Abstract
Latent autoimmune diabetes in adults (LADA) is a heterogeneous disease characterized by autoantibodies against insulin producing pancreatic beta cells and initial lack of need for insulin treatment. The aim of the present study was to investigate if individuals with LADA have an altered gut microbiota relative to non-diabetic control subjects, individuals with type 1 diabetes (T1D), and individuals with type 2 diabetes (T2D). Bacterial community profiling was performed with primers targeting the variable region 4 of the 16S rRNA gene and sequenced. Amplicon sequence variants (ASVs) were generated with DADA2 and annotated to the SILVA database. The gut virome was sequenced, using a viral particle enrichment and metagenomics approach, assembled, and quantified to describe the composition of the viral community. Comparison of the bacterial alpha- and beta-diversity measures revealed that the gut bacteriome of individuals with LADA resembled that of individuals with T2D. Yet, specific genera were found to differ in abundance in individuals with LADA compared with T1D and T2D, indicating that LADA has unique taxonomical features. The virome composition reflected the stability of the most dominant order Caudovirales and the families Siphoviridae, Podoviridae, and Inoviridae, and the dominant family Microviridae. Further studies are needed to confirm these findings.
Subject terms: Type 1 diabetes, Diabetes, Type 1 diabetes, Type 2 diabetes, Microbiology, Bacteria, Virology
Introduction
Diabetes is a heterogeneous disease, characterized by elevated levels of blood glucose. Diabetes might be viewed as a spectrum, ranging from severely insulin deficient type 1 diabetes (T1D), with onset primarily in early childhood, to insulin resistant age-related type 2 diabetes (T2D), with onset primarily in middle-aged individuals and in late adulthood. The spectrum also encompasses latent autoimmune diabetes in adults (LADA), which is rather common, accounting for up to 10% of individuals initially diagnosed with T2D1. Phenotypically, LADA is placed between T1D and T2D, with respect to age-at diagnosis, measures of adiposity, circulating lipid levels, blood pressure, and progression to insulin dependence1–6. Also genetically, LADA seems to be an intermediate between T1D and T2D5,7–11. However, the contribution from T1D -associated genomic variants in the predisposition to LADA seems to be stronger than the contribution from T2D-associated variants, likely in part due to a considerable difference in effect sizes associated with T1D- and T2D-associated variants, respectively. The overlaps between diabetic subtypes can make the classification of adult diabetes challenging. Hence, improved understanding of the pathophysiology of LADA, and identification of biomarkers that could help distinguish LADA from T1D and T2D could be of clinical value. One strategy to shed light on these issues might be characterization of the human gut microbiome.
The human gut microbiome composition is shaped by multiple factors including host genetics, age, diet, environment, lifestyle, and external factors such as the use of medication, particularly antibiotics and metformin12,13. During the last two decades, the role of the gut microbiota in health and disease, particularly the bacteriome, has gained increasing attention. The gut microbiome has multiple functions for host metabolism, immunity, and brain biology14–16. Hence, growing evidence, from both animal and human studies, suggests that compositional, taxonomical, and functional changes in the gut bacteriome are linked to both T1D and T2D17–19. Gut-bacteriome abnormalities have been shown to affect the immune system and the gut permeability, suggested to trigger the autoimmune reaction leading to T1D20,21, and to disrupt the energy-generation and expenditure balance, which influence the risk of T2D22. Fang et al. 23 compared the gut bacterial microbiome between individuals with LADA, T1D, and T2D. This comparison indicated that these diabetic subtypes might differ in gut bacterial composition. Furthermore, Hu et al. 24 used metagenomics to compare the gut microbiome between individuals with adult-onset T1D and T2D including DNA viruses in the analyses and also found signatures differentiating the two groups. The intestinal virome plays an essential role in modulating the gut bacteriome since its core is composed of bacteriophages. The balance between bacteria and virus populations is critical for a healthy state of the overall gut microbiome25. Finding associations between the viral community and diseases is, however, a challenge due to the high variability across individuals. Yet, recent studies have shed some light on the role of the virome both in T1D and T2D26,27.
The aim of this study was to explore the intestinal bacterial and viral microbiome of individuals with LADA and compare findings with microbiome features of individuals with T1D and T2D as well as with non-diabetic control subjects.
Results
The individuals with LADA were intermediary between T1D and T2D with respect to adiposity-related traits and measures of glucose homeostasis (Table 1).
Table 1.
Overview of study population and quality of data parameters.
| Controls | Type 1 diabetes | LADA | Type 2 diabetes | |
|---|---|---|---|---|
| Sex | ||||
| Male | 44 | 17 | 37 | 44 |
| Female | 26 | 13 | 23 | 26 |
| Age (year) | 61 ± 10 | 52 ± 7 | 64 ± 10 | 61 ± 10 |
| BMI (kg/m2) | 27 ± 4 | 27 ± 5 | 29 ± 5 | 31 ± 6 |
| Metformin treatment | ||||
| Yes | 0 | 0 | 48 | 47 |
| No | 70 | 30 | 12 | 23 |
| Waist circumference (cm) | 93 ± 11 | 98 ± 13 | 103 ± 14 | 107 ± 14 |
| Hip circumference (cm) | 101 ± 8 | 106 ± 8 | 107 ± 10 | 108 ± 10 |
| Waist/hip ratio | 0.92 ± 0.08 | 0.92 ± 0.08 | 0.96 ± 0.09 | 1.00 ± 0.08 |
| HbA1c (mmol/l) | 38.5 ± 3.9 | 73.3 ± 8.8 | 63.1 ± 13.1 | 51.6 ± 13.9 |
| Fasting plasma glucose (mmol/l) | 5.8 ± 0.5 | 9.7 ± 4.2 | 8.8 ± 3.0 | 8.5 ± 2.7 |
| Bacteriome sequencing depth (Mill. annotated Reads) | 0.136 ± 0.026 | 0.150 ± 0.028 | 0.128 ± 0.024 | 0.129 ± 0.023 |
| Viral sequencing depth (Mill. mapped Reads) | 4.295 ± 2.747 | 3.932 ± 2.831 | 5.936 ± 2.718 | 5.803 ± 3.187 |
| Cell count (Bill. cells/g) | 20.4 ± 6.8 | 18.7 ± 5.7 | 18.9 ± 5.5 | 19.6 ± 6.1 |
Numerical values except counts are provided as means ± st. dev.
