Abstract
The order Eurotiales is diverse and includes species that impact our daily lives in many ways. In the past, its taxonomy was difficult due to morphological similarities, which made accurate identification of species difficult. This situation improved and stabilised with recent taxonomic and nomenclatural revisions that modernised Aspergillus, Penicillium and Talaromyces. This was mainly due to the availability of curated accepted species lists and the publication of comprehensive DNA sequence reference datasets. This has also led to a sharp increase in the number of new species described each year with the accepted species lists in turn also needing regular updates. The focus of this study was to review the 160 species described between the last list of accepted species published in 2020 until 31 December 2022. To review these species, single-gene phylogenies were constructed and GCPSR (Genealogical Concordance Phylogenetic Species Recognition) was applied. Multi-gene phylogenetic analyses were performed to further determine the relationships of the newly introduced species. As a result, we accepted 133 species (37 Aspergillus, two Paecilomyces, 59 Penicillium, two Rasamsonia, 32 Talaromyces and one Xerochrysium), synonymised 22, classified four as doubtful and created a new combination for Paraxerochrysium coryli, which is classified in Xerochrysium. This brings the number of accepted species to 453 for Aspergillus, 12 for Paecilomyces, 535 for Penicillium, 14 for Rasamsonia, 203 for Talaromyces and four for Xerochrysium. We accept the newly introduced section Tenues (in Talaromyces), and series Hainanici (in Aspergillus sect. Cavernicolarum) and Vascosobrinhoana (in Penicillium sect. Citrina). In addition, we validate the invalidly described species Aspergillus annui and A. saccharicola, and series Annuorum (in Aspergillus sect. Flavi), introduce a new combination for Dichlaena lentisci (type of the genus) and place it in a new section in Aspergillus subgenus Circumdati, provide an updated description for Rasamsonia oblata, and list excluded and recently synonymised species that were previously accepted. This study represents an important update of the accepted species lists in Eurotiales.
Taxonomic novelties: New sections: Aspergillus section Dichlaena Visagie, Kocsubé & Houbraken. New series: Aspergillus series Annuorum J.J. Silva, B.T. Iamanaka, Frisvad. New species: Aspergillus annui J.J. Silva, M.H.P. Fungaro, Frisvad, M.H. Taniwaki & B.T. Iamanaka; Aspergillus saccharicola J.J. Silva, Frisvad, M.H.P. Fungaro, M.H. Taniwaki & B.T. Iamanaka. New combinations: Aspergillus lentisci (Durieu & Mont.) Visagie, Malloch, L. Kriegsteiner, Samson & Houbraken; Xerochrysium coryli (Crous & Decock) Visagie & Houbraken.
Citation: Visagie CM, Yilmaz N, Kocsubé S, Frisvad JC, Hubka V, Samson RA, Houbraken J (2024). A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Studies in Mycology 107: 1–66. doi: 10.3114/sim.2024.107.01
Keywords: Accepted species list, Aspergillaceae, DNA barcodes, new taxa, nomenclature, Penicillaginaceae, phylogenetic species concept, Thermoascaceae, Trichocomaceae
INTRODUCTION
Eurotiales is one of the most diverse orders of fungi and includes genera such as Aspergillus, Penicillium, Paecilomyces and Talaromyces. Species identification in these speciose genera has been very difficult in the past. Recent taxonomic and nomenclatural studies have modernised the morphology-based classifications to the extent that these genera now have one of the most modern taxonomies of all fungi. The basis and main driving force for this are the so-called ‘accepted species lists’. Nomenclators such as MycoBank (https://www.mycobank.org/) list more than 3 000 names of Eurotiales. However, many of these names belong to other genera, were considered synonyms of accepted species, or remain unrecognisable because old descriptions were insufficient for recognition and/or no material is available. Knowledge of a genus at a given time was traditionally published in monographs that contained descriptions for all species and keys for their identification. Pitt (1980), in his monograph on Penicillium and its associated sexual (teleomorphic) genera Eupenicillium and Talaromyces, published a review of the recognised names and listed synonyms and excluded names that he considered indeterminate. This overview was later extended to the ‘Names in Current Use’ published by Pitt & Samson (1993) for Trichocomaceae and updated by Pitt et al. (2000). These lists were based on the morphological species concept used at the time, and did not include comments on taxonomy. The main aim of these lists was to record information on species that were ‘accepted’ in these genera, and the purpose was not to formally conserve or reject names as allowed for by the International Code of Nomenclature for algae, fungi, and plants (ICNafp; Turland et al. 2018). The authors of these lists were aware that taxonomy may change as species concepts evolve, and when new concepts were adopted, old names may be accepted in the future. This happened when Houbraken & Samson (2011) revised the taxonomy of Trichocomaceae and classified the species into three families: Aspergillaceae (Aspergillus, Hamigera, Leiothecium, Monascus, Penicilliopsis, Penicillium, Phialomyces, Sclerocleista, Warcupiella and Xeromyces), Thermoascaceae (Byssochlamys/Paecilomyces and Thermoascus) and Trichocomaceae (Rasamsonia, Sagenomella, Talaromyces, Thermomyces, and Trichocoma). They proposed the adoption of Aspergillus and Penicillium over their associated sexual genera, pre-empting the move to a single name nomenclature for fungi (McNeill et al. 2012), and reclassified several other sexual and asexual genera (e.g. Chromocleista, Eupenicillium, Eladia, Hemicarpenteles, Torulomyces and Thysanophora were considered synonymous with Penicillium). These changes were mainly based on phylogenetic analyses, which have become standard practise to study the relationships between and within these genera.
In the following years, accepted species lists for Aspergillus, Penicillium and Talaromyces were published (Samson et al. 2014, Visagie et al. 2014, Yilmaz et al. 2014), representing the first modern lists for fungi to incorporate DNA sequence data into decision-making. Several recommendations were made, from how to work with these genera or describe new species to the procedures required to identify strains more precisely, including the use of the recommended DNA barcode markers β-tubulin (BenA; for Penicillium and Talaromyces) or calmodulin (CaM; for Aspergillus). These recommendations were supported by the metadata associated with each name entry, including authority, citation, MycoBank number, type, ex-type, subgeneric classification and GenBank accession numbers for DNA sequences obtained from ex-type cultures. As with previous lists (Pitt & Samson 1993, Pitt et al. 2000), excluded names were not formally rejected. This approach, together with the wealth of reference data released, resulted in a strong backbone for these genera on which taxonomic revisions of specific groups could be built. It also facilitated the description of new species and their comparison with close relatives, leading to the description of many new species. For this reason, Houbraken et al. (2020) updated the accepted species lists of Aspergillus (increased from 339 to 446 species), Penicillium (increased from 354 to 483 species), Talaromyces (increased from 88 to 171 species) and expanded the list to include other Eurotiales (but excluding Elaphomycetaceae). Houbraken et al. (2020) also reintroduced a series classification in Aspergillus and Penicillium. This taxonomic rank provides information on what functional characters the species might have and is useful in phenotype-based identification. In addition, the current series classification makes it even easier than before to compare putative new species and their close relatives. Since 2020, 160 new species were described in Eurotiales. The focus of this study was to review these and provide comments and opinions on them.
MATERIALS AND METHODS
Phylogenetic analyses
Phylogenies were calculated for all species described since Houbraken et al. (2020), with datasets compiled to represent the genera, sections and/or series these belong to. Datasets (see Table 1 & Suppl. Table S1) were assembled using DNA reference sequences obtained from NCBIs GenBank nucleotide database (https://www.ncbi.nlm.nih.gov/genbank/) and included the internal transcribed spacer rDNA region (ITS), 28S large subunit (LSU), beta-tubulin (BenA), calmodulin (CaM), and RNA polymerase II second largest subunit (RPB2).
Table 1.
List of species described since Houbraken et al. (2020), and reviewed in the current study.
| Species | Strain | Status | Subgenus | Section | Series | Country | Substrate | ITS | BenA | CaM | RPB2 | LSU | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aspergillus agricola | A2-A | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987060 | - | - | Singh et al. (2020) | |
| BC09-F | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987065 | - | - | Singh et al. (2020) | ||
| C3-J | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987059 | - | - | Singh et al. (2020) | ||
| E13-L | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987064 | - | - | Singh et al. (2020) | ||
| EC37-C | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987066 | - | - | Singh et al. (2020) | ||
| J11-B | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987062 | - | - | Singh et al. (2020) | ||
| J11-C | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987063 | - | - | Singh et al. (2020) | ||
| J15-H | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987061 | - | - | Singh et al. (2020) | ||
| NRRL 66869 | T | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987053 | - | - | Singh et al. (2020) | |
| NRRL 66870 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987054 | - | - | Singh et al. (2020) | ||
| NRRL 66871 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987055 | - | - | Singh et al. (2020) | ||
| NRRL 66872 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987056 | - | - | Singh et al. (2020) | ||
| NRRL 66873 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987057 | - | - | Singh et al. (2020) | ||
| Sanpatong22 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987068 | - | - | Singh et al. (2020) | ||
| Sukhothai 19 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987067 | - | - | Singh et al. (2020) | ||
| TXA35-K | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987058 | - | - | Singh et al. (2020) | ||
| Ubon3 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987069 | - | - | Singh et al. (2020) | ||
| Aspergillus alboluteus | CBS 145854 | Circumdati | Flavipedes | Spelaei | USA | Air (indoor) | MW448664 | MW478498 | MW478512 | MW478533 | - | Sklenář et al. (2021) | |
| CBS 145855 | T | Circumdati | Flavipedes | Spelaei | USA | Air (outdoor) | MW448663 | MW478497 | MW478511 | MW478532 | - | Sklenář et al. (2021) | |
| CBS 145859 | Circumdati | Flavipedes | Spelaei | USA | Airconditioner | MW448662 | MW478496 | MW478510 | MW478531 | - | Sklenář et al. (2021) | ||
| CBS 147065 | Circumdati | Flavipedes | Spelaei | Nigeria | Unknown | MW448666 | MW478500 | MW478514 | MW478535 | - | Sklenář et al. (2021) | ||
| CCF 5849 | Circumdati | Flavipedes | Spelaei | USA | Storage room | MW448665 | MW478499 | MW478513 | MW478534 | - | Sklenář et al. (2021) | ||
| CMW 56637 | Circumdati | Flavipedes | Spelaei | Botswana | Soil (bat cave) | MW480881 | MW480789 | MW480707 | MW480791 | - | Sklenář et al. (2021) | ||
| Aspergillus alboviridis | CBS 142665 | T | Circumdati | Flavipedes | Spelaei | Spain | Dung (herbivor) | LT798909 | LT798936 | LT798937 | LT798938 | - | Sklenář et al. (2021) |
| Aspergillus annui | IBT 36122 | T | Circumdati | Flavi | Annuorum | Brazil | Sweet paprika | OP691228 | ON529842 | ON529841 | ON529843 | - | Silva et al. (2022) |
| IBT 36123 | Circumdati | Flavi | Annuorum | Brazil | Sweet paprika | - | ON643012 | ON643060 | ON642964 | - | Silva et al. (2022) | ||
| IBT 36124 | Circumdati | Flavi | Annuorum | Brazil | Sweet paprika | - | - | - | - | - | Silva et al. (2022) | ||
| Aspergillus arizonicus | CCF 5341 | T | Fumigati | Fumigati | Neoglabri | USA | Air (hospital) | OK322364 | OK334128 | OK334127 | OK334129 | - | Crous et al. (2021b) |
| Aspergillus banksianus | FRR 6047 | T | Fumigati | Fumigati | Brevipedes | Australia | Soil under Banksia integrifolia | MH280013 | MT 184780 | MT184786 | MT184792 | - | Crous et al. (2020b) |
| Aspergillus barbosae | CBS 145863 | T | Circumdati | Terrei | Terrei | Brazil | Indoor environment | LR536042 | LR031377 | LR031392 | LR031407 | - | Barbosa et al. (2018) |
| URM 5870 | Circumdati | Terrei | Terrei | Brazil | Industrial castor cake | LR536043 | LR031378 | LR031393 | LR031408 | - | Barbosa et al. (2018) | ||
| URM 7011 | Circumdati | Terrei | Terrei | Brazil | Soil | LR536041 | LR031376 | LR031391 | LR031406 | - | Barbosa et al. (2018) | ||
| Aspergillus burnettii | CBS 146237 | T | Circumdati | Flavi | Alliacei | Australia | Soil | MK429758 | MT211761 | MT211762 | MT211763 | - | Gilchrist et al. (2020) |
| Aspergillus curvatus | EMCCN2213 | T | Circumdati | Circumdati | Steyniorum | Egypt | Water (alkaline lake) | MN006961 | - | - | - | - | Al-Bedak (2020a) |
| Aspergillus gaarensis | AUMC 11046 | T | Circumdati | Circumdati | Steyniorum | Egypt | Soil (lake) | MN648408 | - | - | - | - | Al-Bedak (2020b) |
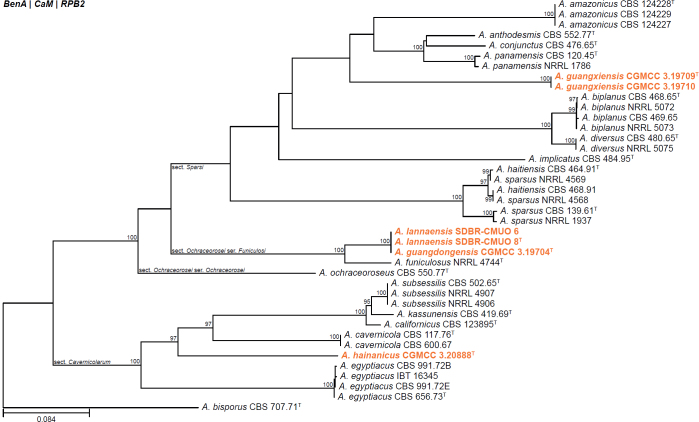
| Aspergillus guangdongensis | CGMCC 3.19704 | T | Nidulantes | Ochraceorosei | Funiculosi | China | Soil | MN640760 | MN635246 | MN635257 | MN635269 | - | Sun et al. (2022a) |
| Aspergillus guangxiensis | CGMCC 3.19709 | T | Nidulantes | Sparsi | Conjuncti | China | Soil | MN640765 | MN635251 | MN635262 | MN635274 | - | Sun et al. (2022a) |
| CGMCC 3.19710 | Nidulantes | Sparsi | Conjuncti | China | Soil | MN640766 | MN635252 | MN635263 | MN635275 | - | Sun et al. (2022a) | ||
| Aspergillus hainanicus | CGMCC 3.20888 | T | Nidulantes | Cavernicolarum | Hainanici | China | Soil | OM414846 | OM475626 | OM475630 | OM475634 | - | Wang & Zhuang (2022b) |
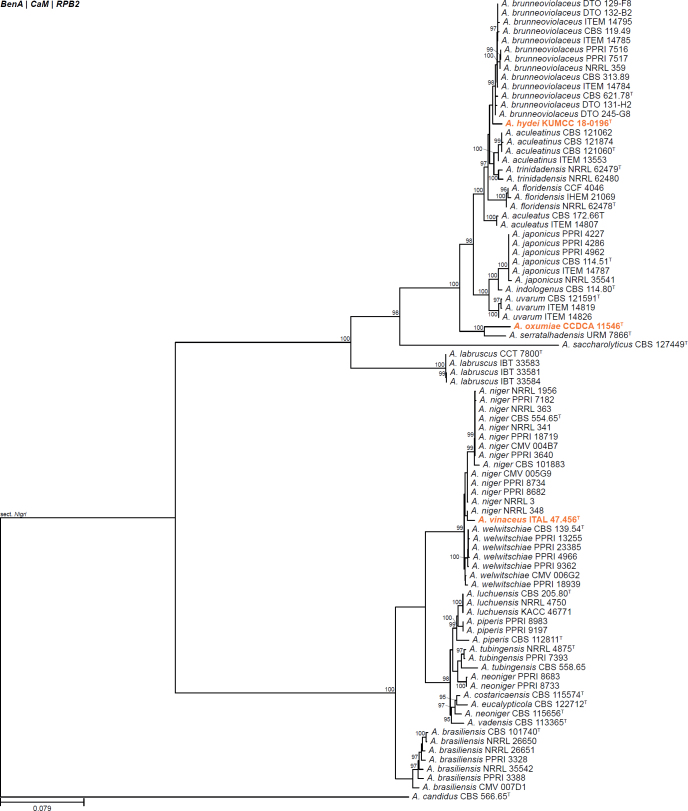
| Aspergillus hydei | KUMCC 18-0196 | T | Circumdati | Nigri | Japonici | China | Air (outdoor) | MT152332 | MT161679 | MT178247 | MT384370 | - | Doilom et al. (2020) |
| Aspergillus inusitatus | CBS 147044 | T | Circumdati | Flavipedes | Spelaei | Tunisia | Soil | MW448669 | MW478502 | MW478517 | MW478542 | - | Sklenář et al. (2021) |
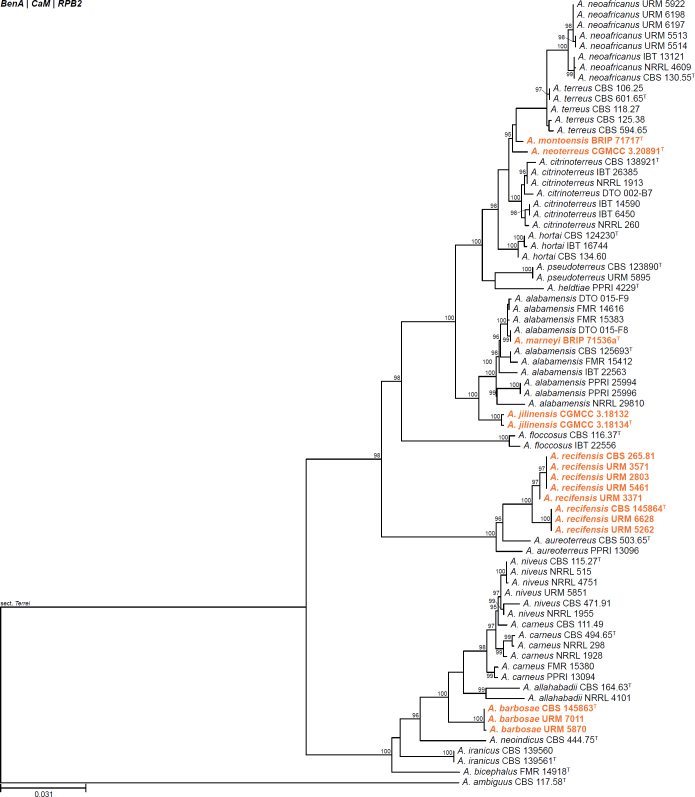
| Aspergillus jilinensis | CGMCC 3.18132 | Circumdati | Terrei | Terrei | China | Soil | KX443223 | KX443161 | KX443192 | - | - | Huang et al. (2020) | |
| CGMCC 3.18134 | T | Circumdati | Terrei | Terrei | China | Soil | KX443224 | KX443162 | KX443193 | - | - | Huang et al. (2020) | |
| Aspergillus kumbius | FRR 6049 | T | Circumdati | Circumdati | Sclerotiorum | Australia | Soil (pasture) | MT179307 | MT184782 | MT184788 | MT184794 | - | Crous et al. (2020b) |
| Aspergillus lannaensis | SDBR-CMUO 6 | Nidulantes | Ochraceorosei | Funiculosi | Thailand | Soil | - | MW219782 | MW219780 | MW219784 | - | Boonmee et al. (2021) | |
| SDBR-CMUO 8 | T | Nidulantes | Ochraceorosei | Funiculosi | Thailand | Soil | - | MW219783 | MW219781 | MW219785 | - | Boonmee et al. (2021) | |
| Aspergillus lanuginosus | NRRL4610 | T | Circumdati | Flavipedes | Spelaei | Haiti | Soil | EF669604 | EU014080 | EF669562 | EF669646 | - | Sklenář et al. (2021) |
| Aspergillus lebretii | URM 8450 | Cremei | Cremei | Wentiorum | Brazil | Air (outdoor) | ON862927 | OP672381 | OP290539 | OP290510 | - | Alves et al. (2022b) | |
| URM 8451 | T | Cremei | Cremei | Wentiorum | Brazil | Air (outdoor) | ON862928 | OP672382 | OP290540 | OP290511 | - | Alves et al. (2022b) | |
| Aspergillus lentisci | CBS 150189 | T | Circumdati | Dichlaena | - | Portugal | Pistacia leaf | OR142402 | OR145977 | OR145992 | OR146003 | OR142413 | Present study |
| DTO 426-F1 | Circumdati | Dichlaena | - | Portugal | Pistacia leaf | OR142405 | OR145976 | OR145991 | OR146002 | OR142414 | Present study | ||
| DTO 426-F2 | Circumdati | Dichlaena | - | Portugal | Pistacia leaf | OR142404 | OR145975 | OR145990 | OR146001 | OR142415 | Present study | ||
| DTO 426-F3 | Circumdati | Dichlaena | - | Portugal | Pistacia leaf | OR142409 | OR145978 | OR145993 | OR146004 | OR142416 | Present study | ||
| Aspergillus limoniformis | CGMCC 3.19323 | T | Polypaecilum | Polypaecilum | Canini | China | Bat guano | MK329066 | MK336093 | - | MK335972 | - | Zhang et al. (2020) |
| LC12610 | Polypaecilum | Polypaecilum | Canini | China | Bat guano | MK329067 | MK336094 | - | MK335973 | - | Zhang et al. (2020) | ||
| Aspergillus luteorubrus | CBS 146723 | T | Fumigati | Fumigati | Fennelliarum | Australia | Soil | MT179305 | MT 184781 | MT184787 | MT184793 | - | Crous et al. (2020b) |
| Aspergillus magnus | UAMH 1324 | T | Circumdati | Candidi | Candidi | Canada | Mouse | ON156376 | ON164570 | ON164619 | ON164517 | - | Glässnerová et al. (2022) |
| Aspergillus malvicolor | CBS 146724 | T | Circumdati | Circumdati | Sclerotiorum | Australia | Soil under Arachis hypogaea | MT179308 | MT184784 | MT184790 | MT184796 | - | Crous et al. (2020b) |
| Aspergillus marneyi | BRIP 71536a | T | Circumdati | Terrei | Terrei | Australia | Crown of Medicago sativa | OL691080 | OL741659 | - | OL741656 | - | Tan & Shivas (2022) |
| Aspergillus montoensis | BRIP 71717 | T | Circumdati | Terrei | Terrei | Australia | Root of Vigna radiata | OK441076 | OK533535 | - | OK509073 | - | Tan et al. (2021) |
| Aspergillus nanangensis | CBS 146238 | T | Circumdati | Janorum | Janorum | Australia | Soil | MK979278 | MT184783 | MT184789 | MT184795 | - | Crous et al. (2020b) |
| Aspergillus neoterreus | CGMCC 3.20891 | T | Circumdati | Terrei | Terrei | China | Soil | OM414849 | OM475629 | OM475633 | OM475637 | - | Wang & Zhuang (2022b) |
| Aspergillus neotritici | CBS 129260 | Circumdati | Candidi | Candidi | USA | Soil | ON156397 | ON164591 | ON164632 | ON164541 | - | Glässnerová et al. (2022) | |
| CBS 129307 | Circumdati | Candidi | Candidi | Unknown | Soil | ON156398 | ON164592 | ON164633 | ON164542 | - | Glässnerová et al. (2022) | ||
| CBS 133055 | Circumdati | Candidi | Candidi | Japan | Unknown | ON156395 | ON164587 | ON164628 | ON164537 | - | Glässnerová et al. (2022) | ||
| CBS 266.81 | Circumdati | Candidi | Candidi | India | Triticum aestivum grains | LT626958 | EU076293 | EU076305 | MN969098 | - | Glässnerová et al. (2022) | ||
| CCF 1649 | Circumdati | Candidi | Candidi | Czech Republic | Flour | FR733810 | LT627024 | FR751427 | LT627025 | - | Glässnerová et al. (2022) | ||
| CCF 3314 | Circumdati | Candidi | Candidi | Czech Republic | Air (outdoor) | FR733812 | LT627022 | FR751426 | LT627023 | - | Glässnerová et al. (2022) | ||
| CCF 3853 | T | Circumdati | Candidi | Candidi | Czech Republic | Human toenail | FR727136 | FR775327 | HE661598 | LT627021 | - | Glässnerová et al. (2022) | |
| CCF 4030 | Circumdati | Candidi | Candidi | Czech Republic | Vermicompost | FR733814 | LT627018 | FR751425 | LT627019 | - | Glässnerová et al. (2022) | ||
| CCF 4653 | Circumdati | Candidi | Candidi | Czech Republic | Human toenail | HG915890 | HG916674 | HG916677 | LT627020 | - | Glässnerová et al. (2022) | ||
| CCF 4658 | Circumdati | Candidi | Candidi | Czech Republic | Human toenail | HG915891 | HG916675 | HG916676 | LT627026 | - | Glässnerová et al. (2022) | ||
| CCF 4914 | Circumdati | Candidi | Candidi | USA | Air (hospital) | ON156392 | ON164556 | ON164605 | ON164503 | - | Glässnerová et al. (2022) | ||
| CCF 6202 | Circumdati | Candidi | Candidi | USA | Air (house) | ON156396 | ON164588 | ON164629 | ON164538 | - | Glässnerová et al. (2022) | ||
| CCF 6397 | Circumdati | Candidi | Candidi | Czech Republic | Human abdominal cavity | ON156394 | ON164589 | ON164630 | ON164539 | - | Glässnerová et al. (2022) | ||
| IBT 12659 | Circumdati | Candidi | Candidi | USA | Soil (kangaroo rat burrow) | ON156393 | ON164557 | ON164606 | ON164504 | - | Glässnerová et al. (2022) | ||
| Aspergillus okavangoensis | CMW 56636 | T | Circumdati | Flavipedes | Flavipedes | Botswana | Soil (bat cave) | MW480880 | MW480788 | MW480706 | MW480790 | - | Visagie et al. (2021) |
| Aspergillus oxumiae | CCDCA11546 | T | Circumdati | Nigri | Japonici | Brazil | Soil under Agave sisalana | MN431160 | - | MN531842 | MN521389 | - | Crous et al. (2020b) |
| Aspergillus phialiformis | CGMCC 3.19314 | T | Polypaecilum | Polypaecilum | Canini | China | Rock | MK329068 | MK336095 | - | MK335974 | - | Zhang et al. (2020) |
| LC12537 | Polypaecilum | Polypaecilum | Canini | China | Rock | MK329069 | MK336096 | - | MK335975 | - | Zhang et al. (2020) | ||
| Aspergillus phialosimplex | CGMCC 3.19637 | T | Polypaecilum | Polypaecilum | Canini | China | Plant debris | MK329070 | MK336097 | - | MK335976 | - | Zhang et al. (2020) |
| LC12625 | Polypaecilum | Polypaecilum | Canini | China | Animal faeces | MK329071 | MK336098 | - | MK335977 | - | Zhang et al. (2020) | ||
| LC12658 | Polypaecilum | Polypaecilum | Canini | China | Plant root | MK329072 | MK336099 | - | MK335978 | - | Zhang et al. (2020) | ||
| Aspergillus gilianyuensis | CGMCC 3.20889 | T | Nidulantes | Nidulantes | Versicolores | China | Soil | OM414847 | OM475627 | OM475631 | OM475635 | - | Wang & Zhuang (2022b) |
| Aspergillus recifensis | CBS 145864 | T | Circumdati | Terrei | Nivei | Brazil | Soil | LR536036 | LR031370 | LR031385 | LR031400 | - | Barbosa et al. (2018) |
| URM 2803 | Circumdati | Terrei | Nivei | Brazil | Bird food | LR536040 | LR031375 | LR031390 | LR031405 | - | Barbosa et al. (2018) | ||
| URM 3371 | Circumdati | Terrei | Nivei | Brazil | Bird food | - | KR051530 | - | - | - | Barbosa et al. (2018) | ||
| URM 3571 | Circumdati | Terrei | Nivei | Brazil | Water from tank | LR536039 | LR031373 | LR031388 | LR031403 | - | Barbosa et al. (2018) | ||
| URM 5262 | Circumdati | Terrei | Nivei | Brazil | Soil (rhizosphere of Croton sp) | LR536037 | LR031371 | LR031386 | LR031401 | - | Barbosa et al. (2018) | ||
| URM 5461 | Circumdati | Terrei | Nivei | Brazil | Water from pool | LR536038 | LR031372 | LR031387 | LR031402 | - | Barbosa et al. (2018) | ||
| URM 6628 | Circumdati | Terrei | Nivei | Brazil | Soil | LR536035 | LR031369 | LR031384 | LR031399 | - | Barbosa et al. (2018) | ||
| Aspergillus rouenensis | CBS 149067 | T | Polypaecilum | Polypaecilum | Salinarum | France | Quercus bore dust of Xestobium rufovillosum | ON603782 | ON605641 | ON653193 | ON653194 | - | Crous et al. (2022) |
| CBS 149068 | Polypaecilum | Polypaecilum | Salinarum | France | Quercus bore dust of Xestobium rufovillosum | - | ON605642 | - | - | - | Crous et al. (2022) | ||
| Aspergillus saccharicola | IBT 36125 | Circumdati | Flavi | Flavi | Brazil | Sugarcane | - | ON642978 | ON643026 | ON642930 | - | Silva et al. (2022) | |
| IBT 36126 | T | Circumdati | Flavi | Flavi | Brazil | Sugarcane | OP611470 | ON529845 | ON529844 | ON529846 | - | Silva et al. (2022) | |
| IBT 36127 | Circumdati | Flavi | Flavi | Brazil | Sugarcane | - | ON642982 | ON643030 | ON642934 | - | Silva et al. (2022) | ||
| Aspergillus sakultaensis | AUMC 13885 | T | Circumdati | Flavipedes | Flavipedes | Egypt | Water | MK391495 | - | - | - | - | Zhori et al. (2020) |
