Abstract
Foamy viruses (spumaretroviruses) represent a retroviral genus which exhibits unusual features relating it to pararetroviruses. Previously, we reported the existence of a protein species harboring Env, Bel, and Bet epitopes in human foamy virus (HFV)-infected cells (M. L. Giron, F. Rozain, M. C. Debons-Guillemin, M. Canivet, J. Périès, and R. Emanoil-Ravier, J. Virol. 67:3596–3600, 1993). Here, we identify this protein as a 160-kDa Env-Bet fusion glycoprotein (gp160) translated from an mRNA species harboring a highly conserved splice site which deletes the membrane anchor domain of Env and fuses the env open reading frame with that of bel1/bet. While gp160 and Bet proteins were both secreted into the supernatant, only Bet was taken up by recipient cells. Since Bet plays a key role in the switch from lytic to chronic infection, secretion of Bet and gp160, together with cellular uptake of Bet, could be highly relevant for both immune response and development of HFV infection in vivo.
Foamy viruses (FV), or spumaviruses, are complex retroviruses known to induce persistent infection in their hosts without causing apparent disease (22, 25). Some properties set the FV apart from all other retroviruses, such as the formation of a specific pol mRNA and the presence of large amounts of double-stranded viral DNA in the extracellular virion (27); in these two features, FV resemble pararetroviruses. Another property peculiar to the FV is their budding into the endoplasmic reticulum (ER) rather than from the plasma membrane. This is attributed to the presence of a dilysine ER-sorting motif in the cytoplasmic domain of the Env protein (6, 7). Regulatory genes are located between the 3′ end of the env gene and the 3′ long terminal repeat and are under the control of an internal promoter (11). In human foamy virus (HFV), these regulatory genes are bel1, bel2, bel3 and bet. The nuclear transactivator Bel1 is essential for viral replication (15), while Bet, a fusion protein between Bel1 and Bel2, plays an important role in the establishment and control of viral persistence, in vitro as well as in vivo (19, 20).
In a previous work, we have characterized viral polypeptides produced during HFV infection (5). A specific env monoclonal antibody (B4) immunoprecipitated four viral glycoproteins from HFV-infected cells: gp160, gp130, gp70-80, and gp48 (5). We demonstrated that gp70-80 and gp48 correspond to the surface (SU) and transmembrane (TM) mature Env glycoproteins, respectively, and that the gp130 polypeptide represents the Env precursor. However, the gp160 protein was immunoprecipitated by the Env-specific monoclonal antibody as well as by a specific anti-Bet antiserum, raising the issue of the existence of a putative Env-Bet polypeptide.
The present study further characterizes this gp160 protein and demonstrates that it represents a fusion protein between the env and bel regions derived from a spliced viral mRNA. The evolutionarily conserved splice deletes the transmembrane anchor of the TM Env protein as well as the ER retention motif and fuses the env open reading frame (ORF) with those of bel or bet. An Env-Bet fusion protein represents the major form detected during HFV infection, although reverse transcription-PCR (RT-PCR) experiments demonstrate the existence of an env-bel1 transcript. While both gp160 and Bet proteins are secreted into the supernatant, Bet appears to be the only protein taken up by naive recipient cells. Secretion of viral proteins harboring regulatory or structural domains or both could have major implications for the biology of these viruses in an in vivo context both for immune response and virus-cell interactions.
MATERIALS AND METHODS
Cells and virus.
Mycoplasma-free HFV stocks were grown on U373-MG cells, a human neural cell line maintained in Dulbecco’s modified Eagle’s medium supplemented with nonessential amino acids, sodium pyruvate, and 10% fetal calf serum. COS-6, a simian cell line, and BHK21, a hamster cell line, were maintained in the same medium. Virus stocks were subjected to titer determination by the end-point dilution method on U373-MG cells as described previously (20).
Transfection experiments.
COS-6 cells were transfected with Lipofectin reagent (Gibco, BRL) as specified by the manufacturer. At 48 h posttransfection, the cells were lysed in lysis buffer and proteins were studied by immunoprecipitation.
Protein analysis.
