18, MIP-1β, and IL-2.78 IL-6 is the mediator of the acute phase response and induces hepatic synthesis and secretion. Increases in circulating glycoprotein concentrations have been shown to correlate with clinically relevant acute phase proteins such as CRP,79 where in our study, the r was only equal to 0.45 (Figure S10). Previous studies have shown GlycA concentrations to be directly associated with endotoxemia, which also was correlated with higher levels of VLDL, while being inversely associated with HDL and LDL particle diameter.80 We found HDL4 to be lower in ICU patients, who went on to develop sepsis or septic shock. Mokkala et al. reported an association between intestinal permeability and GlycA.81 Elevated GlycA is associated with chronic infections and has also been found to be predictive of future hospitalization for acute infection,78 which may be indicative that GlycA plays a role in susceptibility to infection.
Figure 3.
Box plots of healthy controls (black), patients with no sepsis (blue), patients with sepsis (yellow), and patients with septic shock (red) for the glycoproteins and SPC within 48 h of admission into ICU. Mann–Whitney-U tests between the groups are shown above the corresponding plots. (A) GlycA (B) GlycB, (C) SPC1, (D) SPC2, (E) SPC3, (F) SPC/Glyc.
Figure 4.
Box plots of healthy controls (black), patients with no sepsis (blue), patients with sepsis (yellow), and patients with septic shock (red) for the glycoproteins and SPC 48 h post-collection of the first blood sample at admission in ICU. Mann–Whitney-U tests between the groups are shown above the corresponding plots. (A) GlycA, (B) GlycB, (C) SPC1, (D) SPC2, (E) SPC3, (F) SPC/Glyc.
We found a significant difference in GlycA and GlycB levels in sepsis and septic shock patients at admission compared to controls. By 48 h post-first collection, non sepsis, sepsis, and septic shock patients could not be differentiated by GlycA or GlycB (Figures 3 and 4). On the other hand, the plasma Supramolecular Phospholipid Composite, particularly SPC1, showed a more sustained response over the first 48 h in ICU, being lower in ICU patients than healthy controls and of the patients in ICU (Figures 3C and 4C). Significant p-values were obtained when comparing SPC1 in the no sepsis ICU patients with sepsis patients (p = 5.2 × 10–4) and no sepsis ICU patients with septic shock patients (p = 5.0 × 10–4). This confirms the findings of model 1 in this study, where HDL4 is prognostic in determining those patients with sepsis and septic shock in comparison to those without at admission into ICU.
The contrasting patterns between the supramolecular phospholipid cluster (specifically SPC1) and the acute-phase glycoproteins (GlycA and GlycB) may suggest that the glycoproteins reflect acute inflammatory crises, whereas depletion of SPC1 reflects a more reactive response in differentiating ICU patients who did or did not have sepsis. Since neopterin, which was identified as differentiating between ICU patients who did/did not have sepsis or septic shock, is also a known inflammatory marker, we assessed whether plasma neopterin concentrations were correlated with GlycA, GlycB, or SPC and found no significant correlation. The lack of coherence among these three sets of inflammatory markers would suggest that their different temporal patterns are consistent with distinct inflammatory stages or processes. Both GlycA and neopterin have been found to independently outperform CRP in terms of predicting early infection and inflammation82 although some studies have reported contrasting findings.83
Conclusions
Sepsis and septic shock remain a significant economic burden and are associated with high morbidity and mortality. Fast diagnosis of these conditions would lead to clinically advantageous outcomes. We have shown that nonsepsis, sepsis, and septic shock ICU patients have differential metabolic signatures, which can be utilized to diagnose patients in a time-efficient manner. While a total of 1128 lipoproteins, lipids, amino acid and tryptophan metabolites, and inflammatory markers were measured here, we have demonstrated that of these 15 parameters can be used to correctly stratify patients into the correct clinical outcome with high accuracy. Using these metabolites, lipids, lipoproteins, and inflammatory markers as a diagnostic for sepsis and septic shock could potentially reduce the mortality rates as diagnosis can be made within an hour of blood collection.
There are several limitations within this study; first, the sample size is small; however, we have demonstrated a proof of concept (with ROC for the models being 0.81 and 0.85), to further validate with a larger cohort. It should also be noted that time of onset of infection and sepsis is not known, and blood collections were only completed on admission into ICU. In addition, only clinical metadata such as CRP is provided at the first blood collection. Therefore, no modeling longitudinally was completed for each patient.
Acknowledgments
We thank Oscar Millet from CICBiogune for providing the healthy control samples used within this study. We thank the Department of Jobs, Tourism, Science and Innovation, Government of Western Australian Premier’s Fellowship and the ARC Laureate Fellowship funding for E.H. and the MRFF for funding the Australian National Phenome Centre for this work.
Data Availability Statement
The data that support the findings of this study are available at DOI: 10.5281/zenodo.10669068.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00803.