| Aspergillus sibiricus | CBS 143307 | T | Fumigati | Fumigati | Unilaterales | Russia | Soil (cole mine) | MG587008 | MG722970 | MG722971 | MG710809 | - | Iliushin (2022) |
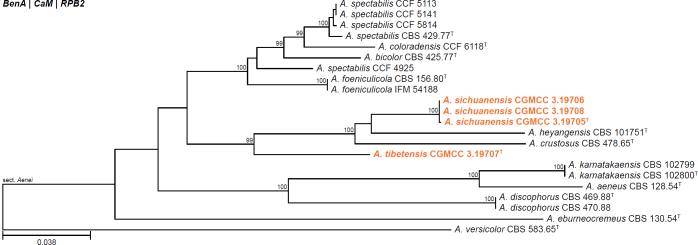
| Aspergillus sichuanensis | CGMCC 3.19705 | T | Nidulantes | Aenei | Aenei | China | Soil | MN640761 | MN635247 | MN635258 | MN635270 | - | Sun et al. (2022a) |
| CGMCC 3.19706 | Nidulantes | Aenei | Aenei | China | Soil | MN640762 | MN635248 | MN635259 | MN635271 | - | Sun et al. (2022a) | ||
| CGMCC 3.19708 | Nidulantes | Aenei | Aenei | China | Soil | MN640764 | MN635250 | MN635261 | MN635273 | - | Sun et al. (2022a) | ||
| Aspergillus sigarelli | CBS 141579 | T | Nidulantes | Usti | Calidousti | China | Cigarette | MN640758 | MN635244 | MN635255 | MN635267 | - | Sun et al. (2020c) |
| Aspergillus telluris | CGMCC 3.19701 | T | Polypaecilum | Polypaecilum | Canini | China | Soil | MN640767 | MN635253 | MN635264 | MN635276 | - | Sun et al. (2022a) |
| CGMCC 3.19702 | Polypaecilum | Polypaecilum | Canini | China | Soil | MN640768 | MN635254 | MN635265 | MN635277 | - | Sun et al. (2022a) | ||
| CGMCC 3.19703 | Polypaecilum | Polypaecilum | Canini | China | Soil | MN640769 | MN635243 | MN635266 | MN635278 | - | Sun et al. (2022a) | ||
| Aspergillus tenebricus | CBS 147048 | T | Circumdati | Candidi | Candidi | South Africa | Soil | ON156389 | ON164584 | ON164623 | ON164532 | - | Glässnerová et al. (2022) |
| CBS 147376 | Circumdati | Candidi | Candidi | Australia | Soil | ON156390 | ON164585 | ON164624 | ON164533 | - | Glässnerová et al. (2022) | ||
| DTO 440-E2 | Circumdati | Candidi | Candidi | Australia | Soil | ON156391 | ON164586 | ON164625 | ON164534 | - | Glässnerová et al. (2022) | ||
| Aspergillus tibetensis | CGMCC 3.19707 | T | Nidulantes | Aenei | Aenei | China | Soil | MN640763 | MN635249 | MN635260 | MN635272 | - | Sun et al. (2022a) |
| Aspergillus toxicus | A34-N | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987102 | - | - | Singh et al. (2020) | |
| BG14-F | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987108 | - | - | Singh et al. (2020) | ||
| BRG3458A | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987090 | - | - | Singh et al. (2020) | ||
| BRG3458H | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987095 | - | - | Singh et al. (2020) | ||
| BRG3458J | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987096 | - | - | Singh et al. (2020) | ||
| BRG5138J | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987097 | - | - | Singh et al. (2020) | ||
| CR10-G | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987105 | - | - | Singh et al. (2020) | ||
| CR20-D | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987098 | - | - | Singh et al. (2020) | ||
| CR24-F | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987103 | - | - | Singh et al. (2020) | ||
| D16-J | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987099 | - | - | Singh et al. (2020) | ||
| D25-A-S | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987100 | - | - | Singh et al. (2020) | ||
| E21-B | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987101 | - | - | Singh et al. (2020) | ||
| EC24-C | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987106 | - | - | Singh et al. (2020) | ||
| EC49-L | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987107 | - | - | Singh et al. (2020) | ||
| J15-B | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987104 | - | - | Singh et al. (2020) | ||
| K44-K | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987085 | - | - | Singh et al. (2020) | ||
| K849-B | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987086 | - | - | Singh et al. (2020) | ||
| NRRL 66868 | T | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987092 | - | - | Singh et al. (2020) | |
| NRRL 66897 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987091 | - | - | Singh et al. (2020) | ||
| NRRL 66899 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987093 | - | - | Singh et al. (2020) | ||
| NRRL 66900 | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987094 | - | - | Singh et al. (2020) | ||
| TX04A5-B | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987089 | - | - | Singh et al. (2020) | ||
| TX07CB73-I | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987087 | - | - | Singh et al. (2020) | ||
| TXLaFeria2-F | Circumdati | Flavi | Flavi | USA | Soil (maize field) | - | - | MN987088 | - | - | Singh et al. (2020) | ||
| Aspergillus vinaceus | ITAL 47.456 | T | Circumdati | Nigri | Nigri | Brazil | Grapes (Vitis labrusca) | MN575692 | MN583579 | MN583580 | MN583581 | - | Silva et al. (2020) |
| Aspergillus xishaensis | CGMCC 3.20890 | T | Circumdati | Flavipedes | Flavipedes | China | Soil | OM414848 | OM475628 | OM475632 | OM475636 | - | Wang & Zhuang (2022b) |
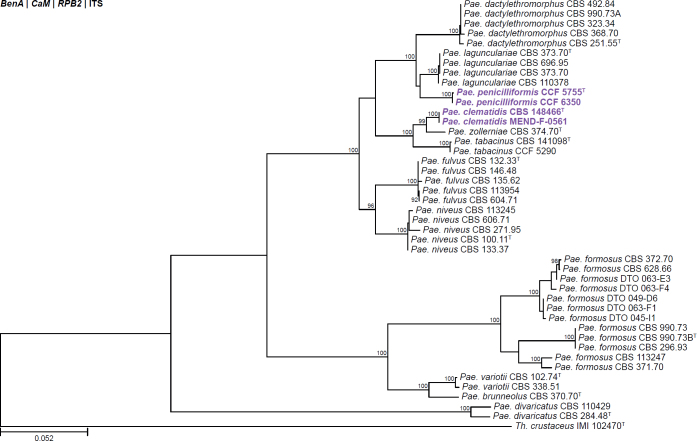
| Paecilomyces clematidis | CBS 148466 | T | - | - | - | Czech Republic | Root of Clematis | MZ923760 | MZ927740 | MZ927738 | OL332316 | - | Spetik et al. (2022) |
| MEND-F-0561 | - | - | - | Czech Republic | Root of Clematis | MZ923761 | MZ927741 | MZ927739 | OL332317 | - | Spetik et al. (2022) | ||
| Paecilomyces penicilliformis | CCF 5755 | T | - | - | - | USA | Air (pharmacy) | LR679769 | LR679768 | LR778299 | - | - | Crous et al. (2020b) |
| CCF 6350 | - | - | - | USA | Juice (peach-mango) | LR736038 | LR778163 | LR778165 | - | - | Crous et al. (2020b) | ||
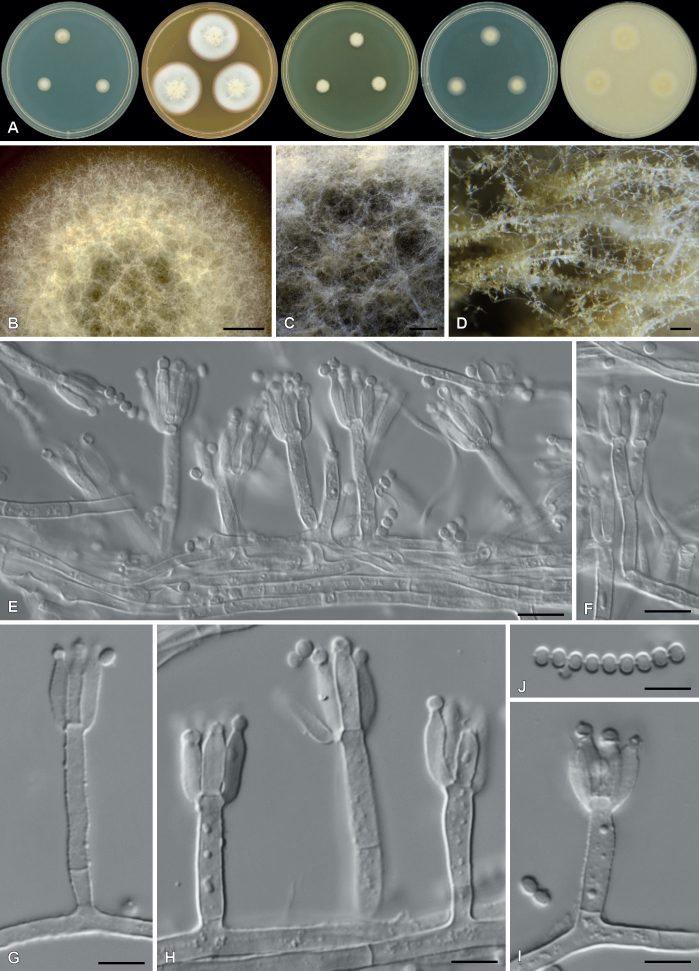
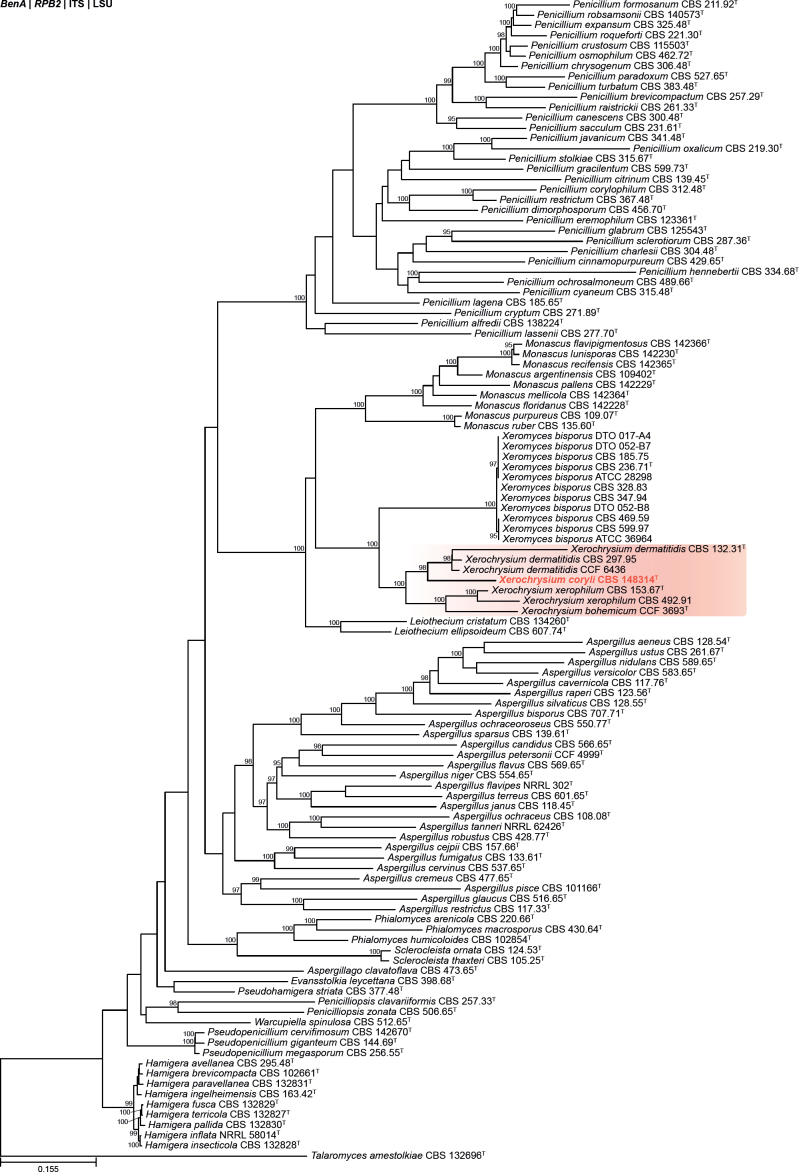
| Paraxerochrysium coryli | CBS 148314 | T | - | - | - | Belgium | Hazelnut (Corylus avellana) | OK664748 | OK651216 | - | OK651178 | OK663787 | Crous et al. (2021b) |
| Penicillium allaniae | BRIP 74886a | T | Aspergilloides | Exilicaulis | Restrict! | Australia | Soil | OP903476 | OP921959 | OP921957 | OP921958 | OP925816 | Tan & Shivas (2022) |
| BRIP 74899 | Aspergilloides | Exilicaulis | Restrict! | Australia | Unknown | OP903475 | OP921956 | OP921954 | OP921955 | Tan & Shivas (2022) | |||
| Penicillium allsoppiae | CBS 138943 | T | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140830 | JX140992 | JX157384 | KP016895 | - | Visagie & Yilmaz (2022) |
| CBS 138945 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140822 | JX141004 | JX157399 | KP016910 | - | Visagie & Yilmaz (2022) | ||
| CN 086C6 | Penicillium | Canescentia | Canescentia | South Africa | Soil | MW364385 | MW357820 | MW357831 | MW357840 | - | Visagie & Yilmaz (2022) | ||
| CN 086C7 | Penicillium | Canescentia | Canescentia | South Africa | Soil | MW364386 | MW357821 | MW357832 | MW357841 | - | Visagie & Yilmaz (2022) | ||
| CN 086C8 | Penicillium | Canescentia | Canescentia | South Africa | Soil | MW364387 | MW357822 | - | MW357842 | - | Visagie & Yilmaz (2022) | ||
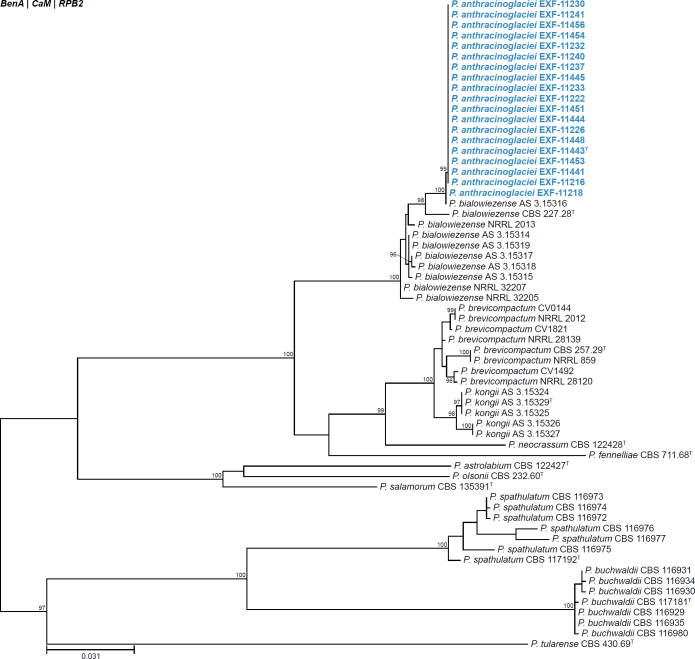
| Penicillium anthracinoglaciei | EXF-11216 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080468 | MT080527 | MT080509 | - | Perini et al. (2023) | |
| EXF-11218 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080469 | MT080528 | MT080510 | - | Perini et al. (2023) | ||
| EXF-11222 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080472 | MT080531 | MT080511 | - | Perini et al. (2023) | ||
| EXF-11226 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Supraglacial water | - | MT080475 | MT080534 | MT080512 | - | Perini et al. (2023) | ||
| EXF-11230 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Supraglacial water | - | MT080479 | MT080538 | MT080508 | - | Perini et al. (2023) | ||
| EXF-11232 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Supraglacial water | - | MT080481 | MT080540 | MT080513 | - | Perini et al. (2023) | ||
| EXF-11233 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Supraglacial water | - | MT080482 | MT080541 | MT080514 | - | Perini et al. (2023) | ||
| EXF-11237 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080483 | MT080542 | MT080515 | - | Perini et al. (2023) | ||
| EXF-11240 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080485 | MT080544 | MT080516 | - | Perini et al. (2023) | ||
| EXF-11241 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Clear ice | - | MT080486 | MT080545 | MT080517 | - | Perini et al. (2023) | ||
| EXF-11443 | T | Penicillium | Brevicompacta | Brevicompacta | Greenland | Dark ice | - | MT080493 | MT080552 | MT080519 | - | Perini et al. (2023) | |
| EXF-11444 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Dark ice | - | MT080494 | MT080553 | MT080520 | - | Perini et al. (2023) | ||
| EXF-11445 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Dark ice | - | MT080495 | MT080554 | MT080521 | - | Perini et al. (2023) | ||
| EXF-11448 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080498 | MT080557 | MT080522 | - | Perini et al. (2023) | ||
| EXF-11451 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Cryoconite | - | MT080501 | MT080560 | MT080523 | - | Perini et al. (2023) | ||
| EXF-11453 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Clear ice | - | MT080503 | MT080562 | MT080524 | - | Perini et al. (2023) | ||
| EXF-11454 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Clear ice | - | MT080504 | MT080563 | MT080525 | - | Perini et al. (2023) | ||
| EXF-11456 | Penicillium | Brevicompacta | Brevicompacta | Greenland | Clear ice | - | MT080506 | MT080565 | MT080526 | - | Perini et al. (2023) | ||
| Penicillium aquadulcis | CNUFC JT1301 | T | Aspergilloides | Citrina | Westlingiorum | Republic of Korea | Water | OK356194 | QK105100 | OK105102 | - | - | Nguyen et al. (2021b) |
| CNUFC JT1302 | Aspergilloides | Citrina | Westlingiorum | Republic of Korea | Water | OK356195 | OK105101 | OK105103 | - | - | Nguyen et al. (2021b) | ||
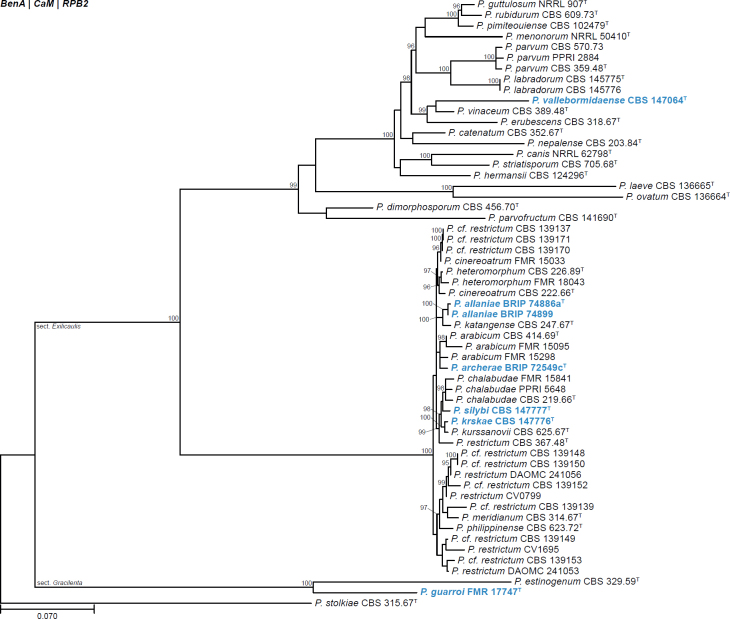
| Penicillium archerae | BRIP 72549c | T | Aspergilloides | Exilicaulis | Restricti | Australia | Soil | OP903477 | OP921961 | - | OP921960 | - | Tan & Shivas (2022) |
| Penicillium aspericonidium | CBS 141832 | T | Aspergilloides | Charlesia | Indica | Australia | Soil | MT309657 | MT302240 | - | MT302224 | - | Sun et al. (2021) |
| Penicillium ausonanum | FMR 16948 | T | Aspergilloides | Lanata-Divaricata | Dalearum | Spain | Fluvial sediment | LR655808 | LR655809 | LR655810 | LR655811 | - | Torres-Garcia et al. (2022) |
| Penicillium barbosae | URM 7705 | T | Aspergilloides | Sclerotiorum | Adametziorum | Brazil | Soil | MW191494 | MG452818 | MW183245 | LR898886 | - | Ramos et al. (2021) |
| URM 7824 | Aspergilloides | Sclerotiorum | Adametziorum | Brazil | Soil | MW191495 | MG452819 | MW183246 | LR898887 | - | Ramos et al. (2021) | ||
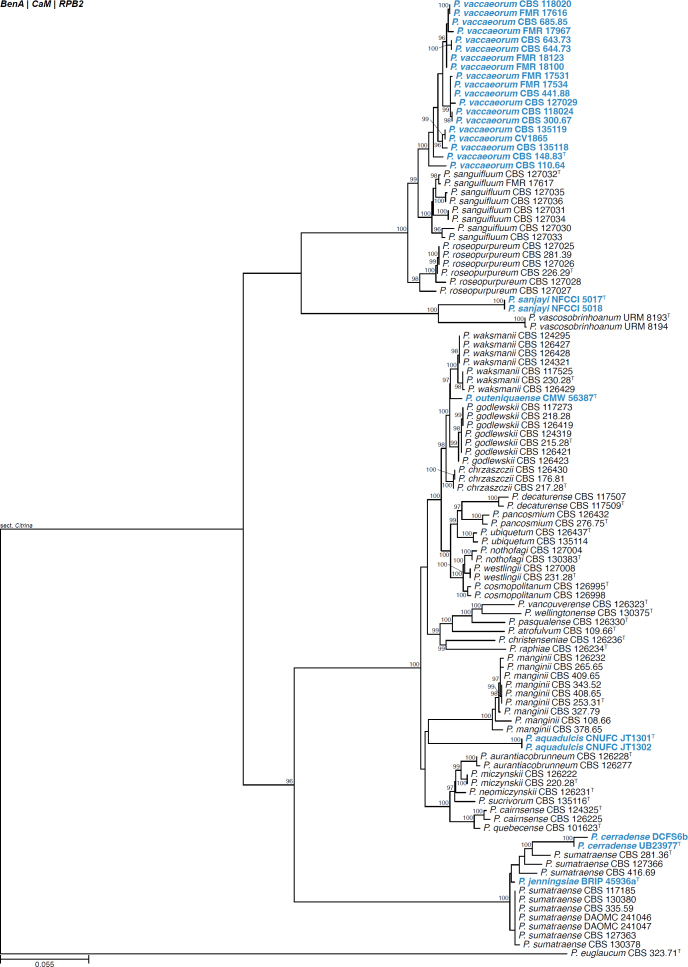
| Penicillium cerradense | UB23977 | T | Aspergilloides | Citrina | Sumatraensia | Brazil | Soil | MT006126 | MT416533 | MT416534 | MT416532 | - | Andrade et al. (2021) |
| DCFS6b | Aspergilloides | Citrina | Sumatraensia | Brazil | Soil | MT006127 | MT416536 | MT416537 | MT416535 | - | Andrade et al. (2021) | ||
| Penicillium claroviride | CMW 56197 | T | Penicillium | Canescentia | Atroveneta | South Africa | Soil | MT949909 | MT957414 | MT957456 | MT957482 | - | Visagie & Yilmaz (2022) |
| CMW 56198 | Penicillium | Canescentia | Atroveneta | South Africa | Soil | MT949910 | MT957415 | MT957457 | MT957483 | - | Visagie & Yilmaz (2022) | ||
| Penicillium doidgeae | CBS 138947 | T | Penicillium | Canescentia | Atroveneta | South Africa | Mite from Protea repens infructescens | JX140804 | JX141006 | JX157413 | KP016915 | - | Visagie & Yilmaz (2022) |
| CBS 138948 | Penicillium | Canescentia | Atroveneta | South Africa | Mite from Protea repens infructescens | JX140805 | JX141007 | JX157414 | KP016916 | - | Visagie & Yilmaz (2022) | ||
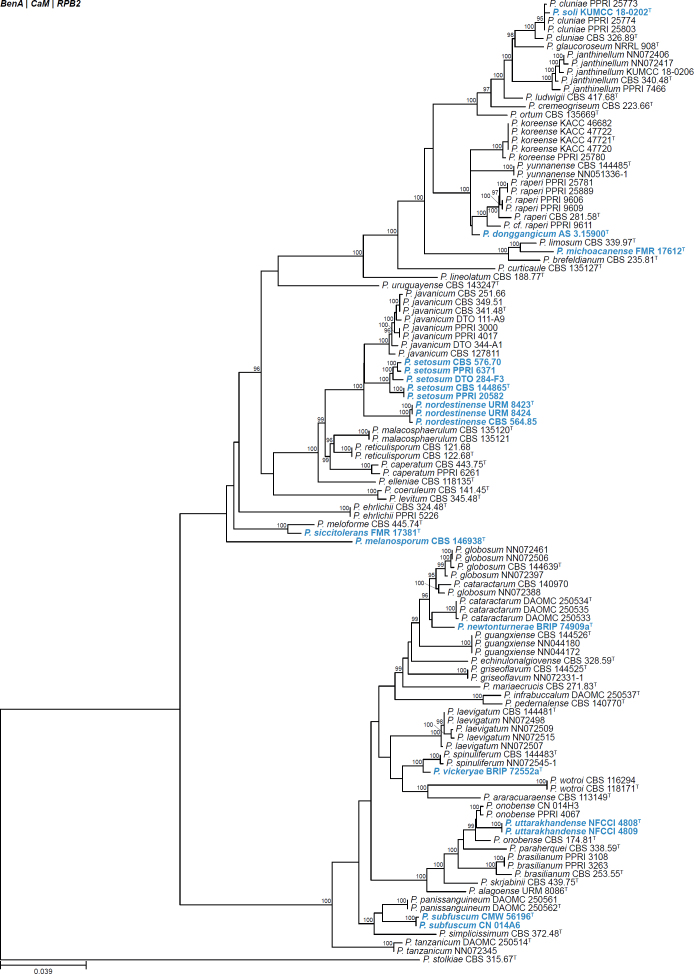
| Penicillium donggangicum | AS 3.15900 | T | Aspergilloides | Lanata-Divaricata | Janthinella | China | Soil | MW946996 | MZ004914 | MZ004918 | MW979253 | - | Xu et al. (2022) |
| Penicillium eickerii | CBS 138939 | T | Penicillium | Canescentia | Canescentia | South Africa | Mite from Protea repens infructescens | JX140824 | JX140979 | JX157365 | KP016876 | - | Visagie & Yilmaz (2022) |
| CBS 138940 | Penicillium | Canescentia | Canescentia | South Africa | Bract from Protea repens infructescens | JX140825 | JX140980 | JX157366 | KP016877 | - | Visagie & Yilmaz (2022) | ||
| Penicillium elizabethiae | NRRL917 | T | Penicillium | Canescentia | Canescentia | Scotland | Soil | KP016840 | KJ866964 | KJ867021 | KP016918 | - | Visagie & Yilmaz (2022) |
| Penicillium ezekielii | CBS 149115 | T | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Indonesia | Zea mays kernels | ON723772 | ON920778 | ON920781 | ON920784 | ON911289 | Tan et al. (2022) |
| CBS 149114 | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Indonesia | Zea mays kernels | ON723771 | ON920777 | ON920780 | ON920783 | ON911288 | Tan et al. (2022) | ||
| DTO 463-A7 | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Nigeria | Oryza sativa kernels | ON723773 | ON920779 | ON920782 | - | ON911290 | Tan et al. (2022) | ||
| Penicillium ferraniaense | CBS 147594 | Aspergilloides | Sclerotiorum | Sclerotiorum | Italy | Compost | MW694952 | MW689337 | MW689339 | MW689341 | - | Crous et al. (2021a) | |
| CBS 147595 | T | Aspergilloides | Sclerotiorum | Sclerotiorum | Italy | Compost | MW694951 | MW689336 | MW689338 | MW689340 | - | Crous et al. (2021a) | |
| Penicillium fusiforme | CBS 250.66 | T | Aspergilloides | Charlesia | Fellutana | The Netherlands | Unknown | MT309668 | MT302253 | MT302220 | MT302236 | - | Sun et al. (2021) |
| Penicillium gercinae | URM 8348 | T | Aspergilloides | Ramigena | Georgiensia | Brazil | Soil | MW648591 | MW646389 | MW646391 | MW646393 | - | Alves et al. (2022a) |
| Penicillium guarroi | FMR 17747 | T | Aspergilloides | Gracilenta | Estinogena | Spain | Fluvial sediment | LR814139 | LR814134 | LR814140 | LR814145 | - | Torres-Garcia et al. (2022) |
| Penicillium hepuense | AS 3.16039 | T | Aspergilloides | Lanata-Divaricata | Oxalica | China | Soil | MW946994 | MZ004912 | MZ004916 | MW979254 | - | Xu et al. (2022) |
| AS 3.16040 | Aspergilloides | Lanata-Divaricata | Oxalica | China | Soil | MW946995 | MZ004913 | MZ004917 | MW979255 | - | Xu et al. (2022) | ||
| Penicillium irregulare | FMR 17859 | T | Penicillium | Canescentia | Canescentia | Spain | Fluvial sediment | LR814181 | LR814144 | LR814151 | LR814182 | - | Torres-Garcia et al. (2022) |
| Penicillium jenningsiae | BRIP 45936a | T | Aspergilloides | Citrina | Sumatraensia | Australia | Compost | - | OL741657 | - | OL741660 | - | Tan & Shivas (2022) |
| Penicillium jiaozhouwanicum | AS 3.16027 | Aspergilloides | Lanata-Divaricata | Oxalica | China | Soil | OM203537 | OM220087 | OM220088 | OM220089 | - | Xu et al. (2022) | |
| AS 3.16038 | T | Aspergilloides | Lanata-Divaricata | Oxalica | China | Soil | MW946993 | MZ004911 | MZ004915 | MW979252 | - | Xu et al. (2022) | |
| Penicillium kalander | CMW 56202 | T | Aspergilloides | Sclerotiorum | Sclerotiorum | South Africa | Soil | MT949914 | MT957421 | MT957461 | MT957487 | - | Visagie & Yilmaz (2022) |
| CMW 56203 | Aspergilloides | Sclerotiorum | Sclerotiorum | South Africa | Soil | MT949915 | MT957422 | MT957462 | MT957488 | - | Visagie & Yilmaz (2022) | ||
| CMW 56204 | Aspergilloides | Sclerotiorum | Sclerotiorum | South Africa | Soil | MT949916 | MT957423 | MT957463 | MT957489 | - | Visagie & Yilmaz (2022) | ||
| CMW 56205 | Aspergilloides | Sclerotiorum | Sclerotiorum | South Africa | Soil | MT949917 | MT957424 | MT957464 | MT957490 | - | Visagie & Yilmaz (2022) | ||
| CMW 56390 | Aspergilloides | Sclerotiorum | Sclerotiorum | South Africa | Soil | MT949918 | MT957425 | MT957465 | MT957491 | - | Visagie & Yilmaz (2022) | ||
| Penicillium krskae | CBS 147776 | T | Aspergilloides | Exilicaulis | Restricta | Austria | Air (indoor) | MW794123 | MW774594 | MW774595 | MW774593 | - | Labuda et al. (2021) |
| Penicillium limae | URM 7706 | T | Aspergilloides | Sclerotiorum | Adametziorum | Brazil | Soil | MW191493 | MG452820 | MW183244 | LR898888 | - | Ramos et al. (2021) |
| Penicillium linzhiense | Z863 | T | Penicillium | Canescentia | Canescentia | China | Soil | MT461156 | MT 461157 | MT461162 | - | - | Liang et al. (2021) |
| Penicillium longiconidiophorum | CBS 141831 | T | Aspergilloides | Charlesia | Phoenicea | Madagascar | Soil | MT309669 | MT302254 | MT302221 | MT302237 | - | Sun et al. (2021) |
| DTO 092-C6 | Aspergilloides | Charlesia | Phoenicea | Madagascar | Soil | MT309670 | MT302255 | MT302222 | MT302238 | - | Sun et al. (2021) | ||
| Penicillium mattheeae | CMW 56195 | Aspergilloides | Aspergilloides | Saturniiormia | South Africa | Soil | MT949905 | MT957409 | MT957452 | MT957478 | - | Visagie & Yilmaz (2022) | |
| CMW 56388 | T | Aspergilloides | Aspergilloides | Saturniiormia | South Africa | Soil | MT949904 | MT957408 | MT957451 | MT957477 | - | Visagie & Yilmaz (2022) | |
| CMW 56633 | Aspergilloides | Aspergilloides | Saturniiormia | South Africa | Soil | MT949906 | MT957410 | MT957453 | MT957479 | - | Visagie & Yilmaz (2022) | ||
| Penicillium melanosporum | CBS 146938 | T | Aspergilloides | Lanata-Divaricata | Janthinella | Spain | Soil | LR655192 | LR655196 | LR655200 | LR655204 | - | Rodriguez-Andrade et al. (2021) |
| Penicillium michoacanense | FMR 17612 | T | Aspergilloides | Lanata-Divaricata | Janthinella | Mexico | Soil | LR655194 | LR655198 | LR655202 | LR655206 | - | Rodriguez-Andrade et al. (2021) |
| Penicillium neoherguei | CBS 148692 | T | Aspergilloides | Sclerotiorum | Hergueorum | USA | White mushroom sporocarp | MW341222 | OL840853 | OL840855 | MW349119 | - | Crous et al. (2022) |
| Penicillium newtonturnerae | BRIP 74909a | T | Aspergilloides | Lanata-Divaricata | Simplicissima | Australia | Soil | OP903478 | OP921964 | OP921962 | OP921963 | OP925817 | Tan & Shivas (2022) |
| Penicillium nordestinense | CBS 564.85 | Aspergilloides | Lanata-Divaricata | Janthinella | Brazil | Pollen samples inside nests of (Melipona scutellaris) | OV312015 | MH846596 | MH846609 | MH846584 | - | Barbosa et al. (2022) | |
| URM 8423 | T | Aspergilloides | Lanata-Divaricata | Janthinella | Brazil | Pollen samples inside nests of (Melipona scutellaris) | OV265270 | OV265324 | OV265272 | OM927721 | - | Barbosa et al. (2022) | |
| URM 8424 | Aspergilloides | Lanata-Divaricata | Janthinella | Brazil | Pollen samples inside nests of (Melipona scutellaris) | OV265271 | OV265337 | OV265273 | OM927721 | - | Barbosa et al. (2022) | ||
| Penicillium outeniguaense | CMW 56387 | T | Aspergilloides | Citrina | Westlingiorum | South Africa | Soil | MT949903 | MT957405 | MT957450 | MT957476 | - | Visagie & Yilmaz (2022) |
| Penicillium poederi | CBS 147622 | T | Aspergilloides | Torulomyces | Torulomyces | Iceland | Volcanic soil | MF611757 | MF611760 | MF611763 | MF611766 | - | Kirchmair et al. (2022) |
| CBS 147623 | Aspergilloides | Torulomyces | Torulomyces | Iceland | Volcanic soil | MF611758 | MF611761 | MF611764 | MF611767 | - | Kirchmair et al. (2022) | ||
| CBS 147624 | Aspergilloides | Torulomyces | Torulomyces | Iceland | Volcanic soil | MF611759 | MF611762 | MF611765 | MF611768 | - | Kirchmair et al. (2022) | ||
| Penicillium pole-evansii | CBS 138946 | T | Penicillium | Canescentia | Atroveneta | South Africa | Bract from Protea repens infructescens | JX140831 | JX141005 | JX157412 | KP016911 | - | Visagie & Yilmaz (2022) |
| Penicillium rotoruae | CBS 145838 | T | Aspergilloides | Lanata-Divaricata | Rolfsiorum | New Zealand | Pinus radiata timber on ground | MN315103 | MN315104 | MN315102 | MT240842 | - | O’Callahan et al. (2020) |
| Penicillium saanichanum | DAOMC 251850 | T | Aspergilloides | Cinnamopurpurea | Idahoensia | Canada | House dust | KY469059 | KY469096 | KY469020 | MN795070 | - | Crous et al. (2020a) |
| Penicillium sanjayi | NFCCI 5017 | T | Aspergilloides | Citrina | Vascosobrinhoana | India | Soil | MZ571358 | MZ558484 | MZ558492 | MZ558482 | - | Ashtekar et al. (2022) |
| NFCCI 5018 | Aspergilloides | Citrina | Vascosobrinhoana | India | Soil | MZ571359 | MZ558485 | MZ558493 | MZ558483 | - | Ashtekar et al. (2022) | ||
| Penicillium scottii | CBS 138935 | Penicillium | Canescentia | Canescentia | South Africa | Air (outdoor) | JX140823 | JX140977 | JX157351 | KP016863 | - | Visagie & Yilmaz (2022) | |
| CBS 138937 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140826 | JX140978 | JX157355 | KP016867 | - | Visagie & Yilmaz (2022) | ||
| CBS 138941 | Penicillium | Canescentia | Canescentia | South Africa | Air (outdoor) | JX140827 | JX140981 | JX157371 | KP016882 | - | Visagie & Yilmaz (2022) | ||
| CBS 138944 | Penicillium | Canescentia | Canescentia | South Africa | Bract from Protea repens infructescens | JX140820 | JX141002 | JX157396 | KP016907 | - | Visagie & Yilmaz (2022) | ||
| CBS 138951 | T | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140812 | JX140991 | JX157383 | KP016894 | - | Visagie & Yilmaz (2022) | |
| CV0939 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140814 | JX140994 | JX157386 | KP016897 | - | Visagie & Yilmaz (2022) | ||
| IBT 31903 | Penicillium | Canescentia | Canescentia | South Africa | Bract from Protea repens infructescens | JX140821 | JX141003 | JX157397 | KP016908 | - | Visagie & Yilmaz (2022) | ||
| IBT 31904 | Penicillium | Canescentia | Canescentia | South Africa | Soil | KP016833 | JX140995 | JX157387 | KP016898 | - | Visagie & Yilmaz (2022) | ||
| IBT 31906 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140815 | JX140996 | JX157388 | KP016899 | - | Visagie & Yilmaz (2022) | ||
| IBT 31907 | Penicillium | Canescentia | Canescentia | South Africa | Soil | KP016832 | JX140988 | JX157378 | KP016889 | - | Visagie & Yilmaz (2022) | ||
| IBT 31908 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140816 | JX140997 | JX157389 | KP016900 | - | Visagie & Yilmaz (2022) | ||
| IBT 31909 | Penicillium | Canescentia | Canescentia | South Africa | Soil | KP016834 | JX140998 | JX157390 | KP016901 | - | Visagie & Yilmaz (2022) | ||
| IBT 31910 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140817 | JX140999 | JX157391 | KP016902 | - | Visagie & Yilmaz (2022) | ||
| IBT 31911 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140818 | JX141000 | JX157392 | KP016903 | - | Visagie & Yilmaz (2022) | ||
| IBT 31912 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140808 | JX140984 | JX157374 | KP016885 | - | Visagie & Yilmaz (2022) | ||
| IBT 31913 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140811 | JX140990 | JX157382 | KP016893 | - | Visagie & Yilmaz (2022) | ||
| IBT 31914 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140813 | JX140993 | JX157385 | KP016896 | - | Visagie & Yilmaz (2022) | ||
| IBT 31915 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140807 | JX140982 | JX157372 | KP016883 | - | Visagie & Yilmaz (2022) | ||
| IBT 31916 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140809 | JX140985 | JX157375 | KP016886 | - | Visagie & Yilmaz (2022) | ||
| IBT 31917 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140810 | JX140987 | JX157377 | KP016888 | - | Visagie & Yilmaz (2022) | ||
| IBT31918 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140828 | JX140986 | JX157376 | KP016887 | - | Visagie & Yilmaz (2022) | ||
| IBT 31919 | Penicillium | Canescentia | Canescentia | South Africa | Soil | KP016831 | JX140983 | JX157373 | KP016884 | - | Visagie & Yilmaz (2022) | ||