Bacterial alpha and beta diversity
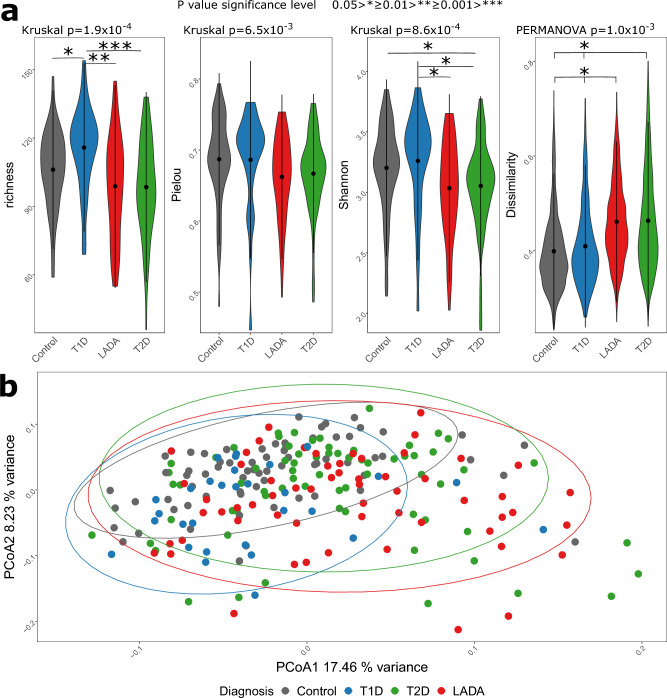
In analyses of the bacterial gut microbiota alpha-diversity and beta-diversity metrics were compared between the diagnostic groups to investigate if individuals with LADA had a distinct compositional profile. For all alpha-diversity measures, richness (p = 1.9 × 10−4), Pielous evenness (p = 6.5 × 10−3), and Shannon diversity (p = 8.6 × 10−4), a significant difference was detected across groups using the Kruskal–Wallis test. Individuals with LADA resembled individuals with T2D across all alpha-diversity measures, and the difference was most pronounced between T1D and the remaining groups, mostly driven by differences in richness (Fig. 1a). Comparison of beta-diversity revealed a significant difference in bacterial composition between diagnostic groups, based on differences in centroid position (PERMANOVA p = 1.0 × 10−3) and dispersion (betadisper p = 4.5 × 10−4) (Fig. 1a). A visual evaluation based on the beta-diversity in the PCoA plot revealed that individuals with T2D and LADA clustered, and differed the most compared with T1D (Fig. 1b). This observation was also evident from the pairwise PERMANOVA, where T2D and LADA were the only groups that did not differ significantly from each other (Fig. 1a). To control for the possible confounding of metformin treatment, we ran additional analyses removing metformin treated individuals with LADA (n = 48) and T2D (n = 47). In these analyses, with a much reduced sample size, individuals with LADA and T2D resembled controls both with respect to alpha-diversity and beta-diversity (Supplementary Fig. 1a,b).
Figure 1.
Exploration of differences between diagnostic groups investigating alpha and beta-diversity parameters (a), and by PCoA (b). Alpha-diversity was visualized with violin plots and included richness, Pielou’s evenness, and Shannon diversity. A non-parametric overall comparison of diagnostic groups was performed using the Kruskal–Wallis test. If significant, a follow-up pairwise comparison of groups was performed with the Mann–Whitney test and p-values were Bonferroni corrected. Bray–Curtis dissimilarity calculated from Hellinger transformed total sum scaled data was used as beta-diversity measure and the violin plot represented the within group variation. PERMANOVA was used to compare all diagnostic groups and follow-up pairwise comparison of diagnostic groups where p-values were Bonferroni corrected. PCoA were used to visualize dissimilarities of all samples. Variance explained by the first two axes were included in their labels. Violin plots of beta-diversity represented within diagnostic group dissimilarity.
