Abstract
Purpose
To perform a prospective epigenome-wide association study of DNA methylation (DNAm) and 28-year proliferative diabetic retinopathy (PDR) incidence in type 1 diabetes (T1D).
Design
Prospective observational cohort study.
Participants
The Pittsburgh Epidemiology of Diabetes Complications (EDC) study of childhood-onset (< 17 years) T1D.
Methods
Stereoscopic fundus photographs were taken in fields 1, 2, and 4 at baseline, 2, 4, 6, 8, 16, 23, and 28 years after DNAm measurements. The photos were graded using the modified Airlie House System. In those free of PDR at baseline (n = 265; mean T1D duration of 18 years at baseline), whole blood DNAm (EPIC array) at 683 597 CpGs was analyzed in Cox models for time to event. Associations between significant CpGs and clinical risk factors were assessed; genetic variants associated with DNAm were identified (methylation quantitative trait loci [meQTLs]). Mendelian randomization was used to examine evidence of causal associations between DNAm and PDR. Post hoc regional and functional analyses were performed.
Main Outcome Measures
Proliferative diabetic retinopathy was defined as the first instance of a grade of ≥ 60 in at least 1 eye or pan-retinal photocoagulation for PDR. Follow-up time was calculated from the study visit at which DNAm data were available (baseline) until PDR incidence or censoring (December 31, 2018 or last follow-up).
Results
PDR incidence was 53% over 28-years’ follow-up. Greater DNAm of cg27512687 (KIF16B) was associated with reduced PDR incidence (P = 6.3 × 10−9; false discovery rate [FDR]: < 0.01); 113 cis-meQTLs (P < 5 × 10−8) were identified. Mendelian randomization analysis using the sentinel meQTL as the instrumental variable supported a potentially causal association between cg27512687 and PDR. Cg27512687 was also associated with lower pulse rate and albumin excretion rate and higher estimated glomerular filtration rate, but its association with PDR remained independently significant after adjustment for those factors. In regional analyses, DNAm of FUT4, FKBP1A, and RIN2 was also associated with PDR incidence.
Conclusions
DNA methylation of KIF16B, FUT4, FKBP1A, and RIN2 was associated with PDR incidence, supporting roles for epigenetic regulation of iron clearance, developmental pathways, and autophagy in PDR pathogenesis. Further study of those loci may provide insight into novel targets for interventions to prevent or delay PDR in T1D.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: DNA methylation, Epigenetics, Proliferative diabetic retinopathy, Retina, Type 1 diabetes
Despite the widespread adoption of intensive insulin therapy and subsequent improvements in glycemic control over the past 25 to 30 years, proliferative diabetic retinopathy (PDR) remains common in people with type 1 diabetes (T1D). By 30 years’ duration of T1D, prevalence of PDR is estimated to be > 30%.1 Proliferative diabetic retinopathy is a leading cause of vision loss; thus, preventing its occurrence or slowing its progression is imperative. While there is a strong relationship between higher glycemic exposure and PDR,2 even people with hemoglobin A1c (HbA1c) at or below the current clinical target of 7% can develop it.3 Thus, there remains a critical need for novel markers to identify people at increased risk for PDR. Such markers may also improve understanding of the pathophysiology of PDR and aid in finding new intervention targets to prevent or delay its development.
DNA methylation (DNAm) provides a link between genetic risk and environmental or lifestyle exposures and its study may reveal insights into novel mechanistic pathways to complex diseases like PDR. Furthermore, DNAm has potential to be pharmacologically modified.4 To our knowledge, there has only been 1 epigenome-wide association study (EWAS) of PDR in T1D to date.5 In that cross-sectional study comparing 28 PDR cases to 30 controls, with DNAm measured in whole blood, the authors identified CpGs in loci related to inflammation, retinal development, oxidative stress, and other diabetes complications. However, their study had limited data on clinical risk factors and was unable to adjust for important confounders of DNAm-outcome associations such as smoking and blood cell composition. The importance of incorporating known clinical risk factors to discern the potential independent contribution of DNAm to T1D complication risk is apparent from more recent reports. For example, although a full EWAS of PDR has not been performed in the Diabetes Complications and Control Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications study, the investigators found DNAm at HbA1c-associated candidate CpG cg19693031 explained 41% of the association between HbA1c and PDR.6 Additionally, in the Pittsburgh Epidemiology of Diabetes Complications (EDC) study of childhood-onset T1D, the same cohort examined in the current analysis, we observed that associations between DNAm and cardiovascular disease were only modestly attenuated after adjustment for traditional cardiometabolic risk factors, suggesting epigenetic regulation of the identified loci may make an independent contribution to future cardiovascular disease risk.7
Given the prior evidence that DNAm may influence the risk of T1D complications, we hypothesized that DNAm is prospectively associated with risk of PDR independent of traditional risk factors, including HbA1c, in T1D. To test our hypothesis, we performed an EWAS of 28-year PDR incidence and examined whether the identified associations remained significant after adjustment for traditional clinical risk factors. We additionally sought to identify genetic variants associated with DNAm at significant CpGs and examined functional data to elucidate potential pathophysiologic pathways underlying PDR.