| IBT 31922 | Penicillium | Canescentia | Canescentia | South Africa | Mite from Protea repens infructescens | JX140819 | JX141001 | JX157394 | KP016905 | - | Visagie & Yilmaz (2022) | ||
| IBT 31953 | Penicillium | Canescentia | Canescentia | South Africa | Soil | JX140829 | JX140989 | JX157379 | KP016890 | - | Visagie & Yilmaz (2022) | ||
| Penicillium setosum | CBS 144865 | T | Aspergilloides | Lanata-Divaricata | Janthinella | India | Withania somniiera | KT852579 | MF184995 | MH105905 | - | - | Barbosa et al. (2022) |
| CBS 576.70 | Aspergilloides | Lanata-Divaricata | Janthinella | Mexico | Soil | - | MH846595 | MH846608 | MH846583 | - | Barbosa et al. (2022) | ||
| DTO 284-F3 | Aspergilloides | Lanata-Divaricata | Janthinella | India | Withania somniiera | - | MH846594 | MH846607 | MH846582 | - | Barbosa et al. (2022) | ||
| PPRI 20582 | Aspergilloides | Lanata-Divaricata | Janthinella | South Africa | Insect | MK450718 | MK451255 | MK451649 | - | - | Barbosa et al. (2022) | ||
| PPRI 6371 | Aspergilloides | Lanata-Divaricata | Janthinella | South Africa | Scarabid larvae | MK450717 | MK 451227 | MK451648 | MK450852 | - | Barbosa et al. (2022) | ||
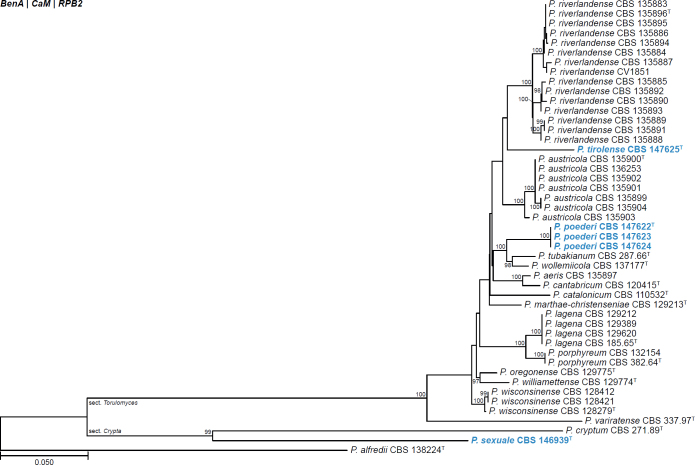
| Penicillium sexuale | CBS 146939 | T | Aspergilloides | Crypta | Crypta | Spain | Soil | LR655195 | LR655199 | LR655203 | LR655207 | - | Rodriguez-Andrade et al. (2021) |
| Penicillium siccitolerans | FMR 17381 | T | Aspergilloides | Lanata-Divaricata | Janthinella | Spain | Soil | LR655193 | LR655197 | LR655201 | LR655205 | - | Rodriguez-Andrade et al. (2021) |
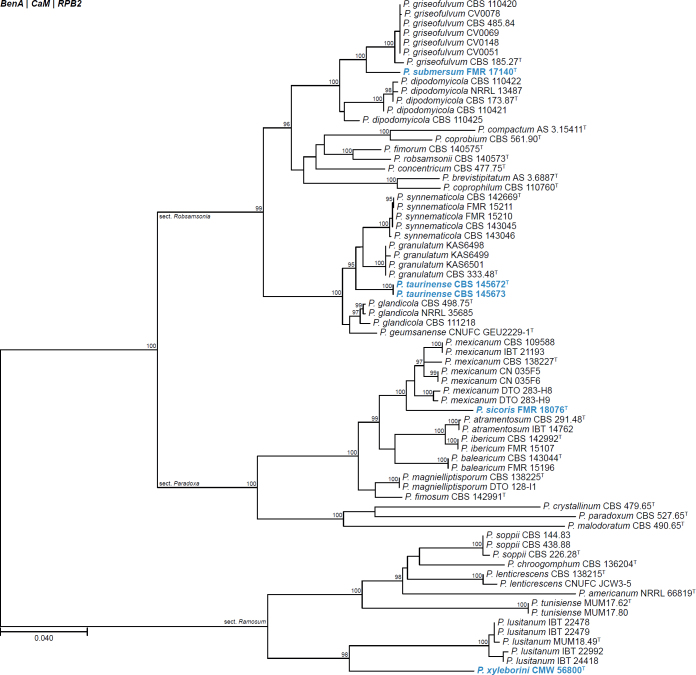
| Penicillium sicoris | FMR 18076 | T | Penicillium | Paradoxa | Atramentosa | Spain | Fluvial sediment | LR884497 | LR884494 | LR884496 | LR884495 | - | Torres-Garcia et al. (2022) |
| Penicillium silybi | CBS 147777 | T | Aspergilloides | Exilicaulis | Restricta | USA | Milk thistle (Silybum marianum) | KF367458 | MW774592 | MW774591 | AB860248 | - | Labuda et al. (2021) |
| Penicillium soli | KUMCC 18-0202 | T | Aspergilloides | Lanata-Divaricata | Janthinella | China | Soil | MT152337 | MT161681 | MT178249 | MT384372 | - | Doilom et al. (2020) |
| Penicillium stangiae | URM 8347 | T | Aspergilloides | Lanata-Divaricata | Dalearum | Brazil | Soil | MW648590 | MW646388 | MW646390 | MW646392 | - | Alves et al. (2022a) |
| Penicillium subfuscum | CMW 56196 | T | Aspergilloides | Lanata-Divaricata | Simplicissima | South Africa | Soil | MT949907 | MT957412 | MT957454 | MT957480 | - | Visagie & Yilmaz (2022) |
| CN014A6 | Aspergilloides | Lanata-Divaricata | Simplicissima | South Africa | Soil | MW329997 | MW340969 | MW340970 | MW340971 | - | Visagie & Yilmaz (2022) | ||
| Penicillium submersum | FMR 17140 | T | Penicillium | Robsamsonia | Urticicola | Spain | Fluvial sediment | LR814194 | LR814187 | LR814188 | LR814195 | - | Torres-Garcia et al. (2022) |
| Penicillium taurinense | CBS 145672 | T | Penicillium | Robsamsonia | Glandicolarum | Italy | Chestnut mill | MF595981 | MF595977 | MF595979 | MT253108 | - | Crous et al. (2020b) |
| CBS 145673 | Penicillium | Robsamsonia | Glandicolarum | Italy | Chestnut mill | MF595982 | MF595978 | MF595980 | - | - | Crous et al. (2020b) | ||
| Penicillium tealii | BRIP 72734c | T | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Australia | Dead spider | OP101639 | OP039547 | - | OP039546 | - | Tan et al. (2022) |
| BRIP 72735b | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Australia | Dead spider | OP101642 | OP039553 | - | OP039552 | - | Tan et al. (2022) | ||
| BRIP 72742b | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Australia | Dead spider | OP101643 | OP039554 | - | OP039546 | - | Tan et al. (2022) | ||
| BRIP 72731b | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Australia | Dead spider | OP101641 | OP039551 | - | OP039555 | - | Tan et al. (2022) | ||
| BRIP 72732b | Aspergilloides | Cinnamopurpurea | Jiangxiensia | Australia | Dead spider | OP101640 | OP039549 | - | OP039559 | - | Tan et al. (2022) | ||
| Penicillium tirolense | CBS 147625 | T | Aspergilloides | Torulomyces | Torulomyces | Austria | Sporocarp of Serpula lacrymans | MW145398 | MW143069 | MW143068 | MW143067 | - | Kirchmair et al. (2022) |
| Penicillium tolerans | BRIP 64090a | T | Aspergilloides | Aspergilloides | Sclerotiorum | Australia | Soil | OK639006 | OL741658 | - | - | - | Tan & Shivas (2022) |
| Penicillium ucsense | CBS 146492 | T | Aspergilloides | Lanata-Divaricata | Oxalica | Brazil | Intestinal tract of Anobium punctatum larva | OM914583 | ON024157 | ON024158 | ON024159 | - | Lenz et al. (2022) |
| Penicillium ulleungdoense | KACC 48990 | T | Aspergilloides | Sclerotiorum | Sclerotiorum | Republic of Korea | Root of Phedimus takesimensis | MN640087 | MN737487 | MN745074 | MN756007 | - | Choi et al. (2020) |
| KACC 48991 | Aspergilloides | Sclerotiorum | Sclerotiorum | Republic of Korea | Root of Sedum oryzifolium | MN640088 | MN737488 | MN745075 | MN756008 | - | Choi et al. (2020) | ||
| KACC 48992 | Aspergilloides | Sclerotiorum | Sclerotiorum | Republic of Korea | Root of Aster spathulifolius | MN640089 | MN737489 | MN745076 | MN756009 | - | Choi et al. (2020) | ||
| Penicillium umkhoba | CMW 56199 | Aspergilloides | Sclerotiorum | Hergueorum | South Africa | Soil | MT949911 | MT957416 | MT957458 | MT957484 | - | Visagie & Yilmaz (2022) | |
| CMW 56200 | T | Aspergilloides | Sclerotiorum | Hergueorum | South Africa | Soil | MT949912 | MT957417 | MT957459 | MT957485 | - | Visagie & Yilmaz (2022) | |
| CMW 56201 | Aspergilloides | Sclerotiorum | Hergueorum | South Africa | Soil | MT949913 | MT957418 | MT957460 | MT957486 | - | Visagie & Yilmaz (2022) | ||
| Penicillium uttarakhandense | NFCCI 4808 | T | Aspergilloides | Lanata-Divaricata | Simplicissima | India | Soil (garden) | MN967315 | MN972443 | MN972445 | MN972447 | - | Crous et al. (2021a) |
| NFCCI 4809 | Aspergilloides | Lanata-Divaricata | Simplicissima | India | Soil (garden) | MN967316 | MN972444 | MN972446 | MN972448 | - | Crous et al. (2021a) | ||
| Penicillium vaccaeorum | CBS 110.64 | Aspergilloides | Citrina | Roseopurpurea | Turkey | Soil | - | JN606829 | JN606533 | - | - | Torres-Garcia et al. (2022) | |
| CBS 118020 | Aspergilloides | Citrina | Roseopurpurea | Canada | Ants | - | JN606832 | JN606536 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 118024 | Aspergilloides | Citrina | Roseopurpurea | Canada | Ants | - | JN606833 | JN606537 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 127029 | Aspergilloides | Citrina | Roseopurpurea | Argentina | Soil | - | JN606814 | JN606544 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 135118 | Aspergilloides | Citrina | Roseopurpurea | South Africa | Soil | JX140867 | JX141019 | JX141510 | MN418449 | - | Torres-Garcia et al. (2022) | ||
| CBS 135119 | Aspergilloides | Citrina | Roseopurpurea | South Africa | Soil | JX140865 | JX141020 | JX141511 | MK461491 | - | Torres-Garcia et al. (2022) | ||
| CBS 148.83 | T | Aspergilloides | Citrina | Roseopurpurea | Spain | Soil | JN617689 | JN606835 | JN606543 | JN606614 | - | Torres-Garcia et al. (2022) | |
| CBS 300.67 | Aspergilloides | Citrina | Roseopurpurea | The Netherlands | Soil | - | JN606841 | JN606561 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 441.88 | Aspergilloides | Citrina | Roseopurpurea | Chile | Soil | - | JN606846 | JN606568 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 643.73 | Aspergilloides | Citrina | Roseopurpurea | Canada | Soil | - | JN606853 | JN606576 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 644.73 | Aspergilloides | Citrina | Roseopurpurea | Canada | Soil | - | JN606854 | JN606577 | - | - | Torres-Garcia et al. (2022) | ||
| CBS 685.85 | Aspergilloides | Citrina | Roseopurpurea | Spain | Soil | JN617711 | JN606855 | JN606578 | - | - | Torres-Garcia et al. (2022) | ||
| CV1865 | Aspergilloides | Citrina | Roseopurpurea | South Africa | Soil | JX140866 | JX141021 | JX157340 | MK461547 | - | Torres-Garcia et al. (2022) | ||
| FMR 17531 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | LR814213 | LR814203 | LR814204 | - | - | Torres-Garcia et al. (2022) | ||
| FMR 17534 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | OU375272 | OU375168 | OU375273 | - | - | Torres-Garcia et al. (2022) | ||
| FMR 17616 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | LR814217 | LR814212 | LR814218 | - | - | Torres-Garcia et al. (2022) | ||
| FMR 17967 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | LR814235 | LR814226 | LR814227 | - | - | Torres-Garcia et al. (2022) | ||
| FMR 18100 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | LR814241 | LR814234 | LR814242 | - | - | Torres-Garcia et al. (2022) | ||
| FMR 18123 | Aspergilloides | Citrina | Roseopurpurea | Spain | Fluvial sediment | LR814273 | LR814265 | LR814264 | - | - | Torres-Garcia et al. (2022) | ||
| Penicillium vallebormidaense | CBS 147064 | T | Aspergilloides | Exilicaulis | Erubescentia | Italy | Compost | MT316359 | MW115862 | MW115863 | MW115864 | - | Crous et al. (2020a) |
| Penicillium vickeryae | BRIP 72552a | T | Aspergilloides | Lanata-Divaricata | Simplicissima | Australia | Soil | OP903479 | OP921966 | - | OP921965 | - | Tan & Shivas (2022) |
| Penicillium vietnamense | VTCC 930029 | T | Aspergilloides | Charlesia | Indica | Vietnam | Sea water | MT102836 | MT230561 | ON209438 | MT222288 | - | Nguyen & Pham (2022) |
| Penicillium xyleborini | CMW 56800 | T | Penicillium | Ramosum | Soppiorum | South Africa | Beetle (Xyleborinus saxesenii) | MW504356 | MW480817 | MW480823 | MW480824 | - | Visagie & Yilmaz (2022) |
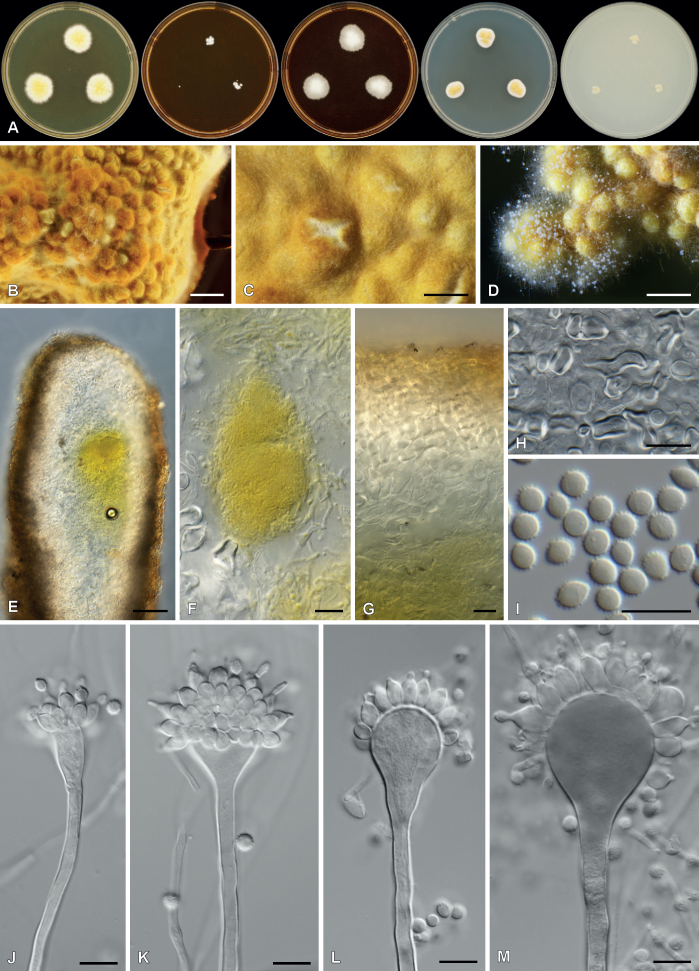
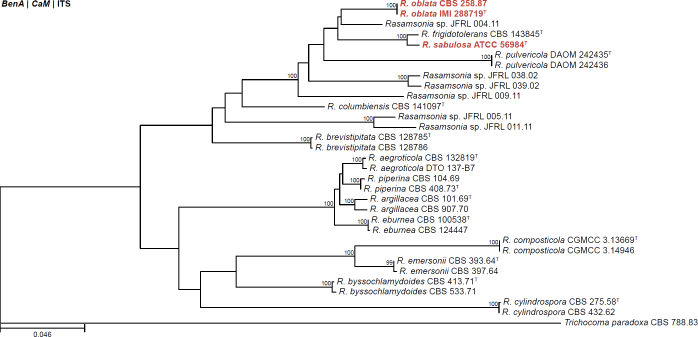
| Rasamsonia oblata | IMI 288719 | T | - | - | - | Australia | Spoiled baby food | LC546718 | LC546729 | LC546740 | - | - | Yanai et al. (2020) |
| CBS 258.87 | T | - | - | - | Australia | Spoiled baby food | OR142403 | OR145988 | OR145994 | - | - | Present study | |
| Rasamsonia sabulosa | ATCC 56984 | T | - | - | - | Australia | Spoiled fruit juice | LC546720 | LC546726 | LC546742 | - | - | Yanai et al. (2020) |
| Talaromyces africanus | CBS 147340 | T | - | Trachyspermi | - | South Africa | House dust | OK339610 | OK338782 | OK338808 | OK338833 | - | Pyrri et al. (2021) |
| Talaromyces albisclerotius | CBS 141839 | T | - | Trachyspermi | - | China | Soil | MN864276 | MN863345 | MN863322 | MN863334 | - | Sun et al. (2020b) |
| Talaromyces aspriconidius | CBS 141835 | T | - | Talaromyces | - | China | Soil | MN864274 | MN863343 | MN863320 | MN863332 | - | Sun et al. (2020b) |
| Talaromyces atkinsoniae | BRIP 72528a | T | - | Talaromyces | - | Australia | Gills of Marasmius crinisequi | OP059084 | OP087524 | - | OP087523 | - | Tan et al. (2022) |
| Talaromyces aureolinus | AS 3.15864 | - | Talaromyces | - | China | Soil | MK837954 | MK837938 | MK837946 | MK837962 | - | Wei et al. (2021) | |
| AS 3.15865 | - | Talaromyces | - | China | Soil | MK837953 | MK837937 | MK837945 | MK837961 | - | Wei et al. (2021) | ||
| NM6-1 | - | Talaromyces | - | China | Soil | MN059095 | MN059093 | MN059094 | MN059096 | - | Wei et al. (2021) | ||
| Talaromyces bannicus | AS 3.15862 | T | - | Talaromyces | - | China | Soil | MK837955 | MK837939 | MK837947 | MK837963 | - | Wei et al. (2021) |
| Talaromyces brevis | CBS 141833 | T | - | Talaromyces | - | China | Soil | MN864269 | MN863338 | MN863315 | MN863328 | - | Sun et al. (2020b) |
| CBS 118436 | - | Talaromyces | - | Marocco | Soil | MN864271 | MN863340 | MN863317 | MN863330 | - | Sun et al. (2020b) | ||
| DTO 307-C1 | - | Talaromyces | - | Turkey | Soil | MN864270 | MN863339 | MN863316 | MN863329 | - | Sun et al. (2020b) | ||
| Talaromyces calidominioluteus | CBS 113167 | - | Trachyspermi | - | Unknown | Air (indoor) | OK339611 | OK338785 | OK338816 | OK338836 | - | Pyrri et al. (2021) | |
| CBS 147313 | T | - | Trachyspermi | - | The Netherlands | Melon (imported from Brazil) | OK339612 | OK338786 | OK338817 | OK338837 | - | Pyrri et al. (2021) | |
| CBS 147341 | - | Trachyspermi | - | Iran | Grapevine | OK339602 | OK338788 | KU711896 | OK338839 | - | Pyrri et al. (2021) | ||
| CBS 147342 | - | Trachyspermi | - | Thailand | House dust | OK339600 | KP330045 | OK338815 | OK338835 | - | Pyrri et al. (2021) | ||
| DTO 265-H8 | - | Trachyspermi | - | Iran | Grapevine | OK339601 | OK338787 | KU711894 | OK338838 | - | Pyrri et al. (2021) | ||
| DTO 266-A5 | - | Trachyspermi | - | Iran | Grapevine | OK339603 | OK338789 | KU711900 | OK338840 | - | Pyrri et al. (2021) | ||
| DTO 269-H1 | - | Trachyspermi | - | Thailand | House dust | KJ775721 | KJ775214 | - | - | - | Pyrri et al. (2021) | ||
| DTO 269-H4 | - | Trachyspermi | - | Thailand | House dust | KJ775722 | KJ775215 | - | - | - | Pyrri et al. (2021) | ||
| DTO 270-A1 | - | Trachyspermi | - | Thailand | House dust | KJ775728 | KJ775221 | - | - | - | Pyrri et al. (2021) | ||
| DTO 270-C3 | - | Trachyspermi | - | Thailand | House dust | KJ775733 | KJ775226 | - | - | - | Pyrri et al. (2021) | ||
| DTO 390-E9 | - | Trachyspermi | - | Nigeria | Cocoa beans | MN788104 | MN787900 | MN787896 | OK338847 | - | Pyrri et al. (2021) | ||
| DTO 390-F1 | - | Trachyspermi | - | Nigeria | Cocoa beans | MN788103 | MN787901 | MN787895 | OK338848 | - | Pyrri et al. (2021) | ||
| DTO 390-I9 | - | Trachyspermi | - | Nigeria | Cocoa beans | MN788115 | MN787911 | MN787885 | OK338849 | - | Pyrri et al. (2021) | ||
| DTO 391-A5 | - | Trachyspermi | - | Nigeria | Cocoa beans | MN788111 | MN787914 | MN787883 | OK338850 | - | Pyrri et al. (2021) | ||
| Talaromyces cavernicola | URM 8448 | T | - | Talaromyces | - | Brazil | Air (cave) | ON862935 | OP672383 | OP290543 | OP290515 | - | Alves et al. (2022b) |
| URM 8449 | - | Talaromyces | - | Brazil | Air (cave) | ON862936 | OP672384 | OP290544 | OP290516 | - | Alves et al. (2022b) | ||
| Talaromyces chongqingensis | CBS 270.35 | - | Trachyspermi | - | China | Soil | OK339609 | OK338781 | OK338807 | OK338832 | - | Zhang et al. (2021b) | |
| CGMCC 3.20482 | T | - | Trachyspermi | - | China | Soil | MZ358001 | MZ361343 | MZ361350 | MZ361357 | - | Zhang et al. (2021b) | |
| CS26-63 | - | Trachyspermi | - | China | Soil | MZ358002 | MZ361344 | MZ361351 | MZ361358 | - | Zhang et al. (2021b) | ||
| CS26-73 | - | Trachyspermi | - | China | Soil | MZ358003 | MZ361345 | MZ361352 | MZ361359 | - | Zhang et al. (2021b) | ||
| CS26-75 | - | Trachyspermi | - | China | Soil | MZ358004 | MZ361346 | MZ361353 | MZ361360 | - | Zhang et al. (2021b) | ||
| NRRL 1064 | - | Trachyspermi | - | China | Soil | KM066172 | KM066129 | Zhang et al. (2021b) | |||||
| Talaromyces gaditanus | CBS 104.71 | - | Trachyspermi | - | The Netherlands | Tulip | OK339614 | OK338792 | OK338820 | OK338852 | - | Pyrri et al. (2021) | |
| CBS 144771 | - | Trachyspermi | - | The Netherlands | Sputum of cystic fibroses patient | OK339616 | OK338794 | OK338822 | OK338842 | - | Pyrri et al. (2021) | ||
| CBS 169.81 | T | - | Trachyspermi | - | Spain | Air | MH861318 | OK338775 | OK338802 | OK338827 | - | Pyrri et al. (2021) | |
| CBS 442.89 | - | Trachyspermi | - | Denmark | Soil | OK339615 | OK338793 | OK338821 | OK338853 | - | Pyrri et al. (2021) | ||
| CBS 444.89 | - | Trachyspermi | - | Denmark | Cranberry (imported from USA) | OK339597 | OK338776 | OK338803 | OK338828 | - | Pyrri et al. (2021) | ||
| CBS 996.72 | - | Trachyspermi | - | The Netherlands | Jute sugar bag | MH860641 | OK338774 | OK338813 | OK338826 | - | Pyrri et al. (2021) | ||
| CBS 138.84 | - | Trachyspermi | - | Spain | Apple (Malus sylvestris) | OK339604 | OK338791 | OK338819 | OK338851 | - | Pyrri et al. (2021) | ||
| Talaromyces germanicus | CBS 147314 | T | - | Trachyspermi | - | Germany | Indoor environment | OK339619 | OK338799 | OK338812 | OK338845 | - | Pyrri et al. (2021) |
| Talaromyces ginkgonis | CGMCC 3.20698 | T | - | Talaromyces | - | China | Fruit of Ginkgo biloba | OL638158 | OL689844 | OL689846 | OL689848 | - | Wang & Zhuang (2022a) |
| Talaromyces gwangjuensis | CNUFC WT19-1 | T | - | Purpurei | - | Republic of Korea | Freshwater | MK766233 | MZ318448 | - | MK912174 | - | Nguyen et al. (2021a) |
| CNUFC WT19-2 | - | Purpurei | - | Republic of Korea | Freshwater | MK766234 | MZ318449 | - | MK912175 | - | Nguyen et al. (2021a) | ||
| Talaromyces haitouensis | AS 3.16101 | T | - | Talaromyces | - | China | Soil | MZ045695 | MZ054634 | MZ054637 | MZ054631 | - | Han et al. (2021) |
| Talaromyces koreanus | CNUFC YJW2-13 | T | - | Helici | - | Republic of Korea | Freshwater | MZ315100 | MZ318450 | MZ332529 | MZ332533 | - | Nguyen et al. (2021a) |
| CNUFC YJW2-14 | - | Helici | - | Republic of Korea | Freshwater | MZ315101 | MZ318451 | MZ332530 | MZ332534 | - | Nguyen et al. (2021a) | ||
| Talaromyces nanjingensis | CCTCC-M-2012167 | T | - | Talaromyces | - | China | Soil | MW130720 | MW147759 | MW147760 | MW147762 | - | Sun et al. (2022b) |
| Talaromyces peaticola | CGMCC 3.18620 | T | - | Trachyspermi | - | China | Soil (peat) | MF135613 | MF284705 | MF284703 | MF284704 | - | Tian et al. (2021) |
| CGMCC 3.18767 | - | Trachyspermi | - | China | Soil (peat) | MF960857 | MF960859 | MF960861 | MF960863 | - | Tian et al. (2021) | ||
| CGMCC 3.18768 | - | Trachyspermi | - | China | Soil (peat) | MF960858 | MF960860 | MF960862 | MF960864 | - | Tian et al. (2021) | ||
| Talaromyces penicillioides | AS 3.15822 | T | - | Talaromyces | - | China | Soil | MK837956 | MK837940 | MK837948 | MK837964 | - | Wei et al. (2021) |
| Talaromyces pernambucoensis | URM 6894 | T | - | Trachyspermi | - | Brazil | Soil | LR535947 | LR535945 | LR535946 | LR535948 | - | Crous et al. (2019) |
| Talaromyces phuphaphetensis | TBRC 16281 | T | - | Trachyspermi | - | Thailand | Soil (cave) | ON692803 | ON706960 | ON706962 | ON706964 | - | Nuankaew et al. (2022) |
| Talaromyces pulveris | CBS 146831 | T | - | Purpurei | - | France | Bore dust of deathwatch beetle (Xestobium rufovillosum) infesting floorboards (Quercus wood) | MW175345 | MW173136 | MW173099 | MW173115 | - | Crous et al. (2020a) |
| Talaromyces resedanus | CBS 181.71 | T | - | Subinflati | - | Seychelles | Soil | MN864280 | MN863349 | MN863326 | MN969214 | - | Sun et al. (2020b) |
| Talaromyces rosarhiza | GUCC 190040.1 | T | - | Talaromyces | - | China | Endophyte of Rosa roxburghii | MZ221603 | MZ333143 | MZ333137 | MZ333141 | - | Zhang et al. (2021a) |
| GUCC 197011.1 | - | Talaromyces | - | China | Endophyte of Rosa roxburghii | MZ221604 | MZ333144 | MZ333138 | MZ333142 | - | Zhang et al. (2021a) | ||
| Talaromyces rufus | CBS 141834 | T | - | Talaromyces | - | China | Soil | MN864272 | MN863341 | MN863318 | MN863331 | - | Sun et al. (2020b) |
| DTO 274-C5 | - | Talaromyces | - | Republic of Korea | Soil | MN864273 | MN863342 | MN863319 | - | - | Sun et al. (2020b) | ||
| Talaromyces samsonii | CBS 137.84 | T | - | Trachyspermi | - | Spain | Apple (Malus sylvestris) | MH861709 | OK338798 | OK338824 | OK338844 | - | Pyrri et al. (2021) |
| CBS 147356 | - | Trachyspermi | - | The Netherlands | Soil | OK339598 | OK338777 | OK338804 | OK338829 | - | Pyrri et al. (2021) | ||
| CBS 147357 | - | Trachyspermi | - | Greece | Air (indoor) | OK339599 | OK338778 | OK338805 | OK338830 | - | Pyrri et al. (2021) | ||
| Talaromyces santanderensis | HF05 | T | - | Talaromyces | - | Colombia | Soil (cacao field) | OP082331 | OP067657 | OP067656 | OP067655 | - | Guerra Sierra et al. (2022) |
| Talaromyces satunensis | TBRC 16246 | T | - | Trachyspermi | - | Thailand | Soil (cave) | ON692804 | ON706961 | ON706963 | - | - | Nuankaew et al. (2022) |
| Talaromyces saxoxalicus | MUM20.30 | T | - | Purpurei | - | Portugal | Limestone walls | MT039882 | MT052003 | - | MT052004 | - | Trovao et al. (2021) |
| Talaromyces shilinensis | CGMCC 3.20699 | T | - | Talaromyces | - | China | Rotten twig | OL638159 | OL689845 | OL689847 | OL689849 | - | Wang & Zhuang (2022a) |
| Talaromyces sparsus | AS 3.15880 | T | - | Talaromyces | - | China | Soil | MK837958 | MK837942 | MK837950 | MK837966 | - | Wei et al. (2021) |
| Talaromyces teleomorphus | CNUFC YJW2-5 | T | - | Helici | - | Republic of Korea | Freshwater | MZ315102 | MZ318452 | MZ332531 | MZ332535 | - | Nguyen et al. (2021a) |
| CNUFC YJW2-6 | - | Helici | - | Republic of Korea | Freshwater | MZ315103 | MZ318453 | MZ332532 | MZ332536 | - | Nguyen et al. (2021a) | ||
| Talaromyces tenuis | CBS 141840 | T | - | Tenues | - | China | Soil | MN864275 | MN863344 | MN863321 | MN863333 | - | Sun et al. (2020b) |
| Talaromyces wushanicus | CGMCC 3.20481 | T | - | Talaromyces | - | China | Soil | MZ356356 | MZ361347 | MZ361354 | MZ361361 | - | Zhang et al. (2021b) |
| CS17-04 | - | Talaromyces | - | China | Soil | MZ356357 | MZ361348 | MZ361355 | MZ361362 | - | Zhang et al. (2021b) | ||
| CS17-06 | - | Talaromyces | - | China | Soil | MZ356358 | MZ361349 | MZ361356 | MZ361363 | - | Zhang et al. (2021b) | ||
| Talaromyces yunnanensis | KUMCC 18-0208 | T | - | Talaromyces | - | China | Soil | MT152339 | MT161683 | MT178251 | - | - | Doilom et al. (2020) |
| Talaromyces zhenhaiensis | AS 3.15693 | - | Talaromyces | - | China | Soil | KY007094 | KY007110 | KY007102 | KY112592 | - | Han et al. (2021) | |
| AS 3.16102 | T | - | Talaromyces | - | China | Soil | MZ045697 | MZ054636 | MZ054639 | MZ054633 | - | Han et al. (2021) |
Datasets were aligned in MAFFT v. 7.490 (Katoh & Standley 2013) using the G-INS-i option. For multigene phylogenies, datasets were concatenated in Geneious Prime v. 2023.0.1 (Biomatters, NZ). Partitionfinder v. 2.1.1 (Lanfear et al. 2017) was used to select the partitioning schemes and nucleotide substitution models for each alignment, with exons, introns, and codon positions treated as independent datasets. Maximum Likelihood phylogenies were computed in IQ-TREE v. 2.1.3 (Minh et al. 2020) with support in nodes calculated using UFBoot (Hoang et al. 2018) ultrafast bootstrapping with 1 000 replicates, as implemented in IQ-TREE. Phylogenies were visualised in TreeViewer v. 2.0.1 (https://treeviewer.org) and further edited for publication in Affinity Publisher v. 2.0.3 [Serif (Europe) Ltd, Nottingham, UK]. Here we use a phylogenetic species concept and apply Genealogical Concordance Phylogenetic Species Recognition (GCPSR; Taylor et al. 2000).
Morphology
Descriptions were made following the recommendations of Samson et al. (2014) and Visagie et al. (2014). Strains were inoculated at three-equidistant points onto Czapek yeast autolysate agar (CYA), CYA with 5 % NaCl (CYAS), CYA with 20 % sucrose (CY20S), malt extract agar (MEA), yeast extract sucrose agar (YES), dichloran 18 % glycerol agar (DG18), oatmeal agar (OA), and Creatine sucrose agar (CREA). These were incubated at 25 °C for 7 d, with additional CYA plates incubated at 20, 30 and 37 °C. A subset of plates were incubated for longer periods to observe possible sexual states. After incubation, the species were characterised based on their growth rates, colony characteristics and microscopic features. Colonies were photographed using a Sony A6400 camera and a Sony SEL50M28 lens (Tokyo, Japan). Microscopic observations were made using a Nikon SMZ25 stereomicroscope (Tokyo, Japan) and Zeiss AXIO Imager.A2 compound microscope (Carl Zeiss CMP, Göttingen, Germany), both equipped with Nikon DS-Ri2 cameras and using Nikon Elements D v. 5.11 software. The photoplates were created in Affinity Photo v. 2.0.3 [Serif (Europe), Nottingham, UK].
