Abstract
While protein kinases have been successfully targeted in a variety of diseases, protein phosphatases remain an underutilized therapeutic target, in part because of incomplete characterization of their effects on signaling networks. The PH domain Leucine-rich repeat Protein Phosphatase (PHLPP) is a relatively new player in the cell signaling field, and increasing roles in controlling the balance between cell survival, proliferation, and apoptosis are being increasingly identified. While originally characterized for its tumor-suppressive function in deactivating the pro-survival kinase Akt, recent evidence suggests that PHLPP may have an opposing role in promoting survival. This review summarizes the current knowledge of PHLPP, and highlights emerging functions as both a tumor suppressor and an oncogene. Understanding the context-dependent functions of PHLPP is essential for appropriate therapeutic intervention.
Keywords: PHLPP, Akt, PKC, Phosphatase, Phosphorylation, Cancer, Transcription
1. INTRODUCTION
The PH domain Leucine-rich repeat Protein Phosphatase (PHLPP) belongs to the protein phosphatase magnesium/manganese (PPM)-dependent ‘shrub’ in the human protein phosphatome [1] (Figure 1A). Originally identified in a rational search for phosphatases that would suppress growth factor signaling by dephosphorylating a key regulatory site on Akt, the hydrophobic motif [2], the repertoire of PHLPP substrates has rapidly expanded in the past few years. Indeed, PHLPP is emerging as a central regulator of diverse biologies, not only by acutely controlling the activity and stability of kinases, but also by suppressing processes such as inflammatory signaling via its dephosphorylation of transcription factors. This review provides an overview of PHLPP, highlighting recent findings on both its tumor suppressive and oncogenic roles in cancer and inflammatory signaling.
Figure 1. Phylogeny and Domain Architecture of PHLPP Phosphatases.
(A) PHLPP1 and PHLPP2 are PP2C-type Ser/Thr protein phosphatases, belonging to the protein phosphatase magnesium/manganese (PPM)-dependent shrub of the protein phosphatase phylogenetic tree [115]. (B) Domain structure of ancestral and mammalian PHLPP. Yeast CYR1 contains a Gα domain, 26 Leucine rich repeats (LRR), a PP2C phosphatase domain (PP2C) fused to adenylate cyclase (AC), and C-terminal cyclase-associated protein 1 binding domain (CAP). Mammalian PHLPP retains the LRR domain (18 in PHLPP1 and 19 in PHLPP2) and PP2C phosphatase domain in addition to a PH domain (PH), N-terminal extension (NTE) containing a bipartite Nuclear Localization Signal (NLS), and C-terminal PDZ ligand. Scale bar denotes 100 amino acids (a.a.).
2. DOMAINS OF PHLPP
The PHLPP family comprises two genes that share the same domain composition and 41% overall amino acid identity in humans (Figure 1B). PHLPP is evolutionarily conserved in yeast as Cyr1 [3], which curiously contains a C-terminal adenylate cyclase domain in addition to the Leucine-rich repeat (LRR) and PP2C domains of PHLPP [4] (Figure 1). The regulatory modules of PHLPP are located on the same polypeptide, distinguishing it from other Ser/Thr phosphatases, which typically consist of separate phosphatase and regulatory modules encoded by multiple genes [5].
2.1. N-Terminal Extension
The N-terminus of PHLPP1 (but not PHLPP2) has an approximately 50 kDa unstructured extension containing a bipartite Nuclear Localization Signal (NLS) that regulates the nuclear entry of PHLPP1 [6]. This NLS is opposed by a Nuclear Exclusion Signal (NES) at the C-terminal end of the LRR segment of the protein [6]. The PHLPP1 NLS is necessary for its nuclear function in controlling transcription (see Section 3.4).
2.2. PH Domain
The PH domains of PHLPP isozymes have weak homology to the canonical PH domain sequence [7] and binding studies reveal that the module has a ‘borderline’ affinity for phosphoinositides [8]. The PHLPP PH domain was acquired later in evolution compared to its other domains and is only found in mammalian PHLPP genes, suggesting that the spectrum of PHLPP substrates has broadened during evolution as more regulatory modules were added to the phosphatase. The PH domain is necessary for the effective interaction and dephosphorylation of protein kinase C (PKC) by PHLPP in cells (see Section. 3.2.) [9], suggesting that PHLPP regulation of PKC evolved after its role in controlling Akt activity.
2.3. LRR Domain
The LRR domains are characterized by multiple repeats of leucine residues at invariant positions. The LRR domain of PHLPP1 is composed of 18 repeats [10] and was reported to mediate the interaction of PHLPP1 with the nucleotide-free form of K-Ras in the brain, blocking K-Ras nucleotide binding and downstream activation of ERK [11]. Interestingly, the LRR domain of the yeast homolog, Cyr1, which contains 26 repeats [12], has also been reported to bind Ras in yeast [13], although with a different functional outcome: yeast Cyr1 interacts with the GTP-bound form of Ras to promote its activation [14].
2.4. PP2C Domain
The phosphatase domain of the PHLPP family members belongs to the PPM ‘shrub’ of Ser/Thr protein phosphatases (Figure 1A). This shrub houses the well-characterized PP2Cα (PPM1A), whose structure has been solved [15], and the recently identified Rab-GTPase phosphatase PPM1H, which counteracts LRRK2 signaling [16]. A hallmark of PP2C phosphatases is their resistance to common Ser/Thr phosphatase inhibitors such as okadaic acid (OA) and microcystin [17, 18]. The insensitivity of PHLPP isozymes to OA provides a useful tool to distinguish between PHLPP-specific activity and that of other non-PP2C phosphatases [2, 19, 20]. The catalytic activity of the PHLPP PP2C domain is relatively low (on the order of one reaction per second towards phospho-peptide substrates [20]), suggesting that stoichiometric association with substrates via its protein interaction modules is an important determinant for substrate dephosphorylation in cells.
2.5. PDZ Ligand
The last four carboxyl-terminal residues of PHLPP1 (DTPL) and PHLPP2 (DTAL) encode Type 1 PDZ (PSD-95/Disc-large/ZO-1) ligands. In the case of PHLPP1, its PDZ ligand is crucial for dephosphorylation of Akt in cells, as well as its pro-apoptotic and tumor-suppressive effects in a glioblastoma xenograft model [2]. The PDZ ligands of PHLPP1 and PHLPP2 also bind the scaffold, Na+/H+ exchanger regulatory factor 1 (NHERF1) (see Section 4.1.), an interaction that localizes the phosphatases near the membrane where they can dephosphorylate Akt [21]. The PHLPP PDZ ligand is conserved in lower organisms such as C. elegans and Drosophila, suggesting that antagonizing Akt signaling is an evolutionarily conserved function of PHLPP.
3. PHLPP SUBSTRATES
3.1. Hydrophobic Motif Phosphatase
The hydrophobic motif is a phosphorylation site on the C-terminal segment of a large number of AGC kinases, including Akt and PKC, that serves as a regulatory switch for kinase function (Figure 2, green square on PKC, Akt, and S6K) [22]. Originally identified in PKC and S6K [23], this phosphorylation site entered the limelight upon its acceptance as a marker for the activation state of Akt [24]. Highlighting the importance of this phosphorylation site, the hydrophobic motif is among the most frequently mutated post-translational modification sites among AGC kinases in human cancer [25, 26]. This site remains the best characterized substrate of the PHLPP [27].
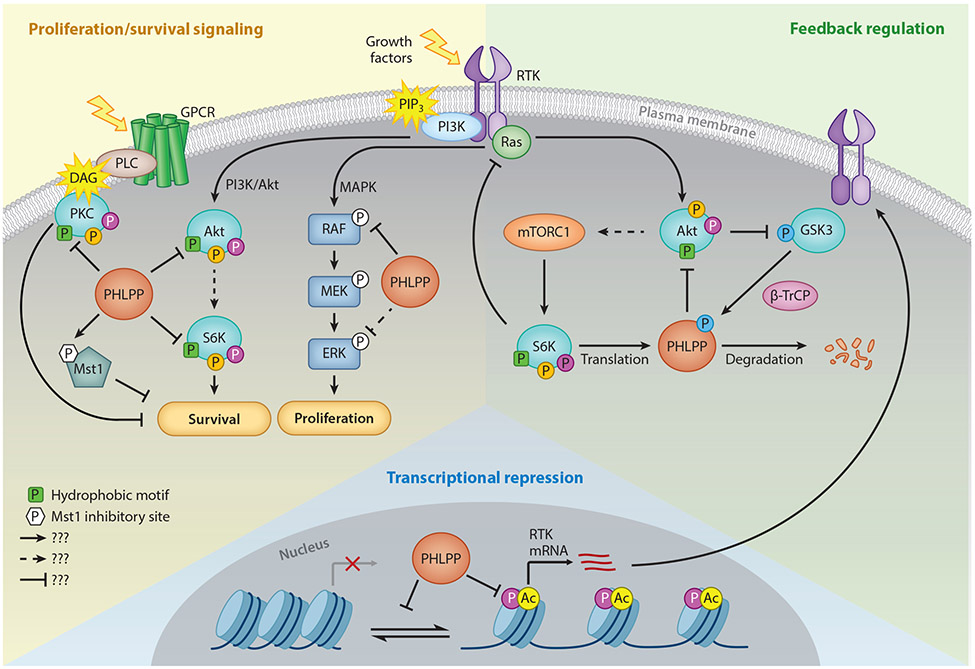
Figure 2. PHLPP Function in Growth Factor Signaling.