For immunoprecipitation assays, acutely HFV-infected cells or transfected cells (107 cells) were labeled with [35S]methionine-cysteine (50 μCi/ml; 1,245 Ci/mmol specific activity; Dupont NEN) for different times in minimal essential medium lacking methionine-cysteine and supplemented with 5% fetal calf serum. The cells were lysed in 50 mM Tris-HCl (pH 7.4)–100 mM NaCl–5 mM MgCl2–1% Triton X-100–0.5% deoxycholate, 0.05% sodium dodecyl sulfate [SDS]–3 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. After centrifugation, the supernatant was collected and immunoprecipitated with a rabbit anti-whole-virus antiserum as described previously (5). For immunoprecipitation assays with supernatants of transfected cells, 75 μCi of [35S]methionine-cysteine per ml was used in minimal essential medium lacking methionine-cysteine without fetal calf serum, as previously described (23).
The antibodies (Ab) used were serum from HFV-infected rabbits (20), a mouse monoclonal Ab (D11) against the Bet protein, a mouse monoclonal Ab (B4) against the SU domain of the Env protein, and rabbit polyclonal anti-Bel1 and anti-Bel2 antisera (kindly provided by R. M. Flügel), all at a 1/100 dilution.
Peptide mapping and acid treatment.
U373-MG-infected cells were immunoprecipitated with the rabbit anti-HFV polyclonal antiserum. After immunoprecipitation and polyacrylamide gel electrophoresis (PAGE), the slab gel was rinsed with water and dried without Amplify. After autoradiography, the bands corresponding to Bet, gp160, and gp130 were cut out of the gel and placed on a second SDS-polyacrylamide slab gel. The proteins were digested with 5 μg of V8 protease (Worthington, Freehold, N.J.) in a stacking gel essentially as described previously (3). For the acid treatment, the same procedure was used to collect the gp160 and gp130 bands, which were resuspended in 100 μl of glycine-acetate buffer (pH 4) and treated for 1 h at 37°C in the presence of 0.2 mM phenylmethylsulfonyl fluoride and 10 U of aprotinin. Then 100 μl of 2× electrophoresis buffer was added and the samples were analyzed by SDS-PAGE.
Generation of eukaryotic expressing vectors.
The Bet-expressing plasmid was obtained by blunting the AatII-SalI (nucleotides [nt] 9341 to 12034 from the infectious clone, pHSRV13) fragment with the Klenow enzyme and subsequent subcloning into the unique SmaI site of the pSG5M-expressing vector.
To study the formation of the Env-Bel fusion proteins, plasmid p2EB was constructed by cutting pHSRV13 with BanI and SalI. The insert was blunted with the Klenow enzyme and subcloned in the pSG5M-expressing vector at the SmaI site of the polylinker. This plasmid contains the second ATG (nt 6526 on the HFV map) found at the 5′ end of the env gene. Plasmid p1EB, which expresses the Env protein from the first ATG (nt 6495), was created by inserting the KpnI-SpeI insert fragment obtained by PCR (direct, ATT TTG GTA CCA TCT TGG CAA C; reverse, TTG TGG AAT ACT AGT CAT ATT TAC; the KpnI and the SpeI sites are underlined) into p2EB at the KpnI-SpeI sites, replacing the 5′ end of this latter plasmid. Deletion mutants with mutations in the pEB plasmids were established by cutting with BamHI (nt 9675 from HFV map) and subsequent religation, leading to plasmids p1EnvΔBel and p2EnvΔBel, in which the bel region is missing.
Point mutants were obtained with the QuickChange site-directed mutagenesis kit (Stratagene). The conservative mutation site on the donor splice site at the 3′ end of env was obtained with the following primers: direct, AAG GAA TTG GCA ACT TTT TAT; reverse: ATA AAA AGT TGC CAA TTC CTT (the point mutation is underlined). The NheI-XmnI insert containing the mutation was sequenced and subcloned in the same sites of the parental plasmid. The resulting plasmids, p1EctB and p2EctB, were used in transfection experiments. Plasmids p1EBΔRGD and p2EBΔRGD were constructed by the same procedure with the following primers: direct, CCT TAT GGA GAT GGG GGT GAT GCA; reverse, TGC ATC ACC CCC ATC TCC ATA AGG, to insert the codon mutation agg→ggg (Arg→Gly) into the RGD motif in the bet gene on p1EB and p2EB, respectively. The BglII-BglII insert was sequenced and recloned into the parental vector.