RESULTS
Phylogenetic analyses
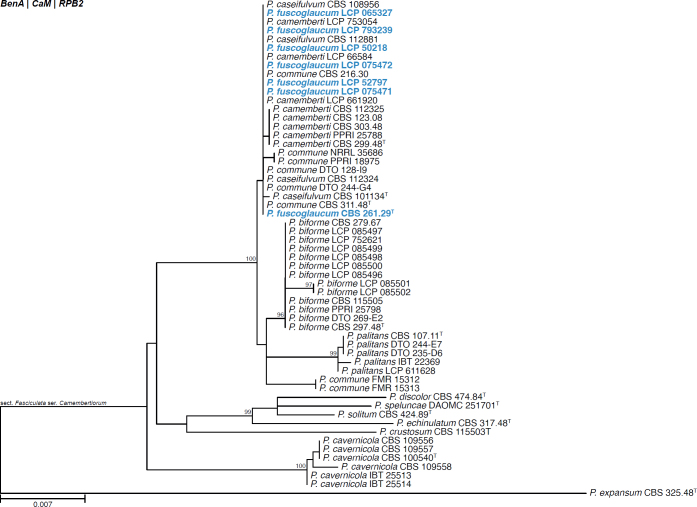
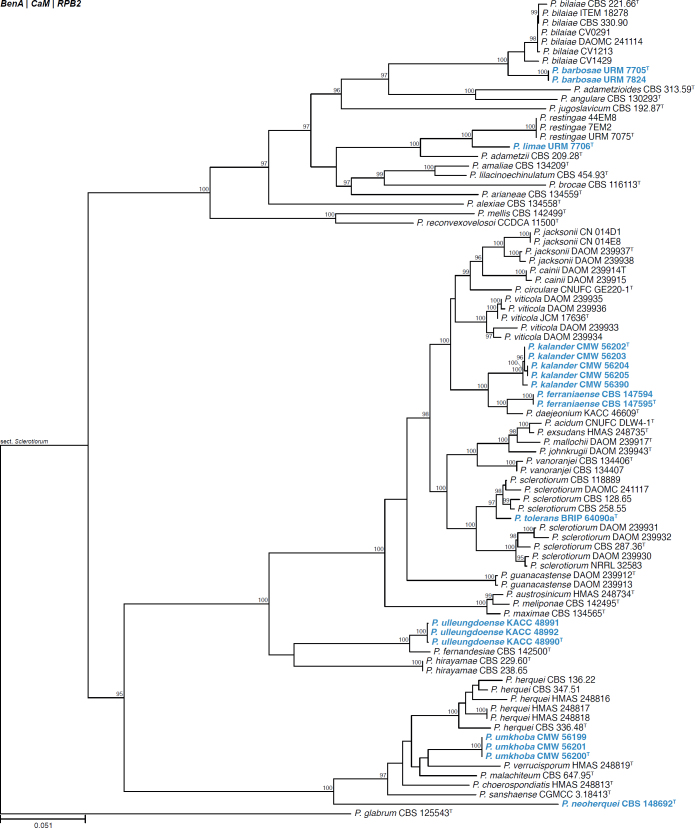
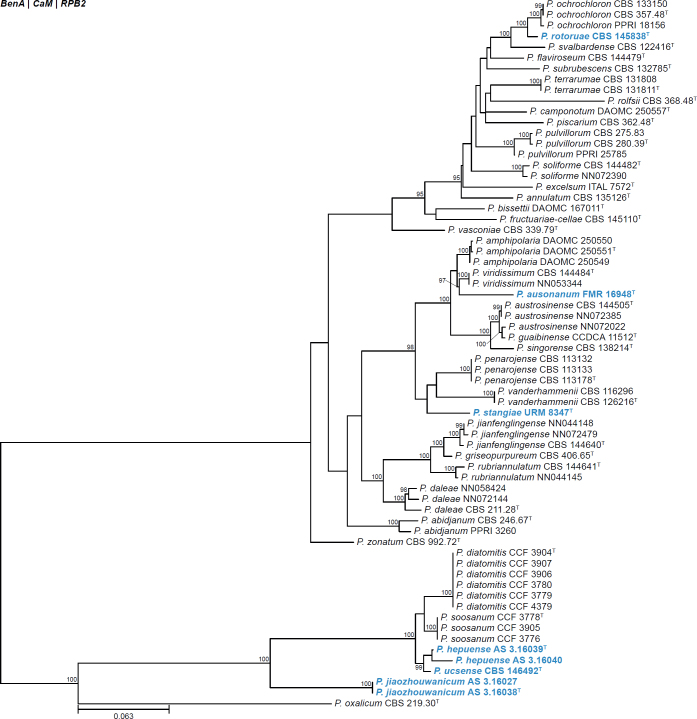
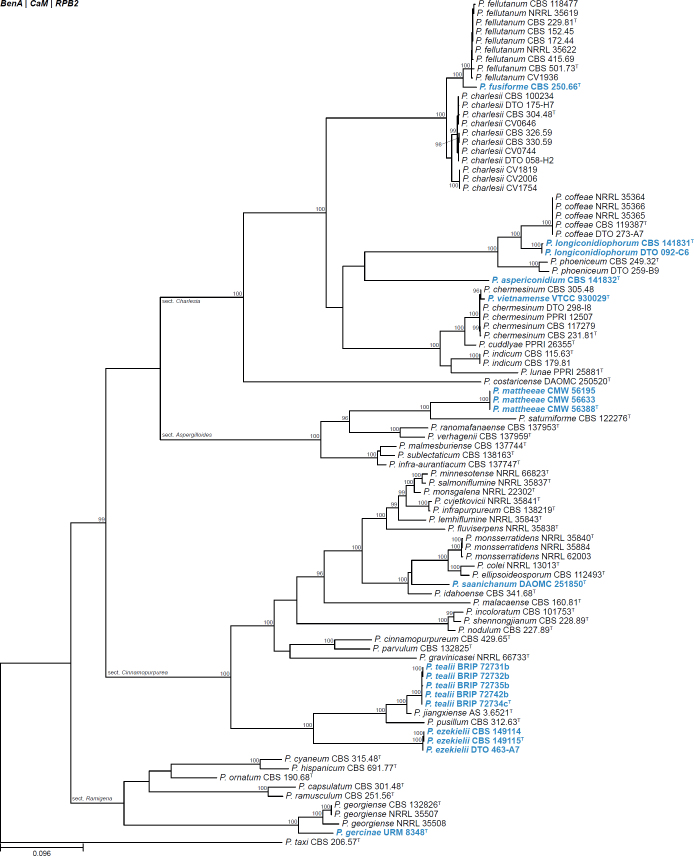
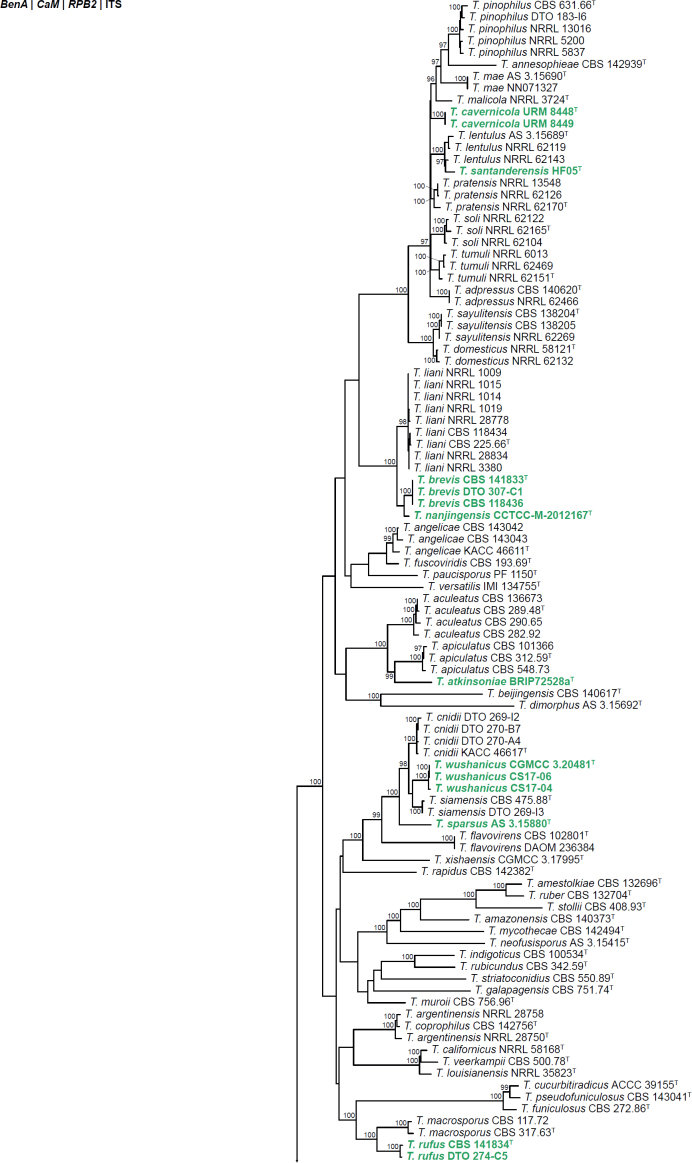
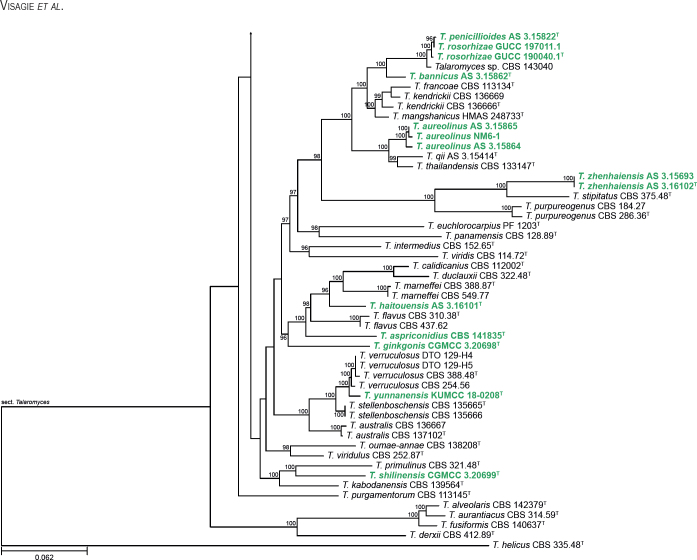
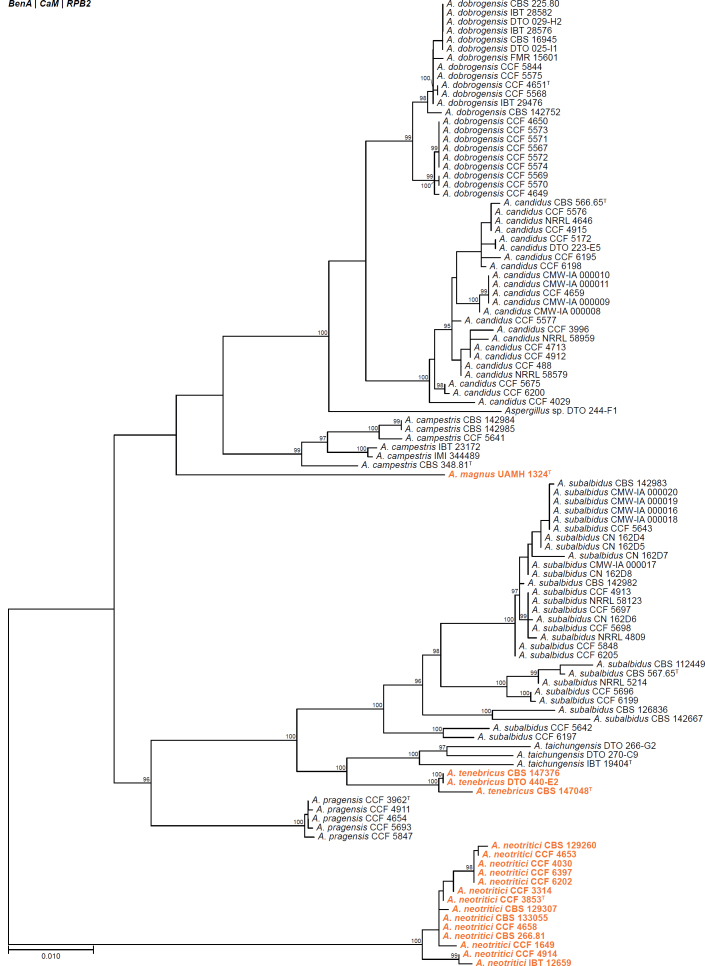
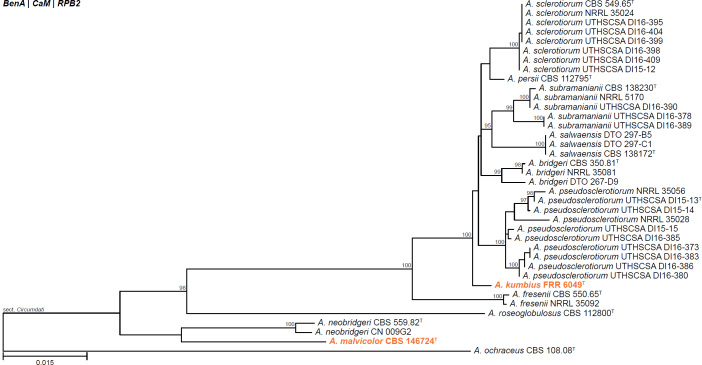
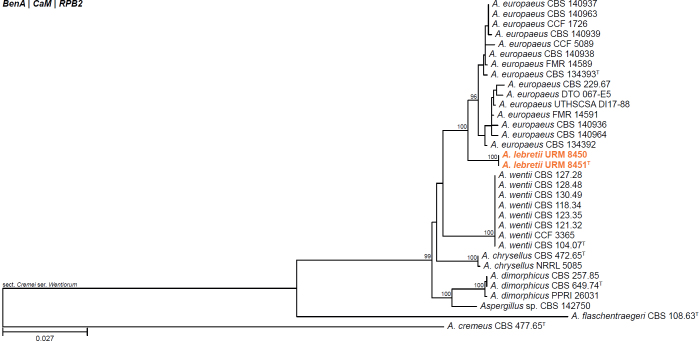
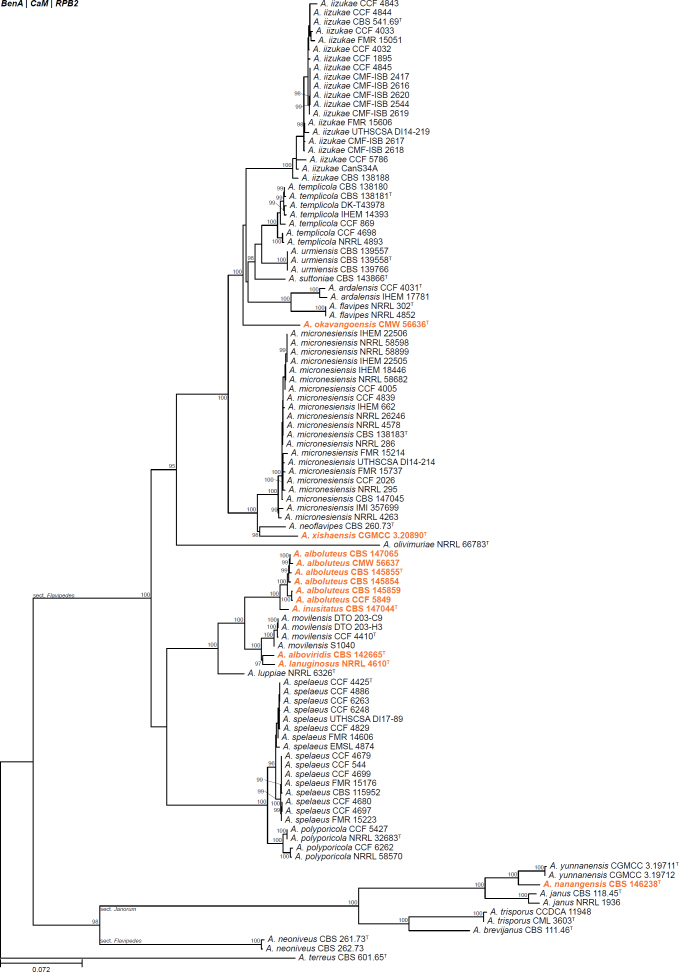
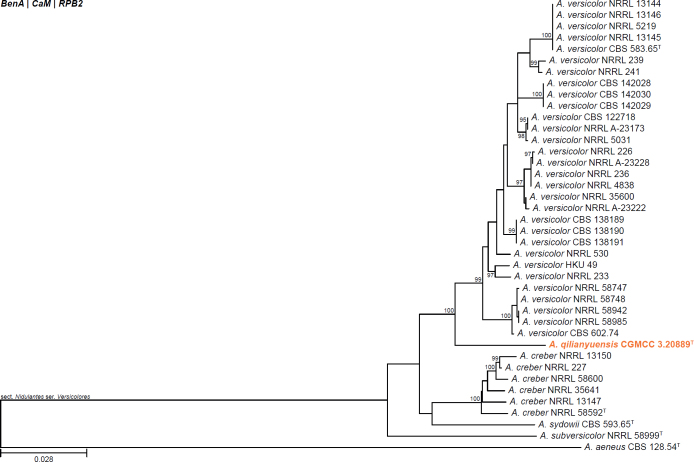
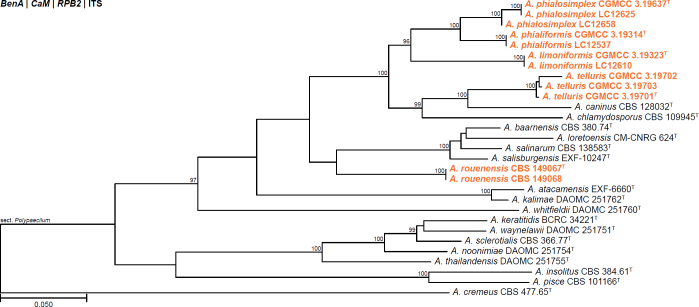
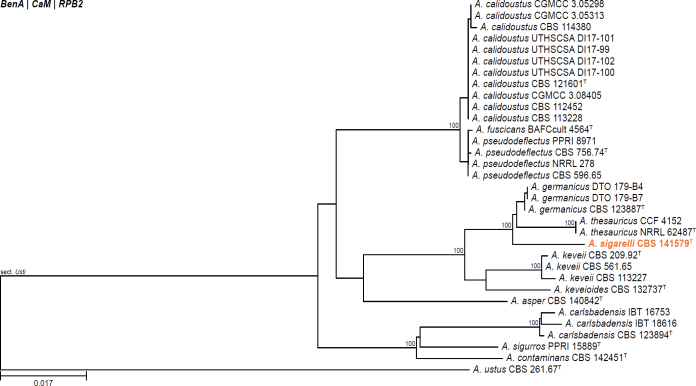
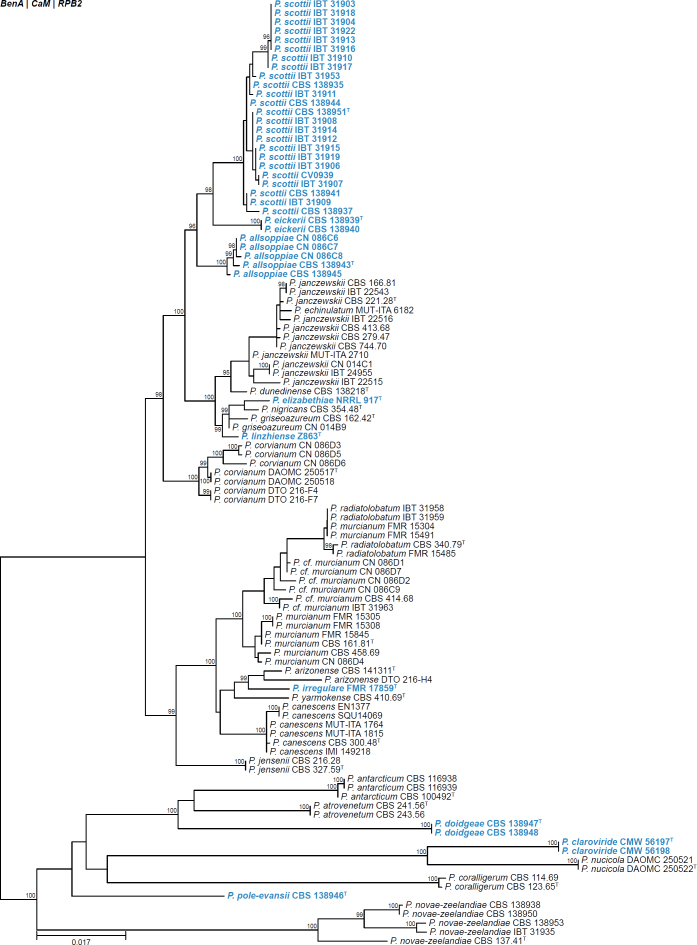
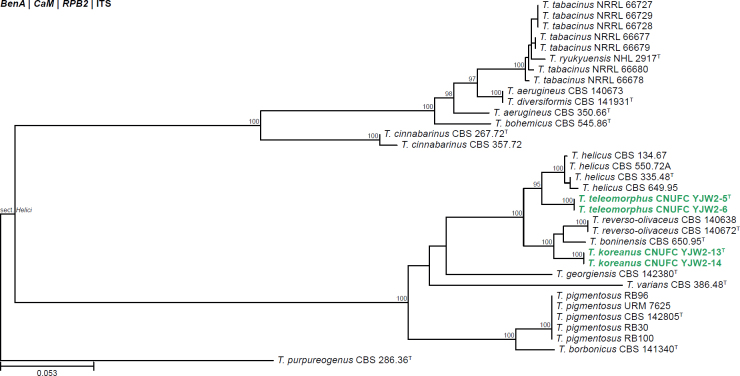
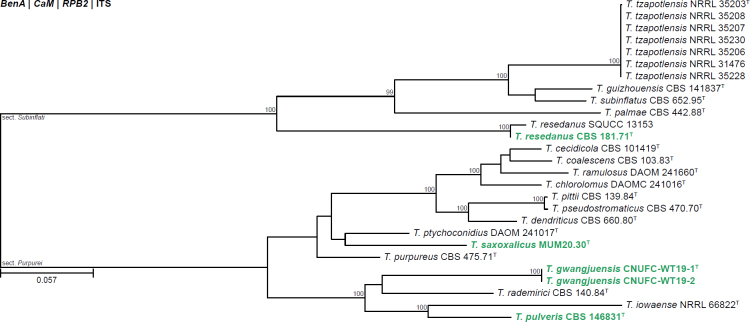
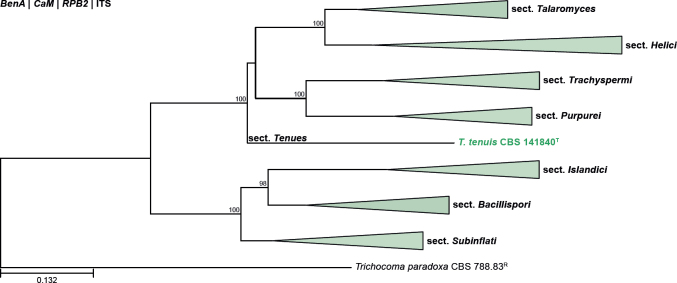
All alignments, partitioning schemes, model tests, and phylogenetic trees were deposited in the University of Pretoria research data repository hosted on Figshare (doi: 10.25403/UPresearchdata.23723277). The phylogenies largely confirmed the novelty of the newly introduced species, with some exceptions commented on using notes in the species list below (Figs 1–33, Suppl. Figs S1–S30).
Fig. 1.
Phylogenetic tree of Aspergillus section Aenei series Aenei based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. versicolor. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S1.
Fig. 33.
Phylogenetic tree of Talaromyces section Trachyspermi, based on a concatenated dataset of BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to T. purpureus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T.
TAXONOMY
Here we present new additions and changes to the list of accepted species published in Houbraken et al. (2020) and provide an overview of the 160 species described (51 Aspergillus, three Emericella, one Neosartorya, two Paecilomyces, one Paraxerochrysium, 64 Penicillium, two Rasamsonia, 35 Talaromyces and one Xerochrysium) and the new genus Paraxerochrysium. At the continental level, species were described from Africa (n = 28), Asia (n = 57), Europe (n = 36), North America (n = 15), Oceania (n = 21) and South America (n = 19).
These originated from 42 countries with most coming from China (n = 39), Australia (n = 20), South Africa (n = 16), Brazil (n = 16), Spain (n = 12), and the USA (n = 8). Species were described from a wide range of substrates, mainly soil (n = 91), plant material (n = 15), food and feed (n = 14), air (n = 12) and indoor environments (n = 8). Of the 160, 22 were classified as synonyms and four as doubtful species. Ten were invalidly described. Two of these names are validated below, the remaining eight are considered synonyms or doubtful species. We consider the four combinations introduced by Pitt & Hocking (2022) contradictory to our view on Aspergillus and the sexual (teleomorphic) genera associated with it (Samson et al. 2014, Kocsube et al. 2016). We also document species that were previously accepted but then subsequently reduced as synonyms due to various taxonomic revisions. Finally, species accepted in Houbraken et al. (2020) for which no DNA sequence data are available are now listed as doubtful. Taking these changes into account, we currently accept 453 Aspergillus, 12 Paecilomyces, 535 Penicillium, 14 Rasamsonia, 203 Talaromyces and four Xerochrysium species.
Aspergillus series Annuorum J.J. Silva, Iamanaka & Frisvad ser. nov. MycoBank MB 849339. Fig. 6 & Suppl. Fig. S6.
Fig. 6.
Phylogenetic tree of Aspergillus section Flavi series Alliacei and Flavi based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by superscript R. See Suppl. Fig. S6.
Synonym: Aspergillus ser. Annuorum [nom. inval. Art. 40.1 (Shenzhen)] J.J. Silva, Fungaro, Frisvad, Taniwaki & Iamanaka, J. Fungi 8 (no 1279): 14. 2022. MycoBank MB 845971.
In: Aspergillus subgen. Circumdati sect. Flavi.
Typus: Aspergillus annui J.J. Silva, Fungaro, Frisvad, Taniwaki & Iamanaka [MB 849336].
Etymology: Named after Aspergillus annui.
Description: See Silva et al. (2022).
Aspergillus annui J.J. Silva, Fungaro, Frisvad, Taniwaki & Iamanaka, sp. nov. MycoBank MB 849336. Fig. 6 & Suppl. Fig. S6.
Synonym: Aspergillus annui [nom. inval. Art. 40.7 & Art. 40.8 (Shenzhen)] J.J. Silva, Fungaro, Frisvad, Taniwaki & Iamanaka, J. Fungi 8 (no 1279): 14. 2022. MycoBank MB 845969.
In: Aspergillus subgen. Circumdati sect. Flavi ser. Annuorum.
DNA barcodes: ITS = OP691228; BenA = ON529842; CaM = ON529841; RPB2 = ON529843.
Etymology: The specific epithet refers to the substrate from which it was isolated, paprika pepper (Capsicum annuum).
Typus: Brazil, São Paulo State, São Paulo City, 23°35’29.7”S 46°40’52.1”W, sweet paprika pepper (Capsicum annuum), 6 Apr. 2017, isol. C.A. Yasumura (holotype IBT 36122 preserved as a metabolically inactive culture, culture ex-type 365-IT-PPK = IBT 36122).
Description: See Silva et al. (2022).
Notes: This species was invalidly described because Silva et al. (2022) cited two holotypes and failed to mention that the holotype was metabolically inactive. This also means that series Annuorum introduced for A. annui is invalid. Here we validate both the species and the series.
Aspergillus saccharicola J.J. Silva, Frisvad, Fungaro, Taniwaki & Iamanaka, sp. nov. MycoBank MB 849338. Fig. 6 & Suppl. Fig. S6.
Synonym: Aspergillus saccharicola [nom. inval. Art. 40.7 & Art. 40.8 (Shenzhen)] J.J. Silva, Frisvad, M.H.P. Fungaro, M.H. Taniwaki & B.T. Iamanaka, J. Fungi 8 (no 1279): 16. 2022. MycoBank MB 845970.
In: Aspergillus subgen. Circumdati sect. Flavi ser. Flavi.
DNA barcodes: ITS = OP611470; BenA = ON529845; CaM = ON529844; RPB2 = ON529846.
Etymology: The specific epithet refers to the substrate from which it was isolated, sugarcane (Saccharum officinarum).
Typus: Brazil, São Paulo State, São Paulo City, 23°35’29.7”S 46°40’52.1”W, sugarcane (Saccharum officinarum), 14 Sep. 2011, isol. B.T. Imanaka (holotype IBT 36126 preserved as a metabolically inactive culture, culture ex-type 117-IT-SGC = IBT 36126).
Description: See Silva et al. (2022).
Notes: This species was invalidly described because Silva et al. (2022) cited two holotypes and failed to mention that the holotype was metabolically inactive. Here we validate the species.
Aspergillus section Dichlaena Visagie, Kocsubé & Houbraken, sect. nov. MycoBank MB 849488. Fig. 14.
Fig. 14.
Phylogenetic tree showing the relationship of Dichlaena within Aspergillus based on a concatenated dataset of BenA, CaM, RPB2, RPB1, ITS and LSU. Strains of recently described species are shown in bold coloured text. The tree was rooted to Trichocoma paradoxa. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by superscript R.
In: Aspergillus subgenus Circumdati.
Etymology: Named after the genus Dichlaena.
Typus: Aspergillus lentisci (Durieu & Mont.) Visagie, Malloch, L. Kriegsteiner, Samson & Houbraken [MB 849485].
Description: We follow the Malloch & Cain (1972) description of Dichlaena and apply this to the section.
Notes: The genus Dichlaena is synonymised with Aspergillus. Aspergillus lentisci (bas. Dichlaena lentisci) strains resolve as a distinct clade, with section Petersoniorum (subg. Circumdati) members being the closest relatives (Fig. 14). Aspergillus lentisci strains are strongly xerophilic, similar to Aspergillus sections Aspergillus and Restricti, while section Petersoniorum species also grow well on low water activity media. We thus introduce this as a new section. The section is based on the single species, Aspergillus lentisci. The sexual state for this xerophilic species has not been observed in culture, but was documented in detail by Malloch & Cain (1972) who observed it from leaves, twigs and dung. Three other species have been described in Dichlaena. Dichlaena bovina [MB 329872] was considered a synonym of Thielavia bovina [MB 240118] by Malloch & Cain (1973), while we consider Dichlaena indica [MB 127024] and Dichlaena pterodontis [MB 312952] doubtful species with no material available for study.
Aspergillus lentisci (Durieu & Mont.) Visagie, Malloch, L. Kriegsteiner, Samson & Houbraken comb. nov. MycoBank MB 849485. Figs 34 & 35.
Fig. 34.
Aspergillus lentisci in nature. A, B. Ascomata on dried leaf material. C, D. Ascomata observed under light microscope. E. Outer cell layer of ascomata. F. Ascospores released from ascomata. G. Ascospores. Scale bars = 10 µm.
Fig. 35.
Aspergillus lentisci in culture. A. Colonies on, from left to right, MY50G, MY20, MY40, DG18 and OA. B–D. Colony overview on DG18 showing sclerotia and conidiophores (D). E, F. Section of sclerotia showing potential early development of asci and ascomata. G. Section of sclerotia showing its wall structure. H. Hülle cells from sclerotia. I. Conidia. J–M. Conidiophores. Scale bars: B = 2 mm; C, D = 500 µm; E = 50 µm; F–H = 20 µm; I–M = 10 µm.
Basionym: Dichlaena lentisci Durieu & Mont., Expl. Sci. l’Algérie 1 (11): 405. 1849. MycoBank MB 249716.
In: Aspergillus subgen. Circumdati sect. Dichlaena
DNA barcodes: ITS = OR142402; BenA = OR145977; CaM = OR145992; RPB2 = OR146003; LSU = OR142413.
Typus: Durieu & Mont., Expl. Sci. l’Algérie 1 (11): 405, tab. 22 bis, fig. 2, lectotype designated here, MBT10014202. Portugal, Aljezur, Pistacia leaf, Apr. 2019, coll. L. Kriegsteiner, isol. R.A. Samson [epitype designated here, CBS H-25292 (dried culture of CBS 150189), MBT10014196, culture ex-epitype CBS 150189 = DTO 426-E9].
Additional strains examined: Portugal, Aljezur, Pistacia leaf, Apr. 2019, coll. L. Kriegsteiner, isol. R.A. Samson, cultures DTO 426-F1, DTO 426-F2, and DTO 426-F3.
Colony diam, 25 °C (7 d, in mm): CYA no growth; MEA no growth; OA no growth; YES no growth; MY20 no growth; MY40 microcolonies–6; MY50G 5–10. After 14 d (in mm): CYA no growth; MEA no growth; OA microcolonies–6; YES no growth; MY20 no growth to microcolonies; MY40 9–17; MY50G 13–20; DG18 5–13; CREA no growth.
Colony characters (25 °C, 14 d): DG18, colonies floccose; mycelial areas white to yellowish; ascocarp bearing stromata light yellow to orange yellow to greyish yellow (4A4–6–B6), globose, sclerotiod, 600–1 200(−1 900) µm; sporulation absent to sparse, becoming moderate with prolonged incubation, white to cream; exudate sometimes present, clear; soluble pigment brown, inconspicuous; reverse yellowish white (4A2) to light brown to yellowish brown to brown (5D7–E8–6E8). MY50G, colonies floccose; mycelial areas white to yellowish; ascocarp bearing stromata light yellow to orange yellow to greyish yellow (4A4–6–B6), globose, sclerotiod, 600–1 200(−1 900) µm; sporulation sparse, white to cream; exudate absent; soluble pigment absent; reverse yellowish white (4A2) to greyish orange (5B6). MY40, colonies floccose; mycelial areas white to yellowish; sporulation absent; exudate absent; soluble pigment absent.
Micromorphology: Conidial heads radiate; conidiophores uniseriate; stipes hyaline, minor proportion pigmented, smooth, 200–830 × 4–8 μm; vesicles subglobose, sometimes clavate, phialides cover 50–75 % of head, 11–22(−32) μm wide; phialides ampulliform, 6.5–13 × 2.5–4(−4.5) μm; conidia globose to subglobose, some tapering towards one end, rough to echinulate, 3.5–5 × 3–4.5 (4.25 ± 0.27 × 3.88 ± 0.27, n = 50) μm, length/width 0.91 ± 0.1. Ascomata not produced in culture, in nature fide Malloch & Cain (1972), ‘arising from coiled or twisted initials at one to three points in the stroma, completely filling the stroma at maturity, irregular in shape, colorless to pale yellow; stroma tissue hard and stony, of two types when young; stromal cells of the outer tissue pale yellow, very thick-walled, pyriform to globose, often nearly open at one end, about 5–13 µm in diameter; stromal cells of the inner tissue hyaline, very thick-walled (but thinner-walled than those of the outer tissue), irregular in shape, usually elongated, often greater than 40 µm length, up to 15 µm broad; ascocarp peridium hyaline to pale yellow, consisting of a filamentous tissue with a membranous “backing”; asci irregularly disposed, ellipsoidal, short-stipitate, eight-spored, evanescent, 9 × 7 µm [fide Von Höhnel (1910)]; ascospores oblate, hyaline, smooth, with an inconspicuous equatorial band, 3.0–4.2 × 2.0–3.5 µm’.
Rasamsonia oblata (Pitt & A.D. Hocking) Yanai & Udagawa, Jap. J. Mycol. 61: 93. 2020. MycoBank MB 836491. Figs 28, 36 & Suppl. Fig. S26.
Fig. 28.
Phylogenetic tree of Rasamsonia, based on a concatenated dataset of BenA, CaM and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to Trichocoma paradoxa. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S26.
Fig. 36.
Rasamsonia oblata. A. Colonies on, from left to right, CYA, MEA, YES, DG18 and OA. B–D. Colony overview on MEA (B) and OA (C, D). E–I. Conidiophores. J. Conidia. Scale bars: B = 2.5 mm; C, D = 1 mm; E–J = 10 µm.
Basionym: Penicillium oblatum Pitt & A.D. Hocking, Mycologia 77: 810. 1985. MycoBank MB 104603.
DNA barcodes: ITS = LC546718; BenA = LC546729; CaM = LC546740.
Typus: Australia, New South Wales, Sydney, from spoiled baby food, 1979, isol. A.D. Hocking (holotype FRR 2234, culture ex-type CBS 258.87 = IMI 288719 = NBRC 33091).
Colony diam, 25 °C (7 d, in mm): CYA 20 °C 8–9; CYA 8–9; CYA 30 °C 10–12; CYA 37 °C 4–5; CYAS no growth; CREA no growth; DG18 14–15; MEA 29–32; OA 22–23; YES 9–10.
Colony characters: CYA 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white; texture funiculose; sporulation very sparse, conidia en masse not determined; soluble pigments absent; exudates absent; reverse pale yellow (3A2). MEA 25 °C, 7 d: Colonies deep, plane; margins subsurface, wide; mycelia white; texture funiculose; sporulation sparse to moderately dense, conidia en masse Brown to Dark Blonde (5D4); soluble pigments absent; exudates absent. YES 25 °C, 7 d: Colonies deep, plane; margins moderately deep, narrow, entire; mycelia white; texture funiculose; sporulation very sparse, conidia en masse not determined; soluble pigments absent; exudates absent; reverse pale yellow (4A2). DG18 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white; texture funiculose; sporulation very sparse, conidia en masse not determined; soluble pigments absent; exudates absent; reverse pale yellow (3A2). OA 25 °C, 7 d: Colonies low, plane; margins subsurface, wide; mycelia white; texture funiculose; sporulation sparse to moderately dense, conidia en masse Brown to Dark Blonde (5D4); soluble pigments absent; exudates absent.
