Abstract
Adeno-associated virus (AAV) integrates very efficiently into a specific site (AAVS1) of human chromosome 19. Two elements of the AAV genome are sufficient: the inverted terminal repeats (ITRs) and the Rep78 or Rep68 protein. The incorporation of the AAV integration machinery in nonviral delivery systems is of great interest for gene therapy. We demonstrate that purified recombinant Rep68 protein is functionally active when directly delivered into human cells by using the polycationic liposome Lipofectamine, promoting the rescue-replication of a codelivered ITR-flanked cassette in adenovirus-infected cells and its site-specific integration in noninfected cells. The sequencing of cloned virus-host DNA junctions confirmed that lipofected Rep68 protein triggers site-specific integration at the same sites in chromosome 19 already characterized in cells latently infected with AAV.
Adeno-associated virus (AAV) is a defective parvovirus with a single-stranded DNA genome of 4.7 kb comprising two open reading frames coding for nonstructural (Rep) and structural (Cap) proteins, and the entire genome is flanked by two identical 145-base inverted terminal repeats (ITRs) (3). AAV replicates only in cells coinfected by a helper virus such as adenovirus (Ad); in the absence of coinfection, the virus integrates stably into a defined region, called AAVS1, of human chromosome 19 (q13.4-qter) (9, 16, 17, 27, 29).
The mechanism of AAV integration has been partially elucidated. The two hairpinned ITRs are the minimal elements required for integration; in fact, recombinant AAV vectors lacking the rep and cap genes still integrate into the human genome, albeit not specifically in chromosome 19 (6, 15, 39). The efficiency of ITR-dependent integration is presently unknown (39). Integration into the AAVS1 site also requires the presence of the viral Rep78 or Rep68 protein (6, 23). Rep78 and Rep68 are expressed from unspliced and spliced transcripts, respectively, initiated at the common p5 promoter (3). The two proteins have similar biochemical properties: they bind to a specific DNA sequence (the Rep binding site [RBS]) present in the AAV ITRs and the p5 promoter, nick the terminal resolution site (trs) in the ITRs in a strand- and site-specific manner, and have an ATP-dependent helicase activity (11, 12). The first step in site-specific integration is postulated to be the formation of a complex, mediated by Rep78 or Rep68, between the AAV ITR and an RBS which has been identified within the AAVS1 site (37). Subsequently, integration is presumed to proceed through a replication-mediated recombination process, in which several template switches occur during synthesis of new DNA strands; the entire process requires the activity of the cellular factors involved in DNA replication and repair (22).
The use of elements of AAV to drive site-specific integration of exogenous DNA sequences delivered with nonviral systems is of great interest for gene therapy (6, 21). In fact, one of the major problems associated with nonviral delivery, the transient expression of the transgene due to the rapid degradation of delivered DNA, might be overcome by utilizing viral components that facilitate integration of the transgene into the host genome. The feasibility of this approach has been demonstrated recently: expression vectors for either Rep78 or Rep68 can promote the integration into the AAVS1 site of an ITR-flanked transgene delivered either in the same plasmid or in a cotransfected plasmid (1, 31, 34). The advantages of nonviral delivery are that no potentially replicating viruses are introduced and that, in theory, there would be no size constraint for the integrating sequence. Cationic liposomes currently represent one of the most promising tools for nonviral delivery in gene therapy, and a great deal of work is being devoted to enhancing their efficacy in vivo (5, 8, 18, 19, 24, 35). Although they are used mainly for introducing DNA, they can also deliver proteins or peptides in vitro and in vivo (2, 7, 13, 14, 20, 30, 40). As a first step toward the assembling of an “ideal” nonviral particle incorporating the AAV integration machinery, we thus decided to test the hypothesis that functionally active (i.e., integration-competent) Rep78 or Rep68 recombinant protein could be delivered into cultured cells by using cationic liposomes.
