Abstract
Primary glioblastoma(pGBM) is the most malignant tumor of the central nervous system. Radiotherapy, chemotherapy and surgical treatment have little effect on the survival of pGBM patients. The prognosis is often poorly once the tumor recurs. It is urgent to develop new therapies for patients. In recent years, studies have been clarified that miRNA have a powerful regulating effect on the genes. However, the main group of miRNAs in regulating long-term survival specific related genes of pGBM is still unclear. Given that the survival period of most glioma patients is relatively short, studying long-term survival patients with pGBM is of great value for this disease. Our study aim to identify key miRNAs with long-term survival related genes present in pGBM and uncover their potential mechanisms. The gene expression profiles of GSE53733, GSE15824, GSE30563, GSE50161 were obtained from the Gene Expression Omnibus database. Firstly, samples were divided into 3 groups according to its survival time and each group compare to the normal control group. Then we obtained differential expression genes (DEGs) with a long-term survival specific (LTSDEGs) and a short-term survival specific DEGs (STSDEGs). Next, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were conducted with LTSDEGs and STSDEGs together. Moreover, we used the UALCAN database to verify LTSDEGs and STSDEGs, and obtained long-term verified survival specific DEGs(LTVSDEGs) and short-term verified survival specific DEGs(STVSDEGs). Finally, we established the predicted key miRNAs-LTVSDEGs interaction network. The protein expressions of the top 4 LTVSDEGs were verified in the HPA database with immunohistochemical staining. In total, we found 260 genes changed in LTSDEGs and 822 genes changed in STSDEGs. GO and KEGG results shown that the major changes are focused on tumor metabolism. 9 LTVSDEGs and 18 STVSDEGs were verified in UALCAN database. As for protein expression verification in top 4 LTVSDEGs, ZNF630, BLVRB and RPA3 were verified, while TPBG was not detected. We obtained 59 key miRNA from the predicted key miRNAs-LTVSDEGs interaction network. 25 key miRNAs were verified using GSE90603. Finally, we constructed the key miRNAs-LTVSDEGs network using a Sankey diagram, including 25 miRNAs and 7 LTVSDEGs. In conclusion, our study shows that there is a close relationship between metabolic changes and survival in pGBM. Besides, we established a key miRNAs-LTVSDEGs network for pGBM, which could be the key path in prolonging the life of pGBM patients.
Keywords: RNA-seq, Glioblastoma, Microarray chips, Overall survival, Long-term survival
1. Introduction
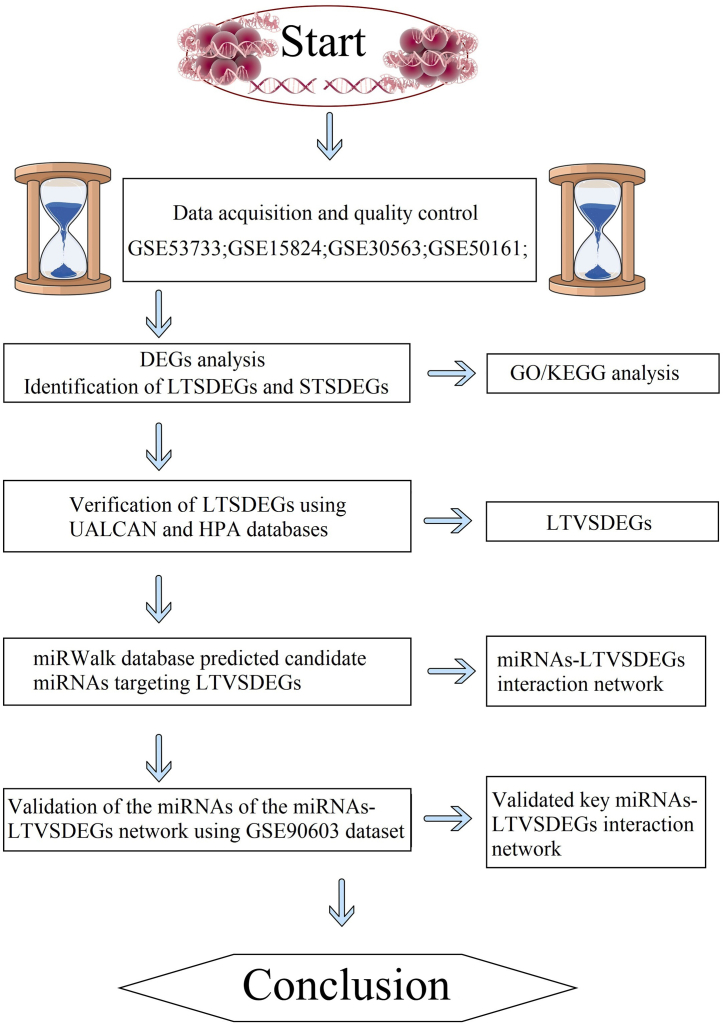
Glioma is the most common type of tumor in the central nervous system [1]. pGBM is one of the most malignant type of glioma [2,3]. The frequently-used methods of clinical treatment include radiotherapy, chemotherapy and surgical treatment [2,3]. Despite current clinical interventions, including radiotherapy, chemotherapy, and surgery, failing to substantially enhance patient survival rates, the disease remains a substantial societal and healthcare burden, with approximately 77,000 new diagnoses reported annually in the United States and Europe [[2], [3], [4], [5]]. Unfortunately, the five-year survival rate for patients with pGBM is dismally low at < 3% [6,7]. While numerous studies have explored the relationship between microRNAs (miRNAs) and functional genes in pGBM, the identification of a significant miRNA network linked to survival-related genes, particularly in the context of long-term survival, remains elusive [[8], [9], [10], [11]]. Fortunately, advancements in gene microarray technology and the establishment of comprehensive network databases have facilitated the collection and analysis of extensive clinical data. Leveraging these resources, our present study aims to extract valuable information from diverse microarray datasets through bioinformatics analysis. The ultimate goal is to unveil a pivotal miRNA network influencing survival-related genes in pGBM. This research seeks to offer novel targets and strategies for the clinical treatment of pGBM. The overall analysis process of this project is shown in Fig. 1.
Fig. 1.
A flow chart displaying the screening process in this study. DEGs, differentially expressed genes; LTSDEGs, long-term survival specific differentially expressed genes; STSDEGs, short-term survival specific differentially expressed genes ; LTVSDEGs; long-term verified survival specific differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
2. Materials and methods
2.1. Microarray data
GSE53733, GSE15824, GSE30563 and GSE50161 were sourced from the GEO database(www.ncbi.nlm.nih.gov/geo/). All datasets were based on the Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). In total, 18 normal control samples obtained from GSE15824 (3 samples), GSE30563 (13 samples) and GSE50161 (2 samples), while 70 pGBM samples selected from GSE53733.The detail information is shown in Table S1. Based on data from the population of GBM patients, long-term survival is uncommon, and the median overall survival (OS) remains less than 12 months. Only a small percentage (3–5%) of GBM patients survive for more than 36 months. Therefore, pGBM samples in our study were divided into three groups: short-term survival group (ST; OS ≤ 12 months), intermediate survival group (IT; 12 months < OS < 36 months), and long-term survival group (LT; OS ≥ 36 months). Our grouping criteria are consistent with previous studies. pGBM samples included 16 ST samples, 31 IT samples, and 23 LT samples.
2.2. Data quality control
The “Affy” R package was employed for gene microarray quality control [12]. RNA degradation plots were generated using R to assess the uniformity and quality of the RNA extraction. A uniform curve with minimal error indicated good RNA quality, providing a solid foundation for the subsequent merging of the four datasets.
2.3. Identification of DEGs
Differentially expressed genes (DEGs) between 70 pGBM samples and 18 normal control samples were conducted by using “limma” R package [13]. The Affy package [12]and Robust Multichip Average [14]algorithm were applied to identify DEGs. We identified the long-term survival differentially expressed genes (LTDEGs) by comparing the LT group with the normal group. Similarly, the intermediate survival differentially expressed genes (ISDEGs) were obtained by comparing the IS group with the normal group, and the short-term survival differentially expressed genes (STDEGs) were identified by comparing the ST group with the normal group. P < 0.05 and |Fold Change(FC)|> 2 were considered statistically significant. Heatmaps of top 10 regulated DEGs of each comparative group were generated using Morpheus online software (Morpheus, https://software.broadinstitute.org/morpheus).
2.4. Venn diagram analysis for the DEGs
A Venn diagram was generated using three previous DEGs results (LTDEGs, ITSDEGs, and STDEGs). Online tool “calculate and draw custom Venn diagrams” was utilized for drawing the diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). Among the venn diagram, we determined long-term survival specific differentially expressed genes (LTSDEGs) by excluding all overlapping genes from the LTDEGs. Similarly, the intermediate survival specific differentially expressed genes (ITSDEGs) were identified by excluding all overlapping genes from the ITDEGs, and the short-term survival specific differentially expressed genes (STSDEGs) were obtained by excluding all overlapping genes from the STDEGs.
2.5. GO and pathway enrichment analyses
Gene Ontology (GO) analysis, a widely recognized method for annotating genes and gene products, was employed to identify characteristic biological attributes of transcriptome data or high-throughput genome [15,16]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/) database, a well-known resource for the systematic analysis of gene functions in biological pathways, was also utilized [17]. The gene lists of LTSDEGs and STSDEGs were together mapped to the relevant biological annotation using the DAVID online database (https://david.ncifcrf.gov/) [18,19]. DAVID is an easy online tool for gene functional analysis, offering the functionality to perform simultaneous GO and KEGG analysis. P < 0.05 was set as the threshold to indicate a statistically significant difference.
2.6. Verification of survival related genes
The LTSDEGs and the STSDEGs were put together into the UALCAN online database(http://ualcan.path.uab.edu) [20] to verify the authenticity of genes, and obtained long-term verified survival specific differentially expressed genes (LTVSDEGs) and short-term verified survival specific differentially expressed genes (STVSDEGs). All survival curves were conducted using the UALCAN database. The UALCAN database contains data on 31 types of cancer. The algorithm construction of UALCAN was based on the TCGA level 3 RNA-seq database. The LTSDEGs and STSDEGs were uploaded to explore their expression and prognostic value in GBM using the UALCAN database. Genes with insufficient data were automatically eliminated. P < 0.05 was considered to indicate a statistically significant difference.