Micromorphology: Conidiophores monoverticillate, small proportion biverticillate; stipes rough walled, some finely roughened, nonvesiculate, 10–28 × 2.5–3 μm; metulae 2–3 per stipe, 9.5–14 × 2.5–3 μm; phialides acerose, in verticils of 3–6, smooth to roughened, 7–8.5(−9.5) × 2–3 μm; conidia smooth, broadly ellipsoid, some subglobose, 2–2.5 × 1.5–2 μm (2.14 ± 0.1 × 1.67 ± 0.1), average width/length = 0.78, n = 50, connected in chains with distinct connectors, connected on short end.
Notes: Pitt & Hocking (1985a) described Penicillium oblatum and referred to it as an interface species, noting that its ‘rough-walled stipes bearing appressed monoverticillate penicilli and its brown, often oblate conidia distinguish it from other Penicillia’, but that its conidial and phialide shape preclude it from Paecilomyces or Geosmithia. Both Samson et al. (2011) and Yilmaz et al. (2014) mentioned that the ex-type strain (CBS 258.87) was contaminated and was not representative of the original description. Recently, Yanai et al. (2020) analysed sequences generated for P. oblatum (ex-type strains IMI 288719 and NBRC 33091) and P. sabulosum (ex-type strain ATCC 56984; CBS 261.87 is contaminated), showed that they belong to Rasamsonia, and provided new combinations as R. oblata and R. sabulosa. They also provided new morphological descriptions in Japanese. We were able to purify CBS 258.87 and provide an English description above based on this strain.
Xerochrysium coryli (Crous & Decock) Visagie & Houbraken, comb. nov. MycoBank MB 849335. Fig. 16.
Fig. 16.
Phylogenetic tree showing the relationship of Xerochrysium coryli (bas. Paraxerochrysium coryli) within Aspergillaceae based on a concatenated dataset of BenA, RPB2, ITS and LSU. Strains of recently described species are shown in bold coloured text. The tree was rooted to Talaromyces amestolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T.
Basionym: Paraxerochrysium coryli Crous & Decock, Persoonia 47: 261. 2021. MycoBank MB 841830.
DNA barcodes: ITS = OK664748; BenA = OK651216; RPB2 = OK651178.
Typus: Belgium, Louvain-la-Neuve, from dry hazelnut (Corylus avellana), Feb. 2021, C. Decock (holotype CBS H-24853, culture ex-type CBS 148314 = CPC 41272 = MUCL 58103).
Description: See Crous et al. (2021b).
Notes: A multigene phylogenetic analysis of Aspergillaceae based on BenA, RPB2, ITS and LSU showed that Paraxerochrysium coryli resolves as a unique species inside Xerochrysium (Fig. 16). We thus synonymise Paraxerochrysium with Xerochrysium and provide a new combination here.
Current Eurotiales families, genera and subgeneric classifications (* indicates recently introduced sections or series)
-
Aspergillaceae
Aspergillago
-
Aspergillus
-
subgenus Aspergillus
-
section Aspergillus
series Aspergillus, Chevalierorum, Leucocarpi, Rubri, Tamarindosolorum, Teporium and Xerophili
-
section Restricti
series Halophilici, Penicillioides, Restricti and Vitricolarum
-
-
subgenus Circumdati
-
section Candidi
series Candidi
-
section Circumdati
series Circumdati, Sclerotiorum and Steyniorum
section Dichlaena*
-
section Flavi
series Alliacei, Annuorum*, Avenacei, Bertholletiarum, Coremiiformes, Flavi, Kitamyces, Leporum and Nomiarum
-
section Flavipedes
series Flavipedes, Neonivei, Olivimuriarum and Spelaei
-
section Janorum
series Janorum
-
section Nigri
series Carbonarii, Heteromorphi, Homomorphi, Japonici and Nigri
-
section Petersoniorum
series Petersoniorum
-
section Robusti
series Robusti
-
section Tannerorum
series Tannerorum
-
section Terrei
series Ambigui, Nivei and Terrei
-
-
subgenus Cremei
-
section Cremei
series Arxiorum, Brunneouniseriati, Cremei, Inflati, Pulvini and Wentiorum
-
-
subgenus Fumigati
-
section Cervini
series Acidohumorum and Cervini
-
section Clavati
series Clavati
-
section Fumigati
series Brevipedes, Fennelliarum, Fumigati, Neoglabri, Spathulati, Thermomutati, Unilaterales and Viridinutantes
-
section Vargarum
series Vargarum
-
-
subgenus Nidulantes
-
section Aenei
series Aenei
-
section Bispori
series Bispori
-
section Cavernicolarum
series Cavernicolarum, Egyptiaci and Hainanici*
-
section Nidulantes
series Aurantiobrunnei, Multicolores, Nidulantes, Speluncei, Stellati, Unguium and Versicolores
-
section Ochraceorosei
series Funiculosi and Ochraceorosei
-
section Raperorum
series Raperorum
-
section Silvatici
series Silvatici
-
section Sparsi
series Biplani, Conjuncti, Implicati and Sparsi
-
section Usti
series Calidousti, Deflecti, Monodiorum and Usti
-
-
subgenus Polypaecilum
-
section Polypaecilum
series Canini, Kalimarum, Noonimiarum, Polypaecilum, Salinarum and Whitfieldiorum
-
-
Evansstolkia
Hamigera
Leiothecium
Monascus
Paraxerochrysium
Penicilliopsis
-
Penicillium
-
subgenus Aspergilloides
-
section Alfrediorum
series Alfrediorum
-
section Aspergilloides
series Fortuita, Glabra, Hoeksiorum, Improvisa, Kiamaensia, Livida, Longicatenata, Pinetorum, Quercetorum, Saturniformia, Spinulosa, Sublectatica, Thiersiorum, Thomiorum and Verhageniorum
-
section Charlesia
series Costaricensia, Fellutana, Indica and Phoenicea
-
section Cinnamopurpurea
series Cinnamopurpurea, Idahoensia, Jiangxiensia and Nodula
-
section Citrina
series Citrina, Copticolarum, Euglauca, Gallaica, Paxillorum, Roseopurpurea, Sheariorum, Sumatraensia, Vascosobrinhoana* and Westlingiorum
-
section Crypta
series Crypta
-
section Eremophila
series Eremophila
-
section Exilicaulis
series Alutacea, Citreonigra, Corylophila, Erubescentia, Lapidosa and Restricta
-
section Gracilenta
series Angustiporcata, Estinogena, Gracilenta and Macrosclerotiorum
-
section Griseola
series Griseola
-
section Inusitata
series Inusitata
-
section Lanata-Divaricata
series Dalearum, Janthinella, Oxalica, Rolfsiorum and Simplicissima
-
section Lasseniorum
series Lasseniorum
-
section Ochrosalmonea
series Ochrosalmonea
-
section Ramigena
series Georgiensia and Ramigena
-
section Sclerotiorum
series Adametziorum, Herqueorum and Sclerotiorum
-
section Stolkia
series Stolkia
-
section Thysanophora
series Thysanophora
-
section Torulomyces
series Torulomyces
-
-
subgenus Penicillium
-
section Brevicompacta
series Brevicompacta, Buchwaldiorum, Olsoniorum and Tularensia
-
section Canescentia
series Atroveneta and Canescentia
-
section Chrysogena
series Aethiopica, Chrysogena, Crustacea, Goetziorum and Persicina
-
section Eladia
series Eladia
-
section Fasciculata
series Camembertiorum, Corymbifera, Gladioli, Verrucosa and Viridicata
-
section Formosana
series Formosana
-
section Osmophila
series Osmophila and Samsoniorum
-
section Paradoxa
series Atramentosa and Paradoxa
-
section Penicillium
series Clavigera, Digitata, Italica, Penicillium and Sclerotigena
-
section Ramosum
series Lanosa, Raistrickiorum, Scabrosa, Soppiorum and Virgata
-
section Robsamsonia
series Claviformia, Glandicolarum, Robsamsonia and Urticicola
-
section Roquefortorum
series Roquefortorum
-
section Turbata
series Turbata
-
-
Phialomyces
Pseudohamigera
Pseudopenicillium
Sclerocleista
Warcupiella
Xerochrysium
Xeromyces
-
incertae sedis
Dendrosphaera
-
Penicillaginaceae
Penicillago
-
Thermoascaceae
Paecilomyces
Thermoascus
-
Trichocomaceae
Acidotalaromyces
Ascospirella
Rasamsonia
Sagenomella
-
Talaromyces
section Bacillispori
section Helici
section Islandici
section Purpurei
section Subinflati
section Talaromyces
section Tenues*
section Trachyspermi
Thermomyces
Trichocoma
List of species described since Houbraken et al. (2020)
Aspergillus agricola [nom. inval. Art. 40.8 (Shenzhen)] P. Singh, K.A. Callicott, M.J. Orbach & P.J. Cotty, Front. Microbiol. 11, 1236: 7. 2020. [MB 830377]. — Type: NRRL 66869 (holotype). Ex-type: NRRL 66869 = CR9-G = A2400. Infragen. class.: subgen. Circumdati sect. Flavi ser. Flavi. DNA barcodes: ITS = n.a.; BenA = n.a.; CaM = MN987053; RPB2 = n.a.
Synonym of: Aspergillus flavus Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809. [MB 209842].
Notes: This species was invalidly described. The CaM phylogeny resolved this species within A. flavus (Suppl. Fig. S6). We therefore consider it a synonym of A. flavus and choose not to validate this species.
Aspergillus alboluteus F. Sklenar, Jurjević, Ezekiel, Houbraken & Hubka, Stud. Mycol. 99 (no. 100120): 19. 2021. [MB 839382]. — Type: PRM 952200 (holotype). Ex-type: CBS 145855 = EMSL 2420 = CCF 5695 = IFM 66815. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Spelaei. DNA barcodes: ITS = MW448663; BenA = MW478497; CaM = MW478511; RPB2 = MW478532.
Aspergillus alboviridis J.P.Z. Siqueira, Gene, F. Sklenar & Hubka, Stud. Mycol. 99 (no. 100120): 19. 2021. [MB 821808]. — Type: CBS H-23128 (holotype). Ex-type: CBS 142665 = FMR 15175 = CCF 6049 = IFM 66819. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Spelaei. DNA barcodes: ITS = LT798909; BenA = LT798936; CaM = LT798937; RPB2 = LT798938.
Aspergillus annui J.J. Silva, Fungaro, Frisvad, Taniwaki & Iamanaka, published here. [MB 849336]. — Type: IBT 36122 (holotype). Ex-type: 365-IT-PPK = IBT 36122. Infragen. class.: subgen. Circumdati sect. Flavi ser. Annuorum. DNA barcodes: ITS = OP691228; BenA = ON529842; CaM = ON529841; RPB2 = ON529843.
Notes: This species was invalidly described in Silva et al. (2022) and is validated above.
Aspergillus arizonicus Jurjević, Glässnerová, Yaguchi & Hubka, Persoonia 47: 273. 2021. [MB 841359]. — Type: PRM 954611 (holotype). Ex-type: CCF 5341 = CBS 148476 = IFM 66805 = EMSL 2204. Infragen. class.: subgen. Fumigati sect. Fumigati ser. Neoglabri. DNA barcodes: ITS = OK322364; BenA = OK334128; CaM = OK334127; RPB2 = OK334129.
Aspergillus banksianus Pitt, Persoonia 44: 355. 2020. [MB 835223]. — Type: DAR 85042 (holotype). Ex-type: FRR 6047 = MST FP2248. Infragen. class.: subgen. Fumigati sect. Fumigati ser. Brevipedes. DNA barcodes: ITS = MH280013; BenA = MT184780; CaM = MT184786; RPB2 = MT184792.
Aspergillus barbosae A.C.R. Barros-Correia, R.N. Barbosa, Houbraken & Souza-Motta, Mycol. Prog. 19: 892. 2020. [MB 830077]. — Type: URM 93046 (holotype). Ex-type: URM 5930 = CBS 145863. Infragen. class.: subgen. Circumdati sect. Terrei ser. Terrei. DNA barcodes: ITS = LR536042; BenA = LR031377; CaM = LR031392; RPB2 = LR031407.
Aspergillus burnettii Pitt, Fungal Genet. Biol. 143 (no. 103435): 5. 2020. [MB 835453]. — Type: DAR 84902 (holotype). Ex-type: CBS 146237 = FRR 5400 = MST FP2249. Infragen. class.: subgen. Circumdati sect. Flavi ser. Alliacei. DNA barcodes: ITS = MK429758; BenA = MT211761; CaM = MT211762; RPB2 = MT211763.
Synonym of: Aspergillus alliaceus Thom & Church, Aspergilli: 163. 1926. [MB 256402].
Notes: BenA and CaM phylogenies resolved A. burnettii inside A. alliaceus, while RPB2 resolved it on an unsupported distinct branch (Suppl. Fig. S6). As a result, we consider A. burnetti a synonym of A. alliaceus.
Aspergillus chiangmaiensis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12, 705896: 6. 2021. [MB 830887]. — Type: SDBR-CMUI4 (holotype). Ex-type: TBRC 10407. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = MK457198; BenA = MK457200; CaM = MK457199; RPB2 = MK457203.
Synonym of: Aspergillus tubingensis Mosseray, La Cellule 43: 245. 1934. [MB 255209] (Bian et al. 2022).
Notes: Khuna et al. (2021) introduced A. chiangmaiensis based on DNA sequences that are of low quality and the species was subsequently considered a synonym of A. tubingensis by Bian et al. (2022).
Aspergillus curvatus Al-Bedak & Moubasher, Asian J. Mycol. 3: 327. 2020. [MB 557788]. — Type: AUMC 11038 (holotype). Ex-type: EMCCN 2213. Infragen. class.: subgen. Circumdati sect. Circumdati ser. Steyniorum. DNA barcodes: ITS = MN006961; BenA = n.a.; CaM = n.a.; RPB2 = n.a.
Notes: Doubtful species. This species was described with only an ITS sequence. Based on sequence alignments, the ITS sequence is potentially of low-quality. We obtained a culture of the ex-type strain but this unfortunately represented a different species. As we could not confirm the status of A. curvatus, we consider it a doubtful species (Suppl. Fig. S4). Two identifiers were cited in the protologue. IF 557788 (= MB 557788) was listed first and is thus the one that is permanently associated with the name. MycoBank MB 831990 should not be used.
Aspergillus gaarensis [nom. inval. Art. 40.7 (Shenzhen)] Al-Bedak & Moubasher, Stud. Fungi 5: 61. 2020. [MB 833223]. — Type: AUMC 11046 = EMCCN 2221 (holotype). Ex-type: AUMC 11046 = EMCCN 2221. Infragen. class.: subgen. Circumdati sect. Circumdati ser. Steyniorum. DNA barcodes: ITS = MN648408; BenA = n.a.; CaM = n.a.; RPB2 = n.a.
Notes: Doubtful species. This species was invalidly described with only an ITS sequence to support it. However, based on sequence alignments it was potentially a low-quality sequence. We obtained a culture of the ex-type strain but this unfortunately represented a different species. As we could not confirm the status of A. gaarensis, we consider it a doubtful species (Suppl. Fig. S4) and choose not to validate this species.
Aspergillus guangdongensis B.D. Sun, X.Z. Jiang & A.J. Chen, J. Fungi 8 (no 1205): 4. 2022. [MB 837898]. — Type: HMAS 248373 (holotype). Ex-type: CGMCC 3.19704 = MN 014767. Infragen. class.: subgen. Nidulantes sect. Ochraceorosei ser. Funiculosi. DNA barcodes: ITS = MN640760; BenA = MN635246; CaM = MN635257; RPB2 = MN635269.
Synonym of: Aspergillus lannaensis Suwannar., S. Khuna & Lumyong, Fungal Divers. 111: 145. 2021. [MB 838058].
Notes: Phylogenies resolved A. lannaensis and A. guangdongensis in the same clade (Fig. 3 & Suppl. Fig. S3) and we consider them synonyms with A. lannaensis having priority over A. guangdongensis.
Fig. 3.
Phylogenetic tree of Aspergillus sections Cavernicolarum, Ochraceorosei and Sparsi based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. bisporus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S3.
Aspergillus guangxiensis B.D. Sun, X.Z. Jiang & A.J. Chen, J. Fungi 8 (no 1205): 9. 2022. [MB 837899]. — Type: HMAS 248372 (holotype). Ex-type: CGMCC 3.19709 = MN 115635. Infragen. class.: subgen. Nidulantes sect. Sparsi ser. Conjuncti. DNA barcodes: ITS = MN640765; BenA = MN635251; CaM = MN635262; RPB2 = MN635274.
Aspergillus hainanicus X.C. Wang & W.Y. Zhuang, J. Fungi 8 (225): 9. 2022. [MB 570967]. — Type: HMAS 247855 (holotype). Ex-type: CGMCC 3.20888. Infragen. class.: subgen. Nidulantes sect. Cavernicolarum ser. Hainanici. DNA barcodes: ITS = OM414846; BenA = OM475626; CaM = OM475630; RPB2 = OM475634.
Aspergillus hydei Doilom, Front. Microbiol. 11 (no. 585215): 4. 2020. [MB 557860]. — Type: MFLU 20-0430 (holotype). Ex-type: KUMCC 18-0196. Infragen. class.: subgen. Circumdati sect. Nigri ser. Japonici. DNA barcodes: ITS = MT152332; BenA = MT161679; CaM = MT178247; RPB2 = MT384370.
Notes: The phylogenetic trees are not well-resolved (Fig. 10 & Suppl. Fig. S10), and further studies including additional strains and/or extended datasets are needed. Here, we give benefit of doubt to the publisher.
Fig. 10.
Phylogenetic tree of Aspergillus section Nigri series Japonici and Nigri based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. candidus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S10.
Aspergillus inusitatus F. Sklenar, C. Silva Pereira, Houbraken & Hubka, Stud. Mycol. 99: 20. 2021. [MB 839383]. — Type: PRM 954606 (holotype). Ex-type: CBS 147044 = DTO 121-G5 = CCF 6552. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Spelaei. DNA barcodes: ITS = MW448669; BenA = MW478502; CaM = MW478517; RPB2 = MW478542.
Aspergillus jilinensis X.Z. Jiang, Mycoscience 61: 207. 2020. [MB 819490]. — Type: HMAS 247010 (holotype). Ex-type: CGMCC 3.18134. Infragen. class.: subgen. Circumdati sect. Terrei ser. Terrei. DNA barcodes: ITS = KX443224; BenA = KX443162; CaM = KX443193; RPB2 = n.a.
Aspergillus kumbius Pitt, Persoonia 44: 359. 2020. [MB 835225]. — Type: DAR 85044 (holotype). Ex-type: FRR 6049 = MST FP2250. Infragen. class.: subgen. Circumdati sect. Circumdati ser. Sclerotiorum. DNA barcodes: ITS = MT179307; BenA = MT184782; CaM = MT184788; RPB2 = MT184794.
Aspergillus lannaensis Suwannar., S. Khuna & Lumyong, Fungal Divers. 111: 145. 2021. [MB 838058]. — Type: SDBR-CMUO 8 (holotype). Ex-type: SDBR-CMUO 8. Infragen. class.: subgen. Nidulantes sect. Ochraceorosei ser. Funiculosi. DNA barcodes: ITS = n.a.; BenA = MW219783; CaM = MW219781; RPB2 = MW219785.
Aspergillus lanuginosus F. Sklenar & Hubka, Stud. Mycol. 99: 20. 2021. [MB 839384]. — Type: PRM 954608 (holotype). Ex-type: NRRL 4610 = IMI 350352 = CCF 4551 = IFM 66818. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Spelaei. DNA barcodes: ITS = EF669604; BenA = EU014080; CaM = EF669562; RPB2 = EF669646.
Aspergillus lebretii V.C.S. Alves, J.D.P. Bezerra & R.N. Barbosa, Fungal Syst. Evol. 10: 144. 2022. [MB 846119]. — Type: URM 95150 (holotype). Ex-type: URM 8451 = FCCUFG 09. Infragen. class.: subgen. Cremei sect. Cremei ser. Wentiorum. DNA barcodes: ITS = ON862928; BenA = OP672382; CaM = OP290540; RPB2 = OP290511.
Aspergillus lentisci (Durieu & Mont.) Visagie, Malloch, L. Kriegsteiner, Samson & Houbraken, published here. [MB 849485]. Basionym: Dichlaena lentisci Durieu & Mont., Expl. Sci. l’Algérie 1: 405. 1849. [MB 249716]. — Type: Durieu & Mont., Expl. Sci. l’Algérie 1 (11): 405, tab. 22 bis, fig. 2 (lectotype), CBS H-25292 (epitype). Ex-epitype: CBS 150189. Infragen. class.: subgen. Circumdati sect. Dichlaena ser. Dichlaena. DNA barcodes: ITS = OR142402; BenA = OR145977; CaM = OR145992; RPB2 = OR146003.
Aspergillus limoniformis Z.F. Zhang & L. Cai, Fungal Divers. 106: 77. 2021. [MB 556394]. — Type: HMAS 248014 (holotype). Ex-type: CGMCC 3.19323 = LC126098. Infragen. class.: subgen. Polypaecilum sect. Polypaecilum ser. Canini. DNA barcodes: ITS = MK329066; BenA = MK336093; CaM = n.a.; RPB2 = MK335972.
Aspergillus luteorubrus Pitt, Persoonia 44: 361. 2020. [MB 835226]. — Type: DAR 85045 (holotype). Ex-type: CBS 146723 = FRR 5427 = MST FP2246. Infragen. class.: subgen. Fumigati sect. Fumigati ser. Fennelliarum. DNA barcodes: ITS = MT179305; BenA = MT184781; CaM = MT184787; RPB2 = MT184793.
Synonym of: Aspergillus fennelliae Kwon-Chung & S.J. Kim, Mycologia 66: 629. 1974. [MB 309218].
Notes: Even though the multigene phylogeny resolves A. luteorubrus basal to A. fennelliae, both the BenA and RPB2 phylogenies places it intermixed with strains of the latter (Fig. 8 & Suppl. Fig. S8). We thus consider it a synonym.
Fig. 8.
Phylogenetic tree of Aspergillus section Fumigati based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. clavatus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by superscript R. See Suppl. Fig. S8.
Aspergillus magnus Glässnerová & Hubka, Stud. Mycol. 102: 38. 2022. [MB 844202]. — Type: PRM 956934 (holotype). Ex-type: UAMH 1324 = IBT 14560. Infragen. class.: subgen. Circumdati sect. Candidi ser. Candidi. DNA barcodes: ITS = ON156376; BenA = ON164570; CaM = ON164619; RPB2 = ON164517.
Aspergillus malvicolor A.D. Hocking, Persoonia 44: 363. 2020. [MB 835227]. — Type: DAR 85046 (holotype). Ex-type: CBS 146724 = FRR 2383 = MST FP2244. Infragen. class.: subgen. Circumdati sect. Circumdati ser. Sclerotiorum. DNA barcodes: ITS = MT179308; BenA = MT184784; CaM = MT184790; RPB2 = MT184796.
Aspergillus marneyi Y.P. Tan, Bishop-Hurley, E. Lacey & R.G. Shivas, Index Austral. Fungi 3: 1. 2022. [MB 900143]. — Type: BRIP 71536a (holotype). Ex-type: BRIP 71536a. Infragen. class.: subgen. Circumdati sect. Terrei ser. Terrei. DNA barcodes: ITS = OL691080; BenA = OL741659; CaM = n.a.; RPB2 = OL741656.
Synonym of: Aspergillus alabamensis Balajee, Baddley, Frisvad & Samson, Eukar. Cell 8: 720. 2009. [MB 543648].
Notes: Phylogenies resolve A. marneyi inside the A. alabamensis clade (Fig. 12 & Suppl. Fig. S12). We thus consider it a synonym.
Fig. 12.
Phylogenetic tree of Aspergillus section Terrei series Nivei and Terrei based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. ambiguus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S12.
Aspergillus montoensis Y.P. Tan, Bishop-Hurley, S.M. Thompson & R.G. Shivas, Index Fungorum 503: 1. 2021. [MB 558828]. — Type: BRIP 64553a (holotype). Ex-type: BRIP 64553a = CBS 149107. Infragen. class.: subgen. Circumdati sect. Terrei ser. Terrei. DNA barcodes: ITS = OK441076; BenA = OK533535; CaM = n.a.; RPB2 = OK509073.
Notes: Both the ITS and BenA sequences are identical to A. terreus, but RPB2 resolves it distinct from A. terreus as a close relative of A. citrinoterreus (Fig. 12 & Suppl. Fig. S12). Further studies including additional strains and/or extended datasets are needed. Here, we give benefit of doubt to the publisher.
Aspergillus nanangensis Pitt, Persoonia 44: 365. 2020. [MB 836001]. — Type: DAR 84903 (holotype). Ex-type: CBS 146238 = FRR 6048 = MST FP2251. Infragen. class.: subgen. Circumdati sect. Janorum ser. Janorum. DNA barcodes: ITS = MK979278; BenA = MT184783; CaM = MT184789; RPB2 = MT184795.
Aspergillus neoterreus X.C. Wang and W.Y. Zhuang, J. Fungi 8 (3, no. 225): 10. 2022. [MB 570968]. — Type: HMAS 247856 (holotype). Ex-type: CGMCC 3.20891. Infragen. class.: subgen. Nidulantes sect. Terrei ser. Terrei. DNA barcodes: ITS = OM414849; BenA = OM475629; CaM = OM475633; RPB2 = OM475637.
Notes: The phylogenetic trees are not well-resolved (Fig. 12 & Suppl. Fig. S12), and further studies including additional strains and/or extended datasets are needed. Here, we give benefit of doubt to the publisher.
Aspergillus neotritici Glässnerová & Hubka, Stud. Mycol. 102: 38. 2022. [MB 844204]. — Type: PRM 956940 (holotype). Ex-type: CCF 3853 = IBT 32725. Infragen. class.: subgen. Circumdati sect. Candidi ser. Candidi. DNA barcodes: ITS = FR727136; BenA = FR775327; CaM = HE661598; RPB2 = LT627021.
Notes: This new name was introduced for the invalid A. tritici [nom. inval. Art. 8.1, Art. 8.4, Art 40.1, Art. 40.4 (Shenzhen)] by Glässnerová et al. (2022).
Aspergillus okavangoensis Visagie & Nkwe, Fungal Syst. Evol. 8: 85. 2021. [MB 840269]. — Type: PREM 63212 (holotype). Ex-type: CBS 147420 = CMW 56636 = CN073A3 = DN24. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Flavipedes. DNA barcodes: ITS = MW480880; BenA = MW480788; CaM = MW480706; RPB2 = MW480790.
Aspergillus oxumiae C.N. Figueiredo, L.S. Sales, Y.F. Figueiredo, J.P. Andrade & J.T. De Souza, Persoonia 44: 357. 2020. [MB 832766]. — Type: HURB 22369 (holotype). Ex-type: CCDCA 11546 = UFLA115. Infragen. class.: subgen. Circumdati sect. Nigri ser. Japonici. DNA barcodes: ITS = MN431160; BenA = MN521388; CaM = MN531842; RPB2 = MN521389.
Aspergillus pakistanicus [nom. inval. Art. 40.7 & Art. 40.8 (Shenzhen)] Abrar, Mughal & S. Sarwar, Appl. Ecol. Environm. Res. 18: 3797. 2020. [MB 830877]. — Type: AA100717 (holotype). Ex-type: AA100717. Infragen. class.: subgen. Circumdati sect. Flavi ser. Flavi. DNA barcodes: ITS = n.a.; BenA = n.a.; CaM = n.a.; RPB2 = n.a.
Notes: The species was invalidly described. Only an SSU (18S small subunit nrDNA) sequence is available for this species (GenBank MK371711). Even though it is unique, there are very little SSU data available for Aspergillus, and we therefore decided not to validate the species here.
Aspergillus phialiformis Z.F. Zhang & L. Cai, Fungal Divers. 106: 79. 2021. [MB 556395]. — Type: HMAS 248017 (holotype). Ex-type: CGMCC 3.19314 = LC12536. Infragen. class.: subgen. Polypaecilum sect. Polypaecilum ser. Canini. DNA barcodes: ITS = MK329068; BenA = MK336095; CaM = n.a.; RPB2 = MK335974.
Aspergillus phialosimplex Z.F. Zhang & L. Cai, Fungal Divers. 106: 79. 2021. [MB 556396]. — Type: HMAS 248007 (holotype). Ex-type: CGMCC 3.19637 = LC12578. Infragen. class.: subgen. Polypaecilum sect. Polypaecilum ser. Canini. DNA barcodes: ITS = MK329070; BenA = MK336097; CaM = n.a.; RPB2 = MK335976.
Aspergillus pseudopiperis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12, 705896: 6. 2021. [MB 830888]. — Type: SDBR-CMUI1 (holotype). Ex-type: TBRC 10408. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = n.a.; BenA = MK457194; CaM = MK457193; RPB2 = MK457196.
Synonym of: Aspergillus tubingensis Mosseray, La Cellule 43: 245. 1934. [MB 255209] (Bian et al. 2022).
Notes: Khuna et al. (2021) introduced this species based on DNA sequences that are of low quality and the species was subsequently considered a synonym of A. tubingensis by Bian et al. (2022).
Aspergillus pseudotubingensis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12, 705896: 9. 2021. [MB 830889]. — Type: SDBR-CMUO2 (holotype). Ex-type: TBRC 10409. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = n.a.; BenA = MK457206; CaM = MK457205; RPB2 = MK457208.
Notes: Doubtful species. Khuna et al. (2021) introduced this species based on DNA sequences that are of low quality and its taxonomic position was subsequently considered doubtful by Bian et al. (2022).
Aspergillus qilianyuensis X.C. Wang & W.Y. Zhuang, J. Fungi 8 (225): 12. 2022. [MB 570969]. — Type: HMAS 247857 (holotype). Ex-type: CGMCC 3.20889. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = OM414847; BenA = OM475627; CaM = OM475631; RPB2 = OM475635.
Aspergillus recifensis A.C.R. Barros-Correia, R.N. Barbosa, Houbraken & Souza-Motta, Mycol. Prog. 19: 894. 2020. [MB 830081]. — Type: URM 93050 (holotype). Ex-type: URM 6605 = CBS 145864. Infragen. class.: subgen. Circumdati sect. Terrei ser. Nivei. DNA barcodes: ITS = LR536036; BenA = LR031370; CaM = LR031385; RPB2 = LR031400.
Aspergillus rouenensis Crous & Decock, Persoonia 48: 289. 2022. [MB 844262]. — Type: CBS H-24987. Ex-type: CBS 149067 = CPC 41585 = MUCL 58110. Infragen. class.: subgen. Polypaecilum sect. Polypaecilum ser. Salinarum. DNA barcodes: ITS = ON603782; BenA = ON605641; CaM = ON653193; RPB2 = ON653194.
Aspergillus saccharicola J.J. Silva, Frisvad, M.H.P. Fungaro, M.H. Taniwaki & B.T. Iamanaka, published here. [MB 849338]. — Type: IBT 36126 (holotype). Ex-type: 117-IT-SGC = IBT 36126. Infragen. class.: subgen. Circumdati sect. Flavi ser. Flavi. DNA barcodes: ITS = OP611470; BenA = ON529845; CaM = ON529844; RPB2 = ON529846.
Notes: This species was invalidly described in Silva et al. (2022) and is validated above.
Aspergillus sakultaensis [nom. inval. Art. 40.8 (Shenzhen)] Al-Bedak, Zohri & Abdel-Kareem, J. Environm. Stud. 20: 23. 2020. [MB 831480]. — Type: UMC 13885 (holotype). Ex-type: AUMC 13885. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Flavipedes. DNA barcodes: ITS = MK391495; BenA = n.a.; CaM = n.a.; RPB2 = n.a.
Synonym of: Aspergillus templicola Visagie, Hirooka & Samson, Stud. Mycol. 78: 103. 2014. [MB 809191].
Notes: Aspergillus sakultaensis was described with only an ITS sequence to support it, but this is identical to A. templicola (Suppl. Fig. S7). As a result, we consider it a synonym of the latter.
Aspergillus sibiricus V.A. Iliushin, Phytotaxa 531: 68. 2022. [MB 841752]. — Type: LE F-341005 (holotype). Ex-type: CBS 143307. Infragen. class.: subgen. Fumigati sect. Fumigati ser. Unilaterales. DNA barcodes: ITS = MG587008; BenA = MG722970; CaM = MG722971; RPB2 = MG710809.
Aspergillus sichuanensis B.D. Sun, X.Z. Jiang & A.J. Chen, J. Fungi 8 (no 1205): 10. 2022. [MB 837900]. — Type: HMAS 248374 (holotype). Ex-type: CGMCC 3.19706 = MN 18437. Infragen. class.: subgen. Nidulantes sect. Aenei ser. Aenei. DNA barcodes: ITS = MN640762; BenA = MN635248; CaM = MN635259; RPB2 = MN635271.
Aspergillus sigarelli B.D. Sun, Houbraken, A.J. Chen and Samson, Int. J. Syst. Evol. Microbiol. 70: 9. 2020. [MB 833129]. — Type: CBS H-22725 (holotype). Ex-type: CBS 141579 = DTO 348-D4 = CGMCC 3.03936. Infragen. class.: subgen. Nidulantes sect. Usti ser. Calidousti. DNA barcodes: ITS = MN640758; BenA = MN635244; CaM = MN635255; RPB2 = MN635267.
Aspergillus telluris B.D. Sun, X.Z. Jiang & A.J. Chen, Phytotaxa 455: 146. 2020. [MB 834494]. — Type: HMAS 248375 (holotype). Ex-type: CGMCC 3.19701. Infragen. class.: subgen. Polypaecilum sect. Polypaecilum ser. Canini. DNA barcodes: ITS = MN640767; BenA = MN635253; CaM = MN635264; RPB2 = MN635276.
Aspergillus tenebricus Houbraken, Glässnerová & Hubka, Stud. Mycol. 102: 45. 2022. [MB 844203]. — Type: PRM 957108 (holotype). Ex-type: CBS 147048 = DTO 337-H7. Infragen. class.: subgen. Circumdati sect. Candidi ser. Candidi. DNA barcodes: ITS = ON156389; BenA = ON164584; CaM = ON164623; RPB2 = ON164532.
Aspergillus tibetensis B.D. Sun, X.Z. Jiang & A.J. Chen, J. Fungi 8 (no 1205): 12. 2022. [MB 837901]. — Type: HMAS 248371 (holotype). Ex-type: CGMCC 3.19707 = MN 110445. Infragen. class.: subgen. Nidulantes sect. Aenei ser. Aenei. DNA barcodes: ITS = MN640763; BenA = MN635249; CaM = MN635260; RPB2 = MN635272.
Aspergillus toxicus [nom. inval. Art. 40.8 (Shenzhen)] P. Singh, K.A. Callicott, M.J. Orbach & P.J. Cotty, Front. Microbiol. 11, 1236: 10. 2020. [MB 832486]. — Type: NRRL 66898 (holotype). Ex-type: NRRL 66898 = A5-B-S = A2406. Infragen. class.: subgen. Circumdati sect. Flavi ser. Flavi. DNA barcodes: ITS = n.a.; BenA = n.a.; CaM = MN987092; RPB2 = n.a.
