Abstract
Hepatitis B virus DNA was extracted from serial serum samples of a hepatitis B surface antigen-negative patient with antibodies to the core protein as the only marker of an infection with hepatitis B virus. This patient showed no symptoms of hepatic injury. Sequencing of the amplified viral DNA demonstrated multiple amino acid changes clustering in surface-exposed regions of the surface protein. Synthesis and association of the middle (M) and small (S) surface proteins could be shown in vitro. The variant surface antigens were recognized neither by monoclonal antibodies to the surface antigen nor by the vaccinee’s sera. Consequences for hepatitis B surface antigen testing and vaccine development are discussed.
The variability of the hepatitis B virus (HBV) is reflected by the occurrence of at least six genotypes (24). Moreover, a number of mutants and variants for nearly all regions of the genome were described previously, and some of them are thought to be related to different courses of the infection (see reference 5 and references therein).
Escape mutants induced by active or passive immunization with amino acid changes resulting in the loss of the group-specific determinant called a of HBV surface antigen (HBsAg) have been reported by several authors (8, 10, 12, 17, 22, 23, 25, 35). An amino acid insertion between positions 122 and 123 in combination with the glycine-145-to-arginine substitution (known to be responsible for the majority of immune escape variants described above) was reported for an HBV isolate of an HBsAg-negative vaccinated patient with fulminant HBV infection (7). Insertions in this region are also found in HBV isolates of HBsAg-negative patients with chronic liver injury (15, 34).
Combining these data with those from studies concerning antigenicity and secretion of surface antigen variants, the major hydrophilic region of HBsAg may be separated into five functional areas related to the antigenic effect of variants and their selection pressure, indicated as HBs1 to -5 (6).
In this work, HBV DNA was amplified by PCR from sera of an HBsAg-negative patient with no hepatic injury. Patient F was a male renal dialysis patient with pharmacogenic renal failure. Serum samples taken at different time points (F1, September 1992; F2, June 1993; F3, March 1994; F4, March 1995; and F5, September 1995) were tested for hepatitis virus serologic parameters and human immunodeficiency virus. No markers of infection with hepatitis A virus, hepatitis C virus, and human immunodeficiency virus were detected. All sera tested negative for HBsAg, but high titers of anti-HBV core protein (HBc) were detectable by routine diagnostic testing (Amerlite HBsAg assay [Ortho Diagnostic Systems, Neckargemünd, Germany], Eti Mak 3 [Sorin Biomedica, Saluggia, Italy], and Amerlite anti-HBc assay). Anti-HBs antibodies were present in very low levels in samples F1, F2, and F4 by an in-house radioimmunoassay. Sera tested for anti-hepatitis delta virus immunoglobulin G antibodies (F1 and F5) with ETI-AB-DELTAK-2 (Sorin Biomedica) were also positive for this parameter. HBV DNA could be amplified by diagnostic PCR in all samples. DNA was extracted from 200 μl of serum treated with proteinase K-sodium dodecyl sulfate (SDS). Serum was incubated for 2 h at 56°C in a final volume of 500 μl with 2.5 mg of proteinase K per ml in a mixture of 10 mM Tris-HCl (pH 8.3), 100 mM NaCl, 5 mM EDTA, and 0.5% (wt/vol) SDS. After phenol-chloroform extraction and ethanol precipitation, DNA was resuspended in 50 μl of 10 mM Tris-HCl (pH 8.3). Alternatively, HBV DNA extraction was carried out with the QIAamp blood kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA was eluted with 50 μl of 10 mM Tris-HCl (pH 8.3). A DNase digestion was carried out prior to extraction procedures, in order to distinguish packaged, DNase-resistant viral DNA from free, DNase-sensitive, viral DNA. As viral DNA was extracted from serum samples, no cellular DNA was present in DNA preparations.