RT-PCR and PCR experiments.
For RT-PCR analysis, total cellular RNAs from infected or transfected cell pellets were extracted with an RNA extraction kit (Bioprobe Systems). RT-PCR experiments were performed with the Access RT-PCR system (Promega). Briefly, 500 ng of total RNA was used as the template for the synthesis of the first-strand cDNA for 45 min at 48°C in the presence of avian myeloblastosis virus reverse transcriptase. After denaturation at 94°C, the synthesis of the second strand and enzymatic amplifications were carried out with Tfl DNA for 40 cycles of 94°C for 45 s, 54°C for 45 s, and 72°C for 1 min. The primers used were as follows: direct, GAT TAC CAC ATT TGG TTG GAAT (nt 9091 to 9112 on the HFV map [primer A]); reverse, GTT TTG GAC CTT CTG AGC A (nt 10001 to 10019 [primer B]).
Southern blots, prepared by standard procedures (24), were hybridized overnight with an [α-32P]dCTP-labeled probe at 42°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS–5× Denhardt’s solution–50% formamide–100 μg of denatured salmon sperm DNA per ml. Washing was performed in 0.1× SSC–0.1% SDS buffer at 60°C for 30 min twice. Plasmid pHSRV13 was used as a probe for PCR hybridization.
DNA sequencing.
DNA inserts or PCR products subcloned in the pGEM-Easy vector (Promega) were sequenced with the ThermoSequenase kit (United States Biochemicals) as specified by the manufacturer.
RESULTS
A gp160 protein is immunoprecipitated with anti-Bel1, anti-Bel2, anti-Bet, and anti-Env antisera in HFV-infected cells.
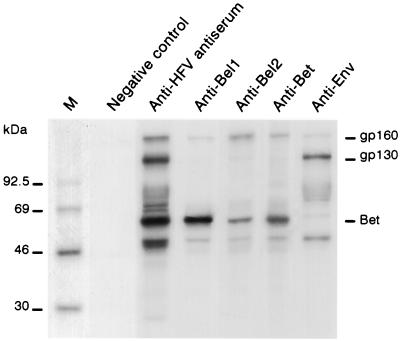
Protein extracts from U373-MG cells infected with 1 PFU/cell for 4 days were immunoprecipitated with a set of different Ab raised against HFV proteins. An antiserum obtained from HFV-infected rabbits and recognizing all the HFV proteins gave rise to the classical pattern previously described (5), corresponding to structural as well as regulatory viral gene products. The B4 monoclonal Ab raised against SU Env protein precipitated four distinct glycoproteins: the gp130 Env precursor, SU as a smear between 70 and 80 kDa, the 48-kDa TM, and a glycoprotein at 160 kDa (5). Interestingly, the use of rabbit polyclonal anti-Bel1 and anti-Bel2 Abs as well as a mouse monoclonal anti-Bet (D11) Ab precipitated the same gp160 polypeptide, strongly suggesting that this protein species harbors both Env and Bel/Bet epitopes (Fig. 1).
FIG. 1.
Identification of Env and Bel gene products by immunoprecipitation of protein extracts from HFV-infected cells. M, molecular mass markers. Rabbit anti-HFV polyclonal antiserum on protein extracts from noninfected cells (negative control) or from infected cells (positive control) were used as controls. Rabbit polyclonal anti-Bel1 and anti-Bel2 Abs and mouse monoclonal anti-Bet (D11) and anti-SU-Env (B4) Abs were used to detect the gp160 band.