Schematic showing some of the substrates and signaling pathways regulated by PHLPP. In general, PHLPP suppresses proliferative and survival pathways through direct dephosphorylation of Akt, S6K1, Mst1, and Raf (red sector). PHLPP dephosphorylates the hydrophobic motif (green square with P) of Akt and S6K1 to inactivate them, while PHLPP activates the pro-apoptotic kinase, Mst1, via removal of an inhibitory phosphorylation (white hexagon with P). PHLPP also dephosphorylates PKC isozymes on the hydrophobic motif, which destabilizes them and reduces their cellular levels. Filled in circles on Akt, S6K1, and PKC denote other key phosphorylation sites, including the PDK-1 site on the activation loop (magenta circle) and mTORC2-regulated turn motif (orange circle). In addition, PHLPP suppresses the transcription and hence steady-state protein levels of RTKs such as the EGFR, thus dampening PI3K survival and ERK proliferative pathways, by an epigenetic mechanism (blue sector). PHLPP itself is under feedback regulation by components of the PI3K pathway (tan sector). High Akt activity increases PHLPP protein levels by suppressing GSK3β-dependent degradation. Furthermore, high mTORC1 activity increases S6K1-dependent translation of PHLPP (left). Moreover, S6K1 activity also activates a negative feedback loop that suppresses Akt activity.
3.1.1. Akt
Akt is acutely activated by reversible phosphorylation at two sites, the activation loop (Thr308 in Akt1; pink circle on Akt in Figure 2) and the hydrophobic motif (Ser473 in Akt1), following activation of the PI3-kinase pathway [24]. Signaling is terminated by dephosphorylation of the activation loop, catalyzed by PP2A family members, and of the hydrophobic motif, catalyzed by PHLPP (Figure 2). The PHLPP-mediated dephosphorylation inhibits Akt-triggered apoptosis and suppresses tumor growth [2, 28]. PHLPP1 specifically interacts with and dephosphorylates Akt2, modulating the phosphorylation of HDM2 and glycogen synthase kinase (GSK)-3α to prevent p53 degradation; whereas PHLPP2 specifically interacts with and dephosphorylates Akt1, inhibiting the phosphorylation of p27 to block cell cycle progression [28]. Both PHLPP isozymes interact with and dephosphorylate Akt3, inhibiting the phosphorylation of GSK-3β and TSC2 to suppress cell survival [28]. The amplified activity of Akt following loss of PHLPP stimulates the well-characterized negative feedback loop from ribosomal protein S6 kinase 1 (S6K1), such that knock-down of both PHLPP1 and PHLPP2 does not have an additive effect on Akt activity compared to knock-down of only one PHLPP isozyme, unless S6K1 is inhibited [28]. The PHLPP-mediated dephosphorylation of Akt is also conserved in Drosophila [2]. Interestingly, a common polymorphism in the substrate-binding region of the phosphatase domain of PHLPP2, L1016S, impairs the ability of PHLPP to dephosphorylate Akt in cells [29], without impairing other biological functions [30].
3.1.2. PKC
In contrast to Akt, PKC is constitutively phosphorylated at the hydrophobic motif (Ser660 in PKCβII) [31]. Furthermore, phosphorylation of this site does not control the acute activity of the kinase, rather it controls the chronic stability of the enzyme. Phosphorylation at this site occurs shortly after biosynthesis by intramolecular autophosphorylation, one of a series of phosphorylation events in the ‘maturation’ of PKC; phosphorylation of the hydrophobic motif followed PDK-1-catalyzed phosphorylation of the activation loop (Figure 2, magenta circle on PKC) and mTORC2-dependent phosphorylation of the turn motif (Figure 2; orange circle on PKC). Phosphate at the hydrophobic motif is necessary for PKC to adopt an autoinhibited conformation that is resistant to degradation, and loss of phosphate at this site results in the degradation of the enzyme. Because PHLPP controls the phosphorylation state of this site, it plays a critical role in setting the cellular levels of PKC. The steady-state levels of both conventional (Ca2+-dependent) and novel (Ca2+-independent) PKC isozymes are regulated by PHLPP [9]. While extensive biochemical and cellular studies have clearly established that PKC lacking phosphate at this site is unstable and degraded, this finding was recently validated in human patient samples: analysis of over 5,000 patient tumor samples from 19 different cancers revealed a 1:1 correlation between PKC levels and phosphorylation at the hydrophobic motif [32]. Thus, if PKC is not phosphorylated at the hydrophobic motif, it is degraded. As mentioned in Section 2.2., the cellular recognition of PKC by PHLPP depends on the PH domain of PHLPP, a module that is dispensable for PHLPP to dephosphorylate Akt in cells, and is independent of the PDZ ligand, a motif that is necessary for PHLPP to dephosphorylate Akt in cells. Furthermore, co-immunoprecipitation studies revealed that PHLPP1 recognizes PKC when it is in an “open” conformation and not when it is in a stable and autoinhibited conformation [32]. This conformational sensing of PKC was recently shown to provide a “quality control” step in the maturation of PKC, whereby newly-synthesized PKC must ultimately become phosphorylated to adopt the autoinhibited and stable conformation which ultimately is reversibly activated in response to second-messengers [32]. PHLPP1 was shown to bind newly-synthesized PKC to ensure that any aberrant enzyme that cannot be autoinhibited is dephosphorylated at the hydrophobic motif and shunted to degradation. Accordingly, cancer-associated mutations in the PKCβ autoinhibitory pseudosubstrate segment prevent autoinhibition and are subject to PHLPP1-catalyzed dephosphorylation and degradation [32]. Thus, PHLPP1 binds to both newly-synthesized PKC and activated PKC to enact the dephosphorylation and degradation of improperly folded or aberrantly active enzymes [32]. PKC quality control by PHLPP1 presents a potentially ubiquitous mechanism by which PKC is lost in cancer. Compromised PKC quality control, conversely, may enable PKC gain-of-function effects that drive neurodegeneration [31].
3.1.3. S6K1
S6K1 is another AGC kinase family member that is a direct substrate of PHLPP (Figure 2), establishing a role for PHLPP in regulating protein translation initiation and cell size. Activation of S6K1 downstream of mTOR is controlled by growth factors, nutrients, and energy balance. Full activation of S6K1 requires phosphorylation on both the hydrophobic motif (Thr389), mediated by mTORC1, and the activation loop (Thr229), by PDK-1 [33]. Consistent with its effect on other substrates from the AGC kinase family, PHLPP selectively dephosphorylates S6K1 on its hydrophobic motif in vitro, and overexpression studies have revealed that PHLPP reduces S6K1 phosphorylation in cells independently of Akt dephosphorylation [34]. Functionally, PHLPP depletion resulted in increased phosphorylation of ribosomal protein S6 (rpS6) and increased binding of rpS6 to the translation initiation complex, correlating with increased cell size, protein content, and rate of cap-dependent translation [34]. As noted in Section 3.1., PHLPP depletion also activates the S6K1-mediated negative feedback regulation of Akt.
3.2. Mst1
PHLPP can also induce apoptosis in cancer cells independently of its regulation of Akt and PKC. Specifically, it binds and dephosphorylates Mst1 at an inhibitory site, Thr387 (Figure 2), leading to its activation and induction of apoptosis through p38 and JNK [35]. Mst1 is crucial for apoptosis and is frequently implicated in tumor growth [36]. Therefore, PHLPP and Mst1 synergize to promote a much greater increase in apoptosis than through either protein alone. Furthermore, Akt and Mst1 are reciprocally inhibitory [37, 38]. PHLPP may thus also induce apoptosis by indirect activation of Mst1 via inhibition of Akt activity.