PCR was carried out with 5 to 10 μl of the extracted DNA in a mixture of 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 2.5 mM MgCl2; 0.05% (wt/vol) gelatin; 0.25 mM (each) dATP, dCTP, dGTP, and dTTP; 90 nM (each) sense and antisense primer; and 0.5 U of Taq DNA polymerase (Boehringer, Mannheim, Germany), in a final volume of 50 μl. For diagnostic PCR, primer pairs 1394-23f/1701-20r and 1425-23f/1673-24r were used for first- and second-round amplification, respectively. Amplification was performed for 40 cycles with denaturation at 95°C for 20 s, annealing at 60°C for 20 s, and extension at 72°C for 20 s. In all sera, there were about 102 to 103 genome equivalents per ml, as estimated by PCR end-point titration (data not shown). Positive results were confirmed by amplification with primer pairs 2360-20f/477-23r and 2382-21f/433-22r for first- and second-round amplification with 20 s at 95°C, 20 s at 60°C, and 1 min at 72°C for 35 cycles.
For amplification of the viral pre-S regions and the S gene of wild-type control R2 (reference serum 2 of Eurohep standard, subtype ay [11]) and samples F2, F3, and F4, primers 2045-23f/1303-21r or 2694-20f/1303-21r were used for first-round amplification. The second PCR was carried out with primer pair 2724-25f/1276-22r. PCR was performed for 35 cycles; each cycle included denaturation at 95°C for 20 s, annealing at 50°C for 20 s, and extension at 72°C for 2 min. The sequences of oligonucleotides used for PCR amplification are listed in Table 1.
TABLE 1.
Sequences of oligonucleotides used for PCR amplifications and sequencinga
| Primer, position | Sequence 5′→3′ | Use |
|---|---|---|
| 412-22f | CCTGCTGCTATGCCTCATCTTC | Sequencing |
| 433-22r | GAAGATGAGGCATAGCAGCAGG | PCR and sequencing |
| 477-23r | GGACAAACGGGCAACATACCTTG | PCR |
| 685-20f | GCCATTTGTTCAGTGGTTCG | Sequencing |
| 720-21r | GTGGGGGAAAGCCCTACGAAC | Sequencing |
| 1276-22r | CCGCAGTATGGATCGGCAGAGG | PCR and sequencing |
| 1303-21r | TGCGAGCAAAACAAGCGGCTA | PCR |
| 1394-23f | CCAACTGGATCCTGCGCGGGACG | Diagnostic PCR |
| 1425-23f | TTACGTCCCGTCGGCGCTGAATC | Diagnostic PCR |
| 1673-24r | GAGTCCAAGAGTCCTCTTATGTAA | Diagnostic PCR |
| 1701-20r | CCTCAAGGTCGGTCGTTGAC | Diagnostic PCR |
| 2045-23f | TCACCTCACCATACNGCACTCAG | PCR |
| 2694-20f | GGCATTAAACCTTATTATCC | PCR |
| 2360-20f | CGAGGCAGGTCCCCTAGAAG | PCR |
| 2382-21f | GAACTCCCTCGCCTCGCAGAC | PCR |
| 2724-25f | GTTAATCATTACTTCCAAACYAGAC | PCR and sequencing |
| 2850-22r | GTAGCTCTTGTTCCCAAGAATA | Sequencing |
| 3100-35fb | GGCATACTACAAGCTTTGCCAGCAA ATCCGCCTCC | PCR |
| 3207-21f | GGCCATGCAGTGGAAC/TTCCAC | Sequencing |
| 3221-21r | TTCCACTGCATGGCCTGAGGA |
Sequences are numbered with regard to sequence hvhepb (18) starting at the EcoRI restriction site. Numbers after the dashes specify the length of the oligonucleotides; f means forward, or sense, orientation, and r means reverse, or antisense, orientation.
The HindIII restriction site is underlined.
Nucleotide sequences were determined for both strands with the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer, Weiterstadt, Germany) on an ABI 373A DNA sequencer (Applied Biosystems, Weiterstadt, Germany) with PCR primers and internal oligonucleotides (Table 1). Sequence analysis was performed with the program package GCG (Wisconsin Sequence Analysis Package, version 8.1; Genetics Computer Group, Madison, Wis.). Within each sample, identical sequences were determined for the main virus strains with repetition of DNA isolation, PCR, and sequencing. Wild-type sequences, which might coexist with the variant ones in samples F2 to -4, were not found in any of the PCR products and clones characterized but may have existed below the detection limit of the assay.