2.7. Verification of top 4 LTVSDEGs protein expression
We use immunohistochemical staining database HPA (www.proteinatlas.org) [21]to verify the top 4 LTVSDEGs protein expression in human high-grade glioma. We compared the expression of each protein in both normal brain tissue and tumor tissue.
2.8. Establishment of the predicted key miRNAs-LTVSDEGs interaction network
Our miRNA network analysis was performed using the online microRNA database miRWalk (http://mirwalk.umm.uni-heidelberg.de/) [22]. The miRWalk database is a project of the Medical Research Center, a core scientific facility of the Medical Faculty Mannheim of the University of Heidelberg. Cytoscape software (version 3.4) [23] was used to conduct network visualization graphic analysis. We applied a filter to identify a significant LTVSDEGs-miRNAs network according to the degree between LTVSDEGs and their related miRNAs. The standard of node degree threshold was set between 7 and infinity, inclusive, ensuring that the threshold degree was higher than the lowest degree of LTVSDEGs. Subsequently, we excluded all LTVSDEGs from the selected nodes, establishing a miRNA-LTVSDEGs interaction network.
2.9. Validation of the key miRNAs and construction of the key miRNAs-LTVSDEGs network
The differential expression of the biomarkers was further confirmed in the testing dataset GSE90603 from the GEO database. The “limma” R package was applied to identify the DEGs between patients with pGBM and normal group. The “limma”, “pheatmap” and “ggplot2” R packages were used to create volcano plots, heatmaps and boxplot, respectively. The thresholds were |logFC| >0.263 (|FC| > 1.2) and P < 0.05. ROC curves of training and testing cohorts were computed and visualized by the “pROC” R package. P < 0.05 was considered statistically significant. The validated key miRNAs-LTVSDEGs interaction network was displayed with Sankey diagram function by the “gglluvial” R packages.
3. Results
3.1. Microarray chip quality control
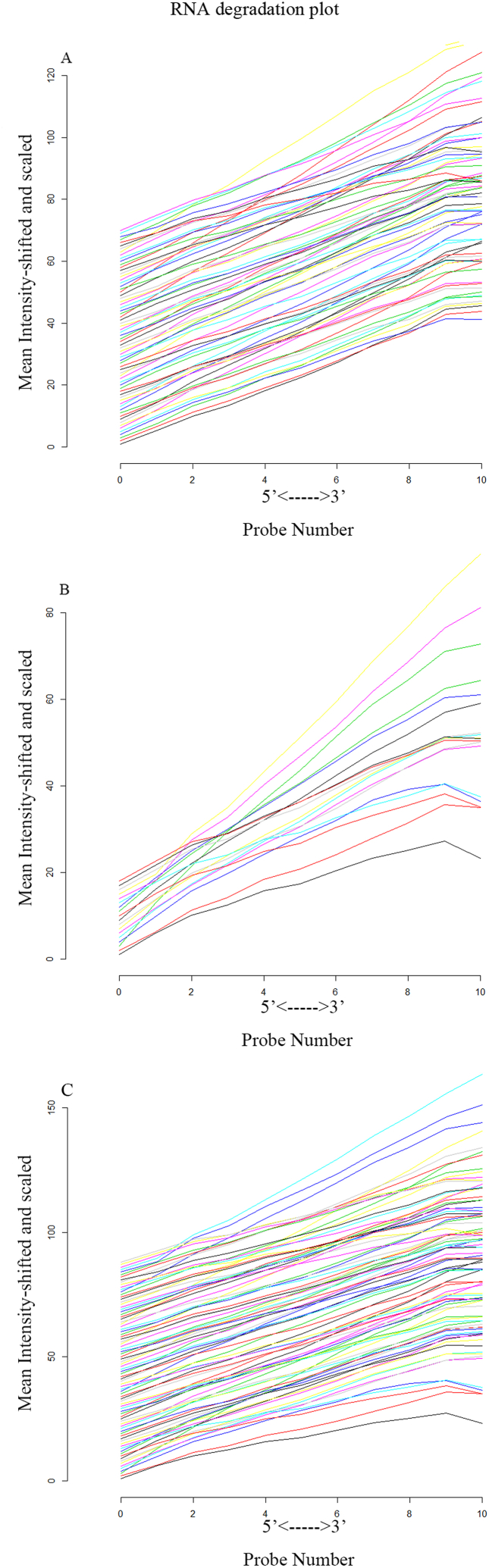
The 70 pGBM samples (including LT, IT, and ST groups) were all extracted from GSE53733 and the 18 normal samples were obtained from the GSE15824, GSE30563 and GSE50161. Affy package in R software was utilized to perform quality control on all CEL files. The RNA degradation plot showed a good consistency in the slope of the line, signifying successful merging of the pGBM groups and the normal group (Fig. 2A–C). Additionally, all samples exhibited a lower numerical value at the 5’ end (Fig. 2A–C).
Fig. 2.
Data quality control of Microarray data.(A) The RNA degradation plot of 18 normal samples from GSE15824, GSE30563 and GSE50161; (B) The RNA degradation plot of 70 pGBM samples from GSE53733; (C) The RNA degradation plot of 18 normal and 70 pGBM samples.
3.2. Identification of DEGs
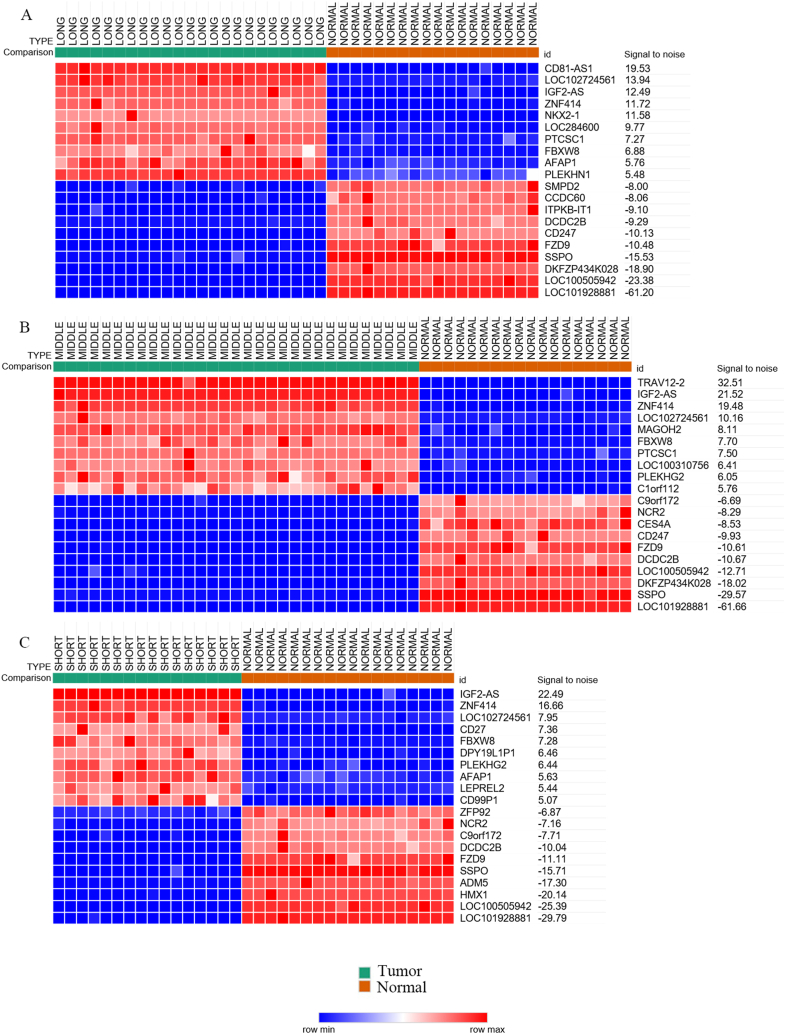
By using limma package of the R software, three comparations were conducted between the LT group and the normal group, the ST group and the normal group, and the IT group and the normal group. The thresholds were |logFC| >2 and P < 0.05. Then, 5035 LTDEGs, 5840 ITDEGs and 5698 STDEGs were identified, and the detail information was displayed in Table 1. Heatmaps of three group of top 10 regulated DEGs were generated using Morpheus online software. The results were shown in Fig. 3A–C.
Table 1.
DEGs between tumor groups and normal group.
| Group | Up-regulated genes | Down-regulated genes | Total account |
|---|---|---|---|
| Long-term vs normal | 2187 | 2848 | 5035 |
| Intermediate-term vs normal | 3107 | 2733 | 5840 |
| Short-term vs normal | 2099 | 3599 | 5698 |
Fig. 3.
Heatmaps of top 10 regulated DEGs of each comparison group. red: up-regulation; blue: down-regulation. The value of expression intensity is based on the gene expression level analysis by R software. (A) Comparison of long-term survival tumor group with normal control group. (B)Comparison of intermediate-term survival tumor group with normal control group. (C)Comparison of short-term survival tumor group with normal control group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Venn diagram of three comparative groups
Venn diagram of three comparative groups showed 3987 genes change in common. There were 260 LTSDEGs genes, 822 ITDEGs and 740 STDEGs in the venn diagram (Fig. 4). Genes that were up-regulated and down-regulated in specific long-term survival group (LTSDEGs) and specific short-term survival group (STSDEGs) were counted separately. The results are displayed in Table .2.
Fig. 4.