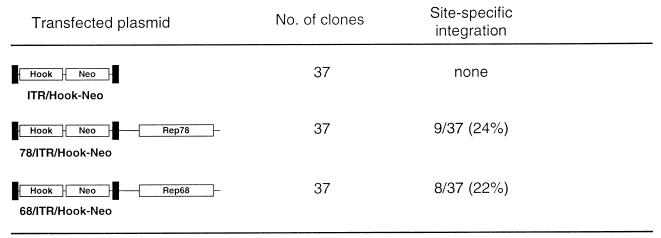
Before determining whether a functional recombinant Rep78 or Rep68 could be lipofected into cells, we tested whether the two genes promote site-specific integration with the same efficiency. Three different plasmids were constructed. The first construct, ITR/Hook-Neo (Fig. 1), contains an “integration element,” represented by the expression cassette for the neomycin resistance gene (Neo) and the expression cassette for the membrane-bound single-chain antibody, both cloned between the AAV ITRs contained in vector pSub201 (28). From ITR/Hook-Neo, two plasmids, called 78/ITR/Hook-Neo and 68/ITR/Hook-Neo, were derived; these contain, outside the ITR-flanked cassettes and under the control of the cytomegalovirus (CMV) enhancer/promoter element, cDNAs for Rep78 and Rep68, respectively (Fig. 1). Western blotting performed with whole-cell extracts from transiently transfected HeLa cells confirmed that only Rep78 and Rep68 were produced from plasmids 78/ITR/Hook-Neo and 68/ITR/Hook-Neo, respectively (data not shown).
FIG. 1.
Site-specific integration in HeLa cell clones. Transfected plasmids are schematically represented. Black boxes represent the AAV ITRs flanking the Hook and Neo expression cassettes. The Neo gene was derived from plasmid PRc/RSV, and the Hook gene was obtained from plasmid pHook-1 (both from Invitrogen). The individual cDNAs coding for Rep78 and Rep68 were obtained by site-directed mutagenesis of the Rep open reading frame (nucleotides 320 to 2252 of the AAV genome [33]) as described elsewhere (34). Integration at the AAVS1 site was assessed by DNA hybridization analysis with AAVS1 and neo-specific probes as described in the text.
Plasmid ITR/Hook-Neo and its two Rep-expressing derivatives were transfected into HeLa cells by the calcium phosphate technique: following 14 to 18 days of growth in selective medium containing 700 μg of G418/ml, 37 single-cell-derived neomycin-resistant clones were isolated and expanded. Genomic DNA was extracted, digested with the restriction enzyme BamHI, which has no recognition site in the transgene and in the subregion of AAVS1 in which the great majority of integration events are detected (17, 39), and analyzed by Southern blotting using probes specific for AAVS1 and for the neo gene. Two criteria were adopted to monitor site-specific integration: the detection, by using the AAVS1 probe, of an upshifted band with respect to the normal AAVS1 site present in the parental cell line and the cohybridization of the same band with the Neo probe. According to this scoring system, no site-specific integration was detected in cells transfected with plasmid ITR/Hook-Neo; in contrast, integration in AAVS1 was evident in clones derived from cells transfected with the Rep-expressing plasmids. As summarized in Fig. 1, similar percentages of site-specific integration were observed for constructs expressing Rep78 and those expressing Rep68 (24% for 78/ITR/Hook-Neo and 22% for 68/ITR/Hook-Neo). This demonstrates that the two Rep proteins mediate integration in the AAVS1 site with similar efficiencies. We thus decided to investigate integration using the smaller of the two proteins, Rep68.
