Abstract
The long-term evolution of the hepatitis C virus hypervariable region (HVR) and flanking regions of the E1 and E2 envelope proteins have been studied in a cohort of women infected from a common source of anti-D immunoglobulin. Whereas virus sequences in the infectious source were relatively homogeneous, distinct HVR variants were observed in each anti-D recipient, indicating that this region can evolve in multiple directions from the same point. Where HVR variants with dissimilar sequences were present in a single individual, the frequency of synonymous substitution in the flanking regions suggested that the lineages diverged more than a decade previously. Even where a single major HVR variant was present in an infected individual, this lineage was usually several years old. Multiple lineages can therefore coexist during long periods of chronic infection without replacement. The characteristics of amino acid substitution in the HVR were not consistent with the random accumulation of mutations and imply that amino acid replacement in the HVR was strongly constrained. Another variable region of E2 centered on codon 60 shows similar constraints, while HVR2 was relatively unconstrained. Several of these features are difficult to explain if a neutralizing immune response against the HVR is the only selective force operating on E2. The impact of PCR artifacts such as nucleotide misincorporation and the shuffling of dissimilar templates is discussed.
More than 80% of individuals infected by hepatitis C virus (HCV) become chronically infected (25), with outcomes varying from persistent asymptomatic infection to chronic hepatitis, cirrhosis, or hepatocellular carcinoma. This phenomenon distinguishes HCV from other members of the Flaviviridae such as yellow fever virus, dengue virus, or pestiviruses, which do not normally establish persistent infections, but the reason for this difference is unclear.
One possible mechanism for the establishment of persistent infections by HCV relates to the most variable portion of the HCV genome, the hypervariable region (HVR) at the NH2 terminus of the envelope protein E2 (9, 49). Nucleotide sequence analysis has revealed that many different HVR variants can be present within an infected individual (15, 46), that the relative proportion of each variant can change over time (8, 13), and that variation within the HVR tends to accelerate as disease progresses, with few substitutions occurring during acute infection (26, 30, 35, 51). These observations are consistent with the idea that amino acid substitution of the HVR allows variants to evade neutralizing immune responses, thus leading to persistent infection.
This theory is supported by the observation that antibody to the HVR is produced in the majority of viremic individuals (36, 54), suggesting that the HVR is a major immunogenic domain of E2. Antibodies can be specific for different HVR variants (22), and new specificities develop after the emergence of new dominant HVR variants (1, 16, 39). The resolution of acute infection has been associated with an early antibody response to the HVR (1), specifically to the NH2 terminus (53), whereas antibodies directed against the COOH terminus of the HVR coexist with the virus in chronically infected individuals. Protection against infection with HCV has been demonstrated in chimpanzees by using a hyperimmune serum against the HVR (7), although no protection was conferred against some HVR variants present in the challenge inoculum. There is also evidence that in the absence of an antibody response to the HVR, variation of the HVR is reduced (19, 52). In an experimentally infected chimpanzee, variation of the HVR occurred only after a delay of 6 years and after the appearance of anti-HVR antibodies (48).
However, direct evidence for neutralization of HCV by anti-HVR antibodies has been difficult to obtain in the absence of an efficient in vitro culture system. Anti-HVR antibodies produced during acute infection have been shown to block viral attachment to tissue culture cells (54), but in most cases anti-HVR antibodies seem to coexist with the HVR variants that they recognize and are frequently cross-reactive with epidemiologically unrelated HVR sequences (22, 36, 53, 54). Alternative explanations for the generation of diversity in the HVR are that different HVR variants have tropisms for particular tissues (37) or that this region is simply less functionally constrained than other parts of the genome (40). Some of the variation observed in individuals infected with multiple HVR variants might be preexisting in the infectious source, and different variants might become dominant as infection progresses and different foci of infection become active. In each of these explanations, variation of the HVR would be an effect of persistent infection rather than a cause. Finally, the importance of the cellular immune response in selecting HVR variants is uncertain. The cytotoxic T-lymphocyte response in the hepatic parenchyma has been reported to recognize variable regions of the envelope and nonstructural proteins (17). Proliferation studies have shown that peripheral CD4+ T lymphocytes from chronically infected patients recognize the carboxyl terminus of core and less frequently E1, E2, and NS3 (21).
Our understanding of the HVR in HCV infection has also been hampered by a lack of information about the general characteristics of HVR evolution. For example, although there have been many studies of HVR evolution within a single infected individual or following a transmission event, there have been few studies in which virus evolution has been studied in parallel in several individuals infected from the same source (10). In addition, the extent of virus heterogeneity in the infectious source is often unknown, although this could clearly influence the complexity of the virus population during persistent infection. Two extreme scenarios following infection from a homogeneous source are (i) that different HVR variants appear in each infected individual or (ii) that functional constraints and shared selection pressures lead to the emergence of similar variants in different individuals.
This work aims to address these questions by studying virus evolution in a cohort of Irish women, infected in 1977 from a batch of anti-D immunoglobulin contaminated with HCV (32, 33). Sequence analysis of virus E1 and NS5B genes revealed that different anti-D recipients were all infected with a subtype 1b virus which was more similar to virus sequences present in an infective batch of immunoglobulin than to unrelated subtype 1b sequences (32, 44). Analysis of individual virus genomes by limiting dilution revealed relatively little variation of NS5B sequences within the infective batch (44). This is consistent with there being a single implicated donor who was probably acutely infected (6). All the infectious batches were manufactured with plasma collected over a period of 10 days from the implicated donor 2 months after first becoming jaundiced and who at that time had an incomplete pattern of serological reactivity consistent with acute infection. This cohort also has the advantage that it comprises women of a similar age group with the same duration of infection and who are not coinfected with other viruses or suffering from other chronic diseases. We have therefore been able to study the evolution of the HVR in parallel in different individuals and to investigate the types of constraints placed on its variation.
MATERIALS AND METHODS
Samples.
Plasma and serum samples were obtained in 1994 from Irish women who had been exposed to an HCV-contaminated batch of anti-D immunoglobulin in 1977 and were anti-HCV positive and HCV RNA positive for the 5′ noncoding region by reverse transcription-PCR. Archived samples of batches 238 and 250 were reconstituted with 2 ml DEPC-treated water and made up to 7.5 ml with RPMI medium. Virus was collected by ultracentrifugation at 100,000 g for 90 min.
Reverse transcription-PCR.
Virus RNA was extracted either from 0.1 ml of plasma or serum or, for four samples, from 0.5 to 2 ml by centrifugation at 100,000 × g at 4°C for 90 min. Virus RNA was extracted by incubation with proteinase K-polyadenylic acid-sodium dodecyl sulfate as reported previously (12). Synthesis of cDNA was carried out with 5 μl of extracted RNA and 10 U of avian myeloblastosis virus reverse transcriptase (Promega) in 20 μl of buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 5 mM dithiothreitol, 50 mM KCl, 0.05 μg of bovine serum albumin per μl, 15% dimethyl sulfoxide (DMSO), 600 μM each dGTP, dATP, dTTP, and dCTP, 1.5 μM primer, and 10 U of RNasin (Promega). A fragment containing the COOH terminus of E1 and an NH2-terminal region of E2 were amplified with primers 2174 (5′-TTCATCCAYGTRCASCCRAACCA-3′, antisense, positions 1645 to 1667 numbered from the AUG initiation codon) and 2173 (5′-CAYCGNATGGCNTGGGAYATGATG-3′, sense, positions 946 to 969) for the first round of PCR and primers 8914 (5′-CGGGATCCGGGTGCTCACTGGGGAGTCCTGGCGGGC-3′, sense, positions 1048 to 1074 incorporating a BamHI site) and 2070 (5′-GGAATTCGTGAARCARTACACYGGRCCRCANAC-3′, antisense, positions 1504 to 1529 incorporating an EcoRI site) for the second round of PCR. Optimized conditions for PCR amplification of the HVR involved 5 μl of cDNA and 30 cycles, each consisting of 0.6 min at 94°C, 0.7 min at 50°C, and 1.5 min at 72°C, with a final extension period of 6.5 min at 72°C. Reactions were carried out with 0.4 U of Taq DNA polymerase (Promega) in 50 μl of buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), Triton X-100, 1.5 mM MgCl2, 30 μM each dGTP, dATP, dTTP, and dCTP, and 0.25 μM each of the outer primers. For the second round of PCR, 1 μl of PCR product was transferred to a fresh tube containing 100 μl of reaction mix and subjected to 30 cycles of PCR as before.