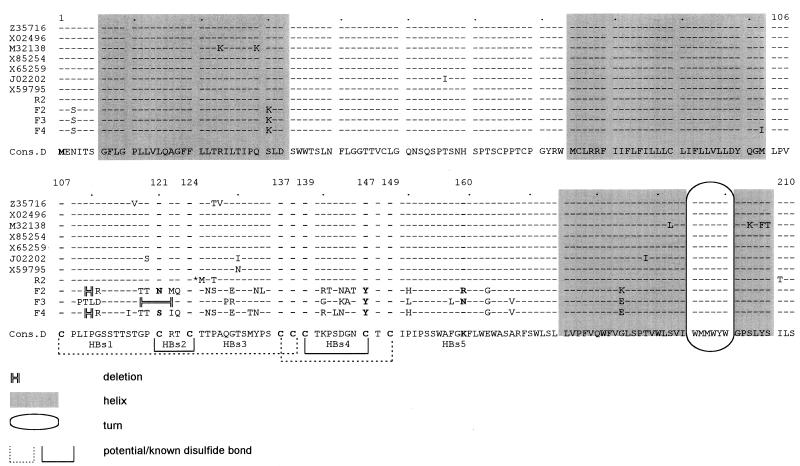
There were no remarkable nucleotide changes in the pre-S regions of isolates F2 to -4, but base changes and in-frame deletions were found within the S gene. Figure 1 shows an alignment of the variant amino acid sequences deduced from the nucleotide sequences with published HBV sequences and the wild-type control R2 sequence. Most of the nucleotide changes found with isolates F2, F3, and F4 clustered in the major hydrophilic region of the S gene coding for surface-exposed regions of the S protein. Regions facing the inner side of the HBV particle and regions where transmembrane helices are predicted showed little or no variations. There were various exchanges for nearly all epitopes defined by Carman (6). Deletions were located in HBs1 (and part of HBs2), where amino acids 110 and 111 (isolates F2 and F4) or 119 to 122 (isolate F3) were missing. Nucleotides deleted in the case of one isolate were exchanged and led to amino acid changes in the case of the other isolate. Carman et al. (6, 7) postulated an interaction of a section preceding amino acid 124 with the region between positions 139 and 147, resulting in an epitope cluster. These authors described the influence of insertions upstream of amino acid 124 on the binding of antibodies directed to the epitope between amino acids 139 and 147. The deletion found in isolate F3, as well as amino acid changes described for F2 and F4, is in the same region where other authors (7, 15, 34) found the amino acid insertions; a hot spot for insertions in this area is assumed (15). The results achieved in this work also argue for an immunologic importance of amino acids 118 to 123. The deletions found in isolates F2 and F4, as well as amino acid changes described for F3, are located somewhat more toward the N terminus. Nevertheless, they might alter the postulated immunologically important epitope. In region HBs3, two amino acid exchanges were found with isolate F3, and five amino acids were exchanged in isolates F2 and F4, where an additional N-glycosylation site is created at position 126. Three or four amino acids were exchanged in HBs4, and in all isolates, cysteine 147 was replaced by tyrosine. For the structure of the group determinant a, this cysteine does not seem to be as important as C107 (21), while lack of C121, which is exchanged in F2 and F4 and deleted in F3, as well as of C147 leads to distortion of the subtype y determinant. In isolate F2, asparagine 146, the N-glycosylation site, was replaced by threonine, but an alternative N-glycosylation site occurred at position 144. Within epitope HBs5, at amino acid 160, which appears to be crucial for r/w subtype determinant expression, there was remarkable variability. Isolate F2 had arginine at this position, while F3 had asparagine and F4 had lysine.
FIG. 1.
Alignment of deduced HBsAg amino acid sequences of isolates R2, F2, F3, and F4 with published HBV sequences, genotype D (GenBank/EMBL accession numbers are given for all published sequences). Topological predictions are based on the data of Chen et al. (9) and Persson and Argos (27). Functional areas of the major hydrophilic region of HBsAg are indicated as HBs1 to -5, referring to the work of Carman (6). The asterisk indicates methionine 125, which is observed as well in some local HBV isolates (unpublished data) and might therefore represent local variation. Cons.D, consensus of published genotype D sequences. All cysteine positions of the HBs epitopes and position 160, which is crucial for r/w subtype determinant expression, are given in bold type.
