Multicellular animals are a diverse lot, with widely varied body plans and lifestyles. One feature they share, however, is a nearly universal reliance on sexual reproduction for species propagation. Humans have long been fascinated by human sex differences and formal theories on how human sex is determined date at least to Aristotle (in De Generatione Animalium, ca. 335 BCE). However, it is only in the past couple of decades that the genetic and molecular programs responsible for generating the two sexes have been understood in any detail. Sex, it turns out, can be established by many very different and fast-evolving mechanisms, but often these involve a conserved class of transcriptional regulators, the DM domain proteins.
Making sexes: determination and differentiation
Sexual reproduction in multicellular animals requires, at a minimum, male and female gametes. Indeed, these specialized haploid cells are how we define the sexes: in a given species individuals with big gametes are females and those with small gametes are males. Individuals that can make both kinds are hermaphrodites, and may be self-fertile or cross-fertile with other individuals. Gametes in most animal species are made in a specialized organ, the gonad. Before sexual reproduction can take place, sexual development must occur. That is, a mechanism is needed to decide which sex a given embryo will adopt — sex determination — as well as mechanisms to control subsequent development of those parts of the embryo that differ between sexes — sexual differentiation. The final result is individuals that can differ remarkably not just in their gametes and gonads but in many aspects of their anatomy, physiology, and behavior — think of the tail of the male peacock, milk production in female mammals, or the courtship rituals of the male bowerbird. Even though these sexual dimorphisms are essential to the propagation of the species, they can be so extreme that in some cases it is difficult to recognize that their bearers are in fact members of the same species. Some sexually dimorphic traits are essential for reproduction or have obvious benefits to reproductive fitness. However, many sexually dimorphic characters seem antithetical to natural selection, which greatly troubled Darwin (“The sight of a feather in a peacock’s tail, whenever I gaze at it, makes me sick!”). The prevalence of these seemingly disadvantageous traits led to Darwin’s second great insight, the theory of sexual selection based on “the advantage which certain individuals have over other individuals of the same sex and species solely in respect of reproduction,” proposing that these traits provide a competitive advantage in mating. In thinking about the molecular basis of sexually dimorphic traits and how they evolve it helps to be mindful of the distinctive selection mechanisms shaping them.
Many paths lead to sexual dimorphism
Despite its near universality, sex determination is controlled by quite different mechanisms in different species. Broadly speaking, sex can be determined two ways: genetically (genotypic sex determination or GSD), where the chromosomal composition determines an individual’s sex at fertilization; or environmentally (environmental sex determination or ESD), where conditions encountered during development determine an individual’s sex. These two categories can be further subdivided based on the precise mechanisms involved. In some GSD species, for example, the male is the heterogametic sex, that is, the gender with two different sex chromosomes. This includes the familiar XX/XY sex-determining mechanism in humans and other mammals where the presence of a Y chromosome initiates male development. Alternatively, as in birds, snakes and butterflies, the female can be the heterogametic sex; this is termed a ZZ/ZW system.
Just as GSD is composed of several distinct mechanisms, ESD can involve a variety of environmental stimuli. Perhaps the best-known examples of ESD involve temperature-dependent sex determination, or TSD, where temperature during a critical window of embryonic development influences the sex of the offspring. A common TSD pattern, seen in alligators and crocodiles and some turtle and lizard species, for example, involves females developing at both low and high incubation temperatures while males are produced at intermediate temperatures. Other environmental variables that can act as sex-determining mechanisms include: the proximity of conspecifics, as is found in the echiurid marine worm, Bonellia viridis, where planktonic larvae that settle in isolation become females whereas larvae that settle near females become males; and photoperiod, as seen in some populations of the brackish water shrimp, Gammarus duebeni, where males are produced on long days and females are produced on short days.
