Abstract
Childhood environments are critical in shaping cognitive neurodevelopment. With the increasing availability of large-scale neuroimaging datasets with deep phenotyping of childhood environments, we can now build upon prior studies that have considered relationships between one or a handful of environmental and neuroimaging features at a time. Here, we characterize the combined effects of hundreds of inter-connected and co-occurring features of a child’s environment (“exposome”) and investigate associations with each child’s unique, multidimensional pattern of functional brain network organization (“functional topography”) and cognition. We apply data-driven computational models to measure the exposome and define personalized functional brain networks in pre-registered analyses. Across matched discovery (n=5139, 48.5% female) and replication (n=5137, 47.1% female) samples from the Adolescent Brain Cognitive Development study, the exposome was associated with current (ages 9–10) and future (ages 11–12) cognition. Changes in the exposome were also associated with changes in cognition after accounting for baseline scores. Cross-validated ridge regressions revealed that the exposome is reflected in functional topography and can predict performance across cognitive domains. Importantly, a single measure capturing a child’s exposome could more accurately and parsimoniously predict cognition than a wealth of personalized neuroimaging data, highlighting the importance of children’s complex, multidimensional environments in cognitive neurodevelopment.
Keywords: Cognition, Functional networks, Development, Environment, Exposome, Topography
Highlights
-
•
Complex childhood environments captured by the “exposome” are related to cognition.
-
•
Changes in the exposome are associated with changes in cognition over two years.
-
•
The exposome is reflected in personalized functional brain network organization.
-
•
Cross-validated ridge regressions trained on the exposome predict future cognition.
1. Introduction
Our minds and brains are highly unique, shaped not just by our genetics (“genome”) but also by our complex network of environmental exposures (“exposome”). In line with this idea is the observation that individual differences in cognition may increase throughout development (Kidd et al., 2018) as environmental exposures and individual experiences continually mold the brain’s functional organization. It is imperative that we characterize how individual differences in cognition emerge during childhood, not only as a window into understanding what makes us unique individuals, but also because individual differences in cognition are associated with socio-economic (Moffitt et al., 2011), physical health (Calvin et al., 2011), and mental health (Shanmugan et al., 2016) outcomes across the lifespan. Understanding cognitive neurodevelopment at the level of the individual requires a characterization of how the unique features of each child’s environment may be reflected in the unique features of each child’s brain.
Many aspects of a child’s environment have been linked with cognitive functioning. The Adolescent Brain Cognitive Development (ABCD) Study (Volkow et al., 2018) (n=11,878) is a large-scale longitudinal study of development with deep phenotyping of multidimensional environmental features and cognition in children from twenty-one sites across the United States. In the ABCD Study, environmental features that are associated with cognition include family dynamics (Thompson et al., 2022) experiences at school (Meredith et al., 2022), socio-economic status (Botdorf et al., 2022), physical activity (Walsh et al., 2018), and stress (Demidenko et al., 2021) among others. Despite the long-recognized importance of the environment in shaping cognitive development, only recently have we been able to leverage data-driven approaches in deeply-phenotyped datasets of this size to capture the multitude of inter-connected features that make up an individual’s environment and experience.
The concept of the “exposome” has been introduced as a way of capturing the totality of co-occurring environmental exposures and experiences (Rappaport, 2011), in contrast to examining single environmental features in isolation. While the first exposome studies primarily focused on associations with physical health (e.g., cancer risk) in adults, more recent work has increasingly focused on mental health outcomes (Guloksuz et al., 2018) including recent studies in children. Factor analytic approaches have made it possible to quantify both a general exposome score as well as sub-factors capturing specific aspects of perinatal, familial, social, and physical environments (Moore et al., 2022), all of which are associated with mental functioning. This approach has revealed that a child’s exposome is associated with psychopathology (Moore et al., 2022, Barzilay et al., 2022, Pries et al., 2022).
Large-scale deeply-phenotyped neuroimaging datasets have also enabled the definition of robust multidimensional features of an individual’s functional brain organization. Studies have highlighted inter-individual heterogeneity in the size, shape and spatial arrangement of functional brain regions across cortex (Glasser et al., 2016, Gordon et al., 2017, Kong et al., 2019, Laumann et al., 2015). Despite this heterogeneity, most human neuroimaging studies still rely on standardized network atlases (Power et al., 2011, Yeo et al., 2011) that are spatially warped to individual brains, smearing away the rich complexity of individual differences. Cognitive functions in particular are supported by spatially-distributed, large-scale networks that tend to have the highest inter-individual heterogeneity in both adults (Gordon et al., 2017) and youth (Cui et al., 2020) compared to other large-scale networks, exacerbating this problem for studies of cognitive development. To overcome this challenge, precision functional mapping techniques have been developed to derive individually-defined networks and map unique patterns of functional topography – the spatial layout of functional brain networks across the cortex. Such personalized functional networks (PFNs) are stable within individuals (Gordon et al., 2017, Keller et al., 2023a) and have been successfully derived from both resting-state and task-based neuroimaging data (Gordon et al., 2017, Cui et al., 2020, Keller et al., 2023a).
Here, we investigate associations among children’s complex, multidimensional environments, unique patterns of functional brain network organization, and cognitive development. To characterize reproducible cross-sectional and longitudinal environment-brain-behavior associations, we leverage ABCD Study® data to conduct our pre-registered analyses (Keller and Barzilay, 2023). We use three previously validated data-driven approaches to reduce dimensionality across rich multivariate data types: 1) we define both general and specific exposome factors using longitudinal exploratory bifactor analysis (Moore et al., 2022); 2) we define personalized functional networks using non-negative matrix factorization (Cui et al., 2020, Keller et al., 2023a, Li et al., 2017); and 3) we use cognitive factors defined by principal components analysis in a previous study (Thompson et al., 2019). Using linear mixed effects models and cross-validated ridge regressions, we relate individual differences in the exposome to unique patterns of PFN topography and cognition across domains. Given the importance of large samples to identify reproducible brain-behavior associations (Marek, Tervo-Clemmens et al., 2022), we replicate our analyses across matched discovery (n=5139, 48.5% female) and replication (n=5137, 47.1% female) samples (Cordova et al., 2021, Feczko et al., 2021). Our findings highlight the importance of capturing multidimensional childhood environments to better understand functional brain network organization and cognition in children on the precipice of the transition into adolescence.
2. Materials and methods
2.1. Participants
Data were drawn from the Adolescent Brain Cognitive Development℠ (ABCD) study (Volkow et al., 2018) baseline sample from the ABCD BIDS Community Collection (ABCC, ABCD-3165) (Feczko et al., 2021), which included n=11,878 children aged 9–10 years old and their parents/guardians collected across 21 sites. Parents and guardians provided written informed consent as part of the ABCD study. Institutional Review Board (IRB) approval was received from the University of California, San Diego and the respective IRBs of each site. Inclusion criteria included being within the desired age range (9–10 years old), English language proficiency in the children, and having the ability to provide informed consent (parent) and assent (child). Exclusion criteria included the presence of severe sensory, intellectual, medical or neurological issues that would have impacted the child’s ability to comply with the study protocol and MRI scanner contraindications. We additionally excluded participants with incomplete data or excessive head motion during scanning. To test the generalizability of our results, we repeated our analyses in discovery (n=5139) and replication samples (n=5137) that were matched across socio-demographic variables (Cordova et al., 2021, Feczko et al., 2021) (Table S1).
2.2. Cognitive assessment
Participants completed a battery of cognitive assessments, including seven tasks from the NIH Toolbox (Picture Vocabulary, Flanker Test, List Sort Working Memory Task, Dimensional Change Card Sort Task, Pattern Comparison Processing Speed Task, Picture Sequence Memory Task, and the Oral Reading Test) as well as two additional tasks (the Little Man Task and the Rey Auditory Verbal Learning Task) (Luciana et al., 2018). To reduce dimensionality, we used scores in three cognitive domains (general cognition, executive function and learning/memory) derived from a prior study (Thompson et al., 2019), downloaded from the ABCD Data Exploration and Analysis Portal (DEAP). Given that not all cognitive tasks were administered across all timepoints, our longitudinal analyses used scores on five cognitive tasks that were assessed at both baseline and two-year timepoints (Picture Vocabulary, Flanker, Picture Sequence Memory, Pattern Comparison and Reading Recognition).
2.3. Definition of exposome factors
We defined both general and specific exposome factors capturing a child’s unique, complex, multidimensional experiences and environment by applying the same approach as in our previous cross-sectional work (Moore et al., 2022) to data collected across multiple longitudinal timepoints. Table S3 shows the results of the exploratory structural equation model (ESEM) with one general factor and six orthogonal sub-factors derived from 354 variables capturing various aspects of a child’s environment. These variables were in multiple formats (continuous, ordinal, and nominal), different lengths (scales used in the ABCD Study® ranged from 2 to 59 items in length), and multiple sources (youth-report, parent-report, geo-coded data, etc.). The first step was to determine whether each scale should be reduced to data-driven summary scores rather than using individual items. This was determined largely by the relative lengths of the scales, where the goal was to avoid having longer scales (those with more items) or variable sets (e.g. 91 geographic variables) “dominate” the exposome factors in the final analysis (see below). Scales were also reduced to a summary score if the scree plot from the inter-item correlations clearly indicated a single factor with an obvious ‘elbow’ after the first eigenvalue; for example, the three-item neighborhood safety scale was reduced to a single score for this reason.