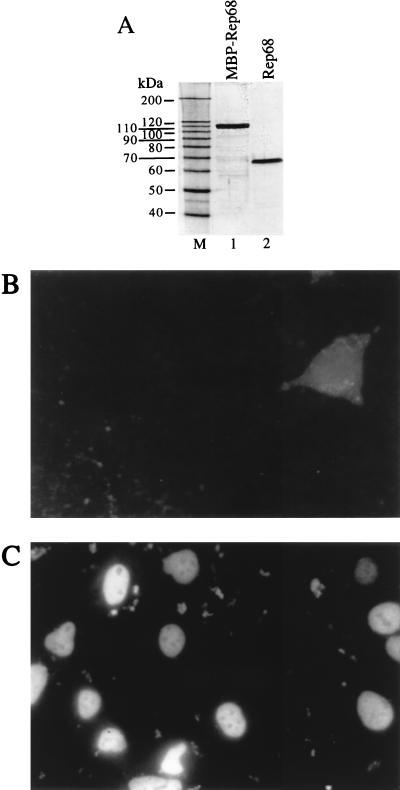
Rep68 protein was expressed in Escherichia coli and purified to homogeneity (Fig. 2). Its functionality was checked by testing its capability to bind to and cleave AAV ITRs and to support AAV DNA replication in vitro (data not shown). To test whether Rep68 could be lipofected in cells, we used Lipofectamine, a commercially available liposome preparation consisting of a 3:1 (wt/wt) mixture of the polycationic lipid 2,3-dioleyloxy-N-[2(sperminecarboxamido)ethyl]-N,N-dimethyl-1-propaminium trifluoroacetate (DOSPA) and the neutral lipid dioleoyl phosphatidylethanolamine (DOPE). Five micrograms of Rep68 was mixed with 10 μg of Lipofectamine in a total volume of 200 μl of Optimem; following 30 min of incubation, 800 μl of Optimem was added and the mixture was layered on 2 × 105 human renal carcinoma 293 cells. In control experiments, cells were incubated with Rep68 alone. After 2 h, the intracellular distribution of Rep68 was visualized by indirect immunofluorescence with an anti-Rep monoclonal antibody. No signal was observed in cells incubated with Rep68 alone; only a small percentage of cells (about 2 to 3%) displayed the faint and diffuse staining shown in Fig. 2B. In contrast, Rep68 was clearly visible in the nuclei of cells transfected with the Rep68–Lipofectamine complex (Fig. 2C); in different experiments, the efficiency of transfection, measured as the percentage of positively stained cells, ranged from 20 to 40%. This result demonstrates that the lipofected protein efficiently enters the cells and undergoes a physiological cytoplasm-to-nucleus transport. A strong signal was also observed at 4 and 8 h posttransfection, while after 1 and 2 days, the number of positive cells strongly decreased. Similar results were obtained with the adenocarcinoma-derived cell line HeLa and the hepatocarcinoma-derived cell lines HuH7 and Hep3B (data not shown). It is worth noting that during the 2 days of these experiments, we did not observe any obvious phenotypic difference between cells (293, HeLa, Hep3B, and HuH7) lipofected with Rep68 protein and cells transfected with a Rep68 expression vector. Experiments were also designed to monitor the stability of delivered protein: Western blotting experiments, performed by probing whole-cell extracts prepared at different times postlipofection with an anti-Rep68 polyclonal serum, demonstrated that lipofected Rep68 undergoes progressive proteolytic degradation according to a kinetics which parallels, and therefore accounts for, the gradual decrease of the nuclear signal observed in immunofluorescence experiments (data not shown).
FIG. 2.
Lipofection of Rep68 protein. (A) Silver staining of a sodium dodecyl sulfate-polyacrylamide gel. Rep68 protein was expressed in E. coli as a fusion protein with maltose binding protein (MBP) as described elsewhere (4) and was partially purified by amylose affinity chromatography (lane 1), cleaved with Factor Xa to remove the maltose-binding moiety, and purified to homogeneity (lane 2) by fast protein liquid chromatography with MonoQ (anion exchange) and Superdex-75 (gel filtration) columns (both from Pharmacia). M, molecular size markers. (B and C) Intracellular localization of lipofected Rep68 protein. Two hours after transfection, 293 cells were washed, fixed in 3% formaldehyde, and permeabilized by treatment with 0.1% Triton X-100. The intracellular location of Rep68 was monitored by sequential incubation with the mouse monoclonal anti-Rep antibody 226.7 (Progen) and a rhodamine-conjugated anti-mouse immunoglobulin G goat polyclonal serum. Shown are results of staining of cells incubated for 2 h with Rep68 alone (B) or with the Rep68–Lipofectamine complex (C).