Table S1: Annotation of the keys used by the Bruker IVDr Lipoprotein Subclass Analysis (B.I.-LISA) method; Figure S1: Area under the ROC and PR curves for model 1 and model 2 using only C-reactive protein; Figure S2: O-PLS-DA and eruption plot for nonsepsis patients versus septic and septic shock patients for the amino acid and tryptophan pathway intermediates; Table S2: O-PLS-DA loadings, Cliff’s delta, and adjusted p-values for the amino acids and tryptophan pathway intermediates for nonseptic versus septic and septic shock; Figure S3: O-PLS-DA for non sepsis patients versus sepsis and septic shock patients for the lipoproteins and eruption plot; Table S3: OPLS loadings, Cliff’s delta, and adjusted p-values for the lipoproteins for no sepsis versus sepsis and septic shock; Figure S4: O-PLS-DA and eruption plot for non sepsis patients versus sepsis and septic shock patients for the glycoproteins and SPC; Table S4: OPLS loadings, Cliff’s delta, and adjusted p-values for the glycoprotein and SPC for no sepsis versus sepsis and septic shock; Figure S5: Box plots of healthy controls, patients with no sepsis, patients with sepsis, and patients with septic shock for the model 1 lipoproteins and neopterin: Figure S6: O-PLS-DA and eruption plot for sepsis patients versus septic shock patients for the lipoproteins; Table S5: OPLS loadings, Cliff’s delta, and adjusted p-values for the lipoproteins of sepsis patients vs septic shock patients at the baseline; Figure S7: O-PLS-DA and eruption plot for sepsis versus septic shock patients for the lipids; Table S6: OPLS loadings, Cliff’s delta, and adjusted p-values for the lipids of the sepsis patients versus septic shock patients; Figure S8: O-PLS-DA for sepsis patients versus septic shock patients for the glycoproteins and SPC; Table S7: OPLS loadings, Cliff’s delta, and adjusted p-values for the glycoproteins and SPC of sepsis patients vs septic shock patients; Figure S9: Box plots of healthy controls, patients with no sepsis, patients with sepsis, and patients with septic shock for the glycoproteins and SPC at 7 days post ICU admission; Figure S10: CRP and GlycA correlation (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Paoli C. J.; Reynolds M. A.; Sinha M.; Gitlin M.; Crouser E. Epidemiology and Costs of Sepsis in the United States - An Analysis Based on Timing of Diagnosis and Severity Level. Critical Care Medicine. 2018, 46, 1889–1897. 10.1097/CCM.0000000000003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins A. M.; Brooker J. E.; Mackie M.; Cooper D. J.; Harris A. H. Health Economic Evaluations of Sepsis Interventions in Critically Ill Adult Patients: A Systemic Review. J. Intensive Care. 2020, 8, 5. 10.1186/s40560-019-0412-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E.; Johnson S. C.; Agesa K. M.; Shackelford K. A.; Tsoi D.; Kievlan D. M.; Colombara D. V.; Ikuta K. S.; Kissoon N.; Finfer S.; Fleischmann-Struzek C.; Machado F. R.; Reinhart K. K.; Rowan K.; Seymour C. W.; Watson R. S.; West T. E.; Marinho F.; Hay S. I.; Lozano R.; Lopez A. D.; Angus D. C.; Murray C. J. L.; Naghavi M. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. The Lancet. 2020, 395, 200–211. 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott H. C.; Angus D. C. Enhancing Recovery From Sepsis A Review. JAMA. 2018, 319, 62–75. 10.1001/jama.2017.17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Poll T.; Van de Veerdonk F. L.; Scicluna B. P.; Netea M. G. The Immunopathology of Sepsis and Potential Therapeutic Targets. Nat Rev Immunol. 2017, 17, 407–420. 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- Mantzarlis K.; Tsolaki V.; Zakynthinos E. Role of Oxidative Stress and Mitochondrial Dysfunction in Sepsis and Potential Therapies. Oxid Med. Cell Longev. 2017, 5985209 10.1155/2017/5985209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince C.; Mayeux P. R.; Nguyen T.; Gomez H.; Kellum J. A.; Ospina-Tascon G. A.; Hernandez G.; Murray P.; De Backer D. The Endothelium in Sepsis. Shock 2016, 45, 259–270. 