Abstract
This report describes the effects of mutating highly conserved residues in the primer grip domain of human immunodeficiency virus type 1 reverse transcriptase (RT) on virus formation and infectivity. Among a series of RT mutant viruses, three (M230A, L234D, and W239A) were found to be noninfectious or very poorly infectious. Our data indicate that these mutations in RT caused severe defects in proviral DNA synthesis. Interestingly, assembly and maturation of mutant virus M230A were similar to those of the wild type, while mutants L234D and W239A showed impaired maturation. The immature morphology of RT mutants L234D and W239A is due at least in part to premature cleavage of the gag-pol precursor, prior to virion budding, indicating that intracellular stability of Pr160gag-pol is of key importance during virus assembly.
During formation of human immunodeficiency virus type 1 (HIV-1), structural proteins, enzymes, envelope glycoproteins, and genomic viral RNA are coordinately assembled at the cell membrane. The internal structural proteins of the virion are encoded by gag and are synthesized in the form of the polyprotein precursor, Pr55gag. The enzymatic components of the virion, which are encoded by pol, are synthesized as components of the larger polyprotein precursor, Pr160gag-pol, which contains both gag- and pol-encoded sequences (37). Synthesis of Pr160gag-pol occurs by occasional ribosomal frameshifting into the overlapping Pol reading frame during translation of gag (14, 38). Pr55gag and Pr160gag-pol are cotranslationally modified by N-terminal attachment of a myristate (3, 11, 25, 29) and transported to the cell membrane, where assembly and budding occur (13, 37). Incorporation of Pr160gag-pol into assembling particles is thought to be mediated through interactions of its N-terminal Gag domain with Pr55gag (13, 28, 34, 35). Although expression of the internal structural proteins alone is sufficient for the formation of noninfectious particles (8, 34), the presence of Pr160gag-pol is required in the virion for maturation, which is an event controlled by the pol-encoded protease (PR) (7, 11, 17, 24). Interestingly, when a truncated recombinant consisting only of gag-PR is expressed, maturation occurs only in a fraction of the resultant particles with a marked reduction in Pr55gag processing (8, 33), suggesting that only full-length gag-pol allows normal particle maturation.
Mature reverse transcriptase (RT) is composed of the p66 and p51 subunits. The p66 subunit is 560 amino acids in length and constitutes the active element of the enzyme (5) with polymerase and RNase H activities, while the p51 subunit is a 440-residue derivative of p66 without the RNase H segment. In p66, a β-sheet (composed of β-strands 12 to 15) located downstream of the polymerase active site is implicated in binding the nucleic acid duplex and its translocation (23). β-Strands 12 and 13 (amino acids 227 to 235) form the primer grip domain responsible for maintaining the primer terminus in an orientation appropriate for nucleophilic attack on the incoming deoxynucleoside triphosphate (15, 16, 23). The HIV-1 RT primer grip domain is highly conserved among several related retroviruses (16, 40), and its implication in RT functions was demonstrated by means of biochemical studies (9, 16, 27, 31).
Highly conserved residues in and around the primer grip motif were mutated (see Fig. 1), and the effects of these mutations on HIV-1 virus structure and replication were examined. Three mutants, namely, M230A, L234D, and W239A (Met230 to Ala, Leu234 to Asp, and Try239 to Ala, respectively) retained our attention (Fig. 1). M230A infectivity was highly attenuated (a decrease of approximately 104-fold compared to the wild type [WT]), and mutants L234D and W239A were found to be noninfectious, while most other mutants (E224A, P225A, F227A, G231A, E233A, and H235A) showed WT or slightly attenuated infectivity.
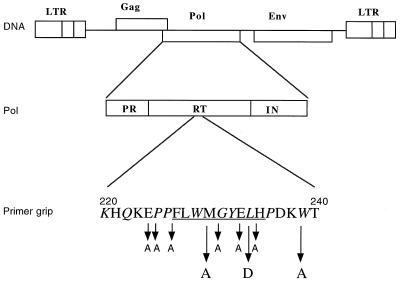
FIG. 1.