Limiting-dilution PCR.
cDNA was diluted until PCR products were derived from single cDNA templates (41). Reaction conditions were as above except that the reaction mix contained 1% DMSO and the second round of PCR was for 40 cycles with primers 2172 (5′-CGGGATCCATGATGMTNAAYTGGTCNCC-3′, sense, positions 964 to 983) and 588 (5′-GGYGSGTARTGCCAGCARTANGG-3′, antisense, positions 1450 to 1472). For sequencing, the second round of PCR was repeated with one of the primers biotinylated. Single-stranded DNA was obtained by binding the PCR product to paramagnetic streptavidin-coated beads (Dynabeads; Dynal) and releasing the unlabelled strand by treatment with alkali. Dideoxynucleotide sequencing was performed with the Sequenase 2.0 enzyme (Amersham) in the presence of 10% DMSO.
Cloning and sequencing of PCR products.
PCR products and plasmid pUC18 were cleaved with both BamHI and EcoRI (Promega) and purified from 0.8% low-melting-point agarose (Gibco BRL). Ligations were performed with 50 ng of vector, 300 ng of PCR insert, and 5 U of T4 DNA ligase (Promega), and the mixtures were incubated at room temperature overnight. Ligation products were transformed into competent TG1 cells and plated on Luria agar plates containing 200 μg of ampicillin per ml, 10 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Plasmid DNA was extracted from transformants producing white colonies by alkali denaturation and sequenced by standard methods with the Sequenase 2.0 enzyme (Amersham) and primers DBS6 (5′-CACTGGGGAGTCCTGGCGGGC-3′, positions 1054 to 1074), S6645 (5′-TGCCARCTNCCRTTGGTRTT-3′, positions 1243 to 1262) and pUC reverse primer (5′-CAGGAAACAGCTATGAC-3′). Nucleotide sequences were entered, aligned, and checked with Simmonic Performance+ software (version 1.0, P. Simmonds).
Phylogenetic analysis.
Phylogenetic analysis was carried out with the Molecular Evolutionary Phylogenetic Analysis (MEGA) version 1.02 package (18). Evolutionary distances were calculated by the Kimura two-parameter method (all sites) or the Jukes-Cantor correction (synonymous sites). Sliding-window analysis of synonymous and nonsynonymous distances was carried out with the program Windows (11).
Previously published sequences have the following references or GenBank accession numbers: 1a sequences, references 15, 27, and 49, no. M62231, M62381, A27609, D10749, L16891, L19371 to L19374, L19376-L19378, L198380, L198381, M74804, M74805, M74808, M74811, M74812, S55848, S72725, U14232, U14233, U14239, U51791 to U51795, and X84079; 1b sequences, references 10 and 15, no. D50483-5, U45476, D30613, M58335, D001171, D10934, S62220, L02836, M96362, and U01214; 2a sequence, no. D00944; 2b sequence, no. D01221; 2c sequence, no. D50409; 3a sequences, references 3 and 5, no. D17763, D14311, Z68743, Z68742, and D28917; 3b sequence, no. D26556; “10a” sequence, no. D63821; 4a sequence, no. Y11604; 5a sequence, no. L29578, and Y13184; 6a sequence, no. Y12083; “11a” sequence, no. D63822.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences reported here are AF056733 to AF056925.
RESULTS
Homogeneity of the infectious source.
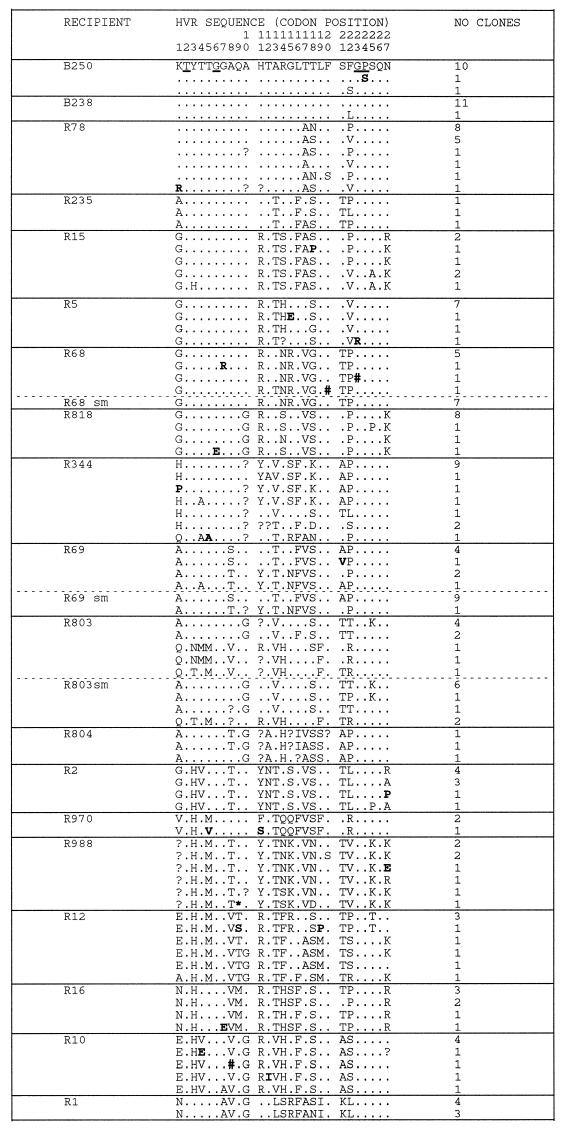
Nucleotide sequence analysis of the E1 and E2 envelope genes (positions 1096 to 1458) from two different infective batches of anti-D immunoglobulin revealed limited variation between virus genomes (Fig. 1, B250 and B238) with a mean evolutionary distance between clones of 0.008. Within the hypervariable region, amino acid sequences were identical in 21 clones and differed by single sporadic amino acid substitutions in 3 different clones (Fig. 2). These and seven other sporadic substitutions observed in the region flanking the HVR are likely to represent artifacts introduced by Taq DNA polymerase during PCR amplification (see Discussion). Irrespective of this interpretation, variation of the E1 and E2 genes, including the hypervariable region, was extremely limited among the virus genomes present in infective batches of anti-D immunoglobulin.