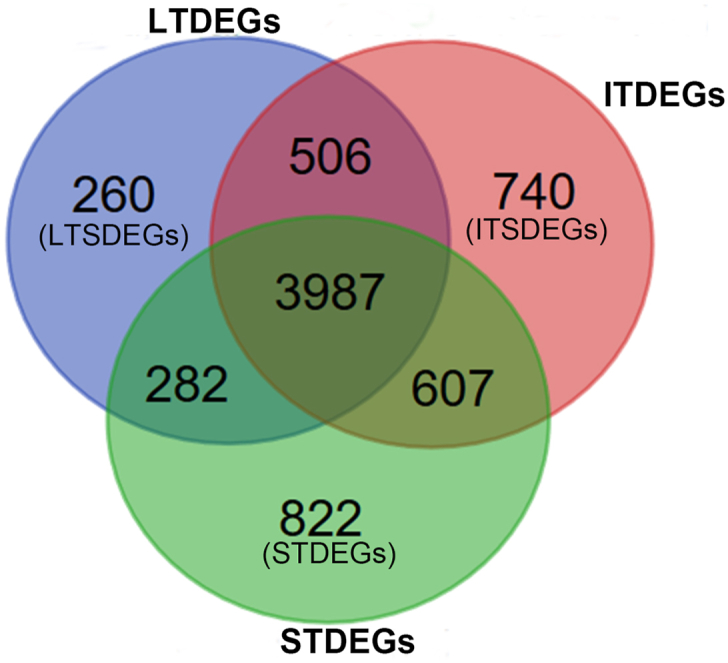
Venn diagram of three comparative groups(LTDEGs, ITSDEGs, and STDEGs). Long-term survival specific differentially expressed genes (LTSDEGs) by excluding all overlapping genes from the LTDEGs. Similarly, the intermediate survival specific differentially expressed genes (ITSDEGs) were identified by excluding all overlapping genes from the ITDEGs, and the short-term survival specific differentially expressed genes (STSDEGs) were obtained by excluding all overlapping genes from the STDEGs.
Table 2.
Statistics of specific DEGs of long-term survival group and short-term survival group of Venn diagram.
| Group | Up-regulated genes | Down-regulated genes | Total account |
|---|---|---|---|
| Specific Long-term survival group | 76 | 184 | 260 |
| Specific Short-term survival group | 149 | 676 | 822 |
3.4. GO term enrichment analysis
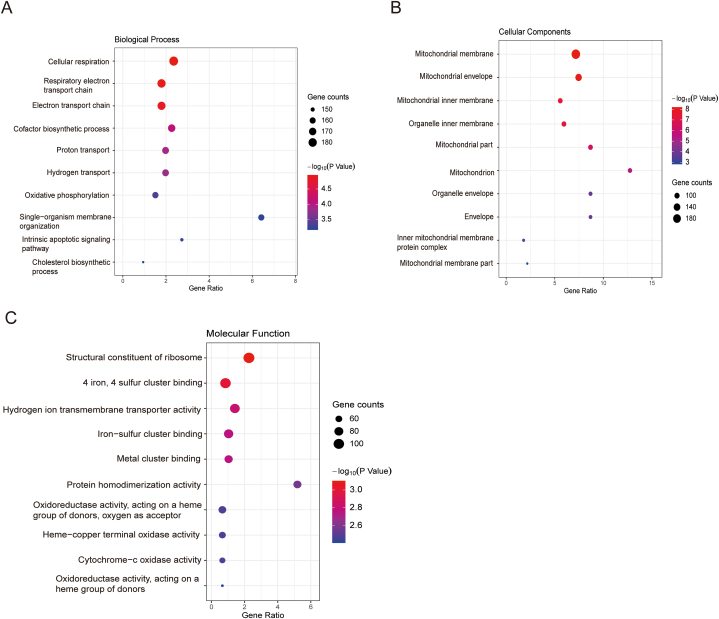
The online software DAVID was used to conduct GO categories and KEGG pathways analysis. All LTSDEGs and STSDEGs were together mapped to the database, and the figures were generated using ggplot2 package of R software. GO analysis indicated that the LTSDEGs and STSDEGs were significantly enriched in biological processes (BP), molecular function(MF) and cell components(CC). Top 10 GO items of each category were shown according to the gene counts. The GO enriched in BP, including cellular respiration, respiratory electron transport chain and electron transport chain (Fig. 5A). In addition, with regards to CC, GO were enriched in mitochondrial membrane, mitochondrial envelope and mitochondrial inner membrane (Fig. 5B). As for MF, GO were enriched in structural constituent of ribosome, 4 iron, 4 sulfur cluster binding and hydrogen ion transmembrane transporter activity (Fig. 5C).
Fig. 5.
Top 10 GO analysis of LTSDEGs and STSDEGs together with GBM. (A)Biological processes; (B)Cell components; (C) Molecular function.
3.5. KEGG pathway analysis
All LTSDEGs and STSDEGs were together enriched pathways in KEGG database. A total of 12 pathways were enriched, with a P value cutoff of <0.05 set as a statistically significant threshold. The top 3 pathways of analysis identified were Huntington's disease, Oxidative phosphorylation and Metabolic pathways. Results are displayed in full in Table .3.
Table 3.
KEGG pathway analysis of LTSDEGs and STSDEGs.
| Pathway ID | Name | Gene count | % | P value | Genes |
|---|---|---|---|---|---|
| hsa05016 | Huntington's disease | 24 | 2.26 | 1.18E-04 | NDUFB4, CREB3, SLC25A5, NDUFA9, NDUFA7, COX8A, CYCS, COX4I1, NDUFC1, SOD1, VDAC2, POLR2C, DCTN4, COX6C, NDUFA11, PPIF, SDHB, EP300, GNAQ, ATP5C1, COX6B1, CREB3L4, PLCB1, NDUFS1 |
| hsa00190 | Oxidative phosphorylation | 19 | 1.79 | 1.40E-04 | NDUFB4, COX10, NDUFA9, ATP6AP1, NDUFA7, COX8A, COX4I1, NDUFC1, ATP6V1G1, COX6C, NDUFA11, ATP6V1C1, SDHB, ATP6V1E2, ATP5C1, COX6B1, ATP6V0A1, ATP5I, NDUFS1 |
| hsa01100 | Metabolic pathways | 90 | 8.49 | 2.48E-04 | IMPA1, GNPDA2, NT5C3B, HMGCR, COX10, ATP6AP1, SAT2, SYNJ1, LSS, PMVK, PDHB, CKB, GOT2, MTHFD2, ST3GAL5, ENOPH1, PDHA1, PLCB1, PTDSS1, ATP5I, AGPAT2, NDUFS1, MINPP1, PIK3C2B, POLE, CYCS, COX4I1, NDUFC1, PFKM, MAN1A1, LPIN1, NDUFA11, COQ6, COX6C, ATP6V1C1, MAN2A2, NME5, GLUL, ADO, SQLE, ATP5C1, PLA2G7, AOC3, NDUFB4, GCNT2, MVD, ENPP1, GLUD2, ST8SIA1, UPP1, UROS, ATP6V1G1, POLR2C, HADHB, GCH1, DCT, MUT, DGKB, PLA2G12A, COX6B1, ALDH4A1, GALE, UGT8, GALNT12, GAD1, DNMT3A, POLR3F, NOS1, MOCS2, ST6GAL2, MSMO1, POLR3H, NDUFA9, MAOA, MGAT4C, NDUFA7, COX8A, MCAT, EPHX2, FDPS, IDH3B, AMPD2, AMPD3, TST, SDHB, ATP6V1E2, ATP6V0A1, DPM3, GAMT, IDI1 |
| hsa05010 | Alzheimer's disease | 19 | 1.79 | 0.0024 | BID, NDUFB4, NOS1, NDUFA9, NDUFA7, COX8A, CYCS, COX4I1, BAD, NDUFC1, COX6C, NDUFA11, SDHB, GNAQ, RYR3, ATP5C1, COX6B1, PLCB1, NDUFS1 |
| hsa05012 | Parkinson's disease | 16 | 1.51 | 0.0060 | NDUFB4, NDUFA9, SLC25A5, NDUFA7, COX8A, CYCS, COX4I1, NDUFC1, VDAC2, COX6C, NDUFA11, PPIF, SDHB, ATP5C1, COX6B1, NDUFS1 |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 8 | 0.75 | 0.0132 | BID, SLC1A2, NOS1, GRIA2, MAPK12, CYCS, BAD, SOD1 |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 15 | 1.42 | 0.0228 | BID, NDUFB4, NDUFA9, NDUFA7, CYCS, COX8A, COX4I1, NDUFC1, DDIT3, COX6C, NDUFA11, SDHB, COX6B1, NDUFS1, PIK3R1 |
| hsa03050 | Proteasome | 7 | 0.66 | 0.239 | PSMB10, PSMA1, PSMD14, PSMC6, PSMB6, PSMD12, PSMD8 |
| hsa00900 | Terpenoid backbone biosynthesis | 5 | 0.47 | 0.0240 | MVD, HMGCR, FDPS, PMVK, IDI1 |
| hsa04728 | Dopaminergic synapse | 13 | 1.23 | 0.0311 | CREB3, MAOA, PPP2R5D, GRIA4, GNAQ, MAPK12, GRIA2, GNG10, CREB3L4, PPP2R5E, GNB3, PPP2R2B, PLCB1 |
| hsa00330 | Arginine and proline metabolism | 7 | 0.66 | 0.0416 | GOT2, NOS1, MAOA, SAT2, ALDH4A1, GAMT, CKB |
| hsa01130 | Biosynthesis of antibiotics | 18 | 1.70 | 0.0447 | MSMO1, MVD, HMGCR, FDPS, IDH3B, LSS, PFKM, AMPD2, AMPD3, PDHB, HADHB, GOT2, NME5, SDHB, SQLE, PLA2G7, PDHA1, IDI1 |
3.6. Verification of survival related genes by UALCAN database
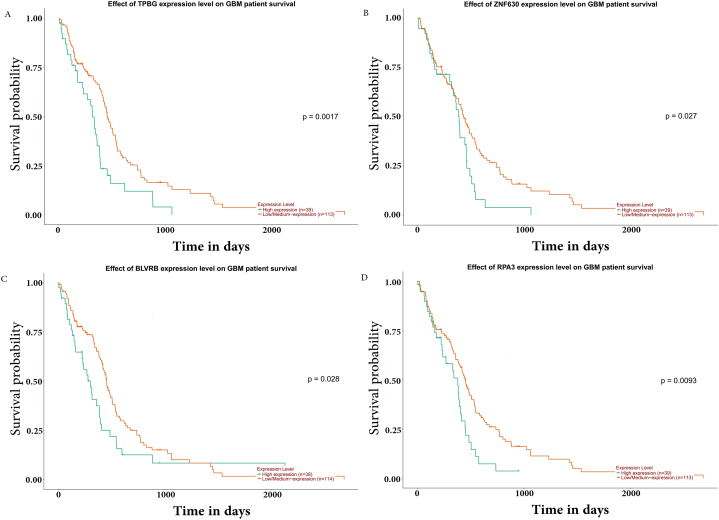
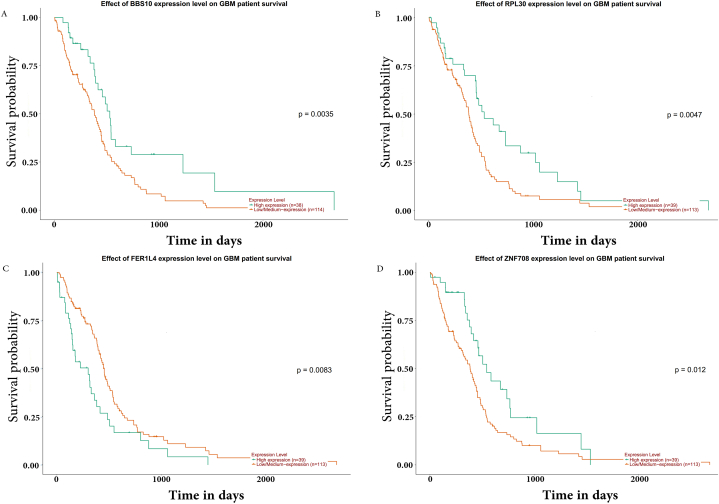
We upload all LTSDEGs and STSDEGs to the UALCAN to verify survival related genes. After analyzing gene expression level and its effect on the survival curve,we identified 9 genes in the LTVSDEGs and 18 genes in the STVSDEGs. Results are displayed in full in Table .4. Top 4 genes of each group were shown in Fig. 6A–D and Fig. 7A–D, based on their P values.