The traditional view that divides sex-determining mechanisms strictly into GSD and ESD is being challenged by evidence that both GSD and ESD can coexist in the same species. Sex determination in two lizard species exemplifies the false dichotomy between GSD and ESD. The bearded dragon Pogona vitticeps and the skink Bassiana duperreyi both possess sex chromosomes — ZZ/ZW and XX/XY systems, respectively — yet genotypic sex can be overridden at extreme incubation temperatures, resulting in individuals with a mismatch between genotype and sexual phenotype (e.g., ZZ female bearded dragons and XX male skinks). Similarly, infection by the symbiotic bacterium Wolbachia can override GSD in a variety of insects, and depletion of oocytes can cause female-to-male sex reversal in some fish species (e.g., Oryzias and Danio). GSD and ESD may be better viewed, therefore, as extreme points along a continuum, with sex determination being more influenced by genetic factors in some species and by environmental factors in others.
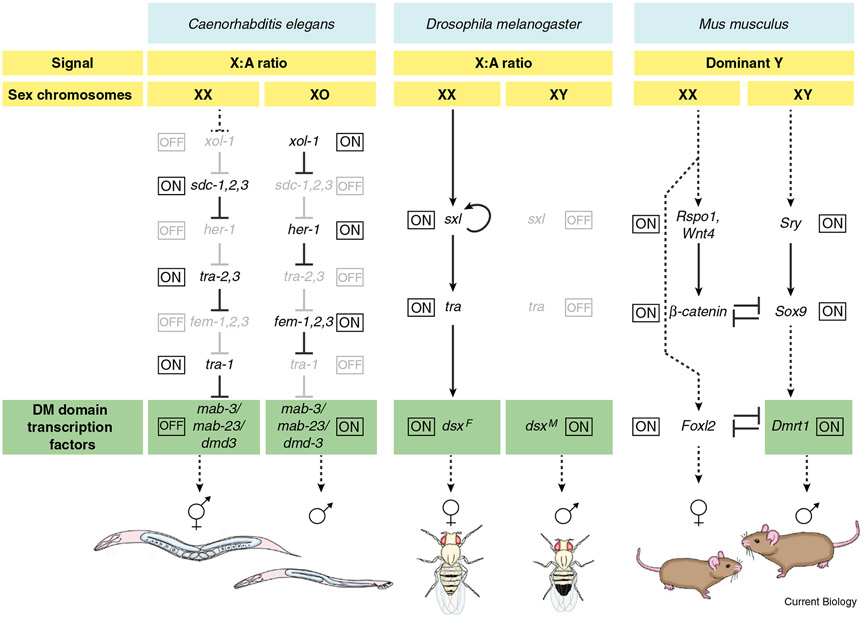
Categorizing GSD species simply into male and female heterogamety obscures surprising variation in the underlying genetic mechanisms. A comparison of the male heterogametic systems of mice, roundworms and fruit flies illustrates this point (Figure 1). The mammalian Y chromosome contains the dominant male determinant sex-determining region on Y (Sry), a transcriptional regulator that antagonizes a Wnt/β-catenin pathway whose activity promotes female development. This permits expression of the related gene Sry-box 9 (Sox9), whose activity is necessary to initiate male development. Sex determination in the fruit fly, Drosophila melanogaster, is determined by the ratio of X chromosomes to autosomes (X:A ratio) or possibly by the number of X chromosomes, currently a point of some controversy. An XX embryo shows transient early expression of the splicing regulator Sex-lethal (Sxl) and lifelong production of functional Sxl ultimately triggers production of the female-specific isoform of doublesex (dsx) via a cascade of sex-specific alternative mRNA splicing. An XY embryo has lower Sxl expression, resulting in the subsequent production of the male-determining splice form of dsx. The Drosophila Y chromosome is not involved in sex determination but contains genes required for male fertility; an XO fly, therefore, is male but sterile. Sex determination in the roundworm Caenorhabditis elegans is initiated by the X:A ratio, but unlike Drosophila there is no Y chromosome. An X:A ratio of 1 (XX) initiates an inhibitory signal transduction pathway resulting in elevated activity of the transcriptional regulator transformer 1 (tra-1) and development into a hermaphrodite (anatomically female but able to make sperm and oocytes). An X: A ratio of 0.5 (XO, the result of rare spontaneous chromosome nondisjunction during meiosis) results in low tra-1 activity and male development. The C. elegans dose-sensing system is exquisitely sensitive: experiments using induced polyploids (worms with one or more extra sets of chromosomes) showed that embryos can reliably distinguish between X:A ratios of 0.67 and 0.75 to become males and hermaphrodites, respectively.