3.3. MAPK Pathway
Early studies reported that PHLPP1 directly interacts with K-Ras via its LRR domain to negatively regulate the Ras-Raf-MEK-ERK pathway involved in memory formation in the hypothalamus [11, 39]. Additionally, a subsequent study reported a positive feedback loop whereby oncogenic H-Ras can inhibit PHLPP1 expression in MCF-10A and HCT116 cells, leading to activation of the p38 pathway and subsequent inhibition of anoikis, a caspase-dependent cell death program [40]. PHLPP was also reported to reduce ERK phosphorylation (Figure 2), as knock-down of PHLPP1 was shown to increase ERK phosphorylation in hippocampal cultures [41]. Additionally, PHLPP1 and PHLPP2 interact directly with the proto-oncogene Raf1 and dephosphorylate Raf1 in vitro on Ser338 (Figure 2), thereby inhibiting its activity and contributing to the negative regulation of MAPK signaling [42].
PHLPP also suppresses ERK signaling by a mechanism unrelated to any of its previously characterized functions as a phosphatase for kinases: by suppressing the transcription of receptor tyrosine kinases (RTKs) to reduce their steady-state levels (Figure 2) [30]. Specifically, nuclear PHLPP opposes histone phosphorylation and acetylation, modifications associated with open chromatin, to reduce the expression of many RTKs including the epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), and insulin receptor (INSR) [30]. This function of PHLPP depends on its nuclear localization and is accompanied by direct binding of PHLPP to chromatin [30]. PHLPP1 and PHLPP2 were also shown to interact with and dephosphorylate c-Jun transcription factor at Ser63/73, inhibiting its transcriptional activity [43]. As AP-1 proteins such as c-Jun are responsible for enhancer-mediated EGFR expression in EGFR-driven cancers such as glioblastoma and head and neck squamous cell carcinoma, it is plausible that PHLPP also reduces EGFR levels through inactivation of its transcription factors [44]. Therefore, PHLPP-mediated suppression of MAPK signaling proceeds both through direct inactivation of pathway components, such as dephosphorylation of Raf1, and also by transcriptional processes involving chromatin silencing and inactivation of transcription factors that downregulate growth factor receptor expression.
3.4. STAT1
One of the most recently identified substrates of PHLPP is the transcription factor Signal Transducer and Activator of Transcription 1 (STAT1) (Figure 3) [6]. Members of this family of transcription factors are recruited from the cytosol to cytokine-bound receptors, where they are phosphorylated on a key Tyr reside (Tyr701 on STAT1) by Janus Kinases (JAKs); this event promotes their dimerization and nuclear entry where they bind to specific promoter sequences and initiate gene transcription [45]. They also transduce signals from type 1 and II interferons following ligation of interferon receptors, resulting ultimately in the transcription of inflammatory genes. Their transcriptional activity is enhanced by phosphorylation of a key regulatory Ser (Ser727 for STAT1) and it is this site that is opposed by PHLPP1. Biochemical studies have revealed that nuclear PHLPP1 binds STAT1 to specifically dephosphorylate Ser727 with no effect on Tyr701 [6]. STAT1 dephosphorylation by PHLPP1 mediates resolution of the inflammatory response by profoundly altering the transcriptome. Analysis of differentially expressed genes in bone marrow-derived macrophages from Phlpp1−/− treated with the major LPS component KLA revealed enrichment of STAT transcription factor motifs in 46% of the upregulated genes (92 of 199 genes upregulated compared to WT) [6]. This regulation of STAT1 cements a role of PHLPP1 in suppressing inflammatory signaling.
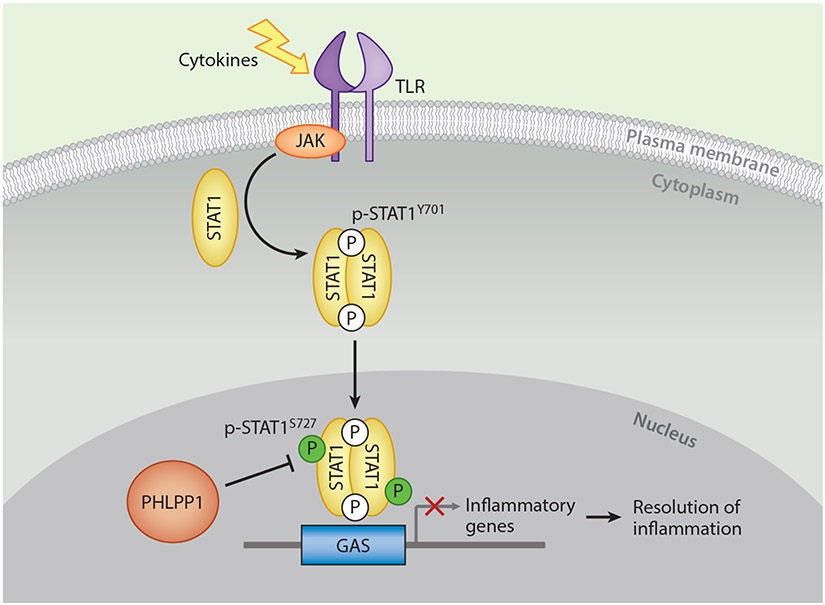
Figure 3. Emerging Roles for PHLPP in Inflammatory Signaling.
Binding of IFNγ to interferon receptors activates inflammatory transcriptional programs. Among the mediators of this inflammatory response are components of the JAK/STAT pathway: phosphorylation of STAT1 transcription factors on Tyr701 by Janus kinases (JAKs) induces STAT1 dimerization, nuclear translocation, and activation to promote transcription of target genes. This activity is enhanced by phosphorylation on Ser727. PHLPP1 opposes STAT1-mediated transcription to promote resolution of the inflammatory response by directly desphosphorylating Ser727 to tune STAT1 activity [6].
3.5. Other Substrates
The number of proteins identified to interact with PHLPP is steadily increasing, particularly with the surge in unbiased proteomics approaches. In this regard, the recent analysis of the “human interactome” by Mann and colleagues identified PHLPP1 as binding six interrogated proteins in HeLa cells: protein phosphatase 2 regulatory subunit B″α (PPP2R3A), the mitotic checkpoint kinase BUB1, mitogen activated kinase MAPK8/JNK1, the histone deacetylase HDAC1, the TATA-binding protein associated factor (TAF15), and the transcriptional regulator protein MAX [46]. Identification of novel PHLPP interactors additionally elucidates the subcellular localizations where PHLPP functions. For example, Rac1 was shown to recruit PHLPP1 to lysosomes to promote chaperone-mediated autophagy through suppression of Akt [47]. In more targeted analyses, PHLPP2 was reported to interact with the tumor suppressor PML (promyelocytic leukemia) in the nucleus, an interaction that promotes the dephosphorylation of nuclear Akt. This association is inhibited in many cancers by deregulated protein kinase CK2, which phosphorylates PML and shunts it to degradation, thus dismantling the PHLPP phosphatase platform that allows access to Akt [48]. This demonstrates that PHLPP is capable of synergizing with other tumor suppressor proteins to inhibit Akt activity. PHLPP2 was also shown to suppress NF-κB signaling, a pro-inflammatory signaling pathway activated in many cancers, by disrupting the Bcl10-MALT1 ubiquitin-ligase complex that enables IKKβ phosphorylation and subsequent NF-κB activation [49]. Identification of interactors that localize PHLPP1 and PHLPP2 to the nucleus will further contribute to their expanding repertoire of nuclear functions, including core transcriptional programs in disease contexts (discussed further in Section 6.).
4. REGULATION OF PHLPP
4.1. Protein Scaffolds
Protein scaffolds are likely to play a key role in controlling specificity and efficacy in downstream signaling by PHLPP. The quantitative analysis of the human interactome in HeLa cells reported intracellular concentrations of PHLPP1 and PHLPP2 as approximately 1 nM each, with substrates such as Akt1 at 30 nM [46]. This underscores the importance of subcellular targeting to position PHLPP near its relevant targets. The key role of spatial organization in dictating the function of PHLPP is illustrated by the finding that loss of the PHLPP1 PDZ ligand abolishes its recognition of Akt2 in cells, whereas loss of the PH domain abolishes its recognition of PKC [9]. Positioning PHLPP near its substrates affords tight control, essential for the regulation of enzymes such as Akt, in which the correct balance between phosphorylated and dephosphorylated states is essential for homeostasis.