Bacterial abundance
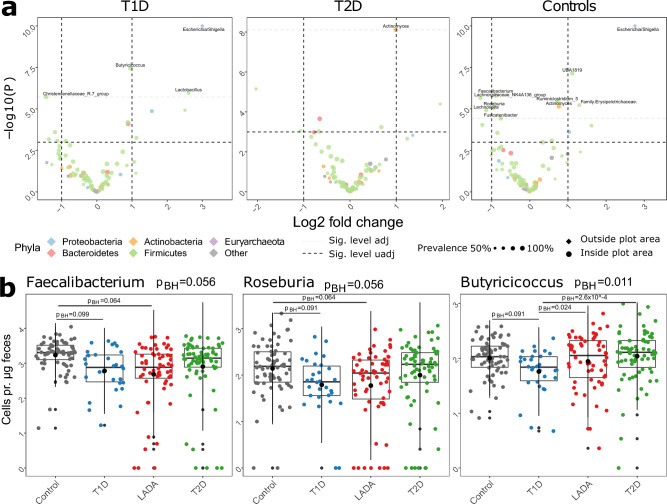
The abundance of a number of bacterial genera was significantly different when comparing individuals with LADA with the three remaining diagnostic groups (Fig. 2a). The absolute abundance of one genus differed significantly between individuals with LADA and T2D (Actinomyces: pBH = 0.025). Ten genera differed significantly between individuals with LADA and the control group, where Escherichia was the most significant (Escherichia/Shigella: pBH = 1.6 × 10−5). Four genera differed when comparing individuals with LADA and T1D (Escherichia/Shigella: pBH = 3.2 × 10−6, Butyricicoccus: pBH = 0.024, Lactobacillus: pBH = 0.068, Christensenellaceae_R.7_group: pBH = 0.068) (Fig. 2a,b, Supplementary Fig. 2). Hence, supporting the observation of individulas with LADA being more similar to T2D than T1D. When analyzing short-chain fatty acid producers, including Faecalibacterium, Clostridium, Roseburia, Anaerostipes, Bifidobacterium, Butyricicoccus, and Akkermansia, we found that two of these genera were less abundant in individuals with LADA compared with non-diabetic control subjects (Roseburia: pBH = 0.064, Faecalibacterium: pBH = 0.064) (Supplementary File 1). Faecalibacterium was the most abundant genus in control subjects, but not in any of the diabetes groups (Fig. 2b, Supplementary Fig. 3). With respect to other butyrate producers, the abundance of Butyricicoccus was lower in individuals with T1D relative to both LADA (pBH = 0.024) and T2D (pBH = 2.6 × 10−4). All of the above-mentioned genera were significantly different in the likelihood ratio test (Fig. 3, Supplementary Table 1).
Figure 2.
Differences in specific genera observed between diagnostic groups, Volcano plot showing results from the differential abundance analysis (a), and boxplots of selected genera (b). Differential abundance tests were performed with cell counts as normalization factors in DESeq2 performing both likelihood ratio test, comparing all diagnostic groups, and a Wald test to make pairwise comparisons between diagnostic groups. p-values in the Wald test were Benjamini–Hochberg corrected.
Figure 3.
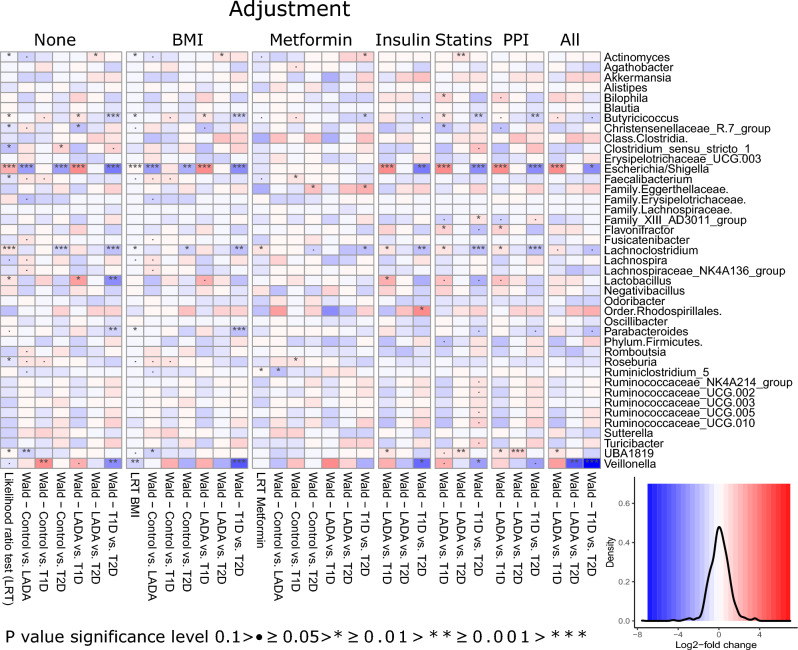
Heatmap of DESeq test results comparing across all groups using the likelihood ratio test implementation (LRT) and comparing diagnostic groups pairwise using Wald test. Analysis included adjustment for BMI, use of anti-diabetic medication (insulin, statins, PPI), and all of the mentioned covariates. Adjustment for metformin was analogous to the overall analysis scheme by removing samples from individuals treated with metformin. The first specified diagnostic group is the reference for the DESeq test in the Wald test. A density plot of the effect sizes (log2fold change) is included with the same color key used in the heatmap. Genera with the lowest p-value in the LRT without adjustment were included, but Log2fold changes and p-values from all genera comparison are provided in Supplementary Table 1. The heatmap was generated with the R package pheatmap with layout adjustments performed in inkscape.
When controlling for metformin treatment, by removing individuals with LADA and T2D treated with metformin, the number of significantly differentially abundant genera was limited to Ruminiclostridium_5 (pBH = 0.018) when comparing individuals with LADA with the control group (Supplementary Fig. 2a, Supplementary Table 1). Comparing the differential abundance of genera after correcting for BMI and anti-diabetic medication (insulin, statins, and proton pump inhibitors (PPI)), revealed in general consistent signatures. Of note, removing individuals treated with metformin, the differences observed for Escherichia/Shigella and UBA1819 disappeared (Fig. 3, Supplementary Table 1).
Prediction of bacterial function
The functional prediction of the bacterial gut microbiota revealed similar patterns in PCoA analyses across groups. However, relatively large differences were observed for the evenness estimates between the diagnostic groups resulting in a higher Shannon diversity in individuals with T2D and LADA relative to control subjects and individuals with T1D. In general, a higher dissimilarity within individual groups of individuals with diabetes was observed relative to control subjects (Supplementary Fig. 4a,b, Supplementary File 2).
Compared with control subjects, 12 KEGG orthologs were significantly more abundant in individuals with LADA. Ten of these orthologs were also significantly more abundant in individuals with LADA compared with T1D. These more abundant orthologs were mainly involved in xenobiotics biodegradation and metabolism and lipid metabolism. Other highlighted pathways included two involved in metabolism of terpenoids, where the putative abundance of the function geraniol degradation was both significantly higher in individuals with LADA and T2D relative to T1D and control subjects. Individuals with LADA and T2D also had higher abundance of carotenoid biosynthesis when compared with control subjects. Furthermore, abundance of pathways involved in retinol metabolism was significantly higher in individuals with LADA relative to control subjects. No KEGG orthologs differed significantly between individuals with LADA and T2D (Supplementary Fig. 5a,b, Supplementary File 2).