Figure 1. Sex determination pathways in diverse model organisms.
Variations in an XX/XY GSD system. Differences among species are evident throughout all stages of the sex determination hierarchy, although each of the pathways converge on conserved downstream regulators in the DM domain gene family that are essential for male development. For simplicity and clarity, in each gene network shown a number of peripheral, relatively minor, or sex non-specific regulators have been omitted. Regulatory interactions (solid lines) are meant to indicate the regulatory logic of each pathway but do not necessarily imply direct regulation. Dashed lines indicate temporal relationships.
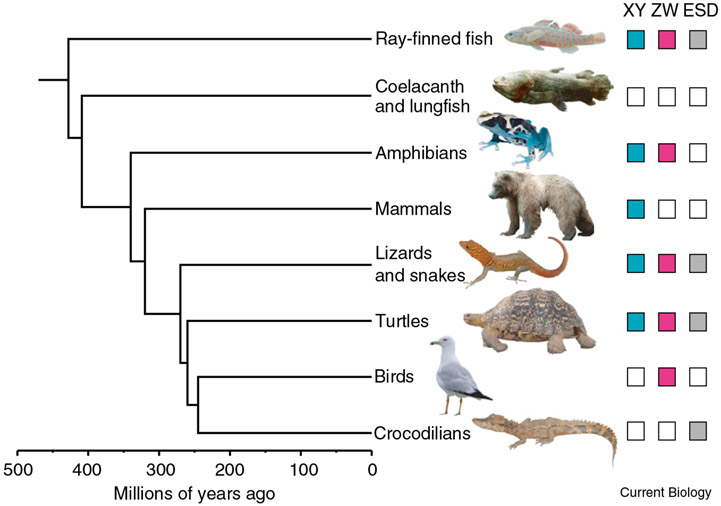
The great diversity in how sex is determined indicates that transitions among sex-determining mechanisms have occurred repeatedly across the tree of life. These transitions are clearly illustrated when sex-determining mechanisms are displayed in a phylogenetic context (Figure 2). Some transitions are ancient and have resulted in relatively stable sex determining mechanisms across all members of a clade. Thus, an XX/XY system regulated by SRY is found in virtually all therian mammals; a ZZ/ZW system is present in all birds, including ratites (e.g., ostriches and emus); and TSD occurs in all crocodilians. In other cases transitions have occurred among closely related species or even among populations within a single species. For example both ZZ/ZW and XX/XY systems are found among different populations of the Japanese frog Rana rugosa, with female heterogamety evolving at least twice independently within the species. Comparing divergent sex-determining systems among closely related species will be particularly useful for learning how these transitions can occur.
Figure 2. Frequent changes in sex-determining mechanisms among major clades of bony vertebrates.
Colored boxes indicate known sex-determining mechanisms for each clade. Lineages with more than one colored box possess multiple species with different sex-determining mechanisms. Some lineages, e.g. birds, mammals and crocodilians, show great stability of sex-determining mechanisms over long periods of time while other lineages, e.g. ray-finned fish, amphibians, lizards and snakes, and turtles, comprise species with diverse mechanisms, implying more frequent evolutionary changes within those groups. Sex-determining mechanisms are unknown in lungfish and coelacanths.
The extreme lability of sex-determining mechanisms was beautifully demonstrated in C. elegans by genetic experiments performed by Jonathan Hodgkin, who showed that simple loss- and gain-of-function mutations in seven of the core nematode sex-determining genes could be used to create a wide variety of stable sex-determining mechanisms, including XX/XY, ZZ/ZW, and environmental mechanisms. Because the different genes that were altered are located on different chromosomes, it was possible to turn each autosome into a sex chromosome, and because these genes encode many different types of proteins, it appears that essentially any type of molecule should be able to control sex. Indeed, use of an amber nonsense allele (a UAG translational stop codon) introduced into the sex-determining gene transformer 3 (tra-3) allowed an amber-suppressor tRNA, which permits read-through of the stop codon, to serve as the primary determinant of sex. These laboratory manipulations strongly suggest that rapid and dramatic transitions in sex-determining mechanism should occur in nature and that they might result from relatively simple loss- or gain-of-function mutations in key sex-regulatory genes.