Several scaffolds that bind PHLPP have been identified. FKBP51 binds both PHLPP isozymes to promote the specific suppression of Ser473 phosphorylation and inhibition of Akt activity [50, 51], a function that is deregulated in diverse diseases from prostate cancer to mitochondrial dysfunction and neurodegeneration [51-54]. The scaffold Scribble binds the PHLPP1 C-terminal segment (but not the PDZ ligand) via its own LRR to localize the phosphatase near the plasma membrane for effective regulation of Akt [55]; knock-down in DLD1 colon cancer cells results in a reduction of PHLPP1 at the membrane fraction and higher accumulation in the cytoplasm of, leading to an increase in Akt Ser473 phosphorylation. Another scaffold protein that localizes PHLPP to the plasma membrane is NHERF1. This tandem PDZ domain scaffold coordinates two suppressors of PI3K signaling, PTEN and PHLPP, to allow effective suppression of Akt signaling. PTEN binds PDZ1 of NHERF1 [56] and PHLPP2 binds PDZ2, whereas PHLPP1 has been reported to bind both PDZ domains. Deregulated PI3K signaling is prevalent in glioblastoma, and the tumor suppressive complex of PTEN-NHERF1-PHLPP1 is frequently disabled in high-grade glioblastoma tumors, correlating with enhanced Akt activation and low patient survival [21].
4.2. Transcriptional Regulation
A number of studies have reported on the transcriptional regulation of PHLPP. In chondrocytes, the binding of histone deacetylase 3 (HDAC3) to Smad binding elements in the PHLPP1 promoter was shown to suppress the transcription of PHLPP1, resulting in decreased PHLPP1 protein levels, increased Akt activity, and chondrocyte hypertrophy. This association of HDAC3 with the PHLPP1 promoter was released by TGFβ treatment, indicating that PHLPP1 transcription is enhanced by this cytokine [57]. HDAC3 was also shown to bind the PHLPP1 promoter in breast cancer cell lines: in this study, the long noncoding RNA Xist (X inactive-specific transcript) decreased HDAC3 recruitment to the PHLPP1 promoter, increasing histone acetylation on the promoter and subsequent expression of PHLPP1 mRNA to increase PHLPP1 protein levels and suppress Akt phosphorylation [58]. Another study showed that DNA hypermethylation of the PHLPP1 gene leads to suppression of PHLPP1 transcription in melanoma and promotes Akt activation [59]. The region in the PHLPP1 promoter that is involved in transcriptional activation of PHLPP1 is between −64 and −3 and contains a consensus binding site for the transcription factor Sp1. Indeed, deletion of the Sp1-binding site or Sp1 knock-down inhibited transcription of PHLPP1 [59], and lipolysaccharide (LPS) was shown to negatively regulate PHLPP expression in macrophages by an Sp1-dependent mechanism [60]. Consistent with promoter methylation regulating PHLPP transcription, high PHLPP1 mRNA levels in the cartilage from osteoarthritis patients correlated with reduced PHLPP1 promoter methylation [61]. In this study, the expression of PHLPP1 was shown to be induced in chondrocytes by inhibition of cytosine methylation or treatment with pro-inflammatory cytokines that can induce DNA demethylation, IL6 and TNFα [61]. A high degree of PHLPP1 methylation at the end of exon 1 has also been reported to strongly correlate with low levels of PHLPP1 protein [62]. Lastly, the transcription of both PHLPP isozymes has been reported to be inhibited by Cullin 4B (CUL4B), the scaffold protein of the CRL4B-E3 ligase complex, which was reported to bind the PHLPP promoter and monoubiquitinate H2AK119, thus repressing transcription [63].
The mRNA levels of CYR1, the yeast homolog of PHLPP1 which also has an adenylate cyclase module, are sensitive to glucose availability. As yeast exhaust their glucose supply, they reach a growth plateau before a diauxic shift to utilize other carbon sources. At this point, Cyr1 mRNA and protein levels decrease, resulting in decreased cAMP levels [64]. Whether mammalian PHLPP mRNA is also regulated by glucose remains to be explored.
4.3. Post-Translational Regulation
The steady-state levels of PHLPP protein are sensitive to Akt activity by both translational and post-translational mechanisms (Figure 2). First, Gao and coworkers established that a key downstream effector of Akt, mTORC1, promotes the translation of PHLPP1 and PHLPP2 mRNA in colon and breast cancer cells [65]. This regulation of protein translation can be independent of Akt function, as Akt-independent mechanisms to activate mTORC1, such as amino acid or glucose starvation, can also enhance PHLPP protein expression. This provides an additional negative feedback loop to suppress Akt activity when mTORC1 activity is elevated, complementing the well-known negative feedback loop mediated by S6K1 suppressing PIP3 generation. In this case, Akt itself is directly kept in check by higher PHLPP activity. Secondly, cellular levels of PHLPP1 protein are enhanced via a phosphodegron that is negatively regulated by Akt activity [66]. Activated Akt phosphorylates and inactivates GSK-3β, inhibiting GSK-3β-dependent phosphorylation at sites in the PP2C domain of PHLPP1. This in turn prevents ubiquitination by the β-TrCP-containing Skp-Cullin 1-F-box protein (SCF) complex (SCFβ-TrCP) and subsequent proteasomal degradation. This second feedback mechanism ensures that high Akt activity is counterbalanced by high PHLPP. SGT1, which was reported to be over-expressed in gastric cancer, was also reported to enhance the binding between PHLPP1 and β-TrCP, therefore promoting PHLPP1 degradation and amplified Akt signaling [67]. This feedback loop is lost in aggressive glioblastoma due to altered localization of β-TrCP from the cytoplasm (in normal brain tissue) to the nucleus (in astrocytoma cell lines), spatially separating it from PHLPP1, uncoupling the levels of a negative regulator PHLPP from the level of its substrate, phosphorylated Akt [68]. Thus, high Akt activity ramps up the amount of PHLPP protein in the cells by 1] increasing protein translation and 2] preventing protein degradation.
PHLPP protein translation is inhibited by various microRNAs (miRNA), including miR224, miR-17~92 cluster, and miR375. These bind the 3’-UTR region of PHLPP mRNA, leading to decreased PHLPP protein expression and consequently, enhanced Akt activation [69-71]. Interestingly, many miRNAs, including miR-17~92, are located in chromosomal regions that are amplified in diverse cancers [72], and it was shown that suppression of miR-17~92 increased PHLPP2 protein levels to promote terminal differentiation of myeloid leukemia cells [73].
PHLPP has also been reported to bind several deubiquitinating enzymes (DUBs) that deubiquitinate and stabilize it, such as ubiquitin-specific protease (USP)1 [74], USP46 [75], USP12, and WDR48, an activator of several DUBs [76]. Loss of such DUBs has been observed in several cancers and is correlated with decreased PHLPP expression levels and amplified Akt phosphorylation [75]. A recent study reported the formation of a protein complex between PHLPP1, PHLPP2, Akt, the Fanconi anemia complementation group proteins FANCI and FANCD2, and the DUBs USP1 and UAF1 [77]. In response to DNA damage, the interaction between FANCI, Akt and PHLPP1 is reduced, resulting in an increase of Akt phosphorylation and activity [77]. This identifies yet another complex that positions Akt near its negative regulator PHLPP to control its activity.
PHLPP is replete with predicted post-translational modification sites, and many have been identified by unbiased phosphoproteomic approaches [78], yet surprisingly few have been characterized. The coming years are likely to see a significant increase in our understanding of how post-translational modifications regulate the stability, activity, and subcellular location of PHLPP.
5. ROLE OF PHLPP IN CANCER
5.1. Tumor Suppressive Role
PHLPP is an accepted tumor suppressor based on animal models and analysis of human tumors. Loss of PHLPP1 causes neoplasia and, when co-deleted with PTEN, carcinoma in mouse prostate [79]. Samples from metastatic prostate tumors reveal frequent co-deletion of PTEN and PHLPP1 genes compared to samples from primary tumors. Interestingly, PHLPP2 levels increase following co-deletion of PTEN and PHLPP1 in an mTOR-dependent manner, similar to the observed increase in p53 levels. Therefore, it is not surprising that late stage metastatic prostate samples from patients also show a high rate of co-deletion of PHLPP1/2 and TP53 genes with PTEN to further break this feedback between loss of TP53 and PTEN and enhanced PHLPP2 levels [79]. PHLPP levels are reduced or lost in many other different cancers (as reviewed in [80]). In chronic lymphocytic leukemia (CLL), with 13q14 deletion, PHLPP mRNA is lower or undetectable in more than 50% of cases [81]. In colon cancer, the expression of both PHLPP isozymes is decreased [82]. In glioblastoma cell lines, the mRNA of PHLPP1 is 40% lower compared with that of normal astrocytoma cell lines [68]. In melanoma, where the PI3K/Akt pathway is activated in up to 70% of cases, PHLPP1, but not PHLPP2, mRNA and protein levels are reduced as a result of hypermethylation of the PHLPP1 gene [59]. The chromosomal regions of PHLPP1 and PHLPP2 are adjacent to other tumor suppressor genes where a frequent loss of heterozygosity occurs in tumors: PHLPP1 is on 18q21.33, one of the most frequently lost regions in colon cancer [83, 84], and PHLPP2 is on 16q22.3, a region frequently lost in breast [85], ovarian [86], liver [87], kidney [88] and prostate [89] tumors. Therefore, low levels of PHLPP can serve as a biomarker in these cancer contexts.