Ultimately, twelve scales were reduced in this data-driven manner: youth-report School Risk and Protective Factors Survey (four sub-scores), youth-report Family Environment Scale (two sub-scores), parent-report Family Environment Scale (two sub-scores), youth-report Parent Monitoring Survey (one sub-score), parent-report Neighborhood Safety/Crime Survey (one sub-score), parent-report Community Risk and Protective Factors (one sub-score), parent-report Mexican-American Cultural Values Scale (four sub-scores), parent-report Parental Rules on Substance Use (one sub-score), parent-report Sports and Activities Involvement Questionnaire (three sub-scores), Traumatic Brain Injury sum scores (one sub-score), youth-report Youth Substance Use Attitudes Questionnaire (one sub-score), and youth-reported hours of screen time on various devices (one sub-score). Additionally, the address-/geocode-based measures of the neighborhood and state environment were reduced to eight sub-scores. The above “pre-reduction” steps were conducted using exploratory factor analysis (EFA), where the number of factors to extract was determined by a combination of interpretability and subjective evaluation of the scree plot. Table S2 shows the sub-scales resulting from the above analyses, along with the items composing them. Note that there are 30 sub-scales, while only 29 were used; sub-scale “dry_heat” was dropped from analyses because of difficulty in interpretation and unbalanced representation of humidity and temperature indicators. Temperature was nonetheless accounted for by the “traditional_south_and_midwest” sub-score, which included a count of the number of “extreme heat days” experienced in a given year. Analyses in this “pre-reduction” step were conducted using the psych package (Revelle, 2019) in R. Note that, in addition to the 29 sub-scales created in this step, the final analysis (below) included parent education, household income, and a binary variable indicating whether the child’s parents were married, for a total of 32 variables.
For the final analysis of 32 variables we used an exploratory structural equation model (ESEM) (Asparouhov and Muthén, 2009) using bifactor rotation (BI-CF-QUARTIMAX) accounting for clustering by families, stratified by site, and constraining factor loadings to be equal across time points (ensuring longitudinal measurement invariance). Note that ‘stratification’ in this sense is a technical term ensuring Mplus considers site stratification when estimating standard errors; it does not affect model parameter estimates. Critically, the model was constrained to have the same configuration, loadings, and intercepts (with unconstrained factor means) across time points. If this constraint were unrealistic–i.e. if the exposome models differed across time points–this would be reflected in the model fit indices, providing an embedded check on the assumption of measurement invariance across time (age). The number of factors to extract was determined by a combination of interpretability and model fit, where fit was assessed using the comparative fit index (CFI; >0.90 acceptable), root mean-square error of approximation (RSMEA; <0.08 acceptable), and standardized root mean-square residual (SRMR; <0.08 acceptable) (Hu and Bentler, 1999). Analyses were conducted using Mplus version 8.4 (Muthén and Muthén, 2017). Table S3 shows the results of the ESEM with one general factor and six orthogonal sub-factors using 32 exposome variables. Fit of the model is acceptable, with CFI = 0.94, RMSEA = 0.026 ± 0.001, and SRMR = 0.032. The general factor reflects the general exposome (somewhat analogous to a first principal component), with the strongest indicators relating to socioeconomic status (household income = 0.780; neighborhood poverty = −0.695; parent education = 0.680) and weakest indicator related to neighborhood characteristics associated with retirement (-0.009).
In addition to a general exposome factor score for each participant, the bifactor model provided six sub-factors capturing specific dimensions of a child’s environment: School, Family Values, Family Turmoil, Dense Urban Poverty, Extracurriculars and Screen Time. These sub-factors are necessarily orthogonal to one another as well as orthogonal to the general exposome factor (Figure S1). The first specific factor comprises school involvement, enjoyment, and performance, as well as some weaker influences of parental monitoring and youth-reported family turmoil. The second specific factor comprises all sub-scales of the Mexican American Cultural Values Scale (MACVS), which captures many aspects of family values, centrality, and culture. The third specific factor relates to family turmoil from the points-of-view of both the parents and youth, as well as a weak relation to substance abuse risk in that area. The fourth specific factor captures several aspects of the youth’s neighborhood, especially poverty, density, safety, and pollution. The fifth specific factor comprises extracurricular activities and traumatic brain injuries (TBIs) (possibly related, as TBIs and extracurricular activities are weakly correlated in the ABCD Study at baseline assessment (r(11,847)=0.073, p=2.54 ×10−15) and two-year follow-up (r(10,269)=0.040, p=6.56 ×10−5)). Finally, the sixth specific factor relates to screen time and (weakly) to peer deviance.
Beyond the model fit indices described above, bifactor models have specific reliability indices useful for evaluating the relative strengths of the factors, appropriateness and reliability of scores, etc (Rodriguez et al., 2016). These indices are shown in Table S4. Thorough discussion of these bifactor-specific metrics is beyond the present scope, but the most important for the present purposes is factor determinacy (Grice, 2001). Determinacy is an indication of how representative factor scores are of the factors from which they were derived. For example, note that the inter-factor correlations in Figure S1 are not exactly 0 despite the factors being modeled as orthogonal; this is due to slight indeterminacy of the factors, and is always seen when scores are calculated from factors. The key value for our present purposes is the Factor Determinacy for the general exposome factor, which is 0.89. This value is beyond the conventionally used minimum of 0.80 recommended for score determinacy, suggesting the general factor score used in the present study is sufficiently determined.
2.4. Quantification of longitudinal change in cognition and in exposome
We computed difference scores for both cognitive task performance and the general exposome factor score to quantify change from baseline assessment (9–10 years old) to two-year follow up (11–12 years old). Due to the phenomenon of regression to the mean (Galton, 1886), change scores will always be negatively correlated with their starting values. Therefore, when relating a change score to another variable of interest, it is unclear whether that relationship is driven by the change itself or simply the starting point of that change score. To account for this, it is common to include the starting value as a covariate in any analysis involving a change score (Moore et al., 2017); however, the correlation between the change score and the starting score is nuisance collinearity in the analysis. To account for this, the baseline scores can be regressed out of the change scores, where the residuals of that analysis represent the “true” change controlling for regression to the mean. Any relationship of that “true” change with another variable can be interpreted as purely involving the change (not the baseline). For each measure, we therefore first subtracted baseline scores from two-year follow up scores and then regressed out the baseline scores to compute residualized difference scores to account for regression to the mean. We then compared the residualized change in exposome score with the residualized change in cognitive performance using linear regressions with correction for multiple comparisons.
2.5. Image processing
Imaging acquisition for the ABCD Study® has been described elsewhere (Feczko et al., 2021). As previously described (Feczko et al., 2021), the ABCD-BIDS Community Collection (ABCC; Collection 3165) from which we drew our data was processed according to the ABCD-BIDS pipeline. This pipeline includes distortion correction and alignment, denoising with Advanced Normalization Tools (ANTS88), FreeSurfer89 segmentation, surface registration, and volume registration using FSL FLIRT rigid-body transformation. Processing was done according to the DCAN BOLD Processing (DBP) pipeline which included the following steps: 1) de-meaning and de-trending of all fMRI data with respect to time; 2) denoising using a general linear model with regressors for signal and movement; 3) bandpass filtering between 0.008 and 0.09 Hz using a 2nd order Butterworth filter; 4) applying the DBP respiratory motion filter (18.582–25.726 breaths per minute), and 5) applying DBP motion censoring (frames exceeding an FD threshold of 0.2 mm or failing to pass outlier detection at +/- 3 standard deviations were discarded). Motion censoring was applied for all functional runs, including both rest and task. Following preprocessing, we concatenated the time series data for both resting-state scans and three task-based scans (Monetary Incentive Delay Task, Stop-Signal Task, and Emotional N-Back Task) as in prior work (Cui et al., 2020, Keller et al., 2023a) to maximize the available data for our analyses. After removing participants who failed to pass ABCD quality control for their T1 or resting-state fMRI scan, we additionally excluded participants with fewer than 600 TRs remaining after motion censoring for the concatenated time series, accounting for both resting-state and task-based fMRI data quality. Head motion (mean fractional displacement) is also included as a covariate in all analyses.
2.6. Regularized non-negative matrix factorization
As previously described (Cui et al., 2020, Keller et al., 2023a, Li et al., 2017), we used non-negative matrix factorization (NMF) to derive individualized functional networks. NMF identifies networks by positively weighting connectivity patterns that covary, leading to a highly specific and reproducible parts-based representation. As NMF requires nonnegative input, we re-scaled the data by shifting time courses of each vertex linearly to ensure all values were positive.25 As in prior work, to avoid features in greater numeric ranges dominating those in smaller numeric range, we further normalized the time course by its maximum value so that all the time points have values in the range of [0,1]. For this study, we used identical parameter settings as in prior validation studies (Cui et al., 2020, Keller et al., 2023a, Li et al., 2017).
To facilitate group-level interpretations of individually-defined PFNs, we used a group consensus atlas from a previously published study in an independent dataset (Cui et al., 2020) as an initialization for individualized network definition. In this way, we also ensured spatial correspondence across all subjects. Details regarding the derivation of this group consensus atlas can be found in previous work (Cui et al., 2020, Keller et al., 2023a). Briefly, group-level decomposition was performed multiple times on a subset of randomly selected subjects and the resulting decomposition results were fused to obtain one robust initialization that is highly reproducible. Next, inter-network similarity was calculated and normalized-cuts (Cai et al., 2011) based spectral clustering method was applied to group the PFNs into 17 clusters. For each cluster, the PFN with the highest overall similarity with all other PFNs within the same cluster was selected as the most representative. The resulting group-level network loading matrix V was transformed from fsaverage5 space to fslr space using Connectome Workbench (Marcus et al., 2011), and thus the resultant matrix had 17 rows and 59,412 columns. Each row of this matrix represents a functional network, while each column represents the loadings of a given cortical vertex.
Using the previously-derived group consensus atlas (Cui et al., 2020) as a prior to ensure inter-individual correspondence and a data locality regularization term to improve robustness to imaging noise, we derived each individual’s specific network atlas using NMF based on the acquired group networks (17 ×59,412 loading matrix) as initialization and each individual’s specific fMRI times series. See (Li et al., 2017) for optimization details. This procedure yielded a loading matrix V (17 ×59,412 matrix) for each participant, where each row is a PFN, each column is a vertex, and the value quantifies the extent each vertex belongs to each network. In other words, each of the 17 loading values at a given vertex represents the extent to which that vertex exhibits group membership to each of the networks. This probabilistic (soft) definition can be converted into discrete (hard) network definitions for display purposes by labeling each vertex according to its highest loading. Split-half reliability of PFN loadings was found to be high in prior work (Keller et al., 2023a), indicating excellent reliability.