Table 4.
Survival related genes of LTVSDEGs and STVSDEGs.
| Group | Gene Name | p value |
|---|---|---|
| LTVSDEG | BLVRB | 0.028 |
| LTVSDEG | FAM110C | 0.047 |
| LTVSDEG | FOSB | 0.03 |
| LTVSDEG | MGST1 | 0.041 |
| LTVSDEG | RPA3 | 0.0093 |
| LTVSDEG | SLC2A3 | 0.047 |
| LTVSDEG | TPBG | 0.0017 |
| LTVSDEG | ULBP2 | 0.029 |
| LTVSDEG | ZNF630 | 0.027 |
| STVSDEG | ASCC1 | 0.022 |
| STVSDEG | ATP1F1 | 0.015 |
| STVSDEG | BAALC | 0.025 |
| STVSDEG | BBS10 | 0.0035 |
| STVSDEG | CDC5L | 0.049 |
| STVSDEG | CHRNB2 | 0.049 |
| STVSDEG | EP300 | 0.014 |
| STVSDEG | FER1L4 | 0.0083 |
| STVSDEG | GLUD2 | 0.013 |
| STVSDEG | KCNT2 | 0.016 |
| STVSDEG | MFF | 0.034 |
| STVSDEG | PFN2 | 0.04 |
| STVSDEG | PPM1L | 0.042 |
| STVSDEG | RNF115 | 0.041 |
| STVSDEG | RPL30 | 0.0047 |
| STVSDEG | SF3A1 | 0.015 |
| STVSDEG | ZNF708 | 0.012 |
| STVSDEG | ZNRF3 | 0.037 |
Fig. 6.
Top 4 genes related to survival of LTVSDEGs. (A)TPBG; (B)ZNF630; (C)BLVRB; (D)RPA3.
Fig. 7.
Top 4 genes related to survival of STVSDEGs. (A) BBS10; (B)RPL30; (C)FER1L4; (D)ZNF708.
3.7. Verification of top 4 LTVSDEGs by HPA database
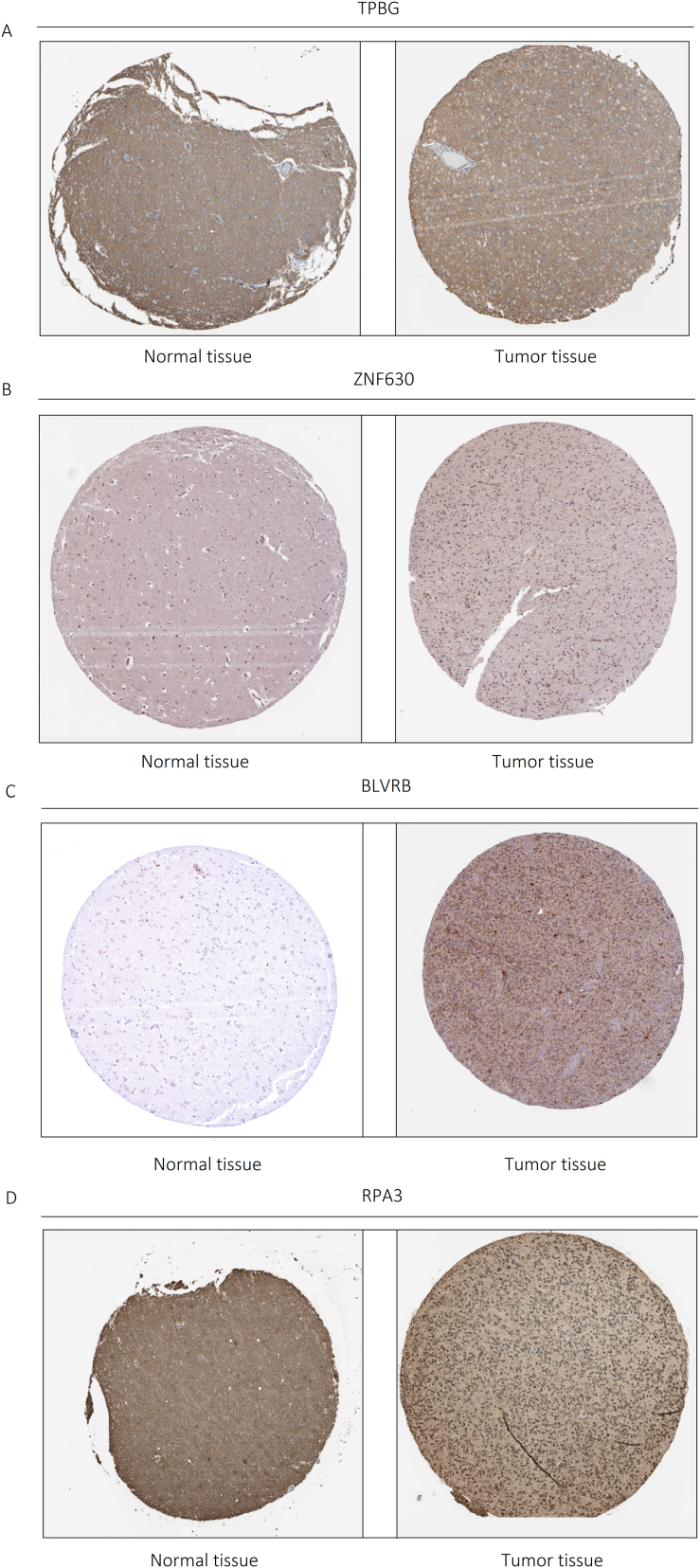
The expression of Top 4 LTVSDEGs were verified in the HPA http://www.proteinatlas.orgonline database. We compared the intensity of immunohistochemical staining between normal tissue and tumor tissue. ZNF630, BLVRB and RPA3 were verified, while TPBG was not detected. Results were displayed in Fig. 8A–D.
Fig. 8.
Protein verification by Immunohistochemical staining. Image from The Human Protein Atlas(http://www.proteinatlas.org) (A)TPBG(https://www.proteinatlas.org/ENSG00000146242-TPBG/tissue/cerebral+cortex#img;https://www.proteinatlas.org/ENSG00000146242-TPBG/pathology/glioma#img); (B)ZNF630(https://www.proteinatlas.org/ENSG00000221994-ZNF630/tissue/cerebral+cortex#img;https://www.proteinatlas.org/ENSG00000221994-ZNF630/pathology/glioma#img); (C) BLVRB(https://www.proteinatlas.org/ENSG00000090013-BLVRB/tissue/cerebral+cortex#img;https://www.proteinatlas.org/ENSG00000090013-BLVRB/pathology/glioma#img); (D)RPA3(https://www.proteinatlas.org/ENSG00000106399-RPA3/tissue/cerebral+cortex#img;https://www.proteinatlas.org/ENSG00000106399-RPA3/pathology/glioma#img).
