Abstract
Previous studies have shown that single amino acid changes in the amino-terminal matrix (MA) domain, p17, of the human immunodeficiency virus type 1 Gag precursor Pr55, can abrogate virion particle assembly. In the three-dimensional structure of MA such mutations lie in a single helix spanning residues 54 to 68, suggesting a key role for this helix in the assembly process. The fundamental nature of this involvement, however, remains poorly understood. In the present study, the essential features of the MA helix required for virus assembly have been investigated through the analysis of a further 15 site-directed mutants. With previous mutants that failed to assemble, residues mapped as critical for assembly were all located on the hydrophobic face of the helix and had a key role in stabilizing the trimeric interface. This implies a role for the MA trimer in virus assembly. We support this interpretation by showing that purified MA is trimeric in solution and that mutations that prevent virus assembly also prevent trimerization. Trimerization in solution was also a property of a larger MA-capsid (CA) Gag molecule, while under the same conditions CA only was a monomer. These data suggest that Gag trimerization driven by the MA domain is an intermediate stage in normal virion assembly and that it relies, in turn, on an MA conformation dependent on the hydrophobic core of the molecule.
The matrix protein (MA) of human immunodeficiency virus (HIV), p17, has been ascribed a number of biological functions. In the late stage of the viral life cycle, as part of the Pr55 Gag precursor, MA contributes sequences that are necessary for targeting the Gag precursor to the plasma membrane (3, 31, 39, 42) and for the incorporation, by the assembling virion, of the major surface glycoprotein gp160 (8, 11, 37, 38). Some studies have suggested that MA is dispensable for a form of virus assembly (10, 21, 36), but a larger number of studies suggest a role for MA in normal virus assembly (5, 6, 12, 28, 35). One study has reported that simian immunodeficiency virus (SIV) MA alone is capable of the formation of virus-like particles (VLP) (15). Together, these findings indicate that while there are powerful assembly signals in other regions of Gag, MA appears to have a key role in the process of authentic virus assembly. Aside from mutations in the amino terminus of MA, some of which prevent myristylation, the region of MA found to play a role during the normal assembly process has been localized by site-directed mutagenesis studies to a discrete region between amino acids 54 and 68 (7, 12, 14, 28). Moreover, peptide inhibition of virion assembly has been observed with peptides derived from an overlapping sequence (5, 30). The importance of this region in MA is also highlighted by the conservation of amino acid sequence between residues 54 and 70 among HIV type 1 (HIV-1), HIV-2, and SIV (14). The recently reported crystal structures of HIV and SIV MAs reveal the molecule to be a trimer (16, 33) in which residues 54 to 68 form a discrete alpha-helix (helix 4) suggested to provide an essential spar within the molecule, precisely spacing the residues involved in trimer contact (33). Alteration of the conformation of helix 4 could, therefore, be the molecular basis by which the mutations in this region that prevent assembly, at Gly56, Cys57, Leu64, and Ile60 (5, 12, 14, 28), exert their phenotype. However, while the MA trimer is present in the crystal structure and supports the finding of a threefold axis of symmetry in the structure of Gag within the budding virus (29), it has yet to be observed in solution (25), preventing a direct test of the hypothesis that it forms an essential assembly intermediate. Here, to address these issues, we extend our previous study (28) to examine the ability of a further 12 single-residue and 3 double-residue mutations of helix 4 to produce Gag VLP and identify only one face of helix 4 as critical for assembly. We also show that following expression and purification of soluble forms of Gag, wild-type MA and a larger MA-capsid (CA) protein sediment as trimers in solution, a property not shared with those mutants that fail to assemble (referred to hereafter in this work as assembly-negative mutants). These data provide a possible explanation for the role of the MA domain within Pr55 during virus assembly.
Expression of helix 4 mutants and VLP formation.