2.7. Linear mixed-effects models
We used linear mixed effects models (“lme4” package in R) to assess associations between exposome factors and cognitive performance while accounting for both fixed and random predictors. All models included fixed effects parameters for age and biological sex and random effects for family (accounting for siblings) and site. Sensitivity analyses tested whether results held with the inclusion of measures of socio-economic status (household income and parental education), psychiatric medication use (ADHD medications, Antidepressants, or Antipsychotics) assessed using the PhenX instrument and coded as in our previous work (Shoval et al., 2021), or across stratifications by biological sex and parent-reported racial identification.
2.8. Ridge regression models
To uncover associations between the full multivariate pattern of PFN functional topography and each participant’s general exposome factor score, we trained linear ridge regression models using nested two-fold cross validation as in our previous work (Cui et al., 2020, Keller et al., 2023a). These models were trained on concatenated network loading matrices across all PFNs. Independent network models were also trained on loadings from specific networks. All models included covariates for age, sex, site, and motion (mean FD) that were all regressed out separately in the training and testing sets prior to training.
Ridge regression models were trained and tested in our matched discovery and replication samples (Cordova et al., 2021, Feczko et al., 2021) using nested two-fold cross-validation (2 F-CV), with outer 2 F-CV estimating the generalizability of the model and the inner 2 F-CV determining the optimal tuning parameter (λ) for the ridge regression. For the inner 2 F-CV, one subset was selected to train the model under a given λ value in the range [1, 10, 100, 500, 1000, 5000, 10,000, 15,000, 20,000], and the remaining subset was used to test the model. This procedure was repeated 2 times such that each subset was used once as the testing dataset, resulting in two inner 2F-CV loops in total. For each λ value, the correlation r between the observed and predicted outcome as well as the mean absolute error (MAE) were calculated for each inner 2F-CV loop, and then averaged across the two inner loops. The sum of the mean correlation r and reciprocal of the mean MAE was defined as the inner prediction accuracy, and the λ with the highest inner prediction accuracy was chosen as the optimal λ (Cui et al., 2020).
To ensure that our matched discovery and replication sample selection procedure did not bias our results, we performed repeated random cross-validation over 100 iterations, each time randomly splitting the sample and repeating the nested 2F-CV procedure to generate a distribution of prediction accuracies for each model. Furthermore, we used permutation testing to generate null distributions for both the primary models and the repeated random cross-validation models by randomly shuffling the outcome variable. To ensure that our results were not overfit as a result of leakage across samples by the general exposome factor outcome variables derived in the whole sample, we also trained ridge regression models with the general exposome factor derived by two independent longitudinal bifactor analyses in the discovery and replication samples separately. Repeating our main analyses with these new predictive models, we find nearly identical results as shown in Figure S2.
To investigate associations among the exposome, PFN topography, and cognition, we trained three additional ridge regression models using the same procedure as above: Model 1 (“Exp-Factor”) used only a participant’s general exposome score; Model 2 (“PFN Topography”), reported in our prior work (Keller et al., 2023a), used each participant’s multivariate pattern of personalized functional network topography; and Model 3 (“Exp-Factor + PFN Topography”) used a combination of the features in the first and second model types, hypothesized to perform best by capitalizing on both shared and unique variance across features. Note that the model training procedure remained the same even for the single-variable model to allow the slope of the association between the general exposome factor and cognition to vary with the value of the ridge constraint during nested cross-validation. We also trained models to predict cognitive performance two years later (11–12 years old), covarying for baseline cognitive performance. Models were compared by assessing the Akaike Information Criterion (AIC) (Akaike, 1998) and Bayesian Information Criterion (BIC) (Schwarz, 1978), which (unlike measures like R2) consider the number of features the model is trained on, penalizing more complex models.
3. Results
3.1. A general measure of the exposome is associated with individual differences in cognition
We first characterized associations between a child’s complex, multidimensional environment (“exposome”) and their cognitive abilities. To do so, we derived a measurement of each child’s exposome using multilevel (clustered) exploratory factor analysis with a bifactor rotation (Jennrich and Bentler, 2011). To investigate both cross-sectional and longitudinal associations, we derived exposome scores using data from multiple timepoints, accounting for clustering by family, stratification by site, and ensuring measurement invariance across time. We used a bifactor approach (Reise et al., 2010) because, based on prior work (Moore et al., 2022), we anticipated that a general exposome factor would capture variance across dimensions of a child’s complex environment (e.g., neighborhood, family, school, etc.; Fig. 1a). As depicted in Table S3, many of the variables loading most strongly on the general exposome factor were those capturing dimensions of socio-economic status (SES; e.g., household income, parental education and marital status, neighborhood poverty, children’s involvement in extracurricular sports/activities, neighborhood safety, crowding and crime), with positive associations between general exposome scores and SES measures (Figure S3).
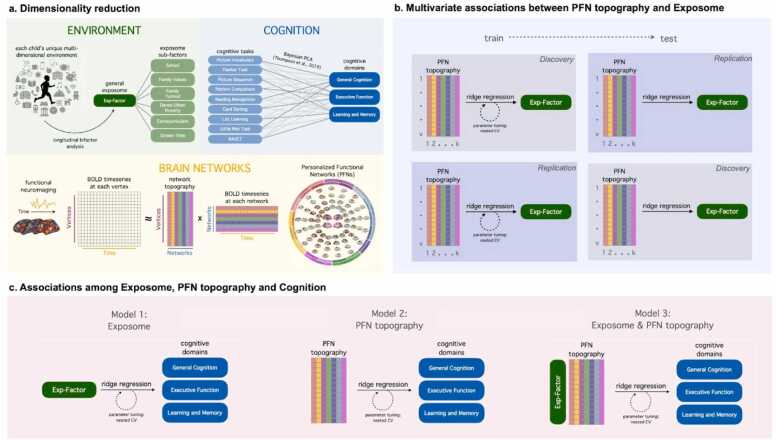
Fig. 1.
Summary of Methodological Approach. (A) We aimed to define broad factors describing a child’s environment, cognition, and brain network organization by reducing high-dimensional data with three unsupervised machine-learning methods: longitudinal bifactor analysis, principal components analysis, and non-negative matrix factorization. (B) To investigate whether children’s general exposome is encoded in their multivariate patterns of PFN topography, we trained ridge regression models using two-fold cross-validation across our matched discovery and replication samples. To ensure that model performance was not influenced by the choice of split, we also performed repeated random cross-validation using multiple random splits. (C) To investigate associations among the exposome, PFN topography, and cognition, we trained three models: Model 1 (“Exposome”) used only a participant’s general exposome score; Model 2 (“PFN Topography”) used each participant’s multivariate pattern of PFN topography; and Model 3 (“Exposome + PFN Topography”) used both exposome scores and PFN topography.
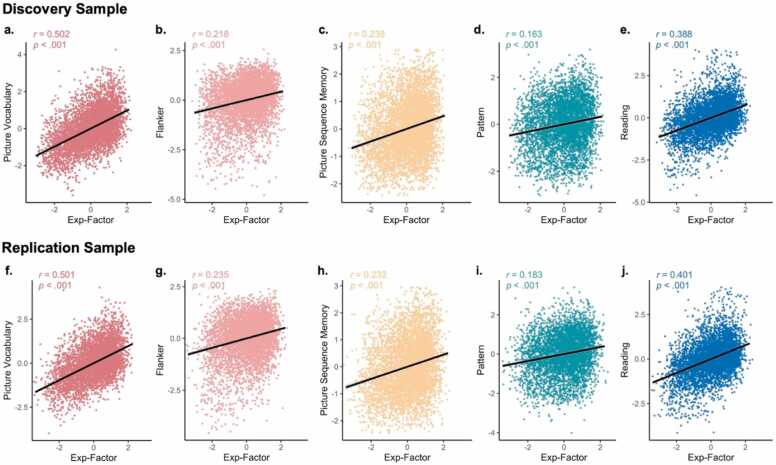
To investigate associations between exposome factor scores and cognition at baseline (9–10 years old) and two-year follow-up (11–12 years old), we estimated cross-sectional and longitudinal linear mixed effects models. All models accounted for family structure and ABCD Study site as random effects as well as age and biological sex as fixed effects. Given that the general exposome factor and six exposome sub-factors are necessarily orthogonal (Figure S1), we included all seven factors together. Across demographically-matched discovery (n=5139, 48.5% female) and replication (n=5137, 47.1% female) samples, the general exposome factor was significantly associated with cognition on all five cognitive tasks at baseline (Fig. 2) and survived Bonferroni correction for multiple comparisons (discovery: ꞵs=0.12–0.50, all pbonf<.001; replication: ꞵs=0.15–0.48, all pbonf<.001; Table 1). While the majority of exposome sub-factor scores were not consistently associated with cognition, we did find a few specific associations that were significant across both samples: between Family Values and Picture Vocabulary scores (discovery: ꞵ=-0.10, pbonf=1.91 ×10−11; replication: ꞵ=-0.10, pbonf=2.26 ×10−11), between Screen Time and Picture Sequence Memory (discovery: ꞵ=-0.12, pbonf=4.26 ×10−7; replication: ꞵ=-0.08, pbonf=0.001) and between Screen Time and Reading Recognition (discovery: ꞵ=-0.08, pbonf=0.001; replication: ꞵ=-0.06, pbonf=0.013). Notably, the associations between the general exposome factor and all domains of cognition also remained significant in longitudinal models predicting cognition two years later while accounting for baseline cognition, which is known to be a strong predictor of future cognitive performance, across both the discovery (n=2763, 47.9% female) and replication (n=2739, 47.1% female) samples (discovery: ꞵs=0.08–0.24, all pbonf<.001; replication: ꞵs=0.11–0.22, all pbonf<.001; Table S5).