10.1097/SHK.0000000000000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Feng Y. w.; Yao Y. m. Potential Therapy Strategy: Targeting Mitochondrial Dysfunction in Sepsis. Military Med. Res. 2018, 5, 41. 10.1186/s40779-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasyluk W.; Zwolak A. Metabolic Alterations in Sepsis. J Clin Med. 2021, 10, 2412. 10.3390/jcm10112412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Sun Y.; Teng S.; Li K. Prediction of Sepsis Mortality Using Metabolite Biomarkers in the Blood: a Meta-analysis of Death-Related Pathways and Prospective Validation. BMC Med. 2020, 18, 83. 10.1186/s12916-020-01546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H.; Wang H.; Wang B.; Liu X.; Gao H.; Xu M.; Zhao H.; Deng X.; Lin D. Systemic Metabolic Changes of Traumatic Critically Ill Patients Revealed by an NMR-Based Metabonomic Approach. J.Proteome Res. 2009, 8, 5423–5430. 10.1021/pr900576y. [DOI] [PubMed] [Google Scholar]
- Tesfai A.; MacCallum N.; Kirkby N. S.; Gashaw H.; Gray N.; Want E.; Quinlan G. J.; Mumby S.; Leiper J. M.; Paul-Clark M.; Ahmetaj-Shala B.; Mitchell J. A. Metabolomic Profiling of Amines in Sepsis Predicts Changes in NOS Canonical Pathways. PLOS ONE. 2017, 12, e0183025 10.1371/journal.pone.0183025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She H.; Du Y.; Du Y.; Tan L.; Yang S.; Luo X.; Li Q.; Xiang X.; Lu H.; Hu Y.; Liu L.; Li T. Metabolomics and Machine Learning Approaches for Diagnostic and Prognostic Biomarkers Screening in Sepsis. BMC Anesthesiol. 2023, 23, 367. 10.1186/s12871-023-02317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsolus I. L.; Sweeney T. E.; Liesenfeld O.; Ledeboer N. A.; Kraft C. S. Diagnosing and Managing Sepsis by Probing the Host Response to Infection: Advances, Opportunities, and Challenges. J. Clin. Microbiol. 2019, 25, e00425–19. 10.1128/JCM.00425-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder A. L.; Gupta N.; Lulaj E.; Furgiuele M.; Hidalgo I.; Jones M. P.; Jolly T.; Gennis P.; Birnbaum A. Predictors of Early Progression to Severe Sepsis or Shock Among Emergency Department Patients with Nonsevere Sepsis. Int. J. Emerg. Med. 2016, 9, 10. 10.1186/s12245-016-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. L. C.; Mason A. J.; Gordon A. C.; Brett S. J. Are Large Randomised Controlled Trials in Severe Sepsis and Septic Shock Statistically Disadvantaged by Repeated Inadvertent Underestimates of Required Sample Size?. BMJ Open. 2018, 8, e020068 10.1136/bmjopen-2017-020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong J. Z.; Koh Z. X.; Samsudin M. I.; Fook-Chong S.; Liu N.; Ong M. E. H. Validation of the Mortality in Emergency Department Sepsis (MEDS) Score in a Singaporean Cohort. Medicine (Baltimore). 2019, 98, e16962 10.1097/MD.0000000000016962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaka F.; EthmaneAbouElMaali C.; Cytron M. A.; Fowler K.; Javaux V. M.; O’Brien J. Predicting Mortality of Patients with Sepsis: A Comparison of APACHE II and APACHE III Scoring Systems. J. Clin. Med. Res. 2017, 9, 907–910. 10.14740/jocmr3083w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugar S.; Choudhary C.; Duggal A. Sepsis and Septic Shock: Guideline-based Management. Cleveland Clinic Journal of Medicine. 2020, 87, 53–64. 10.3949/ccjm.87a.18143. [DOI] [PubMed] [Google Scholar]
- Mok K.; Christian M. D.; Nelson S.; Burry L. Time to Administration of Antibiotics Among Inpatients with Severe Sepsis or Septic Shock. Can. J. Hosp. Pharm. 2014, 67, 213–219. 10.4212/cjhp.v67i3.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew Clayton T.; Lindon J. C.; Cloarec O.; Antti H.; Charuel C.; Hanton G.; Provost J. P.; Le Net J. L.; Baker D.; Walley R. J.; Everett J. R.; Nicholson J. K. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006, 440, 1073–1077. 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- Clayton T. A.; Baker D.; Lindon J. C.; Everett J. R.; Nicholson J. K. Pharmacometabonomic Identification of a Significant Host-Microbiome Metabolic Interaction Affecting Human Drug Metabolism. PNAS. 2009, 106, 14728–14733. 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett J. R.; Loo R. L.; Pullen F. S. Pharmacometabonomics and Personalized Medicine. Annals of Clinical Biochemistry. 2013, 50, 523–545. 10.1177/0004563213497929. [DOI] [PubMed] [Google Scholar]