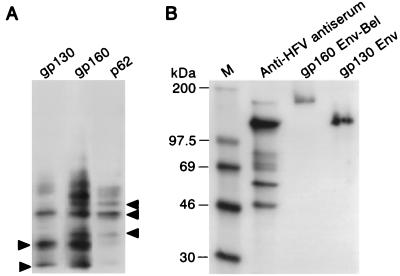
To confirm that the gp160 protein presented both Env and Bel/Bet epitopes, peptide-mapping experiments were performed. After immunoprecipitation and autoradiography, the bands corresponding to gp130, gp160, and Bet proteins were cut out from the gel and placed on a second SDS-polyacrylamide slab gel. The proteins were then digested with V8 protease as previously described (3). Figure 2A shows that several polypeptides are shared by gp130 and gp160 and others are shared by gp160 and Bet. These results are consistent with the presence of both Env and Bel/Bet sequences in the 160-kDa glycoprotein.
FIG. 2.
The gp160 band does not represent a protein complex. (A) Peptide mapping of gp130, gp160, and Bet. Several bands are shared by gp130 and gp160 and others are shared by gp160 and Bet (arrows). (B) Low-pH treatment of gp160 and gp130. M, molecular mass markers. The apparent molecular masses of the two proteins are not modified after 1 h of incubation at pH 4.
One hypothesis was to assume that the 160-kDa band could constitute a complex between the gp130 Env precursor (or a major part of it) and the Bel or Bet proteins. This putative complex might remain stable during SDS-PAGE migration, as previously described for homodimers of the Env precursor of human immunodeficiency virus type 2 (16). In this case, although the complex was resistant to 1% SDS and reducing agents, the authors succeeded in obtaining protein dissociation after acid treatment. Thus, after immunoprecipitation with the rabbit polyclonal anti-HFV antiserum, SDS-PAGE migration, and autoradiography, the gp130 and gp160 bands were extracted from the gel and incubated for 1 h at pH 4. After neutralization, the proteins were subjected to SDS-PAGE on a second gel (see Materials and Methods). As shown in Fig. 2B, acid treatment also failed to dissociate the gp160. Moreover, in protein extracts prepared in the presence of 1% SDS or under highly reducing conditions, no dissociation of the gp160 protein was detected (data not shown).
Altogether, these results strongly suggest that the 160-kDa band is not a complex formed between Env and Bel/Bet proteins but a glycoprotein harboring Bel, Bet, and Env sequences.
An evolutionarily conserved splicing event generates the Env-Bel fusion protein.
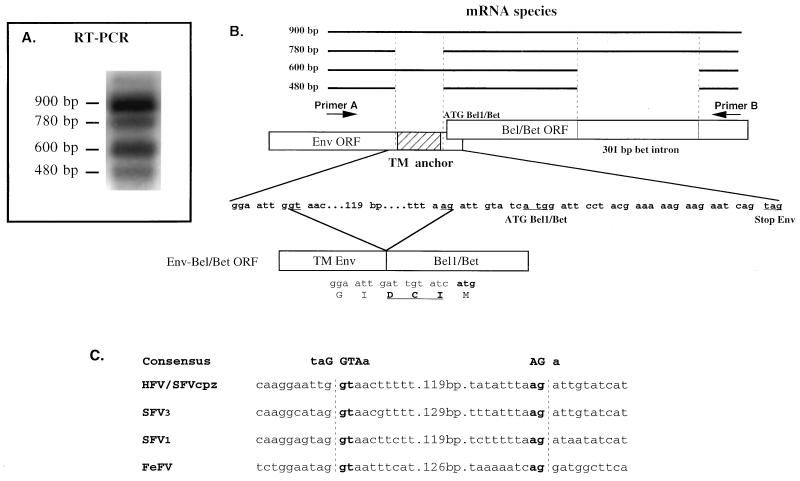
The presence of a high-molecular-weight fusion Env-Bel/Bet glycoprotein could be accounted for by the existence of a multiply spliced mRNA (derived from the long terminal repeat) which would either suppress the stop codon in the env ORF or, indirectly, change the ORF of the env gene upstream of the stop codon, leading to the production of an Env-Bel or Env-Bet fusion product. To assess this possibility, RT-PCR experiments were performed with specific primers which flank the end of the env gene and the bel region (Fig. 3A). After 4 days of infection of U373-MG cells with wild-type HFV (0.5 PFU/cell), total RNAs were extracted and RT-PCR was performed as described in Materials and Methods. With the primers used, we were able to amplify four different RNA species at approximately 900, 780, 600, and 480 bp (Fig. 3A). PCR products were cloned, and three independent clones of each amplification were sequenced. The 900-bp species represents the full-length RNA. The 600-bp species corresponds to the previously described 301-bp splice generating the Bet mRNA also harbored by ΔHFV (12, 21). Interestingly, the 780-bp species corresponds to a 119-bp splice (nt 9307 to 9425) flanked by typical donor and acceptor sites, as previously described (12). This 119-bp deletion results in an ORF which deletes the transmembrane domain of the Env protein and changes the env ORF in its 3′ end by suppressing the stop codon by fusing in frame the env ORF to the bel/bet one. Note that three amino acids (Asp-Cys-Ile) are inserted between Env and Bel (Fig. 3B). These splice sites are highly conserved among the spumaretroviruses sequenced to date (8, 9, 13, 14), even in the remotely related feline foamy virus (26) (Fig. 3C), suggesting an important role of this specific splicing event in the biology of this retroviral family. The 780-bp RNA could encode a putative Env-Bel1 product, while the 480-bp species, which harbors the 119-bp as well as the 301-bp splice, generates an Env-Bet fusion protein. Note that this latter RNA species and the full-length RNA are the most abundant species detected by RT-PCR.