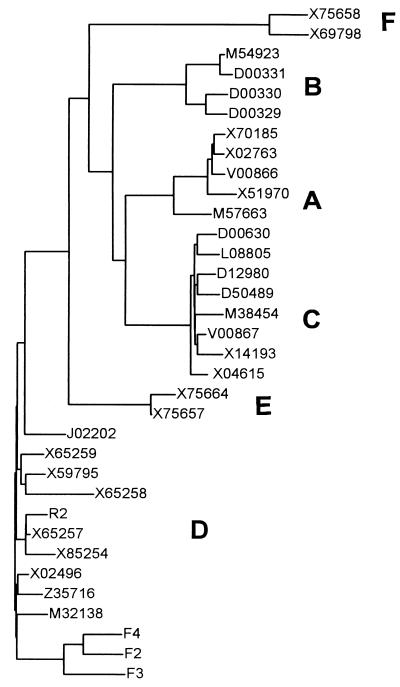
Assuming a hepadnaviral mutation rate for nucleotide substitutions concerning genes coding for structural proteins of between 1.75 × 10−5 (C gene; nonsynonymous substitution) and 7.62 × 10−5 (pre-S gene; synonymous substitution) substitutions per site per year (26), it is unlikely that isolate F4 emerged from F3 and F3 emerged from F2 in only 1 year. Thus, the predominant strains at each time point must have been coexisting at a very low level with the other strains in previous sera but were undetectable. Isolates F2, F3, and F4 form a distinct cluster in phylogenetic analysis (Fig. 2), so all may have emerged from a common ancestor, which might have infected the patient several years before. The closer relationship between F2 and F4 underlines this theory. Unfortunately, information is not available concerning the date of infection, and amplification of the whole S gene was unsuccessful with samples F1 and F5. Therefore, these presumptions cannot be confirmed.
FIG. 2.
Phylogenetic tree based on the nucleotide sequences of the pre-S/S genes of 30 published human HBV genomes and 4 isolates characterized in this work. Accession numbers are given for all published HBV isolates. The unrooted dendrogram was obtained with Growtree with uncorrected distances and neighbor joining. Genotypes A to F are indicated. Isolate R2, F2, F3, and F4 are all genotype D. Isolates F2 to -4 form a distinct cluster apart from all other HBV genomes. There is also high variability among isolates F2 to -4, which is striking, as all these HBV sequences were obtained from sera of the same patient within only 2 years.
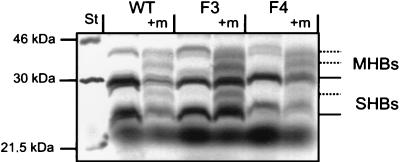
For in vitro expression, the pre-S2 regions and variant S genes of isolates F3 and F4 and the corresponding genome segment of wild-type HBV isolate R2 were inserted into expression vector pcDNAI (Invitrogen). First-round amplification products were reamplified with primers 3100-35f (containing a HindIII restriction site) and 1276-22r, and the HindIII/Nsp75241 fragments were cloned into the corresponding site of the vector (31). The resulting plasmids were named pcDNAF3, pcDNAF4, and pcDNAR2. In vitro expression with rabbit reticulocyte lysate (Promega) showed production of proteins of the expected size of glycosylated and nonglycosylated M and S proteins (MHBs and SHBs) (Fig. 3). Expressing pcDNAF4, the 27-kDa band corresponding to once-glycosylated S protein was missing, but double-glycosylated S protein due to use of the second glycosylation site found at amino acid 126 (Fig. 1) might be overlapping with unglycosylated M protein with a size of about 30 kDa. So in general, it could be demonstrated that the mutations of the S gene do not prevent the formation of the corresponding proteins.
FIG. 3.