5.1.1. Suppression of Akt signaling.
The best characterized mechanism by which PHLPP acts as a tumor suppressor is through inhibition of Akt activity (Figure 2). An abundance of studies have shown a correlation between reduced levels of PHLPP and increased Ser473 phosphorylation of Akt in different cancer cells, such as metastatic breast cancer [90], colon and glioblastoma cell lines [2, 82], CLL B-cells [91], and melanoma [59]. Over-expression of PHLPP in cancer cells leads to inhibition of tumor growth, increased apoptosis, and decreased Akt Ser473 phosphorylation [28, 82, 91]. Interestingly, 30% of the population has a heterozygous polymorphism in the PP2C domain of PHLPP2 in which a Leu residue is substituted with Ser at position 1016. As noted in Section 3.1., the Ser1016 variant of PHLPP2 has decreased activity towards Akt both in vitro and in cells, resulting in reduced apoptosis. In heterozygous individuals the loss of the wild-type Leu1016 allele contributes to higher grade breast cancer: in pair-matched high grade breast cancer samples, only the Ser allele is retained from heterozygous patients [29]. The same Leu1016Ser polymorphism in PHLPP2 was also reported in ovarian tumors, presenting more frequently in mucinous compared to serous tumors, with loss of heterozygosity in one case [92].
5.1.2. Suppression of RTK Signaling
PHLPP functions as a tumor suppressor through additional mechanisms that are independent of its function as the Akt hydrophobic motif phosphatase; notably, PHLPP also controls growth factor signaling by suppressing the transcription of RTKs. As discussed in Section 3.3., loss of PHLPP1 or PHLPP2 increases the transcription and steady-state protein levels of a number of RTKs, including EGFR, leading to amplified growth factor signaling [30]. This epigenetic regulation of RTK levels by PHLPP could provide one mechanism to account for the frequent upregulation of RTKs in cancer. Other studies have shown that PHLPP can act as tumor suppressor independently of Akt, PKC, and ERK pathways. Over-expression of PHLPP in Panc1 pancreatic cancer cells decreased colony numbers by more than 50% as assessed by anchorage-independent growth, but did not affect Akt or ERK phosphorylation or total levels of PKC [35]. Rather, the mechanism by which PHLPP induced apoptosis was by dephosphorylation at an inhibitory site of the pro-apoptotic kinase, Mst1. PHLPP2 was also shown to interact with IKKβ, leading to inhibition of its phosphorylation and subsequent activation of a major signaling pathway in cancer, the NF-κB pathway [49]. Additionally, PHLPP overexpression was reported to upregulate the expression and phosphorylation of p21 and p27, negative regulators of cell cycle progression [59, 82]. In turn, PHLPP1 expression is positively controlled by p27 [93], the loss of which is strongly associated with colorectal cancer progression [94]. Thus, PHLPP suppresses survival signaling by multiple mechanisms.
5.1.3. Suppression of Inflammation
PHLPP has been shown to be important for immunological processes. Regulatory T cells (Treg) are activated through either the T cell receptor or IL-2 and have a reduced capacity to activate the PI3K/Akt pathway compared to conventional T cells, which is necessary for Treg cell development and function [95]. PHLPP1 and PHLPP2 levels are upregulated in Treg cells, and PHLPP1 knock-out mice have impaired Treg cell development and suppressive capacity [96]. Furthermore, PHLPP1 was shown to be essential for the induction of Treg cells, as inhibition of both PHLPP and PTEN phosphatases results in decreased mitochondrial membrane potential in blood-derived Treg cells [97]. These data suggest a role for PHLPP in the regulation of immunological tolerance.
Another role for PHLPP was recently reported in mediating the resolution of the innate immune response [6]. Phlpp1−/− mice either challenged with E. coli or injected with LPS were protected from sepsis-induced death. This suggest that inhibition of PHLPP would be beneficial as an adjuvant therapy in antibiotic treatment of gram-negative sepsis. While the mechanism for this protection awaits elucidation, studies with bone marrow-derived macrophages identified PHLPP1 as a major regulator opposing STAT1-mediated transcription of inflammatory genes (see Section 3.4). PHLPP resolution of the inflammatory response may also play a role in chronic inflammatory disorders that predispose individuals to cancer such as inflammatory bowel disorder and nonalcoholic steatohepatitis (NASH). Intriguingly, Phlpp1/Phlpp2 double knock-out mice were protected against dextran sulfate sodium (DSS)-induced colitis due to an Akt-dependent reduction in apoptosis of intestinal epithelial cells [98]. Protection against acute colitis was also found to be mediated by PHLPP-deficient neutrophils, which exhibited enhanced immune homeostasis and elevated phosphorylation of Akt, ERK, and STAT1 [99]. These findings suggest that PHLPP enacts coordinated effects on inflammatory processes through modulation of distinct cell types. Additionally, PHLPP itself is regulated by immune signaling, as PHLPP1 and PHLPP2 protein were shown to be downregulated after 24 hours of LPS treatment [98]. Conversely, the induction of PHLPP2 in prostate cancer upon loss of PTEN and p53 is dependent upon the pro-inflammatory cytokine IL-6 [100]. Thus, further studies are required to understand the signaling pathways underlying PHLPP regulation of the immune system in various cell types and disease contexts.
5.2. Oncogenic Role
Could PHLPP also function as an oncogene in certain contexts? Recent evidence indicates that both PHLPP1 and PHLPP2 negatively regulate two tumor suppressors, PKC and Myc (Figure 4).
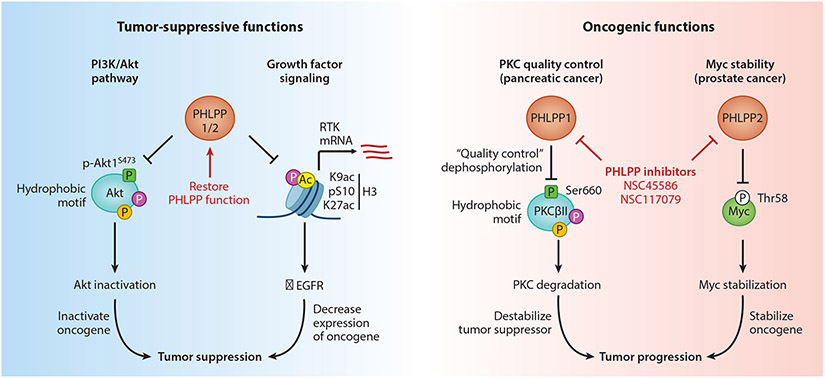
Figure 4. Pharmacological Targeting of PHLPP in Cancer.
Despite the discovery of PHLPP as a bona fide tumor suppressor by opposing PI3K and growth factor signaling (left), complex roles in cancer involving novel oncogenic functions present therapeutic opportunities for PHLPP inhibitors (right). In pancreatic cancer, PHLPP1 suppresses levels of the tumor suppressor PKC through quality control dephosphorylation of the hydrophobic motif, where the loss of PKC correlates inversely with patient survival [32]. In prostate cancer, PHLPP2 stabilization of oncogenic Myc is critical for tumor progression and metastatic disease [106]. Thus, future studies should explore the potential therapeutic benefit of PHLPP inhibition in these specific contexts.
5.2.1. PKC
Recent analysis of cancer-associated mutations in PKC genes has revealed that they are generally loss-of-function, suggesting that this family of kinases has a tumor-suppressive role [101]. Consistent with this, high levels of PKC protein confer improved survival in diverse cancers [102]. Furthermore, genome-editing to correct mutant PKCβII in a colon cancer cell line revealed not only that the mutant PKC was dominant-negative, but that the PKCβII was haploinsufficient with respect to cell growth in 3D: two alleles encoding wild-type PKCβII were considerably more effective at suppressing anchorage-independent growth of the cancer cell line compared with one allele [101]. As discussed in Section 3.1.2., dephosphorylation of the hydrophobic motif of PKC by PHLPP is a key determinant in setting the steady-state levels of PKC family members. While there are many mechanisms to control the steady-state levels of PKC, PHLPP1 is the dominant regulator in pancreatic cancer [32]. Analysis of protein levels from 105 patient tumors revealed an inverse correlation between PHLPP1 levels and PKC levels [32]. Furthermore, patients with high levels of PKC hydrophobic motif phosphorylation and low levels of PHLPP1 had greatly improved survival compared to patients with low PKC and high PHLPP1 protein levels [32]. PKC’s tumor-suppressive function was originally elucidated in a K-Ras-driven colorectal cancer xenograft model, and one mechanism by which PKC may act as a checkpoint in tumor progression is through attenuation of K-Ras signaling [101, 103]. Thus, in cancers that rely less heavily on Akt signaling, PHLPP1 may function as an oncogene through negative regulation of PKC, particularly in the context of oncogenic K-ras, which is frequently mutated in pancreatic cancer (Figure 4) [104, 105].