FIG. 3.
(A) RT-PCR experiments on total RNA from HFV-infected cells. Four distinct classes of RNAs, which correspond to the four RNA species of 900, 780, 600, and 480 bp, are amplified. Plasmid pHSRV13 was used as a probe for PCR hybridization. (B) Schematic representation of the different mRNA species which can be detected between primer A and primer B. Note that three new amino acids are created by the splicing event (Asp-Cys-Ile in boldface type). The dilysine ER retention motif is also depicted in the env ORF. (C) Sequence comparisons of known and putative splice sites among different sequenced FV. The donor and acceptor splice sites flank the transmembrane anchor sequence of the Env TM protein and fuse the env ORF with the transactivator one. SFVcpz, simian FV from chimpanzee; FeFV, feline FV.
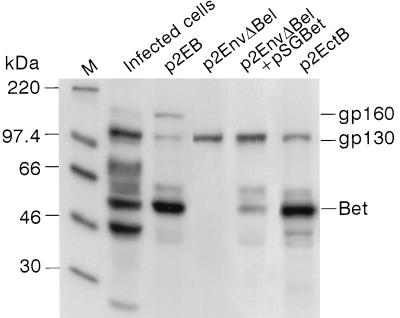
To obtain more insights into the formation of the gp160 protein, p1EB and p2EB, harboring the entire env and bel ORFs, as well as the 3′ LTR, under the transcriptional control of a simian virus 40 early promoter were constructed (see Materials and Methods). p1EB contains the first ATG (nt 6495 on the HFV map) of the env ORF, while p2EB harbors only the second initiator ATG codon (nt 6526). Since similar results were obtained with the two constructs, we report only those obtained with p2EB. After transfection of p2EB into COS-6 cells, we were able to immunoprecipitate not only the 130-kDa Env precursor and the Bet protein but also the 160-kDa glycoprotein (Fig. 4). Moreover, upon transfection of the p2EnvΔBel plasmid (a mutant of p2EB with the entire bel and bet gene sequences deleted), the 130-kDa Env protein was clearly detected by immunoprecipitation, but neither Bet nor gp160 was detected. Furthermore, cotransfection of p2EnvΔBel and a Bet-expressing vector (pSGBet) did not lead to the reappearance of the gp160 band, confirming our first biochemical results that gp160 is not a complex between gp130 and Bet (Fig. 4).
FIG. 4.
Identification of the gp160 protein in pEB-transfected cells. Immunoprecipitation was performed with the rabbit anti-HFV antiserum. M, molecular mass markers. Cells were transfected with p2EB, p2EnvΔBel, p2EnvΔBel plus pSGBet, and p2EctB. Mutation of the splice acceptor site abolishes the formation of gp160. Note the absence of the gp160 species but the detection of Bet in p2EctB-transfected cells.