In vitro expression of middle and small surface proteins. Coupled eukaryotic in vitro transcription and translation reactions were carried out with expression plasmids driving the synthesis of wild-type (WT) and variant (F3 and F4) middle and small surface proteins with the T7 RNA polymerase promoter. All reactions were also carried out with canine microsomal membranes (+m) for posttranslational modifications. [35S]methionine-radiolabelled proteins were analyzed by SDS-gel electrophoresis and autoradiography. The S proteins were expressed as unglycosylated and once-glycosylated peptides, with the exception of F4, for which once-glycosylated S protein could not be detected. Double-glycosylated S protein with a predicted size of about 30 kDa might be overlapping with the band seen for nonglycosylated M protein. The M proteins showed an additional double-glycosylated form due to an additional glycosylation site in the pre-S2 domain. Lane St, molecular mass markers. All autoradiograms were scanned with a Mustek Twain hand scanner and modified with program iPhoto Plus (version 1.1; U-Lead Systems, Inc.).
For cell culture transfection and immunoprecipitation experiments, inserts of the pcDNA clones were subcloned in vector pSV33H (3). The resulting plasmids carried the 3′ noncoding portion of adw2val (32) in addition to the variant pre-S2 region and S gene. Vector pSV33H was used as a wild-type control. COS7 cells were grown in six-well plates and handled as described by Bruss and Thomssen (2). Transfection was carried out by lipofection (DOTAP; Boehringer Mannheim) with 2 μg of plasmid DNA per well according to the manufacturer’s instructions.
In vitro labelling of expressed proteins and immunoprecipitation were carried out as described previously (2). With antibody Q19/10 binding to an MHBs-specific, glycan-dependent N-terminal epitope in the pre-S2 domain (14), precipitation of cell-culture-expressed intracellular protein as well as secreted variant small surface protein together with MHBs was possible. This coprecipitation of middle-sized surface proteins with the S protein due to the formation of mixed dimers was described previously for wild-type proteins (33). So, one can presume at least secretion and aggregation of the expressed variant surface proteins, and particle formation might also be possible.
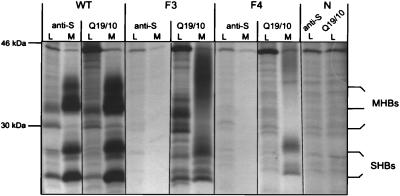
In contrast, the variant proteins expressed by vectors pSVF3 and -F4 were not recognized by the polyclonal anti-HBsAg antibody (Dako, Hamburg, Germany), or by human antisera (Fig. 4 and 5).
FIG. 4.
Immunoprecipitation of middle and small surface proteins expressed in cell culture. Expression plasmids driving the synthesis of wild-type (WT) and variant (F3 and F4) M and S proteins by the simian virus 40 promoter were transiently transfected into COS7 cells. HBV surface proteins were immunoprecipitated with antibodies directed to HBsAg (anti-S) and glycosylated pre-S2 (Q19/10) from cell lysates (L) and medium (M) after radioactive pulse-labelling and a chase of 24 h. Variants F3 and F4 were not recognized by anti-S antibody, but expression and secretion of variant M and S proteins could be demonstrated with the anti-pre-S2 antibody. Instead of distinct bands for the M proteins, F3 and F4 showed a diffuse protein smear of 33 to 40 kDa in cell culture medium. Lane N (negative control), untransfected cells.
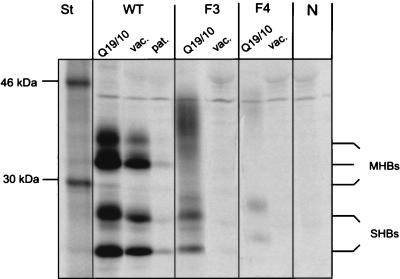
FIG. 5.
Immunoprecipitation of M and S proteins with human antisera. Wild-type (WT) and variant (F3 and F4) M and S proteins were expressed in COS7 cells, radioactively pulse-chase-labelled, and immunoprecipitated from cell culture medium with anti-pre-S2 antibody (Q19/10), vaccinee sera (vac.), and sera of an anti-HBs-positive patient after HBV infection (pat.). Immunoprecipitation was not very efficient with patient sera after HBV infection; only sera with high titers of anti-HBs after vaccination were suitable for this experiment. In contrast to the wild-type control, variants F3 and F4 were not recognized by human antisera. Lane N, immunoprecipitation of wild-type proteins with normal human serum; lane St, molecular mass markers.