5.2.2. Myc
Discerning the expanding roles of PHLPP2 in cancer presents an even more complex picture. Despite the fact that elevation of Akt signaling resulting from PHLPP1 and PTEN deletion is critical for the onset, progression, and outcome of prostate cancer, PHLPP2 levels become elevated in prostate cancer metastasis upon co-deletion of Pten and Tp53, correlating with decreased Akt phosphorylation on Ser473 [100]. This apparent paradox was resolved by the finding that PHLPP2 dephosphorylates Myc on Thr58 to promote the stabilization of Myc in advanced prostate cancer [106]. Therefore, although capable of suppressing Akt activation that drives the disease, PHLPP2 is required for prostate cancer progression by stabilizing Myc [106] (Figure 4). Supporting a role of PHLPP2-mediated dephosphorylation of Myc on Thr58 in oncogenesis, a recurrent Myc T58A mutation observed in Burkitt’s lymphoma was sufficient to induce lymphoma and enhanced lethality in adoptive transfer of mouse hematopoietic stem cells [107]. The potential role of PHLPP2 as an oncogene provides one mechanism for the paradoxical “Akt-off” state in tumors with PI3K pathway alterations [108], which may be applicable to many apparently PI3K-driven malignancies. Whether tumor Akt status can be utilized to predict the function of PHLPP as oncogenic or tumor-suppressive is an attractive hypothesis.
6. PHARMACOLOGICAL TARGETING OF PHLPP IN CANCER
Resistance to chemotherapy is a major challenge in cancer therapy. Thus, information on the expression levels of certain biomarkers has tremendous therapeutic potential in outcome prediction and in matching available kinase inhibitors with the right patient or designing personalized medicines. PHLPP is a good candidate to be such a biomarker in cancer. For example, re-expressing PHLPP significantly increases the sensitivity of colon cancer cells to PI3K inhibition [82]. Therefore, determining the expression levels of PHLPP in cancer may help to predict which patients will respond better to PI3K and Akt inhibitors. Similarly, PHLPP expression predicts the prognosis of lung adenocarcinoma patients and PHLPP levels correlate with the latency of acquired resistance to EGFR inhibitors [109, 110]. Determining PHLPP expression could also be beneficial in determining the response to rapamycin treatment in certain cancer cell lines that develop resistance to rapamycin-mediated growth inhibition. In rapamycin-treated cells, PHLPP translation is inhibited, indicating that PHLPP may play a role in the induction of chemoresistance. Indeed, overexpression of PHLPP proteins significantly enhanced the inhibitory effect of rapamycin on growth of these cancer cells [65]. Furthermore, hypoxia is a common feature of tumors that plays a critical role in promoting resistance to chemotherapy [111]. Hypoxia also inhibits mTOR and cap-dependent translation, which are upstream of PHLPP and positively regulate its expression. Indeed, downregulation of PHLPP contributes to hypoxia-induced chemoresistance in colon cancer cells, dependent upon hypoxia-inducible factor 1α (HIF1α) [112]. Conversely, in cancers where PHLPP has oncogenic functions, as described previously in pancreatic [32] and prostate [106] cancers, PHLPP levels may correlate inversely with sensitivity to treatment or with patient outcome. Thus, determining the levels of PHLPP and other biomarkers in tumors may not only aid in the identification of critical dependencies of specific tumors, but may also facilitate personalized cancer treatments and minimize off-target effects.
The choice of the right strategy to target PHLPP may have important consequences in different cancers. Drugs that stabilize PHLPP or enhance its activity would be generally beneficial in cancers driven by deregulated growth factor signaling (Figure 4, left). In this pathway, PHLPP opposes cell survival and cell growth through transcriptional repression of growth factor receptors and by suppressing the activity of the downstream kinases Akt and S6K. In contrast, cancers driven by oncogenes such as K-Ras, which are sensitive to suppression by PKC, would benefit from restoring PKC by inhibiting PHLPP function (Figure 4, right).
In the case of Akt-driven cancers, designing drugs to stimulate the activity of PHLPP is more challenging. PHLPP is commonly deleted in cancer, making drug design impossible in such cases. However, targeting many of the mechanisms that regulate PHLPP itself could stabilize the amount of protein in the cell or redirect it to relevant substrates. In this regard, a recent study identified γ-tocopherol as a molecule that binds the PH domains of Akt and PHLPP1, promoting their co-recruitment to the plasma membrane and thus enhancing the dephosphorylation of Akt on Ser473 by PHLPP1 [113]. Moreover, the authors reported that tumor growth in a xenograft mouse model was suppressed upon oral administration of the compound, suggesting suppression of tumor growth attributable to PHLPP1 inhibition of Akt [113].
PP2C phosphatases are largely refractory to commonly used small molecule inhibitors such as calyculin A and OA. However, several small molecule inhibitors selective for PHLPP over the related PP2Cα were identified in a chemical and virtual screen of the NCI repository of inhibitors against the phosphatase activity of the purified phosphatase domains of PHLPP1 and PHLPP2 [114]. Biochemical and cellular assays validated two structurally diverse compounds that were identified from the screen: these selectively inhibit PHLPP in vitro, increase Akt signaling in cells, and suppress apoptosis. The two compounds, NSC 117079 and 45586, were predicted to be uncompetitive inhibitors of PHLPP that bind the hydrophobic cleft near the active site and interact with one of the Mn2+ ions. The compounds are not specific for PHLPP1 vs PHLPP2 but are selective for PHLPP compared to other phosphatases tested. Both compounds show potential therapeutic promise for diseases in which cell survival pathways are suppressed, however the IC50 values towards Akt in cells are high, approximately 30 and 70 μM for 117079 and 45586, respectively. Optimizing PHLPP inhibitors may be beneficial to target emerging oncogenic functions of PHLPP such as PHLPP1-mediated degradation of PKC in pancreatic cancer [32] and PHLPP2-mediated stabilization of Myc in prostate cancer [106] (Figure 4, right).
PHLPP plays a central role in cellular homeostasis, with enhanced activity generally promoting apoptosis and reduced activity promoting survival. Pharmacologically targeting PHLPP is encumbered by its broadening roles in controlling many diverse, and often opposing, aspects of cell signaling. Optimally designing drugs to target PHLPP therapeutically in particular tissues and cell types for specific diseases, while challenging, will have enormous therapeutic potential.
7. SUMMARY
Since the discovery of PHLPP as the hydrophobic motif phosphatase of Akt in 2005 [2], PHLPP has moved quickly into the spotlight of tumor suppressor studies. The last few years have unveiled many new targets and mechanisms of function in different pathologies for this relative late-comer to the phosphatome. Numerous studies have cemented a central role for PHLPP in suppression of survival pathways and induction of apoptosis; however, it is clear that PHLPP performs multifaceted and often opposing roles, as evidenced by the recently-elucidated oncogenic functions in pancreatic and prostate cancer and complex role in the immune system. The coming years are likely to see an increase in our understanding of PHLPP regulation in normal and pathological conditions, which will be essential for the development of therapies to treat specific cancers and further establish the importance of PHLPP as a biomarker in cancer.
ACKNOWLEDGEMENTS
We thank members of the Newton lab for helpful comments. This work was supported by NIH R35 GM122523 to A.C.N. T.R.B. was supported by the PhRMA Foundation Pre Doctoral Fellowship in Pharmacology/Toxicology (#20183844) and the UCSD Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752). K.C.K. was supported by the UCSD Training Program in Biochemistry of Growth Regulation and Oncogenesis (NIH/NCI T32 CA009523).