Methods
Study Population
Data were from the Pittsburgh EDC study, a prospective cohort study of childhood-onset (<17 years old) T1D (n = 658).8 Participants were followed 1986 to 1988 to 2016 to 2018. DNA was collected at study visits between 1988 and 1998, with 86% of the DNA specimens collected at the 1988 to 1990 visit, 9% at the 1990 to 1992 visit, and the remaining 5% between 1992 and 1998. A diagram detailing the derivation of the total analytic sample of n = 411 participants is shown in Figure S1 (available at https://www.aaojournal.org). Research protocols were approved by the University of Pittsburgh institutional review board (approval #19040065). All participants provided written informed consent, and the research adhered to the tenets of the Declaration of Helsinki.
DNAm Arrays, Quality Control, and Data Processing
The methylation arrays, methylation data quality control (QC) and data processing have previously been described in detail.7 Briefly, high molecular weight DNA was isolated from whole blood-derived leukocytes. DNAm was assayed using Illumina Infinium MethylationEPIC BeadChip arrays (Illumina).9 We implemented QC in 2 stages: first using a standard QC pipeline in minfi v1.32.010 and then using a second pipeline to confirm and expand QC in SeSAMe v.1.8.10,11 both in R v4.1.0 (R Core Team 2021). Of the 865 918 probes on the EPIC array, we dropped a previously published curated exclusion set12 of 95 923 and an additional set of 72 868 poor quality probes with detection rate < 95% in all samples, resulting in a final analytic set of 683 597 probes mapped to autosomal chromosomes. The final methylation fraction β values for analysis were generated using SeSAMe11 as previously described.7 For each methylation probe we excluded β values > ±3 standard deviations from the mean to remove extreme outliers prior to analysis. Cell type composition was estimated using the estimateCellCounts2 function from the R package FlowSorted.Blood.EPIC v1.5.2.13
Assessment of PDR and Clinical Risk Factors
Stereoscopic fundus photographs were taken in fields 1, 2, and 4 using a Zeiss camera (Carl Zeiss) at each study visit. The photos were graded at the Fundus Photography Reading Center, University of Wisconsin, Madison, using the modified Airlie House System.14 PDR was defined as the first instance of a grade of ≥60 in at least 1 eye or pan-retinal photocoagulation for PDR. Follow-up time was calculated from the study visit at which DNAm data were available (baseline) until complication incidence or censoring (December 31, 2018 or last follow-up).
Each participant’s clinical risk factor data were taken from the same study visit when their DNAm data were available. Details regarding ascertainment of the clinical measures have been published previously.7 Fasting blood samples were obtained to measure HbA1, lipids, and serum creatinine. HbA1 values were converted to DCCT-aligned HbA1c values using a regression equation derived from duplicate assays (DCCT HbA1c: 0.14 + 0.83 [EDC HbA1]).15 Total cholesterol and triglycerides were determined enzymatically and high-density lipoprotein (HDL) cholesterol was determined using a modified precipitation technique.16 Non-HDL cholesterol was calculated by subtracting HDL cholesterol from total cholesterol. Height and weight were measured using standard methods to calculate body mass index. Blood pressure was measured according to the Hypertension Detection and Follow-Up protocol with a random-zero sphygmomanometer.17 Hypertension (HTN) was defined as blood pressure > 140/90 or reported use of blood pressure lowering medication for indication of HTN or high blood pressure. Pulse rate (beats per minute) was determined by palpitating the radial pulse for 30 seconds and multiplying by 2. To assess albuminuria, 3 timed urine specimens (24-hour, overnight, and 4-hour) were collected during the 2 weeks before each study visit. Albumin excretion rate (AER) was calculated for each specimen; the median of the 3 AER was used in analysis. Serum creatinine was measured using an Ectachem 400 Analyzer (Eastman Kodak Co.). Glomerular filtration rate was estimated (estimated glomerular filtration rate [eGFR]) using the Chronic Kidney Disease Epidemiology Collaboration creatinine equation.18 Smoking and insulin regimen were self-reported via questionnaire. Insulin dose was calculated as total insulin units per day divided by body weight (kg).
PDR EWAS and Clinical Risk Factor Associations
After excluding prevalent cases of PDR at baseline, 265 were eligible for analysis (Figure S1, available at https://www.aaojournal.org). A time-to-event EWAS for PDR incidence was performed using Cox regression. Each CpG probe β-value was modeled as the main independent variable, adjusting for T1D duration, sex, pack years of smoking, cell type composition variables, plate/run number, well position, green CpC to TpC bisulfite score, and DNA extraction method. Because the identification of genetic variants associated with DNAm was a prespecified aim of our study, the first 2 ancestry principal components based on GWAS data 19 were also included as covariates. CpGs with a Benjamini-Hochberg false discovery rate (FDR) < 0.05 were considered statistically significant. The EDC is an exclusively childhood-onset (<17 years) T1D cohort; thus, age and T1D duration are highly correlated (r = 0.86; P < 0.0001). Because T1D duration is the exposure of greater interest in the current analysis, the results we present were adjusted for T1D duration only. However, results remained the same in alternative models adjusting for age instead of T1D duration. Because the conventional λ can overestimate test statistic inflation in EWAS, we used the bacon method (λ.bacon) developed specifically for EWAS 20 to assess evidence of inflation.
We assessed cross-sectional associations between significant CpGs and continuous baseline clinical risk factors using linear regression. Risk factors were HbA1c, body mass index, HDL cholesterol, non-HDL cholesterol, triglycerides, systolic blood pressure (SBP) and diastolic blood pressure, HTN, pulse rate, AER, and eGFR. For each significant CpG, we re-fit the corresponding Cox model, adjusting for the identified CpG-associated clinical risk factors, to obtain risk factor independent estimates of DNAm-PDR associations. CpG x risk factor interaction terms were also assessed. We also assessed whether significant CpGs were associated with longitudinal risk factors over the subsequent 28 years using linear mixed models. Models of the form were fit for each significant CpG probe, where Y is the postbaseline longitudinal risk factor for subject s at time t, S0s is the subject-specific random intercept offset, and est is the subject-specific error term. Models were adjusted for T1D duration, sex, pack years of smoking, and cell type composition (denoted as β1X1 through βjXj in the equation). Model residuals were plotted and visually examined to assess fit.