To identify the features of helix 4 that were essential for assembly, 11 single-amino acid, and 3 double-amino acid changes as indicated in Table 1 were introduced into the HIV-1 MA in the context of the Gag precursor Pr55. Mutant gag genes were cloned into the baculovirus expression vector pAcCL29-1 (22) for expression of Gag VLP in recombinant baculoviruses (2, 13, 19). In addition, each mutant MA domain was rescued from the Pr55 precursor by PCR and cloned into the Escherichia coli expression vector pGEX2T (34) for the expression and purification of soluble MA antigen as described previously (28). The ability of each mutation to support VLP formation was assayed by (i) detection of particulate antigen in the supernatant of infected Spodoptera frugiperda (Sf9) cells at 36 to 48 h postinfection by sucrose gradient fractionation and Western blotting and (ii) direct visualization of VLP formation by electron microscopy (EM) of thin sections of infected cells as described elsewhere (17, 19, 40). The ability of mutant MA antigen to oligomerize with wild-type MA was assayed by protein overlay blotting (18) as modified by Morikawa et al. (28). Finally, the ability of each mutant to incorporate Env antigen was assayed after coinfection of each Gag mutant virus with a recombinant baculovirus expressing HIV-1 gp160 (27, 37) followed by sucrose density fractionation of the VLP and Western blotting for both Gag and Env antigens. Of all the mutations, only the double mutation Leu-to-Ala at position 61/68 (Leu61Ala/Leu68Ala) prevented Gag VLP formation, a phenotype that was matched by an inability of the isolated MA domain to interact with the wild-type MA protein in vitro (Table 1). All mutations that allowed VLP development also incorporated the envelope glycoprotein gp160 in keeping with the direct mapping of residues concerned with Env incorporation to a more amino-terminal region of MA (11, 39).
TABLE 1.
Analysis of mutations in the 54 to 68 region of HIV MAa
| Mutation | Gag VLP | MA-MA interaction |
|---|---|---|
| S54A | + | + |
| E55A | + | + |
| C57Sb | − | − |
| R58A | + | + |
| Q59A | + | + |
| I60A | + | + |
| L61A | + | + |
| Q63A | + | + |
| L64Ab | − | − |
| Q65A | + | + |
| P66A | + | + |
| S67A | + | + |
| L68A | + | + |
| R58A/Q65A | + | + |
| Q59A/P66A | + | + |
| L61A/L68A | − | − |
| Y79A | − | ND |
Mutations were analyzed for properties of Gag VLP formation and the ability to incorporate Env antigen and MA oligomerization. We found no intermediate results in these assays and no evidence by EM for altered structures associated with any of the particle-competent mutants characterized. +, positive result; −, negative result; ND, not done.
Data from Morikawa et al. (28).
A predicted distal mutation also affects VLP assembly.
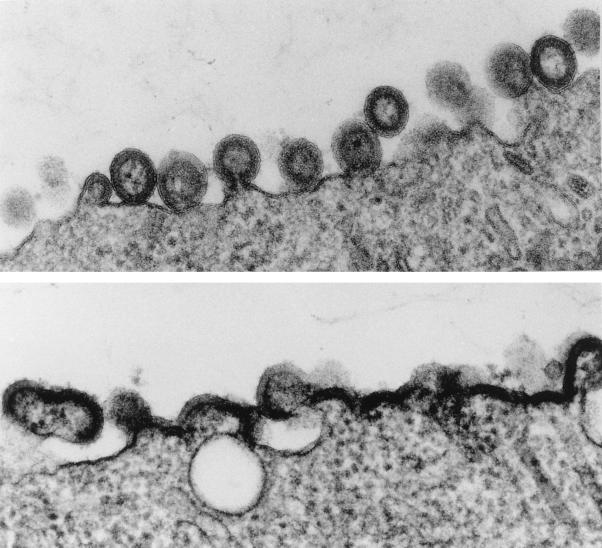
Helix 4 forms one boundary of a 5-Å-radius hydrophobic core within MA centered on Ile60 (33), and the cumulative mutants identified to date as essential for assembly were all hydrophobic in character and oriented toward the MA interior. To test if other residues marking out the hydrophobic core of MA may be required for assembly, we identified in the structure a key tyrosine residue, Tyr79, present on the long central helix 5, whose side chain interfaced with those hydrophobic residues identified by our mutational studies as essential for assembly. Accordingly, Tyr79 was mutated to Ala and the effect on VLP formation was assessed as before. Generally, residues in helix 5 have little or no effect on virus assembly (12), but in our analysis mutation of Tyr79 led to near abolition of antigen release into the media and to gross deformation of particle morphology at the surface of the expressing cells (Fig. 1). Thus, the hydrophobic core of MA between helices 4 and 5 generated by the residues Cys57, Leu61, Leu64, Leu68, and Tyr79 appeared to be a crucial component of the MA conformation required for virus assembly.