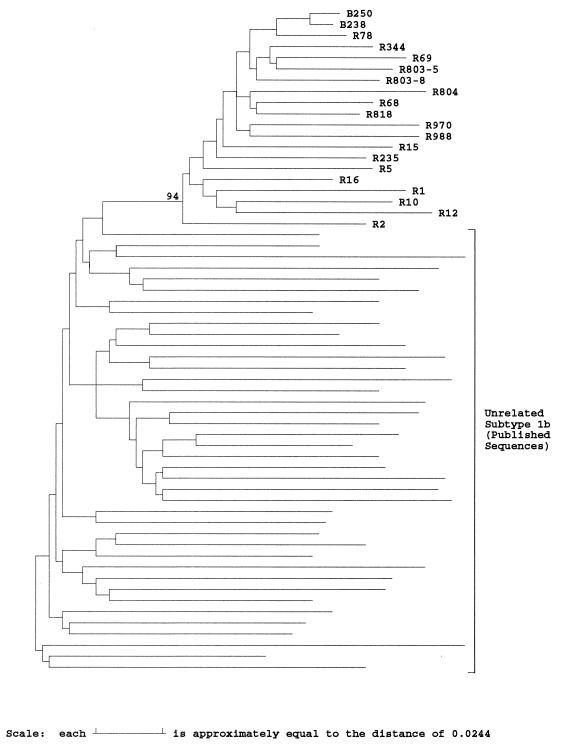
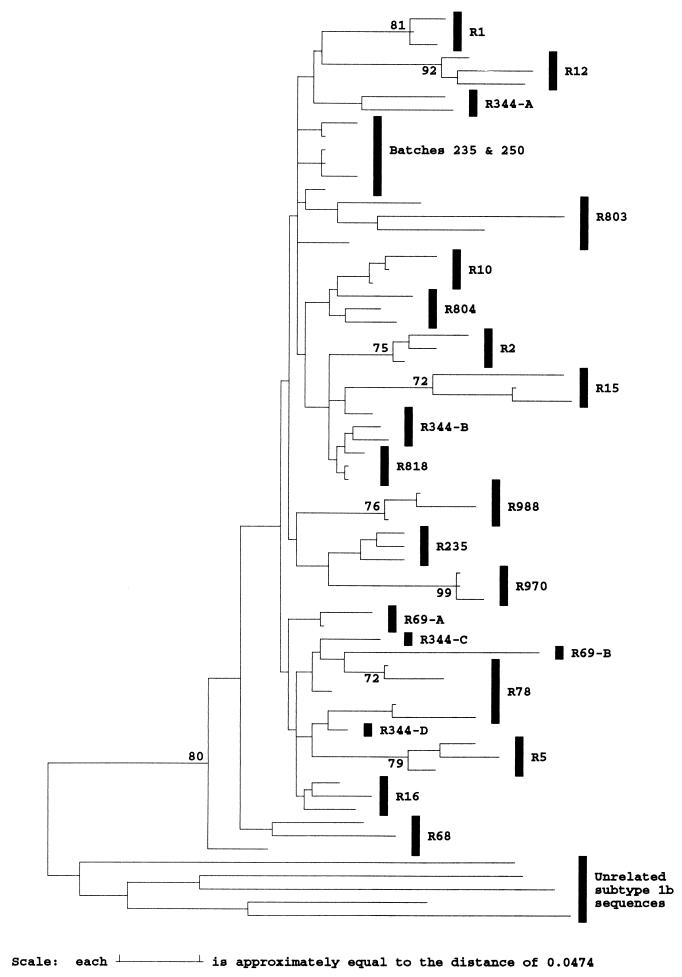
FIG. 1.
Phylogenetic analysis of E1 and E2 sequences from anti-D recipients and subtype 1b isolates. Evolutionary distances between representative sequences from two anti-D immunoglobulin batches (B), 17 anti-D recipients (R), and 40 epidemiologically unrelated subtype 1b sequences in the E1/E2 region (nucleotide positions 1096 to 1458) were used to construct a neighbor-joining tree. The bootstrap support (100 replicates) for the group of sequences from anti-D recipients is indicated.
FIG. 2.
Phylogenetic analysis of E1/E2 sequences from anti-D recipients and infectious batches. Distinct HVR amino acid sequences (codons 1 to 27 of E2) found in cloned virus sequences from two anti-D immunoglobulin batches and 17 recipients are compared with the most frequent sequence variant present in the batches (identities shown by ., sequence ambiguities shown by ?, a stop codon shown by ∗, and single nucleotide deletions shown by #). Sequences derived from single cDNA molecules by direct sequence analysis of PCR products obtained at limiting dilution are indicated by sm. The number of clones or direct sequences that share a given HVR sequence is indicated. Amino acids underlined in the batch sequence are strongly conserved in the entire data set, while sporadic amino acid substitutions are indicated by boldface type.
Variation among anti-D recipients.
Much greater variation was observed between E1 and E2 sequences from 17 different anti-D recipients sampled 17 years after exposure (mean evolutionary distance, 0.12). Phylogenetic analysis confirmed that all sequences had a common branch that separated them from 40 epidemiologically unrelated subtype 1b sequences and that this branch was observed in 94% of bootstrap resampling replicates (Fig. 1). Since virus sequences in the infectious source were relatively homogeneous, differences between virus sequences in different individuals have mostly arisen during the 17 years of separate evolution from their common source.
Analysis of the frequency of substitution across the region sequenced revealed that nonsynonymous substitutions that result in amino acid alterations were concentrated at the NH2 terminus of the E2 gene, corresponding to the HVR (data not shown). In addition, there was considerable variability between sequences from different anti-D recipients for codons 55 to 65 of E2 and to a lesser extent around codon 93, corresponding to HVR2 (15). A similar pattern of variability was observed among epidemiologically unrelated subtype 1b sequences (data not shown).
Sporadic nonsynonymous substitutions, defined as substitutions occurring in only 1 of the 153 clones sequenced, were evenly distributed throughout the regions sequenced. Of the 80 sporadic substitutions, 3 resulted in termination codons; in addition, there were 5 sporadic single-nucleotide deletions. The frequency of these sporadic changes is consistent with their origin as nucleotide misincorporation during PCR (see Discussion). Other nonsynonymous substitutions were present in two or more clones and presumably represent segregating polymorphisms present in different virus genomes.
Synonymous substitutions were relatively evenly distributed throughout the region sequenced except for the region of E1 immediately preceding the HVR. The frequency of synonymous substitutions between different anti-D recipients, excluding the HVR (mean, 0.154), was higher than previously documented for the same cohort in an adjoining region of E1 (0.053) or NS5B (0.037) (44). This trend is also observed upon comparison of complete genome sequences of epidemiologically unrelated subtype 1b isolates (45).
Variation of the HVR.
The diversity of HVR amino acid sequences within individual anti-D recipients between the 3 to 19 different clones sequenced was generally quite limited (Fig. 2 and 3). For 12 recipients, a single major HVR variant was present along with minor variants that differed at no more than two positions. In three recipients (R78, R15 and R69), two major variants were present, although these differed from each other at only 2 or 3 positions, while in two recipients (R12 and R803), there were two major variants that differed from each other at 7 or 12 positions. Finally, in R344, four distinct variants were present, differing from each other at 5 to 10 positions.
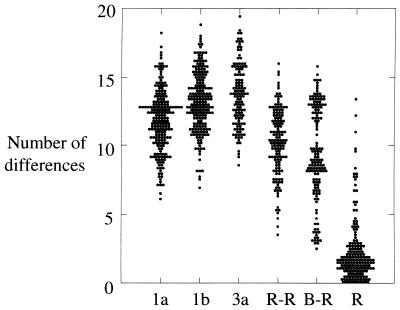
FIG. 3.
Scatter plot of the number of amino acid differences between different groups of HVR sequences. The number of amino acid substitutions in the HVR (positions 383 to 408) was calculated for sequences of viruses of genotypes 1a (n = 58), 1b (n = 46) or 3a (n = 16), among representative sequences from different anti-D recipients (R-R), between these sequences and the consensus sequence of the infectious source (B-R), and within individual anti-D recipients (R).
Much greater divergence was observed between different anti-D recipients. No two recipients had the same HVR sequence (Fig. 2), and the number of amino acid differences between the variants infecting different recipients (mean, 9.8) was almost as great as that between epidemiologically unrelated viruses within subtype 1a, 1b, or 3a (means, 12.0, 13.2, and 13.3, respectively [Fig. 3]). This diversity was not present among virus sequences isolated from the two infectious batches (Fig. 2) and so must represent divergent evolution during the 17 years of chronic infection. Surprisingly, the divergence between HVR sequences in different anti-D recipients and that present in the infectious batch was only slightly less (9.3 differences) than that between different recipients (9.8 differences), despite the period of divergence being only half as long. This anomaly may reflect the existence of constraints within the HVR that result in saturation of substitutions after relatively short periods (see below). In addition, distances between the batch and recipients were distributed into two separate peaks, one having a mean of 13 differences and the other having a mean of 7 differences, reflecting the lack of variation at the NH2 terminus among several recipients (Fig. 2).