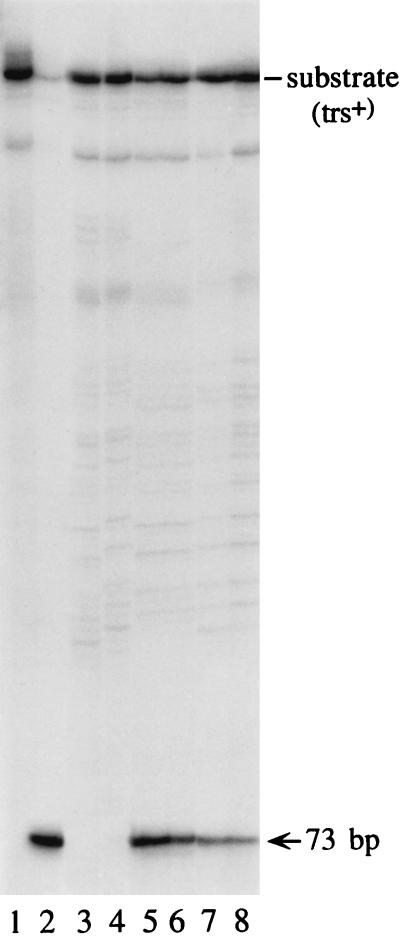
To test whether intranuclear Rep68 retained its biochemical properties, nuclear extracts were prepared 2 h after lipofection of 293 cells with Rep68 and incubated in the standard trs endonuclease reaction with labelled AAV ITR as described elsewhere (32). As a control, extracts from cells lipofected with the expression vector pCMVrep68, containing the cDNA for Rep68 cloned downstream of the CMV enhancer/promoter, were tested. Figure 3 shows that a site- and strand-specific nicking activity was detected only in the nuclear extracts of cells either lipofected with Rep68 (lanes 7 and 8) or transfected with the Rep68 expression vector (lanes 5 and 6): no specific activity was detected in untransfected cells (lanes 3 and 4).
FIG. 3.
Endonuclease reaction with nuclear extracts of cells lipofected with Rep68 protein. Ten thousand counts per minute of trs+ hairpin substrate that had been 5′-end labelled with 32P, prepared as described elsewhere (32), was incubated for 1 h in endonuclease assay buffer (32) with 1 and 3 μg of nuclear extracts from untransfected cells (lanes 3 and 4, respectively), cells lipofected with the Rep68 expression vector pCMVrep68 (lanes 5 and 6), or cells lipofected with Rep68 protein (lanes 7 and 8). A standard endonuclease reaction was carried out (32). The reaction was terminated by treatment with proteinase K, phenol extraction, and ethanol precipitation; samples were then resolved on an 8% polyacrylamide sequencing gel. The position of the 73-base product of the nicking reaction is indicated on the right. In lane 1, the starting trs+ substrate was loaded. Lane 2 shows the results of a control experiment with 10 ng of purified Rep68. Nuclear extracts were prepared as described elsewhere (36).