FIG. 1.
Thin sections of Sf9 cells expressing wild-type Pr55 (top panel) or Pr55 bearing the Tyr79Ala change in the MA domain (bottom panel). Compared to the wild type, Tyr79Ala produced deformed, only partly budded particles and an accumulation of Gag antigen at the membrane typical of other virus assembly mutants previously described (reference 17 and references therein).
Purified Gag antigen is trimeric in solution.
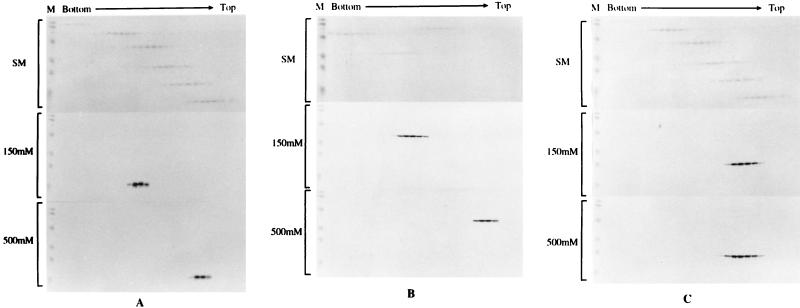
A role for MA in virus assembly based upon its capacity to oligomerize was suggested by the discovery of the MA trimer in the crystal structure and by EM observation of plasma membrane-associated Gag assemblies (29). However, although plausible, there has been no independent evidence that MA exists as a trimer in solution or that the trimer plays an essential role in normal virus assembly, and the solution structures of MA reported have been monomeric (23, 25). In addition, the determined structures of isolated MA may not reflect the form of MA that exists within the Pr55 precursor, in which context all assembly mutations to date have been mapped. In order to readdress the question of the role of MA in the Gag oligomeric form, the coding regions for HIV-1 MA, CA, and MA-CA were cloned into the expression vector pTrcHisA (Invitrogen Corp.) and protein representing each of these Gag domains was produced as a histidine-tagged fusion protein in E. coli. Fusion proteins were purified under conditions of either low (150 mM NaCl) or high (500 mM NaCl) salt and then subjected to sedimentation analysis on 15 to 30% glycerol gradients. Gag antigen was detected by gel and Western blotting and compared to molecular mass markers sedimented in parallel. Under these conditions and at a physiological salt concentration, MA and MA-CA sedimented at calculated molecular masses of 51 and 120 kDa, respectively, the equivalent of a trimeric form of each antigen. Purified CA by contrast sedimented at 25 kDa, the size of the monomer. When Gag antigens were purified under high-salt conditions, all antigens migrated at molecular masses equivalent to the monomeric form (Fig. 2). These data suggest that (i) the MA-CA Gag molecule is a trimer in solution, (ii) trimerization is dependent on the MA domain, not CA, and (iii) the trimeric association of MA and MA-CA is dependent on ionic strength.
FIG. 2.
Sedimentation profiles of purified Gag antigens on glycerol gradients. Purified soluble Gag antigens were sedimented through 15 to 30% glycerol gradients made in 20 mM Tris (pH 7.4)–100 mM NaCl–1 mM dithiothreitol–0.5 mM EDTA at 48,000 rpm in an SW50 rotor and 4°C for 40 h (for analysis of MA and CA) or for 27 h (for analysis of MA-CA). Molecular mass markers in the gradients were provided by high (analysis of MA-CA)- or low (analysis of MA and CA)-molecular-mass calibration kits (Pharmacia), are shown in the upper section of each panel in all cases, and are marked SM (sedimentation markers). The high-molecular-mass range consisted of the following: catalase, 4 × 58 kDa = 232 kDa; lactate dehydrogenase, 4 × 36 kDa = 140 kDa; and serum albumin, 67 kDa. The low-molecular-mass range consisted of the following: phosphorylase b, 94 kDa; serum albumin, 67 kDa; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa; and a-lactalbumin, 14.4 kDa. Gel markers (M) are prestained molecular mass markers (Bio-Rad). Gradients were fractionated from the bottom, and Gag antigen was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. The middle and bottom sections of each panel show an analysis of antigen prepared under low- and high-salt conditions, respectively. (A) MA protein; (B) MA-CA protein; (C) CA protein.