Constraints on HVR variation.
Despite the diversity of HVR amino acid sequences observed among the different anti-D recipients, sequence change was not random but varied depending on the position within the HVR. Six amino acids (positions 2, 6, 20, 23, 24, and 26, numbered from the NH2 terminus of E2) were completely conserved among all HVR sequences from the 17 different anti-D recipients (Fig. 2), except for the occasional serine residue at position 20 and five sporadic substitutions. At the majority of variable positions in the HVR, amino acids were confined to two, three, or four different residues with similar characteristics. At seven positions (positions 4, 7, 8, 10, 13, 17, and 21), almost all amino acids were small uncharged residues (glycine, alanine, serine, threonine, or valine). At another two positions (positions 3 and 11) amino acids were confined to large residues with a dissociable proton (histidine, arginine, or tyrosine), while two other positions (positions 16 and 19) were predominantly large hydrophobic residues (leucine, isoleucine, phenylalanine, or methionine). At four positions, residues were confined to four or fewer amino acids but with dissimilar characteristics (position 5, threonine or methionine; position 9, glutamine, threonine, or methionine; position 12, threonine, asparagine, or alanine; position 27, asparagine, lysine, arginine, or alanine). Finally, at the remaining six positions (positions 1, 14, 15, 18, 22, and 25) five or more amino acids occurred, again with no discernible pattern. At three of these positions (positions 1, 18, and 22), the amino acid residue present in the HVR sequence from the infectious batch was present in 10% or fewer of the clones sequenced from anti-D recipients.
Characteristics of substitution in the HVR.
The observation that amino acid substitutions at some positions in the HVR were confined to particular amino acids might be related to the ease with which these substitutions could be produced from the nucleotide sequence of virus in the infectious batch. There is a 3- to 6-fold bias against transversion substitutions (A or G↔C or U) and toward transition substitutions (A↔G or C↔U) in the HCV genome as a whole (4, 28, 30), rising to 16-fold at the third position of codons (45), where confounding effects of constraints on amino acid substitution are weakest. A change of the tyrosine at position 3 of the HVR to histidine only requires a single transition mutation, while a change to isoleucine, threonine, alanine, or glycine would require two independent transversion mutations. Hence, a change to histidine at position 3 might be expected to occur more frequently than changes requiring one or more transversions.
The validity of this explanation was investigated by comparing the nucleotide sequence of the HVR from the infectious batches with that present in clones from 17 different recipients. Over the HVR as a whole, the ratio of transitions to transversions was lower (median, 2.3) than for the flanking region (median, 3.7) or for the HCV genome as a whole (median, 4.3) (45). At nonsynonymous sites in the HVR, there were 179 transition substitutions compared to 122 transversions (Fig. 4), similar to the ratio (1.4) observed at nonsynonymous sites elsewhere in the genome (45), suggesting that change is not random.
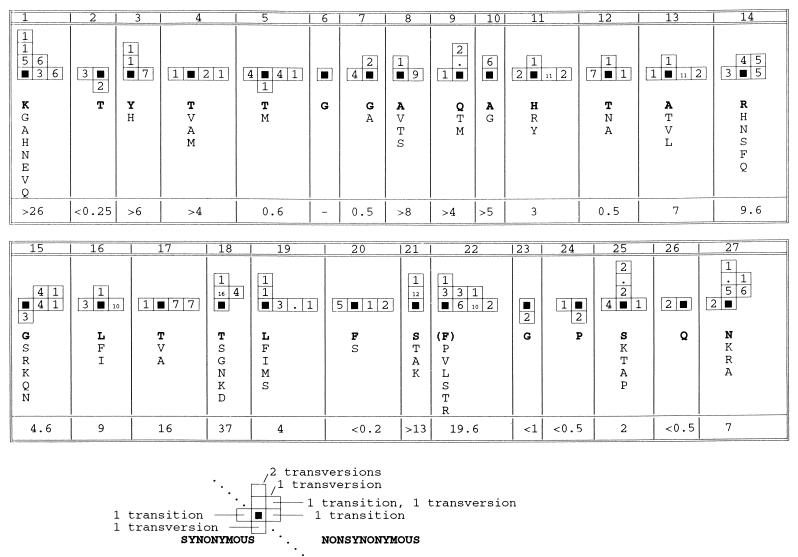
FIG. 4.
Pattern of nucleotide substitution within the HVR. The number and type of substitutions occurring in different recipients at each codon of the HVR are summarized schematically. The solid black squares represent the nucleotide sequence of the codon found in the infectious batch at each position. Synonymous transition substitutions are represented by boxes on the horizontal axis to the left of the origin, while synonymous transversion substitutions are represented by boxes below the origin on the vertical axis. Similarly, nonsynonymous substitutions are shown to the right of the origin if they are transitions and above the origin if they are transversions. Where a codon contains multiple substitutions, this is indicated by adding the individual vectors, so that a nonsynonymous substitution produced by two transversion substitutions is indicated by two boxes on the vertical axis above the origin (see the example below the figure). The number of times each combination of substitutions was observed is indicated by the number in the corresponding box; identical substitutions occurring in different recipients were considered independent events, while sporadic substitutions were ignored. The range of amino acid residues observed at each codon is indicated, with the residue found in the infectious batch in boldface type. The ratio of the number of nonsynonymous to synonymous substitutions between the batch sequence and representative sequences from each recipient is indicated for each codon.
Considering each codon separately (Fig. 4), at some sites the pattern of substitution was consistent with the random accumulation of mutations, since both synonymous and nonsynonymous substitutions occurred, with transitions outnumbering transversions (codons 4, 5, 11, 12, 13, 14, 16, 17, and 20). Hence, the observation that amino acid replacements at positions 4, 11, and 13 were limited to residues with similar biochemical properties might instead reflect the biased pattern of substitution during replication of the HCV genome. However, one or more of the potential nonsynonymous transition mutations did not occur at codons 12 (isoleucine absent), 14 (cysteine), 16 (proline), 17 (methionine), and 20 (leucine), suggesting the presence of negative selection against certain amino acid replacements at these sites. Another class of sites included those where there were more transversion mutations than expected, in some cases multiple substitutions, even though one or more of the three possible single transition mutations was not observed (codons 1, 3, 7, 9, 10, 15, 18, 21, 25, and 27). This pattern of substitution implies selection for or against particular amino acid replacements rather than random fixation or selection for change per se. The most extreme example was position 18, where although three different double mutations occurred, no single transition substitutions were observed, and where three different transversion substitutions occurred but did not include either of the two potential synonymous transversions. All three of the potential transition substitutions were observed in at least one of the 150 clones sequenced at only three codons. There were no substitutions at codon 6 in any of the clones sequenced, while only synonymous substitutions were observed at codons 2, 23, 24, and 26, implying strong negative selection against amino acid replacement at these positions.
Synonymous transition substitutions did not occur at 11 of the 27 codons in the HVR, although other transitions or transversions were observed at all codons except codon 6. This deficiency may be partly due to a bias against U or A at the third position of codons (45), since for 10 of these 11 codons the third position was G or C. In contrast, of the 16 codons at which a synonymous transition substitution was observed, 6 had U or A at the third position, similar to the relative frequency over the HCV genome as a whole (45).
Another way of investigating the characteristics of substitution in the HVR is to compare distances between the batch sequence and representative sequences from different anti-D recipients at synonymous (dS) and nonsynonymous (dN) sites. Over the HVR as a whole, the ratio of dN to dS was 2.0, or 2.5 if the six completely conserved sites were excluded. This compares with an average of 1.13 for the next 76 codons of E2 and less than 0.25 for comparisons between complete genomes (45). Comparing codons individually, the number of nonsynonymous substitutions was more than twice the number of synonymous substitutions at 18 positions and lower ratios were observed only at the 6 invariant codons and at codons 5, 7, and 12, which were the next most strongly conserved (Fig. 4).