To directly prove that the 160-kDa protein was generated from a 119-bp splice event at the 3′ end of the env gene, a conservative point mutation was generated on the 5′ donor splice site, changing a GT into CT (nt 9307 and 9308 on the HFV map), leading to the p2EctB plasmid. As expected, after transfection of COS-6 cells with this construct, the Env precursor and the Bet protein were both immunoprecipitated by the rabbit polyclonal anti-HFV antiserum but the gp160 glycoprotein was no longer immunoprecipitated (Fig. 4). Moreover, RT-PCR experiments performed on total RNA extracted from p2EctB-transfected cells did not detect the 780- and 480-bp bands (data not shown). These results directly demonstrate that gp160 is translated from an mRNA harboring the 119-bp splice in the env gene. Note that in our immunoprecipitation assays, we failed to detect cleavage products of the gp130 or gp160 protein from the pEB-transfected COS-6 cells (Fig. 4).
The fusion protein is secreted into the supernatant.
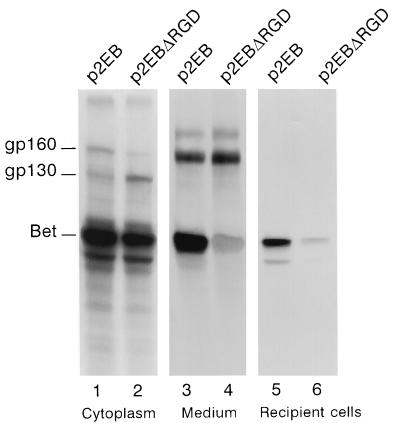
Since the gp160 protein possesses a signal peptide from the Env protein but lacks its transmembrane anchor domain and the dilysine ER retention motif, we wondered whether it could be secreted in the extracellular compartment by the cellular secretory pathways. Therefore, we transfected COS-6 cells with p2EB and labeled the transfected cells with [35S]Met-Cys-containing medium 48 h posttransfection. After overnight labeling, we immunoprecipitated the cytoplasmic extracts of transfected cells or filtered culture supernatant (0.45-μm-pore-size filter [Corning]) with the rabbit anti-HFV antiserum and performed SDS-PAGE. Although gp160, gp130, and Bet were immunoprecipitated in transfected cells, only gp160 and Bet were detected in the supernatant (Fig. 5, lane 3). This result has been confirmed by the use of the D11 monoclonal Ab directed against Bet (data not shown). The absence of the gp130 protein in the acellular culture medium and the use of p2EB instead of the lytic wild-type virus strongly suggest that the detection of Bet and gp160 in the cell culture supernatant is due not to cell lysis but, rather, to their secretion from the transfected cells. However, in HFV-infected BHK21 cells, gp160 and Bet were also found in the culture medium (together with cleavage products from gp160) prior to cell lysis (data not shown).
FIG. 5.
Secretion and uptake of Env-Bet and Bet proteins. Cytoplasmic extracts from cells transfected with p2EB (lane 1), p2EBΔRGD (lane 2) and supernatants from p2EB (lane 3)- and p2EBΔRGD (lane 4)-transfected cells are shown. The upper (200-kDa) band probably represents a cellular protein. After 18 h of labeling, filtered culture medium from transfected cells was incubated with naive recipient COS-6 cells for 4 h and subsequent immunoprecipitations were performed with the rabbit anti-HFV antiserum on cytoplasmic extracts. Lane 5, extracts from p2EB-transfected cells; lane 6, extracts from p2EBΔRGD-transfected cells.
Of the total HFV proteins immunoprecipitated in infected cells, 5 to 15% were found in the supernatant exclusively as Bet and gp160. We wondered whether these proteins could be internalized by naive recipient cells. To test this hypothesis, COS-6 cells were transfected with the p2EB vector. At 48 h posttransfection, the cells were labeled for 2, 6, 16, or 18 h and acellular supernatants were incubated with 106 recipient COS-6 cells at 37°C for 2, 4 or 6 h. After intensive washes of the cell layer with phosphate-buffered saline (to avoid detection of viral proteins adsorbed onto the plasma membrane), the cells were collected and HFV proteins were immunoprecipitated with the rabbit polyclonal anti-HFV antiserum. Approximately 10 to 20% of secreted labeled viral proteins were taken up in 4 h. As shown in Fig. 5, Bet is clearly taken up whereas gp160 is undetectable. However, such failure to detect gp160 in recipient cells could be a function of the sensitivity of our immunoprecipitation assay. Furthermore, some Bet was found in nuclear extracts from recipient cells, demonstrating that Bet is internalized and transported and not simply adsorbed on the plasma membrane (data not shown). The specificity, the in vivo relevance, and the putative cellular partners of the uptake mechanism will be the focus of future studies.