Cell culture supernatant of cells transfected with the variant pre-S2/S sequences was negative in two separate tests with anti-HBsAg monoclonal antibodies and in a polyclonal HBsAg assay (Table 2), and so the virus variants described in this work are both diagnostic and immune escape variants (as they were not recognized by the vaccinee’s sera). With some escape variants described by other authors, diminished reactivity with different HBsAg test systems is described elsewhere (7, 23). The changeover of diagnostic test systems to polyclonal assays might be necessary. However, the variants described in this work were not reactive in a polyclonal HBsAg assay at all. Inclusion of antibodies specific for pre-S domains (such as Q19/10) in HBsAg assays should be kept in mind as well, with the aim of detecting such very unusual variants. However, in contrast to the small surface antigen, only small amounts of middle and large surface proteins are found in sera of HBV-infected patients (13). In rare cases such as those described here, with very low virus titers in addition to unusual serologic patterns, one has to rely on detection of anti-HBc as a marker of such cryptic HBV infection (16). Active viral replication has to be proved by virus-specific DNA amplification.
TABLE 2.
Results of different HBsAg assaysa
| Sample | Monoclonal 1 (int.) | Monoclonal 2 (abs.) | Polyclonal (cpm) |
|---|---|---|---|
| F3 | φ (0.095) | φ (0.015) | φ (267) |
| F4 | φ (0.055) | φ (0.028) | φ (317) |
| Pos. control | + (110; ≈150 ng/ml) | + (3.94; ≈150 ng/ml) | + (4,578) |
| Neg. control | φ (0.036) | φ (0.028) | φ (247) |
| Cutoff | 1 | 0.050 | 557 |
Cell culture supernatants of transfected COS cells were diluted 10-fold and tested for the presence of HBsAg 48 h posttransfection. Values are given as intensity (int.) and absorbance (abs.) for the monoclonal assays (Amerlite HBsAg assay [Ortho Diagnostic Systems]; and Eti Mak 3 [Sorin Biomedica]), and in counts per minute for the polyclonal HBsAg test (AUSRIA II-125; Abbott Laboratories, North Chicago, Ill.). F3, F4, Pos. control, and Neg. control indicate supernatants of cells transfected with pSVF3, pSVF4, and pSV33H and untransfected cells, respectively. Cutoff, cutoff of the respective assay. φ, negative; +, positive.
In contrast to the patients bearing similar HBV variants reported by several authors (7, 15, 34), patient F showed no clinical signs of hepatic injury. The virus might be replication competent, but virus multiplication was strongly decreased. Those very low virus titers might be responsible for the failure of HBsAg detection, independently of substitutions concerning the surface antigen (1, 20, 28). As patient F was also positive for antibodies against hepatitis delta virus, suppression of HBV replication and HBsAg negativity due to interference with hepatitis delta virus is possible (4, 19). The influence of the mutations on viral polymerase functions has to be considered as well, but deletions and most of the amino acid changes for the reverse transcriptase are located in a region of the viral polymerase where deletions and/or exchanges are tolerable (29). Whether the asymptomatic course of the patient’s HBV infection is related to the changes in the HBsAg or to the low virus titer, possibly caused by the same mutations, can only be answered by further infection experiments.
The epidemiology of variant viral strains such as F2, F3, and F4 remains unclear. The small number of works reporting massive changes within the surface antigen, even in studies with sera from patients with unusual HBV serologic parameters (1, 20, 28), implies a low prevalence of these variants.
Recently, persistence of HBV for decades after patients’ recovery from acute viral hepatitis was described as a common event (30). Thus, the occurrence and outcome of these unusual variants might also reflect a mechanism for maintaining the viral persistence.
At the moment, there is no vaccine in use protecting against infection with HBV escape mutants. Any future vaccine ought to comprise protective pre-S epitopes in addition to HBsAg, for example.
In conclusion, we have shown in this work the occurrence of unusual HBV immune and diagnostic variants in sera from an HBsAg-negative patient. The structure of the major antigenic determinants was severely affected by multiple amino acid exchanges, which did not prevent oligomerization and secretion of envelope proteins. Before conclusions concerning routine HBsAg testing and vaccine strategies can be drawn, the infectivity and pathogenicity of these variants have to be studied.