- Ruffieux H.; Hanson A. L.; Lodge S.; Lawler N. G.; Whiley L.; Gray N.; Nolan T. H.; Bergamaschi L.; Mescia F.; Turner L.; de Sa A.; Pelly V. S.; The Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) BioResource COVID-19 Collaboration;; Kotagiri P.; Kingston N.; Bradley J. R.; Holmes E.; Wist J.; Nicholson J. K.; Lyons P. A.; Smith K. G. C.; Richardson S.; Bantug G. R.; Hess C. A Patient-centric Modeling Framework Captures Recovery from SARS-CoV-2 Infection.. Nature Immunol. 2023, 24, 349–358. 10.1038/s41590-022-01380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; A Patient-centric Modeling Framework Captures Recovery from SARS-CoV-2 Infection. Nature Immunology 2023, 24, 349–358. [DOI] [PMC free article] [PubMed]
- Lodge S.; Lawler N. G.; Gray N.; Masuda R.; Nitschke P.; Whiley L.; Bong S. H.; Yeap B. B.; Dwivedi G.; Spraul M.; Schaefer H.; Gil-Redondo R.; Embade N.; Millet O.; Holmes E.; Wist J.; Nicholson J. K. Integrative Plasma Metabolic and Lipidomic Modelling of SARS-CoV-2 Infection in Relation to Clinical Severity and Early Mortality Prediction. Int. J. Mol. Sci. 2023, 24, 11614. 10.3390/ijms241411614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda R.; Lodge S.; Whiley L.; Gray N.; Lawler N.; Nitschke P.; Bong S. H.; Kimhofer T.; Loo R. L.; Boughton B.; Zeng A. X.; Hall D.; Schaefer H.; Spraul M.; Dwivedi G.; Yeap B. B.; Diercks T.; Bernardo-Seisdedos G.; Mato J. M.; Lindon J. C.; Holmes E.; Millet O.; Wist J.; Nicholson J. K. Exploration of Human Serum Lipoprotein Supramolecular Phospholipids Using Statistical Heterospectroscopy in n-Dimensions (SHY-n): Identification of Potential Cardiovascular Risk Biomarkers Related to SARS-CoV-2 Infection. Anal. Chem. 2022, 94, 4426–4436. 10.1021/acs.analchem.1c05389. [DOI] [PubMed] [Google Scholar]
- Nitschke P.; Lodge S.; Kimhofer T.; Masuda R.; Bong S. H.; Hall D.; Schäfer H.; Spraul M.; Pompe N.; Diercks T.; Bernardo-Seisdedos G.; Mato J. M.; Millet O.; Susic D.; Henry A.; El-Omar E. M.; Holmes E.; Lindon J. C.; Nicholson J. K.; Wist J. J-Edited DIffusional Proton Nuclear Magnetic Resonance Spectroscopic Measurement of Glycoprotein and Supramolecular Phospholipid Biomarkers of Inflammation in Human Serum. Anal. Chem. 2022, 94, 1333–1341. 10.1021/acs.analchem.1c04576. [DOI] [PubMed] [Google Scholar]
- Litton E.; Anstey M.; Broadhurst D.; Chapman A.; Currie A.; Ferrier J.; Gummer J.; Higgins A.; Lim J.; Manning L.; Myers E.; Orr K.; Palermo A. M.; Paparini A.; Pellicano S.; Raby E.; Rammohan A.; Regli A.; Richter B.; Salman S.; Strunk T.; Waterson S.; Weight D.; Wibrow B.; Wood F. Early and Sustained Lactobacillus Plantarum Probiotic Therapy in Critical Illness: The Randomised, Placebo-Controlled, Restoration of Gut Microflora in Critical Illness Trial (ROCIT). Intensive Care Med. 2021, 47, 307–315. 10.1007/s00134-020-06322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona A. C.; Jiménez B.; Schäfer H.; Humpfer E.; Spraul M.; Lewis M. R.; Pearce J. T. M.; Holmes E.; Lindon J. C.; Nicholson J. K. Precision High-Throughput Proton NMR Spectroscopy of Human Urine, Serum, and Plasma for Large-Scale Metabolic Phenotyping. Anal. Chem. 2014, 19, 9887–9894. 10.1021/ac5025039. [DOI] [PubMed] [Google Scholar]
- Lodge S.; Nitschke P.; Kimhofer T.; Wist J.; Bong S-H.; Loo R. L.; Masuda R.; Begum S.; Richards T.; Lindon J. C.; Bermel W.; Reinsperger T.; Schafer H.; Spraul M.; Holmes E.; Nicholson J. K. Diffusion and Relaxation Edited Proton NMR Spectroscopy of Plasma Reveals a High-Fidelity Supramolecular Biomarker Signature of SARS-CoV-2 Infection. Anal. Chem. 2021, 93, 3976–3986. 10.1021/acs.analchem.0c04952. [DOI] [PubMed] [Google Scholar]