Abbreviations
- DUB
Deubiquitinating enzyme
- GSK
Glycogen synthase kinase
- HDAC3
Histone deacetylase 3
- IFNγ
Interferon γ
- LPS
Lipolysaccharide
- LRR
Leucine-rich repeat
- Mst1
Mammalian sterile 20–like kinase 1
- NHERF1
Na+/H+ exchanger regulatory factor 1
- NTE
N-terminal extension
- PDZ
PSD-95/Disc-large/ZO-1
- PHLPP
PH domain leucine-rich repeat protein phosphatase
- PI3K
Phosphoinositide 3-kinase
- PKC
Protein kinase C
- PTEN
Phosphatase and tensin homolog
- RA
Ras association
- RTK
Receptor tyrosine kinase
- S6K1
Ribosomal protein S6 kinase 1
- STAT1
Signal Transducer and Activator of Transcription 1
- TLR
Toll-like receptor
LITERATURE CITED
- 1.Chen MJ, Dixon JE, and Manning G, Genomics and evolution of protein phosphatases. Sci Signal, 2017. 10(474). [DOI] [PubMed] [Google Scholar]
- 2.Gao T, Furnari F, and Newton AC, PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell, 2005. 18(1): p. 13–24. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto K, et al. , Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A, 1982. 79(7): p. 2355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shima F, et al. , Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation. Mol Cell Biol, 2000. 20(1): p. 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Serine/threonine phosphatases: mechanism through structure. Cell, 2009. 139(3): p. 468–84. [DOI] [PubMed] [Google Scholar]
- 6.Cohen Katsenelson K, et al. , PHLPP1 counter-regulates STAT1-mediated inflammatory signaling. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park WS, et al. , Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell, 2008. 30(3): p. 381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon MA, Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp, 2007(74): p. 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao T, Brognard J, and Newton AC, The Phosphatase PHLPP Controls the Cellular Levels of Protein Kinase C. Journal of Biological Chemistry, 2008. 283: p. 6300–6311. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu K, et al. , SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett, 1999. 458(3): p. 363–9. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu K, et al. , Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J Biol Chem, 2003. 278(17): p. 14920–5. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka T, Broek D, and Wigler M, DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell, 1985. 43(2 Pt 1): p. 493–505. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki N, et al. , Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc Natl Acad Sci U S A, 1990. 87(22): p. 8711–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field J, et al. , Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science, 1990. 247(4941): p. 464–7. [DOI] [PubMed] [Google Scholar]
- 15.Das AK, et al. , Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. Embo J, 1996. 15(24): p. 6798–809. [PMC free article] [PubMed] [Google Scholar]
- 16.Berndsen K, et al. , PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fjeld CC and Denu JM, Kinetic analysis of human serine/threonine protein phosphatase 2Calpha. J Biol Chem, 1999. 274(29): p. 20336–43. [DOI] [PubMed] [Google Scholar]
- 18.Cohen P and Cohen PT, Protein phosphatases come of age. J Biol Chem, 1989. 264(36): p. 21435–8. [PubMed] [Google Scholar]
- 19.Gao T, Brognard J, and Newton AC, The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem, 2008. 283(10): p. 6300–11. [DOI] [PubMed] [Google Scholar]
- 20.Sierecki E and Newton AC, Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat protein phosphatase. Biochemistry, 2014. 53(24): p. 3971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina JR, et al. , PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene, 2012. 31(10): p. 1264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton AC, Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J, 2003. 370(Pt 2): p. 361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton AC, Protein Kinase C: Structural and Spatial Regulation by Phosphorylation, Cofactors, and Macromolecular Interactions. Chem. Rev, 2001. 101: p. 2353–2364. [DOI] [PubMed] [Google Scholar]
- 24.Manning BD and Cantley LC, AKT/PKB signaling: navigating downstream. Cell, 2007. 129(7): p. 1261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang LC, et al. , Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci Rep, 2018. 8(1): p. 6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayascas JR and Alessi DR, Regulation of Akt/PKB Ser473 phosphorylation. Mol Cell, 2005. 18(2): p. 143–5. [DOI] [PubMed] [Google Scholar]
- 27.Grzechnik AT and Newton AC, PHLPPing through history: a decade in the life of PHLPP phosphatases. Biochem Soc Trans, 2016. 44(6): p. 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brognard J, et al. , PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell, 2007. 25(6): p. 917–31. [DOI] [PubMed] [Google Scholar]
- 29.Brognard J, et al. , Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. J Biol Chem, 2009. 284(22): p. 15215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes G, et al. , Pleckstrin homology domain leucine-rich repeat protein phosphatases set the amplitude of receptor tyrosine kinase output. Proc Natl Acad Sci U S A, 2014. 111(38): p. E3957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton AC, Protein kinase C: perfectly balanced. Crit Rev Biochem Mol Biol, 2018. 53(2): p. 208–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baffi TR, et al. , Protein Kinase C Quality Control by Phosphatase PHLPP1 Unveils Loss-of-Function Mechanism in Cancer. Mol Cell, 2019. 74(2): p. 378–392 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magnuson B, Ekim B, and Fingar DC, Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J, 2012. 441(1): p. 1–21. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, et al. , PHLPP-mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol Cell Biol, 2011. 31(24): p. 4917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao M, et al. , Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol Cell, 2010. 38(4): p. 512–23. [DOI] [PubMed] [Google Scholar]
- 36.Zeng Q and Hong W, The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell, 2008. 13(3): p. 188–92. [DOI] [PubMed] [Google Scholar]
- 37.Cinar B, et al. , The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J, 2007. 26(21): p. 4523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang SW, et al. , Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J Biol Chem, 2007. 282(42): p. 30836–44. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu K, et al. , Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell, 2007. 128(6): p. 1219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason JA, et al. , Oncogenic Ras differentially regulates metabolism and anoikis in extracellular matrix-detached cells. Cell Death Differ, 2016. 23(8): p. 1271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson TC, et al. , PHLPP1 splice variants differentially regulate AKT and PKCalpha signaling in hippocampal neurons: characterization of PHLPP proteins in the adult hippocampus. J Neurochem, 2010. 115(4): p. 941–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. , PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology, 2014. 146(5): p. 1301–12 e1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, et al. , Crucial role of c-Jun phosphorylation at Ser63/73 mediated by PHLPP protein degradation in the cheliensisin a inhibition of cell transformation. Cancer Prev Res (Phila), 2014. 7(12): p. 1270–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jameson NM, et al. , Intron 1-Mediated Regulation of EGFR Expression in EGFR-Dependent Malignancies Is Mediated by AP-1 and BET Proteins. Mol Cancer Res, 2019. 17(11): p. 2208–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmarsh AJ and Davis RJ, Regulation of transcription factor function by phosphorylation. Cell Mol Life Sci, 2000. 57(8–9): p. 1172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hein MY, et al. , A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell, 2015. 163(3): p. 712–23. [DOI] [PubMed] [Google Scholar]
- 47.Arias E, et al. , Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Molecular Cell, 2015. 59: p. 270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee A, Chatterjee U, and Ghosh MK, Activation of protein kinase CK2 attenuates FOXO3a functioning in a PML-dependent manner: implications in human prostate cancer. Cell Death Dis, 2013. 4: p. e543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal NK, et al. , PHLPP2 suppresses the NF-kappaB pathway by inactivating IKKbeta kinase. Oncotarget, 2014. 5(3): p. 815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, et al. , Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res, 2008. 68(17): p. 7050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pei H, et al. , FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell, 2009. 16(3): p. 259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver BS, et al. , Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell, 2011. 19(5): p. 575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang Z, et al. , LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic acids research, 2019. 47: p. 4211–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boonying W, et al. , Pink1 regulates FKBP5 interaction with AKT/PHLPP and protects neurons from neurotoxin stress induced by MPP+. Journal of Neurochemistry, 2019. 150: p. 312–329. [DOI] [PubMed] [Google Scholar]
- 55.Li X, et al. , Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. EMBO Rep, 2011. 12(8): p. 818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi Y, et al. , PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J, 2006. 25(4): p. 910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradley EW, Carpio LR, and Westendorf JJ, Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem, 2013. 288(14): p. 9572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang YS, et al. , Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong L, et al. , Oncogenic suppression of PHLPP1 in human melanoma. Oncogene, 2014. 33(39): p. 4756–66. [DOI] [PubMed] [Google Scholar]