Nucleotide sequence accession numbers.
Accession numbers are AJ003026 for F2, AJ003027 for F3, AJ003028 for F4, and AJ003116 for R2.
Acknowledgments
We gratefully acknowledge Volker Bruss for providing vector pSV33H and Angela Uy for providing the patients’ sera. We thank Richard Walker (University of Glasgow) for critical reading of the manuscript.
REFERENCES
- 1.Bréchot, C., D. Kremsdorf, P. Paterlini, and V. Thiers. 1991. Hepatitis B virus DNA in HBsAg-negative patients. J. Hepatol. 14(Suppl. 4):49–55. [DOI] [PubMed]
- 2.Bruss V, Thomssen R. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J Virol. 1994;68:1643–1650. doi: 10.1128/jvi.68.3.1643-1650.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss V, Lu X, Thomssen R, Gerlich W. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caredda F, Antinori S, Pastecchia C, Coppin P, Palla M, Ponzetto A, Rizzetto M, Moroni M. Incidence of hepatitis delta virus infection in acute HBsAg-negative hepatitis. J Infect Dis. 1989;159:977–979. doi: 10.1093/infdis/159.5.977. [DOI] [PubMed] [Google Scholar]
- 5.Carman W F. Molecular variants of hepatitis B virus. Clin Lab Med. 1996;16:407–428. [PubMed] [Google Scholar]
- 6.Carman, W. F. 1997. The clinical significance of surface antigen variants of hepatitis B virus. J. Viral Hepatitis 4(Suppl. 1):11–20. [DOI] [PubMed]
- 7.Carman W F, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–1407. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 8.Carman W F, Zanetti A R, Karyiannis P, Waters J, Manzillo G, Tanzi E, Zuckermann A, Thomas H C. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Delbrook K, Dealwis C, Mimms L, Mushawar I K, Mandecki W. Discontinuous epitopes of hepatitis B surface antigen derived from a filamentous phage peptide library. Proc Natl Acad Sci USA. 1996;93:1997–2001. doi: 10.1073/pnas.93.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuji H, Moryiama K, Sakamoto N, Kondo T, Yasuda K, Hiraizumi Y, Yamazaki M, Sakaki Y, Okochi K, Nakajima E. Gly 145 to arg substitution in HBs antigen of immune escape mutant of hepatitis B virus. Biochem Biophys Res Commun. 1992;184:1152–1154. doi: 10.1016/s0006-291x(05)80003-8. [DOI] [PubMed] [Google Scholar]
- 11.Gerlich W H, Heermann K H, Thomssen R. Third Eurohep trial on generation of reference samples for hepatitis B virus DNA. In: Baya C, editor. Advances in medical biology. Amsterdam, The Netherlands: IOS Press; 1994. pp. 249–251. [Google Scholar]
- 12.Harrisson T J, Hopes E A, Oon C J, Zanetti A R, Zuckermann A J. Independent emergence of a vaccine-induced escape mutant of hepatitis B virus. J Hepatol. 1991;14:105–107. doi: 10.1016/0168-8278(91)90037-c. [DOI] [PubMed] [Google Scholar]
- 13.Heermann K H, Gerlich W H. Surface proteins of hepatitis B viruses. In: McLachlan A, editor. Molecular biology of hepatitis B virus. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 109–153. [Google Scholar]
- 14.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou J, Karyiannis P, Waters J, Luo K, Liang C, Thomas H C. A unique insertion in the S gene of surface antigen negative hepatitis B virus chinese carriers. Hepatology. 1995;21:273–278. doi: 10.1016/0270-9139(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 16.Jilg W, Siegert E, Zachoval R, Schatzl H. Individuals with antibodies against hepatitis core antigen as the only serological marker for hepatitis B virus infection: high percentage of carriers of hepatitis B and C virus. J Hepatol. 1995;23:14–20. doi: 10.1016/0168-8278(95)80305-x. [DOI] [PubMed] [Google Scholar]
- 17.Karthigesu V D, Allison L M C, Fortuim M, Mendy M, Whitthle H C, Howard C R. A novel hepatitis B virus variant in the sera of immunized children. J Gen Virol. 1994;75:443–448. doi: 10.1099/0022-1317-75-2-443. [DOI] [PubMed] [Google Scholar]
- 18.Köchel, H. G., A. Schüler, S. Lottmann, and R. Thomssen. 1990. Sequence of HBV isolate 991. EMBL database. Accession no. X51970.