- Jimenez B.; Holmes E.; Heude C.; Tolson R. F.; Harvey N.; Lodge S.; Chetwynd A. J.; Cannet C.; Fang F.; Pearce J. T. M.; Lewis M. R.; Viant M. R.; Lindon J. C.; Spraul M.; Schafer H.; Nicholson J. K. Quantitative Lipoprotein Subclass and Low Molecular Weight Metabolite Analysis in Human Serum and Plasma by 1H NMR Spectroscopy in a Multilaboratory Trial. Anal. Chem. 2018, 90, 11962–11971. 10.1021/acs.analchem.8b02412. [DOI] [PubMed] [Google Scholar]
- Lawler N. G.; Gray N.; Kimhofer T.; Boughton B.; Gay M.; Yang R.; Morillon A-C.; Chin S-T.; Ryan M.; Begum S.; Bong S-H.; Coudert J. D.; Edgar D.; Raby E.; Pettersson S.; Richards T.; Holmes E.; Whiley L.; Nicholson J. K. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J Proteome Res. 2021, 20, 2796–2811. 10.1021/acs.jproteome.1c00052. [DOI] [PubMed] [Google Scholar]
- Gray N.; Lawler N. G.; Yang R.; Morillon A-C.; Gay M. C. L.; Bong S-H.; Holmes E.; Nicholson J. K.; Whiley L. A Simultaneous Exploratory and Quantitative Amino Acid and Biogenic Amine Metabolic Profiling Platform for Rapid Disease Phenotyping via UPLC-QToF-MS. Talanta. 2021, 223, 121872 10.1016/j.talanta.2020.121872. [DOI] [PubMed] [Google Scholar]
- Ryan M. J.; Grant-St James A.; Lawler N. G.; Fear M. W.; Raby E.; Wood F. M.; Maker G. L.; Wist J.; Holmes E.; Nicholson J. K.; Whiley L.; Gray N. Comprehensive Lipidomic Workflow for Multicohort Population Phenotyping Using Stable Isotope Dilution Targeted Liquid Chromatography-Mass Spectrometry. J. Proteome Res. 2023, 22, 1419–1433. 10.1021/acs.jproteome.2c00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. J.; Pratt B.; Bose N.; Dubois L. G.; St. John-Williams L.; Perrott K. M.; Ky K.; Kapahi P.; Sharma V.; MacCoss M. J.; Moseley M. A.; Colton C. A.; MacLean B. X.; Schilling B.; Thompson J. W. Alzheimer’s Disease Metabolomics Consortium. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H.; Ji F.; Chen Y.; Cai Z. statTarget: A Streamlined Tool for Signal Drift Correction and Interpretations of Quantitative Mass Spectrometry-based Omics Data. Anal. Chim. Acta 2018, 1036, 66–72. 10.1016/j.aca.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Cliff N. Dominance Statistics: Ordinal Analyses to Answer Ordinal Questions. Psychological Bulleti. 1993, 114, 494–509. 10.1037/0033-2909.114.3.494. [DOI] [Google Scholar]
- Bylesjo M.; Rantalainen M.; Cloarec O.; Nicholson J. K.; Holmes E.; Trygg J. OPLS Discriminant Analysis: Combining the Strengths of PLS-DA and SIMCA Classification. J Chemometrics. 2006, 20, 341–351. 10.1002/cem.1006. [DOI] [Google Scholar]
- Lee S. H.; Park M. S.; Park B. H.; Jung W. J.; Lee I. S.; Kim S. Y.; Kim E. Y.; Jung J. Y.; Kang Y. A.; Kim Y. S.; Kim S. K.; Chang J.; Chung K. S. Prognostic Implications of Serum Lipid Metabolism Over Time During Sepsis. Biomed. Res. Int. 2015, 2015, 789298 10.1155/2015/789298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen H. J.; Heezius E. C. J.; Dallinga G. M.; van Strijp J. A. G.; Verhoef J.; van Kessel K. P. M. Lipoprotein Metabolism in Patients with Severe Sepsis. Crit Care Med. 2003, 31, 1359–1366. 10.1097/01.CCM.0000059724.08290.51. [DOI] [PubMed] [Google Scholar]
- Tanaka S.; Couret D.; Tran-Dinh A.; Duranteau J.; Montravers P.; Schwendeman A.; Meilhac O. High-Density Lipoproteins During Sepsis: From Bench to Bedside. Crit. Care. 2020, 24, 134. 10.1186/s13054-020-02860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasi A.; Franzin R.; Fiorentino M.; Squiccimarro E.; Castellano G.; Gesualdo L. Multifaced Roles of HDL in Sepsis and SARS-CoV-2 Infection: Renal Implications. Int J Mol Sci. 2021, 22, 5980. 10.3390/ijms22115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirillo A.; Catapano A. L.; Norata G. D.. HDL in Infectious Diseases and Sepsis. In: von Eckardstein A.; Kardassis D. (eds) High Density Lipoproteins. Handbook of Experimental Pharmacology; Springer; 2015, 224, 483-508. [DOI] [PubMed] [Google Scholar]