Identification of Methylation Quantitative Trait Loci and Mendelian Randomization
For each significant CpG (FDR< 0.05), we identified methylation quantitative trait loci (meQTLs) via GWAS using existing imputed genotyping array data in the EDC cohort,19 applying a genome-wide significance cut-off of P < 5 × 10−8. Linkage disequilibrium (LD)–based clumping was performed in PLINK v1.90.b6.24 to select the top single nucleotide polymorphisms (SNPs) from the LD block. Results were compared to the Genetics of DNA Methylation Consortium database of meQTLs 21 and the Human Whole Blood meQTL Atlas from the Chronic Renal Insufficiency Cohort.22 We examined evidence of the meQTLs’ effects on gene expression by determining whether the variants were annotated as whole blood expression quantitative trait loci (eQTLs) in the Genotype-Tissue Expression (GTEx) project database (data obtained from the GTEx Portal 30 May 2023).
We used Mendelian randomization (MR) to examine a potential causal association between DNAm and PDR. As large GWAS data for PDR in T1D are limited, 2-sample MR was not feasible, so we performed a 1-sample MR analysis using individual-level data from the EDC cohort in the R package OneSampleMR v0.1.3.23 The causal log odds of PDR associated with each 5% increase in DNAm was estimated using 2-stage predictor substitution estimators with a logit link and the representative SNP as the instrumental variable (IV).24 Mendelian randomization models were adjusted for the same covariates as the main EWAS. Model assumptions were assessed using Hansen J-test.
Differentially Methylated Region and Post Hoc Functional Analyses
Differentially methylated regions (DMRs) were identified using the Enmix-comb method in the ENmix v1.28.8 Bioconductor package for R.25 The P values from the EWAS, Chromosome and CpG start and end positions were provided as inputs to the combp function, using a region size of 1000, bin size of 310, and seed of 0.05. DMRs containing < 3 CpGs were excluded. To account for multiple comparisons, a Šidák value of < 0.05 was considered significant.
Loci containing individual CpGs with FDR < 0.05 or significant DMRs (Šidák value < 0.05) were considered. We performed Gene Set Enrichment Analysis based on gene ontology and Kyoto Encyclopedia of Genes and Genomes pathways using the Database for Annotation, Visualization, and Integrated Discovery 2021.26 We also identified a Reactome Functional Interaction network 27 with clustered modules28 using Cytoscape.29 We performed Reactome pathway analysis on the resulting modules with ≥5 total nodes.
Results
The 28-year incidence of PDR was 52.5% (139 of 265), with a median PDR-free survival time of 16 years after the study baseline (interquartile range 7–28 years). Baseline characteristics overall and by PDR incidence status are in Table 1. The 10 most statistically significant CpGs for PDR are shown in Table 2. Only cg27512687 in KIF16B (P = 6.27 × 10−9; FDR < 0.01) reached the significance threshold of FDR < 0.05. There was no evidence of meaningful inflation or deflation of the EWAS test statistics (λ.bacon = 1.04). To gain additional insight into potential pathways through which DNAm at cg27512687 may influence risk of PDR, we assessed associations between cg27512687 and traditional clinical risk factors (Table S3, available at https://www.aaojournal.org). In cross-sectional analysis, greater DNAm of cg27512687 was inversely associated with pulse rate and ln(AER) and positively associated with eGFR. After adjusting for those risk factors, the cg27512687-PDR association effect size was reduced by 4.3% but remained significant (log[hazard ratio] = −2.68, standard error = 0.50, P = 6.38 × 10−8). There was a significant interaction between cg27512687 and SBP (P = 0.0002) with respect to PDR, such that DNAm of cg27512687 was more strongly protective against PDR with lower SBP. There were no significant interactions between cg27512687 and the other risk factors, including HbA1c. Associations between cg27512687 and subsequent longitudinal risk factors were similar to those observed at baseline, except that cg27512687 in addition to being associated with lower pulse rate and ln(AER) and higher eGFR over follow-up, cg27512687 was also associated with lower SBP (Table S4, available at https://www.aaojournal.org).
Table 1.