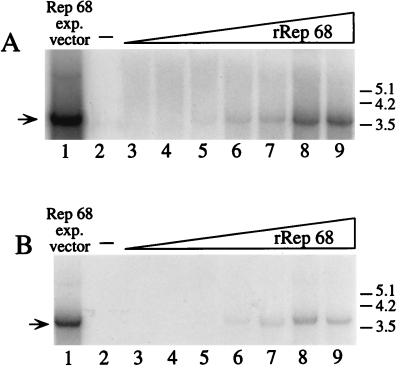
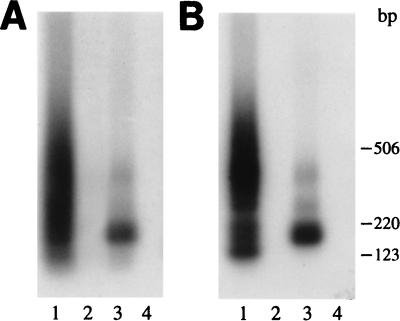
To further verify the functionality of the lipofected Rep68, we tested its capability to stimulate, in Ad-infected cells, the rescue-replication of an ITR-flanked cassette contained in a codelivered recombinant plasmid (28). Two cell lines were used: HeLa and HepG2. In both cases, 7.5 μg of plasmid ITR/Hook-Neo and increasing concentrations of Rep68 were coincubated in 1.8 ml of Optimem with 60 μg of Lipofectamine; after 30 min, 7.2 ml of medium was added and the mixture was layered on 2 × 106 cells which had been infected 2 h before with Ad-2 at a multiplicity of infection of 10. In control experiments, the same plasmid ITR/Hook-Neo was mixed with equivalent amounts of the pCMVrep68 expression vector or a carrier plasmid. After 8 h of incubation, medium was changed and cells were grown for additional 60 h. Low-molecular weight DNA was then isolated according to Hirt’s procedure (10), digested extensively with DpnI to eliminate unreplicated input plasmid DNA (38), and then analyzed by Southern blotting with a neo probe. As shown in Fig. 4, a Rep68-dose-dependent increase in the amount of rescued and replicated DNA was observed in both HeLa (Fig. 4A) and HepG2 cells (Fig. 4B). The signal corresponding to the rescued monomer was clearly detected with 4 μg of Rep68 protein (Fig. 4, lanes 6) and reached a plateau when 16 μg of the protein was lipofected (Fig. 4, lanes 8). As expected, no signal was detected in the absence of Rep (Fig. 4, lanes 2), while strong rescue-replication occurred in cells transfected with the Rep68 expression vector (Fig. 4, lane 1). Similar results were obtained with 293 cells (data not shown).
FIG. 4.
Southern blot analysis of rescue-replication from plasmid ITR/Hook-Neo in Ad-infected cells lipofected with Rep68 protein. Rescue-replication assays were performed in HeLa (A) and HepG2 (B) cells. In both cases, cells were lipofected with 7.5 μg of plasmid ITR/Hook-Neo in combination with either an equivalent amount of pCMVrep68 expression vector (lanes 1) or increasing concentrations (0.5, 1, 2, 4, 8, 16, and 32 μg) of Rep68 protein (lanes 3, 4, 5, 6, 7, 8, and 9, respectively). As a control, DNA was extracted from cells transfected only with the ITR/Hook-Neo plasmid (lanes 2). Equivalent amounts of low-Mr DNA samples isolated at 68 h posttransfection were digested with DpnI, electrophoresed on agarose gels, and analyzed on Southern blots with a neo probe. The arrows on the left indicate the rescued monomer; dimeric forms were also observed after longer exposure (data not shown). Molecular sizes are shown in kilobases.