Viral abundance
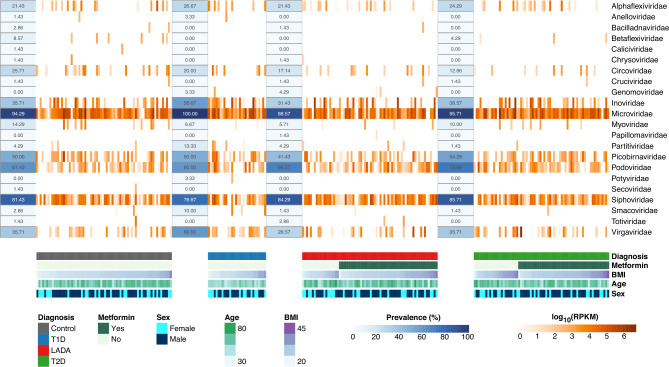
Our analyses of the viral gut microbiota showed that the most represented families were Microviridae (455 sequences), Siphoviridae (325), Picobirnaviridae (263), Podoviridae (199), and Inoviridae (41). The most represented families were also the most prevalent in the different diagnostic groups (Fig. 4). There was no association between the presence of a viral family and the diagnostic group of study participants (Supplementary Table 2). Considering the mean abundance of each viral family, the most abundant were Microviridae (101748 RPKM), Inoviridae (21822 RPKM), Virgaviridae (13683 RPKM), Siphoviridae (10598 RPKM), and Podoviridae (3715 RPKM). Comparing the quantification ranking between different groups (Wilcoxon rank test), we found no difference between diagnostic groups (Supplementary Table 3). The differential abundance analysis revealed Podoviridae as more abundant in individuals with T2D when compared with control subjects (Podoviridae: pBH = 0.023) (Supplementary Table 4). However, considering outliers with extremely high counts, the difference observed might be false positive (Supplementary Fig. 6).
Figure 4.
Heatmap of virome abundance and prevalence. The abundance of 22 taxonomic families is represented in log10(RPKM) and grouped by diagnosis. The prevalence represents the percentage of individuals in one group where the virus could be identified. The sex and usage of metformin are observed as qualitative information, while BMI (kg/m2) and age (years) are pictured as a quantitative colormap. The heatmap was generated with the R package pheatmap with layout adjustments performed in illustrator.
Discussion
We examined the composition, taxonomy, and predicted functional profile of the bacterial gut microbiota, using 16S rRNA gene amplicon sequencing, and the viral gut microbiota, using metagenomics, in individuals with LADA compared with non-diabetic control subjects, and individuals with T1D or T2D. Overall, individuals with LADA were characterized by distinct features in their bacterial gut microbiota resembling those, which have been reported previously23. However, our results also indicated that the bacterial gut microbiota of individuals with LADA was most similar to individuals with T2D, which is in contrast to a previous study showing that the microbiota of individuals with LADA was more similar to individuals with T1D23.
By performing taxonomic and functional analysis of alpha and beta-diversity measures, we showed differences across diagnostic groups, but also revealed that individuals with LADA were most similar to individuals with T2D. Based on bacterial beta-diversity Hu et al. 24, using metagenomics, similarly found adult-onset T1D to be more similar, but still significantly different compared to T2D, relative to healthy cotrols. With respect to the abundance of bacterial genera, we found that Actinomyces was significantly more abundant in LADA compared with T2D and represented the only genus that was significantly different in the comparison of LADA and T2D. This signature was also observed after correcting for BMI, and anti-diabetic medication. Higher abundance of Actinomyces in feces has been associated with gestational diabetes and inflammation of the gut28,29. The link to inflammation provides a speculative explanation of the similarity of the abundance of Actinomyces in LADA and T1D. Despite previous observations that an increase in the abundance of Actinomyces is a marker of obesity and altered bacterial gut microbiota28,30, we did not observe a difference between individuals with T2D and control subjects.
On the bacterial genera level, the abundance of key short-chain fatty acid producers, including Faecalibacterium and Roseburia, have been reported to be relatively depleted in T1D and T2D 19,24,31,32. Here, we found that both genera were less abundant in individuals with LADA compared with the control group, which is in line with previous observations23. However, in contrast to previous observations we observed similar abundance of these short-chain fatty acid producers in LADA and T1D. This was also observed after correcting for BMI or removing individuals treated with metformin. These findings could indicate that lower abundance of these genera is a general change of the gut bacterial microbiota characterizing individuals with diabetes. Moreover, another key butyrate producer, Clostridium, was also observed to have lower abundance among individuals with T2D compared with control subjects, which is in line with previous observations in individuals with pre-diabetes and T2D33,34. The abundance of Clostridium did not differ between individuals with LADA and control subjects. Comparing individuals with LADA with non-diabetic control subjects, we observed a previously described metformin signature12, as Escherichia/Shigella and UBA1819 were significantly more abundant among individuals with LADA, but only in the analysis where we included the metformin treated individuals.