Baseline Characteristics of EDC Study Participants by 28-Year Microvascular Complication Incidence Status
| Proliferative Retinopathy |
||
|---|---|---|
| Yes (n = 139) | No (n = 126) | |
| Age, yrs | 27.7 (7.3) | 26.5 (8.2) |
| Type 1 diabetes duration, yrs | 18.8 (6.4) | 18.3 (7.4) |
| Age at type 1 diabetes onset, yrs | 8.9 (4.0) | 8.2 (4.4) |
| Female sex, % (n) | 46.8% (65) | 49.2% (62) |
| Bachelor’s degree and/or beyond, % (n) | 35.3% (49) | 37.3% (47) |
| HbA1c, % | 9.5 (1.5) | 8.5 (1.4) |
| HbA1c, mmol/mol | 79.9 (16.9) | 69.4 (15.2) |
| Smoking, pack-yrs∗ | 0 (0–2.4) | 0 (0–0) |
| Body mass index, kg/m2 | 24.3 (3.3) | 23.6 (3.3) |
| Insulin dose, insulin units/kg body weight | 0.8 (0.2) | 0.8 (0.3) |
| MDI† or insulin pump use, % (n) | 9.4% (13) | 16.7% (21) |
| Self-monitoring of blood glucose, % (n) | 69.1% (96) | 77.0% (97) |
| Total cholesterol (mg/dl) | 192.9 (42.5) | 172.1 (41.0) |
| HDLc (mg/dl) | 54.5 (12.6) | 53.6 (13.0) |
| Non-HDLc (mg/dl) | 138.4 (41.6) | 118.4 (38.2) |
| Triglycerides (mg/dl)∗ | 81 (56–119) | 67 (51–108) |
| Systolic blood pressure (mmHg) | 113.7 (15.3) | 109.2 (11.7) |
| Diastolic blood pressure (mmHg) | 73.0 (10.0) | 69.6 (8.9) |
| Hypertension, % (n) | 11.5% (16) | 3.2% (4) |
| Pulse rate, bpm | 75.9 (10.5) | 72.8 (10.1) |
| Albumin excretion rate, μg/min∗ | 11.0 (6.6–49.4) | 7.4 (4.7–13.5) |
| Estimated clomerular filtration rate, ml/min/1.73 m2 | 120.5 (26.6) | 120.2 (28.6) |
| White blood cell count, ×109 cells/l | 6.7 (1.9) | 6.6 (2.0) |
EDC = Epidemiology of Diabetes Complications; HbA1c = hemoglobin A1c; MDI = multiple daily injections.
Participants with prevalent PDR at baseline were excluded. Values are mean (SD) unless specified.
Median (p25, p75).
Multiple daily injections (≥ 3 insulin injections per day).
Table 2.
DNA Methylation and 28-Year Incidence of Proliferative Retinopathy in the EDC Cohort: The 10 Most Statistically Significant CpGs Sorted by Ascending P Value
| CpG | Chr | hg38 Position | Location | Gene | Location Relative to Gene | Log (HR) per 5% Methylation | SE | P Value | FDR |
|---|---|---|---|---|---|---|---|---|---|
| cg27512687 | 20 | 16279513 | Open Sea | KIF16B | Body | −2.798 | 0.482 | 6.27E-09 | < 0.01 |
| cg04202206 | 9 | 86282804 | Island | ISCA1 | TSS1500 | −2.436 | 0.481 | 4.00E-07 | 0.14 |
| cg14678509 | 1 | 34532068 | Open Sea | n/a | Intergenic | −0.833 | 0.167 | 6.34E-07 | 0.14 |
| cg19776580 | 22 | 23898936 | S. Shelf | MIF-AS1 | TSS200 | −1.087 | 0.229 | 1.99E-06 | 0.21 |
| cg21665700 | 20 | 2450393 | Open Sea | n/a | Intergenic | −0.901 | 0.191 | 2.51E-06 | 0.21 |
| cg06825886 | 1 | 30775696 | S. Shelf | n/a | Intergenic | −0.713 | 0.152 | 2.64E-06 | 0.21 |
| cg04399632 | 16 | 11743690 | S. Shore | TXNDC11 | TSS1500 | −1.141 | 0.246 | 3.39E-06 | 0.21 |
| cg11843868 | 5 | 158382704 | Open Sea | LOC1019227697 | Body | −0.810 | 0.174 | 3.45E-06 | 0.21 |
| cg20073831 | 6 | 30466458 | Island | n/a | Intergenic | −1.803 | 0.389 | 3.55E-06 | 0.21 |
| cg06644457 | 10 | 133164642 | Island | KNDC1 | Body | −0.532 | 0.115 | 3.81E-06 | 0.21 |
Bolded text indicates CpGs with FDR < 0.05.
Chr = chromosome; EDC = Epidemiology of Diabetes Complications; FDR = false discovery rate; HR = hazard ratio; SE = standard error.
meQTLs and MR
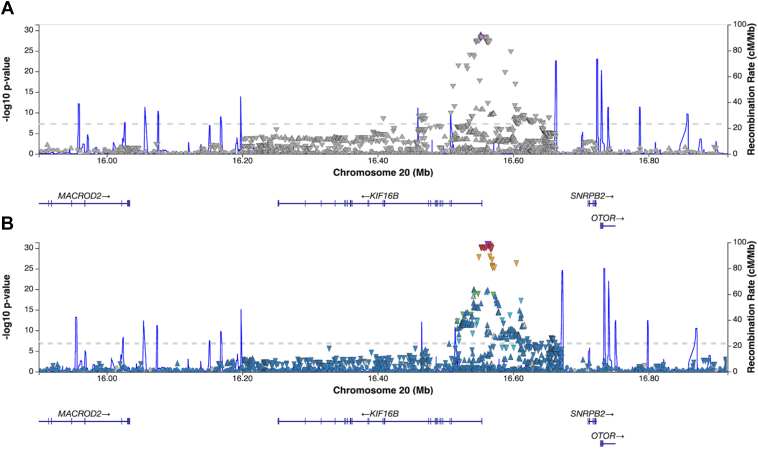
We identified 113 cis variants in the KIF16B region of chromosome 20 that were significantly (P < 5 × 10−8, genomic inflation factor λ = 1.00) associated with DNAm at cg27512687 (Figure 2, panel A and Table S5, available at https://www.aaojournal.org). There were no significant meQTLs for cg27512687 in the Genetics of DNA Methylation Consortium database, but 126 SNPs in the KIF16B region were significant meQTLs for cg27512687 in the Chronic Renal Insufficiency Cohort. Furthermore, 96 of the 113 variants identified as meQTLs in EDC were also annotated as whole blood eQTLs in GTeX (Figure 2, panel B).