Synonym of: Aspergillus minisclerotigenes Vaamonde, Frisvad & Samson, Int. J. Syst. Evol. Microbiol. 58: 733. 2008. [MB 505188].
Notes: This species was invalidly described. The CaM phylogeny resolved this species within the A. minisclerotigenes clade (Suppl. Fig. S6) and we thus consider this species to be a synonym of the latter and, therefore, choose not to validate this species.
Aspergillus vinaceus Ferranti, Iamanaka, Frisvad, O. Puel & J.J. da Silva, J. Fungi 6 (no. 371): 14. 2020. [MB 833399]. — Type: ITAL 47.456 (holotype). Ex-type: ITAL 47.456 = IBT 35556. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = MN575692; BenA = MN583579; CaM = MN583580; RPB2 = MN583581.
Synonym of: Aspergillus niger Tiegh., Ann. Sci. Nat., Bot., ser. 58: 240. 1867. [MB 284309].
Notes: Phylogenies resolved A. vinaceus in the A. niger clade (Fig. 10 & Suppl. Fig. S10), similar to the results found during a recent revision of series Nigri (Bian et al. 2022).
Aspergillus xishaensis X.C. Wang & W.Y. Zhuang, J. Fungi 8 (225): 13. 2022. [MB 570970]. — Type: HMAS 247858 (holotype). Ex-type: CGMCC 3.20890. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Flavipedes. DNA barcodes: ITS = OM414848; BenA = OM475628; CaM = OM475632; RPB2 = OM475636.
Emericella sydowii (Bainier & Sartory) Pitt & A.D. Hocking, Fungi and Food Spoilage. 4th Ed.: 611. 2022. [MB 838069]. Basionym: Sterigmatocystis sydowii Bainier & Sartory, Ann. Mycol. 11: 25. 1913. [MB 197979] — Type: IMI 211384.
Synonym of: Aspergillus sydowii (Bainier & Sartory) Thom & Church, The Aspergilli: 147. 1926. [MB 279636]. — Type: IMI 211384. Ex-type: CBS 593.65 = NRRL 250 = IMI 211384 = NRRL 254. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652450; BenA = EF652274; CaM = EF652362; RPB2 = EF652186.
Notes: Pitt & Hocking (2022) introduced this new combination to accommodate A. sydowii in Emericella. The genus Emericella is considered a synonym of Aspergillus (Samson et al. 2014). We therefore do not accept this new combination and consider it a synonym of Aspergillus sydowii.
Emericella versicolor (Vuill.) Pitt & A.D. Hocking, Fungi and Food Spoilage. 4th Ed.: 611. 2022. [MB 838068]. Basionym: Sterigmatocystis versicolor Vuill., Erreur Dét. Asp. Paras. Homme: 15. 1903. [MB 233198]. — Type: CBS 583.65 (holotype).
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159]. — Type: CBS 583.65. Ex-type: CBS 583.65 = NRRL 238 = ATCC 9577 = IFO 33027 = IMI 229970 = JCM 10258 = QM 7478 = Thom 5519.57 = WB 238. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652442; BenA = EF652266; CaM = EF652354; RPB2 = EF652178.
Notes: Pitt & Hocking (2022) introduced this new combination to accommodate A. versicolor in Emericella. The genus Emericella is considered a synonym of Aspergillus (Samson et al. 2014). We therefore do not accept this new combination and consider this species a synonym of Aspergillus versicolor.
Emericella usta (Bainier) Pitt & A.D. Hocking, Fungi and Food Spoilage. 4th Ed.: 611. 2022. [MB 838032]. Basionym: Sterigmatocystis usta Bainier, Bull. Soc. Bot. France 28: 78. 1881. [MB 536545]. — Type: IMI 211805 (holotype).
Synonym of: Aspergillus ustus (Bainier) Thom & Church, The Aspergilli: 152. 1926. [MB 281216]. — Type: IMI 211805. Ex-type: CBS 261.67 = NRRL 275 = ATCC 1041 = ATCC 16818 = IMI 211805 = QM 7477 = WB 275. Infragen. class.: subgen. Nidulantes sect. Usti ser. Usti. DNA barcodes: ITS = EF652455; BenA = EF652279; CaM = EF652367; RPB2 = EF652191.
Notes: Pitt & Hocking (2022) introduced this new combination to accommodate A. ustus in Emericella. The genus Emericella is considered a synonym of Aspergillus (Samson et al. 2014). We therefore do not accept this new combination and consider this species a synonym of Aspergillus ustus.
Neosartorya clavata (Desm.) Pitt & A.D. Hocking, Fungi and Food Spoilage. 4th Ed.: 611. 2022. [MB 838031]. — Type: IMI 15949 (holotype).
Synonym of: Aspergillus clavatus Desm., Ann. Sci. Nat., Bot., ser. 2: 71. 1834. [MB 211530]. — Type: IMI 15949. Ex-type: CBS 513.65 = NRRL 1 = ATCC 1007 = ATCC 9598 = ATCC 9602 = CECT2674 = DSM 816 = IMI 15949 = LSHBA c .86 = LSHBA c .95 = MIT213 = NCTC 3887 = NCTC 9 = NCTC 978 = NRRL 1656 = QM 1276 = QM 7404 = Thom 107 = WB 1. Infragen. class.: subgen. Fumigati sect. Clavati ser. Clavati. DNA barcodes: ITS = EF669942; BenA = EF669802; CaM = EF669871; RPB2 = EF669730.
Notes: Pitt & Hocking (2022) introduced this new combination to accommodate A. clavatus in Neosartorya. The genus Neosartorya is considered a synonym of Aspergillus (Samson et al. 2014). We therefore do not accept this new combination and consider it a synonym of Aspergillus clavatus.
Paecilomyces clematidis Spetik, Eichmeier, Gramaje & Berraf-Tebbal, Phytotaxa 559: 242. 2022. [MB 843540]. — Type: BRNU 677844 (holotype). Ex-type: CBS 148466 = MEND-F-0560. DNA barcodes: ITS = MZ923760; BenA = MZ927740; CaM = MZ927738; RPB2 = OL332317.
Paecilomyces penicilliformis Jurjević & Hubka, Persoonia 44: 431. 2020. [MB 834874]. — Type: BPI 911216 (holotype). Ex-type: CCF 5755 = CBS 146003 = EMSL 3392. DNA barcodes: ITS = LR679769; BenA = LR679768; CaM = LR778299; RPB2 = n.a.
Paraxerochrysium Crous & Decock, Persoonia 47: 261. 2021. [MB 841829]
Synonym of: Xerochrysium Pitt, SIM 4 (2): 236. 2013. [MB 807003].
Notes: See notes for Xerochrysium coryli above.
Paraxerochrysium coryli Crous & Decock, Persoonia 47: 261. 2021. [MB 841830]. — Type: CBS H-24853 (holotype). Ex-type: CBS 148314 = CPC 41272 = MUCL 58103. DNA barcodes: ITS = OK664748; BenA = OK651216; CaM = n.a.; RPB2 = OK651178.
Notes: See notes for Xerochrysium coryli above.
Penicillium allaniae Y.P. Tan, Bishop-Hurley, Marney, R.G. Shivas, Index Austral. Fungi 3: 6. 2022. [MB 900139]. — Type: BRIP 74886a (holotype). Ex-type: BRIP 74886a. Infragen. class.: subgen. Aspergilloides sect. Exilicaulis ser. Restricta. DNA barcodes: ITS = OP903476; BenA = OP921959; CaM = OP921957; RPB2 = OP921958.
Notes: Since Houbraken et al. (2020), P. allaniae, P. archerae, P. krskae and P. silybi were introduced bringing the number of accepted species in series Restricta to thirteen. However, the series clearly needs a taxonomic revision (Fig. 23 & Suppl. Fig. S21) as noted by Visagie et al. (2016). In the meantime, we accept the four recently introduced species.
Fig. 23.
Phylogenetic tree of Penicillium sections Exilicaulis and Gracilenta based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S21.
Penicillium allsoppiae Visagie, A. Visagie, Frisvad & K. Jacobs, Persoonia 46: 176. 2021. [MB 834426]. — Type: CBS H-22036 (holotype). Ex-type: CBS 138943 = DAOM 241348 = DTO 182-D5 = CV 931. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = JX140830; BenA = JX140992; CaM = JX157384; RPB2 = KP016895.
Penicillium anthracinoglaciei Perini, Frisvad & Zalar, Microbial Ecol. 86: 287. 2022 (2023). [MB 835602]. — Type: EXF11443H (Holotype). Ex-type: EXF-11443 = IBT 34739. Infragen. class.: subgen. Penicillium sect. Brevicompacta ser. Brevicompacta. DNA barcodes: ITS = n.a.; BenA = MT080493; CaM = MT080552; RPB2 = MT080519.
Notes: The phylogenetic relationship between P. anthracinoglaciei and P. bialowiezense is unresolved and needs further study (Fig. 18 & Suppl. Fig. S16).
Fig. 18.
Phylogenetic tree of Penicillium section Brevicompacta based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. tularense. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S16.
Penicillium aquadulcis Hyang B. Lee & T.T.T. Nguyen, Mycobiology 49: 542. 2021. [MB 556953]. — Type: CNUFC HT19009 (holotype). Ex-type: CNUFC JT1301. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Westlingiorum. DNA barcodes: ITS = OK356194; BenA = OK105100; CaM = OK105102; RPB2 = n.a.
Penicillium archerae Y.P. Tan, Bishop-Hurley, R.G. Shivas, Index Austral. Fungi 3: 7. 2022. [MB 900140]. — Type: BRIP 72549c (holotype). Ex-type: BRIP 72549c. Infragen. class.: subgen. Aspergilloides sect. Exilicaulis ser. Restricta. DNA barcodes: ITS = OP903477; BenA = OP921961; CaM = n.a.; RPB2 = OP921960.
Notes: Since Houbraken et al. (2020), P. allaniae, P. archerae, P. krskae and P. silybi were introduced bringing the number of accepted species in series Restricta to thirteen. However, the series clearly needs a taxonomic revision (Fig. 23 & Suppl. Fig. S21) as noted by Visagie et al. (2016). In the meantime, we accept the four recently introduced species.
Penicillium aspericonidium B.D. Sun, A.J. Chen & Houbraken, Mycol. Prog. 20: 1387. 2021. [MB 838211]. — Type: CBS H-22830 (holotype). Ex-type: CBS 141832 = DTO 030-C5. Infragen. class.: subgen. Aspergilloides sect. Charlesia ser. Phoenicea. DNA barcodes: ITS = MT309657; BenA = MT302240; CaM = n.a.; RPB2 = MT302224.
Penicillium ausonanum Torres-Garcia, Gené & Dania García, MycoKeys 86: 119. 2022. [MB 840556]. — Type: CBS H-24781 (holotype). Ex-type: CBS 148237 = FMR 16948. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Dalearum. DNA barcodes: ITS = LR655808; BenA = LR655809; CaM = LR655810; RPB2 = LR655811.
Penicillium barbosae S. Ramos, R. Cruz, R.N. Barbosa & Houbraken, Mycol. Prog. 20: 828. 2021. [MB 837908]. — Type: URM 94474 (holotype). Ex-type: URM 7705. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Adametziorum. DNA barcodes: ITS = MW191494; BenA = MG452818; CaM = MW183245; RPB2 = LR898886.
Penicillium cerradense Cruvinel, P.O. Magalh. & Pinho, Sci. Rep. 11 (no. 17861): 2. 2021. [MB 835241]. — Type: UB23977 (holotype). Ex-type: DCFS6a. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Westlingiorum. DNA barcodes: ITS = MT006126; BenA = MT416533; CaM = MT416534; RPB2 = MT416532.
Synonym of: Penicillium sumatraense [as ‘sumatrense’] Szilvinyi, Archiv. Hydrobiol. 14 Suppl. 6: 535. 1936. [MB 319297].
Notes: Penicillium cerradense belongs to the P. sumatraense clade and is thus considered a synonym (Fig. 21 & Suppl. Fig. S19).
Fig. 21.
Phylogenetic tree of Penicillium section Citrina based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. euglaucum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S19.
Penicillium claroviride Visagie & Yilmaz, Mycologia 115: 90. 2022. [MB 844184]. — Type: PREM 63221 (holotype). Ex-type: CMW 56197 = CBS 147458 = CN014D2. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Atroveneta. DNA barcodes: ITS = MT949909; BenA = MT957414; CaM = MT957456; RPB2 = MT957482.
Penicillium doidgeae Visagie, Frisvad & K. Jacobs, Persoonia 46: 176. 2021. [MB 834427]. — Type: CBS H-22038 (holotype). Ex-type: CBS 138947 = IBT 31950 = DAOM 241107 = DTO 183-G7 = CV 2189. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Atroveneta. DNA barcodes: ITS = JX140804; BenA = JX141006; CaM = JX157413; RPB2 = KP016915.
Penicillium donggangicum L. Wang, PeerJ 10 (e13224): 8. 2022. [MB 841518]. — Type: HMAS 350265 (holotype). Ex-type: AS 3.15900. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = MW946996; BenA = MZ004914; CaM = MZ004918; RPB2 = MW979253.
Penicillium eickeri Visagie, Frisvad & K. Jacobs, Persoonia 46: 179. 2021. [MB 834428]. — Type: CBS H-22034 (holotype). Ex-type: CBS 138939 = IBT 31921 = DAOM 241352 = DTO 181-G3 = CV 475. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = JX140824; BenA = JX140979; CaM = JX157365; RPB2 = KP016876.
Penicillium elizabethiae Visagie & Frisvad, Persoonia 46: 179. 2021. [MB 834432]. Basionym: Penicillium echinatum E. Dale, Ann. Mycol. 24: 137. 1926. (nom. illegit. Art. 53.1; non Rivolta (1873). [MB 505484]. — Type: CBS H-22052 (holotype). Ex-type: NRRL 917 = MUCL 29170 = IBT 21955 = DTO 189-B8. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = KP016840; BenA = KJ866964; CaM = KJ867021; RPB2 = KP016918.
Notes: Penicillium elizabethiae was introduced for the illegitimate P. echinatum E. Dale.
Penicillium ezekielii Houbraken & Oyedele, Persoonia 49: 309. 2022. [MB 844770]. — Type: CBS H-25015 (holotype). Ex-type: CBS 149115 = DTO 065-D2. Infragen. class.: subgen. Aspergilloides sect. Cinnamopurpurea ser. Jiangxiensia. DNA barcodes: ITS = ON723772; BenA = ON920778; CaM = ON920781; RPB2 = ON920784.
Penicillium ferraniaense Houbraken & Di Piazza, Persoonia 46: 491. 2021. [MB 839119]. — Type: CBS H-24757 (holotype). Ex-type: CBS 147595 = DTO 400-D8. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Sclerotiorum. DNA barcodes: ITS = MW694951; BenA = MW689336; CaM = MW689338; RPB2 = MW689340.
Penicillium fuscoglaucum Biourge, La Cellule 33: 128. 1923. [MB 265659]. — Type: Belgium, source and collection date unknown, P. Biourge [Biourge, La Cellule 33: Pl. col. I. dr., n°4; Pl. noire II, fig. 9. 1923, lectotype designated here, MBT10014260); CBS H-15446 (dried culture), epitype designated here, MBT10014261]. Ex-epitype: CBS 261.29 = IMI 092259 = LSHB P81 = MUCL 28651 = NRRL 892 = CBS 122423 = DTO 461-D2. Infragen. class.: subgen. Penicillium sect. Fasciculata ser. Camembertiorum. DNA barcodes: ITS = MH855062; BenA = OR206420; CaM = OR206421; RPB2 = OR206422.
Notes: Giraud et al. (2010) and Ropars et al. (2020) considered P. fuscoglaucum as a distinct species, closely related to P. camemberti, and suggested that P. caseifulvum (CBS 101134T) is a variety, P. camemberti var. ‘caseifulvum’. Following Frisvad & Samson (2004), we disagree with the idea of introducing varieties in Penicillium. We accept P. camemberti, P. caseifulvum and P. commune, as suggested in the taxonomy of Frisvad & Samson (2004), but also tentatively accept P. fuscoglaucum and P. biforme following the taxonomic insights of Ropars et al. (2020). The taxonomies of Frisvad & Samson (2004) and Ropars et al. (2020) are not congruent and a taxonomic revision is therefore needed to resolve relationships and species boundaries within the clade (see Fig. 19 & Suppl. Fig. S17), noting that P. camemberti, and maybe also P. caseifulvum, are domesticated species related to cheese production (Bodinaku et al. 2019, Ropars et al. 2020).
Fig. 19.
Phylogenetic tree of Penicillium section Fasciculata series Camembertiorum based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. expansum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S17.
Penicillium fusiforme B.D. Sun, A.J. Chen & Houbraken, Mycol. Prog. 20: 1389. 2021. [MB 838212]. — Type: CBS H-22840 (holotype). Ex-type: CBS 250.66 = DTO 035-D7. Infragen. class.: subgen. Aspergilloides sect. Charlesia ser. Fellutana. DNA barcodes: ITS = MT309668; BenA = MT302253; CaM = MT302220; RPB2 = MT302236.
Penicillium gercinae A.L. Alves, A.C.S. Santos, R.N. Barbosa, C.M. Souza-Motta, R.F.R. Melo, P.V. Tiago, Acta Bot. Brasil 36 (e2022abb0006): 10. 2022. [MB 841260]. — Type: URM 94476 (holotype). Ex-type: URM 8348. Infragen. class.: subgen. Aspergilloides sect. Ramigena ser. Georgiensia. DNA barcodes: ITS = MW648591; BenA = MW646389; CaM = MW646391; RPB2 = MW646393.
Penicillium guarroi Torres-Garcia, Gené & Dania García, MycoKeys 86: 121. 2022. [MB 840567]. — Type: CBS H-24782 (holotype). Ex-type: CBS 148238 = FMR 17747. Infragen. class.: subgen. Aspergilloides sect. Gracilenta ser. Estinogena. DNA barcodes: ITS = LR814139; BenA = LR814134; CaM = LR814140; RPB2 = LR814145.
Penicillium hepuense L. Wang, PeerJ 10 (e13224): 11. 2022. [MB 841525]. — Type: HMAS 350263 (holotype). Ex-type: AS 3.16039 = TT2-4X3. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Oxalica. DNA barcodes: ITS = MW946994; BenA = MZ004912; CaM = MZ004916; RPB2 = MW979254.
Penicillium irregulare Torres-Garcia, Gené & Dania García, MycoKeys 86: 125. 2022. [MB 840558]. — Type: CBS H-24783 (holotype). Ex-type: CBS 148240 = FMR 17859. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = LR814181; BenA = LR814144; CaM = LR814151; RPB2 = LR814182.
Penicillium jenningsiae Y.P. Tan, Bishop-Hurley, E. Lacey & R.G. Shivas, Index Austral. Fungi 3: 8. 2022. [MB 900141]. — Type: BRIP 45936a (holotype). Ex-type: BRIP 45936a. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Sumatraensia. DNA barcodes: ITS = n.a.; BenA = OL741657; CaM = n.a.; RPB2 = OL741660.
Synonym of: Penicillium sumatraense [as ‘sumatrense’] Szilvinyi, Archiv. Hydrobiol. 14 Suppl. 6: 535. 1936. [MB 319297].
Notes: Penicillium jenningsiae belongs to the P. sumatraense clade and is thus considered a synonym. (Fig. 21 & Suppl. Fig. S19)
Penicillium jiaozhouwanicum L. Wang, PeerJ 10 (e13224): 13. 2022. [MB 841531]. — Type: HMAS 350262 (holotype). Ex-type: AS 3.16038 = 0801H2-2. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Oxalica. DNA barcodes: ITS = MW946993; BenA = MZ004911; CaM = MZ004915; RPB2 = MW979252.
Penicillium kalander Visagie & Yilmaz, Mycologia 115: 95. 2022. [MB 844185]. — Type: PREM 63223 (holotype). Ex-type: CMW 56202 = CN014E1. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Sclerotiorum. DNA barcodes: ITS = MT949914; BenA = MT957421; CaM = MT957461; RPB2 = MT957487.
Penicillium krskae Labuda, Kubátová, C. Schüller & J. Strauss, J. Fungi 7 (7, no. 557): 7. 2021. [MB 839112]. — Type: PRM 955188 (holotype). Ex-type: CBS 147776 = BiMM-F280 = CCF 6561. Infragen. class.: subgen. Aspergilloides sect. Exilicaulis ser. Restricta. DNA barcodes: ITS = MW794123; BenA = MW774594; CaM = MW774595; RPB2 = MW774593.
Notes: Since Houbraken et al. (2020), P. allaniae, P. archerae, P. krskae and P. silybi were introduced bringing the number of accepted species in series Restricta to thirteen. However, the series clearly needs a taxonomic revision (Fig. 23 & Suppl. Fig. S21) as noted by Visagie et al. (2016). In the meantime, we accept the four recently introduced species.
Penicillium limae S. Ramos, R. Cruz, C. Souza-Motta & N. Tinti, Mycol. Prog. 20: 831. 2021. [MB 837909]. — Type: URM 94475 (holotype). Ex-type: URM 7706. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Adametziorum. DNA barcodes: ITS = MW191493; BenA = MG452820; CaM = MW183244; RPB2 = LR898888.
Penicillium linzhiense H-K. Wang & R. Jeewon, Front. Cell. Infect. Microbiol. 10 (no 6045044): 4. 2021. [MB 838576]. — Type: CCTCC-M 2019870 (holotype). Ex-type: CCTCC-M 2019870. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = MT461156; BenA = MT461157; CaM = MT461162; RPB2 = n.a.
Penicillium longiconidiophorum B.D. Sun, A.J. Chen & Houbraken, Mycol. Prog. 20: 1389. 2021. [MB 838213]. — Type: CBS H-22829 (holotype). Ex-type: CBS 141831 = DTO 088-C1. Infragen. class.: subgen. Aspergilloides sect. Charlesia ser. Phoenicea. DNA barcodes: ITS = MT309669; BenA = MT302254; CaM = MT302221; RPB2 = MT302237.
Penicillium mattheeae Visagie & Yilmaz, Mycologia 115: 97. 2022. [MB 844186]. — Type: PREM 63219 (holotype). Ex-type: CMW 56388 = CBS 147415 = CN014C5. Infragen. class.: subgen. Aspergilloides sect. Aspergilloides ser. Saturniformia. DNA barcodes: ITS = MT949904; BenA = MT957408; CaM = MT957451; RPB2 = MT957477.
Penicillium melanosporum Rodr.-Andr., Cano & Stchigel, J. Fungi 7 (2, no. 126): 5. 2021. [MB 835938]. — Type: CBS H-24465 (holotype). Ex-type: CBS 146938 = FMR 17424. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = LR655192; BenA = LR655196; CaM = LR655200; RPB2 = LR655204.
Penicillium michoacanense Rodr.-Andr., Cano & Stchigel, J. Fungi 7 (2, no. 126): 8. 2021. [MB 835940]. — Type: CBS H-24467 (holotype). Ex-type: FMR 17612. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = LR655194; BenA = LR655198; CaM = LR655202; RPB2 = LR655206.
Penicillium neoherquei Labuda, Kubátová, Nebesářová, Oberlies & Raja, Persoonia 48: 339. 2022. [MB 842267]. — Type: PRM 956035 (holotype). Ex-type: CBS 148692 = CCF 6604. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Herqueorum. DNA barcodes: ITS = MW341222; BenA = OL840853; CaM = OL840855; RPB2 = MW349119.
Penicillium newtonturnerae Y.P. Tan, Bishop-Hurley, Marney & R.G. Shivas, Index Austral. Fungi 3: 10. 2022. [MB 900142]. — Type: BRIP 74909a (holotype). Ex-type: BRIP 74909a. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Simplicissima. DNA barcodes: ITS = OP903478; BenA = OP921964; CaM = OP921962; RPB2 = OP921963.
Penicillium nordestinense J.E.F. Santos & R.N. Barbosa, Acta Bot. Brasil 36 (e2021abb0390): 4. 2022. [MB 845495]. — Type: URM 83558 (holotype). Ex-type: URM 8423. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = OV265270; BenA = OV265324; CaM = OV265272; RPB2 = OM927721.
Penicillium outeniquaense Visagie & Yilmaz, Mycologia 115: 97. 2022. [MB 844187]. — Type: PREM 63218 (holotype). Ex-type: CMW 56387 = CBS 147414 = CN014C2. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Westlingiorum. DNA barcodes: ITS = MT949903; BenA = MT957405; CaM = MT957450; RPB2 = MT957476.
Penicillium poederi Kirchm. & Neuh., Fungal Syst. Evol. 10: 92. 2022. [MB 845496]. — Type: IBF2017/0007 (holotype). Ex-type: CBS 147622 = SF014017 = Bq214. Infragen. class.: subgen. Aspergilloides sect. Torulomyces ser. Torulomyces. DNA barcodes: ITS = MF611757; BenA = MF611760; CaM = MF611763; RPB2 = MF611766.
Penicillium pole-evansii Visagie, Frisvad & K. Jacobs, Persoonia 46: 179. 2021. [MB 834429]. — Type: CBS H-22037 (holotype). Ex-type: CBS 138946 = IBT 31929 = DAOM 241106 = DTO 183-D5 = CV 1758. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Atroveneta. DNA barcodes: ITS = JX140831; BenA = JX141005; CaM = JX157412; RPB2 = KP016911.
Penicillium rotoruae O’Callahan & Vaidya, Curr. Microbiol. 77: 4131. 2020. [MB 834084]. — Type: NZFS 4797 (holotype). Ex-type: CBS 145838 = NMI V19/026738. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Rolfsiorum. DNA barcodes: ITS = MN315103; BenA = MN315104; CaM = MN315102; RPB2 = MT240842.
Penicillium saanichanum Visagie, Assabgui & Seifert, Persoonia 45: 373. 2020. [MB 835962]. — Type: DAOM 745787 (holotype). Ex-type: DAOMC 251850 = KAS 6184. Infragen. class.: subgen. Aspergilloides sect. Cinnamopurpurea ser. Idahoensia. DNA barcodes: ITS = KY469059; BenA = KY469096; CaM = KY469020; RPB2 = MN795070.
Penicillium sanjayi Rajeshk., Visagie, N. Ashtekar & Yilmaz, Mycol. Prog. 21 (4, no. 42): 5. 2022. [MB 840642]. — Type: AMH 10349 (holotype). Ex-type: NFCCI 5017. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Vascosobrinhoana. DNA barcodes: ITS = MZ571358; BenA = MZ558484; CaM = MZ558492; RPB2 = MZ558482.
Notes: The identifier “MB 840643” cited in Ashtekar et al. (2022) is that of the series Vascosobrinhoana introduced in the same publication, but the species is nevertheless validly published (Art. F.5.6 San Juan Chapter F) (May et al. 2019) since “MB 840642” was issued by MycoBank prior to the publication of the name.
Penicillium scottii Visagie, Frisvad & K. Jacobs, Persoonia 46: 182. 2021. [MB 834430]. — Type: CBS H-22040 (holotype). Ex-type: CBS 138951 = IBT 31905 = DTO 185-F8 = CV 930. Infragen. class.: subgen. Penicillium sect. Canescentia ser. Canescentia. DNA barcodes: ITS = JX140812; BenA = JX140991; CaM = JX157383; RPB2 = KP016894.
Penicillium setosum T.K. George, Houbraken, L. Mathew & M.S. Jisha, Acta Bot. Brasil 36 (e2021abb0390): 6. 2022. [MB 842377]. — Type: CBS H-24872 (holotype). Ex-type: CBS 144865 = DTO 455-G4 = WSR 62 = MCC 1370 = NCFT NO 8222.16 = AMH-9974. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = KT852579; BenA = MF184995; CaM = MH105905; RPB2 = n.a.
Notes: In the original description (George et al. 2018), P. setosum was invalidly described because it was not stated that the holotype was preserved as a metabolically inactive culture. The name was subsequently validated by Barbosa et al. (2022).
Penicillium sexuale Rodr.-Andr., Cano & Stchigel, J. Fungi 7 (2, no. 126): 10. 2021. [MB 835941]. — Type: CBS H-24468 (holotype). Ex-type: CBS 146939 = FMR 17380. Infragen. class.: subgen. Aspergilloides sect. Crypta ser. Crypta. DNA barcodes: ITS = LR655195; BenA = LR655199; CaM = LR655203; RPB2 = LR655207.
Penicillium siccitolerans Rodr.-Andr., Cano & Stchigel, J. Fungi 7 (2, no. 126): 7. 2021. [MB 835939]. — Type: CBS H-24466 (holotype). Ex-type: FMR 17381. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = LR655193; BenA = LR655197; CaM = LR655201; RPB2 = LR655205.
Penicillium sicoris Torres-Garcia, Gené & Dania García, MycoKeys 86: 127. 2022. [MB 840559]. — Type: CBS H-24784 (holotype). Ex-type: CBS 148241 = FMR 18076. Infragen. class.: subgen. Penicillium sect. Paradoxa ser. Atramentosa. DNA barcodes: ITS = LR884497; BenA = LR884494; CaM = LR884496; RPB2 = LR884495.
Penicillium silybi Labuda, Kubátová, Raja & Oberlies, J. Fungi 7 (7, no. 557): 9. 2021. [MB 839113]. — Type: PRM 955189 (holotype). Ex-type: CBS 147777 = G85 = CCF 6562. Infragen. class.: subgen. Aspergilloides sect. Exilicaulis ser. Restricta. DNA barcodes: ITS = KF367458; BenA = MW774592; CaM = MW774591; RPB2 = AB860248.
Notes: Since Houbraken et al. (2020), P. allaniae, P. archerae, P. krskae and P. silybi were introduced bringing the number of accepted species in series Restricta to thirteen. However, the series clearly needs a taxonomic revision (Fig. 23 & Suppl. Fig. S21) as noted by Visagie et al. (2016). In the meantime, we accept the four recently introduced species.
Penicillium soli Doilom, C.F. Liao & D. Pem, Front. Microbiol. 11 (no. 585215): 13. 2020. [MB 557862]. — Type: MFLU 20-0432 (holotype). Ex-type: KUMCC 18-0202. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Janthinella. DNA barcodes: ITS = MT152337; BenA = MT161681; CaM = MT178249; RPB2 = MT384372.
Synonym: Penicillium cluniae [nom. inval. Art. 40.7 (Shenzhen)] Quintan., Avances en Alimentation y Mechora Animal 30: 174. 1990. [MB 130240].
Notes: Phylogenies resolved P. soli in the P. cluniae clade (Fig. 24 & Suppl. Fig. S22). The exception was RPB2. Pairwise comparisons showed that the ex-type sequences of both species were identical for BenA, but that there was some variation between them for CaM and RPB2. This suggests that they belong to the same species. Penicillium cluniae was previously included in the accepted species lists (Visagie et al. 2014, Houbraken et al. 2020), but was invalidly described by Quintanilla (1990) because no collection or herbarium was specified with the “1532” type designation. Consequently, we reduce P. cluniae as a synonym of P. soli.
Fig. 24.
Phylogenetic tree of Penicillium section Lanata-Divaricata series Janthinella and Simplicissima, based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S22.
Penicillium stangiae A.L. Alves, A.C.S. Santos, R.N. Barbosa, C.M. Souza-Motta, R.F.R. Melo, P.V. Tiago, Acta Bot. Brasil 36: e2022abb0006: 10. 2022. [MB 841261]. — Type: URM 94477 (holotype). Ex-type: URM 8347. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Dalearum. DNA barcodes: ITS = MW648590; BenA = MW646388; CaM = MW646390; RPB2 = MW646392.
Penicillium subfuscum Visagie & Yilmaz, Mycologia 115: 99. 2022. [MB 844188]. — Type: PREM 63220 (holotype). Ex-type: CMW 56196 = CBS 147455 = CN014C9. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Simplicissima. DNA barcodes: ITS = MT949907; BenA = MT957412; CaM = MT957454; RPB2 = MT957480.
Penicillium submersum Torres-Garcia, Gené & Dania García, MycoKeys 86: 129. 2022. [MB 840560]. — Type: CBS H-24785 (holotype). Ex-type: CBS 148242 = FMR 17140. Infragen. class.: subgen. Penicillium sect. Robsamsonia ser. Urticicola. DNA barcodes: ITS = LR814194; BenA = LR814187; CaM = LR814188; RPB2 = LR814195.
Penicillium taurinense S. Prencipe, Houbraken & D. Spadaro, Persoonia 44: 435. 2020. [MB 834715]. — Type: CBS H-24332 (holotype). Ex-type: CBS 145672 = DTO 333-B8 = CAS16. Infragen. class.: subgen. Penicillium sect. Robsamsonia ser. Glandicolarum. DNA barcodes: ITS = MF595981; BenA = MF595977; CaM = MF595979; RPB2 = MT253108.
Penicillium tealii Y.P. Tan, Bishop-Hurley, Marney & R.G. Shivas, Persoonia 49: 307. 2022. [MB 845000]. — Type: BRIP 72734c (holotype). Ex-type: BRIP 72734c. Infragen. class.: subgen. Aspergilloides sect. Cinnamopurpurea ser. Jiangxiensia. DNA barcodes: ITS = OP101639; BenA = OP039547; CaM = n.a.; RPB2 = OP039546.
Penicillium tirolense Kirchm., Embacher & Neuh., Fungal Syst. Evol. 10: 96. 2022. [MB 845496]. — Type: IBF2019/0162 (holotype). Ex-type: CBS 147625 = SF014017. Infragen. class.: subgen. Aspergilloides sect. Torulomyces ser. Torulomyces. DNA barcodes: ITS = MW145398; BenA = MW143069; CaM = MW143068; RPB2 = MW143067.
Penicillium tolerans Y.P. Tan, Bishop-Hurley, E. Lacey, Grice & R.G. Shivas, Index Austral. Fungi 3: 12. 2022. [MB 900151]. — Type: BRIP 64090a (holotype). Ex-type: BRIP 64090a. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Sclerotiorum. DNA barcodes: ITS = OK639006; BenA = OL741658; CaM = n.a.; RPB2 = n.a.
Synonym of: Penicillium sclerotiorum J.F.H. Beyma, Zentralbl. Bakteriol. Parasitenk., Abt. 2 96: 418. 1937. [MB 277708].
Notes: Species classified in Penicillium section Sclerotiorum were reviewed in Rivera & Seifert (2011) and Visagie et al. (2013), both who accepted a broad concept of P. sclerotiorum. Phylogenetically, the strains considered to belong to P. sclerotiorum can be divided into two clades, but without a taxonomic revision of the species, we consider P. tolerans to be a synonym of the former. (Fig. 27 & Suppl. Fig. S25)
Fig. 27.
Phylogenetic tree of Penicillium section Sclerotiorum, based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. glabrum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S25.
Penicillium ucsense A. Lenz & Houbraken, Antonie van Leeuwenhoek 115: 9. 2022. [MB 839721]. — Type: CBS H-24331 (holotype). Ex-type: CBS 146492 = DTO 426-B1 = IOC 4717 = 2HH. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Oxalica. DNA barcodes: ITS = OM914583; BenA = ON024157; CaM = ON024158; RPB2 = ON024159.
Synonym of: Penicillium hepuense L. Wang, PeerJ 10 (e13224): 11. 2022. [MB 841525].
Notes: Phylogenies resolved P. ucsense and P. hepuense in the same clade (Fig. 25 & Suppl. Fig. S23). Both were introduced in 2022, but P. hepuense was published first (2022/05/06 vs 2022/06/09) and thus has priority over P. ucsense.