- Barker G.; Leeuwenburgh C.; Brusko T.; Moldawer L.; Reddy S. T.; Guirgis F. W. Lipid and Lipoprotein Dysregulation in Sepsis: Clinical and Mechanistic Insights into Chronic Critical Illness. J Clin Med. 2021, 10, 1693. 10.3390/jcm10081693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P. A.; Berbee J. F. P.; Rensen P. C. N.; Kitchens R. L. Apolipoprotein A-II Augments Monocyte Responses to LPS by Suppressing the Inhibitory Activity of LPS-Binding Protein. Innate Immun. 2008, 14, 365–374. 10.1177/1753425908099171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. D.; Ramos R. A.; Tobias P. S.; Ulevitch R. J.; Mathison J. C. CD14, a Receptor for Complexes of Lipopolysaccharide (LPS) and LPS Binding Protein. Science. 1990, 249, 1431–1433. 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Schumann R. R.; Leong S. R.; Flaggs G. W.; Gray P. W.; Wright S. D.; Mathison J. C.; Tobias P. S.; Ulevitch R. J. Structure and Function of Lipopolysaccharide Binding Protein. Science. 1990, 249, 1429–1431. 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- Cockerill G. W.; Rye K. A.; Gamble J. R.; Vadas M. A.; Barter P. J. High-Density Lipoproteins Inhibit Cytokine Induced Expression of Endothelial Cell Adhesion Molecules. Arteriosclerosis, Thrombosis and Vascular Biology. 1995, 15, 1987–1994. 10.1161/01.ATV.15.11.1987. [DOI] [PubMed] [Google Scholar]
- Cirstea M.; Walley K. R.; Russell J. A.; Brunham L. R.; Genga K. R.; Boyd J. H. Decreased High-Density Lipoprotein Cholesterol Level is an Early Prognostic Marker for Organ Dysfunction and Death in Patients with Suspected Sepsis. J Crit Care. 2017, 38, 289–294. 10.1016/j.jcrc.2016.11.041. [DOI] [PubMed] [Google Scholar]
- Toth P. R. High-Density Lipoprotein and Cardiovascular Risk. Circulation. 2004, 109, 1809–1812. 10.1161/01.CIR.0000126889.97626.B8. [DOI] [PubMed] [Google Scholar]
- Femlak M.; Gluba-Brzózka A.; Ciałkowska-Rysz A.; Rysz J. The Role and Function of HDL in Patients with Diabetes Mellitus and the Related Cardiovascular Risk. Lipids Health Disease 2017, 16, 207. 10.1186/s12944-017-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda R.; Wist J.; Lodge S.; Kimhofer T.; Hunter M.; Hui J.; Beilby J. P.; Burnett J. R.; Dwivedi G.; Schlaich M. P.; Bong S-H.; Loo R. L.; Holmes E.; Nicholson J. K.; Yeap B. B. Plasma Lipoprotein Subclass Variation in Middle-Aged and Older Adults: Sex-stratified Distributions and Associations with Health Status and Cardiometabolic Risk Factors. Journal of Clinical Lipidology. 2023, 17, 677–687. 10.1016/j.jacl.2023.06.004. [DOI] [PubMed] [Google Scholar]
- Lodge S.; Nitschke P.; Kimhofer T.; Coudert J. D.; Begum S.; Bong S-H.; Richards T.; Edgar D.; Raby E.; Spraul M.; Schaefer H.; Lindon J. C.; Loo R. L.; Holmes E.; Nicholson J. K. NMR Spectroscopic Windows on the Systemic Effects of SARS-CoV-2 Infection on Plasma Lipoproteins and Metabolites in Relation to Circulating Cytokines. J Proteome Res 2021, 20, 1382–1396. 10.1021/acs.jproteome.0c00876. [DOI] [PubMed] [Google Scholar]
- Lodge S.; Nitschke P.; Loo R. L.; Kimhofer T.; Bong S. H.; Richards T.; Begum S.; Spraul M.; Schaefer H.; Lindon J. C.; Holmes E.; Nicholson J. K. Low Volume in Vitro Diagnostic Proton NMR Spectroscopy of Human Blood Plasma for Lipoprotein and Metabolite Analysis: Application to SARS-CoV-2 Biomarkers. J. Proteome Res. 2021, 20, 1415–1423. 10.1021/acs.jproteome.0c00815. [DOI] [PubMed] [Google Scholar]
- Masuda R.; Lodge S.; Nitschke P.; Spraul M.; Schaefer H.; Bong S. H.; Kimhofer T.; Hall D.; Loo R. L.; Bizkarguenaga M.; Bruzzone C.; Gil-Redondo R.; Embade N.; Mato J. M.; Holmes E.; Wist J.; Millet O.; Nicholson J. K. Integrative Modeling of Plasma Metabolic and Lipoprotein Biomarkers of SARS-CoV-2 Infection in Spanish and Australian COVID-19 Patient Cohorts. J. Proteome Res. 2021, 20, 4139–4152. 10.1021/acs.jproteome.1c00458. [DOI] [PubMed] [Google Scholar]