Figure 2.
LocusZoom plots of genetic variants significantly associated with methylation of cg27512687 in the Epidemiology of Diabetes Complications cohort (panel A) and variants annotated as eQTLs in GTEx in the same genomic region (panel B).
As the identified significant cis-meQTLs/eQTLs support a possible functional role for cg27512687 DNAm on PDR development, we performed an exploratory MR analysis to assess evidence of a causal association between cg27512687 and PDR. After clumping, 13 SNPs were selected as representative of the 113 variants identified as meQTLs in the KIF16B LD block. Of those, rs35834087 was the most strongly associated with cg27512687 (P = 2.97 × 10−29), and thus was selected as the IV. The MR estimated odds ratio for PDR associated with each 5% methylation of cg27512687 was 0.029 (95% confidence interval: 0.002, 0.330), suggesting a significant causal protective effect.
DMRs and Post Hoc Functional Analyses
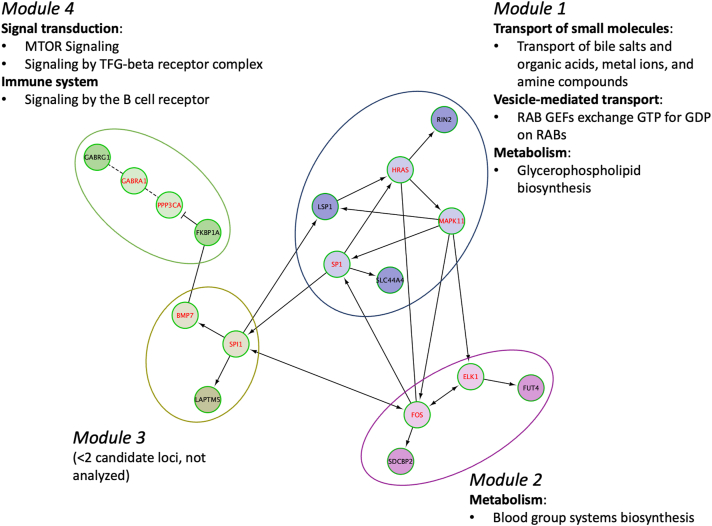
Detailed results of the DMR analyses are shown in Table 6. Based on a Šidák value < 0.05, we identified 17 significant DMRs for PDR, 15 of which were annotated to a gene(s), including ACY3, CDK2AP1, FKBP1A, FUT4, GABRG1, GCSAML, LAPTM5, LINC00028, LINC00649, LSP1, NOS1AP, RIN2, SDCBP2, SDHAP3, SLC44A4, and VSTM5. Those loci and KIF16B were further examined in functional analyses. There were no significantly enriched gene ontology terms or Kyoto Encyclopedia of Genes and Genomes pathways; however, we identified a Reactome Functional Interaction network comprising 4 modules in which a total of 7 subpathways were significantly enriched within transport of small molecules, vesicle-mediated transport, metabolism, signal transduction, and immune system top-level pathways (Figure 3 and Table S7, available at https://www.aaojournal.org).
Table 6.
Significant Differentially Methylation Regions Associated with Proliferative Retinopathy Incidence in the EDC Cohort
| Chr | hg38 Start Position | hg38 End Position | # CpGs in the DMR | Location | Gene | Location Relative to Gene | P Value | FDR | Šidák Value |
|---|---|---|---|---|---|---|---|---|---|
| 11 | 67650487 | 67650895 | 9 | Open Sea | ACY3 | TSS200, TSS1500, 5'UTR | 4.49E-13 | 1.21E-11 | 7.52E-10 |
| 11 | 93850343 | 93850808 | 7 | Island, S. Shore | VSTM5 | TSS200, TSS1500, 1stExon | 2.22E-09 | 2.00E-08 | 3.26E-06 |
| 6 | 31180555 | 31180890 | 14 | Island, S. Shore | Intergenic | n/a | 4.37E-09 | 2.95E-08 | 8.92E-06 |
| 5 | 1594561 | 1594619 | 4 | Island | SDHAP3 | TSS200 | 1.50E-09 | 2.00E-08 | 1.77E-05 |
| 7 | 1505886 | 1506184 | 4 | S. Shore | Intergenic | n/a | 9.36E-09 | 3.61E-08 | 2.15E-05 |
| 11 | 94545241 | 94545438 | 6 | Island | FUT4 | 1stExon | 1.32E-08 | 4.46E-08 | 4.59E-05 |
| 20 | 31485596 | 31485774 | 5 | S. Shore | LINC00028 | TSS200 | 2.23E-08 | 6.70E-08 | 8.58E-05 |
| 20 | 1336956 | 1337103 | 3 | Open Sea | FKBP1A-SDCBP2; SDCBP2-AS1 | Body | 3.56E-08 | 9.62E-08 | 0.0002 |
| 4 | 46124049 | 46124357 | 6 | Open Sea | GABRG1 | TSS200, TSS1500 | 1.03E-07 | 2.52E-07 | 0.0002 |
| 11 | 1890948 | 1890957 | 3 | Open Sea | LSP1 | 3'UTR | 7.81E-09 | 3.51E-08 | 0.0006 |
| 20 | 19889335 | 19889574 | 3 | Open Sea | RIN2 | TSS200, 1stExon, 5'UTR | 2.17E-07 | 4.53E-07 | 0.0006 |
| 6 | 31878992 | 31879252 | 8 | Open Sea | SLC44A4 | TSS200, TSS1500, 1stExon, 5'UTR | 2.56E-07 | 4.93E-07 | 0.0007 |
| 1 | 30758367 | 30758575 | 3 | Open Sea | LAPTM5 | TSS1500 | 2.18E-07 | 4.53E-07 | 0.0007 |
| 1 | 162366828 | 162367088 | 4 | N. Shore | NOS1AP | Body | 4.63E-07 | 7.36E-07 | 0.0012 |
| 12 | 123268081 | 123268370 | 7 | N. Shore | CDK2AP1 | 5'UTR, TSS200, 1stExon, Body | 7.01E-07 | 1.05E-06 | 0.0016 |
| 21 | 33948292 | 33948364 | 3 | Open Sea | LINC00649 | TSS1500, Body | 4.01E-07 | 6.85E-07 | 0.0038 |
| 1 | 247518279 | 247518480 | 5 | Island | GCSAML | TSS200, 1stExon, 5'UTR | 6.53E-06 | 9.28E-06 | 0.0220 |
Chr = chromosome; DMR = differentially methylated region; EDC = Epidemiology of Diabetes Complications; FDR = false discovery rat.