A picture emerges of pathways whose logic and individual components can exchange quickly during evolution, with a fluidity that is possible because the only essential function of a sex determination switch is to make different individuals develop as different sexes. Phylogenetic studies suggest that the downstream ‘business’ end of a sex-determination pathway does tend to be more stable than the triggering mechanism at the top; in insects for example, tra and dsx play widely conserved roles in sex determination while Sxl does not.
When and where sex is determined
The initial sex determination decision needs eventually to result in sexual dimorphism throughout the body, from brain to tail. There is, however, considerable diversity in where sex determination is initiated and how the decision is transmitted to the cells that require sex-specific development. A familiar example is that of mammals, where Sry acts exclusively in the fetal gonads and then gonadal sex hormones convey the sex determination decision to the rest of the body; this explains why surgical removal of the male gonad during fetal development in mammals causes XY embryos to develop as females. Manipulations of gonadal sex hormones in other vertebrates also have confirmed the critical importance of the gonad in sexual development. However, animals such as roundworms and fruit flies lack gonadal sex hormones and in these creatures the sex determination decision is more spatially distributed, with the sex chromosome complement acting directly in cells throughout the body. Even these examples are generalizations, however: there is good evidence that the sex chromosomes also act directly on the fetal mammalian brain to cause sexually dimorphic gene expression, and secreted signaling molecules do play important roles in sexual dimorphism of both flies and roundworms.
In some species sex determination mechanisms seem to differ between the gonad and other tissues. In birds, for example, the Z-linked doublesex and mab-3 related transcription factor 1 (Dmrt1) genes are critical for gonadal sex, but the sex chromosomes act via a separate mechanism to control the sex of non-gonadal tissues. This is graphically illustrated by bilateral gynandromorphs, chimaeric birds in which one side is predominantly ZZ and the other ZW (Figure 3). In these birds one half appears physically male and the other female, a pattern that cannot involve Dmrt1, which is not expressed outside the urogenital system, and also cannot be explained solely by gonadal sex hormone levels, which should be similar in both sides. Similarly, in marsupials, like eutherian mammals, the Y-linked Sry gene controls gonadal sex, but the number of X chromosomes determines whether a pouch or scrotum develops. An XO kangaroo therefore is a female with ovaries but has a scrotum instead of a pouch. An XXY kangaroo, on the other hand, is a male with testes, but with a pouch — perhaps the prototypical ‘man-purse’ (Figure 3).
Figure 3. Animals of mixed-sex genotypes showing both male and female sexual traits.
(A) Bilateral gynandromorph chicken. Right side has cells that are predominantly ZW, while left side cells are predominantly ZZ. Consequently, female characteristics, e.g. small wattle and small leg spur, are expressed on the bird’s right side (brown plumage) while male characteristics, e.g. large wattle, large leg spur and greater muscle mass, are expressed on the bird’s left side (gold and white plumage). Photo courtesy of Dr. Michael Clinton, The Roslin Institute, The University of Edinburgh. (B) “Vincent”, an XXY gray kangaroo (Macropus giganteus) showing both male and female characteristics. The black arrow indicates the pouch, which is determined by the number of X chromosomes, while the white arrow indicates the penis, which along with the testes is determined by the presence of a Y chromosome. Photo courtesy of Professor D.W. Cooper, University of New South Wales.
What are the genes that control sex?
Clearly a number of different mechanisms can trigger sex determination, but how similar are the downstream gene networks that respond to these triggers and actually do the work? Our current understanding of the genetic control of sex determination mainly derives from two intertwined strands of research: studies of human patients whose genetic and physical sex are discordant, and molecular genetic analysis in model organisms, mainly fruit flies, roundworms, and mice.