Schematic representation of the HIV-1 genome with localization of the RT primer grip domain in Pol. Amino acids constituting the β-strands 12 and 13 are underlined. Conserved residues (compared with HIV-2, simian immunodeficiency virus, feline immunodeficiency virus, and equine infectious anemia virus) are indicated in italics. The three substitutions (M230A, L234D, and W239A) analyzed here are emphasized by long arrows. The SphI/EcoRI fragment of the pNL4-3 HIV-1 molecular clone (1) was inserted into M13mp18 for use as a target for site-directed mutagenesis as described elsewhere (21, 26). Mutations L234D and W239A were obtained with oligonucleotides 5′-CAGGATGGTCTTCATAACCC-3′ and 5′-GGCTGTACTGTCGCTTTATCAGG-3′, respectively. Underlined nucleotides represent the changes responsible for the amino acid substitutions. Mutation M230A was introduced into the plasmid pNL4-3 by replacing the EcoRV-KpnI fragment (positions 2979 to 3830 of the plasmid) with the homologous EcoRV-KpnI fragment (positions 2561 to 3412 of HIV-1 strain Lai) from the plasmid pRTM230A (23, 39). Other substitutions are indicated by short arrows and correspond to E224A, P225A, F227A, G231A, E233A, and H235A.
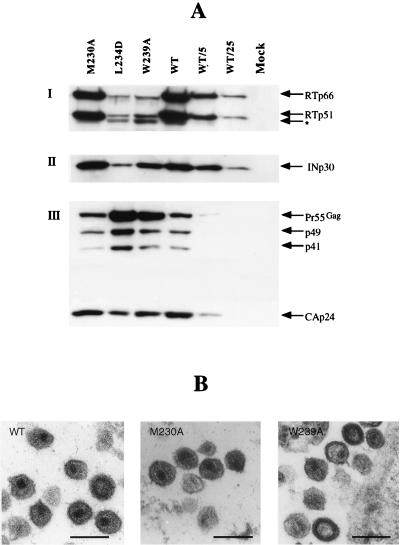
The protein content of viral particles was analyzed by Western blotting of virions harvested from the supernatant of transfected HeLa P4 cells (6) with antibodies directed against the capsid protein (CAp24), RT, or integrase (IN). Virus production between the mutants and the WT was consistently similar (data not shown). Figure 2A shows that the protein pattern for the mutant M230A was similar to that of the WT, since RT and IN were present in virions at WT levels, and the Gag precursor was processed normally; the ratio of Pr55gag to CAp24 for M230A was identical to that for the WT. It was unexpected that mutants L234D and W239A presented abnormalities, since, for example, RT and IN were decreased by approximately 20-fold in L234D particles and 10-fold in W239A particles compared to WT (Fig. 2A, I and II). The gag precursor Pr55gag was found to be processed, but less extensively than it was in WT virus, with a relatively high Pr55gag-to-CAp24 ratio (Fig. 2A, III), suggesting that mutations L234D and W239A affect the maturation process. An additional protein of approximately 45 kDa was detected for L234D and W239A with the anti-RT serum (Fig. 2A), which may have resulted from abnormal protease cleavage. Figure 2B shows a morphological comparison between WT and mutant viruses. WT particles were of a typical mature shape, with a well-defined conical electron-dense core. M230A mutant viruses exhibited a shape similar to that of WT particles, whereas mutants L234D (not shown) and W239A revealed a heterogeneous population of particles with immature or aberrant morphology. These observations are consistent with the Western blotting data, in that the aberrant morphology reflects abnormal viral protein content.
FIG. 2.
Structure of RT mutant virions. (A) Protein content of RT mutant virions. Western blotting of virions isolated from transfected HeLa P4 cell supernatants (6) was performed with anti-RT (I) or anti-INp30 (II) polyclonal antiserum or anti-CAp24 (III) monoclonal antibodies. Membrane was stripped (0.1 M glycine [pH 3.0] for 90 min and then 2% sodium dodecyl sulfate for 30 min) between immunodetections, and proteins were revealed by enhanced chemiluminescence (Amersham). WT/5 and WT/25, 1/5 and 1/25 of the normalized WT sample, respectively; ∗, processed form of RT observed for mutants L234D and W239A. Data are representive of three independent experiments. (B) Electron microscopy analysis. Transfected HeLa P4 cells were fixed and processed for 70-nm thin-section electron microscopy (Philips CM 120 transmission electron microscope [CMEABG]). Representative virions produced by HeLa P4 cells after transfection with WT, mutant M230A, or mutant W239A DNA are shown. Bars, 200 nm.