This data suggests that for the majority of positions in the HVR the restricted pattern of amino acid substitution observed among the anti-D recipient cohort is not simply a consequence of the biased pattern of nucleotide substitution in the HCV genome. Instead, there is evidence for positive selection for particular amino acid replacements at the majority of positions and for negative selection against amino acid replacement at other positions.
Variation of the regions flanking the HVR.
While the COOH-terminal 18 residues of the E1 polypeptide were highly conserved with only one site variable, 33 of the 76 positions in E2 following the HVR were polymorphic. All but three of these sites were also polymorphic in a set of 40 epidemiologically unrelated subtype 1b sequences, and in the majority of cases the same amino replacements occurred. Four potential N-linked glycosylation sites at positions 34 to 35, 40 to 42, 47 to 49, and 65 to 67 were conserved except for eight sporadic substitutions and three segregating polymorphisms, while cysteine residues at positions 46, 69, 76, and 103 were also conserved except for four sporadic substitutions. A cluster of these conserved residues between positions 34 to 49 and immediately following the HVR coincides with the region of lowest variability in E2. All but two of the anti-D recipient had distinct sequences in the region between positions 51 and 63, which also differed from that present in the infective batch, while for HVR-2 several of the anti-D recipients had identical sequences, although these differed from the infectious batch sequence by one to four substitutions.
Origin of HVR variants within an individual.
Phylogenetic analysis of sequences from anti-D recipients, excluding the HVR, produced distinct groupings of sequences for most recipients, seven of which were supported by bootstrap resampling of replicates of synonymous sites (Fig. 5) or of all sites (data not shown). Two separate groups of sequences were observed for R69, while four different groups were observed for R344, some of which were supported by bootstrap resampling. These groups correspond to the groups of distinct HVR variants detected within these recipients. Less extreme subgroupings of flanking region sequences were observed for R12, R15, R78, R344, and R803, and these groups of sequences corresponded to the groups of HVR variants present in these individuals (Fig. 2). Exceptions to this segregation were a clone from R12 that grouped separately from three clones from which it differed in the HVR by only two sporadic substitutions, two clones from R78 that grouped with clones bearing HVR sequences differing at two sites, and one clone from R69 that grouped with clones differing in their HVR sequence at four positions rather than with clones differing at only two positions. These exceptions may have arisen through template shuffling during PCR (see Discussion).
FIG. 5.
Phylogenetic analysis of the HVR flanking region at synonymous sites. Evolutionary distances between sequences at synonymous sites between positions 1096 to 1150 and 1231 to 1458 were calculated with the Jukes-Cantor correction for representative sequences from different anti-D recipients (R), two infectious batches (238 and 250), and five unrelated subtype 1b sequences and used to construct a neighbor-joining tree. Sequences from the same recipient are grouped by vertical bars, except where these group separately on the tree, in which case a suffix indicates the HVR group to which the sequences belong. Bootstrap values of 70% or more (500 replicates) are indicated.
Substitutions at synonymous sites flanking the HVR might be expected to have little effect on virus viability and to accumulate linearly with time. In support of this possibility, evolutionary distances at synonymous sites between flanking region sequences from different recipients averaged 0.16 compared to 0.105 between recipients and the infectious batch, consistent with the greater period of separation between virus sequences in different recipients. Using the rate of accumulation of substitutions between different recipients to estimate the time of divergence between distinct HVR variants within a single recipient gives times of 8.5 to 9.5 years for the variants in R12, 15 years for the variants in R803, and 11 to 15 years for the variants in R344. Similar times of divergence (10 to 16 years) are implied for the variant groups in R15, R69, and R78 that differed at two or three positions. Estimated times of divergence were lower in individuals in whom a single HVR variant was detected, with times of 2 years or less in four recipients, less than 5 years in a further five recipients, and less than 9 years in a further two recipients (mean, 3.3). These times of divergence were shorter than those estimated within groups of sequences with similar HVR sequences for recipients infected with two or more HVR variant groups (1.7 to 10.2 years; mean, 5.9).
DISCUSSION
Artifacts and the interpretation of HVR variation.
We have found evidence for two types of artifact among our sequence data, both of which probably arose during PCR amplification. First, 90 sporadic nonsynonymous substitutions were observed in the sequence data set, 3 of which produced termination codons while there were 5 deletions of a single base pair. Although these might represent minor sequence variants in the virus population, their frequency is consistent with their origin during PCR because of nucleotide misincorporation by Taq DNA polymerase, equivalent to an error rate of 3.5 × 10−5 per nucleotide per cycle of amplification, within the documented range (2 × 10−5 to 20 × 10−5) (42). We tested this interpretation for three different recipients by direct sequencing of 7 to 10 PCR products obtained at limiting dilution of virus cDNA. Since these PCR products are expected to be derived from a single cDNA molecule, errors introduced by Taq during PCR would be visible as a heterogeneity on the sequence gel only if they occurred in the first cycle of PCR but would otherwise be diluted out (41). Only 2 sporadic nonsynonymous substitutions were observed among 27 sequences obtained at limiting dilution, while more than 10 would be expected from the same number and length of cloned sequences. Similar frequencies of sporadic substitution were observed in several other studies of the diversity of the HVR within infected individuals (42) suggesting that diversity in this part of the genome may be lower than was previously suggested.
Another way in which artifacts can be generated during PCR is through shuffling of templates due to the annealing of incompletely extended products to heterologous templates (23). Evidence for this type of artifact comes from the rare cases in which polymorphisms in the HVR did not segregate with those in the flanking region. These exceptions are unlikely to represent the products of convergent evolution since polymorphisms in the flanking region switched as a large block from being typical of one HVR variant group to being typical of another. Sequences of this type could also result from recombination in vivo, but apart from one report (15), there is no evidence for recombination between HCV genomes (34, 43). In addition, when 7 to 10 virus sequences were obtained at limiting dilution for three anti-D recipients (R68, R69, and R803), conditions which should allow the detection of in vivo recombinants but prevent the shuffling of dissimilar templates during PCR, the HVR variant groups segregated with the same flanking region polymorphisms as for the sequences derived from cloned PCR products (data not shown).
The HVR has multiple evolutionary pathways.
HVR sequences from each anti-D recipient were distinct and were almost as different from each other as were epidemiologically unrelated subtype 1b sequences (Fig. 3). This diversification occurred although all individuals were infected from a common source of limited diversity (Fig. 2) that would have presented the same antigenic stimulus upon first infection. This observation implies that the HVR of HCV is not constrained to follow a particular sequence of substitution in response to selective pressures imposed during infection, as typified by the sequential evolution of the influenza A virus hemagglutinin from year to year (2). Instead, it appears that although amino acid replacements are highly constrained (see below), the HVR can evolve in multiple directions from a given starting point.
Sequence change of the HVR is not random.
Despite the widespread acceptance of the idea that changes within the HVR are driven by antibody-mediated selection (8, 19, 47, 48, 50, 53), direct evidence in support of this possibility has been difficult to obtain in the absence of an in vitro culture system (54). Alternative explanations include the possibility that variation of the HVR is involved in cell tropism, as suggested for the V3 loop of human immunodeficiency virus, or that this region is unconstrained so that nonsynonymous substitutions occur in this region at the higher rate normally observed for synonymous substitutions (45).
Because of the strong bias against transversion substitutions in the HCV genome, this last explanation implies that the amino acid replacements should be those that can be produced by transition substitutions. Our study provides an opportunity to test this possibility because the HVR of virus in the infectious source was homogeneous, so that HVR substitutions observed in different anti-D recipients can be assumed to have arisen by independent events. At the majority of codons in the HVR, the pattern of substitution was inconsistent with the random accumulation of substitutions in the HVR (Fig. 4); instead, amino acid replacements were generally limited to particular sets of amino acids with shared biochemical properties or were completely conserved (Fig. 6). In addition, evolutionary distances between the batch sequence and sequences present in different recipients at nonsynonymous sites were twice those at synonymous sites, compared with ratios of less than 0.25 elsewhere in the virus genome. The bias toward nonsynonymous substitution in the HVR is even stronger than suggested by this ratio, since several positions were completely conserved and only certain types of amino acid replacement occurred at others. These observations are inconsistent with the possibility that the HVR evolves by random drift and suggest instead that substitutions are influenced by both negative and positive selection.