- Harbaum L.; Ghataorhe P.; Wharton J.; Jimenez B.; Howard L. S. G.; Gibbs J. S. R.; Nicholson J. K.; Rhodes C. J.; Wilkins M. R. Reduced Plasma Levels of Small HDL Particles Transporting Fibrinolytic Proteins in Pulmonary Arterial Hypertension. Thorax. 2019, 74, 380–389. 10.1136/thoraxjnl-2018-212144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Parker G.; Gaddam R. R.; Moltchanova E.; Carr A.; Shaw G.; Chambers S.; Gieseg S. P. Oxidative Stress and Immune Cell Activation Quantification in Sepsis and Non-sepsis Critical Care Patients by Neopterin/7,8-dihydroneopterin Analysis. Pteridines. 2020, 31, 68–82. 10.1515/pteridines-2020-0015. [DOI] [Google Scholar]
- Ploder M.; Neurauter G.; Spittler A.; Schroecksnadel K.; Roth E.; Fuchs D. Serum Phenylalanine in Patients Post Trauma and with Sepsis Correlate to Neopterin Concentrations. Amino Acids. 2008, 35, 303–307. 10.1007/s00726-007-0625-x. [DOI] [PubMed] [Google Scholar]
- Eisenhut M. Neopterin in Diagnosis and Monitoring of Infectious Diseases. J. Biomark. 2013, 2013, 196432 10.1155/2013/196432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar B.; Spisarova M.; Bartouskova M.; Krcmova L. K.; Javorska L.; Studentova H. Neopterin as a Biomarker of Immune Response in Cancer Patients. Ann Transl Med. 2017, 5, 280. 10.21037/atm.2017.06.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrak V.; Secnik P.; Skrakova M. Neopterin Screening of Blood Donations. Pteridines. 2006, 17, 105–106. 10.1515/pteridines.2006.17.4.105. [DOI] [Google Scholar]
- Zhang X.; Chen Q.; Ni S.; Xiang Z.; Zhou X.; Huang Y. Serum Neopterin and its Significance as Biomarker in Differentiation of Mods From Sepsis. Pteridines. 2018, 29, 201–205. 10.1515/pteridines-2018-0018. [DOI] [Google Scholar]
- Fisgin N. T.; Aliyazicioglu Y.; Tanyel E.; Coban A. Y.; Ulger F.; Zivalioglu M.; Esen S.; Leblebicioglu H. The Value of Neopterin and Procalcitonin in Patients with Sepsis. South Med J. 2010, 103, 216–219. 10.1097/SMJ.0b013e3181cf11a1. [DOI] [PubMed] [Google Scholar]
- Oweira H.; Lahdou I.; Daniel V.; Hofer S.; Mieth M.; Schmidt J.; Schemmer P.; Opelz G.; Mehrabi A.; Sadeghi M. Early Post-transplant Neopterin Associated with One Year Survival and Bacteremia in Liver Transplant Recipients. Hum. Immunol. 2016, 77, 115–120. 10.1016/j.humimm.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Helan M.; Malaska J.; Tomandl J.; Jarkovsky J.; Helanova K.; Benesova K.; Sitina M.; Dastych M.; Ondrus T.; Pavkova Goldbergova M.; Gal R.; Lokaj P.; Tomandlova M.; Parenica J. Kinetics of Biomarkers of Oxidative Stress in Septic Shock: A Pilot Study. Antioxidants 2022, 11, 640. 10.3390/antiox11040640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G.; Leeuwenburgh C.; Brusko T.; Moldawer L.; Reddy S. T.; Guirgis F. W. Lipid and Lipoprotein Dysregulation in Sepsis: Clinical and Mechanistic Insights into Chronic Critical Illness. J Clin Med. 2021, 10, 1693. 10.3390/jcm10081693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkou A.; Mouzaki A.; Siagris D.; Ravani I.; Gogos C. A. Serum Lipid Profile, Cytokine Production, and Clinical Outcome in Patients with Severe Sepsis. J Crit Care 2014, 29, 723–727. 10.1016/j.jcrc.2014.04.018. [DOI] [PubMed] [Google Scholar]
- Ferrario M.; Brunelli L.; Su F.; Herpain A.; Pastorelli R. The Systemic Alterations of Lipids, Alanine-Glucose Cycle and Inter-Organ Amino Acid Metabolism in Swine Model Confirms the Role of Liver in Early Phase of Septic Shock. Front Physiol. 2019, 10, 11. 10.3389/fphys.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer S.; Giamarellos-Bourboulis E. J.; Pelekanou A.; Marioli A.; Baziaka F.; Tsangaris I.; Bauer M.; Kiehntopf M. Metabolite Profiles in Sepsis: Developing Prognostic Tools Based on the Type of Infection. Crit Care Med. 2016, 44, 1649–1662. 10.1097/CCM.0000000000001740. [DOI] [PubMed] [Google Scholar]