The data shown in Fig. 2 suggest that in mutant viruses L234D and W239A, the level of pol-encoded products is much inferior to that of the WT virus. This observation led us to suspect a defect in incorporation of the Pr160gag-pol precursor into virions and/or a lack of the precursor in the cytoplasm of transfected cells. We thus immunoprecipitated viral proteins from transfected and radiolabeled HeLa P4 cells and from viral particles in the supernatant. In Fig. 3A, virion proteins were revealed by a mixture of anti-RT polyclonal and anti-CAp24 monoclonal antibodies. In confirmation of data in Fig. 2A, mutant M230A showed a Pr55gag-to-CAp24 ratio similar to that of the WT, whereas mutants L234D and W239A showed a higher Pr55gag-to-CAp24 ratio, indicating an incomplete maturation process. Figure 3B shows cytoplasmic viral proteins immunoprecipitated by using the anti-RT polyclonal antiserum alone (lanes 1 to 4) or a mixture of two anti-CAp24 monoclonal antibodies (lanes 5 to 8). While the levels of Pr55gag were similar for all samples, in cells transfected with the L234D plasmid the amount of Pr160gag-pol was about 20% of that of the WT (lanes 3 and 7). For the W239A mutant, the level of Pr160gag-pol was also inferior to the WT level (about 50% [data not shown]). Importantly, an additional fragment of approximately 120 kDa (also present for the WT, but only faintly) was found in significant amounts in samples L234D (Fig. 3B, lane 3) and W239A (not shown), possibly indicating an early cleavage between CA and NC. To determine if this cleavage was due to viral PR activity, a specific HIV protease inhibitor, Palinavir (22), was used during radiolabeling of transfected cells. As shown in Fig. 3C, the 120-kDa fragment disappeared from cells transfected with L234D plasmid DNA upon the addition of Palinavir. These data suggest that for mutants L234D and W239A, Pr160gag-pol precursor is cleaved prematurely within the cell, and this may at least in part explain the low level of RT and IN enzymes in mutant viral particles observed in Fig. 2A. Assuming that there is a similar decrease of viral protease levels in the mutant virions, this might explain the relatively immature morphology observed for mutants W239A (Fig. 2B) and L234D (not shown).
FIG. 3.
Analysis of viral proteins in cells and virions by radiolabeling and immunoprecipitation. (A) Virion proteins (from cell supernatant) were immunoprecipitated with a mixture of one anti-RT polyclonal serum and two anti-CAp24 monoclonal antibodies. Lanes: 1, mock-transfected cells; 2, WT pNL4 virion proteins; 3, 4, and 5, RT mutant virion proteins with the indicated mutations. The positions of RTp66, CAp24, and the gag precursor (Pr55gag) are indicated. (B) Viral proteins in cells. Lanes 1 to 4 show proteins immunoprecipitated with the anti-RT polyclonal serum. Lane 1, mock-transfected cells; lanes 2 and 3, transfection with plasmids M230A and L234D, respectively; lane 4, WT pNL4-3 transfection. Lanes 5 to 8 correspond to proteins immunoprecipitated with a mixture of two anti-CAp24 monoclonal antibodies. Lane 5, mock-transfected cells; lanes 6 and 7, transfection with plasmids M230A and L234D, respectively; lane 8, WT pNL4-3 transfection. (C) Viral proteins in cells were immunoprecipitated with a mixture of the anti-RT polyclonal serum and two anti-CAp24 monoclonal antibodies following treatment with Palinavir (22), an HIV PR inhibitor, used at 10 μM during radiolabeling of transfected cells to block PR activity. Lanes: 1, mock-transfected cells; 2 and 3, transfection with WT and mutant L234D plasmids, respectively. Data are representive of three independent experiments.
HIV-1 PR is activated upon dimerization of the PR subunits on separate gag-pol precursors, followed by autocatalytic cleavage and release of a free, functional dimer (18–20). Dimerization of PR as part of the Pr160gag-pol precursor may require the cooperation of several additional dimerization sites within the polyprotein. Previous reports show that deletions in RT or IN domains or truncation of C-terminal segments of Pr160gag-pol can directly affect gag-pol dimerization, leading to impaired virus maturation (4, 10, 32). The premature processing of Pr160gag-pol in the case of L234D and W239A mutants is prevented in the presence of the specific viral protease inhibitor Palinavir (Fig. 3) and is thus not likely due to cellular protease activity. The p2/NC site, which is located between CAp24 and NCp7 in Pr160gag-pol, is the first cleavage site during gag-pol processing (30). We thus assume that the 120-kDa protein seen with mutants L234D and W239A (Fig. 3B and C) lacks MA and CA domains, in support of which we do not detect it using anti-CAp24 antibodies (Fig. 3B). Since the CA domain in Pr160gag-pol is required for its incorporation into viral particles (34, 35), the prematurely processed 120-kDa form would be left behind in the cell, resulting in the production of virions with low levels of the viral enzymes.