Purified Gag antigen encoding assembly-negative mutants is monomeric.
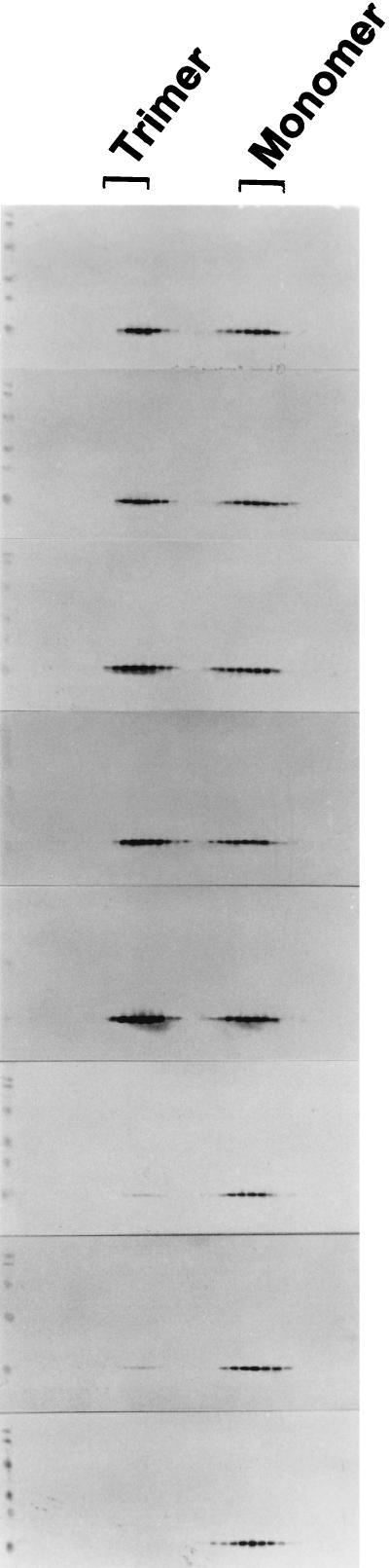
To establish that the finding that Gag antigen is trimeric in solution is relevant to assembly, MA from several of the mutants analyzed in Table 1 was similarly prepared and analyzed under low-salt conditions under which the trimer was normally observed. In these experiments the MAs derived from the assembly-competent mutations Arg58Ala, Gln59Ala, Gln63Ala, Pro66Ala, and Gln59Ala/Pro66Ala showed a trimer and monomer profile, suggesting that the encoded MA domains were competent for trimerization, although the trimer/monomer ratio was reduced compared to that of the wild type. This indicates the presence of a fraction of unassembled Gag, although it is insufficient to prevent the formation of VLP. In the assembly-defective MAs Cys57Ser, Leu64Ala, and Leu61Ala/Leu68Ala mutants, the Gag sedimentation profile was shifted essentially to that of a monomer (Fig. 3), providing a direct link between the failure of the MA domain to undergo trimerization and the ability to form VLP.
FIG. 3.
Sedimentation analysis of assembly-defective and -competent Gag mutant MAs. The MA domain of several Gag mutants was rescued by PCR for expression as a purified MA domain by using pGEX expression as described previously (28). The antigen was prepared under conditions of low salt throughout and analyzed by glycerol velocity gradients under the same conditions described in the legend to Fig. 2. The panels show the analysis of MA antigen prepared from (top to bottom) Arg58Ala, Gln59Ala, Gln63Ala, Pro66Ala, Gln59Ala/Pro66Ala, Cys57Ser, Leu64Ala, and Leu61Ala/Leu68Ala mutants. Prestained markers (Bio-Rad) are visible in the leftmost lane of each gel. A trace of trimer (less than 5% by gel scan) was present in panels containing Cys57Ser and Leu64Ala, but none was visible in Leu61Ala/Leu68Ala mutants.