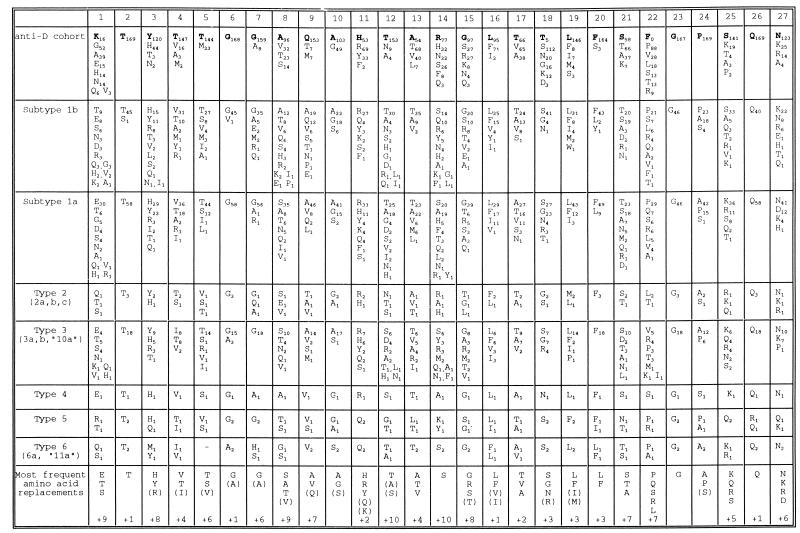
FIG. 6.
Frequency of amino acid replacements within the HVR. At each position in the HVR, the frequency with which amino acid replacement is observed among different groups of sequences is indicated by a subscript. For the anti-D cohort, the amino acid present in the infectious batch is indicated by a boldface letter and sporadic substitutions are omitted. Other groups of sequences were obtained from GenBank or from published sources. The bottom row gives the most frequent amino acid replacements at each site for the whole data set, with less common replacements enclosed in parentheses and the number of additional residues observed indicated on the bottom row.
This interpretation is supported by the observation that very similar constraints on amino acid replacement were observed among a collection of 46 epidemiologically unrelated sequences of subtype 1b (Fig. 6). Five of the six positions that were strongly conserved among the anti-D recipients were also conserved among collections of sequences from other virus genotypes. At codons where replacements among the anti-D recipients were limited to particular groups of amino acids with shared characteristics, the same restrictions were observed among sequences from other genotypes. For example, replacements at positions 4, 5, 13, 17, and 21 were usually small hydrophobic residues while replacements at codons 16, 19, and 20 were typically either leucine, phenylalanine, or isoleucine. Similar observations have been made for sequences from HCV isolates of undefined genotypes (37, 38). At several positions where seven or more amino acid replacements were observed (codons 4, 8, 9, 12, 14, 21, 22, and 25), the most frequent amino acid replacement differed between genotypes although the general characteristics of the amino acids were maintained.
These constraints are reflected in the overall amino composition of the HVR, which differs consistently from that of the entire E2 gene or the complete HCV polyprotein. For the collections of HVR sequences from genotypes 1a, 1b, and 3a, cysteine and tryptophan were almost completely absent, while there was marked underrepresentation of aspartic and glutamic acids and, to a lesser extent, of leucine, isoleucine, and proline. Phenylalanine, glycine, and threonine were overrepresented, although this is difficult to interpret since these amino acids were strongly conserved at one or more codons. While basic residues occurred at about the same frequency as for E2 as a whole, it was noticeable that they were common only at the most variable sites, where seven or more different amino acid replacements were observed in the collections of sequences.
There was also a bias in the pattern of substitution observed within the HVR among the anti-D recipient cohort. While all recipients had several substitutions in the region from codons 10 to 22, several recipients had no or very few substitutions from codons 2 to 9. This observation is consistent with a study of another cohort of anti-D recipients in which reactivity to the COOH terminus of the HVR was more frequent among chronically infected recipients than was reactivity to the NH2 terminus (53). Several other studies have also mapped linear antibody epitopes to the COOH terminus of the HVR in both acutely (14, 36, 37) and chronically (50) infected patients, as well as in experimentally infected chimpanzees (47). Despite these observations, comparison of HVR sequences from viruses of different genotypes provided no evidence for greater heterogeneity at COOH-terminal positions within the HVR (Fig. 6).
Hypervariable codons outside the HVR.
Substitutions between sequences from anti-D recipients or between epidemiologically unrelated subtype 1b isolates did not occur evenly in the regions flanking the HVR but were concentrated in two regions centered on codons 60 and 93 (HVR-2). Substitutions occurring in the region around codon 60 tended to occur between a restricted set of amino acids with common characteristics, such as lysine-arginine (codon 63), alanine-serine (codons 57 and 66), or glutamine-asparagine-histidine-tyrosine (codons 51 and 62). Some of these groups of amino acids were also observed at one or more positions in the HVR, and the extent of variability at individual codons was also similar. In contrast, no common characteristic could be discerned for the substitutions occurring at any of codons 93 to 97, and replacements included nonpolar, polar, and charged residues, suggesting that this region is subject to different constraints and/or selection pressures. Elsewhere in E2, substitutions were either rare or confined to closely related pairs of amino acids such as aspartate-glutamate and isoleucine-valine.
Phylogenetic implications of constraints on sequence change in E2.
Because the HVR of E2 is the most variable region of the HCV genome, it has been used by many groups to investigate the phylogenetic relationship between virus sequences in different infected individuals. However, the observation that sequence change of the HVR is not random means that similarity between sequences may not always reflect their true phylogenetic relationship since substitutions may be convergent and will quickly become saturated. For example, although all anti-D recipients were infected from the same source, most of the sequences obtained 17 years after infection were as divergent from each other (mean, 9.8 differences) as from the infectious batch (mean, 9.3) (Fig. 3). This degree of divergence is smaller than between epidemiologically unrelated subtype 1b sequences (mean, 13.9), but only because few substitutions occurred in the NH2-terminal region of the HVR in some recipients. HVR sequences from different recipients were as different from the consensus HVR sequence of subtype 1b isolates (10.3 differences) as was the sequence of the infectious batch (11 differences). Phylogenetic analysis of HVR amino acid sequences from anti-D recipients failed to provide bootstrap support for a common grouping, even if flanking regions were included (data not shown), although their grouping was supported by 80% of bootstrap replicates if only synonymous sites were considered (Fig. 5). Despite the limited time of divergence between HVR sequences in different anti-D recipients, at 21 of 27 codons the amino acids observed among the cohort accounted for more than 75% of the range observed among epidemiologically unrelated subtype 1b sequences. Only at position 9 did replacements in the cohort account for less than half of the subtype 1b sequences (Fig. 6).
These observations imply that amino acid substitutions in the HVR become saturated within relatively short periods of divergence and suggest that this region may not always be reliable for the investigation of transmission events. For example, in one study of vertical transmission, the HVR from an infected infant differed at five to seven positions from the virus sequences present in the mother (51) but epidemiologically unrelated subtype 1a and 1b sequences sometimes differ to the same extent (Fig. 3). Similarly, two patients of an HCV-infected surgeon were infected with variants differing at only one position in the HVR from one of the five variants present in the surgeon but in other patients there were three, four, or five differences (5). This situation was not clarified by analysis of a 188-nucleotide fragment including the HVR, since in some cases distances between sequences from the surgeon and his infected patients were equivalent to those between some epidemiologically unrelated controls. In both of these examples, virus sequences were obtained relatively close to the time of transmission, and greater problems of interpretation can be expected in cases where samples from the implicated donor and recipient are separated by several years or where the implicated source is infected with a complex mixture of variants.