Fig. 2.
A general measure of exposome is significantly associated with cognition across cognitive tasks. We characterized a child’s unique, complex and multidimensional exposome using a general exposome factor (“Exp-Factor”) derived from longitudinal bifactor analysis. The general exposome factor is significantly associated with cognitive performance across all five tasks assessed across both the discovery (A-E) and replication (F-J) samples. One outlier was excluded from Panel A for visualization purposes (Picture Vocabulary score less than −4) but this participant was retained for all statistical analyses.
Table 1.
The general exposome factor is significantly associated with cognition at ages 9–10. Across all five cognitive tasks and across both the discovery and replication sub-samples, the general exposome factor (Exp-Factor) is positively associated with cognition. These effects held with the inclusion of all six orthogonal exposome sub-factors as covariates and survived Bonferroni correction for multiple comparisons. Note that random effects for site and family are also included as covariates in these models.
|
Picture Vocabulary |
Flanker |
Picture Sequence Memory |
Pattern Comparison |
Reading Recognition |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | β | Std. Error | t | pbonf | β | Std. Error | t | pbonf | β | Std. Error | t | pbonf | β | Std. Error | t | pbonf | β | Std. Error | t | pbonf |
| Discovery Sample | ||||||||||||||||||||
| Intercept | -0.02 | 0.04 | -0.65 | 1.00 | -0.06 | 0.03 | -1.81 | 7.08 ×10−1 | -0.01 | 0.03 | -0.34 | 1.00 | 0.05 | 0.04 | 1.29 | 1.00 | -0.02 | 0.05 | -0.35 | 1.00 |
| Age | 0.23 | 0.01 | 19.88 | 9.62 ×10−84 | 0.18 | 0.01 | 13.22 | 2.76 ×10−38 | 0.11 | 0.01 | 8.25 | 1.93 ×10−15 | 0.22 | 0.01 | 16.48 | 1.64 ×10−58 | 0.21 | 0.01 | 16.80 | 1.07 ×10−60 |
| Sex | 0.07 | 0.02 | 2.81 | 4.96 ×10−2 | 0.05 | 0.03 | 1.87 | 6.21 ×10−1 | -0.12 | 0.03 | -4.27 | 1.99 ×10−4 | -0.13 | 0.03 | -4.72 | 2.40 ×10−5 | -0.00 | 0.03 | -0.14 | 1.00 |
| Exp-Factor | 0.50 | 0.01 | 36.06 | 4.95 ×10−253 | 0.20 | 0.02 | 13.33 | 7.06 ×10−39 | 0.22 | 0.01 | 14.85 | 6.82 ×10−48 | 0.12 | 0.02 | 8.06 | 9.72 ×10−15 | 0.43 | 0.01 | 28.99 | 2.90 ×10−170 |
| School | -0.02 | 0.01 | -1.78 | 7.47 ×10−1 | -0.01 | 0.01 | -0.57 | 1.00 | 0.02 | 0.01 | 1.06 | 1.00 | 0.03 | 0.01 | 2.03 | 4.27 ×10−1 | -0.00 | 0.01 | -0.18 | 1.00 |
| Family Values | -0.10 | 0.01 | -7.06 | 1.91 ×10−11 | 0.02 | 0.02 | 1.49 | 1.00 | -0.03 | 0.02 | -2.10 | 3.60 ×10−1 | 0.01 | 0.02 | 0.67 | 1.00 | -0.01 | 0.01 | -0.63 | 1.00 |
| Family Turmoil | 0.01 | 0.01 | 0.87 | 1.00 | 0.01 | 0.01 | 0.46 | 1.00 | 0.00 | 0.01 | 0.13 | 1.00 | -0.02 | 0.01 | -1.17 | 1.00 | 0.01 | 0.01 | 0.58 | 1.00 |
| Dense Urban Poverty | 0.01 | 0.01 | 0.49 | 1.00 | -0.01 | 0.01 | -0.97 | 1.00 | -0.01 | 0.01 | -0.47 | 1.00 | -0.04 | 0.02 | -2.40 | 1.64 ×10−1 | 0.05 | 0.02 | 3.23 | 1.25 ×10−2 |
| Extracurriculars | -0.02 | 0.02 | -1.00 | 1.00 | 0.01 | 0.02 | 0.64 | 1.00 | 0.02 | 0.02 | 0.92 | 1.00 | -0.01 | 0.02 | -0.32 | 1.00 | -0.01 | 0.02 | -0.45 | 1.00 |
| Screen Time | -0.03 | 0.02 | -1.62 | 1.00 | -0.03 | 0.02 | -1.57 | 1.00 | -0.12 | 0.02 | -5.49 | 4.26 ×10−7 | -0.04 | 0.02 | -2.03 | 4.27 ×10−1 | -0.08 | 0.02 | -3.82 | 1.35 ×10−3 |
| Replication Sample | ||||||||||||||||||||
| Intercept | -0.05 | 0.03 | -1.37 | 1.00 | -0.04 | 0.04 | -1.10 | 1.00 | 0.01 | 0.03 | 0.19 | 1.00 | 0.03 | 0.04 | 0.90 | 1.00 | -0.02 | 0.05 | -0.44 | 1.00 |
| Age | 0.25 | 0.01 | 22.01 | 1.06 ×10−101 | 0.16 | 0.01 | 12.20 | 9.41 ×10−33 | 0.11 | 0.01 | 8.09 | 7.69 ×10−15 | 0.22 | 0.01 | 16.05 | 1.39 ×10−55 | 0.24 | 0.01 | 19.45 | 2.22 ×10−80 |
| Sex | 0.07 | 0.02 | 2.89 | 3.87 ×10−2 | 0.02 | 0.03 | 0.62 | 1.00 | -0.11 | 0.03 | -3.95 | 7.99 ×10−4 | -0.11 | 0.03 | -4.03 | 5.62 ×10−4 | 0.04 | 0.03 | 1.67 | 9.58 ×10−1 |
| Exp-Factor | 0.48 | 0.01 | 36.31 | 4.80 ×10−256 | 0.22 | 0.02 | 14.76 | 2.54 ×10−47 | 0.22 | 0.01 | 14.78 | 1.85 ×10−47 | 0.15 | 0.02 | 9.62 | 9.75 ×10−21 | 0.43 | 0.01 | 29.58 | 1.12 ×10−176 |
| School | -0.02 | 0.01 | -1.25 | 1.00 | 0.00 | 0.01 | 0.23 | 1.00 | 0.01 | 0.01 | 0.76 | 1.00 | 0.06 | 0.01 | 4.03 | 5.66 ×10−4 | -0.01 | 0.01 | -1.09 | 1.00 |
| Family Values | -0.10 | 0.01 | -7.03 | 2.26 ×10−11 | -0.02 | 0.02 | -1.55 | 1.00 | -0.01 | 0.02 | -0.74 | 1.00 | -0.02 | 0.02 | -1.54 | 1.00 | -0.02 | 0.01 | -1.26 | 1.00 |
| Family Turmoil | -0.01 | 0.01 | -1.15 | 1.00 | -0.00 | 0.01 | -0.25 | 1.00 | 0.01 | 0.01 | 0.95 | 1.00 | -0.01 | 0.01 | -0.68 | 1.00 | -0.01 | 0.01 | -0.54 | 1.00 |
| Dense Urban Poverty | -0.00 | 0.01 | -0.32 | 1.00 | -0.02 | 0.02 | -1.53 | 1.00 | -0.04 | 0.01 | -2.70 | 6.94 ×10−2 | 0.00 | 0.02 | 0.11 | 1.00 | 0.02 | 0.01 | 1.39 | 1.00 |
| Extracurriculars | -0.01 | 0.02 | -0.68 | 1.00 | 0.01 | 0.02 | 0.52 | 1.00 | 0.00 | 0.02 | 0.08 | 1.00 | -0.00 | 0.02 | -0.11 | 1.00 | -0.02 | 0.02 | -1.02 | 1.00 |
| Screen Time | -0.06 | 0.02 | -3.22 | 1.28 ×10−2 | -0.03 | 0.02 | -1.28 | 1.00 | -0.08 | 0.02 | -3.79 | 1.50 ×10−3 | -0.07 | 0.02 | -3.07 | 2.13 ×10−2 | -0.06 | 0.02 | -3.21 | 1.33 ×10−2 |
Sensitivity analyses revealed that the general exposome factor, which broadly captures multidimensional socio-economic disparities and co-occurring environmental features, could explain additional variance in cognitive abilities over and above the effects of two commonly used measures of socio-economic status (household income and parental education) (Table S6). This suggests that, although these standard measures of socio-economic status are included in our operationalization of the exposome, the general exposome factor captures additional variance in cognitive abilities beyond what is captured by these measures alone. The general exposome factor was also significantly associated with individual differences in cognition over and above the effects of psychiatric medication use (Table S7). Stratified analyses by racial identification and biological sex revealed consistent positive associations between the general exposome factor and cognition that were significant for nearly all cognitive tasks in all groups after correction for multiple comparisons (Table S8).
To determine whether longitudinal changes in the exposome are associated with changes in cognition, we fit linear mixed-effects models with change in exposome as the independent variable and change in cognition as the dependent variable. All measures of change over time accounted for baseline measurements and models were adjusted for age, family, site, and biological sex across the full sample (n=5632). We found that change in the exposome was significantly associated with change in cognitive performance in three out of the five cognitive tasks assessed after multiple comparisons correction (Picture Vocabulary: ꞵ=0.18, pbonf<.001; Picture Sequence Memory: ꞵ=0.13, pbonf<.001; Reading Recognition: ꞵ=0.11, pbonf<.001; Flanker: ꞵ=0.06, pbonf=0.316; Pattern Comparison: ꞵ=0.05, pbonf=0.640; Table S9).