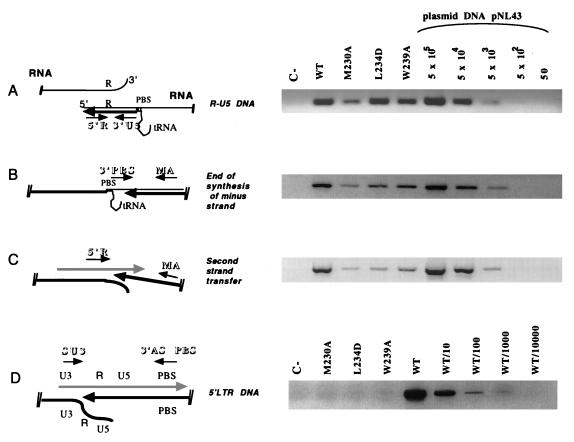
To investigate the defect of the RT mutants at the level of reverse transcription, we used a PCR-based method to analyze the level of proviral DNA synthesis in newly infected cells (2, 36). After confirming that the levels of virus production were similar for mutants and the WT, the same volumes of WT and mutant viral supernatants were used to infect HeLa P4 cells. Cells were incubated with viral supernatant for 18 h, and then a Hirt supernatant was prepared (12). DNA from 105 cells infected with 1 ml of viral supernatant was used for PCR analyses. In order to monitor early and late viral DNA synthesis, four PCR primer pairs were used (Fig. 4): 5′R/3′U5 for R-U5 DNA corresponding to the earliest stage of reverse transcription (strong-stop cDNA) (Fig. 4A), 3′PBS/MA for extended minus-strand DNA (Fig. 4B), 5′R/MA for plus-strand DNA synthesized just after the second strand transfer (Fig. 4C), and SU3/ASPBS SU3 for synthesis of the 5′ LTR DNA (Fig. 4D). The primer pair 5′check/3′check was used to ensure that pNL4-3 plasmid DNA was absent from the samples (not shown). R-U5 DNA synthesis was similar to that for the WT for the mutants L234D and W239A. In contrast, for the mutant M230A, R-U5 DNA synthesis was decreased to 20% of WT levels (Fig. 4A). Very pronounced decreases in minus-strand DNA and plus-strand DNA synthesis were observed for all three mutants (Fig. 4B and C), implying that mutations in the primer grip domain of RT impair the reverse transcription process. No amplification of 5′ long terminal repeat (LTR) was detected for any of the three mutants suggesting that the end of proviral DNA synthesis was very inefficient upon mutating the primer grip of RT.
FIG. 4.
Early and late phases of proviral DNA synthesis analyzed by PCR. HeLa P4 cells were infected for 18 h with WT or mutant viral supernatant. Extrachromosomal DNA was prepared by the method described by Hirt (12), and a fraction (corresponding to 105 cells infected with 1 ml of viral supernatant) was subjected to each PCR amplification with alternative primer pairs for determination of early and late steps of reverse transcription. (The localization of each primer pair relative to the viral genome is schematically represented on the left of each panel.) The primer pairs used were 5′R/3′U5 (5′R, 5′-GGTCTCTCTGGTTAGACCA-3′; 3′U5, 5′-CTGCTAGAGATTTTCCACAC-3′) for R-U5 DNA corresponding to the earliest stage of reverse transcription (A); 3′PBS/MA (3′PBS, 5′-ACTTGAAAGCGAAAGTAAAGC-3′; and MA, 5′-GGTCTCTCTGGTTAGACCA-3′) for extended minus-strand DNA (B); 5′R/MA (5′R, 5′-GGTCTCTCTGGTTAGACCA-3′; MA, 5′-GGTCTCTCTGGTTAGACCA-3′) for plus-strand DNA synthesized just after second-strand transfer (C); and SU3/ASPBS (SU3, 5′-GCACCATCCAAAGGTCAGTGG-3′; ASPBS, 5′-CTCCTCTGGCTTTACTTTCGC-3′) to detect synthesis of the 5′ LTR DNA. The absence of contaminating pNL4-3 plasmid in the samples was confirmed by using primers localized within the flanking (5′check) and the pUC (3′check) sequences of the plasmid (not shown). PCR products were resolved on a 1.5% agarose gel and were visualized by ethidium bromide staining. For PCR amplifications A, B, and C, serial dilutions of the plasmid pNL4-3 (corresponding to 50 to 5 × 105 copies) were used as a positive control and a basis for quantification. Since pNL4-3 contains a hybrid provirus with sequence mismatches between the 5′ LTR and the 3′ LTR, the primer pair 5′SU3/3′ASPBS can amplify only synthesized viral DNA and not the plasmid. Consequently, we used dilutions of the WT sample as the control set for the 5′ LTR proviral DNA amplification (D). In the schematic diagrams, RNA is represented by thin lines, minus-strand DNA is indicated by thick black lines, and plus-strand DNA is indicated by hatched thick lines. Arrows indicate the directions of synthesis, while small arrows represent primers.