Variation in functional potential of the bacterial gut microbiome might provide mechanistic insight into the differentiation of the diabetes entities. In Hu et al. 24 Eubacterium rectale represented a key short-chain acid producer depleted in adult-onset T1D. However, in this study the genus was observed in few samples and did not meet inclusion criteria for differential abundance analyses, highlighting, that despite not being a key microbiome member in the present study the functional profile is conserved. The differences in community members might represent cohort differences, but could also be due to methodology and differences in the diabetes phenotypes compared in the studies. In the present study functional insights were based on predictions from the 16S rRNA gene sequencing data. The abundance of a number of KEGG orthologs involved in lipid metabolism and xenobiotics biodegradation and metabolism, differed comparing individuals with LADA with T1D or control subjects, whereas no differences in abundance of KEGG orthologs were observed between LADA and T2D. Moreover, the abundance of the functional pathway geraniol degradation was significantly higher in LADA compared with T1D and control subjects, which is in line with a previous study showing that Geraniol has antimicrobial properties and is associated with improved glucose homeostasis in diabetes induced in rats35. The pathways carotenoid biosynthesis and retinol metabolism were significantly higher in LADA compared with control subjects, which might be explained by the insulin sensitizing effects of carotenoids and retinoids36,37. No significant differences in functional pathways were observed between individuals with LADA and T2D, which might indicate that some of the underlying pathways linked to the gut bacteria are similar in LADA and T2D. These findings related to functional differences in the bacterial gut microbiota of LADA, T1D, and T2D should be investigated further in studies applying metagenomics sequencing in a larger sample size with well matched diagnostic groups. Also, the interplay between the host and the gut bacterial potential to modify or synthesize these bioactive compounds have not been explored.
Taxonomic and functional investigations into the microbiome have been associated with the development of T1D, T2D, and maturity-onset diabetes of the young. Nevertheless, findings have been somewhat inconsistent due to differences in methodology and heterogeneous populations19,32,38,39. Metformin treatment, a common first-line medication prescribed to individuals diagnosed with LADA and T2D, is a confounding variable in gut microbiota studies12,40,41. Attempting to rule out a possible impact of metformin, we repeated all analyses after removal of individuals treated with metformin. This subgroup analysis indicated that LADA and T2D individuals resembled controls, when comparing alpha and beta-diversity measures of their gut microbiota. However, removal of such a large group of samples consequently resulted in a loss of statistical power, and therefore, might introduce type 2 errors. The inclusion of metformin treated individuals was based on a pragmatic approach, as exclusion of metformin treated T2D individuals would challenge patient recruitment immensely and potentially also lead to selection bias. Besides metformin treatment, it would also have been relevant to take other possible confounders into account, including dietary habits. However, even though the diagnostic groups were collected as part of different studies, we consider our study sample to be relatively homogeneous in terms of lifestyle, environment, and ethnicity, as all individuals were recruited from the same part of the country and all individuals were of Danish ancestry. Statistical power might also be an issue in the comparison to the smaller group of individuals with T1D, which possibly explains the relatively few differentially abundant bacterial genera observed in the comparison with individuals with LADA, where we tentatively expected to find greater differences based on the comparisons of alpha and beta-diversity measures. In the study, bacterial-cell counting was performed, which was used to obtain absolute abundances that benefit quantitative analysis.
The non-redundant virome dataset permitted us to explore the gut environment and health and disease status associations. Although we were able to classify a fraction of the viruses into families (13.7%), most of the phages could not be classified, due to the current incompleteness of phage reference databases. The human virome is largely unknown and description of the virome in association with diabetes is sparse and lacking in the context of LADA24,42. The human gut virome may be considered an individual signature25. The most prevalent families in our study including Microviridae, Siphoviridae, and Podoviridae have been reported as prevalent and abundant in other studies including a range of different cohorts25,43,44. In particular, the family Podoviridae, has been related to systemic lupus erythematosus45, but has not been associated with T2D46,47. Even though we did not find significant associations across the diagnostic groups, this study recognizes the relevance of assessing the microbial environment in a broader context by assessing both the bacterial and viral community. Using metagenomics possibly including metabolomics has the potential to improve the functional understanding of the mechanism underlying the link between microbiota and diabetes subtypes23,24.
In conclusion, the present study characterized both the bacterial and viral gut microbiota across diabetic subtypes. We showed that the bacterial gut microbiota of individuals with LADA was most similar to that of individuals with T2D, but there were also great similarities with individuals with T1D. We identified unique taxonomical features in individuals with LADA, and also the signature of lower abundance of short-chain fatty acid producing bacteria, which is shared across all the diabetic subtypes. We did not observe clear virome signatures associating with the diagnostic groups. Our findings may contribute to the understanding of the gut microbiota-related pathophysiology underlying LADA, but we did not identify clear gut microbial biomarkers that can distinguish LADA from T1D or T2D. To gain further information about the potential role of the gut microbiota in LADA pathophysiology it would be relevant in future studies to assess a larger group of individuals with LADA, and at the same time include additional phenotypic information to facilitate analysis of the potential effect of disease duration, anti-diabetic treatment, dietary habits, as well as type and level of autoantibodies.
Methods
Study participants
The study comprised 60 individuals diagnosed with LADA, 70 individuals diagnosed withT2D, 30 individuals diagnosed with T1D, and 70 non-diabetic control subjects (Table 1). Individuals with LADA were recruited from Steno Diabetes Center Copenhagen. LADA was defined by; age at onset above 30 years, presence of glutamic acid decarboxylase autoantibodies (GADA), and preserved beta-cell function indicated by clinical diagnosis, treatment without insulin during the first year after diagnosis, or fasting C-peptide above 300 pmol/l.
To match inividuals with LADA on age and sex, 70 individuals with T2D were selected from the MicrobDiab study sample48, where all individuals had been diagnosed with diabetes within five years of the clinical visit, and were white Europeans between 35 and 75 years of age. Similarly, 70 matched non-diabetic control subjects were selected from the DanFunD cohort49. From the MetaHIT study we obtained 30 eligible individuals with T1D12,50. In all cohorts participants treated with antibiotics within three weeks before sample collection and examination were excluded. All included individuals were collected from the greater Copenhagen area and were all of Danish ancestry.