- 19.Krogsgaard K, Kryger P, Aldersvile J, Andersson P, Sorentsen T I, Nielsen J O the Copenhagen Hepatitis Acuta Programme. Delta-infection and suppression of hepatitis B virus replication in chronic HBsAg carriers. Hepatology. 1987;7:42–45. doi: 10.1002/hep.1840070110. [DOI] [PubMed] [Google Scholar]
- 20.Liang T J, Blum H E, Wands J R. Characterization and biological properties of a hepatitis B virus isolated from a patient without hepatitis B virus serologic markers. Hepatology. 1990;12:204–212. doi: 10.1002/hep.1840120205. [DOI] [PubMed] [Google Scholar]
- 21.Mangold C M T, Unckell F, Werr M, Streeck R E. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology. 1995;211:535–543. doi: 10.1006/viro.1995.1435. [DOI] [PubMed] [Google Scholar]
- 22.McMahon G, Ehrlich P H, Moustafa Z, McCarthy L, Dottavio D, Tolpin M D, Nadler P I, Östberg L. Genetic alterations in the gene encoding the major HBsAg: DNA and immunological analysis of recurrent HBsAg derived from monoclonal antibody-treated liver transplant patients. Hepatology. 1992;15:757–766. doi: 10.1002/hep.1840150503. [DOI] [PubMed] [Google Scholar]
- 23.Ni F, Fang D, Gan R, Li Z, Duan S, Xu Z. A new immune escape mutant of hepatitis B virus with an asp to ala substitution in aa145 of the envelope major protein. Res Virol. 1995;146:397–407. doi: 10.1016/0923-2516(96)80899-5. [DOI] [PubMed] [Google Scholar]
- 24.Norder H, Couroucé A M, Magnius L. Complete genomes, phylogenetic relatedness and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto H, Yano K, Nozaki Y, Matsui A, Miyazaki H, Yamamoto K, Tsuda F, Machida A, Mishira S. Mutations within the S gene of hepatitis B virus transmitted from mothers to babies immunized with hepatitis B immune globulin and vaccine. Pediatr Res. 1992;32:264–268. doi: 10.1203/00006450-199209000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Orito E, Mizokami M, Ina Y, Moriyama E N, Kameshima N, Yamamoto M, Gojobori T. Host independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–193. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 28.Preisler-Adams S, Schlayer H J, Peters T, Hettler F, Gerok W, Rasenack J. Sequence analysis of hepatitis B virus DNA in immunologically negative infection. Arch Virol. 1993;133:385–396. doi: 10.1007/BF01313777. [DOI] [PubMed] [Google Scholar]
- 29.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehermann B, Ferrari C, Pasquinelli C, Chisari F V. The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med. 1996;2:1104–1108. doi: 10.1038/nm1096-1104. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Valenzuela P, Quiroga M, Zalvidar J, Gray P, Ruttner W. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. In: Fields B N, Jaenisch R, Fox C F, editors. Animal virus genetics. New York, N.Y: Academic Press; 1980. pp. 57–70. [Google Scholar]
- 33.Wunderlich G, Bruss V. Characterization of early hepatitis B virus surface protein oligomers. Arch Virol. 1996;141:1191–1205. doi: 10.1007/BF01718824. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto K, Horikita M, Tsuda F, Itoh K, Akahane Y, Yotsumoto S, Okamoto H, Miyakawa Y, Mayumi M. Naturally occurring escape mutants of hepatitis B virus with various mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J Virol. 1994;68:2671–2676. doi: 10.1128/jvi.68.4.2671-2676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y Y, Nordenfelt E, Hansson B G. Increasing heterogeneity of the ‘a’ determinant of HBsAg found in the presumed late phase of chronic hepatitis B virus infection. Scand J Infect Dis. 1996;28:9–15. doi: 10.3109/00365549609027142. [DOI] [PubMed] [Google Scholar]