Limited diversity within persistently infected individuals.
A surprising finding of this study was that sequence diversity of the HVR was quite limited within chronically infected anti-D recipients, with the majority of recipients being infected with a single major variant. This observation does not simply reflect the recognition of sporadic substitutions as Taq errors in this study, and it contrasts with several previous reports in which multiple divergent HVR variants were found to cocirculate in chronically infected individuals (15, 20, 24, 29). One explanation for this difference may be the relatively homogeneous nature of the infectious source in comparison to other studies, where individuals were often infected with large quantities of virus from potentially heterogeneous sources. For example, an individual infected by blood transfusion from a chronically infected individual might be expected to become infected with the full range of variants present at that time. Another explanation for the limited diversity observed in our study is that the anti-D recipients formed a relatively homogeneous cohort, since they were all women of a similar age and ethnic background, and did not have the complicating effects of factors such as coinfection with other parenterally transmitted viruses, nonviral chronic diseases such as hemophilia or renal failure, or symptomatic chronic liver disease. For example, increased variation of the HVR has been observed in individuals coinfected with HIV (38).
Long-term coexistence of multiple HCV lineages.
Of the 17 anti-D recipients, 3 were infected with multiple HVR variants differing from each other at five or more positions, while variant groups differed by two or three positions in three further recipients. Phylogenetic analysis of sequences flanking the HVR produced groupings of sequences that matched those defined by HVR sequences, suggesting that distinct HVR variants represent different lineages and that convergent evolution is uncommon. Using the extent of divergence at synonymous sites in the region flanking the HVR to estimate the time of divergence of HVR variant lineages gives times ranging between 8 and 16 years (mean, 12.1). In contrast, virus sequences in individuals infected with a single HVR variant group were more closely related to each other, with a mean estimated time of divergence of 3.3 years. These observations imply that HVR variant sequences can be stable for several years and that variants with divergent HVR sequences can coexist in an infected individual while following separate evolutionary pathways.
Nature of selection on HVR variants.
Several of our observations on the long-term evolution of the HVR in individuals infected from a common source cannot be explained if immune system-mediated neutralization is the only selective force on variants with different HVR sequences. First, variation was unequally distributed within the HVR, with some positions being invariant and others being highly variable and usually limited to particular amino acid replacements. This pattern of variation does not simply result from the bias toward transition substitutions in the HCV genome. The pattern is also not due to the homogeneity of the infectious source or the limited period of divergence between sequences, since similar restrictions on amino acid replacement are observed among epidemiologically unrelated sequences of subtype 1b or of more distantly related virus genotypes (Fig. 6). Instead these observations imply that there is strong negative selection against some amino acid substitutions in the HVR while at the majority of codons there is selection for conservative amino acid replacements rather than selection for change per se. In support of this idea, the amino acid present in the infectious batch of anti-D at codons 1, 18, and 22 was rare (<5%) among HVR sequences of other subtype 1b isolates and these codons were the most frequently substituted (>90% of clones) among the anti-D recipient cohort.
A second finding that is difficult to explain by immune selection is that divergent HVR variants infecting a single anti-D recipient belonged to different lineages that had coexisted for 8 to 15 years. These multiple lineages would be unlikely to survive in the face of a neutralizing immune response since an effective response against one variant should lead to a dramatic shift in the composition of the virus population. With repeated cycles of neutralization and immune system escape, it might be expected that one of the variants would be eliminated. Multiple lineages could coexist if convergent evolution led to similar HVR sequences appearing in different lineages, but we have found no evidence for this possibility.
Finally, considerable diversity in the region flanking the HVR was observed even between HVR variants with similar sequences, with 7 of 11 lineages being estimated to have arisen 2 years or more ago and 3 of these to have arisen more than 4 years ago. These estimates imply that selection for new HVR variants is an infrequent event rather than a continuous process and are difficult to reconcile with the observation that antibody to the HVR is produced in the majority of infected individuals (36, 54), that antibody specific for an HVR variant can be present before the variant appears (16, 36, 52), that it can coexist with the variant (37), or that increased variation of the HVR is observed in patients coinfected with human immunodeficiency virus (38).
Together, these observations are consistent with there being strong selection for the maintenance of certain (unknown) properties of the HVR at the same time as there is intermittent positive selection for amino acid replacement. Further work is required to clarify the properties of this region that produce constraints on its evolution, as well as on the nature of the selective forces that are responsible for amino acid replacement. For example, sequence change in the HVR could result from selection against peptides capable of binding to host major histocompatibility complex alleles. This type of selection, which has been documented for human immunodeficiency virus (31), might explain the long-term coexistence of HVR variant lineages within an infected individual, since HVR variants containing peptide epitopes incapable of binding to class I or class II alleles would not be subject to further immune selection. This hypothesis could be tested by investigating the association between particular HVR substitutions and HLA type in this study group.
ACKNOWLEDGMENTS
J.M. and D.B.S. were supported by a grant from the Wellcome Trust. C.C. was supported by a study fellowship from the University of Rome “La Sapienza”. P.S. is a Darwin Fellow.
We are grateful to Anders Widell for making available his collection of subtype 1a HVR sequences.
REFERENCES
- 1.Allander T, Beyene A, Jacobson S H, Grillner L, Persson M A A. Patients infected with the same hepatitis C virus strain display different kinetics of the isolate-specific antibody response. J Infect Dis. 1997;175:26–31. doi: 10.1093/infdis/175.1.26. [DOI] [PubMed] [Google Scholar]
- 2.Bean W J, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster R G. Evolution of the H3 influenza virus haemagglutinin from human and nonhuman hosts. J Virol. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driesel G, Wirth D, Stark K, Baumgarten R, Sucker U, Schreier E. Hepatitis C virus (HCV) genotype distribution in German isolates: studies on the sequence variability in the e2 and NS5 region. Arch Virol. 1994;139:379–388. doi: 10.1007/BF01310799. [DOI] [PubMed] [Google Scholar]
- 4.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b—sensitivity to interferon is conferred by amino acid substitutions in the NS5a region. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteban J I, Gomez J, Martell M, Cabot B, Quer J, Camps J, Gonzalez A, Otero T, Moya A, Esteban R, Guardia J. Transmission of hepatitis C virus by a cardiac surgeon. N Engl J Med. 1996;334:555–560. doi: 10.1056/NEJM199602293340902. [DOI] [PubMed] [Google Scholar]
- 6.Expert Group. Report of the Expert Group on the Blood Transfusion Board. Pn 1538. Dublin, Ireland: Stationary Office; 1995. [Google Scholar]
- 7.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higashi Y, Kakumu S, Yoshioka K, Wakita T, Mizokami M, Ohba K, Ito Y, Ishikawa T, Takayanagi M, Nagai Y. Dynamics of genome change in the E2/NS1 region of hepatitis C virus in vivo. Virology. 1993;197:659–668. doi: 10.1006/viro.1993.1641. [DOI] [PubMed] [Google Scholar]