In subsequent experiments we used a nested-set PCR assay to monitor the capability of recombinant Rep68 to promote site-specific integration. Sixteen micrograms of Rep was lipofected along with 7.5 μg of ITR/Hook-Neo into HeLa cells as described above. In control experiments, cells were lipofected with 7.5 μg of plasmid ITR/Hook-Neo alone or in combination with 7.5 μg of the Rep68 expression vector pCMVrep68. After 48 h, cells were collected, and genomic DNA was extracted and used as a template for a PCR using primers designed to flank the ITR-AAVS1 junction (a scheme of the assay is shown in Fig. 5C). A first amplification was carried out with one ITR-specific primer (p1, 5′-GTAGCATGGCGGGTTAATCA-3′) and one AAVS1-specific primer (p2, 5′-GCGCGCAGAAGCCAGTAGAGC-3′) by using AmpliTaq Gold (Perkin-Elmer) polymerase according to the manufacturer’s instructions in a total volume of 50 μl. The reaction proceeded for 25 cycles (1 min at 94°C, 1 min at 60°C, and 2 min at 72°C). Ten microliters of the amplification product was then used as a template for a second round of PCR, performed exactly like the first one but using the ITR-specific primer p3 (5′-TTAACTACAAGGAACCCCTAGTGATGG-3′) and the AAVS1-specific primer p4 (5′-GATAGACCAGACCTGAGCTATGGGAG-3′). The amplification product from the second round was run on an agarose gel, blotted, and hybridized with AAVS1-derived as well as ITR-derived probes; signals detected with both probes were considered to be derived from specific amplification of ITR-AAVS1 junctions and therefore were scored as true integration events. Figure 5 shows that no signal was detected in untransfected cells (Fig. 5A and B, lanes 4) or those transfected with the ITR/Hook-Neo plasmid alone (Fig. 5A and B, lanes 2); in contrast, positive signals, i.e., site-specific integration events, were observed in cells lipofected with ITR/Hook-Neo and the Rep68 protein (Fig. 5A and B, lanes 3). As expected, integration was also observed in cells transfected with the ITR-flanked cassette and the Rep68 expression vector (Fig. 5A and B, lanes 1). Similar results were obtained with 293 and HuH7 cells and when Rep78 expression vectors were used (data not shown). In cells transfected with the Rep expression vector, the positive signal appeared as a smear on the agarose gel, in line with the observation that site-specific integration can take place in a region spanning at least 500 bp of human chromosome 19, and therefore the amplified ITR-AAVS1 junctions are expected to be heterogeneous in size (38). The positive signal obtained with lipofected Rep68 had a reproducibly different pattern: in this case, one to three major bands (depending on the experiment) were observed in the context of a faint smear. This might indicate that the protein-mediated integration is clustered in a more limited number of sites within AAVS1; more likely, however, the result might reflect a lower intracellular level of Rep68 when the protein, instead of an expression vector, is delivered.
FIG. 5.
Lipofected Rep68 protein mediates site-specific integration into AAVS1. (A and B) Southern blot analysis of PCR amplification products generated from ITR-AAVS1 junctions. (A) Hybridization with the AAVS1 probe; (B) hybridization with the ITR probe. PCRs were carried out by using as a template the genomic DNA isolated from HeLa cells 48 h after lipofection with plasmid ITR/Hook-Neo alone (lanes 2) or in combination with the pCMVrep68 expression vector (lanes 1) or with Rep68 protein (lanes 3). Control reactions were performed with DNA extracted from untransfected cells (lanes 4). Amplification products were blotted onto nylon membranes; the same filter was probed first with a 32P-labelled AAVS1-specific probe (spanning nucleotides 210 to 1140 of the published AAVS1 sequence [17]) and then, after stripping, with an ITR-specific probe, excised as an MscI-PvuII fragment from plasmid pSub201 (28). (C) Sequence analysis of ITR-AAVS1 junctions. At the top is a schematic representation of the PCR assay. The sequence of one strand of the ITRs in plasmid ITR/Hook-Neo, in the “flop” orientation (3, 27), is shown; the nucleotide numbering is relative to the right end of the AAV genome (33). Letters (D, A, B, B′, C, C′, and A′) indicate palindromic sequences. Junctions obtained with the Rep68 proteins (r68-1 through -4) and with the pCMVrep68 expression vector (p68-1 through -6) are shown below the PCR assay representation. The numbers of the last evident cellular and viral nucleotides are given. AAVS1 breakpoints are based on the published AAVS1 sequence (17). Overlapping sequences between the ITR and AAVS1 are underlined. Insertions between the ITR and AAVS1 breakpoints are boldfaced.