Figure 3.
Reactome Functional Interaction network of loci with significant CpGs or differentially methylated regions associated with 28-year proliferative retinopathy incidence in the Epidemiology of Diabetes Complications cohort. Candidate loci are indicated in black font (red font indicates linker genes used only to construct the network). Categories of significantly enriched (false discovery rate < 0.05) Reactome pathways for each module are noted. Solid line = involved in same reaction as inputs or are components of a shared complex, → = activator or catalyst, --| = inhibitor, dashed line = predicted interaction.
Discussion
In this prospective EWAS of PDR incidence in T1D, we observed that greater methylation of cg27512687 in KIF16B (also known as SNX23) was associated with decreased PDR incidence independent of established clinical risk factors, including HbA1c. These findings suggest epigenetic regulation of KIF16B may provide insight into novel pathways to PDR in T1D. In addition, we identified meQTLs for cg27512687 which were validated in an external diabetes cohort and annotated in GTEx as eQTLs in a wide variety of vascular tissues, neural tissues, and whole blood, supporting a possible functional role of cg27512687 DNAm in PDR development. The results of our MR analysis provide additional supporting evidence of a causal association between cg27512687 DNAm and PDR. In addition to cg27512687, we also identified several genomic regions where DNAm was associated with PDR. Those regions include FUT4, FKBP1A, and RIN2, genes with prior evidence of biologically plausible roles in PDR development, including retinal development,30 mTOR-depdendent autophagy,31 and VEGF signaling,32 respectively.
The KIF16B locus encodes Kinesin Family Member 16B, which is involved in receptor recycling and degradation, intracellular transport, and microtubule formation.33 There is evidence KIF16B is required for transport of basolateral transferrin receptor (TfR) from common recycling endosomes to apical recycling endosomes in the retinal pigment epithelium, thus it is likely KIF16B plays a role in preventing iron accumulation in the retina.34 Iron accumulation leads to oxidative damage and inflammation, both of which are involved in the underlying pathogenesis of PDR.35 Increased DNAm at gene bodies is generally associated with increased gene expression.36 Thus, the direction of the association we observed (i.e., greater methylation of cg27512687 in the KIF16B gene body is associated with lower risk of PDR) is consistent with the hypothesis that cg27512687 DNAm may increase KIF16B expression, facilitating increased TfR transport and greater iron removal from the retina, subsequently reducing PDR risk. Indeed, there are prior experimental data demonstrating transferrin protects against retinal degeneration.37 The relationship between cg27512687 and PDR in our study was independent of HbA1c; therefore, DNAm of KIF16B may protect against PDR regardless of glycemic exposure in T1D. Furthermore, our observation that DNAm at cg27512687 was more strongly protective against PDR in those with lower SBP suggests that protection could be offset by the deleterious effects of HTN. Because the concordance between DNAm in peripheral blood and retinal tissue is unclear,38 our study examining peripheral blood DNAm cannot demonstrate a causal role for cg27512687 DNAm PDR. However, our findings suggest further study of epigenetic regulation of KIF16B in retinal tissue is warranted.
In regional analyses, we identified several PDR-associated DMRs, some of which are annotated to genes that have prior animal and cell data supporting biologically plausible roles in retinopathy. They include FUT4, which encodes a protein that catalyzes synthesis of CD15, a cell surface marker expressed on photoreceptor precursors,30 thus raising the possibility that epigenetic regulation of prenatal photoreceptor development may affect future risk of PDR. We also observed associations with DNAm of the FKBP1A region, a gene that regulates autophagy via the mTOR signaling pathway. FKBP1A expression has been shown to be reduced in the retinal pigment epithelium of PDR cases vs. healthy controls,31 supporting the hypothesis that mTOR-dependent autophagy is a key mechanism underlying retinal degeneration.39 Finally, we observed associations with DNAm of RIN2, which is involved in angiogenesis and plays a critical role in VEGF signaling.32 Altogether, our observations suggest epigenetic regulation of specific genes involved in photoreceptor development, autophagy, and angiogenesis may contribute to PDR pathogenesis in T1D, supporting further study of the identified loci.