- 9.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 10.Hohne M, Schreier E, Roggendorf M. Sequence variability in the env-coding region of hepatitis C virus isolated from patients infected during a single source outbreak. Arch Virol. 1994;137:25–34. doi: 10.1007/BF01311170. [DOI] [PubMed] [Google Scholar]
- 11.Ina Y. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput Appl Biosci. 1994;10:11–12. doi: 10.1093/bioinformatics/10.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis L M, Watson H G, McOmish F, Peutherer J F, Ludlam C A, Simmonds P. Frequent reinfection and reactivation of hepatitis C virus genotypes in multitransfused hemophiliacs. J Infect Dis. 1994;170:1018–1022. doi: 10.1093/infdis/170.4.1018. [DOI] [PubMed] [Google Scholar]
- 13.Kao J H, Chen P J, Lai M Y, Wang T H, Chen D S. Quasispecies of hepatitis C virus and genetic drift of the hypervariable region in chronic type C hepatitis. J Infect Dis. 1995;172:261–264. doi: 10.1093/infdis/172.1.261. [DOI] [PubMed] [Google Scholar]
- 14.Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J Virol. 1994;68:4776–4784. doi: 10.1128/jvi.68.8.4776-4784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–123. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 16.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region-1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koziel M J, Dudley D, Wong J T, Dienstag J, Houghton M, Ralston R, Walker B D. Intrahepatic cytotoxic T lymphocytes specific for hepatitis-c virus in persons with chronic hepatitis. J Immunol. 1992;149:3339–3344. [PubMed] [Google Scholar]
- 18.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. Philadelphia: Pennsylvania State University; 1993. [Google Scholar]
- 19.Kumar U, Monjardino J, Thomas H C. Hypervariable region of hepatitis C virus envelope glycoprotein (e2 NS1) in an agammaglobulinemic patient. Gastroenterology. 1994;106:1072–1075. doi: 10.1016/0016-5085(94)90770-6. [DOI] [PubMed] [Google Scholar]
- 20.Kurosaki M, Enomoto N, Marumo F, Sato C. Evolution and selection of hepatitis C virus variants in patients with chronic hepatitis C. Virology. 1994;205:161–169. doi: 10.1006/viro.1994.1631. [DOI] [PubMed] [Google Scholar]
- 21.LerouxRoels G, Esquivel C A, Deleys R, Stuyver L, Elewaut A, Philippe J, Desombere I, Paradijs J, Maertens G. Lymphoproliferative responses to hepatitis C virus core, E1, E2, and NS3 in patients with chronic hepatitis C infection treated with interferon alfa. Hepatology. 1996;23:8–16. doi: 10.1002/hep.510230102. [DOI] [PubMed] [Google Scholar]
- 22.Lesniewski R R, Boardway K M, Casey J M, Desai S M, Devare S G, Leung T K, Mushahwar I K. Hypervariable 5′-terminus of hepatitis C virus E2/NS1 encodes antigenically distinct variants. J Med Virol. 1993;40:150–156. doi: 10.1002/jmv.1890400213. [DOI] [PubMed] [Google Scholar]
- 23.Meyerhans A, Vartanian J P, Wain Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, Kaneshige T, Igimi H, Kamada T, Uchida K. Hepatitis C viral complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology. 1995;108:789–795. doi: 10.1016/0016-5085(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 25.Muller R. The natural history of hepatitis C: clinical experiences. J Hepatol. 1996;24:52–54. [PubMed] [Google Scholar]
- 26.Nakazawa T, Kato N, Ootsuyama Y, Sekiya H, Fujioka T, Shibuya A, Shimotohno K. Genetic alteration of the hepatitis C virus hypervariable region obtained from an asymptomatic carrier. Int J Cancer. 1994;56:204–207. doi: 10.1002/ijc.2910560210. [DOI] [PubMed] [Google Scholar]
- 27.Odeberg J, Yun Z B, Sonnerborg A, Uhlen M, Lundeberg J. Dynamic analysis of heterogeneous hepatitis C virus populations by direct solid-phase sequencing. J Clin Microbiol. 1995;33:1870–1874. doi: 10.1128/jcm.33.7.1870-1874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the e2/NS1 protein of hepatitis-c virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto H, Kojima M, Okada S-I, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2 year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 31.Phillips R E, Rowland Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R M, Rizza C R, McMichael A J. Human immunodeficiency virus genetic variation that can escape cytotoxic T-cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 32.Power J P, Lawlor E, Davidson F, Holmes E C, Yap P L, Simmonds P. Molecular epidemiology of an outbreak of infection with hepatitis C virus in recipients of anti-D immunoglobulin. Lancet. 1995;345:1211–1213. doi: 10.1016/s0140-6736(95)91993-7. [DOI] [PubMed] [Google Scholar]
- 33.Power J P, Lawlor E, Davidson F, Yap P L, Kenny-Walsh E, Whelton M J, Walsh T J. Hepatitis C viraemia in recipients of Irish intravenous anti-D immunoglobulin. Lancet. 1994;344:1166–1167. doi: 10.1016/s0140-6736(94)90679-3. [DOI] [PubMed] [Google Scholar]
- 34.Prescott L E, Berger A, Pawlotsky J M, Conjeevaram P, Pike I, Simmonds P. Sequence analysis of hepatitis C virus variants producing discrepant results with two different genotyping assays. J Med Virol. 1997;53:237–244. [PubMed] [Google Scholar]
- 35.Sakamoto N, Enomoto N, Kurosaki M, Marumo F, Sato C. Sequential change of the hypervariable region of the hepatitis C virus genome in acute infection. J Med Virol. 1994;42:103–108. doi: 10.1002/jmv.1890420119. [DOI] [PubMed] [Google Scholar]
- 36.Scarselli E, Cerino A, Esposito G, Silini E, Mondelli M U, Traboni C. Occurrence of antibodies reactive with more than one variant of the putative envelope glycoprotein (gp70) hypervariable region 1 in viremic hepatitis C virus-infected patients. J Virol. 1995;69:4407–4412. doi: 10.1128/jvi.69.7.4407-4412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiya H, Kato N, Ootsuyama Y, Nakazawa T, Yamauchi K, Shimotohno K. Genetic alterations of the putative envelope proteins encoding region of the hepatitis C virus in the progression to relapsed phase from acute hepatitis: humoral immune response to hypervariable region 1. Int J Cancer. 1994;57:664–670. doi: 10.1002/ijc.2910570509. [DOI] [PubMed] [Google Scholar]
- 38.Sherman K E, Andreatta C, Obrien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23:688–694. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith D B, McAllister J, Casino C, Simmonds P. Virus ’quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 43.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P L, Simmonds P, Conradie J D, Neill A G S, Dusheiko G M, Kew M C, Crookes R, Koshy A, Lin C K, Lai C, Murraylyon I M, Elguneid A, Gunaid A A, Yemen T, Yemen S, Mutimer D, Ahmed M, Nuchprayoon C, Tanprasert S, Preston F E, Makris M, Chuansumrit A, Mahasandana C, Pritchard D, Riley E, Greenwood B M, Saeed A A, Alrasheed A M, Saleh M G, McFarlane I, Tibbs C, Williams R, Power J, Lawlor E, Kiyokawa H. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 44.Smith D B, Pathirana S, Davidson F, Lawlor E, Power J, Yap P L, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- 45.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka T, Kato N, Nakagawa M, Ootsuyama Y, Cho M J, Nakazawa T, Hijikata M, Ishimura Y, Shimotohno K. Molecular cloning of hepatitis C virus genome from a single Japanese carrier: sequence variation within the same individual and among infected individuals. Virus Res. 1992;23:39–53. doi: 10.1016/0168-1702(92)90066-i. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore E E, Peterson D A, Mishiro S. A structurally flexible and antigenically variable n-terminal domain of the hepatitis C virus e2/NS1 protein—implication for an escape from antibody. Virology. 1993;195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 48.van Doorn L J, Capriles I, Maertens G, Deleys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region e2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M, Han J H. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 50.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, et al. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner A J, Thaler M M, Crawford K, Ching K, Kansopon J, Chien D Y, Hall J E, Hu F, Houghton M. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J Virol. 1993;67:4365–4368. doi: 10.1128/jvi.67.7.4365-4368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshioka K, Higashi Y, Tanaka K, Aiyama T, Takayanagi M, Okumura A, Iwata K, Nagai Y, Kakumu S. Deficiency of antibody response to hypervariable region of hepatitis C virus in patients with chronic hepatitis C. J Hepatol. 1996;24:649–657. doi: 10.1016/s0168-8278(96)80259-5. [DOI] [PubMed] [Google Scholar]
- 53.Zibert A, Kraas W, Meisel H, Jung G, Roggendorf M. Epitope mapping of antibodies directed against hypervariable region 1 in acute self-limiting and chronic infections due to hepatitis C virus. J Virol. 1997;71:4123–4127. doi: 10.1128/jvi.71.5.4123-4127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]