- Baisted D. J.; Robinson B. S.; Vance D. E. Albumin Stimulates the release of Lysophosphatidylcholine from Cultured Rat Hepatocytes. Biochem. J. 1988, 253, 693–701. 10.1042/bj2530693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N.; Matsubara T.; Krausz K. W.; Patterson A. D.; Gonzalez F. J. Disruption of Phospholipid and Bile Acid Homeostasis in Mice with Nonalcoholic Steatohepatitis. Hepatology. 2012, 56, 118–129. 10.1002/hep.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmaenner D. A.; Kleyman A.; Press A.; Bauer M.; Singer M. The Many Roles of Cholesterol in Sepsis: A Review. Am J Resp Crit Care Med. 2022, 205, 388–396. 10.1164/rccm.202105-1197TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.; Xu S.; Zhou B.; Zhou R.; Liu P.; Hui X.; Long Y.; Su L. Dynamic Plasma Lipidomic Analysis Revealed Cholesterol Ester and Amides Associated with Sepsis Development in Critically Ill Patients after Cardiovascular Surgery with Cardiopulmonary Bypass. J Pers Med. 2022, 12, 1838. 10.3390/jpm12111838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grion C. M. C.; Cardoso L. T. Q.; Perazolo T. F.; Garcia A. S.; Barbosa D. S.; Morimoto H. K.; Matsuo T.; Carrilho A. J. F. Lipoproteins and CETP Levels as Risk Factors for Severe Sepsis in Hospitalized Patients. Eur J Clin Invest. 2010, 40, 330–338. 10.1111/j.1365-2362.2010.02269.x. [DOI] [PubMed] [Google Scholar]
- Venancio T. M.; Machado R. M.; Castoldi A.; Amano M. T.; Nunes V. S.; Quintao E. C. R.; Camara N. O. S.; Soriano F. G.; Cazita P. M. CETP Lowers TLR4 Expression Which Attenuates the Inflammatory Response Induced by LPS and Polymicrobial Sepsis. Mediators Inflamm. 2016, 2016, 1784014 10.1155/2016/1784014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokkala K.; Houttu N.; Koivuniemi E.; Sørensen N.; Nielsen H. B.; Laitinen K. GlycA, a Novel Marker for Low Grade Inflammation, Reflects Gut Microbiome Diversity and is More Accurate than High Sensitive CRP in Reflecting Metabolomic Profile. Metabolomics. 2020, 16, 76. 10.1007/s11306-020-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. J.; Raby E.; Whiley L.; Masuda R.; Lodge S.; Nitschke P.; Maker G. L.; Wist J.; Holmes E.; Wood F. M.; Nicholson J. K.; Fear M. W.; Gray N. Nonsevere Burn Induces a Prolonged Systemic Metabolic Phenotype Indicative of a Persistent Inflammatory Response Postinjury. J Prot Res. 2023, 10.1021/acs.jproteome.3c00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S. C.; Würtz P.; Nath A. P.; Abraham G.; Havulinna A. S.; Fearnley L. G.; Sarin A. P.; Kangas A. J.; Soininen P.; Aalto K.; Seppälä I.; Raitoharju E.; Salmi M.; Maksimow M.; Männistö S.; Kähönen M.; Juonala M.; Ripatti S.; Lehtimäki T.; Jalkanen S.; Perola M.; Raitakari O.; Salomaa V.; Ala-Korpela M.; Kettunen J.; Inouye M. The Biomarker GlycA is Associated with Chronic Inflammation and Predicts Long-Term Risk of Severe Infection. Cell Systems. 2015, 1, 293–301. 10.1016/j.cels.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Otvos J. D.; Shalaurova I.; Wolak-Dinsmore J.; Connelly M. A.; Mackey R. H.; Stein J. H.; Tracy R. P. GlycA: A Composite Nuclear Magnetic Resonance Biomarker of Systemic Inflammation. Clinical Chemistry. 2015, 61, 714–723. 10.1373/clinchem.2014.232918. [DOI] [PubMed] [Google Scholar]
- Määttä A. M.; Salminen A.; Pietiäinen M.; Leskelä J.; Palviainen T.; Sattler W.; Sinisalo J.; Salomaa V.; Kaprio J.; Pussinen P. J. Endotoxemia is Associated with an Adverse Metabolic Profile. Innate Immun. 2021, 27, 3–14. 10.1177/1753425920971702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokkala K.; Pellonpera O.; Roytio H.; Pussinen P.; Ronnemaa T.; Laitinen K. Increased Intestinal Permeability, Measured by Serum Zonuli, is Associated with Metabolic Risk Markers in Overweight Pregnant Women. Metabolism. 2017, 69, 43–50. 10.1016/j.metabol.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Hara S.; Sanatani T.; Tachikawa N.; Yoshimura Y.; Miyata N.; Sasaki H.; Kuroda R.; Kamikokuryo C.; Eguchi T.; Niiyama S.; Kakihana Y.; Ichinose H. Comparison of the Levels of Neopterin, CRP and IL-6 in Patients Infected with and without SARS-CoV-2. Heliyon. 2022, 8, e09371 10.1016/j.heliyon.2022.e09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. C. Serum C-reactive Protein and Neopterin Concentrations in Patients with Viral or Bacterial Infection. J Clin Pathol 1991, 44, 596–599. 10.1136/jcp.44.7.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available at DOI: 10.5281/zenodo.10669068.