3.2. Exposome scores are reflected in multivariate patterns of functional brain network topography
We next sought to investigate whether the general exposome is reflected in functional brain organization. To capture inter-individual heterogeneity in the spatial topography of functional brain networks, we defined a unique map of functional brain networks for each child using non-negative matrix factorization (Fig. 1a). Personalized functional network (PFN) topography was defined as each individual’s multivariate pattern of vertex-wise loadings for each of 17 PFNs. To investigate associations between these high-dimensional patterns of PFN topography and each child’s general exposome score, we trained ridge regression models using two-fold cross-validation across matched discovery and replication samples with the ridge parameter tuned by nested cross-validation (Fig. 1b), and only report results from testing our models on unseen data. All models included covariates for age, biological sex, scanning site, and head motion.
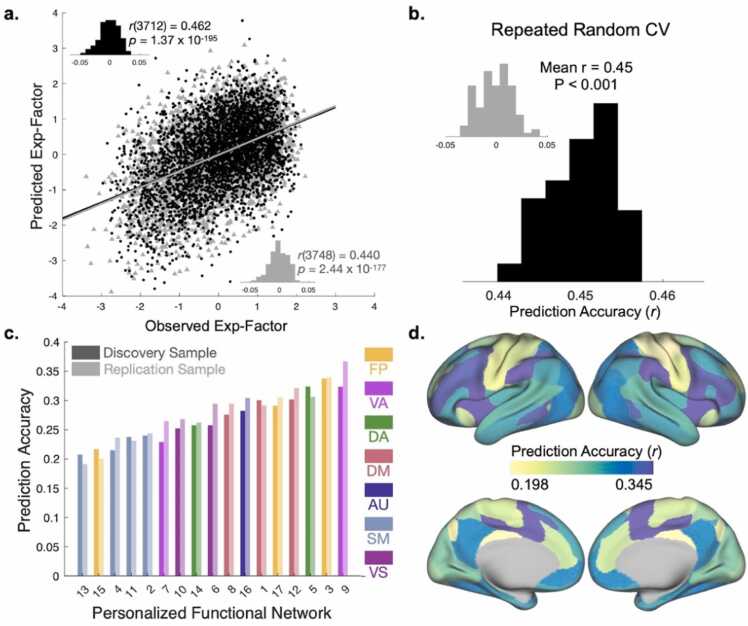
PFN topography was associated with the general exposome factor in unseen data across both the discovery (n=3712) and replication (n=3748) samples, with significant correlations between a child’s observed general exposome factor score and the general exposome factor score estimated by ridge regression (Fig. 3a, discovery: r=0.440, p<0.001, 95% CI: [0.41, 0.47]; replication: r=0.462, p<0.001, 95% CI: [0.44, 0.49]). The accuracy of these models far exceeds the accuracy of models trained to predict a standard continuous measure of socio-economic status (areal deprivation index; discovery: r=0.269, p<0.001, 95% CI: [0.24, 0.30]; replication: r=0.292, p<0.001, 95% CI: [0.26, 0.32]), providing further evidence that the exposome explains more variance in PFN topography patterns than socio-economic status alone. Repeated random cross-validation confirmed that results were not driven by the choice of split (Fig. 3b; mean r = 0.45, p<.001). To further confirm that our results were not driven by leakage across samples, we repeated this training and testing procedure using exposome scores that were generated independently in the discovery and replication samples rather than from the full sample, and found nearly identical results (Figure S2, discovery: r=0.440, p<0.001, 95% CI: [0.41, 0.47]; replication: r=0.460, p<0.001, 95% CI: [0.43, 0.49]). Independent models trained on the functional topography of each PFN highlight variability in prediction accuracies (correlations between true exposome and model-predicted exposome) across networks: fronto-parietal, dorsal and ventral attention networks yielded higher accuracy than sensorimotor networks (Fig. 3c,d). Together, these findings reveal a clear association between a child’s exposome score and their unique multivariate pattern of PFN topography.
Fig. 3.
Exposome scores are reflected in the multivariate pattern of personalized functional brain network topography. (A) Association between observed and predicted general exposome factor scores using two-fold cross-validation (2 F-CV) across both the discovery (black scatterplot) and replication (gray scatterplot) samples. Inset histograms represent null distributions of prediction accuracies with permuted data. (B) Repeated random 2 F-CV provided evidence of stable prediction accuracy across splits of the data, compared with a null distribution with permuted data (inset). (C) Independent network models ranked by prediction accuracy. Note that all p-values associated with prediction accuracies are significant after Bonferroni correction. Numerical assignments for each PFN are consistent with previous work (Cui et al., 2020, Keller et al., 2023a) and can be found in Fig. S4. (D) Prediction accuracy averaged across discovery and replication samples depicted for seventeen cross-validated models trained on each PFN independently. Abbreviations: Exp-Factor: general exposome factor; CV: cross-validation; FP: fronto-parietal; VA: ventral attention; DA: dorsal attention; DM: default mode; AU: auditory; SM: somatomotor; VS: visual.
3.3. Exposome scores and functional topography are associated with cognition
To compare multivariate associations among exposome scores, personalized functional brain network topography and cognitive functioning, we trained three types of linear ridge regression models to predict three domains of cognition (general cognition, executive function, and learning/memory). The first model type (“Exp-Factor”) used only a participant’s general exposome score, while the second model type (“PFN Topography”), reported in our prior work (Keller et al., 2023a), used each participant’s multivariate pattern of PFN topography. The third model type (“Exp-Factor + PFN Topography”) used a combination of exposome and PFN topography, hypothesized to perform best by capitalizing on both shared and unique variance across features. All three models performed well (Table 2), with the highest accuracy for predictions of general cognition and lower accuracy for learning/memory and executive function.
Table 2.
Comparison of models relating exposome and PFN topography to cognitive functioning across domains. We trained linear ridge regression models to predict three domains of cognition (General Cognition, Executive Function, and Learning/Memory). The first model type (“Exp-Factor”) used only a participant’s general exposome score, while the second model type (“PFN Topography”) used each participant’s multivariate pattern of PFN topography. The third model type (“Exp-Factor + PFN Topography”) used both exposome scores and PFN topography. Correlations between true cognitive performance and model-generated cognitive performance (r) were significant for all model types. Model comparison using the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) reveals that the Exp-Factor model is the most parsimonious.
|
Discovery |
Replication |
|||||||
|---|---|---|---|---|---|---|---|---|
| Prediction Accuracy | r | p | AIC | BIC | r | p | AIC | BIC |
| General Cognition | ||||||||
| Exp-Factor | 0.42 | 2.65 ×10−148 | -248.8198 | -8.0250 | 0.46 | 1.17 ×10−179 | -454.2790 | -7.8809 |
| PFN Topography | 0.41 | 3.05 ×10−146 | 2.0198 ×106 | 8.2493 ×106 | 0.45 | 3.85 ×10−174 | 2.0196 ×106 | 8.2267 ×106 |
| Exp-Factor + PFN Topography | 0.44 | 2.44 ×10−166 | 2.0196 ×106 | 8.2491 ×106 | 0.48 | 3.30 ×10−194 | 2.0195 ×106 | 8.2270 ×106 |
| Executive Function | ||||||||
| Exp-Factor | 0.11 | 8.86 ×10−11 | 1217.1946 | -8.8570 | 0.14 | 7.30 ×10−16 | 1035.4933 | -8.7450 |
| PFN Topography | 0.17 | 1.37 ×10−23 | 2.0210 ×106 | 8.2493 ×106 | 0.16 | 5.48 ×10−22 | 2.0209 ×106 | 8.2267 ×106 |
| Exp-Factor + PFN Topography | 0.17 | 4.41 ×10−24 | 2.0210 ×106 | 8.2491 ×106 | 0.17 | 8.54 ×10−23 | 2.0209 ×106 | 8.2270 ×106 |
| Learning/Memory | ||||||||
| Exp-Factor | 0.25 | 1.35 ×10−50 | 470.3910 | -8.4332 | 0.27 | 6.20 ×10−57 | 386.2874 | -8.3685 |
| PFN Topography | 0.27 | 2.06 ×10−61 | 2.0204 ×106 | 8.2493 ×106 | 0.27 | 2.91 ×10−57 | 2.0204 ×106 | 8.2267 ×106 |
| Exp-Factor + PFN Topography | 0.28 | 3.49 ×10−66 | 2.0203 ×106 | 8.2491 ×106 | 0.28 | 4.92 ×10−63 | 2.0203 ×106 | 8.2270 ×106 |
We compared performance across models using the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC). These indices were selected to account for the substantial differences in the number of features used to train each type of model, balancing the tradeoff between model accuracy and complexity. The general exposome factor model was the most parsimonious (lowest AIC and BIC), reflecting its high accuracy and low complexity. While the full Exposome + PFN Topography model yielded a small increase in accuracy, this benefit was outweighed by the substantial increase in model complexity from a single feature to thousands of features. This result also held when using a much lower-dimensional measure of PFN topography, the total cortical representation of each PFN (Table S10). To determine which model type could best predict future cognitive performance, we trained models on baseline (9–10 years old) general exposome scores and PFN topography to predict cognition assessed two years later (11–12 years old), including baseline cognitive scores as covariates. Again, the general exposome factor models were most parsimonious across all five cognitive tasks (Table S11).
Furthermore, the modest boost in model performance for the full Exp-Factor + PFN topography model compared with the PFN topography and Exp-Factor models indicates that there may be substantial shared variance between the general exposome factor and PFN topography, in line with our observation that general exposome scores are reflected in PFN topography. However, there also appears to be some unique variance explained from each feature type (see Figure S5 for a comparison of prediction accuracy maps across models). To further disentangle the unique patterns of functional network organization associated with cognition beyond what is accounted for by the exposome, we trained an additional model associating the multivariate pattern of PFN topography with general cognition after regressing out the general exposome scores. This model achieved moderate accuracy (discovery: r = 0.240, p<0.001, replication: r = 0.233, p<0.001), suggesting that some features of the multivariate pattern of PFN topography are uniquely associated with cognition, some features are uniquely associated with the general exposome, and some variance is shared between both.