Our mutagenesis studies confirm and extend previous data indicating that the hydrophobic residues of helix 4 (Cys57, Leu61, Leu64, and Leu68) are important for particle assembly (12, 14, 28). In this study, however, single-point mutations to alanine at hydrophobic residues Ile60, Pro66, and Leu68 did not prevent particle assembly. Pro66Ala combined with Gln59Ala also failed to prevent VLP assembly (Table 1). The discrepancy between our results at Ile60 (Ile60Ala) and those of Freed et al. (Ile60Glu [12]) is most likely attributable to the nature of the change made. The double mutation Leu61Ala/Leu68Ala abolished MA oligomerization and VLP assembly and gave a phenotype reminiscent of those of the single mutations Cys57Ser and Leu64Ala reported by us previously (28). When considered together, these mutations are distinguished from those with no effect by virtue of their periodicity (C57-XXX-L61-XX-L64-XXX-L68), consistent with their alignment on one side of helix 4, interfacing with helix 5. That Tyr79Ala, whose side group interfaces helix 4, also abolished VLP assembly, provides an argument that it is selective rather than general hydrophobicity in the core of MA, between helices 4 and 5, that is critical for the conformation necessary for assembly. This interpretation would be consistent with the recent finding that the mutation Cys57Ser causes an altered nuclear magnetic resonance spectrum compared with that of the wild-type molecule (5).
It has been proposed that the conformation of the 310 alpha-helix between residues 66 and 70 of MA is related directly to the process of assembly (24). We suggest that the hydrophobic interface between helices 4 and 5 influences this conformation to provide an indirect link with assembly. A corollary of this interpretation is that VLP-negative mutants should demonstrably fail to trimerize MA. In a sedimentation assay, purified MA-CA existed as a trimer under low-salt concentrations, a property shared by MA analyzed in the same way but not by CA, which sedimented as a monomer in solution. Critically, five assembly-proficient MA mutants showed trimerization-competent profiles similar to those of the wild type, while three nonassembly mutants of MA sedimented essentially as monomers. Wild-type MA trimerization was dependent on salt concentration, providing experimental support for the crystallographic finding that the MA monomer-monomer interactions are weak (33). The finding that CA is a monomer at the protein concentrations used here (∼1 mg/ml) agrees with earlier reports that CA is a monomer at a protein concentration of ∼1 mg/ml but may become dimeric at concentrations of ∼10 mg/ml and higher (9, 26). Similarly, purified CA-nucleocapsid (NC) at concentrations of ∼1 mg/ml has been shown to assemble into higher-order structures (4), including structures reminiscent of partially assembled Gag shells (17). Taken together with our data, this suggests that these domains play a major role in the trimer-trimer interactions necessary to give rise to the higher orders of Gag required for the assembly of the submembrane Gag shell and virus (29, 33).
A plausible order of events might be the assembly of Gag trimers via interactions in the MA domain at low concentrations of protein followed by the higher-order multimerization of Gag via interactions in the CA or CA-NC domain(s) at the high concentrations of protein that would occur when Gag antigen accumulates at the plasma membrane. Direct evidence for a role of CA-NC, in particular the junction p2 peptide, in the overall conformation of Gag (e.g., oligomerization) has been demonstrated by the altered rates of Pr55 cleavage by protease and the reduction of infectious particles when p2 is deleted (32). Deletion of p2 or mutation of the cleavage site between CA and NC also causes a concomitant gross alteration in particle morphology (1, 20). These data are consistent with the effect CA-NC has on the higher order of Gag assembly, although a role for the NC domain itself in the complete process is not ruled out (41).
Acknowledgments
We thank the AIDS reagent repository (Harvey Holmes) for the provision of a number of enabling reagents and David Stuart, ZiHe Rao, and Elizabeth Fry at the Department of Molecular Biophysics, University of Oxford, United Kingdom, for constructive discussions.