Fig. 25.
Phylogenetic tree of Penicillium section Lanata-Divaricata series Dalearum, Oxalica and Rolfsiorum, based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to the series Oxalica clade. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S23.
Penicillium ulleungdoense D.H. Choi & J.G. Kim, Mycobiology 49: 48. 2021. [MB 835474]. — Type: KACC 48990 (holotype). Ex-type: KACC 48990. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Sclerotiorum. DNA barcodes: ITS = MN640087; BenA = MN737487; CaM = MN745074; RPB2 = MN756007.
Penicillium umkhoba Visagie & Yilmaz, Mycologia 115: 101. 2022. [MB 844189]. — Type: PREM 63222 (holotype). Ex-type: CMW 56200 = CBS 147457 = CN014D5. Infragen. class.: subgen. Aspergilloides sect. Sclerotiorum ser. Herqueorum. DNA barcodes: ITS = MT949912; BenA = MT957417; CaM = MT957459; RPB2 = MT957485.
Penicillium uttarakhandense Rajeshk., N. Ashtekar, Visagie, G. Anand & Yilmaz, Persoonia 46: 493. 2021. [MB 834093]. — Type: AMH 10225 (holotype). Ex-type: NFCCI 4808. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Simplicissima. DNA barcodes: ITS = MN967315; BenA = MN972443; CaM = MN972445; RPB2 = MN972447.
Notes: The phylogenetic relationship between P. brasilianum, P. onobense, P. paraherquei and P. skrjabinii is unresolved (Fig. 24 & Suppl. Fig. S22). Further studies including additional strains and/or extended datasets are needed. Here, we give benefit of doubt to the publisher.
Penicillium vaccaeorum Quintan., Mycopathologia 80: 77. 1982. [MB 109999]. — Type: CBS 148.83 (holotype). Ex-type: CBS 148.83 = DTO 009-E2 = CECT 2753. Infragen. class.: subgen. Aspergilloides sect. Citrina ser. Roseopurpurea. DNA barcodes: ITS = JN617689; BenA = JN606835; CaM = JN606543; RPB2 = JN606614.
Notes: Considered a synonym of P. sanguifluum by Houbraken, Frisvad & Samson (2011), this old name was more recently re-introduced by Torres-Garcia et al. (2022).
Penicillium vallebormidaense R.N. Barbosa & J.D.P. Bezerra, Persoonia 45: 371. 2020. [MB 837659]. — Type: CBS H-24527 (holotype). Ex-type: CBS 147064 = DTO 402-H5. Infragen. class.: subgen. Aspergilloides sect. Exilicaulis ser. Erubescentia. DNA barcodes: ITS = MT316359; BenA = MW115862; CaM = MW115863; RPB2 = MW115864.
Penicillium vickeryae Y.P. Tan & R.G. Shivas, Index Austral. Fungi 3: 10. 2022. [MB 900144]. — Type: BRIP 72552a (holotype). Ex-type: BRIP 72552a. Infragen. class.: subgen. Aspergilloides sect. Lanata-Divaricata ser. Simplicissima. DNA barcodes: ITS = OP903479; BenA = OP921966; CaM = n.a.; RPB2 = OP921965.
Penicillium vietnamense [nom. inval. Arts 40.7 & 40.8 (Shenzhen)] V.D. Nguyen & T.T. Pham, Mycobiology 50: 157. 2022. [MB 840587]. — Type: VTCC 930029 (holotype). Ex-type: VTCC 930029 = NTU DW14M = NITIA DW14M. Infragen. class.: subgen. Aspergilloides sect. Charlesia ser. Indica. DNA barcodes: ITS = MT102836; BenA = MT230561; CaM = ON209438; RPB2 = MT222288.
Synonym of: Penicillium chermesinum Biourge, Cellule 33: 284. 1923. [MB 260472].
Notes: This species was invalidly described. Phylogenies resolved P. vietnamense inside P. chermesinum (Fig. 17 & Suppl. Fig. S15), and even though CaM was not yet available for analysis, we consider P. vietnamense a synonym of P. chermesinum. We therefore choose not to validate this species.
Fig. 17.
Phylogenetic tree of Penicillium sections Aspergilloides, Charlesia, Cinnamopurpurea and Ramigena based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. taxi. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S15.
Penicillium xyleborini Visagie & W.J. Nel, Persoonia 47: 349. 2021. [MB 840990]. — Type: PREM 63078 (holotype). Ex-type: CMW 56800 = CN001D4. Infragen. class.: subgen. Penicillium sect. Ramosum ser. Soppiorum. DNA barcodes: ITS = MW504356; BenA = MW480817; CaM = MW480823; RPB2 = MW480824.
Rasamsonia oblata (Pitt & A.D. Hocking) Yanai & Udagawa, Jap. J. Mycol. 61: 93. 2020. [MB 836491]. Basionym: Penicillium oblatum Pitt & A.D. Hocking, Mycologia 77: 810. 1985. [MB 104603]. — Type: FRR 2234. Ex-type: CBS 258.87 = IMI 288719 = NBRC 33091. DNA barcodes: ITS = LC546718; BenA = LC546729; CaM = LC546740; RPB2 = n.a.
Notes: Penicillium oblatum was considered a doubtful species by Yilmaz et al. (2014), because sequence data obtained for the ex-type strain CBS 258.87 (= FRR 2234) indicated it as a close relative of T. dendriticus, but its morphology clearly did not match the original description of Pitt & Hocking (1985a). The species was recently revised based on sequences from IMI 288719 and NBRC 33091 (both ex-type strains), which showed that it represents a unique species in Rasamsonia (Yanai et al. 2020). Penicillium oblatum was described as strictly monoverticillate (Pitt & Hocking 1985a), with some subterminal metulae produced. However, Yanai et al. (2020) description of P. oblatum based on IMI 288719 showed both mono- and biverticillate conidiophores.
Rasamsonia sabulosa (Pitt & A.D. Hocking) Yanai & Udagawa, Jap. J. Mycol. 61: 96. 2020. [MB 836492]. Basionym: Penicillium sabulosum Pitt & A.D. Hocking, Mycologia 77: 810. 1985. [MB 104604]. — Type: FRR 2743. Ex-type: ATCC 56984 = FRR 2743 = IMI 288715. DNA barcodes: ITS = LC546720; BenA = LC546726; CaM = LC546742; RPB2 = n.a.
Notes: Penicillium sabulosum and P. corynephorum (CBS 256.87T) were until recently considered synonyms of P. smithii (CBS 276.83T) (Visagie et al. 2016). Penicillium sabulosum was reviewed by Yanai et al. (2020) using ATCC 56984 and they showed it is a unique Rasamsonia species, and a new combination was subsequently made. Morphologically, P. sabulosum was described as growing slowly on CYA at 25 °C (3–6 mm) (Pitt & Hocking 1985a), while P. corynephorum grows 32–40 mm (Pitt & Hocking 1985b) and P. smithii 30–35 mm (after 2 wk) (Quintanilla 1982). Additionally, P. sabulosum has roughened phialides which was not reported for the other species. Penicillium smithii (section Exilicaulis) remains an accepted species with P. corynephorum its synonym.
Talaromyces africanus Houbraken, Pyrri & Visagie, J. Fungi 7 (11, no. 993): 10. 2021. [MB 841228]. — Type: CBS H-24874 (holotype). Ex-type: CBS 147340 = DTO 179-C5 = KAS 3859. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = OK339610; BenA = OK338782; CaM = OK338808; RPB2 = OK338833.
Talaromyces albisclerotius B.D. Sun, A.J. Chen, Houbraken & Samson, MycoKeys 68: 88. 2020. [MB 833135]. — Type: CBS H-22837 (holotype). Ex-type: CBS 141839 = DTO 340-G5. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = MN864276; BenA = MN863345; CaM = MN863322; RPB2 = MN863334.
Talaromyces aspriconidius B.D. Sun, A.J. Chen, Houbraken & Samson, MycoKeys 68: 90. 2020. [MB 833134]. — Type: CBS H-22833 (holotype). Ex-type: CBS 141835 = DTO 340-F8. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MN864274; BenA = MN863343; CaM = MN863320; RPB2 = MN863332.
Talaromyces atkinsoniae Y.P. Tan, Bishop-Hurley, Bransgr. & R.G. Shivas, Persoonia 49: 323. 2022. [MB 845020]. — Type: BRIP 72528a (holotype). Ex-type: BRIP 72528a. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = OP059084; BenA = OP087524; CaM = n.a.; RPB2 = OP087523.
Talaromyces aureolinus L. Wang, Mycologia 113: 495. 2021. [MB 831405]. — Type: HMAS 248136 (holotype). Ex-type: AS 3.15865. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MK837953; BenA = MK837937; CaM = MK837945; RPB2 = MK837961.
Talaromyces bannicus L. Wang, Mycologia 113: 498. 2021. [MB 831406]. — Type: HMAS 248133 (holotype). Ex-type: AS 3.15862. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MK837955; BenA = MK837939; CaM = MK837947; RPB2 = MK837963.
Talaromyces brevis B.D. Sun, A.J. Chen, Houbraken & Samson, MycoKeys 68: 92. 2020. [MB 833132]. — Type: CBS H-22833 (holotype). Ex-type: CBS 141833. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MN864269; BenA = MN863338; CaM = MN863315; RPB2 = MN863328.
Talaromyces calidominioluteus Houbraken & Pyrri, J. Fungi 7 (11, no. 993): 14. 2021. [MB 841229]. — Type: CBS H-24875 (holotype). Ex-type: CBS 147313 = DTO 052-G3. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = OK339612; BenA = OK338786; CaM = OK338817; RPB2 = OK338837.
Talaromyces cavernicola V.C.S. Alves, J.D.P. Bezerra & R.N. Barbosa, Fungal Syst. Evol. 10: 157. 2022. [MB 846125]. — Type: URM 95155 (holotype). Ex-type: URM 8448 = FCCUFG 11. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = ON862935; BenA = OP672383; CaM = OP290543; RPB2 = OP290515.
Talaromyces chongqingensis X.C. Wang & W.Y. Zhuang, Biology 10: 10. 2021. [MB 570851]. — Type: HMAS 247849 (holotype). Ex-type: CGMCC 3.20482. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = MZ358001; BenA = MZ361343; CaM = MZ361350; RPB2 = MZ361357.
Talaromyces gaditanus (C. Ramírez & A.T. Martínez) Houbraken & Soccio, J. Fungi 7 (11, no. 993): 17. 2021. [MB 841226]. Basionym: Penicillium gaditanum C. Ramírez & A.T. Martínez, Mycopathologia 74: 165. 1981. [MB 112521]. — Type: IJFM 5146 (holotype). Ex-type: CBS 169.81 = DTO 228-B8 = ATCC 42230 = IMI 253792 = VKM F-2188 = IJFM 5146. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = MH861318; BenA = OK338775; CaM = OK338802; RPB2 = OK338827.
Notes: Penicillium gaditanum was considered a synonym of T. minioluteus by Yilmaz et al. (2014). A recent revision of T. minioluteus and related species revealed that P. gaditanum is a distinct species and was therefore combined in Talaromyces as T. gaditanus (Pyrri et al. 2021).
Talaromyces germanicus Houbraken & Pyrri, J. Fungi 7 (11, no. 993): 19. 2021. [MB 841227]. — Type: CBS H-24876 (holotype). Ex-type: CBS 147314 = DTO 055-D1. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = OK339619; BenA = OK338799; CaM = OK338812; RPB2 = OK338845.
Talaromyces ginkgonis W.C. Wang & W.Y. Zhuang, J. Fungi 8 (7, no. 647): 7. 2022. [MB 570954]. — Type: HMAS 247853 (holotype). Ex-type: CGMCC 3.20698. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = OL638158; BenA = OL689844; CaM = OL689846; RPB2 = OL689848.
Talaromyces gwangjuensis Hyang B. Lee & T.T.T. Nguyen, J. Fungi 7 (9, no. 722): 8. 2021. [MB 554801]. — Type: CNUFC HT19191 (holotype). Ex-type: CNUFC WT19-1. Infragen. class.: sect. Purpurei. DNA barcodes: ITS = MK766233; BenA = MZ318448; CaM = n.a.; RPB2 = MK912174.
Talaromyces haitouensis L. Wang, J. Fungi 8 (1, no. 36): 3. 2021. [MB 570868]. — Type: HMAS 350335 (holotype). Ex-type: AS 3.16101 = HR1-7. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MZ045695; BenA = MZ054634; CaM = MZ054637; RPB2 = MZ054631.
Talaromyces koreanus [as ‘koreana’] Hyang B. Lee, J. Fungi 7 (9, no. 722): 9. 2021. [MB 648574]. — Type: CNUFC HT19213 (holotype). Ex-type: CNUFC YJW2-13. Infragen. class.: sect. Helici. DNA barcodes: ITS = MZ315100; BenA = MZ318450; CaM = MZ332529; RPB2 = MZ332533.
Talaromyces nanjingensis X.R. Sun, X.Q. Wu & W. Wei, J. Fungi 8 (2, no. 155): 18. 2022. [MB 837590]. — Type: CCTCC-M 2012167 (holotype). Ex-type: CCTCC-M 2012167. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MW130720; BenA = MW147759; CaM = MW147760; RPB2 = MW147762.
Notes: The phylogenies of T. nanjingensis and its close relatives T. brevis and T. liani proved to be problematic (Fig. 31 & Suppl. Fig. S29). BenA resolved T. nanjingensis and T. liani as close relatives, while T. brevis is distantly related. Based on CaM, T. nanjingensis, T. liani and T. brevis are phylogenetically similar species. Finally, T. nanjingensis and T. brevis share RPB2 sequences, and these species are closely related to T. liani. A taxonomic revision including a broader strain sampling is required to assess this species complex, but for the time being, we accept T. nanjingensis as a distinct species.
Fig. 31.
Phylogenetic tree of Talaromyces section Talaromyces, based on a concatenated dataset of BenA, CaM, RPB2, and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to T. helicus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S29.
Talaromyces peaticola Jian Q. Tian & Jing Z. Sun, Stud. Fungi 6: 396. 2021. [MB 553909]. — Type: HMAS 247296 (holotype). Ex-type: CGMCC 3.18620. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = MF135613; BenA = MF284705; CaM = MF284703; RPB2 = MF284704.
Synonym of: Talaromyces diversus (Raper & Fennell) Samson, Yilmaz & Frisvad, Stud. Mycol. 70: 175. 2011. [MB 560649].
Notes: The BenA phylogeny resolved T. peaticola inside T. diversus (Fig. 33 & Suppl. Fig. S30) and we thus consider it a synonym of the latter.
Talaromyces penicillioides L. Wang, Mycologia 113: 501. 2021. [MB 831407]. — Type: HMAS 248132 (holotype). Ex-type: AS 3.15822. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MK837956; BenA = MK837940; CaM = MK837948; RPB2 = MK837964.
Talaromyces phuphaphetensis Nuankaew, Chuaseehar. & Somrith., J. Fungi 8 (8: no. 825): 7. 2022. [MB 844613]. — Type: BBH 49306 (holotype). Ex-type: TBRC 16281 = CV00299. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = ON692803; BenA = ON706960; CaM = ON706962; RPB2 = ON706964.
Talaromyces pulveris Crous, Persoonia 45: 293. 2020. [MB 837837]. — Type: CBS H-24417 (holotype). Ex-type: CBS 146831 = CPC 38523 = MUCL pd8781 = DTO 432-H1. Infragen. class.: sect. Purpurei. DNA barcodes: ITS = MW175345; BenA = MW173136; CaM = MW173099; RPB2 = MW173115.
Talaromyces resedanus (McLennan & Ducker) A.J. Chen, Houbraken & Samson, MycoKeys 68: 96. 2020. [MB 811695]. Basionym: Penicillium resedanum McLennan & Ducker, Austral. J. Bot. 2: 360. 1954. [MB 302422] — Type: IMI 062877 (holotype). Ex-type: CBS 181.71 = DTO 376-A7 = ATCC 22356 = FRR 578 = IMI 062877 = NRRL 578. Infragen. class.: sect. Subinflati. DNA barcodes: ITS = MN431413; BenA = MN969436; CaM = MN969355; RPB2 = MN969214.
Notes: Although an ITS sequence resolved P. resedanum in Talaromyces, Yilmaz et al. (2014) considered it a doubtful species because the CBS strain was not viable. The strain was subsequently successfully revived and formally combined as Talaromyces resedanus by Sun et al. (2020a). The identifier “MB 302422” cited in Sun et al. (2020a) is that of the basionym (P. resedanum), but the species is still validly published (Art. F.5.6 San Juan Chapter F) (May et al. 2019) because “MB 811695” was issued by MycoBank prior to the publication of the name. Sun et al. (2020a) considered T. omanensis (SQUCC 13153T) as synonym of T. resedanus based on morphology and DNA sequence data, which we follow here.
Talaromyces rosorhizae [as ‘rosarhiza’] [nom. inval. Art. 40.8 (Shenzhen)] H. Zhang & Y.L. Jiang, Biodivers. Data J. 9 (e70088): 11. 2021. [MB 662132]. — Type: GUCC 190040.1 (holotype). Ex-type: GUCC 190040.1. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MZ221603; BenA = MZ333143; CaM = MZ333137; RPB2 = MZ333141.
Synonym of: Talaromyces penicillioides L. Wang, Mycologia 113: 501. 2021. [MB 831407].
Notes: This species was invalidly described. Phylogenies resolved T. rosorhizae and T. penicillioides, both introduced in 2021, in the same clade (Fig. 31 & Suppl. Fig. S29). We therefore choose not to validate this species.
Talaromyces rufus B.D. Sun, A.J. Chen, Houbraken & Samson, MycoKeys 68: 99. 2020. [MB 833133]. — Type: CBS H-22832 (holotype). Ex-type: CBS 141834 = DTO 349-D7 = CGMCC 3.13203. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MN864272; BenA = MN863341; CaM = MN863318; RPB2 = MN863331.
Talaromyces samsonii (Quintan.) Houbraken & Pyrri, J. Fungi 7 (11, no. 993): 25. 2021. [MB 841230]. Basionym: Penicillium samsonii Quintan., Mycopathologia 91: 69. 1985. [MB 105614] — Type: CBS H-24877 (holotype). Ex-type: CBS 137.84 = DTO 304-C3 = DTO 169-G6 = CECT 2772 = IMI 282404 = IMI 327872. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = MH861709; BenA = OK338798; CaM = OK338824; RPB2 = OK338844.
Notes: Penicillium samsonii was considered a synonym of T. minioluteus by Yilmaz et al. (2014). A recent revision of T. minioluteus and related species showed that P. samsonii is a distinct species and therefore combined in Talaromyces as T. samsonii (Pyrri et al. 2021).
Talaromyces santanderensis [nom. inval. Art. 40.8 (Shenzhen)] B.E. Guerra-Sierra & L.A. Arteaga-Figueroa, J. Fungi 8 (10, no. 1042): 5. 2022. [MB 845323]. — Type: CBUDES:UDES:3068 (holotype). Ex-type: HF05. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = OP082331; BenA = OP067657; CaM = OP067656; RPB2 = OP067655.
Synonym of: Talaromyces lentulus X.-Z. Jiang & L. Wang, Sci. Rep. 8 (no. 4932): 3. 2018. [MB 824519].
Notes: This species was invalidly described. All phylogenies resolve T. santanderensis inside the T. lentulus clade (Fig. 31 & Suppl. Fig. S29). We consider T. santaderensis a synonym of T. lentulus and therefore choose not to validate this species.
Talaromyces satunensis Nuankaew, Chuaseehar. & Somrith., J. Fungi 8 (8: no. 825): 8. 2022. [MB 844614]. — Type: BBH 49305 (holotype). Ex-type: TBRC 16246 = CV00055. Infragen. class.: sect. Trachyspermi. DNA barcodes: ITS = ON692804; BenA = ON706961; CaM = ON706963; RPB2 = n.a.
Talaromyces saxoxalicus J. Trovão, F. Soares, I. Tiago & A. Portugal, Int. J. Syst. Evol. Microbiol. 71 (12, no. 5175): 4. 2021. [MB 834608]. — Type: MUM-H 20.30 (holotype). Ex-type: MUM 20.30. Infragen. class.: sect. Purpurei. DNA barcodes: ITS = MT039882; BenA = MT052003; CaM = n.a.; RPB2 = MT052004.
Talaromyces shilinensis X.C. Wang & W.Y. Zhuang, J. Fungi 8 (7, no. 647): 10. 2022. [MB 570955]. — Type: HMAS 247854 (holotype). Ex-type: CGMCC 3.20699. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = OL638159; BenA = OL689845; CaM = OL689847; RPB2 = OL689849.
Talaromyces sparsus L. Wang, Mycologia 113: 501. 2021. [MB 831409]. — Type: HMAS 248135 (holotype). Ex-type: AS 3.16003. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MT077182; BenA = MT083924; CaM = MT083925; RPB2 = MT083926.
Talaromyces teleomorphus [as ‚teleomorpha‘] Hyang B. Lee, Frisvad, P.M. Kirk, H.J. Lim & T.T.T. Nguyen, J. Fungi 7 (9, no. 722): 11. 2021. [MB 648560]. — Type: CNUFC HT19251 (holotype). Ex-type: CNUFC YJW2-5. Infragen. class.: sect. Helici. DNA barcodes: ITS = MZ315102; BenA = MZ318452; CaM = MZ332531; RPB2 = MZ332535.
Talaromyces tenuis B.D. Sun, A.J. Chen, Houbraken & Samson, MycoKeys 68: 86. 2020. [MB 833136]. — Type: CBS H-22838 (holotype). Ex-type: CBS 141840 = DTO 340-G9. Infragen. class.: sect. Tenues. DNA barcodes: ITS = MN864275; BenA = MN863344; CaM = MN863321; RPB2 = MN863333.
Talaromyces wushanicus X.C. Wang & W.Y. Zhuang, Biology 10 (8, no. 745): 15. 2021. [MB 570852]. — Type: HMAS 247848 (holotype). Ex-type: CGMCC 3.20481. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MZ356356; BenA = MZ361347; CaM = MZ361354; RPB2 = MZ361361.
Talaromyces yunnanensis Doilom & C.F. Liao, Front. Microbiol. 11 (no. 585215): 18. 2020. [MB 557863]. — Type: MFLU 20-0434 (holotype). Ex-type: KUMCC 18-0208. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MT152339; BenA = MT161683; CaM = MT178251; RPB2 = n.a.
Talaromyces zhenhaiensis L. Wang, J. Fungi 8 (1, no. 36): 7. 2021. [MB 570869]. — Type: HMAS 350336 (holotype). Ex-type: AS 3.16102 = ZH3-18. Infragen. class.: sect. Talaromyces. DNA barcodes: ITS = MZ045697; BenA = MZ054636; CaM = MZ054639; RPB2 = MZ054633.
Xerochrysium bohemicum Kubátová & Hubka, Persoonia 46: 519. 2021. [MB 839239]. — Type: PRM 954080. Ex-type: CCF 3693 = CBS 147157. DNA barcodes: ITS = MW798184; BenA = n.a.; CaM = n.a.; RPB2 = n.a.
Xerochrysium coryli (Crous & Decock) Visagie & Houbraken, published here. [MB 849335]. Basionym: Paraxerochrysium coryli Crous & Decock, Persoonia 47: 261. 2021. [MB 841830]. — Type: CBS H-24853 (holotype). Ex-type: CBS 148314 = CPC 41272 = MUCL 58103. DNA barcodes: ITS = OK664748; BenA = OK651216; CaM = n.a.; RPB2 = OK651178.
Notes: See notes for Xerochrysium coryli above in the description.
Accepted species published before 2020 not included in Houbraken et al. (2020)
The following species were not included the list published by Houbraken et al. (2020).
Aspergillus coreanus S.B. Hong, Frisvad & Samson, Int. J. Syst. Evol. Microbiol. 56: 485. 2006. [MB 521268]. — Type: CBS 117059 (holotype). Ex-type: CBS 117059 = NRRL 35590 = KACC 41659. Infragen. class.: subgen. Fumigati sect. Fumigati ser. Fumigati. DNA barcodes: ITS = JN943570; BenA = AY870758; CaM = AY870718; RPB2 = n.a.
Species with no or inconclusive sequence data available
The following species were accepted by Houbraken et al. (2020). However, since no sequence data are available for these, we exclude these names until their phylogenetic relationships can be verified.
Aspergillus argenteus [as ‘argentum’] J.N. Rai & H.J. Chowdhery, Kavaka 7: 19. 1980. [MB 116063]. — Type: MLLU 104. Ex-type: unknown.
Aspergillus beijingensis D.M. Li, Y. Horie, Yu X. Wang & R.Y. Li, Mycoscience 39: 299. 1998. [MB 446575]. — Type: CBM FD-285. Ex-type: CBM FD-285.
Aspergillus collembolorum Dörfelt & A.R. Schmidt, Mycol. Res. 109: 956, figs 1–9. 2005. [MB 344420]. — Type: Russia: Kaliningrad (Koenigsberg), in succinum Balticum, in exemplare subordines Entomobryomorpha (Collembola), C. & H. W. Hoffeins (coll. Hoffeins, Hamburg, no. 805, holotypus). Ex-type: unknown.
Aspergillus crassihyphae Wadhwani & N. Mehrotra, Indian Bot. Reporter: 52. 1985. [MB 105070]. — Type: unknown. Ex-type: unknown.
Aspergillus curviformis H.J. Chowdhery & J.N. Rai, Nova Hedwigia 32: 231. 1980. [MB 118396]. — Type: unknown. Ex-type: unknown.
Aspergillus ellipsoideus J.N. Rai & H.J. Chowdhery, Kavaka 7: 17. 1980. [MB 116064]. — Type: MLLU 107. Ex-type: unknown.
Aspergillus maritimus Samson & W. Gams, Adv. Pen. Asp. Syst.: 43. 1986. [MB 114709]. — Type: CBS 186.77. Ex-type: CBS 186.77.
Aspergillus qizutongii D.M. Li, Y. Horie, Yu X. Wang & R.Y. Li, Mycoscience 39: 301. 1998. [MB 446576]. — Type: CBM FD-284. Ex-type: CBM FD-284.
Aspergillus raianus [as ‘raianum’] H.J. Chowdhery, Curr. Sci. 48: 953. 1979. [MB 309239]. — Type: MLLU 110. Ex-type: unknown.
Aspergillus subunguis Wadhwani, Dudeja & M.P. Srivast., Curr. Sci. 53: 443. 1984. [MB 105934]. — Type: IMI 254637. Ex-type: IMI 254637.
Aspergillus tapirirae C. Ram & A. Ram, Atti Reale Accad. Sci. Napoli: 100. 1972. [MB 309245]. — Type: IMUFPe 2175. Ex-type: unknown.
Aspergillus vinosobubalinus Udagawa, Kamiya & Kaori Osada, Trans. Mycol. Soc. Japan 34: 255. 1993. [MB 361186]. — Type: CBM BF-33501. Ex-type: CBM BF-33501.
Aspergillus wangduanlii D.M. Li, Y. Horie, Yu X. Wang & R.Y. Li, Mycoscience 39: 302. 1998. [MB 447107]. — Type: CBM FD-283. Ex-type: CBM FD-283.
Penicilliopsis africana Samson & Seifert, Adv. Pen. Asp. Syst.: 408. 1985. [MB 114759]. — Type: unknown. Ex-type: unknown.
Penicilliopsis pseudocordyceps H.M. Hsieh & Y.M. Ju, Mycologia 9: 541. 2002. [MB 484663]. — Type: HAST (Taiwan) Hsieh & Ju 89112611. Ex-type: BCRC 33730.
Penicillium asymmetricum (Subramanian & Sudha) Houbraken & Samson, Stud. Mycol. 70: 47. 2011. [MB 561963]. Basionym: Thysanophora asymmetrica Subram. & Sudha, Kavaka 13: 88. 1985. [MB 135502]. — Type: unknown. Ex-type: unknown.
Penicillium coniferophilum Houbraken & Samson, Stud. Mycol. 70: 47. 2011. [MB 561968]. Basionym: Thysanophora striatispora G.L. Barron & W.B. Cooke, Mycopathologia et Mycologia Applicata 40 (3-4): 353. 1970. [MB 324607]. — Type: unknown. Ex-type: unknown.
Penicillium glaucoalbidum (Desmazières) Houbraken & Samson, Stud. Mycol. 70: 47. 2011. [MB 561965]. Basionym: Sclerotium glaucoalbidum Desm., Ann. Sci. Nat. Bot. 16: 329. 1851. [MB 212120]. — Type: unknown. Ex-type: unknown.
Penicillium longisporum (W.B. Kendr.) Houbraken & Samson, Stud. Mycol. 70: 47. 2011. [MB 561966]. Basionym: Thysanophora longispora W.B. Kendr., Can. J. Bot. 39 (4): 826. 1961. [MB 340086]. — Type: DAOM 63073. Ex-type: CBS 354.62 = DAOM 63073 = MUCL 4168.
Phialomyces fusiformis G. Delgado & Decock, Mycologia 95: 896. 2003. [MB 489106]. — Type: MUCL 43747. Ex-type: MUCL 43747.
Thermoascus taitungiacus K.Y. Chen & Z.C. Chen, Mycotaxon 60: 226. 1996. [MB 436720]. — Type: TAl-Mycology K-Y Chen 8709-2. Ex-type: unknown.
Species in Houbraken et al. (2020) synonymised afterwards
Aspergillus amoenus M. Roberg, Hedwigia 70: 138. 1931. [MB 250654]. — Type: Munster, isol. ex Berberis sp. fruit, M. Roberg (type locality, this specimen was not deposited into herbarium). Ex-type: NRRL 4838 = CBS 111.32. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652480; BenA = JN853946; CaM = JN854035; RPB2 = JN853824.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus austroafricanus Ž. Jurjević, S.W. Peterson & B.W. Horn, SIM 3: 67. 2012. [MB 800597]. — Type: BPI 880914. Ex-type: CBS 145748 = NRRL 233 = DTO 225-D8. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = JQ301891; BenA = JN853963; CaM = JN854025; RPB2 = JN853814.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus capensis Visagie, Hirooka & Samson, Stud. Mycol. 78: 105. 2014. [MB 809193]. — Type: CBS H-21810. Ex-type: CBS 138188 = DTO 179-E6. Infragen. class.: subgen. Circumdati sect. Flavipedes ser. Flavipedes. DNA barcodes: ITS = KJ775550; BenA = KJ775072; CaM = KJ775279; RPB2 = KP987020.
Synonym of: Aspergillus iizukae Sugiy., J. Fac. Sci. Univ. Tokyo, Sect. 3 9: 390. 1967. [MB 326636] (Sklenář et al. 2021).
Aspergillus costaricensis [as ‘costaricaensis’] Samson & Frisvad, Stud. Mycol. 50: 52. 2004. [MB 369151]. — Type: CBS H-13437. Ex-type: CBS 115574 = IBT 23401 = CECT 20579 = ITEM 7555. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = DQ900602; BenA = FJ629277; CaM = FN594545; RPB2 = HE984361.
Synonym of: Aspergillus tubingensis Mosseray, La Cellule 43: 245. 1934. [MB 255209] (Bian et al. 2022).
Aspergillus cvjetkovicii Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800599]. — Type: BPI 880909. Ex-type: NRRL 227 = CBS 599.65. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652440; BenA = EF652264; CaM = EF652352; RPB2 = EF652176.
Synonym of: Aspergillus creber Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800598] (Sklenář et al. 2022).
Aspergillus fructus Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 70. 2012. [MB 800600]. — Type: BPI 880915. Ex-type: NRRL 239 = CBS 584.65. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652449; BenA = EF652273; CaM = EF652361; RPB2 = EF652185.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus griseoaurantiacus Visagie, Hirooka & Samson, Stud. Mycol. 78: 112. 2014. [MB 809197]. — Type: CBS H-21814. Ex-type: CBS 138191 = DTO 267D8. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = KJ775553; BenA = KJ775086; CaM = KJ775357; RPB2 = KU866988.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus hongkongensis C.C. Tsang, T.W.S. Hui, K.C. Lee, J.H.K. Chen, A.H.Y. Ngan, E.W.T. Tam, J.F.W. Chan, A.L. Wu, M. Cheung, B.P.H. Tse, A.K.L. Wu, C.K.C. Lai, D.N.C. Tsang, T.L. Que, C.W. Lam, K.Y. Yuen, S.K.P. Lau & P.C.Y. Woo, Diagn. Microbiol. Infect. Dis. 84: 130. 2016. [MB 810279]. — Type: NBRC H-13268. Ex-type: CBS 145671 = HKU49 = NBRC 110693 = NCPF 7870 = BCRC FU30360 = DTO 351-C3. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = AB987907; BenA = LC000552; CaM = MN969320; RPB2 = LC000578.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus jensenii Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 70. 2012. [MB 800601]. — Type: BPI 880910. Ex-type: NRRL 58600. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = JQ301892; BenA = JN854007; CaM = JN854046; RPB2 = JN853835.
Synonym of: Aspergillus creber Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800598] (Sklenář et al. 2022).
Aspergillus neoniger Varga, Frisvad & Samson, Stud. Mycol. 69: 16. 2011. [MB 560390]. — Type: CBS H-20630. Ex-type: CBS 115656 = NRRL 62634. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = FJ491682; BenA = FJ491691; CaM = FJ491700; RPB2 = KC796429.
Synonym of: Aspergillus tubingensis Mosseray, La Cellule 43: 245. 1934. [MB 255209] (Bian et al. 2022).
Aspergillus pepii Despot, Kocsubé, Varga & Klarić, Mycol. Prog. 16: 67. 2017. [MB 817073]. — Type: SZMC 23791 (holotype). Ex-type: CBS 142028 = MFBF AV11051B IX = SZMC 22333. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = KU613368; BenA = KU613371; CaM = KU613365; RPB2 = n.a.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus piperis Samson & Frisvad, Stud. Mycol. 50: 57. 2004. [MB 500009]. — Type: CBS H-13434. Ex-type: CBS 112811 = IBT 24630 = IBT 26239 = NRRL 62631. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = EU821316; BenA = FJ629303; CaM = EU163267; RPB2 = KC796427.
Synonym of: Aspergillus luchuensis Inui, Journal of the Faculty of Science, Imperial University of Tokyo 15: 469. 1901. [MB 151291] (Bian et al. 2022).
Aspergillus protuberus Munt.-Cvetk., Mikrobiologia 5: 119. 1968. [MB 326650]. Basionym: Aspergillus versicolor var. protuberus (Munt.-Cvetk.) Kozak., Mycological Papers 161: 139. 1989. [MB 127752]. — Type: CBS 602.74. Ex-type: CBS 602.74 = NRRL 3505 = ATCC 18990 = QM 9804. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652460; BenA = EF652284; CaM = EF652372; RPB2 = EF652196.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus puulaauensis Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 71. 2012. [MB 800602]. — Type: BPI 880911. Ex-type: CBS 145750 = NRRL 35641 = DTO 225-G5. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = JQ301893; BenA = JN853979; CaM = JN854034; RPB2 = JN853823.
Synonym of: Aspergillus creber Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800598] (Sklenář et al. 2022).
Aspergillus tabacinus Nakaz., Y. Takeda, Simo & A. Watan., J. Agric. Chem. Soc. Japan 10: 177. 1934. [MB 539544]. — Type: CBS H-24287. Ex-type: CBS 122718 = NRRL 4791 = IFO 4098 = QM 9766 = WB 4791. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = EF652478; BenA = EF652302; CaM = EF652390; RPB2 = EF652214.