Sex chromosomes were identified in the early 1900s. However, it was not until the late 1950s that the mammalian Y chromosome was discovered to determine male sex, the late 1980s that the sex-determining region of the human Y chromosome was identified through cytogenetic analysis, and the early 1990s that SRY was discovered and confirmed, by human translocations of SRY and mice transgenic for Sry, to be the Y-linked gene that triggers testicular differentiation in males. Sry was found to act through the related gene Sox9 and to oppose a female-promoting regulatory network involving Wnt/β-catenin signaling (Figure 1). At the same time, forward genetic studies in flies and worms were identifying the major players controlling sex in these organisms. Surprisingly, there was initially no overlap among these model systems — though many of the genes controlling sex in flies or roundworms were conserved, their sex-determining roles were not. Thus, sex-determination pathways seemed to lack shared components, unlike many other major developmental regulatory pathways, a near heresy in the age of model organisms. Until the late 1990s, a molecular geneticist therefore could be excused for viewing sex determination much as Darwin initially viewed the peacock’s tail.
A measure of resolution came when the downstream male regulator male abnormal 3 (mab-3) was cloned from C. elegans and found to be related to the insect dsx gene. dsx and mab-3 share a novel DNA-binding motif, the DM domain, which was subsequently found in many metazoan sexual regulators, most notably Dmrt1 in vertebrates. Dmrt1 is expressed in the embryonic gonad of all vertebrates examined and Dmrt1 or a close paralog has been shown to be essential for testicular differentiation in mammals, birds, and fish. Thus, at least one family of regulators acting at the interface of sex determination and sexual differentiation does appear to be deeply conserved.
Studies of Dmrt1 in vertebrates also have suggested that simple gene mutations can drive transitions between sex-determining mechanisms in nature, much like those created by Hodgkin in C. elegans in the laboratory. In three different vertebrate groups it appears that a new sex-determining mechanism resulted from a different mutational event affecting Dmrt1: the avian ZZ/ZW system likely arose from a recessive loss-of-function mutation in Dmrt1; in the medaka fish (Oryzias latipes) a new XX/XY system evolved due to a dominant gain-of-function mutation of Dmrt1 in which the new Dmrt1 allele — Dmy — functions analogously to Sry in mammals; and the African clawed frog (Xenopus laevis) ZZ/ZW system likely arose from a truncation that generated a dominant-negative ovary-determining dmrt1 allele — dm-w — that blocks the masculinizing activity of the autosomal dmrt1 gene. Therian mammals and stickleback fish are the only vertebrate groups so far examined where it is clear that Dmrt1 or a paralog does not act as a sex-linked sex-determining gene, although Dmrt1 still plays a crucial role in male gonadal differentiation and maintenance in these species.
Acquiring and evolving sexual traits
Sexual selection and other selective pressures cause sexually dimorphic traits to evolve rapidly. Thus, even dramatic dimorphisms in one species may be very different or absent in related species. Recent work, mainly in fruit flies, has helped reveal how dimorphic traits arise and adapt during evolution. Two distinct mechanisms, both involving the activity of dsx, have been described. The first involves the ability of dsx to regulate downstream transcriptional targets and relies on variation in the presence or location of DSX binding sites in cis-regulatory elements. In abdominal pigmentation, which is present in males but reduced in females, changes in DSX binding sites have altered the ability of DSX to repress the two bric a brac (bab) genes, and thereby promote strong pigmentation in the male posterior abdomen. The second mechanism involves changes in the expression pattern of dsx itself. Although dsx controls most dimorphic traits, it is not actually expressed in all cells, so that in effect some cells ‘know’ their sex and some do not. This feature has been exploited in the evolution of sex combs, male-specific sensory bristles used in mating that are found on the first pair of legs in some Drosophila species. Sex combs develop as a result of the joint expression of the male-specific dsx isoform and the HOX gene sex combs reduced (Scr) and are present only in species that have evolved an appropriate dsx expression domain in the foreleg.