We also cloned and sequenced those PCR products which not only cohybridized with AAVS1 and ITR probes but also were detected by direct ethidium bromide staining of the gels. Three junctions generated in HeLa cells lipofected with the integration cassette and recombinant Rep68 were sequenced; they are shown in Fig. 5C. The insertion of the ITR-flanked cassette occurred within a 16-bp region of the AAVS1 site mapping at nucleotides 1016 to 1032 of the published AAVS1 sequence (17), while the ITR breakpoints occurred in the A (junctions r68-2 and -3) and B (junction r68-1) regions of the terminal repeat. Six junctions generated in cells lipofected with the integration cassette and the Rep68 expression vector were also sequenced (junctions p68-1 through -6 [Fig. 5C]). In this case, the breakpoints in chromosome 19 were located in a larger region spanning about 500 bp, from nucleotide 842 (junction p68-2) to nucleotide 1295 (junction p68-1) of the AAVS1 sequence. It is worth noting that the location of all these junction breakpoints closely resembles that of the virus-cell junctions characterized in cells latently infected with AAV (29, 39). As for all the AAV-cell junctions analyzed so far, in this case, too, a complete ITR was never detected. In addition, the finding of two junctions, p68-5 and p68-6, containing the ITR in the “flip” orientation, the opposite of the “flop” orientation of the ITRs present in the transfected plasmid (Fig. 5C), strongly supports the hypothesis that plasmid integration, like the integration of wild-type AAV, proceeds through intermediate steps of replication of the plasmid template (22, 33).
The expression time of transgenes delivered via nonviral particles such as liposomes might be prolonged by promoting their integration into the host genome. In addition, the use of AAV elements to specifically target the AAVS1 site in human chromosome 19 should minimize the risk of insertional mutagenesis. In relation to this point, the observation that liposome-delivered Rep68 protein promotes the site-specific integration of a codelivered ITR-flanked cassette is of great interest. In fact, lipofection of the Rep68 protein, instead of Rep78 or Rep68 expression vectors, might temporally limit the activity of the protein until its degradation takes place; this could prevent undesired effects, such as the described rearrangements at the AAVS1 site following the integration event (1, 33, 39), possibly a consequence of prolonged expression of the Rep proteins. Utilizing recombinant Rep68 would also eliminate the potential problem of the integration of the rep gene, which has been reported to take place (34). In addition, lipofection of Rep68 protein might increase the specificity of integration; in fact, the assembling of liposomes containing preformed complexes between Rep68 and AAV ITRs could prevent the random insertion of the integration cassette (39), which, when a Rep78 or Rep68 expression vectors is used, would take place until a sufficient quantity of Rep78 or Rep68 is produced. In this connection, it is worth noting that in AAV particles, the Rep78 protein is covalently linked to the packaged genome and that, following infection of cells, a significant portion of the viral single-stranded DNA remains attached to Rep78 after nuclear translocation (25, 26).
In conclusion, the demonstration that AAV Rep68 protein is functional when lipofected into cells represents a first step toward the construction of nonviral particles able to promote site-specific integration of the desired transgene. Future work will be devoted to optimizing this system and further validating the utility of this approach.
Acknowledgments
We thank Gennaro Ciliberto and Nicola La Monica for critically reading the manuscript. We also gratefully acknowledge Janet Clench for editing the manuscript and M. Emili for graphics work.