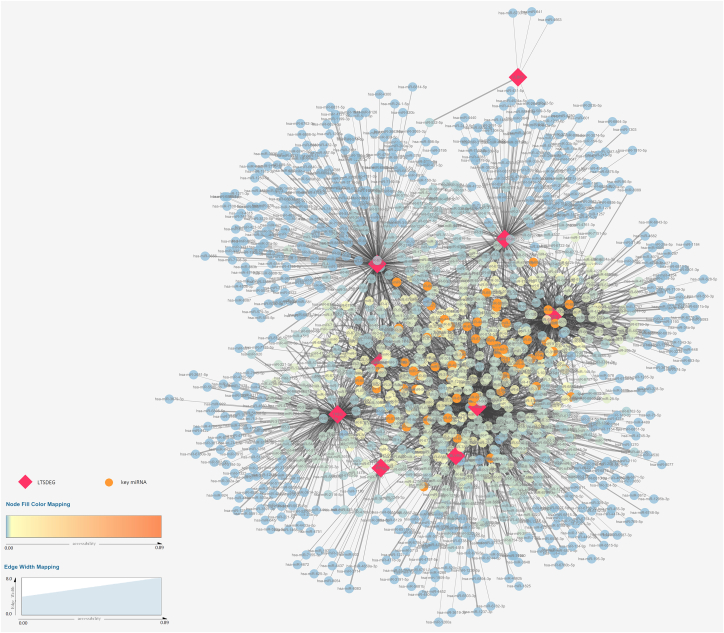
3.8. Establishment of the predicted key miRNAs-LTVSDEGs interaction network
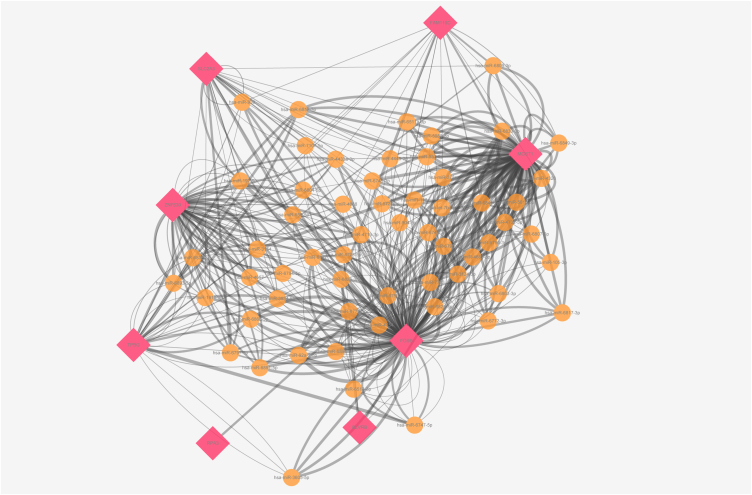
Through the analysis of Cytoscape software, we obtained 59 key miRNAs in total. Results were shown in Table .5. The entire miRNAs-LTVSDEGs interaction network was displayed in figs1 and the key miRNAs-LTVSDEGs interaction network was displayed in Fig. 9.
Table 5.
Key miRNAs in predicted miRNAs-LTVSDEGs interaction network.
| name | Neighborhood Connectivity | Number of Directed Edges |
|---|---|---|
| hsa-miR-1910-3p | 345.75 | 10 |
| hsa-miR-6826-3p | 300.75 | 7 |
| hsa-miR-6127 | 388.67 | 15 |
| hsa-miR-936 | 389.75 | 9 |
| hsa-miR-3124-3p | 439.50 | 10 |
| hsa-miR-6512-3p | 694.00 | 7 |
| hsa-miR-6809-3p | 262.50 | 7 |
| hsa-miR-4688 | 328.00 | 9 |
| hsa-miR-4446-3p | 431.33 | 7 |
| hsa-miR-656-5p | 447.33 | 8 |
| hsa-miR-4433a-3p | 337.75 | 8 |
| hsa-miR-6891-5p | 375.50 | 7 |
| hsa-miR-92a-2-5p | 455.50 | 9 |
| hsa-miR-3934-5p | 337.75 | 7 |
| hsa-miR-378g | 381.75 | 9 |
| hsa-miR-7703 | 439.50 | 10 |
| hsa-miR-6747-5p | 455.50 | 7 |
| hsa-miR-198 | 377.75 | 9 |
| hsa-miR-6804-5p | 312.00 | 8 |
| hsa-miR-105-3p | 439.50 | 7 |
| hsa-miR-6740-3p | 365.33 | 11 |
| hsa-miR-197-5p | 345.75 | 9 |
| hsa-miR-504-5p | 463.50 | 7 |
| hsa-let-7b-3p | 439.50 | 7 |
| hsa-miR-6829-3p | 312.67 | 8 |
| hsa-miR-6794-5p | 375.50 | 7 |
| hsa-miR-6817-3p | 439.50 | 7 |
| hsa-miR-3154 | 288.25 | 7 |
| hsa-miR-6866-3p | 455.50 | 8 |
| hsa-miR-6807-5p | 439.50 | 10 |
| hsa-miR-6852-3p | 201.00 | 7 |
| hsa-miR-6803-3p | 312.67 | 9 |
| hsa-miR-6797-5p | 388.67 | 8 |
| hsa-miR-6777-3p | 376.50 | 7 |
| hsa-miR-4326 | 370.67 | 7 |
| hsa-miR-937-3p | 431.33 | 9 |
| hsa-miR-6795-5p | 439.50 | 8 |
| hsa-miR-6750-3p | 439.50 | 10 |
| hsa-miR-6790-3p | 439.50 | 7 |
| hsa-miR-5088-3p | 439.50 | 8 |
| hsa-miR-7107-5p | 447.33 | 7 |
| hsa-miR-6772-3p | 439.50 | 8 |
| hsa-miR-6893-5p | 230.50 | 9 |
| hsa-miR-4754 | 463.50 | 7 |
| hsa-miR-6748-3p | 431.33 | 9 |
| hsa-miR-6894-5p | 375.50 | 8 |
| hsa-miR-5010-3p | 439.50 | 7 |
| hsa-miR-4725-5p | 270.00 | 8 |
| hsa-miR-4713-3p | 328.00 | 8 |
| hsa-miR-4732-5p | 439.50 | 7 |
| hsa-miR-6849-3p | 370.67 | 7 |
| hsa-miR-4667-3p | 431.33 | 8 |
| hsa-miR-6728-3p | 431.33 | 7 |
| hsa-miR-6841-3p | 285.25 | 8 |
| hsa-miR-4657 | 388.67 | 8 |
| hsa-miR-3605-5p | 455.50 | 7 |
| hsa-miR-6838-3p | 381.33 | 13 |
| hsa-miR-6511a-3p | 431.33 | 7 |
| hsa-miR-6833-3p | 370.67 | 8 |
Fig. 9.
The predicted key miRNAs-LTVSDEGs interaction network.
3.9. Validation of the key miRNAs and construction of the key miRNAs-LTVSDEGs network
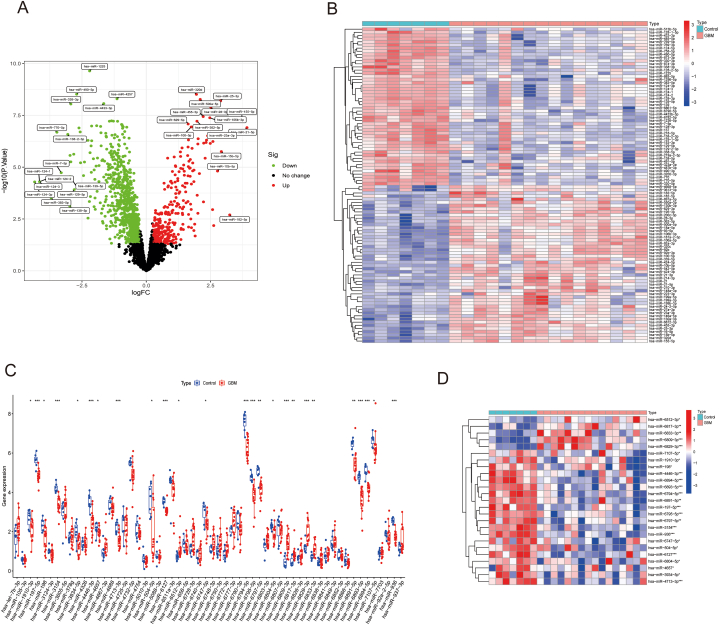
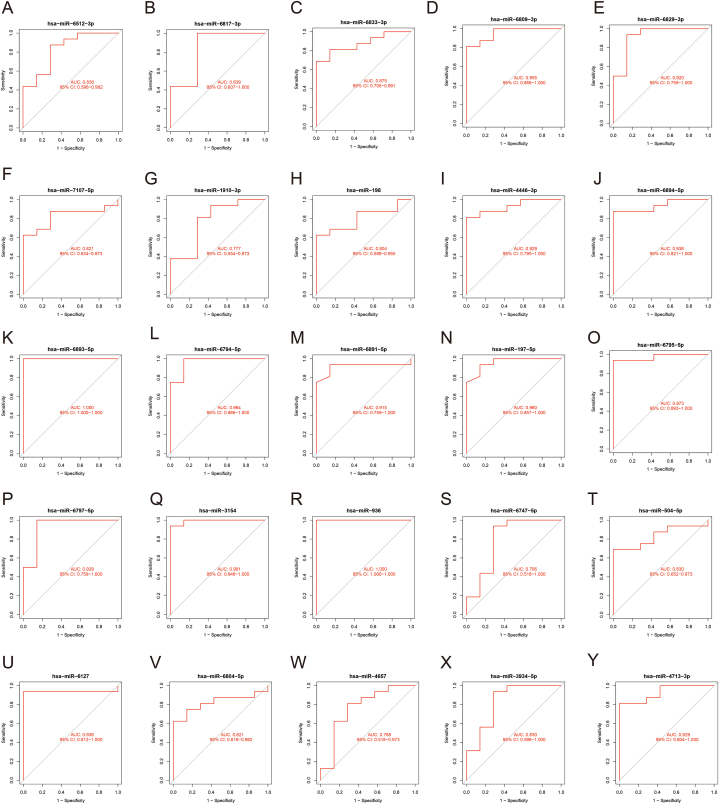
To verify the expression of the key miRNAs, we plotted the overall miRNA expression levels of the GSE90603 via volcano map and heatmap (Fig. 10A–B). The boxplot showed the statistical significance level of these miRNAs(Fig. 10C–D). Compared with Control group, 25 miRNAs exhibited abnormal expression in GBM. Among them, hsa-miR-6512-3p, hsa-miR-6817-3p, hsa-miR-6833-3p, hsa-miR-6809-3p, hsa-miR-6829-3p were up-regulated in GBM, while hsa-miR-7107-5p, hsa-miR-1910-3p, hsa-miR-198, hsa-miR-4446-3p, hsa-miR-6894-5p, hsa-miR-6893-5p, hsa-miR-6794-5p, hsa-miR-6891-5p, hsa-miR-197-5p, hsa-miR-6795-5p, hsa-miR-6797-5p, hsa-miR-3154, hsa-miR-936, hsa-miR-6747-5p, hsa-miR-504-5p, hsa-miR-6127, hsa-miR-6804-5p, hsa-miR-4657, hsa-miR-3934-5p, hsa-miR-4713-3p were down-regulated (Fig. 10C–D). The ROC curve was used to measure predictive utility, and it was discovered that these 25 miRNAs all had a remarkable distinguishing efficiency with a high AUC values (Fig. 11).