The present work was supported by grants from the Ministry of Health and Welfare of Japan and the Medical Research Council of the United Kingdom.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulanger P, Jones I M. Use of heterologous expression systems to study retroviral morphogenesis. Curr Top Microbiol Immunol. 1996;214:237–260. doi: 10.1007/978-3-642-80145-7_8. [DOI] [PubMed] [Google Scholar]
- 3.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon P M, Matthews S, Clark N, Byliss E D, Iourin O, Hockley D J, Kingsman S M, Kingsman A J. Structure-function studies of the human immunodeficiency virus type 1 matrix protein p17. J Virol. 1997;71:3474–3483. doi: 10.1128/jvi.71.5.3474-3483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chazal N, Carrière C, Gay B, Boulanger P. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J Virol. 1994;68:111–122. doi: 10.1128/jvi.68.1.111-122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chazal N, Gay B, Carrière C, Tournier J, Boulanger P. Human immunodeficiency virus type 1 MA deletion mutants expressed in baculovirus-infected cells: cis and trans effects on the Gag precursor assembly pathway. J Virol. 1995;69:365–375. doi: 10.1128/jvi.69.1.365-375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrlich L S, Agresta B E, Carter C A. Assembly of human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fäcke M, Janetzko A, Shoeman R L, Kräusslich H-G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix protein. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez S A, Affranchino J L. Mutational analysis of the conserved cysteine residues in the simian immunodeficiency virus matrix antigen. Virology. 1995;210:501–507. doi: 10.1006/viro.1995.1369. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez S A, Affranchino J L, Gelderblom H, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 16.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric HIV-1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 18.Homann H E, Willenbrink W, Buchholz C J, Neubert W J. Sendai virus protein-protein interactions studied by a protein-blotting protein overlay technique: mapping of domains on NP protein required for binding to P protein. J Virol. 1991;65:1304–1309. doi: 10.1128/jvi.65.3.1304-1309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jowett J B M, Hockley D J, Nermut M, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 20.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livingstone C, Jones I M. Baculovirus expression vectors with single-strand capability. Nucleic Acids Res. 1989;17:2366. doi: 10.1093/nar/17.6.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 24.Massiah M A, Worthylake D, Christensen A M, Sundquist W I, Hill C P, Summers M F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: evidence for conformational changes during viral assembly. Protein Sci. 1996;5:2391–2398. doi: 10.1002/pro.5560051202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, Kingsman S, Kingsman A, Campbell I. Structural similarity between the p17 matrix protein of HIV-1 and interferon gamma. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 26.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossman M G. Crystal structure of the dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 27.Moore J P, McKeating J A, Jones I M, Stephens P E, Clements G, Thomson S, Weiss R A. Characterisation of recombinant gp120 and gp160 from HIV-1: binding to monoclonal antibodies and soluble CD4. AIDS. 1990;4:307–315. doi: 10.1097/00002030-199004000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa Y, Kishi T, Zhang W-H, Nermut M V, Hockley D J, Jones I M. A molecular determinant of HIV particle assembly located in the matrix antigen p17. J Virol. 1995;69:4519–4523. doi: 10.1128/jvi.69.7.4519-4523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Fullerene-like organisation of HIV gag-protein shell in virus-like particles produced by recombinant baculoviruses. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 30.Niedrig M, Gelderblom H R, Pauli G, Marz J, Bickhard H, Wolf H, Modrow S. Inhibition of infectious human immunodeficiency virus type 1 particle formation by Gag protein-derived peptides. J Gen Virol. 1994;75:1469–1474. doi: 10.1099/0022-1317-75-6-1469. [DOI] [PubMed] [Google Scholar]
- 31.Pal R, Reitz M S, Jr, Tschachler E, Gallo R C, Sarngadharan M G, Veronese F D. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses. 1990;6:721–730. doi: 10.1089/aid.1990.6.721. [DOI] [PubMed] [Google Scholar]
- 32.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao Z, Belyeaev A S, Fry E, Roy P, Jones I M, Stuart D S. Crystal structure of simian immunodeficiency virus matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C-T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamshchikov G V, Ritter G D, Vey M, Compans R W. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214:50–58. doi: 10.1006/viro.1995.9955. [DOI] [PubMed] [Google Scholar]
- 38.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan X, Yu X, Lee T-H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]