Analysis of the Bet sequence reveals the presence of an Arg-Gly-Asp (RGD) motif (amino acids 294 to 296), a sequence implicated in complex recognition mechanisms between cells, implying the presence of proteins from the integrin family (fibronectin, laminin, and fibrinogen) at the cell surface (reviewed in references 4 and 18). In some viruses, the presence of a functional RGD motif is required for infection or expression of their cytopathic effects (1, 2, 17). We wondered whether this sequence could be involved in the uptake of Bet. A plasmid (p2EBΔRGD) harboring the GGD motif instead of RGD in Bet was constructed and transfected into COS-6 cells as above. The mutated Bet, produced in the supernatant, was taken up by naive cells in the same way as the wild-type one (Fig. 5, lane 6). Detection of Bet from p2EBΔRGD-transfected cells, both within the cell and in the supernatant, was weaker than from p2EB-transfected cells, possibly reflecting a higher lability of this mutant (Fig. 5). Thus, while Bet and gp160 are secreted, only Bet is taken up by an RGD-independent mechanism.
DISCUSSION
In this report, a new HFV fusion protein species, translated from an mRNA harboring a 119-bp splice at the end of the env ORF, is described. This splice deletes the transmembrane anchor of the Env protein, its dilysine ER retention motif and fuses the env ORF to the bel/bet ORF. The 160-kDa Env-Bet glycoprotein and Bet can both be secreted, but only Bet is taken up by naive recipient cells via an RGD-independent mechanism.
This article describes a new LTR-derived mRNA, harboring a 119-bp intron in the env gene, which, depending upon the presence or absence of the 301-bp intron in the bet gene, can lead to the production of env-bel1 or env-bet mRNA. This 119-bp splice could be harbored by either the genomic, the pol, or the env mRNA. While this very small deletion precludes its detection on a Northern blot, the presence of this splice in p2EB-transfected cells demonstrates that it can indeed be found in env transcripts. The 119-bp intron is located between exon 6 (which harbors the internal promoter) and exon 7 (encoding the bel genes) and is used in the synthesis of bel1 and bet mRNAs from the internal promoter (10–12). The fusion protein detected is an Env-Bet protein, but RT-PCR experiments demonstrate the existence of an env-bel1 transcript (Fig. 3). Interestingly, the different spliced RNAs in Fig. 3A can also be directly amplified by PCR without an RT step on extrachromosomal DNA extracted from HFV-infected cells, as well as from molecular clones generated from chronically infected cells (reference 21 and data not shown). This suggests either that these viral mRNAs are nonspecifically retrotranscribed in infected cells or that these splice events are part of full-length viral DNAs, as is the case of the bet splice in ΔHFV (21).
The fusion protein is inserted into the ER membrane via the peptide signal at the 5′ end of Env but subsequently behaves differently from the wild-type Env and is secreted. We found this protein in supernatants of pEB-transfected cells and HFV-infected BHK21 cells, consistent with the deletion of both the TM anchor and the dilysine ER retention motif. Bet is also secreted and taken up by noninfected cells (Fig. 5). Considering the role of Bet in viral resistance and our demonstration that Bet-expressing stable cell lines are resistant to HFV-induced lysis (23a), uptaken Bet protein could impair viral replication in recipient cells and thus behave as a virokine.
ACKNOWLEDGMENTS
We thank Axel Rethwilm and Dirk Lindemann for communicating results before publication. We warmly thank A. M. Poorters and N. Honoré for technical assistance, D. Sitterlin for help in some experiments, and the Laboratoire Photographique de l’Institut d’Hématologie for the photographic work. We thank Libin Ma (Leiden) for providing pSG5M.