Recently, Wohrl et al. (39) have reported that RT containing the M230A mutation has only a low affinity for dTTP in vitro (72-fold decrease compared to WT). This observation is consistent with the strongly attenuated phenotype of the M230A mutant described herein. Our data show that reverse transcription was severely impaired in cells infected by the M230A virus (Fig. 4), in spite of the presence of WT levels of RT in the virions (Fig. 2). The late stages of reverse transcription were very inefficient and the 5′ end of proviral DNA was not detectable. Thus, RT containing the M230A mutation is poorly functional during reverse transcription in vivo, yet virion morphology and gag processing are little or not affected (Fig. 2). The late phases of reverse transcription in cells infected with mutants L234D and W239A were also severely impaired, and the 5′ end of proviral DNA again was not detected (Fig. 4). However, the level of synthesis of strong-stop cDNA remained high in mutants L234D and W239A (Fig. 4A), considering that virion RT content was only 1/5 to 1/25 of the WT level (Fig. 2A).
Together, these data suggest that the RT primer grip domain delays dimerization of gag-pol precursors to prevent early activation of the protease and gag-pol maturation prior to virus assembly. This emphasizes the central importance of Pr160gag-pol precursor stability during virus assembly. In addition, the results in Fig. 2 and 4 indicate that active RT is probably in large excess in HIV-1 virions.
Acknowledgments
This work was supported by ANRS, SIDACTION and MGEN. S Le G was supported by PHS grant GM 52263. We thank M. Rau for critical reading of the manuscript.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix J L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukovsky A, Gottlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng N, Painter G R, Furman P A. Crosslinking of substrates occurs exclusively to the p66 subunit of heterodimeric HIV-1 reverse transcriptase. Biochem Biophys Res Commun. 1991;174:785–789. doi: 10.1016/0006-291x(91)91486-v. [DOI] [PubMed] [Google Scholar]
- 6.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh M, Jacques P S, Rodgers D W, Ottman M, Darlix J L, Le Grice S F. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 10.Goff S P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquired Immune Defic Syndr. 1990;3:817–831. [PubMed] [Google Scholar]
- 11.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 14.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 15.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacques P S, Wohrl B M, Ottmann M, Darlix J L, Le Grice S F. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J Biol Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 17.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krausslich H G. Genetic analysis and gene expression of human immunodeficiency virus. Curr Opin Genet Dev. 1992;2:82–89. doi: 10.1016/s0959-437x(05)80327-2. [DOI] [PubMed] [Google Scholar]
- 19.Krausslich H G. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc Natl Acad Sci USA. 1991;88:3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krausslich H G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamarre D, Croteau G, Wardrop E, Bourgon L, Thibeault D, Clouette C, Vaillancourt M, Cohen E, Pargellis C, Yoakim C, Anderson P C. Antiviral properties of palinavir, a potent inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1997;41:965–971. doi: 10.1128/aac.41.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb D D, Hutchison C A D, Edgell M H, Farmerie W G, Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J Virol. 1989;63:111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H R, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottmann M, Gabus C, Darlix J L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palaniappan C, Wisniewski M, Jacques P S, Le Grice S F, Fay P J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 28.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J, Morrow C D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell M D, Ghosh M, Jacques P S, Howard K J, Le Grice S F, Levin J G. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 32.Quillent C, Borman A M, Paulous S, Dauguet C, Clavel F. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology. 1996;219:29–36. doi: 10.1006/viro.1996.0219. [DOI] [PubMed] [Google Scholar]
- 33.Ross E K, Fuerst T R, Orenstein J M, O’Neill T, Martin M A, Venkatesan S. Maturation of human immunodeficiency virus particles assembled from the gag precursor protein requires in situ processing by gag-pol protease. AIDS Res Hum Retroviruses. 1991;7:475–483. doi: 10.1089/aid.1991.7.475. [DOI] [PubMed] [Google Scholar]
- 34.Smith A J, Srinivasakumar N, Hammarskjold M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasakumar N, Hammarskjold M L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. . (Editorial.) [DOI] [PubMed] [Google Scholar]
- 38.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 39.Wohrl B M, Krebs R, Thrall S H, Le Grice S F J, Scheidig A J, Goody R S. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J Biol Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 40.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]