4. Discussion
Neurodevelopment does not take place in a vacuum, but amid a variety of environmental exposures and experiences. To characterize reproducible cross-sectional and longitudinal environment-brain-behavior associations, we conducted preregistered analyses (Keller and Barzilay, 2023) in a large-scale dataset of youth. Using validated dimensionality reduction approaches, we defined an individual’s exposome, mapped individual-specific patterns of functional brain organization, and trained cross-validated models to predict cognition from held-out data. We found that individual differences in the exposome are associated with individual differences in current and future cognition and are reflected in children's unique patterns of functional brain network topography. We also found that predictive models trained on a single variable capturing a child’s exposome could predict cognition more parsimoniously than models trained on a wealth of robust, personalized neuroimaging variables. Together, these findings extend prior work identifying associations between the exposome (Rappaport, 2011, Wild, 2005) and a variety of outcomes, including mental health outcomes in children (Moore et al., 2022, Barzilay et al., 2022, Pries et al., 2022), by demonstrating that the exposome explains substantially more variance in youth cognition and functional brain network organization than standard measures of socio-economic status alone.
Studies of the “exposome” (Rappaport, 2011, Wild, 2005) have uncovered associations with a variety of physical and mental health outcomes in adults and, more recently, mental health outcomes in children (Moore et al., 2022, Barzilay et al., 2022, Pries et al., 2022). We have recently described in a cross-sectional analysis that the exposome explains ∼40% of variance in overall psychopathology at early adolescence (Moore et al., 2022). In the present study, we extended this work to investigate cognitive outcomes, revealing that the exposome is also related to individual differences in cognitive functioning across domains. We also extended this work to examine longitudinal outcomes, demonstrating that changes in the exposome are associated with changes in cognition over a two year time period spanning the critical transition from childhood to adolescence. When we compared predictive models trained on a child’s exposome with models trained on a wealth of robust, personalized neuroimaging variables to predict cognition, we found that the single-variable exposome models were just as accurate and more parsimonious than the brain network models. This result highlights the magnitude of the association between the exposome and health as well as the importance of accounting for environmental associations in future studies of cognitive neurodevelopment. This finding also sets the stage for future investigations of further longitudinal timepoints from the ABCD Study to determine whether differences in childhood environments might predispose individuals to different trajectories of functional topography development, leading to individual differences in cognitive functioning. Moreover, given that cognitive impairments may be a risk factor for psychiatric illnesses like depression and anxiety that emerge during adolescence (Larsen and Luna, 2018, Keller et al., 2023b, James et al., 2023, SECTION ON ADOLESCENT HEALTH, COUNCIL ON COMMUNITY PEDIATRICS, COMMITTEE ON ADOLESCENCE, 2019, Wold, 1956, Dalsgaard et al., 2020, Solmi et al., 2022, Kessler et al., 2005, Caspi et al., 2020, Merikangas et al., 2010), our findings point to potential early markers for targeted prevention efforts.
Our observation that the exposome is reflected in patterns of functional brain network topography could suggest that a child’s environment leaves a mark on their neurodevelopment. Prior work has characterized how specific types of experiences or environments such as childhood maltreatment (Teicher et al., 2016) or low socio-economic status (Botdorf et al., 2022) affect brain structure and function. Our results build on this work by examining a general exposome factor derived from many co-occurring environmental features at once, highlighting the utility of capturing potentially important additive effects. Given that the general exposome factor primarily captures co-occurring aspects of wealth and socioeconomic status (e.g., income, education, and neighborhood prosperity), this work contributes to a large body of evidence that supports the promotion of economic prosperity and policies that ensure socioeconomic resources for children (Weissman et al., 2023). Furthermore, our findings provide validation for functional network topography as a potentially useful biomarker of environmental associations with neurodevelopment, complementing studies of activation or functional connectivity. Future studies may further characterize the extent and duration of the association between the exposome and functional network topography.
Adolescence appears to be a particularly crucial sensitive period for the development of higher-order cortices (Larsen and Luna, 2018), including prominently the functional brain networks that support cognition (Keller et al., 2023b). Our observation that multivariate patterns of functional topography in association and attention networks were most strongly related to exposome scores coheres with these prior studies suggesting that these networks might be in a sensitive period of developmental plasticity where brain development might be related to environmental exposures. It has been theorized that sensitive windows for cognitive development may occur opportunistically during adolescence, a period of life when it is crucial to learn to adapt to the environment, though future studies are warranted to investigate the potential causality of environmental influences on brain development. For example, if the environment adaptively shapes the development of attention networks, it may do so by either honing sharp focus amid distractions or heightening broad awareness to monitor for potential threats. Identifying which brain networks (e.g., association networks) appear to most reflect the environment at a given time period during development (e.g. at the transition from childhood to adolescence) may inform future studies investigating causal links between environmental adversity and specific brain circuitry. Such studies may subsequently inform targeted interventions or target prevention efforts more optimally to promote healthy neurodevelopment, particularly during the critical transition from childhood to adolescence when psychiatric illnesses often emerge (Dalsgaard et al., 2020, Solmi et al., 2022, Kessler et al., 2005).
It is difficult to balance specificity, accuracy, and personalization with reproducibility, parsimony, and generalizability. While approaches favoring specificity allow us to deeply characterize each individual, approaches favoring reproducibility allow findings to be more readily understandable and generalizable. In statistical learning, this conundrum is referred to as a tradeoff between model flexibility and interpretability (James et al., 2023), with more complex models tending to overfit to training data. Here, we attempted to balance this tradeoff. We used as much detail as possible to capture each child’s multivariate exposome and defined personalized functional brain networks within individuals, then counterbalanced this detail by using dimensionality reduction to derive broad, generalizable descriptors of environment, brain and behavior. We also maximized reproducibility by pre-registering our analyses (Keller and Barzilay, 2023), using rigorous cross-validation, and leveraging model-selection statistics (AIC and BIC) that balance model flexibility and interpretability.
Our results should be considered in light of several limitations. First, it is challenging to comprehensively capture every possible aspect of a child’s complex, multidimensional environment. Our approach to defining the exposome longitudinally made use of data that were available at multiple timepoints of the ABCD Study®. Although this meant we could make use of a large number of variables capturing a variety of features of each child’s environment, not all aspects of a child’s environment were captured and some aspects may have been better assessed than others. Importantly, the variables available for our study timepoints did not address racial discrimination, which has known effects on development (Trent et al., 2019), nor did they include measures of traumatic life events, though these variables were included in a previous cross-sectional definition of the exposome in this dataset (Moore et al., 2022). Future studies may investigate a wider array of assessments to more fully capture each child’s multidimensional environment. Additionally, our data-driven finding that certain exposome sub-factor scores (Family Values and Screen Time) are associated with specific sub-domains of cognition may provide targets for future hypothesis-driven studies to answer more mechanistic questions than we were able to explore here. Furthermore, given that the ABCD Study® sample is not fully representative of all demographic characteristics in the United States, generalizability may be limited and future studies in more diverse samples are warranted. As the ABCD Study® is observational, we cannot infer causality from exposome to brain/cognition. One important potential mechanistic pathway connecting childhood environments with functional brain network organization that could be explored in future work is stress, in line with a large body of prior research showing the physical and mental health impacts of increased caregiver and child stress (Lupien et al., 2009). Future work may also leverage methods for causal inference to address the question of causality (Wold, 1956) to inform interventions to promote healthy development. Finally, although our study has attempted to focus specifically on environmental exposures, there are likely to be non-environmental (e.g., genetic) effects that are confounded with variables in our model (e.g., through family history of psychiatric illness). Future studies may seek to further characterize the independent and combinatorial effects of the genome, exposome, and functional connectome together on cognitive development.
4.1. Conclusions
Our results highlight the utility of capturing the complex, multidimensional features of a child’s environment to better understand functional brain organization and cognitive development. Our findings build on prior studies investigating associations between individual environmental features and youth cognition by leveraging a multitude of environmental features in a large sample. By attempting to capture the totality of many co-occurring features at once, we were better able to predict cognitive performance in held-out data and found that changes in the environment were associated with changes in cognition over time. Moreover, by defining functional brain networks at the level of the individual rather than by group-average, we demonstrated that childhood environments are reflected in an individual’s unique topographical patterns of functional brain network organization. Future research may build upon this work to more precisely characterize how variability in childhood environments may be associated with specific features or long-term trajectories of cognitive neurodevelopment.
Funding
This study was supported by grants from the National Institutes of Health: R01MH113550 (T.D.S.), (T.D.S.), R01MH120482 (T.D.S.), R01EB022573R37MH125829 (T.D.S. and D.A.F.), K23MH120437 (R.B.), R21MH130797 (R.B.), R01MH123550 (R.T.S.), R01MH112847 (R.T.S.), R01MH123563 (R.T.S.). A.S.K. was supported by a Neuroengineering and Medicine T32 Fellowship from the National Institute of Neurological Disorders and Stroke (5T32NS091006-08), a Neurodevelopment and Psychosis T32 Fellowship from the National Institute of Mental Health (5T32MH019112-32), and a Loan Repayment Award from the National Institute of Mental Health (1L30MH131061-01). A.P. was supported by the Stanford School of Medicine Dean’s Fellowship. Additional support was provided by the Penn-CHOP Lifespan Brain Institute.