Fig. 10.
Validation of the key miRNAs. (A-B)Volcano plot of the differentially expressed miRNAs in the GSE90603 testing dataset; (C) Heatmap of the differentially expressed miRNAs in the GSE90603 testing dataset; (C–D) Boxplot and heatmap of the expression of key miRNAs in the GSE90603 testing dataset.
Fig. 11.
ROC curve analysis of the 25 key miRNAs. A-Y ROC curve analysis of hsa-miR-6512-3p(A), hsa-miR-6817-3p(B), hsa-miR-6833-3p(C), hsa-miR-6809-3p(D), hsa-miR-6829-3p(E), hsa-miR-7107-5p(F), hsa-miR-1910-3p(G), hsa-miR-198(H), hsa-miR-4446-3p(I), hsa-miR-6894-5p(J), hsa-miR-6893-5p(K), hsa-miR-6794-5p(L), hsa-miR-6891-5p(M), hsa-miR-197-5p(N), hsa-miR-6795-5p(O), hsa-miR-6797-5p(P), hsa-miR-3154(Q), hsa-miR-936(R), hsa-miR-6747-5p(S), hsa-miR-504-5p(T), hsa-miR-6127(U), hsa-miR-6804-5p(V), hsa-miR-4657(W), hsa-miR-3934-5p(X) and hsa-miR-4713-3p(Y).
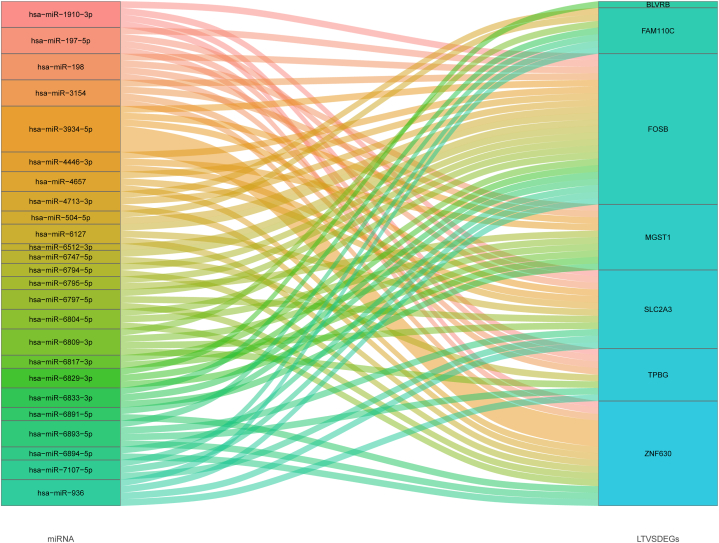
Based on these 25 miRNAs and the predicted key miRNAs-LTVSDEGs interaction network, we finally constructed the key miRNAs-LTVSDEGs network by using Sankey diagram, including 25 miRNAs and 7 LTVSDEGs (Fig. 12).
Fig. 12.
Sankey diagram displaying the key miRNAs-LTVSDEGs interaction network. Each rectangle represents a miRNA or mRNA, and the connection degree is visualized based on the size of the rectangle.
4. Discussion
GBM exhibits a high degree of heterogeneity, and variations in gene expressions play a crucial role in influencing the survival of GBM patients. In clinical practice, the vast majority of patients have a short survival time, and only a very small number of patients can observe long-term survival. Therefore, conducting research on GBM patients with long-term survival is of great clinical importance and scientific significance. In our study, we obtained a specific expression gene sets for long-term survival patients by analyzing the gene transcriptome expression database of clinical long-term survival patients. In recent years, research on miRNA has grown rapidly. The relationship between miRNA and tumors is constantly expounded. Some miRNAs have the potential to serve as diagnostic biomarkers, while others directly act on key genes involved in the disease [24,25]. Additionally, studies indicatedthat different ethnicities may exhibit differential expression of miRNAs in breast cancer [26]. The role of miRNAs in the disease, particularly in tumors, cannot be overlooked. In scientific research practice, there are an excessive number of miRNAs that can affect the specific expression of genes in long-term survival GBM patients, and it is imperative to pay attention to the network of key miRNA interactions. Constructing this network helps us gain a deeper understanding of the interfering mechanism of miRNA in GBM at the level of important gene sets such as LTVSDEGs. This process can identify new targets and miRNA target networks for intervention in GBM.
Current research highlights the crucial role of miRNAs in various aspects of GBM, including growth, cell proliferation, cell cycle regulation, apoptosis, invasion, glioma stem cell behavior, angiogenesis, reversal of temozolomide resistance, and their potential as biomarkers for GBM diagnosis and treatment [[27], [28], [29]]. Despite the rapid development of medical technology in recent years, there is no obvious improvement in the overall survival of the GBM patient. At the same time, there are no research focus on the key miRNAs-LTVSDEGs network so far. Our study not only identified the key LTVSDEGs but also unveiled a key miRNAs-LTVSDEGs network in GBM. The miRNAs within this network may hold the key to influencing critical factors in the survival of cancer patients. This exploration of the miRNA-gene interaction network provides a novel perspective and opens avenues for further research to better understand the intricate mechanisms underlying the survival dynamics of glioblastoma patients.
More concretely, we obtained predicted a key miRNAs-LTVSDEGs network, including 9 key LTVSDEGs and 59 key miRNAs. Also, we obtained 18 STVSDEGs at the same time. We use 2 type of databases to verify our results. Firstly, we used microarray data analysis to filter the specific gene in LT patients and ST patients. Then, we used RNA-seq based database to verify the data and filter out 3 genes in LT patients and 19 genes in ST patiens. Finally, protein expression database was used to verify top 4 LTVSDEGs both in clinical normal tissue and tumor tissue. It is worth noting that in our verification, RPA3, ZNF630 and BLVRB was verified, but TPBG was not detected. Considering the limited number of samples in the database and RNA-seq data, we acknowledge that the possibility of verification is very large when data are abundant enough in the clinical staining database.
On the other hand, we have made a summary of previous studies. A published study shown that polymorphisms in RPA3, combinating with previous reaserches, were associated with glioma developing in Chinese population [30]. Besides, there have been no studies focusing on TPBG, ZNF630 and BLVRB with GBM. Moreover, all LTSDEGs and STSDEGs were together conducted GO and KEGG analysis. Results showed that cellular respiration, respiratory electron transport chain, and electron transport chain were the most biological processes, while mitochondrial membrane, mitochondrial envelope and mitochondrial inner membrane were the most cell components biological processes. Molecular function mainly focused on structural constituent of ribosome, 4 iron, 4 sulfur cluster binding and hydrogen ion transmembrane transporter activity. The KEGG results showed that the process of diseasewas mainly concentrated on Huntington's disease, Oxidative phosphorylation and Metabolic pathways. These findings not only inform us about the concentration of differences in tumors related to metabolic changes but also establish a significant connection with Huntington's disease. As is well-known, Huntington's disease is also a metabolic diseases of the central nervous system. The abnormality of nerve metabolism in Huntington's disease may have same gene alternation with pGBM patient. For example, research demonstrated that mitochondrial SIRT3 was involved in neurodegenerative brain disorders including Huntington's disease, and it also played an important role in glycolytic metabolism of GBM cells [31,32].