Collection and handling of fecal samples were performed according to the same protocol across all studies. The samples were collected at home using a standard stool collection kit. The protocol emphasized antiseptic handling, using sterile containers, and freezing locally at − 20 °C before transferring in cooling bags to a central facility where samples was stored at − 80 °C.
DNA extraction and estimation of bacterial cell load
DNA was extracted from 200 mg of feces using the NucleoSpinSoil kit (Macherey–Nagel, Germany), following the manufacturer’s instructions. Quality control of DNA included measurements of yield, purity, and degree of fragmentation using a Qubit 2.0 fluorometer, a NanoDrop2000 spectrometer, and agarose gel electrophoresis, respectively. Bacterial community profiling was performed with primers (515F/806R) targeting the hypervariable region 4 of the 16S rRNA gene, and sequencing on the Illumina HiSeq platform generating a total of 48,794,462 (MEAN = 203,310 SD = 40,317 Minimum = 86,379) 150 bp paired-end reads. Reads were trimmed, filtered, de-replicated, and merged leaving 32,073,812 (MEAN = 133,641 SD = 25,466 Minimum = 61,161) reads for further processing by generating amplicon sequence variants (ASVs) (nASVs = 6455), and taxonomically annotation to the SILVA database using DADA2 (v1.6.0) in R (v3.4.1)51. Cell counts were obtained by fluorescence-activated Cell Sorting (FACS) and used to obtain absolute numbers of cells, which were used in the differential abundance analyses as described below52. This enabled the quantitative differential abundance analyses setup relative to a classical compositional approach with concurrent loss of power. It was not possible to obtain a cell count from one sample, which was therefore set as the mean of all samples, because the cell counts obtained were not significantly different from each other in the diagnostic groups (p = 0.62) (Table 1).
Analyses of the bacterial gut microbiota
Alpha-diversity metrics included richness, Pielou’s evenness index, and Shannon diversity using the diversity function in the vegan package. Differences in alpha-diversity were compared with a Kruskal–Wallis test and a pairwise follow-up Bonferroni corrected Mann–Whitney test. Beta-diversity calculations were based on Bray–Curtis dissimilarities calculated from Hellinger transformed count data. These were used as input to perform PERMANOVA, heatmap dendrograms, and principal coordinate analysis (PCoA). PERMANOVA was performed using the adonis2 implementation in the vegan package as well as a Bonferroni corrected pairwise PERMANOVA between the diagnostic groups. A heatmap of taxonomic abundances was generated for the 30 most abundant genera standardized to zero mean and unit variance. Sample-based dendrograms were generated from the Bray–Curtis dissimilarity matrix using complete-linkage clustering. The genera-based dendrograms were generated using Pearson product-moment correlation coefficients with complete-linkage clustering. PCoA and canonical correspondence analysis (CCA) plots were generated using the capscale function in vegan unconstrained and constrained, respectively.
Differential abundance tests were performed with cell counts as normalization factors in DESeq2 performing both likelihood ratio test (LRT) comparing all diagnostic groups, and a Wald test to make pairwise comparisons of diagnostic groups53. Only genera present in 50 or more percent of the samples were included. These genera accounted for the vast majority of the reads (MEAN = 88.9% SD = 11.1). The test results reported in the text is with correction for BMI, but the different test results (effect size: log2fold change) were visualized in a heatmap also accounting for the covariates: Insulin, statins, and PPI. However, information was not available for the control group for these covariates. The comparison of LADA with the other diagnostic groups was visualized as volcano plots and selected significant genera as boxplots. Correction of multiple testing were performed using the Benjamini–Hochberg method (BH) with the default significance level p-value < 0.1. In the rest of the analyses a p-value < 0.05 was considered statistically significant.
Metformin was only used as treatment by some of the T2D (n = 47) and LADA (n = 48) individuals and was therefore not represented in the other groups. To account for effects of metformin on gut microbiota, the analyses were performed omitting data from individuals treated with metformin. Test results provided in text is here without the correction for BMI (Supplementary File 1). Metagenomic function of the DADA2 annotated 16S rRNA gene data was predicted using piphillin and the KEGG database54,55. A custom script was used to generate R compatible KEGG brite hierarchies from the online KEGG orthology resource, available at https://github.com/Hansen-Group/KEGGjsonR56. Orthologs classified as human diseases and organismal systems were removed from the downstream analyses. The statistical analyses applied to the taxonomical data described above were performed analogously to investigate differences in functional orthologs (Supplementary File 2).
Statistical analyses of the bacterial gut microbiota were performed in R (v4.1.2) and package specifications and function calls are available in supplementary Files 1 and File 2. Visualization of data was performed with ggplot2 3.3.5, pheatmap 1.0.12 and VennDiagram 1.7.1 with final modifications in Inkscape and illustrator, however, all original plots are available in Supplementary Files 1 and File 2. The count tables of ASVs were aggregated to different taxonomic levels but data presented were at genus level. Analyses were performed including: (1) all individuals, and (2) after exclusion of LADA and T2D individuals treated with metformin (Supplementary File 1).
Processing and analyses of the viral gut microbiota
Separate aliquots of the stool samples were sent for processing according to the NetoVIR protocol, used to enrich viral-like particles and to prepare samples for sequencing57. The virome data was characterized as described in Van Espen et al. 58. In short, raw reads were trimmed to remove low-quality bases and sequencing adapters using Trimmomatic59, reads mapping human reference genome and contaminome using Bowtie2 60 were removed and the remaining reads were per-sample de novo assembled using metaSPAdes 61. The obtained scaffolds longer than 1kb were clustered at 95% identity over 80% sequence length to generate a non-redundant reference set, containing 150,743 scaffolds ranging from 1000 bp long to 301 ,716 bp. The filtered reads were mapped to the non-redundant reference database using BWA 58,62. The number of mapped reads to each contig was used to create an abundance table for statistical analysis. Viral scaffolds (n = 21,678) were identified using a scoring system based on homology to reference viruses at the amino acid and/or genome level, genome structure, the presence of virus-specific genes, and VirSorter category63,64. Eukaryotic viruses were taxonomically classified based on homology to classified reference viruses, while phages (prokaryotic viruses) were classified using vConTACT265.