- 60.Alamuru-Yellapragada NP, et al. , LPS depletes PHLPP levels in macrophages through the inhibition of SP1 dependent transcriptional regulation. Biochemical and Biophysical Research Communications, 2017. 486: p. 533–538. [DOI] [PubMed] [Google Scholar]
- 61.Bradley EW, et al. , Phlpp1 facilitates post-traumatic osteoarthritis and is induced by inflammation and promoter demethylation in human osteoarthritis. Osteoarthritis Cartilage, 2016. 24(6): p. 1021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Hayre M, et al. , Mechanisms and consequences of the loss of PHLPP1 phosphatase in chronic lymphocytic leukemia (CLL). Leukemia, 2012. 26(7): p. 1689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian Y, et al. , The CUL4B/AKT/beta-Catenin Axis Restricts the Accumulation of Myeloid-Derived Suppressor Cells to Prohibit the Establishment of a Tumor-Permissive Microenvironment. Cancer Res, 2015. 75(23): p. 5070–83. [DOI] [PubMed] [Google Scholar]
- 64.Russell M, et al. , Changes in gene expression in the Ras/adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol Biol Cell, 1993. 4(7): p. 757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Stevens PD, and Gao T, mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem, 2011. 286(8): p. 6510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, Liu J, and Gao T, beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol, 2009. 29(23): p. 6192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao G, et al. , SGT1 regulates Akt signaling by promoting beta-TrCP-dependent PHLPP1 degradation in gastric cancer cells. Mol Biol Rep, 2013. 40(4): p. 2947–53. [DOI] [PubMed] [Google Scholar]
- 68.Warfel NA, et al. , Mislocalization of the E3 ligase, beta-transducin repeat-containing protein 1 (beta-TrCP1), in glioblastoma uncouples negative feedback between the pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) and Akt. J Biol Chem, 2011. 286(22): p. 19777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rao E, et al. , The miRNA-17 approximately 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia, 2012. 26(5): p. 1064–72. [DOI] [PubMed] [Google Scholar]
- 70.Liao WT, et al. , microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res, 2013. 19(17): p. 4662–72. [DOI] [PubMed] [Google Scholar]
- 71.Hart M, et al. , Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res, 2014. 12(2): p. 250–63. [DOI] [PubMed] [Google Scholar]
- 72.Calin GA and Croce CM, MicroRNA signatures in human cancers. Nat Rev Cancer, 2006. 6(11): p. 857–66. [DOI] [PubMed] [Google Scholar]
- 73.Yan Y, et al. , Transcription factor C/EBP-β induces tumor-suppressor phosphatase PHLPP2 through repression of the miR-17-92 cluster in differentiating AML cells. Cell Death and Differentiation, 2016. 23: p. 1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhiqiang Z, et al. , USP1 regulates AKT phosphorylation by modulating the stability of PHLPP1 in lung cancer cells. J Cancer Res Clin Oncol, 2012. 138(7): p. 1231–8. [DOI] [PubMed] [Google Scholar]
- 75.Li X, et al. , The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene, 2013. 32(4): p. 471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gangula NR and Maddika S, WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1). J Biol Chem, 2013. 288(48): p. 34545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X, et al. , FANCI is a negative regulator of Akt activation. Cell Cycle, 2016. 15(8): p. 1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hornbeck PV, et al. , PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res, 2012. 40(Database issue): p. D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen M, et al. , Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell, 2011. 20(2): p. 173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Neill AK, Niederst MJ, and Newton AC, Suppression of survival signalling pathways by the phosphatase PHLPP. Febs J, 2013. 280(2): p. 572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ouillette P, et al. , Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res, 2008. 68(4): p. 1012–21. [DOI] [PubMed] [Google Scholar]
- 82.Liu J, et al. , Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene, 2009. 28(7): p. 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goel A, et al. , Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res, 2003. 63(7): p. 1608–14. [PubMed] [Google Scholar]
- 84.Jen J, et al. , Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med, 1994. 331(4): p. 213–21. [DOI] [PubMed] [Google Scholar]
- 85.Rakha EA, et al. , Chromosome 16 tumor-suppressor genes in breast cancer. Genes Chromosomes Cancer, 2006. 45(6): p. 527–35. [DOI] [PubMed] [Google Scholar]
- 86.Patael-Karasik Y, et al. , Comparative genomic hybridization in inherited and sporadic ovarian tumors in Israel. Cancer Genet Cytogenet, 2000. 121(1): p. 26–32. [DOI] [PubMed] [Google Scholar]
- 87.Tsuda H, et al. , Allele loss on chromosome 16 associated with progression of human hepatocellular carcinoma. Proc Natl Acad Sci U S A, 1990. 87(17): p. 6791–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Safford SD, et al. , Fine mapping of Wilms' tumors with 16q loss of heterozygosity localizes the putative tumor suppressor gene to a region of 6.7 megabases. Ann Surg Oncol, 2003. 10(2): p. 136–43. [DOI] [PubMed] [Google Scholar]
- 89.Torring N, et al. , Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer, 2007. 96(3): p. 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiao M, Iglehart JD, and Pardee AB, Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res, 2007. 67(11): p. 5293–9. [DOI] [PubMed] [Google Scholar]
- 91.Suljagic M, et al. , Reduced expression of the tumor suppressor PHLPP1 enhances the antiapoptotic B-cell receptor signal in chronic lymphocytic leukemia B-cells. Leukemia, 2010. 24(12): p. 2063–71. [DOI] [PubMed] [Google Scholar]
- 92.Stemke-Hale K, et al. , Frequency of mutations and polymorphisms in borderline ovarian tumors of known cancer genes. Mod Pathol, 2013. 26(4): p. 544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang D, et al. , The N-terminal region of p27 inhibits HIF-1alpha protein translation in ribosomal protein S6-dependent manner by regulating PHLPP-Ras-ERK-p90RSK axis. Cell Death Dis, 2014. 5: p. e1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hershko DD and Shapira M, Prognostic role of p27Kip1 deregulation in colorectal cancer. Cancer, 2006. 107(4): p. 668–75. [DOI] [PubMed] [Google Scholar]
- 95.Crellin NK, Garcia RV, and Levings MK, Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood, 2007. 109(5): p. 2014–22. [DOI] [PubMed] [Google Scholar]
- 96.Patterson SJ, et al. , Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J Immunol, 2011. 186(10): p. 5533–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen HH, et al. , Immune dysregulation in patients with PTEN hamartoma tumor syndrome: Analysis of FOXP3 regulatory T cells. J Allergy Clin Immunol, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wen YA, et al. , Loss of PHLPP protects against colitis by inhibiting intestinal epithelial cell apoptosis. Biochim Biophys Acta, 2015. 1852(10 Pt A): p. 2013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ran T, et al. , Enhanced neutrophil immune homeostasis due to deletion of PHLPP. Frontiers in Immunology, 2019. 10: p. 2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nowak DG, et al. , MYC Drives Pten/Trp53-Deficient Proliferation and Metastasis due to IL6 Secretion and AKT Suppression via PHLPP2. Cancer Discov, 2015. 5(6): p. 636–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Antal CE, et al. , Cancer-Associated Protein Kinase C Mutations Reveal Kinase's Role as Tumor Suppressor. Cell, 2015. 160(3): p. 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Newton AC and Brognard J, Reversing the Paradigm: Protein Kinase C as a Tumor Suppressor. Trends Pharmacol Sci, 2017. 38(5): p. 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang M-T, et al. , K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell, 2015. 163: p. 1237–1251. [DOI] [PubMed] [Google Scholar]
- 104.Almoguera C, et al. , Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell, 1988. 53(4): p. 549–54. [DOI] [PubMed] [Google Scholar]
- 105.Waters AM and Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med, 2018. 8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nowak DG, et al. , The PHLPP2 phosphatase is a druggable driver of prostate cancer progression. J Cell Biol, 2019. 218(6): p. 1943–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hemann MT, et al. , Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature, 2005. 436(7052): p. 807–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ku SY, et al. , Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science, 2017. 355(6320): p. 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lv D, et al. , High PHLPP expression is associated with better prognosis in patients with resected lung adenocarcinoma. BMC Cancer, 2015. 15: p. 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie Y, et al. , High PHLPP1 expression levels predicts longer time of acquired resistance to EGFR tyrosine kinase inhibitors in patients with lung adenocarcinoma. Oncotarget, 2017. 8: p. 59000–59007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilson WR and Hay MP, Targeting hypoxia in cancer therapy. Nat Rev Cancer, 2011. 11(6): p. 393–410. [DOI] [PubMed] [Google Scholar]
- 112.Wen YA, et al. , Downregulation of PHLPP expression contributes to hypoxia-induced resistance to chemotherapy in colon cancer cells. Mol Cell Biol, 2013. 33(22): p. 4594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan R, et al. , Exploitation of the ability of gamma-tocopherol to facilitate membrane co-localization of Akt and PHLPP1 to develop PHLPP1-targeted Akt inhibitors. J Med Chem, 2015. 58(5): p. 2290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sierecki E, et al. , Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J Med Chem, 2010. 53(19): p. 6899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Min A, et al. , CoralP: Flexible visualization of the human phosphatome. J Open Source Softw, 2019. 4(44). [DOI] [PMC free article] [PubMed] [Google Scholar]