Finally, We obtained 59 key miRNA in key miRNAs-LTVSDEGs interaction network and conducted in-depth validation of this result using the GSE90603 dataset. Among the 25 miRNAs identified, which showed abnormal expression and good AUC value in GBM, and we further constructed the key miRNAs-LTVSDEGs network, including 25 miRNAs and 7 LTVSDEGs. This result indicated that these 25 miRNAs are reliable. The expression levels of these miRNAs are highly correlated with the occurrence and development of diseases, especially the survival period of patients, possibly by affecting the expression levels of LTVSDEGs genes to enable patients to have a longer or shorter survival. Therefore, our analysis shown that the changes in the expression levels of these 25 miRNAs are strongly correlated with the occurrence and development of tumors, which names miR-6512-3p, miR-6817-3p, miR-6833-3p, miR-6809-3p, miR-6829-3p, miR-7107-5p, miR-1910-3p, miR-198, miR-4446-3p, miR-6894-5p, miR-6893-5p, miR-6794-5p, miR-6891-5p, miR-197-5p, miR-6795-5p, miR-6797-5p, miR-3154, miR-936, miR-6747-5p, miR-504-5p, miR-6127, miR-6804-5p, miR-4657, miR-3934-5p, miR-4713-3p. Then, we retrieved the relationship between 25 key miRNA and cancer in the pubmed database. It is speculated that it also has a similar role in GBM and is worth further exploration since a series of studies have confirmed that these miRNAs have a significant impact on the occurrence and development of tumors. These studies are listed as follows: miR-6512-3p has ability as a sponge with genes such as CYTOR, SNHG7 and HOTTIP [[33], [34], [35]]. miR-6817-3p stabilizes Wnt2B through the mechanism of ceRNA combined with AC104041.1, thereby exerting anticancer effects [36]; miR-6833-3p regulates the tumorigenesis effect of colorectal through the LINC-ROR/miR-6833-3p/SMC4 axis [37]; miR-6809-3p, miR-6829-3p, and miR-6804-5p are one of the miRs targeting overexpressed genes in lung cancer [38]; miR-7107-5p and miR-6891-5p are one of the markers identified in endoscopic cholangiopancreatography for the treatment of bile in malignant biliary strictures [39]; miR-1910-3p in exosomes activates NF-kB pathway through MTMR3 to promote proliferation, metastasis and autophagy in breast cancer [40]; The miR198-CUL4B pathway mediates circMFN2 induced glycolysis in ovarian cancer and has sensitization effect in gemcitabine treatment of pancreatic cancer [41,42]; miR-4446-3p was found in the serum of colon adenocarcinoma patients and has diagnostic value [43]; miR-6894-5p promotes gastric cancer progression through the activation of circAFF2 [44]; miR-6794-5p participates in constructing a large-scale microRNA map detection model for esophageal squamous cell carcinoma [45]; miR-197-5p adsorbed by circ0125803 in glioblastoma can lead to tumor progression and it increases the anticancer cytotoxicity of HT1080 fibrosarcoma cells mediated by doxorubicin by reducing drug efflux [46,47]; miR-6795-5p is an important component of LINC01013-miR-6795-5p-FMNL3 axis regulation of liver cancer stem cell characteristics [48]; miR-3154 is one of the potential prognostic biomarkers for cervical cancer and it is one of the potential prognostic biomarkers for cervical cancer [49,50]; miR-936 mediates cell proliferation and invasion in glioma through the miR-936/ERBB4 axis and it mediate the acquisition of non androgen dependent prostate cancer metastasis phenotype when it lossed. It also targeting GPR78 to regulate chemotherapy resistance in non-small cell lung cancer [51]; Down regulation of lincRNA DSCR9 delays breast cancer progression by regulating microRNA-504-5p-dependent G-protein-coupled receptor 65 [52]; The content of miR-6127 significantly increased in extracellular vesicles of metastatic colorectal cancer cell lines intervened by rapamycin [53]; EWI-2, located on the cell membrane and extracellular vesicles of miR-3934-5p, is a key molecule that regulates the distribution of miR-3934-5p and has the function of inhibiting prostate cancer cell metastasis [54]. In addition, miR-4713-3p is the most important miR that affects the expression of EPHX3 gene in head and neck squamous cell carcinoma, often leading to poor prognosis and tumor immune infiltration [55].
However, The complexity of the central nervous system and the presence of physiological disorders pose a significant scientific challenge in traditional drug delivery methods [56].Delivering miRNAs as mimetics or antagonists for GBM treatment is challenging due to their polyanionic properties, moderate in vivo stability, reduced absorption, adverse pharmacokinetic characteristics, high sensitivity to serum nucleases, rapid renal clearance, non-specific biological distribution, and limited ability to cross cell membranes, including the blood-brain barrier (BBB). Therefore, appropriate delivery systems are needed to address the aforementioned challenges to ensure appropriate features and promising results. Therefore, it is crucial to seek an effective and biocompatible delivery system that promotes nucleic acid penetration into specific areas of the brain without degradation [57].At present, the solution to this problem is to nanocrystallize drug formulations. For example, Liu et al. [58] developed polymeric nanoparticles to coload anti-miR-21 and miR-124 mimics into the brain to efficiently treat GBM. Another study used solid lipid nanoparticles to deliver anti-miR-21 and pemetrexed for treating GBM therapy [59]. Teplyuk et al. [60] using the convection-enhanced delivery (CED) technique for the sustained release of anti miR-10b-containing nanoparticles for two weeks, via intracranial osmotic pumps and it mitigated the growth and development of GBM without observing any toxicity.
There are several limitations to the current study. Firstly, gender, age, and others pathological factors of patients were not included in this study, which might be incorporated in future research. Moreover, our present study was based on bioinformatics analysis, and though we employed external datasets for validation, additional in vivo and in vitro research would be necessary to confirm the regulatory relationships in the key miRNAs-LTVSDEGs network of pGBM.
5. Conclusion
Combine microarray database analysis with RNA-seq database verification of the our study provides a comprehensive analysis of survival related DEGs that may be involved in the progress of pGBM. The key miRNAs-LTVSDEGs interaction network provides a set of important miRNA targets for future investigation into the molecular mechanisms and prognostic biomarkers involved in pGBM. With the development of research, more functions of miRNA in pGBM will be clarified and it may be the key node to lengthens the quality of life and survival time.
Funding
The present study was supported by Key Scientific Research Platform and Project of Universities in Guangdong Province (Youth Innovative Talents Project of Education Department of Guangdong Province, 2018GkQNCX085) and the project of Guangzhou Key Laboratory of Child Neurodevelopment (202201020620).
Ethics statement
This study was a bioinformatic analysis and had no ethical implications.
Data availability statement
The datasets generated and/or analyzed during the current study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE53733, GSE15824, GSE30563 and GSE50161. Other datas are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Lingqi Zhou: Writing – review & editing, Resources, Investigation, Formal analysis, Conceptualization. Xuemei Liu: Writing – review & editing, Resources, Investigation, Data curation. Tong Wu: Resources, Investigation, Data curation. Qundi Liu: Investigation, Formal analysis. Meilian Jing: Writing – review & editing, Investigation, Formal analysis. Huahan Li: Writing – review & editing, Formal analysis, Data curation, Conceptualization. Ning Xu: Investigation, Data curation, Conceptualization. Hai Tang: Writing – review & editing, Writing – original draft, Resources, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28439.
Contributor Information
Lingqi Zhou, Email: zhoulq6@mail2.sysu.edu.cn.
Ning Xu, Email: 67781904@qq.com.
Hai Tang, Email: tangh25@mail2.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- 1.Whitfield B.T., Huse J.T. Classification of adult-type diffuse gliomas: impact of the world health organization 2021 update. Brain Pathol. 2022;32(4) doi: 10.1111/bpa.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. Mckinnon, M. Nandhabalan, S.A. Murray, P. Plaha, Glioblastoma: clinical presentation, diagnosis, and management, BMJ Br. Med. J. (Clin. Res. Ed.) 3742021) n1560, 10.1136/bmj.n1560. [DOI] [PubMed]
- 3.Ma R., Taphoorn M., Plaha P. Advances in the management of glioblastoma. J. Neurol. Neurosurg. Psychiatry. 2021;92(10):1103–1111. doi: 10.1136/jnnp-2020-325334. [DOI] [PubMed] [Google Scholar]
- 4.E. Le Rhun, M. Preusser, P. Roth, et al., Molecular targeted therapy of glioblastoma, Cancer Treat. Rev. 802019) 101896, 10.1016/j.ctrv.2019.101896. [DOI] [PubMed]
- 5.A.L. Hung, T. Garzon-Muvdi, M. Lim, Biomarkers and immunotherapeutic targets in glioblastoma, World Neurosurg. 1022017) 494-506, 10.1016/j.wneu.2017.03.011. [DOI] [PubMed]
- 6.Stupp R., Mason W.P., van den Bent M.J., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.Stupp R., Hegi M.E., Mason W.P., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase iii study: 5-year analysis of the eortc-ncic trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 8.B. Xu, J. Mei, W. Ji, et al., Micrornas involved in the egfr pathway in glioblastoma, Biomed. Pharmacother. 1342021) 111115, 10.1016/j.biopha.2020.111115. [DOI] [PubMed]
- 9.N. Mercatelli, S. Galardi, S.A. Ciafre, Micrornas as multifaceted players in glioblastoma multiforme, Int. Rev. Cell Mol. Biol. 3332017) 269-323, 10.1016/bs.ircmb.2017.03.002. [DOI] [PubMed]
- 10.Peng Y., He X., Chen H., et al. Inhibition of microrna-299-5p sensitizes glioblastoma cells to temozolomide via the mapk/erk signaling pathway. Biosci. Rep. 2018;38(5) doi: 10.1042/BSR20181051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Nikaki A., Piperi C., Papavassiliou A.G. Role of micrornas in gliomagenesis: targeting mirnas in glioblastoma multiforme therapy. Expert Opin. Investig. Drugs. 2012;21(10):1475–1488. doi: 10.1517/13543784.2012.710199. [DOI] [PubMed] [Google Scholar]
- 12.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. Affy--analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.I. Diboun, L. Wernisch, C.A. Orengo, M. Koltzenburg, Microarray analysis after rna amplification can detect pronounced differences in gene expression using limma, BMC Genomics 72006) 252, 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed]