REFERENCES
- 1.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang K H, Day C, Walker J, Hyypia T, Stanway G. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 is functionally significant. J Gen Virol. 1992;73:621–626. doi: 10.1099/0022-1317-73-3-621. [DOI] [PubMed] [Google Scholar]
- 3.Cleveland D W, Fischer S G, Kirschner M W, Laemmli U K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1997;252:1102–1106. [PubMed] [Google Scholar]
- 4.D’Souza S E, Ginsberg M H, Plow E F. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem Sci. 1991;16:246–250. doi: 10.1016/0968-0004(91)90096-e. [DOI] [PubMed] [Google Scholar]
- 5.Giron M L, Rozain F, Debons-Guillemin M C, Canivet M, Périès J, Emanoil-Ravier R. Human foamy virus polypeptides: identification of env and bel gene products. J Virol. 1993;67:3596–3600. doi: 10.1128/jvi.67.6.3596-3600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goepfert P A, Shaw K L, Ritter G, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herchenröder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 9.Kupiec J J, Kay A, Hayat M, Ravier R, Périès J, Galibert F. Sequence analysis of the simian foamy virus type 1. Gene. 1991;101:185–194. doi: 10.1016/0378-1119(91)90410-d. [DOI] [PubMed] [Google Scholar]
- 10.Löchelt M, Flügel R M. The molecular biology of human and primate spumaretrovirus. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 11.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome possesses an internal, bel1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muranyi W, Flügel R M. Analysis of splicing pattern of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renshaw R W, Casey J W. Transcriptional mapping of the 3′ end of the bovine syncytial virus genome. J Virol. 1994;68:1021–1028. doi: 10.1128/jvi.68.2.1021-1028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renshaw R W, Gonda M A, Casey J W. Structure and transcriptional status of bovine syncytial virus in cytopathic infections. Gene. 1991;105:179–184. doi: 10.1016/0378-1119(91)90149-6. [DOI] [PubMed] [Google Scholar]
- 15.Rethwilm A. Regulation of foamy virus gene expression. Curr Top Microbiol Immunol. 1995;193:1–24. doi: 10.1007/978-3-642-78929-8_1. [DOI] [PubMed] [Google Scholar]
- 16.Rey M A, Krust B, Laurent A G, Montagnier L, Hovanessian A G. Characterization of human immunodeficiency virus type 2 envelope glycoprotein precursor during processing. J Virol. 1989;63:647–658. doi: 10.1128/jvi.63.2.647-658.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roivainen M, Hyypia T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruoslahti E, Pierschbacher M D. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 19.Saïb A, Koken M, van der Spek P, Périès G, de Thé H. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J Virol. 1995;69:5261–5268. doi: 10.1128/jvi.69.9.5261-5268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saïb A, Neves M, Giron M L, Guillemin M C, Valla J, Peries J, Canivet M. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology. 1997;228:263–268. doi: 10.1006/viro.1996.8383. [DOI] [PubMed] [Google Scholar]
- 21.Saïb A, Périès J, de Thé H. A defective human foamy provirus generated by pregenome splicing. EMBO J. 1993;12:4439–4444. doi: 10.1002/j.1460-2075.1993.tb06129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saïb A, Périès J, de Thé H. Recent insights into the biology of the human foamy virus. Trends Microbiol. 1995;3:173–178. doi: 10.1016/s0966-842x(00)88916-7. [DOI] [PubMed] [Google Scholar]
- 23.Saïb A, Saal F, Giron M L, Valla J, Hojman F, Périès J, Canivet M. Comparative studies on IL-6 production status in HTLV-I chronically infected cell lines derived from different HTLV-I associated pathologies. J Biol Regul Homeostatic Agents. 1993;7:79–84. [PubMed] [Google Scholar]
- 23a.Saïb, A. Unpublished data.
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Weiss R A. A virus in search of a disease. Nature. 1988;333:497–498. doi: 10.1038/333497a0. [DOI] [PubMed] [Google Scholar]
- 26.Winkler I, Bodem J, Haas L, Zemba M, Delius H, Flower R, Flügel R M, Löchelt M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J Virol. 1997;71:6727–6741. doi: 10.1128/jvi.71.9.6727-6741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]