The statistical analysis was performed in the R (v4.0.5). The absolute counts were summarized into viral family levels for differential abundance analysis using DESeq253. Adjusted p-values < 0.1 were considered as significant for discussion. The counts were normalized, calculating the number of reads per kilobase of contig per million reads mapped (RPKM) and used for heatmap visualization. The ranked abundance difference between groups was assessed using the Wilcoxon rank test. The association between viral prevalence and the diagnosis was measured using Fisher's Exact test. All the p-values were adjusted using the FDR method, and p-values < 0.05 were considered significant.
Ethics declarations
All individuals gave written informed consent before participation and studies were approved by the Ethical Committees of the Capital Region of Denmark (MicrobDiab: H-3-2013-102; DanFund: H-3-2011-081 and H-3-2012-0015; MetaHIT: HC-2008-017; LADA: H-16046195). Studies were conducted in accordance with the principles of the Declaration of Helsinki.
Supplementary Information
Acknowledgements
The authors wish to thank Lila Close for processing the fecal samples for virome sequencing.
Author contributions
HV, TH, and MKA were responsible for study concept and design. DH, AL, LVE, TJ, TN, ACA, JM, OP, and MKA were responsible for data collection and curation. CSP, GRF, THH, TH, and MKA were responsible for analysis and interpretation of data. CSP, GRF, TH, and MKA were responsible for drafting and critical revision of the manuscript. TH and MKA obtained funding. CSP, DH, GRF, THH, TK, AL, LVE, TJ, TN, ACA, JM, OP, HV, TH, and MKA contributed to the final version of the manuscript. CSP is the guarantor of this work and, as such, had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
DanFunD was funded by The Lundbeck Foundation (Grant number R155-2013-14070) and Trygfonden (Grant numbers 7-11-0213, 117548). MicrobDiab was funded by Innovation Fund of Denmark (Grant number 0603-00591B) and the Department of Biotechnology, Govt. of India (BT/IN/Denmark/12/VM/2013). MetaHIT was funded from European Community’s Seventh Framework Program (Grant agreement HEALTH-F4-2007-201052). The LADA study and MKA was funded by The Danish Diabetes Academy (Grant ID po0115) and Danish Council for Independent Research (Grant ID DFF-5053-00159). LVE is supported by the Research Foundation Flanders (Fonds voor Wetenschappelijk Onderzoek-FWO-1S25720N). The computational resources were provided by the Flemish Supercomputer Center (VSC) and funded by FWO and the Flemish Government, Department of Economy, Science, and Innovation. Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center, based at the University of Copenhagen, Denmark, and partially funded by an unconditional donation from the Novo Nordisk Foundation (www.cbmr.ku.dk) (Grant number NNF18CC0034900 and NNF23SA0084103).
Data availability
The datasets generated and analysed during the current study are available under accession number PRJEB68363 (https://www.ebi.ac.uk/ena/browser/view/PRJEB68363).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Torben Hansen, Email: torben.hansen@sund.ku.dk.
Mette K. Andersen, Email: metteandersen@sund.ku.dk
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-58985-w.
References
- 1.Tuomi T, et al. The many faces of diabetes: A disease with increasing heterogeneity. Lancet. 2014;383:1084–1094. doi: 10.1016/S0140-6736(13)62219-9. [DOI] [PubMed] [Google Scholar]
- 2.Tuomi T, et al. Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes. 1999;48:150–157. doi: 10.2337/diabetes.48.1.150. [DOI] [PubMed] [Google Scholar]
- 3.Zinman B, et al. Phenotypic characteristics of GAD antibody-positive recently diagnosed patients with type 2 diabetes in North America and Europe. Diabetes. 2004;53:3193–3200. doi: 10.2337/diabetes.53.12.3193. [DOI] [PubMed] [Google Scholar]
- 4.Hawa MI, et al. Metabolic syndrome and autoimmune diabetes: Action LADA 3. Diabetes Care. 2009;32:160–164. doi: 10.2337/dc08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen MK, et al. Latent autoimmune diabetes in adults differs genetically from classical type 1 diabetes diagnosed after the age of 35 years. Diabetes Care. 2010;33:2062–2064. doi: 10.2337/dc09-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z, et al. Frequency, immunogenetics, and clinical characteristics of latent autoimmune diabetes in China (LADA China Study): A nationwide, multicenter, clinic-based cross-sectional study. Diabetes. 2013;62:543–550. doi: 10.2337/db12-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra R, et al. Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med. 2017;15:1. doi: 10.1186/s12916-017-0846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousminer, D. L. et al. First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. in Diabetes Care vol. 41 2396–2403 (American Diabetes Association Inc., 2018). [DOI] [PMC free article] [PubMed]
- 9.Andersen MK, et al. Type 2 diabetes susceptibility gene variants predispose to adult-onset autoimmune diabetes. Diabetologia. 2014;57:1859–1868. doi: 10.1007/s00125-014-3287-8. [DOI] [PubMed] [Google Scholar]
- 10.Ramu D, Perumal V, Paul SFD. Association of common type 1 and type 2 diabetes gene variants with latent autoimmune diabetes in adults: A meta-analysis. J. Diabetes. 2019;11:484–496. doi: 10.1111/1753-0407.12879. [DOI] [PubMed] [Google Scholar]
- 11.Andersen MK. New insights into the genetics of latent autoimmune diabetes in adults. Curr. Diabetes Rep. 2020 doi: 10.1007/s11892-020-01330-y. [DOI] [PubMed] [Google Scholar]
- 12.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vujkovic-Cvijin I, et al. Host variables confound gut microbiota studies of human disease. Nature. 2020;587:448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 15.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;1979(312):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 17.Tai N, Wong FS, Wen L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev. Endocr. Metab. Disord. 2015;16:55–65. doi: 10.1007/s11154-015-9309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durazzo M, Ferro A, Gruden G. Gastrointestinal microbiota and type 1 diabetes mellitus: The state of art. J. Clin Med. 2019;8:1843. doi: 10.3390/jcm8111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurung M, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020 doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts T, et al. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc. Natl. Acad. Sci. USA. 2005;102:2916–2921. doi: 10.1073/pnas.0500178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosi E, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 22.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, et al. Characteristics of the gut microbiota and metabolism in patients with latent autoimmune diabetes in adults: A case-control study. Diabetes Care. 2021;44:2738–2746. doi: 10.2337/dc20-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, J. et al. Distinct signatures of gut microbiota and metabolites in different types of diabetes: A population-based cross-sectional study. EClinicalMedicine. 62, 102132 (2023). [DOI] [PMC free article] [PubMed]