- 14.Irizarry R.A., Hobbs B., Collin F., et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M., Ball C.A., Blake J.A., et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The gene ontology (go) project in 2006 Nucleic Acids Res. 2006;34(Database issue):D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Chandrashekar D.S., Bashel B., Balasubramanya S., et al. Ualcan: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M., Zhang C., Lee S., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352) doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 22.Sticht C., De La Torre C., Parveen A., Gretz N. Mirwalk: an online resource for prediction of microrna binding sites. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F.Y., Zhou C.Y., Liu Y.B., Wang B., Mao L., Li Y. Mir-483 is down-regulated in gastric cancer and suppresses cell proliferation, invasion and protein o-glcnacylation by targeting ogt. Neoplasma. 2018;65(3):406–414. doi: 10.4149/neo_2018_170608N411. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y., Chen F., Shen L., et al. Biomarker micrornas for prostate cancer metastasis: screened with a network vulnerability analysis model. J. Transl. Med. 2018;16(1):134. doi: 10.1186/s12967-018-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard J., Burns P.A., Hughes T.A., et al. Differential expression of micrornas in breast cancers from four different ethnicities. Pathobiology. 2018;85(4):220–226. doi: 10.1159/000488456. [DOI] [PubMed] [Google Scholar]
- 27.Huang S.W., Ali N.D., Zhong L., Shi J. Micrornas as biomarkers for human glioblastoma: progress and potential. Acta Pharmacol. Sin. 2018;39(9):1405–1413. doi: 10.1038/aps.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y.N., Zhang X.H., Wang Y.M., Zhang X., Gu Z. Mir-204 reverses temozolomide resistance and inhibits cancer initiating cells phenotypes by degrading fap-alpha in glioblastoma. Oncol. Lett. 2018;15(5):7563–7570. doi: 10.3892/ol.2018.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G., Chen L., Khan A.A., et al. Mirna-124-3p/neuropilin-1(nrp-1) axis plays an important role in mediating glioblastoma growth and angiogenesis. Int. J. Cancer. 2018;143(3):635–644. doi: 10.1002/ijc.31329. [DOI] [PubMed] [Google Scholar]
- 30.Jin T., Wang Y., Li G., et al. Analysis of difference of association between polymorphisms in the xrcc5, rpa3 and rtel1 genes and glioma, astrocytoma and glioblastoma. Am. J. Cancer Res. 2015;5(7):2294–2300. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G., Fu X.L., Wang J.J., Guan R., Sun Y., Tony T.S. Inhibition of glycolytic metabolism in glioblastoma cells by pt3glc combinated with pi3k inhibitor via sirt3-mediated mitochondrial and pi3k/akt-mapk pathway. J. Cell. Physiol. 2019;234(5):5888–5903. doi: 10.1002/jcp.26474. [DOI] [PubMed] [Google Scholar]
- 32.Anamika, A. Khanna, P. Acharjee, A. Acharjee, S.K. Trigun, Mitochondrial sirt3 and neurodegenerative brain disorders, J. Chem. Neuroanat. 952019) 43-53, 10.1016/j.jchemneu.2017.11.009. [DOI] [PubMed]
- 33.S. Tu, Y. Chen, Y. Feng, et al., Lncrna cytor facilitates osteogenic differentiation of human periodontal ligament stem cells by modulating sox11 via sponging mir-6512-3p, Stem Cells Int. 20232023) 5671809, 10.1155/2023/5671809. [DOI] [PMC free article] [PubMed]
- 34.W. Chen, Snhg7 promotes the osteo/dentinogenic differentiation ability of human dental pulp stem cells by interacting with hsa-mir-6512-3p in an inflammatory microenvironment, Biochem. Biophys. Res. Commun. 5812021) 46-52, 10.1016/j.bbrc.2021.09.081. [DOI] [PubMed]
- 35.Wei H., Xu Z., Chen L., et al. Long non-coding rna paarh promotes hepatocellular carcinoma progression and angiogenesis via upregulating hottip and activating hif-1alpha/vegf signaling. Cell Death Dis. 2022;13(2):102. doi: 10.1038/s41419-022-04505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M., Ding X., Zhang Y., et al. Antisense oligonucleotides targeting lncrna ac104041.1 induces antitumor activity through wnt2b/beta-catenin pathway in head and neck squamous cell carcinomas. Cell Death Dis. 2020;11(8):672. doi: 10.1038/s41419-020-02820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.X. Li, W. Chen, J. Jia, et al., The long non-coding rna-ror promotes the tumorigenesis of human colorectal cancer by targeting mir-6833-3p through smc4, OncoTargets Ther.. 132020) 2573-2581, 10.2147/OTT.S238947. [DOI] [PMC free article] [PubMed]
- 38.Chakraborty S., Nath D. A study on micrornas targeting the genes overexpressed in lung cancer and their codon usage patterns. Mol. Biotechnol. 2022;64(10):1095–1119. doi: 10.1007/s12033-022-00491-3. [DOI] [PubMed] [Google Scholar]
- 39.Kuniyoshi N., Imazu H., Masuzaki R., et al. Diagnostic utility of quantitative analysis of microrna in bile samples obtained during endoscopic retrograde cholangiopancreatography for malignant biliary strictures. PLoS One. 2023;18(8) doi: 10.1371/journal.pone.0289537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B. Wang, J.H. Mao, B.Y. Wang, et al., Exosomal mir-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting mtmr3 and activating the nf-kappab signaling pathway, Cancer Lett.. 4892020) 87-99, 10.1016/j.canlet.2020.05.038. [DOI] [PubMed]
- 41.R. Song, T. Chai, J. Liu, A. Chu, C. Sun, Z. Liu, Knockdown of circmfn2 inhibits cell progression and glycolysis by mir-198/cul4b pathway in ovarian cancer, J. Biochem. Mol. Toxicol. 37 (8) (2023) e23383, 10.1002/jbt.23383.. [DOI] [PubMed]
- 42.Marin-Muller C., Li D., Lu J.M., et al. Nanoparticle-mediated therapy with mir-198 sensitizes pancreatic cancer to gemcitabine treatment through downregulation of vcp-mediated autophagy. Pharmaceutics. 2023;15(8) doi: 10.3390/pharmaceutics15082038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng G., Wang H., Zhang X., et al. Identification and validation of reference genes for qpcr detection of serum micrornas in colorectal adenocarcinoma patients. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bu X., Chen Z., Zhang A., et al. Circular rna circaff2 accelerates gastric cancer development by activating mir-6894-5p and regulating antxr 1 expression. Clin. Res. Hepatol. Gastroenterol. 2021;45(3) doi: 10.1016/j.clinre.2021.101671. [DOI] [PubMed] [Google Scholar]
- 45.Sudo K., Kato K., Matsuzaki J., et al. Development and validation of an esophageal squamous cell carcinoma detection model by large-scale microrna profiling. JAMA Netw. Open. 2019;2(5) doi: 10.1001/jamanetworkopen.2019.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. Tang, F. Liu, D. Huang, et al., Circ0125803 facilitates tumor progression by sponging mir-197-5p and upregulating e2f1 in neuroblastoma, Pathol. Res. Pract. 2332022) 153857, 10.1016/j.prp.2022.153857. [DOI] [PubMed]
- 47.N. Jain, B. Das, B. Mallick, Mir-197-5p increases doxorubicin-mediated anticancer cytotoxicity of ht1080 fibrosarcoma cells by decreasing drug efflux, DNA Repair 1092022) 103259, 10.1016/j.dnarep.2021.103259. [DOI] [PubMed]
- 48.Wang W., Xu S., Di Y., et al. Novel role of linc01013/mir-6795-5p/fmnl3 axis in the regulation of hepatocellular carcinoma stem cell features. Acta Biochim. Biophys. Sin. 2021;53(6):652–662. doi: 10.1093/abbs/gmab040. [DOI] [PubMed] [Google Scholar]
- 49.Zeng Y., Wang K.X., Xu H., Hong Y. Integrative mirna analysis identifies hsa-mir-3154, hsa-mir-7-3, and hsa-mir-600 as potential prognostic biomarker for cervical cancer. J. Cell. Biochem. 2018;119(2):1558–1566. doi: 10.1002/jcb.26315. [DOI] [PubMed] [Google Scholar]
- 50.Wei Y., Wei L., Han T., Ding S. Mir-3154 promotes hepatocellular carcinoma progression via suppressing hnf4alpha. Carcinogenesis. 2022;43(10):1002–1014. doi: 10.1093/carcin/bgac067. [DOI] [PubMed] [Google Scholar]
- 51.Huang X., Wang Y., Chen M., Li G. Mir-936 targets gpr78 and regulates chemotherapy resistance in non-small cell lung cancer by activating the galphaq rho gtpase pathway. Altern. Ther. Health Med. 2023;29(2):58–63. [PubMed] [Google Scholar]
- 52.Li M., Lin C., Cai Z. Downregulation of the long noncoding rna dscr9 (down syndrome critical region 9) delays breast cancer progression by modulating microrna-504-5p-dependent g protein-coupled receptor 65. Hum. Cell. 2023;36(4):1516–1534. doi: 10.1007/s13577-023-00916-4. [DOI] [PubMed] [Google Scholar]
- 53.Tubita V., Segui-Barber J., Lozano J.J., et al. Effect of immunosuppression in mirnas from extracellular vesicles of colorectal cancer and their influence on the pre-metastatic niche. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-47581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu C., Zhang Q., Wang A., et al. Ewi-2 controls nucleocytoplasmic shuttling of egfr signaling molecules and mirna sorting in exosomes to inhibit prostate cancer cell metastasis. Mol. Oncol. 2021;15(5):1543–1565. doi: 10.1002/1878-0261.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Ding, Q. Hong, T. Duan, et al., Mirna-mediated low expression of ephx3 is associated with poor prognosis and tumor immune infiltration in head and neck squamous cell carcinomas, J. Oncol. 20222022) 7633720, 10.1155/2022/7633720. [DOI] [PMC free article] [PubMed]
- 56.D. Bataveljic, M. Milosevic, L. Radenovic, P. Andjus, Novel molecular biomarkers at the blood-brain barrier in als, Biomed Res. Int. 20142014) 907545, 10.1155/2014/907545. [DOI] [PMC free article] [PubMed]
- 57.Ansari M.A., Chung I.M., Rajakumar G., et al. Current nanoparticle approaches in nose to brain drug delivery and anticancer therapy - a review. Curr. Pharm. Design. 2020;26(11):1128–1137. doi: 10.2174/1381612826666200116153912. [DOI] [PubMed] [Google Scholar]
- 58.Y. Liu, M. Zheng, M. Jiao, et al., Polymeric nanoparticle mediated inhibition of mir-21 with enhanced mir-124 expression for combinatorial glioblastoma therapy, Biomaterials 2762021) 121036, 10.1016/j.biomaterials.2021.121036. [DOI] [PubMed]
- 59.Kucukturkmen B., Bozkir A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and mir-21 antisense oligonucleotide to glioblastoma cells. Drug Dev. Ind. Pharm. 2018;44(2):306–315. doi: 10.1080/03639045.2017.1391835. [DOI] [PubMed] [Google Scholar]
- 60.Teplyuk N.M., Uhlmann E.J., Gabriely G., et al. Therapeutic potential of targeting microrna-10b in established intracranial glioblastoma: first steps toward the clinic. EMBO Mol. Med. 2016;8(3):268–287. doi: 10.15252/emmm.201505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE53733, GSE15824, GSE30563 and GSE50161. Other datas are available from the corresponding author on reasonable request.