Our study has many strengths, including the use of data from a well-characterized T1D cohort with long-term follow-up that is epidemiologically representative of the childhood-onset T1D population of Allegheny County, Pennsylvania.40 Importantly, the prospective study design avoids the possibility of reverse causation, which was key limitation of the prior cross-sectional EWAS for PDR in T1D. A further strength is the availability of clinical risk factor data which allowed examination of intermediate phenotypes between DNAm and PDR. Another strength was the use of a cis-meQTL, which was validated in an external diabetes cohort, as the IV in the MR analysis, increasing the biological plausibility of our findings that cg27512687 may play a causal role in PDR development.
Limitations include the use of whole blood for methylation measurement and lack of tissue-specific data. However, DNAm in whole blood is commonly examined in epidemiologic studies such as ours, due to ease of specimen collection and because it facilitates detection of multiple physiologic pathways that lead to complex phenotypes like PDR. The sample size of our study is relatively small, so the results should be validated as more DNAm data become available in T1D cohorts. Because of a lack of available large GWAS for PDR in T1D, we performed a 1-sample MR which carries limitations of potential overfitting and bias if the IV-exposure association is weak; thus, the MR should be replicated in larger studies using a 2-sample approach to validate our findings. Finally, 98% of the EDC cohort is of white/European ancestry, because of the demographics of Allegheny County, Pennsylvania, USA, (< 15% black/African American) and historically lower incidence of T1D among black individuals,41 so our results may not apply to more diverse populations.
Conclusions
Our prospective EWAS provides novel evidence that epigenetic regulation of KIF16B is associated with long-term risk of PDR in T1D, independent of established clinical risk factors, possibly via regulation of TfR transport. In addition, the results of our regional analyses support a role for epigenetic regulation of specific genes involved in development (FUT4), autophagy (FKBP1A), and angiogenesis (RIN2) in PDR pathogenesis. Further study of the identified loci may provide insight into novel targets for interventions to prevent or delay PDR in T1D.
Manuscript no. XOPS-D-23-00298.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s):
R.G.M.: Grant or contracts from any entity - American Diabetes Association (7-23-ICTSWH-19) and National Heart Lung, and Blood Institute (R01HL161879); Honorarium for lecture—American Diabetes Association; Complementary meeting registration as compensation for lecture—American Diabetes Association.
T.C.: Grants or contracts from any entity—R01HL161879, DK094157, HL151328.
The parent EDC study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (Grant #R01-DK034818) and the Rossi Memorial Fund (Pittsburgh, PA). The EDC DNA methylation substudy was supported by the American Diabetes Association (Grant #1-19-JDF-109).
HUMAN SUBJECTS: Human subjects were included in this study. Research protocols were approved by the University of Pittsburgh institutional review board (approval #19040065). All participants provided written informed consent and the research adhered to the tenets of the Declaration of Helsinki.
No animal subjects were used in this study.
Author Contributions:
Research design: Miller, Mychaleckyj, Onengut-Gumuscu, Orchard, Costacou
Data Acquisition and research execution: Miller, Mychaleckyj, Onengut-Gumuscu, Orchard, Costacou
Data analysis and interpretation: Miller, Mychaleckyj, Onengut-Gumuscu, Orchard, Costacou
Manuscript preparation: Miller, Mychaleckyj, Onengut-Gumuscu, Orchard, Costacou
Preliminary data from this manuscript were presented at the American Diabetes Association’s 83rd Scientific Sessions, June 2023, San Diego, CA (Abstract #1461P).
Supplementary Data
References
- 1.Fang M., Echouffo-Tcheugui J.B., Selvin E. Burden of complications in U.S. adults with young-onset type 2 or type 1 diabetes. Diabetes Care. 2020;43:E47–E49. doi: 10.2337/dc19-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein R., Knudtson M.D., Lee K.E., et al. The Wisconsin epidemiologic study of diabetic retinopathy: XXII the twenty-five-year progression of retinopathy in persons with type 1 diabetes. Ophthalmology. 2008;115:1859–1868. doi: 10.1016/j.ophtha.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Krzentowski G., Albert A., Lefebvre P.J. Risk of developing retinopathy in diabetes control and complications trial type 1 diabetic patients with good or poor metabolic control. Diabetes Care. 2001;24:1275–1279. doi: 10.2337/diacare.24.7.1275. [DOI] [PubMed] [Google Scholar]
- 4.Liu X.S., Wu H., Ji X., et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agardh E., Lundstig A., Perfilyev A., et al. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015;13:182. doi: 10.1186/s12916-015-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z., Miao F., Braffett B.H., et al. DNA methylation mediates development of HbA1c-associated complications in type 1 diabetes. Nat Metab. 2020;2:744–762. doi: 10.1038/s42255-020-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller R.G., Mychaleckyj J.C., Onengut-Gumuscu S., et al. DNA methylation and 28-year cardiovascular disease risk in type 1 diabetes: the Epidemiology of Diabetes Complications (EDC) cohort study. Clin Epigenetics. 2023;15:122. doi: 10.1186/s13148-023-01539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orchard T.J., Dorman J.S., Maser R.E., et al. Prevalence of complications in IDDM by sex and duration. Pittsburgh epidemiology of diabetes complications study II. Diabetes. 1990;39:1116–1124. doi: 10.2337/diab.39.9.1116. [DOI] [PubMed] [Google Scholar]