- 25.Shkoporov AN, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26:527–541.e5. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhao G, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. USA. 2017;114:E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, You X, Mai G, Tokuyasu T, Liu C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 2018;6:1–12. doi: 10.1186/s40168-018-0410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Actinomyces and alimentary tract diseases: A review of its biological functions and pathology. BioMed. Res. Int. 2018 doi: 10.1155/2018/3820215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, et al. Alterations of gut microbiota and blood lipidome in gestational diabetes mellitus with hyperlipidemia. Front. Physiol. 2019;10:1015. doi: 10.3389/fphys.2019.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Chierico F, et al. Gut Microbiota markers in obese adolescent and adult patients: Age-dependent differential patterns. Front. Microbiol. 2018;9:1210. doi: 10.3389/fmicb.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leiva-Gea I, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: A case-control study. Diabetes Care. 2018;41:2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 33.Allin KH, et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810–820. doi: 10.1007/s00125-018-4550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 35.Babukumar S, Vinothkumar V, Sankaranarayanan C, Srinivasan S. Geraniol, a natural monoterpene, ameliorates hyperglycemia by attenuating the key enzymes of carbohydrate metabolism in streptozotocin-induced diabetic rats. Pharm. Biol. 2017;55:1442–1449. doi: 10.1080/13880209.2017.1301494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee EJ, Plutzky J. Retinoid metabolism and diabetes mellitus. Diabetes Metab. J. 2012;36:167–180. doi: 10.4093/dmj.2012.36.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roohbakhsh A, Karimi G, Iranshahi M. Carotenoids in the treatment of diabetes mellitus and its complications: A mechanistic review. Biomed. Pharmacother. 2017;91:31–42. doi: 10.1016/j.biopha.2017.04.057. [DOI] [PubMed] [Google Scholar]
- 38.Thingholm LB, et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26:252–264.e10. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siljander H, Honkanen J, Knip M. Microbiome and type 1 diabetes. EBioMedicine. 2019;46:512–521. doi: 10.1016/j.ebiom.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong X, et al. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional chinese herbal formula: A multicenter, randomized, open label clinical trial. mBio. 2018;9:2392–2409. doi: 10.1128/mBio.02392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 42.Fitzgerald CB, et al. Probing the “dark matter” of the human gut phageome: culture assisted metagenomics enables rapid discovery and host-linking for novel bacteriophages. Front. Cell Infect. Microbiol. 2021;11:100. doi: 10.3389/fcimb.2021.616918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garmaeva S, et al. Stability of the human gut virome and effect of gluten-free diet. Cell Rep. 2021;35:109132. doi: 10.1016/j.celrep.2021.109132. [DOI] [PubMed] [Google Scholar]
- 44.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomofuji Y, et al. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Ann. Rheum. Dis. 2022;81:278–288. doi: 10.1136/annrheumdis-2021-221267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang K, et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology. 2021;161:1257–1269.e13. doi: 10.1053/j.gastro.2021.06.056. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q, et al. Enteric phageome alterations in patients with type 2 diabetes. Front. Cell Infect. Microbiol. 2021;10:856. doi: 10.3389/fcimb.2020.575084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez-Silva C, et al. Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Med. 2021;13:1–13. doi: 10.1186/s13073-021-00856-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dantoft TM, et al. Cohort description: The Danish study of Functional Disorders. Clin. Epidemiol. 2017;9:127–139. doi: 10.2147/CLEP.S129335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandeputte D, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 53.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed]
- 54.Iwai S, et al. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11:e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narayan NR, et al. Piphillin predicts metagenomic composition and dynamics from DADA2-corrected 16S rDNA sequences. BMC Genom. 2020;21:56. doi: 10.1186/s12864-019-6427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conceição-Neto N, et al. Modular approach to customise sample preparation procedures for viral metagenomics: A reproducible protocol for virome analysis. Sci. Rep. 2015;5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Espen, L. et al. A previously undescribed highly prevalent phage identified in a danish enteric virome catalog. mSystems6, e0038221 (2021). [DOI] [PMC free article] [PubMed]
- 59.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beller, L. et al. The virota and its transkingdom interactions in the healthy infant gut. Proc. Natl. Acad. Sci. USA119, e2114619119 (2022). [DOI] [PMC free article] [PubMed]
- 64.Roux, S., Enault, F., Hurwitz, B. L. & Sullivan, M. B. VirSorter: Mining viral signal from microbial genomic data. PeerJ3, e985 (2015). [DOI] [PMC free article] [PubMed]
- 65.Bin Jang H, et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available under accession number PRJEB68363 (https://www.ebi.ac.uk/ena/browser/view/PRJEB68363).