Synonym of: Aspergillus versicolor (Vuill.) Tirab., Ann. Bot. (Roma) 7: 9. 1908. [MB 172159] (Sklenář et al. 2022).
Aspergillus tennesseensis Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 73. 2012. [MB 800604]. — Type: BPI 880917. Ex-type: CBS 145752 = NRRL 13150 = DTO 225-F5. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = JQ301895; BenA = JN853976; CaM = JN854017; RPB2 = JN853806. (Sklenář et al. 2022)
Synonym of: Aspergillus creber Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800598] (Sklenář et al. 2022).
Aspergillus venenatus Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 73. 2012. [MB 800605]. — Type: BPI 880916. Ex-type: CBS 145753 = NRRL 13147 = DTO 225-F4. Infragen. class.: subgen. Nidulantes sect. Nidulantes ser. Versicolores. DNA barcodes: ITS = JQ301896; BenA = JN854003; CaM = JN854014; RPB2 = JN853803.
Synonym of: Aspergillus creber Jurjevic, S.W. Peterson & B.W. Horn, SIM 3: 69. 2012. [MB 800598] (Sklenář et al. 2022).
Aspergillus welwitschiae (Bres.) Henn. apud Wehmer, Centralbl. Bakteriol. Parasitenk., 2. Abth. 18: 294. 1907. [MB 490584]. Basionym: Ustilago welwitschiae Bres., Bol. Soc. Brot. 11: 68. 1893. [MB 176748]. — Type: CBS 139.54. Ex-type: CBS 139.54. Infragen. class.: subgen. Circumdati sect. Nigri ser. Nigri. DNA barcodes: ITS = FJ629340; BenA = MN969369; CaM = KC480196; RPB2 = MN969100.
Synonym of: Aspergillus niger Tiegh., Ann. Sci. Nat., Bot. ser. 5 8: 240. 1867. [MB 284309] (Bian et al. 2022).
Penicillium cluniae [nom. inval. Arts 40.7 (Shenzhen)] Quintan., Av. Aliment. Mejora Anim. 30: 174. 1990. [MB 130240].
Synonym of: Penicillium soli Doilom, C.F. Liao & D. Pem, Front. Microbiol. 11 (no. 585215): 13. 2020. [MB 557862] (this paper).
Talaromyces omanensis Halo, Maharachch., Al-Yahyai & Al-Sadi, Phytotaxa 404: 192. 2019. [MB 830302]. — Type: SQU H-106 (holotype). Ex-type: SQUCC 13153. Infragen. class.: sect. Subinflati. DNA barcodes: ITS = MH784402; BenA = MH794502; CaM = MH794503; RPB2 = n.a.
Synonym of: Talaromyces resedanus (McLennan & Ducker) A.J. Chen, Houbraken & Samson, MycoKeys 68: 96. 2020. [MB 811695] (Sun et al. 2020a).
DISCUSSION
In this study, we review Eurotiales species described since the previous accepted species list by Houbraken et al. (2020) up until 31 December 2022. Species were revised based on a phylogenetic species concept and the application of Genealogical Concordance Phylogenetic Species Recognition (GCPSR; Taylor et al. 2000), which is the most widely used species concept in Eurotiales. However, we believe that the polyphasic approach (or consilient concept of species) provides a holistic view of species and therefore consider it important to include morphology and, where possible, extrolite (secondary metabolite) as well as whole-genome sequence data, which will play an important role in the near future. Taxonomists dealing with Eurotiales have historically kept classifications practical and useful to the end user, an approach that has served the community dealing with these important fungi well. In case of doubt about the delimitation of species during this review, we have given preference to the publisher and accepted the name. However, we have classified newly published species as synonyms when the currently available data clearly do not support their novelty, while in cases where species were published without or with poor quality sequence data, we considered them doubtful because their future identification would not be possible. This follows Charles Thom’s idea that a species is only useful if others can identify it (Thom 1954). During this study, several trends were noted where descriptions can go wrong. We discuss these trends in more detail below and provide recommendations to avoid them in the future.
The ICNafp (Turland et al. 2018) governs the naming of fungi and sets out the rules for the valid and legitimate publication of names. Of the 160 new Eurotiales names introduced, 10 were invalid because they either did not comply with Art. 40.7 (the single herbarium, collection or institution in which the type is conserved must be specified), Art. 40.8 (the protologue must include a statement that the culture is preserved in a metabolically inactive state), or in some cases both. In addition to ICNafp, there are other helpful resources to guide valid publication of names. These include ‘The Code Decoded’ (Turland 2019) and several publications on behalf of the International Commission on the Taxonomy of Fungi (ICTF) outlining the best practises for describing a new species (Sigler & Hawksworth 1987, Seifert & Rossman 2010, Aime et al. 2021).
DNA sequence comparisons have become the most important tool for the classification and identification of fungal species. A reliable and complete reference database is crucial for this approach. The openly available and mostly complete ex-type DNA sequence reference sets (Samson et al. 2014, Visagie et al. 2014, Yilmaz et al. 2014, Houbraken et al. 2020) for ITS, BenA, CaM and RPB2 are a great resource. However, as critical as they may be, the ex-type sequences only serve as anchor points for the species, while additional sequences are needed to capture infraspecies variation and delimit species. Bian et al. (2022), Glässnerová et al. (2022) and Sklenář et al. (2022) emphasise the importance of these sequences, which should preferably come from strains isolated from different substrates and (ecological) regions. We therefore recommend not to compare putative new species only with ex-type strains in phylogenies where more extensive data is available as this does not provide information on species boundaries and can lead to incorrect conclusions. An example where this was a problem was the description of Penicillium tolerans as a close relative of P. sclerotiorum (Tan & Shivas 2022). The latter was subject to several taxonomic revisions with phylogenies that placed P. sclerotiorum strains into two closely related clades. However, both Rivera & Seifert (2011) and Visagie et al. (2013) concluded that from a morphological and ecological point of view there is no reason to consider these clades as separate species. In the case of P. tolerans, the ex-type strain (BRIP 64090a) resolves into the clade without the P. sclerotiorum ex-type strain (CBS 287.36) (Fig. 27). The phylogenies presented by Tan & Shivas (2022) did not include a comparison with all closely related sequences and left unmentioned their conclusions about these other ‘P. sclerotiorum’ strains and the reasons why the clade with P. tolerans should be considered distinct. In view of this, we consider P. tolerans a synonym of P. sclerotiorum.
Homology searches are a popular tool to quickly identify strains at the species level and generally works well if you get hits with well curated sequences. If a search does not find highly similar sequences, this may be due to several reasons. The sequence in question could belong to an unsequenced described species, represent an unsequenced genotype of an existing species or indicate a new species. The list of accepted species is very informative because, at least for Eurotiales, we can practically ignore the names not accepted and we have reference sequences of all known species. Dissimilarity higher than 4 %, especially in CaM and RPB2, is a relatively safe indication of new species in Aspergillus (Bian et al. 2022). Nevertheless, the results of homology searches should be interpreted with caution and should not be confused with phylogenetic relatedness. We recommend to always confirm new species or genotypes by phylogenetic analyses and not to rely only on homology search results.
Examples of low-quality sequence data used for species descriptions were noted for several recently described species. This has already been discussed for fungi (Nilsson et al. 2017) and remains problematic, even though careful contig assembly and proper trimming and base-calling should resolve this issue. In our analyses, the ITS alignments generated for Aspergillus sect. Circumdati revealed suspect sequences for both A. curvatus and A. gaarensis. Similarly, the sequences available for the recently published A. pseudopiperis and A. pseudotubingensis were considered to be of low quality (Bian et al. 2022), while Visagie et al. (2021) illustrated the same problem in the descriptions of P. attenuatum, P. ochotense and P. piltunense (Kirichuk, Pivkin & Matveeva 2016). Aspergillus pseudopiperis were considered a synonym of A. tubingensis (Bian et al. 2022), while P. attenuatum, P. ochotense and P. piltunense was considered synonyms of P. antarcticum (Visagie et al. 2021). In most cases, however, low-quality sequences make classification and future identification impossible, so that the species must be considered doubtful.
As mentioned above, the phylogenetic species concept and the application of GCPSR is important for the taxonomy of Eurotiales within our preferred consilient concept of species. GCPSR has largely proven to be a consistent approach to defining species boundaries. In principle, a species is delimited in the narrowest sense at the most recent node that forms consistently between different gene phylogenies (Taylor et al. 2000). In Eurotiales, the four most commonly used gene regions are ITS, BenA, CaM and RPB2, and we recommend that these are at least sequenced when describing new species. Recently, several Eurotiales species were introduced that lacked an ITS sequence, the official DNA barcode for fungi (Schoch et al. 2012). Admittedly, ITS is generally not informative at the species level, but we consider it good practice to sequence ITS at least for the ex-type strain of a new species. Several species have also been introduced without sequence data for all recommended genes. We cannot stress enough how important this is for the taxonomy of Eurotiales at a time when mycologists are struggling with the implications of the ‘Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their utilisation under the Convention on Biological Diversity’ and the interpretation by governments that restricts and, in many cases, prohibits the exchange of strains between mycologists from different countries. The ICNafp (Rec. 8B.1) recommends that strains of new species should be deposited in two recognised public culture or genetic resource collections, preferably in different countries. It is not inconceivable that strain sharing will become even more complicated in the future, making taxonomic revisions more difficult. This is a complex issue, but we recommend looking for ways to enable sharing of strains. Nevertheless, we believe it is critical that new species are described with as much data as possible, not only to prove conclusively that the proposed species are indeed new, but also to avoid a future in which species might be classified as doubtful because no material is available for study or a lack of data on the species renders it unrecognisable, as was the case with the many doubtful species described in the late 1800s, what Pitt (1980) called the ‘dark ages’.
The introduction of species with a single isolate is not ideal, but it is also not considered a problem, provided it is clearly unique based on all available data, which at least includes BenA, CaM and RPB2. Of course, there is always the risk that a species is not new when future data are collected. Currently, 467 of the 1 279 recognised Eurotiales species are known from only the ex-type strain and 219 species for which one additional reference sequence is available in GenBank. For Aspergillus, 143 of 453 accepted species are represented by the ex-type strain only, while an additional strain was sequenced for 93; for Penicillium, 175 of 535 accepted species are represented by the ex-type strain only, while an additional strain was sequenced for 75; and for Talaromyces, 88 of 203 accepted species are represented by the ex-type strain only, while an additional strain was sequenced for 34. Thus, for a considerable proportion of accepted species, we have little knowledge about infraspecific variability and thus also species boundaries, and much remains to be done to capture them. We therefore recommend that when isolating these poorly sequenced species, BenA, CaM and RPB2 sequences be generated and the strains deposited in recognised culture collections or, if this is not possible, the strains be shared with taxonomists working on these genera. Because of the uncertainty that a lack of DNA reference data can cause, we also recommend that workers who are uncertain about the identity of a species ask a taxonomist specialising in these fungi for a second opinion or advice.
Throughout the history of these genera, the way we describe species has evolved, bringing with it many taxonomic changes. The one thing that has remained is the community of collaborators who have adhered to best practises and worked together to create a standard for Eurotiales that serves everyone and provides a solid taxonomic basis for the study of these species. We believe that our recommendations will help to expand this community and provide further stability to the taxonomy of Eurotiales.
Fig. 2.
Phylogenetic tree of Aspergillus section Candidi based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. neotritici. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S2.
Fig. 4.
Phylogenetic tree of Aspergillus section Circumdati series Sclerotiorum based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. ochraceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S4.
Fig. 5.
Phylogenetic tree of Aspergillus section Cremei series Wentiorum based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S5.
Fig. 7.
Phylogenetic tree of Aspergillus sections Flavipedes and Janorum based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. terreus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S7.
Fig. 9.
Phylogenetic tree of Aspergillus section Nidulantes series Versicolores based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. aeneus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S9.
Fig. 11.
Phylogenetic tree of Aspergillus section Polypaecilum based on a concatenated dataset of BenA, CaM, RPB2, and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S11.
Fig. 13.
Phylogenetic tree of Aspergillus section Usti series Calidousti based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. ustus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S13.
Fig. 15.
Phylogenetic tree of Paecilomyces based on a concatenated dataset of BenA, CaM, RPB2 and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to Thermoascus crustaceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S14.
Fig. 20.
Phylogenetic tree of Penicillium section Canescentia based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S18.
Fig. 22.
Phylogenetic tree of Penicillium sections Crypta and Torulomyces based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to P. alfredii. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S20.
Fig. 26.
Phylogenetic tree of Penicillium sections Paradoxa, Ramosum and Robsamsonia, based on a concatenated dataset of BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S24.
Fig. 29.
Phylogenetic tree of Talaromyces section Helici, based on a concatenated dataset of BenA, CaM, RPB2, and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to T. purpureogenus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S27.
Fig. 30.
Phylogenetic tree of Talaromyces sections Purpurei and Subinflati, based on a concatenated dataset of BenA, CaM, RPB2, and ITS. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. See Suppl. Fig. S28.
Fig. 32.
Phylogenetic tree of Talaromyces showing the phylogenetic relationship of the recently introduced section Tenues, based on a concatenated dataset of BenA, CaM, RPB2, and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to Trichocoma paradoxa. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by superscript R. See Suppl. Fig. S30.
Acknowledgments
C.M. Visagie and N. Yilmaz received funding from the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Skłodowska-Curie grant agreement No. 101008129, project acronym “Mycobiomics”. C.M. Visagie was also supported by the University of Pretoria’s University Capacity Development Programme (UCDP) grant. V. Hubka was supported by the Czech Ministry of Health (grant NU21-05-00681). We are grateful to K. Bensch for her advice and guidance, and we thank L. Krieglsteiner (Spraitbach, Germany) and D. Malloch for sharing specimens of Aspergillus lentisci.
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supplementary Material: https://studiesinmycology.org/
Phylogenetic trees of Aspergillus section Aenei series Aenei based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. versicolor. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Candidi series Candidi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. neotritici. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus sections Cavernicolarum, Ochraceorosei and Sparsi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. bisporus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Circumdati series Sclerotiorum based on BenA, CaM, and RPB2, and series Steynii based on ITS. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ochraceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Cremei series Wentiorum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Flavi series Alliacei and Flavi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. avenaceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T. Representative strains are indicated by R.
Phylogenetic trees of Aspergillus section Flavipedes based on BenA, CaM, RPB2 and ITS. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. terreus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Fumigati based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. clavatus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Nidulantes series Versicolores based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. aeneus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Nigri series Japonici and Nigri based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. candidus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Polypaecilum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Terrei series Nivei and Terrei based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ambiguus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Usti series Calidousti based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ustus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Paecilomyces based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to Thermoascus crustaceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Aspergilloides, Charlesia, Cinnamopurpurea and Ramigena based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. taxi. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Brevicompacta based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. tularense (except RPB2 that was midpoint rooted). UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Fasciculata series Camembertiorum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. expansum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Canescentia based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Citrina based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. euglaucum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Crypta and Torulomyces based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. alfredii. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Exilicaulis and Gracilenta based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Lanata-Divaricata series Janthinella and Simplicissima, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Lanata-Divaricata series Dalearum, Oxalica and Rolfsiorum, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to the series Oxalica clade. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Paradoxa, Ramosum and Robsamsonia, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Sclerotiorum, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. glabrum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Rasamsonia, based on BenA, CaM and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to Trichocoma paradoxa. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by R.
Phylogenetic trees of Talaromyces section Helici, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. purpureogenus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Purpurei and Subinflati, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Talaromyces, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. helicus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Trachyspermi, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. purpureus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
List of reference species used for comparisons in this study.
REFERENCES
- Aime MC, Miller AN, Aoki T. et al. (2021). How to publish a new fungal species, or name, version 3.0. SIM 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bedak OA. (2020a). Aspergillus curvatus, a new species in section Circumdati isolated from an alkaline water of Lake Khadra in Wadi-El-Natron, Egypt. Asian Journal of Mycology 3: 325–334. [Google Scholar]
- Al-Bedak OA. (2020b). Aspergillus gaarensis, a new addition to section Circumdati from soil of Lake El-Gaar in Wadi-El-Natron, Egypt. Studies in Fungi 5: 59–65. [Google Scholar]
- Alves AL, Santos ACdS, Barbosa RdN. et al. (2022a). Penicillium gercinae and Penicillium stangiae (Eurotiomycetes, Ascomycota), two new species from soil in Brazil. Acta Botanica Brasilica 36: e2022abb0006. [Google Scholar]
- Alves VCS, Lira RA, Lima JMS. et al. (2022b). Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species. Fungal Systematics and Evolution 10: 139–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade KCR, Fernandes RA, Pinho DB. et al. (2021). Sequencing and characterization of an L-asparaginase gene from a new species of Penicillium section Citrina isolated from Cerrado. Scientific Reports 11: 17861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashtekar N, Rajeshkumar KC, Yilmaz N. et al. (2022). A new Penicillium section Citrina species and series from India. Mycological Progress 21: 42. [Google Scholar]
- Barbosa RdN, Santos JEFd, Bezerra JDP. et al. (2022). Brazilian Atlantic Forest and Pampa Biomes in the spotlight: an overview of Aspergillus, Penicillium, and Talaromyces (Eurotiales) species and the description of Penicillium nordestinense sp. nov. Acta Botanica Brasilica 36: e2021abb0390. [Google Scholar]
- Barbosa RN, Bezerra JDP, Souza-Motta CM. et al. (2018). New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek 111: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, Kusaya Y, Sklenář F. et al. (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology 102: 95–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodinaku I, Shaffer J, Connors AB. et al. (2019). Rapid phenotypic and metabolomic domestication of wild Penicillium molds on cheese. mBio 10: e02445–02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmee S, Wanasinghe DN, Calabon MS. et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111: 1–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DH, You YH, Lee IS. et al. (2020). Penicillium ulleungdoense sp. nov. from Ulleung Island in Korea. Mycobiology 49: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Boers J, Holdom D. et al. (2022). Fungal Planet description sheets: 1383–1435. Persoonia 48: 261–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ. et al. (2019). Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G. et al. (2020a). Fungal Planet description sheets: 1112–1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kolling G. et al. (2021a). Fungal Planet description sheets: 1182–1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž. et al. (2021b). Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi YH. et al. (2020b). Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doilom M, Guo JW, Phookamsak R. et al. (2020). Screening of phosphatesolubilizing fungi from air and soil in Yunnan, China: four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Frontiers in Microbiology 11: 585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174. [Google Scholar]
- George TK, Houbraken J, Mathew L. et al. (2018). Penicillium setosum, a new species from Withania somnifera (L.) Dunal. Mycology 10: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CLM, Lacey HJ, Vuong D. et al. (2020). Comprehensive chemotaxonomic and genomic profiling of a biosynthetically talented Australian fungus, Aspergillus burnettii sp. nov. Fungal Genetics and Biology 143: 103435. [DOI] [PubMed] [Google Scholar]
- Giraud F, Giraud T, Aguileta G. et al. (2010). Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. International Journal of Food Microbiology 137: 204–213. [DOI] [PubMed] [Google Scholar]
- Glässnerová K, Sklenář F, Jurjević Ž. et al. (2022). A monograph of Aspergillus section Candidi. Studies in Mycology 102: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra Sierra BE, Arteaga-Figueroa LA, Sierra-Pelaéz S. et al. (2022). Talaromyces santanderensis: A new Cadmium-tolerant fungus from Cacao soils in Colombia. Journal of Fungi 8: 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han PJ, Sun JQ, Wang L. (2021). Two new sexual Talaromyces species discovered in estuary soil in China. Journal of Fungi 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A. et al. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. (2011). Taxonomy of Penicillium section Citrina. Studies in Mycology 70: 53–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsube S, Visagie CM. et al. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Jiang X, Wu B. et al. (2020). Aspergillus jilinensis sp. nov. and its thermostable alkaline enzymes evaluation. Mycoscience 61: 205–211. [Google Scholar]
- Iliushin VA. (2022). Aspergillus sibiricus (Aspergillaceae, Eurotiales), a novel acid-tolerant species in Aspergillus section Fumigati. Phytotaxa 531: 63–72. [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuna S, Suwannarach N, Kumla J. et al. (2021). Growth enhancement of Arabidopsis (Arabidopsis thaliana) and onion (Allium cepa) with inoculation of three newly identified mineral-solubilizing fungi in the genus Aspergillus section Nigri. Frontiers in Microbiology 12: 705896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmair M, Embacher J, Heimdörfer D. et al. (2022). Penicillium poederi and P. tirolense, two new species of section Torulomyces. Fungal Systematics and Evolution 10: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichuk NN, Pivkin MV, Matveeva TV. (2016). Three new Penicillium species from marine subaqueous soils. Mycological Progress 16: 15–26. [Google Scholar]
- Kocsube S, Perrone G, Magista D. et al. (2016). Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology 85: 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda R, Bacher M, Rosenau T. et al. (2021). Polyphasic approach utilized for the identification of two new toxigenic members of Penicillium section Exilicaulis, P. krskae and P. silybi spp. nov. Journal of Fungi 7: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM. et al. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Lenz AR, Balbinot E, de Abreu FP. et al. (2022). Taxonomy, comparative genomics and evolutionary insights of Penicillium ucsense: a novel species in series Oxalica. Antonie van Leeuwenhoek 115: 1009–1029. [DOI] [PubMed] [Google Scholar]
- Liang L-J, Jeewon R, Dhandevi P. et al. (2021). A novel species of Penicillium with inhibitory effects against Pyricularia oryzae and fungal pathogens inducing Citrus diseases. Frontiers in Cellular and Infection Microbiology 10: 10:604504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloch D, Cain RF. (1972). The Trichocomataceae: Ascomycetes with Aspergillus, Paecilomyces, and Penicillium imperfect states. Canadian Journal of Botany 50: 2613–2628. [Google Scholar]
- Malloch D, Cain RF. (1973). The genus Thielavia. Mycologia 65: 1055–1077. [Google Scholar]
- May TW, Redhead SA, Bensch K. et al. (2019). Chapter F of the International Code of Nomenclature for algae, fungi, and plants as approved by the 11th International Mycological Congress, San Juan, Puerto Rico, July 2018. SIM 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR. et al. (2012). International Code of Nomenclature for algae, fungi and plants (Melbourne Code). Regnum Vegetabile 154. Koeltz Scientific Books, Glashütten, Germany. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O. et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTT, Frisvad JC, Kirk PM. et al. (2021a). Discovery and extrolite production of three new species of Talaromyces belonging to sections Helici and Purpurei from freshwater in Korea. Journal of Fungi 7: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTT, Kwan Noh KJ, Lee HB. (2021b). New species and eight undescribed species belonging to the families Aspergillaceae and Trichocomaceae in Korea. Mycobiology 49: 534–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VD, Pham TT. (2022). Penicillium vietnamense sp. nov., the first novel marine fungi species described from Vietnam with a unique conidiophore structure and molecular phylogeny of Penicillium section Charlesia. Mycobiology 50: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Sánchez-García M, Ryberg MK. et al. (2017). Read qualitybased trimming of the distal ends of public fungal DNA sequences is nowhere near satisfactory. MycoKeys 26: 13–24. [Google Scholar]
- Nuankaew S, Chuaseeharonnachai C, Preedanon S. et al. (2022). Two novel species of Talaromyces discovered in a karst cave in the satun UNESCO Global Geopark of Southern Thailand. Journal of Fungi 8: 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callahan D, Vaidya A, Donaldson L. et al. (2020). Penicillium rotoruae, a new species from an in-ground timber durability test site in New Zealand. Current Microbiology 77: 4129–4139. [DOI] [PubMed] [Google Scholar]
- Perini L, Gostincar C, Likar M. et al. (2023). Interactions of fungi and algae from the Greenland ice sheet. Microbial Ecology 86: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt J, Hocking AD. (2022). Fungi and Food Spoilage. 4th edn. Springer International Publishing. [Google Scholar]
- Pitt JI. (1980). The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, London. [Google Scholar]
- Pitt JI, Hocking AD. (1985a). Interfaces among genera related to Aspergillus and Penicillium. Mycologia 77: 810–824. [Google Scholar]
- Pitt JI, Hocking AD. (1985b). New species of fungi from Indonesian dried fish. Mycotaxon 22: 197–208. [Google Scholar]
- Pitt JI, Samson RA. (1993). Species names in current use in the Trichocomaceae (Fungi, Eurotiales). In: Names in current use in the family Trichocomaceae, Cladoniaceae, Pinaceae, and Lemnaceae (Greuter W. ed.) Koeltz Scientific Books, Königstein, Germany: 13–57. [Google Scholar]
- Pitt JI, Samson RA, Frisvad JC. (2000). List of accepted species and their synonyms in the family Trichocomaceae. In: Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification (Samson RA, Pitt JI. eds). Harwood Academic Publishers, Reading: 9–79. [Google Scholar]
- Pyrri I, Visagie CM, Soccio P. et al. (2021). Re-evaluation of the taxonomy of Talaromyces minioluteus. Journal of Fungi 7: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla JA. (1982). Cuatro nuevas especies de Penicillium aisladas en centeno: P. mariaecrucis, sp. nov., P. castellae, sp. nov., P. cieglerii, sp. nov., y P. smithii, sp. nov. Avances en Alimentation y Mechora Animal 23: 333–343. [Google Scholar]
- Quintanilla JA. (1990). Penicillium cluniae nov. sp. y P. burgense nov. sp., dos nuevas especies aisladas de suelo no cultivado. Avances en Alimentation y Mechora Animal 30: 174–180. [Google Scholar]
- Ramos SMS, Cruz R, Barbosa RdN. et al. (2021). Two new Penicillium section Sclerotiorum species from sugarcane soil in Brazil. Mycological Progress 20: 823–835. [Google Scholar]
- Rivera KG, Seifert KA. (2011). A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Studies in Mycology 70: 139–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta S. (1873). Dei Parassiti Vegetali. 1–592. [Google Scholar]
- Rodriguez-Andrade E, Stchigel AM, Cano-Lira JF. (2021). New xerophilic species of Penicillium from soil. Journal of Fungi 7: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, Didiot E, Rodriguez de la Vega RC. et al. (2020). Domestication of the emblematic white cheese-making fungus Penicillium camemberti and its diversification into two varieties. Current Biology 30: 4441–4453. [DOI] [PubMed] [Google Scholar]
- Samson RA, Visagie CM, Houbraken J. et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78: 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J. et al. (2011). Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S. et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Rossman AY. (2010). How to describe a new fungal species. SIM 1: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler L, Hawksworth DL. (1987). International Commission on the Taxonomy of Fungi (ICTF) Code of Practice for Systematic Mycologists. Mycopathologia 99: 3–7. [DOI] [PubMed] [Google Scholar]
- Silva JJ, Fungaro MH, Wang X. et al. (2022). Deep genotypic species delimitation of Aspergillus section Flavi isolated from brazilian foodstuffs and the description of Aspergillus annui sp. nov. and Aspergillus saccharicola sp. nov. Journal of Fungi 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JJD, Iamanaka BT, Ferranti LS. et al. (2020). Diversity within Aspergillus niger clade and description of a new species: Aspergillus vinaceus sp. nov. Journal of Fungi 6: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Callicott KA, Orbach MJ. et al. (2020). Molecular analysis of S-morphology aflatoxin producers from the United States reveals previously unknown diversity and two new taxa. Frontiers in Microbiology 11: 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Glässnerová K, Jurjević Ž. et al. (2022). Taxonomy of Aspergillus series Versicolores: species reduction and lessons learned about intraspecific variability. Studies in Mycology 102: 53–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Houbraken J. et al. (2021). Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and proposal of four new species. Studies in Mycology 99: 100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetik M, Berraf-Tebbal A, Gramaje D. et al. (2022). Paecilomyces clematidis (Eurotiales, Thermoascaceae): a new species from Clematis root. Phytotaxa 559: 238–246. [Google Scholar]
- Sun B, Luo C, Gerald BF. et al. (2022a). Four new species of Aspergillus subgenus Nidulantes from China. Journal of Fungi 8: 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B-D, Chen AJ, Houbraken J. et al. (2020a). New section and species in Talaromyces. MycoKeys 68: 75–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B-D, Houbraken J, Frisvad JC. et al. (2020b). New species in Aspergillus section Usti and an overview of Aspergillus section Cavernicolarum. International Journal of Systematic and Evolutionary Microbiology 70: 5401–5416. [DOI] [PubMed] [Google Scholar]
- Sun B-D, Huang P-P, Wei H-L. et al. (2020c). Aspergillus telluris, a new soil derived species belonging to Aspergillus subgenus Polypaecilum. Phytotaxa 455: 137–151. [Google Scholar]
- Sun B-D, Visagie CM, Chen AJ. et al. (2021). A taxonomic review of Penicillium section Charlesia. Mycological Progress 20: 1383–1397. [Google Scholar]
- Sun XR, Xu MY, Kong WL. et al. (2022b). Fine identification and classification of a novel beneficial Talaromyces fungal species from masson Pine rhizosphere soil. Journal of Fungi 8: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Bishop-Hurley S, Marney TS. et al. (2021). Nomenclatural novelties. Index Fungorum 503: 1–8. [Google Scholar]
- Tan YP, Bishop-Hurley SL, Shivas RG. et al. (2022). Fungal Planet description sheets: 1436–1477. Persoonia 49: 261–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Shivas RG. (2022). Nomenclatural novelties. Index of Australian Fungi 3: 1–21. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S. et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Thom C. (1954). The evolution of species concepts in Aspergillus and Penicillium. Annals of the New York Academy of Sciences 60: 24–34. [DOI] [PubMed] [Google Scholar]
- Tian JQ, Wang YF, Sun JZ. (2021). Talaromyces peaticola (Aspergillaceae, Eurotiales), a new species from the Zoige wetlands, China. Studies in Fungi 6: 391–399. [Google Scholar]
- Torres-Garcia D, Gene J, Garcia D. (2022). New and interesting species of Penicillium (Eurotiomycetes, Aspergillaceae) in freshwater sediments from Spain. MycoKeys 86: 103–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovao J, Soares F, Tiago I. et al. (2021). Talaromyces saxoxalicus sp. nov., isolated from the limestone walls of the Old Cathedral of Coimbra, Portugal. International Journal of Systematic and Evolutionary Microbiology 71: 005175. [DOI] [PubMed] [Google Scholar]
- Turland NJ. (2019). The Code Decoded: A user’s guide to the International Code of Nomenclature for algae, fungi, and plants. 2nd edn. Pensoft Pulisher, Sofia, Bulgaria. [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR. et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Koeltz Botanical Books, Glashütten, Germany. [Google Scholar]
- Visagie CM, Yilmaz N. (2023). Along the footpath of Penicillium discovery: Six new species from the Woodville Big Tree Forest Trail. Mycologia 115: 87–106. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Frisvad JC, Houbraken J. et al. (2021). A re-evaluation of Penicillium section Canescentia, including the description of five new species. Persoonia 46: 163–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Goodwell M, Nkwe DO. (2021). Aspergillus diversity from the Gcwihaba Cave in Botswana and description of one new species. Fungal Systematics and Evolution 8: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC. et al. (2014). Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Rodriques C. et al. (2013). Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31: 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J. et al. (2016). A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. SIM 7: 75–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Höhnel F. (1910). Fragmente zur Mykologie (X. Mitteilung 475. Dichlaena lentisci). Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften in Wien Mathematisch-Naturwissenschaftliche Classe 119: 399–400. [Google Scholar]
- Wang XC, Zhuang WY. (2022a). New species of Aspergillus (Aspergillaceae) from tropical islands of China. Journal of Fungi 8: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Zhuang WY. (2022b). New species of Talaromyces (Trichocomaceae, Eurotiales) from southwestern China. Journal of Fungi 8: 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Xu X, Wang L. (2021). Four new species of Talaromyces section Talaromyces discovered in China. Mycologia 113: 1–17. [DOI] [PubMed] [Google Scholar]
- Xu KX, Shan XN, Ruan Y. et al. (2022). Three new Penicillium species isolated from the tidal flats of China. PeerJ 10: e13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai M, Maekawa S, Udagawa S-I. (2020). A phylogenetic revision of Penicillium oblatum and P. sabulosum as heat resistant molds. Japanese Journal of Mycology 61: 91–101. [Google Scholar]
- Yilmaz N, Visagie CM, Houbraken J. et al. (2014). Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology 78: 175–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wei TP, Mao YT. et al. (2021a). Ascodesmisrosicola sp. nov. and Talaromycesrosarhiza sp. nov., two endophytes from Rosaroxburghii in China. Biodiversity Data Journal 9: e70088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-F, Zhou S-Y, Eurwilaichitr L. et al. (2020). Culturable mycobiota from karst caves in China II, with descriptions of 33 new species. Fungal Diversity 106: 29–136. [Google Scholar]
- Zhang ZK, Wang XC, Zhuang WY. et al. (2021b). New species of Talaromyces (Fungi) isolated from soil in Southwestern China. Biology 10: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhori A-NA, Al-Bedak OA, Abdel-Kareem MM. (2020). Aspergillus sakultaensis, a new species in section Flavipedes isolated from Sohag Governorate, Egypt. Journal of Environmental Studies 20: 21–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees of Aspergillus section Aenei series Aenei based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. versicolor. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Candidi series Candidi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. neotritici. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus sections Cavernicolarum, Ochraceorosei and Sparsi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. bisporus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Circumdati series Sclerotiorum based on BenA, CaM, and RPB2, and series Steynii based on ITS. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ochraceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Cremei series Wentiorum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Flavi series Alliacei and Flavi based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was rooted to A. avenaceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T. Representative strains are indicated by R.
Phylogenetic trees of Aspergillus section Flavipedes based on BenA, CaM, RPB2 and ITS. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. terreus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Fumigati based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. clavatus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Nidulantes series Versicolores based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. aeneus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Nigri series Japonici and Nigri based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. candidus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Polypaecilum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. cremeus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Terrei series Nivei and Terrei based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ambiguus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Aspergillus section Usti series Calidousti based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to A. ustus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Paecilomyces based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to Thermoascus crustaceus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Aspergilloides, Charlesia, Cinnamopurpurea and Ramigena based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. taxi. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Brevicompacta based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. tularense (except RPB2 that was midpoint rooted). UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Fasciculata series Camembertiorum based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. expansum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Canescentia based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Citrina based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. euglaucum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Crypta and Torulomyces based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. alfredii. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Exilicaulis and Gracilenta based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Lanata-Divaricata series Janthinella and Simplicissima, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. stolkiae. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Lanata-Divaricata series Dalearum, Oxalica and Rolfsiorum, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to the series Oxalica clade. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium sections Paradoxa, Ramosum and Robsamsonia, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. The tree was midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Penicillium section Sclerotiorum, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to P. glabrum. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Rasamsonia, based on BenA, CaM and ITS. Strains of recently described species are shown in bold coloured text. The tree was rooted to Trichocoma paradoxa. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by superscript T. Representative strains are indicated by R.
Phylogenetic trees of Talaromyces section Helici, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. purpureogenus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Purpurei and Subinflati, based on BenA, CaM, and RPB2. Strains of recently described species are shown in bold coloured text. Trees were midpoint rooted. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Talaromyces, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. helicus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
Phylogenetic trees of Talaromyces section Trachyspermi, based on BenA, CaM and RPB2. Strains of recently described species are shown in bold coloured text. Trees were rooted to T. purpureus. UltraFast Bootstrap support values higher than 95 % are shown at relevant branches. Ex-type strains are indicated by T.
List of reference species used for comparisons in this study.