- 9.Moran S., Arribas C., Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aryee M.J., Jaffe A.E., Corrada-Bravo H., et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W., Triche T.J., Laird P.W., Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018;46:e123. doi: 10.1093/nar/gky691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W., Laird P.W., Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45:e22. doi: 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salas L.A., Koestler D.C., Butler R.A., et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19:1–14. doi: 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein R., Klein B.E., Magli Y.L., et al. An alternative method of grading diabetic retinopathy. Ophthalmology. 1986;93:1183–1187. doi: 10.1016/s0161-6420(86)33606-6. [DOI] [PubMed] [Google Scholar]
- 15.Prince C.T., Becker D.J., Costacou T., et al. Changes in glycaemic control and risk of coronary artery disease in type 1 diabetes mellitus: findings from the Pittsburgh Epidemiology of Diabetes Complications Study (EDC) Diabetologia. 2007;50:2280–2288. doi: 10.1007/s00125-007-0797-7. [DOI] [PubMed] [Google Scholar]
- 16.Warnick G.R., Albers J.J. Heparin--Mn2+ quantitation of high-density-lipoprotein cholesterol: an ultrafiltration procedure for lipemic samples. Clin Chem. 1978;24:900–904. [PubMed] [Google Scholar]
- 17.The hypertension detection and follow-up program. Hypertension detection and follow-up program cooperative group. Prev Med. 1976;5:207–215. doi: 10.1016/0091-7435(76)90039-6. [DOI] [PubMed] [Google Scholar]
- 18.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salem R.M., Todd J.N., Sandholm N., et al. Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J Am Soc Nephrol. 2019;30:2000–2016. doi: 10.1681/ASN.2019030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Iterson M., van Zwet E.W., Heijmans B.T., et al. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18:1–13. doi: 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min J.L., Hemani G., Hannon E., et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53:1311–1321. doi: 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng X., Qiu C., Liu H., et al. Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease. Proc Natl Acad Sci U S A. 2020;117:29013–29024. doi: 10.1073/pnas.2005905117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer T., Spiller W., Sanderson E. OneSampleMR: One Sample Mendelian Randomization and Instrumental Variable Analyses, May 5, 2023. https://CRAN.R-project.org/package=OneSampleMR
- 24.Terza J.V., Basu A., Rathouz P.J. Two-stage residual inclusion estimation: addressing endogeneity in health econometric modeling. J Health Econ. 2008;27:531–543. doi: 10.1016/j.jhealeco.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z., Xie C., Taylor J.A., Niu L. ipDMR: identification of differentially methylated regions with interval P-values. Bioinformatics. 2021;37:711–713. doi: 10.1093/bioinformatics/btaa732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman B.T., Hao M., Qiu J., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillespie M., Jassal B., Stephan R., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman M.E.J. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowski J., Welby E., Budinger D., et al. Isolation of human photoreceptor precursors via a cell surface marker panel from stem cell-derived retinal organoids and fetal retinae. Stem Cell. 2018;36:709–722. doi: 10.1002/stem.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N., Wei L., Liu D., et al. Identification and validation of autophagy-related genes in diabetic retinopathy. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.867600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempers L., Wakayama Y., van der Bijl I., et al. The endosomal RIN2/Rab5C machinery prevents VEGFR2 degradation to control gene expression and tip cell identity during angiogenesis. Angiogenesis. 2021;24:695–714. doi: 10.1007/s10456-021-09788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B.J., Chen H., Jiang S.S., et al. PX domain-containing Kinesin KIF16B and microtubule-dependent intracellular movements. J Membr Biol. 2020;253:101–108. doi: 10.1007/s00232-020-00110-9. [DOI] [PubMed] [Google Scholar]
- 34.Perez Bay A.E., Schreiner R., Mazzoni F., et al. The kinesin KIF16B mediates apical transcytosis of transferrin receptor in AP-1B-deficient epithelia. EMBO J. 2013;32:2125–2139. doi: 10.1038/emboj.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhary K., Promsote W., Ananth S., et al. Iron overload accelerates the progression of diabetic retinopathy in association with increased retinal renin expression. Sci Rep. 2018;8:3025. doi: 10.1038/s41598-018-21276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varley K.E., Gertz J., Bowling K.M., et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picard E., Le Rouzic Q., Oudar A., et al. Targeting iron-mediated retinal degeneration by local delivery of transferrin. Free Radic Biol Med. 2015;89:1105–1121. doi: 10.1016/j.freeradbiomed.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Wu J., Liu L.L., Cao M., et al. DNA methylation plays important roles in retinal development and diseases. Exp Eye Res. 2021;211 doi: 10.1016/j.exer.2021.108733. [DOI] [PubMed] [Google Scholar]
- 39.Madrakhimov S.B., Yang J.Y., Kim J.H., et al. mTOR-dependent dysregulation of autophagy contributes to the retinal ganglion cell loss in streptozotocin-induced diabetic retinopathy. Cell Commun Signal. 2021;19:29. doi: 10.1186/s12964-020-00698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller R.G., Secrest A.M., Sharma R.K., et al. Improvements in the life expectancy of type 1 diabetes: the Pittsburgh Epidemiology of Diabetes Complications Study Cohort. Diabetes. 2012;61:2987–2992. doi: 10.2337/db11-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laporte R.E., Tajima N., Dorman J.S., et al. Differences between blacks and whites in the epidemiology of insulin-dependent diabetes mellitus in Allegheny county, Pennsylvania. Am J Epidemiol. 1986;123:592–603. doi: 10.1093/oxfordjournals.aje.a114279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.