CRediT authorship contribution statement
Audrey Luo: Writing – review & editing, Validation. Oscar Miranda-Dominguez: Writing – review & editing, Data curation. Tyler M. Moore: Writing – review & editing, Data curation, Conceptualization. Allyson P. Mackey: Writing – review & editing, Conceptualization. Arielle S. Keller: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Conceptualization. Theodore D. Satterthwaite: Writing – review & editing, Supervision, Conceptualization. Zaixu Cui: Writing – review & editing, Data curation. Damien A. Fair: Writing – review & editing, Supervision, Conceptualization. Alisha Shetty: Writing – review & editing, Visualization, Validation. Kevin Y. Sun: Writing – review & editing. Mārtinš Gatavinš: Writing – review & editing, Conceptualization. Russell T. Shinohara: Writing – review & editing. Elina Visoki: Writing – review & editing, Validation, Data curation, Conceptualization. Hongming Li: Writing – review & editing, Data curation. Audrey Houghton: Writing – review & editing, Data curation. Eric Feczko: Writing – review & editing, Data curation. Ran Barzilay: Writing – review & editing, Supervision, Conceptualization. Yong Fan: Writing – review & editing, Data curation. Adam Pines: Writing – review & editing, Data curation.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Theodore D. Satterthwaite reports financial support was provided by National Institute of Mental Health. Damien A. Fair reports financial support was provided by National Institute of Mental Health. Ran Barzilay reports financial support was provided by National Institute of Mental Health. Russell T. Shinohara reports financial support was provided by National Institute of Mental Health. Arielle S. Keller reports financial support was provided by National Institute of Neurological Disorders and Stroke. Arielle S. Keller reports financial support was provided by National Institute of Mental Health. Ran Barzilay reports a relationship with Taliaz Health that includes: board membership and equity or stocks. Ran Barzilay reports a relationship with Zynerba Pharmaceuticals, Inc that includes: board membership. Russell T. Shinohara reports a relationship with Genentech that includes: consulting or advisory. Russell T. Shinohara reports a relationship with Octave Bioscience that includes: consulting or advisory. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive DevelopmentSM (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9–10 and follow them over 10 years into early adulthood. The ABCD Study® is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD data repository grows and changes over time. The ABCD data used in this report came from [NIMH Data Archive Digital Object Identifier 10.15154/1523041]. DOIs can be found at https://nda.nih.gov/abcd.
All analyses for this study were pre-registered (https://osf.io/2dm9q).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dcn.2024.101370.
Appendix A. Supplementary material
Supplementary material
Data availability
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development Study® (http://abcdstudy.org), held in the NIMH Data Archive (NDA). Only researchers with an approved NDA Data Use Certification (DUC) may obtain ABCD Study data. Code is available at http://github.com/PennLINC/exposome_PFNs_cognition.
References
- Akaike H. In: Selected Papers of Hirotugu Akaike. Parzen E., Tanabe K., Kitagawa G., editors. Springer; 1998. Information Theory and an Extension of the Maximum Likelihood Principle; pp. 199–213. [DOI] [Google Scholar]
- Asparouhov T., Muthén B. Exploratory structural equation modeling. Struct. Equ. Model.: A Multidiscip. J. 2009;16(3):397–438. doi: 10.1080/10705510903008204. [DOI] [Google Scholar]
- Barzilay R., Pries L.-K., Moore T.M., Gur R.E., van Os J., Rutten B.P.F., Guloksuz S. Exposome and trans-syndromal developmental trajectories toward psychosis. Biol. Psychiatry Glob. Open Sci. 2022;2(3):197–205. doi: 10.1016/j.bpsgos.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botdorf M., Dunstan J., Sorcher L., Dougherty L.R., Riggins T. Socioeconomic disadvantage and episodic memory ability in the ABCD sample: contributions of hippocampal subregion and subfield volumes. Dev. Cogn. Neurosci. 2022;57 doi: 10.1016/j.dcn.2022.101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., He X., Han J., Huang T.S. Graph regularized nonnegative matrix factorization for data representation. IEEE Trans. Pattern Anal. Mach. Intell. 2011;33(8):1548–1560. doi: 10.1109/TPAMI.2010.231. [DOI] [PubMed] [Google Scholar]
- Calvin C.M., Deary I.J., Fenton C., Roberts B.A., Der G., Leckenby N., Batty G.D. Intelligence in youth and all-cause-mortality: systematic review with meta-analysis. Int. J. Epidemiol. 2011;40(3):626–644. doi: 10.1093/ije/dyq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A., Houts R.M., Ambler A., Danese A., Elliott M.L., Hariri A., Harrington H., Hogan S., Poulton R., Ramrakha S., Rasmussen L.J.H., Reuben A., Richmond-Rakerd L., Sugden K., Wertz J., Williams B.S., Moffitt T.E. Longitudinal assessment of mental health disorders and comorbidities across 4 decades among participants in the dunedin birth cohort study. JAMA Netw. Open. 2020;3(4) doi: 10.1001/jamanetworkopen.2020.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova, M.M., Doyle, O., Conan, G., Feczko, E., Earl, E., Perrone, A., & Fair, D. (2021, July 2). ABCD Reproducible Matched Samples (ARMS) software. Open Science Framework.
- Cui Z., Li H., Xia C.H., Larsen B., Adebimpe A., Baum G.L., Cieslak M., Gur R.E., Gur R.C., Moore T.M., Oathes D.J., Alexander-Bloch A.F., Raznahan A., Roalf D.R., Shinohara R.T., Wolf D.H., Davatzikos C., Bassett D.S., Fair D.A.…Satterthwaite T.D. Individual variation in functional topography of association networks in youth. Neuron. 2020;106(2):340–353.e8. doi: 10.1016/j.neuron.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard S., Thorsteinsson E., Trabjerg B.B., Schullehner J., Plana-Ripoll O., Brikell I., Wimberley T., Thygesen M., Madsen K.B., Timmerman A., Schendel D., McGrath J.J., Mortensen P.B., Pedersen C.B. Incidence rates and cumulative incidences of the full spectrum of diagnosed mental disorders in childhood and adolescence. JAMA Psychiatry. 2020;77(2):155. doi: 10.1001/jamapsychiatry.2019.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko M.I., Ip K.I., Kelly D.P., Constante K., Goetschius L.G., Keating D.P. Ecological stress, amygdala reactivity, and internalizing symptoms in preadolescence: is parenting a buffer? Cortex. 2021;140:128–144. doi: 10.1016/j.cortex.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E., Conan G., Marek S., Tervo-Clemmens B., Cordova M., Doyle O., Earl E., Perrone A., Sturgeon D., Klein R., Harman G., Kilamovich D., Hermosillo R., Miranda-Dominguez O., Hendrickson T., Juliano A.C., Snider K., Moore L.A., Uriartel J.…Fair D.A. Adolescent brain cognitive development (ABCD) community MRI collection and utilities. BioRxiv. 2021;20 doi: 10.1101/2021.07.09.451638. [DOI] [Google Scholar]
- Galton F. Regression Towards Mediocrity in Hereditary Stature. J. Anthropol. Inst. Great Britain Ireland. 1886;15:246. doi: 10.2307/2841583. [DOI] [Google Scholar]
- Glasser M.F., Coalson T.S., Robinson E.C., Hacker C.D., Harwell J., Yacoub E., Ugurbil K., Andersson J., Beckmann C.F., Jenkinson M., Smith S.M., Van Essen D.C. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536(7615):171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E.M., Laumann T.O., Gilmore A.W., Newbold D.J., Greene D.J., Berg J.J., Ortega M., Hoyt-Drazen C., Gratton C., Sun H., Hampton J.M., Coalson R.S., Nguyen A.L., McDermott K.B., Shimony J.S., Snyder A.Z., Schlaggar B.L., Petersen S.E., Nelson S.M., Dosenbach N.U.F. Precision functional mapping of individual human brains. Neuron. 2017;95(4):791–807.e7. doi: 10.1016/j.neuron.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice J.W. Computing and evaluating factor scores. Psychol. Methods. 2001;6(4):430–450. doi: 10.1037/1082-989X.6.4.430. [DOI] [PubMed] [Google Scholar]
- Guloksuz S., van Os J., Rutten B.P.F. The exposome paradigm and the complexities of environmental research in psychiatry. JAMA Psychiatry. 2018;75(10):985–986. doi: 10.1001/jamapsychiatry.2018.1211. [DOI] [PubMed] [Google Scholar]
- Hu L., Bentler P.M. Cutoff criteria for fit indexes in covariance structure analysis: conventional criteria versus new alternatives. Struct. Equ. Model. 1999;6(1):1–55. doi: 10.1080/10705519909540118. [DOI] [Google Scholar]
- James G., Witten D., Hastie T., Tibshirani R. An introduction to statistical learning with applications in R. Second Edition. Springer; 2023. [Google Scholar]
- Jennrich R.I., Bentler P.M. Exploratory bi-factor analysis. Psychometrika. 2011;76(4):537–549. doi: 10.1007/s11336-011-9218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, A.S., & Barzilay, R. (2023). Associations among exposome factors, personalized functional brain network topography, and cognitive functioning in youth. 〈https://osf.io/2dm9q〉.