Staying committed
Sexual dimorphisms arise throughout much of development in most species and thus sex presumably is determined early. A wide range of experimental approaches have confirmed that this is so, including experiments involving surgical removal of the fetal gonad and conditional deletion of sex determination genes in mammals, temperature shifts in reptiles with TSD, and temperature shifts using conditional alleles of sex-determining genes in worms. In each case sex reversal required intervention during embryonic development and did not alter phenotypic sex after that time (although in flies sexual phenotypes can be altered as late as the pupal stage). It came as a surprise, therefore, when it was shown recently that deleting either of two sex-specific mouse transcription factors — forkhead box L2 (Foxl2) in females or Dmrt1 in males — could cause gonadal cells to reprogram their sex, even in adults. Thus, sex is determined early but not irreversibly, and Foxl2 and Dmrt1 lie at the heart of two opposed maintenance networks that uphold the initial sex determination decision.
The future
Several decades of intensive genetic investigation have provided a detailed view of how sex is determined in several model organisms, including what triggers sex determination, what gene networks respond to the trigger, and how downstream genes integrate sex and pattern to promote sexual differentiation, and have begun to illustrate how sex determination and sexual dimorphism can evolve. New technologies, including inexpensive high-throughput DNA sequencing, RNAi, and other reverse-genetic strategies, herald rapid future progress in our understanding of sex determination and its evolution, making it easier to perform genetic mapping in non-model organisms, to identify and study the function of sex-biased regulatory networks, and to compare sex chromosomes across species. A detailed understanding of how transitions among sex-determining mechanisms can occur is currently lacking, although several theoretical models provide testable hypotheses about how such transitions might be accomplished. Fine-scale phylogeny-based studies will be one way forward and will identify many new models to study. Sex maintenance is another emerging field of study. While we now know that Dmrt1 and Foxl2 are required for maintaining the sex of postnatal gonads in mice, we know little of the gene networks in which they function nor whether sex maintenance occurs in other animals. Darwin’s theories provided the foundation for modern studies of how and why sexual dimorphisms arose and evolved; with the advent of new technologies the near future will certainly provide deeper insights into the molecular basis of these processes.
Further reading
- Bull JJ (1983). Evolution of Sex Determining Mechanisms. (Menlo Park, California: Benjamin Cummings Publishing Company, Inc.) [Google Scholar]
- Darwin C. (1871). The Descent of Man and Selection in Relation to Sex. (London: John Murray.) [Google Scholar]
- Dewing P, Shi T, Horvath S, and Vilain E (2003). Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Mol. Brain. Res 118, 82–90. [DOI] [PubMed] [Google Scholar]
- Graves JAM (2008). Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet 42, 565–586. [DOI] [PubMed] [Google Scholar]
- Herpin A, and Schartl M (2011). Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J. 278, 1010–1019. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. (2002). Exploring the envelope: systematic alteration in the sex-determination system of the nematode Caenorhabditis elegans. Genetics 162, 767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosken DJ, and House CM (2011). Sexual selection. Curr. Biol 21, R62–R65. [DOI] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, and Zarkower D (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, and Zarkower D (2012). Sex and the singular DM domain: insights into sexual regulation, avolution and plasticity. Nat. Rev. Genet 13, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, and Graves JAM (2007). Temperature sex reversal implies sex gene dosage in a reptile. Science 316, 411. [DOI] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, and Baker BS (2010). Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8,e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, and Lovell-Badge R (2009). Sex determination and SRY: down to a wink and a nudge. Trends Genet. 25, 19–29. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, and Kopp A (2011). Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9, e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier A-C, Klugmann C, Klasen C, Holter NI, et al. (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142. [DOI] [PubMed] [Google Scholar]
- Veitia RA (2010). FOXL2 versus SOX9: a lifelong “battle of the sexes”. Bioessays 32, 375–380. [DOI] [PubMed] [Google Scholar]
- Williams TM, and Carroll SB (2009). Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet 10, 797–804. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, and Carroll SB (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, McBride D, Nandi S, McQueen HA, McGrew MJ, Hocking PM, Lewis PD, Sang HM, and Clinton M (2010). Somatic sex identity is cell autonomous in the chicken. Nature 464, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]