REFERENCES
- 1.Balagué C, Kalla M, Zhang W-W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Linden R M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 4.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshnukh H M, Huang L. Liposome and polylysine mediated gene transfer. New J Chem. 1997;21:113–124. [Google Scholar]
- 6.Flotte T R, Carter B J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 7.Gao X, Huang L. Cytoplasmic expression of a reporter gene by co-delivery of T7 RNA polymerase and T7 promoter sequence with cationic liposomes. Nucleic Acids Res. 1993;21:2867–2872. doi: 10.1093/nar/21.12.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X, Huang L. Cationic liposome-mediated gene transfer. Gene Ther. 1995;2:710–722. [PubMed] [Google Scholar]
- 9.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 11.Im D-S, Muzyczka N. The AAV origin-binding protein Rep68 is an ATP dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 12.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep68, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneda Y, Iwai K, Uchida T. Increased expression of DNA cointroduced with nuclear protein in adult liver. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- 14.Kato K, Nakanishi M, Kaneda Y, Uchida T, Okada Y. Expression of hepatitis B virus surface antigen in adult rat liver: cointroduction of DNA and nuclear protein by a simplified liposome method. J Biol Chem. 1993;266:3361–3364. [PubMed] [Google Scholar]
- 15.Kearns W G, Afione S A, Fulmer S B, Pang M G, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated (AAV-CTFR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 16.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, Laughlin S M, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasic D D, Templeton N S. Liposomes in gene therapy. Adv Drug Delivery Rev. 1996;20:221–266. [Google Scholar]
- 19.Ledley F D. Nonviral gene therapy: the promise of genes as pharmaceutical products. Hum Gene Ther. 1995;6:1129–1144. doi: 10.1089/hum.1995.6.9-1129. [DOI] [PubMed] [Google Scholar]
- 20.Lee K-D, Oh Y K, Portnoy D A, Swanson J A. Delivery of macromolecules into cytosol using liposomes containing hemolysin from Listeria monocytogenes. J Biol Chem. 1996;271:7249–7252. [PubMed] [Google Scholar]
- 21.Linden R M, Berns K I. Site-specific integration by adeno-associated virus: a basis for a potential gene-therapy vector. Gene Ther. 1997;4:4–5. doi: 10.1038/sj.gt.3300357. [DOI] [PubMed] [Google Scholar]
- 22.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nabel G J, Nabel E G, Yang Z-H, Fox B A, Plautz G E, Gao X, Huang L, Shu S, Gordon D, Chang A E. Direct gene transfer with DNA-liposome complexes in melanoma: expression, biological activity and lack of toxicity in humans. Proc Natl Acad Sci USA. 1993;90:11307–11311. doi: 10.1073/pnas.90.23.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad K M R, Trempe J P. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology. 1995;214:360–370. doi: 10.1006/viro.1995.0045. [DOI] [PubMed] [Google Scholar]
- 26.Prasad K M R, Zhou C, Trempe J P. Characterization of the Rep78/adeno-associated virus complex. Virology. 1997;229:183–192. doi: 10.1006/viro.1996.8431. [DOI] [PubMed] [Google Scholar]
- 27.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genet Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 28.Samulski R J, Chang L S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study in vitro replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. . (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sells M A, Li J, Chernoff J. Delivery of protein into cells using polycationic liposomes. BioTechniques. 1995;19:72–78. [PubMed] [Google Scholar]
- 31.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycine resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 32.Snyder R O, Im D-S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Templeton N S, Lasic D D, Frederik P M, Strey H H, Roberts D D, Pavlakis G N. Improved DNA:liposome complexes for increased systemic delivery and gene expression. Nature Biotechnol. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 36.Toniatti C, Demartis A, Monaci P, Nicosia A, Ciliberto G. Synergistic transactivation of the human C-reactive protein promoter by transcription factor HNF-1 binding at two distinct sites. EMBO J. 1990;9:4467–4475. doi: 10.1002/j.1460-2075.1990.tb07897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus SV40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoemitsu Y, Kaneda Y, Muraishi A, Yoshizumi T, Sugimachi K, Sueishi K. HVJ (Sendai virus)-cationic liposomes: a novel and potentially effective liposome-mediated technique for gene transfer to the airway epithelium. Gene Ther. 1997;4:631–638. doi: 10.1038/sj.gt.3300463. [DOI] [PubMed] [Google Scholar]