- Keller A.S., Sydnor V.J., Pines A., Fair D.A., Bassett D.S., Satterthwaite T.D. Hierarchical functional system development supports executive function. Trends Cogn. Sci. 2023;27(2):160–174. doi: 10.1016/j.tics.2022.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A.S., Pines A.R., Shanmugan S., Sydnor V.J., Cui Z., Bertolero M.A., Barzilay R., Alexander-Bloch A.F., Byington N., Chen A., Conan G.M., Davatzikos C., Feczko E., Hendrickson T.J., Houghton A., Larsen B., Li H., Miranda-Dominguez O., Roalf D.R.…Satterthwaite T.D. Personalized functional brain network topography is associated with individual differences in youth cognition. Nat. Commun. 2023;14(1):8411. doi: 10.1038/s41467-023-44087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R.C., Berglund P., Demler O., Jin R., Merikangas K.R., Walters E.E. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the national comorbidity survey replication. Arch. Gen. Psychiatry. 2005;62(6):593. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kidd E., Donnelly S., Christiansen M.H. Individual differences in language acquisition and processing. Trends Cogn. Sci. 2018;22(2):154–169. doi: 10.1016/j.tics.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Kong R., Li J., Orban C., Sabuncu M.R., Liu H., Schaefer A., Sun N., Zuo X.N., Holmes A.J., Eickhoff S.B., Yeo B.T.T. Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb. Cortex. 2019;29(6):2533–2551. doi: 10.1093/cercor/bhy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Luna B. Adolescence as a neurobiological critical period for the development of higher-order cognition. Neurosci. Biobehav. Rev. 2018;94:179–195. doi: 10.1016/j.neubiorev.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann T.O., Gordon E.M., Adeyemo B., Snyder A.Z., Joo S.J., Chen M.-Y., Gilmore A.W., McDermott K.B., Nelson S.M., Dosenbach N.U.F., Schlaggar B.L., Mumford J.A., Poldrack R.A., Petersen S.E. Functional system and areal organization of a highly sampled individual human brain. Neuron. 2015;87(3):657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Satterthwaite T.D., Fan Y. Large-scale sparse functional networks from resting state fMRI. NeuroImage. 2017;156:1–13. doi: 10.1016/j.neuroimage.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciana M., Bjork J.M., Nagel B.J., Barch D.M., Gonzalez R., Nixon S.J., Banich M.T. Adolescent neurocognitive development and impacts of substance use: overview of the adolescent brain cognitive development (ABCD) baseline neurocognition battery. Dev. Cogn. Neurosci. 2018;32:67–79. doi: 10.1016/j.dcn.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien S.J., McEwen B.S., Gunnar M.R., Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 2009;10(6):434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- Marcus D., Harwell J., Olsen T., Hodge M., Glasser M., Prior F., Jenkinson M., Laumann T., Curtiss S., Van Essen D. Informatics and data mining tools and strategies for the human connectome project. Front. Neuroinformatics. 2011;5 doi: 10.3389/fninf.2011.00004. 〈https://www.frontiersin.org/articles/10.3389/fninf.2011.00004〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Calabro F.J., Montez D.F., Kay B.P., Hatoum A.S., Donohue M.R., Foran W., Miller R.L., Hendrickson T.J., Malone S.M., Kandala S., Feczko E., Miranda-Dominguez O., Graham A.M., Earl E.A., Perrone A.J., Cordova M., Doyle O., Dosenbach N.U.F. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603(7902):654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith W.J., Cardenas-Iniguez C., Berman M.G., Rosenberg M.D. Effects of the physical and social environment on youth cognitive performance. Dev. Psychobiol. 2022;64(4) doi: 10.1002/dev.22258. [DOI] [PubMed] [Google Scholar]
- Merikangas K.R., He J., Burstein M., Swanson S.A., Avenevoli S., Cui L., Benjet C., Georgiades K., Swendsen J. Lifetime prevalence of mental disorders in U.S. adolescents: results from the national comorbidity survey replication–adolescent supplement (NCS-A) J. Am. Acad. Child Adolesc. Psychiatry. 2010;49(10):980–989. doi: 10.1016/j.jaac.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt T.E., Arseneault L., Belsky D., Dickson N., Hancox R.J., Harrington H.L., Houts R., Poulton R., Roberts B.W., Ross S., Sears M.R., Thomson W.M., Caspi A. A gradient of childhood self-control predicts health, wealth, and public safety. Proc. Natl. Acad. Sci. USA. 2011;108(7):2693–2698. doi: 10.1073/pnas.1010076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T.M., Risbrough V.B., Baker D.G., Larson G.E., Glenn D.E., Nievergelt C.M., Maihofer A., Port A.M., Jackson C.T., Ruparel K., Gur R.C. Effects of military service and deployment on clinical symptomatology: the role of trauma exposure and social support. J. Psychiatr. Res. 2017;95:121–128. doi: 10.1016/j.jpsychires.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T.M., Visoki E., Argabright S.T., Didomenico G.E., Sotelo I., Wortzel J.D., Naeem A., Gur R.C., Gur R.E., Warrier V., Guloksuz S., Barzilay R. Modeling environment through a general exposome factor in two independent adolescent cohorts. Exposome. 2022;2(1) doi: 10.1093/exposome/osac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén L.K., Muthén B.O. Mplus (Version 8) [Computer software] 2017 [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries L.-K., Moore T.M., Visoki E., Sotelo I., Barzilay R., Guloksuz S. Estimating the association between exposome and psychosis as well as general psychopathology: results from the ABCD study. Biol. Psychiatry Glob. Open Sci. 2022;2(3):283–291. doi: 10.1016/j.bpsgos.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport S.M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011;21(1):5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Reise S.P., Moore T.M., Haviland M.G. Bifactor models and rotations: exploring the extent to which multidimensional data yield univocal scale scores. J. Personal. Assess. 2010;92(6):544–559. doi: 10.1080/00223891.2010.496477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle, W. (2019). psych: Procedures for Personality and Psychological Research. (1.9.12) [Computer software]. 〈http://personality-project.org/r〉.
- Rodriguez A., Reise S.P., Haviland M.G. Evaluating bifactor models: calculating and interpreting statistical indices. Psychol. Methods. 2016;21(2):137–150. doi: 10.1037/met0000045. [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann. Stat. 1978;6(2):461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- Shanmugan S., Wolf D.H., Calkins M.E., Moore T.M., Ruparel K., Hopson R.D., Vandekar S.N., Roalf D.R., Elliott M.A., Jackson C., Gennatas E.D., Leibenluft E., Pine D.S., Shinohara R.T., Hakonarson H., Gur R.C., Gur R.E., Satterthwaite T.D. Common and dissociable mechanisms of executive system dysfunction across psychiatric disorders in youth. Am. J. Psychiatry. 2016;173(5):517–526. doi: 10.1176/appi.ajp.2015.15060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval G., Visoki E., Moore T.M., DiDomenico G.E., Argabright S.T., Huffnagle N.J., Alexander-Bloch A.F., Waller R., Keele L., Benton T.D., Gur R.E., Barzilay R. Evaluation of Attention-deficit/hyperactivity disorder medications, externalizing symptoms, and suicidality in children. JAMA Netw. Open. 2021;4(6) doi: 10.1001/jamanetworkopen.2021.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmi M., Radua J., Olivola M., Croce E., Soardo L., Salazar De Pablo G., Il Shin J., Kirkbride J.B., Jones P., Kim J.H., Kim J.Y., Carvalho A.F., Seeman M.V., Correll C.U., Fusar-Poli P. Age at onset of mental disorders worldwide: large-scale meta-analysis of 192 epidemiological studies. Mol. Psychiatry. 2022;27(1):281–295. doi: 10.1038/s41380-021-01161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher M.H., Samson J.A., Anderson C.M., Ohashi K. The effects of childhood maltreatment on brain structure, function and connectivity. Nat. Rev. Neurosci. 2016;17(10):652–666. doi: 10.1038/nrn.2016.111. [DOI] [PubMed] [Google Scholar]
- Thompson R.C., Montena A.L., Liu K., Watson J., Warren S.L. Associations of family distress, family income, and acculturation on pediatric cognitive performance using the NIH toolbox: implications for clinical and research settings. Arch. Clin. Neuropsychol.: Off. J. Natl. Acad. Neuropsychol. 2022;37(4):798–813. doi: 10.1093/arclin/acab082. [DOI] [PubMed] [Google Scholar]
- Thompson W.K., Barch D.M., Bjork J.M., Gonzalez R., Nagel B.J., Nixon S.J., Luciana M. The structure of cognition in 9 and 10 year-old children and associations with problem behaviors: findings from the ABCD study’s baseline neurocognitive battery. Dev. Cogn. Neurosci. 2019;36 doi: 10.1016/j.dcn.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent M., Dooley D.G., Dougé J., Cavanaugh R.M., Lacroix A.E., Fanburg J., Rahmandar M.H., Hornberger L.L., Schneider M.B., Yen S., Chilton L.A., Green A.E., Dilley K.J., Gutierrez J.R., Duffee J.H., Keane V.A., Wallace S.B., SECTION ON ADOLESCENT HEALTH, COUNCIL ON COMMUNITY PEDIATRICS, COMMITTEE ON ADOLESCENCE The Impact of Racism on Child and Adolescent Health. Pediatrics. 2019;144(2) doi: 10.1542/peds.2019-1765. [DOI] [PubMed] [Google Scholar]
- Volkow N.D., Koob G.F., Croyle R.T., Bianchi D.W., Gordon J.A., Koroshetz W.J., Pérez-Stable E.J., Riley W.T., Bloch M.H., Conway K., Deeds B.G., Dowling G.J., Grant S., Howlett K.D., Matochik J.A., Morgan G.D., Murray M.M., Noronha A., Spong C.Y., Weiss S.R.B. The conception of the ABCD study: From substance use to a broad NIH collaboration. Dev. Cogn. Neurosci. 2018;32:4–7. doi: 10.1016/j.dcn.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J.J., Barnes J.D., Cameron J.D., Goldfield G.S., Chaput J.-P., Gunnell K.E., Ledoux A.-A., Zemek R.L., Tremblay M.S. Associations between 24 h movement behaviours and global cognition in US children: a cross-sectional observational study. Lancet Child Adolesc. Health. 2018;2(11):783–791. doi: 10.1016/S2352-4642(18)30278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D.G., Hatzenbuehler M.L., Cikara M., Barch D.M., McLaughlin K.A. State-level macro-economic factors moderate the association of low income with brain structure and mental health in U.S. children. Nat. Commun. 2023;14(1):2085. doi: 10.1038/s41467-023-37778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C.P. Complementing the genome with an “Exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Wold H. Causal inference from observational data: a review of end and means. J. R. Stat. Soc. Ser. A (Gen.) 1956;119(1):28–61. doi: 10.2307/2342961. [DOI] [Google Scholar]
- Yeo B.T.T., Krienen F.M., Sepulcre J., Sabuncu M.R., Lashkari D., Hollinshead M., Roffman J.L., Smoller J.W., Zöllei L., Polimeni J.R., Fischl B., Liu H., Buckner R.L. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development Study® (http://abcdstudy.org), held in the NIMH Data Archive (NDA). Only researchers with an approved NDA Data Use Certification (DUC) may obtain ABCD Study data. Code is available at http://github.com/PennLINC/exposome_PFNs_cognition.