Abstract
Purpose
We describe the implementation of CYP2D6-focused pharmacogenetic testing to guide opioid prescribing in a quaternary care, nonprofit pediatric academic medical center.
Summary
Children are often prescribed oral opioids after surgeries, for cancer pain, and occasionally for chronic pain. In 2004, Cincinnati Children’s Hospital Medical Center implemented pharmacogenetic testing for CYP2D6 metabolism phenotype to inform codeine prescribing. The test and reports were updated to align with changes over time in the testing platform, the interpretation of genotype to phenotype, the electronic health record, and Food and Drug Administration (FDA) guidance. The use of the test increased when a research project required testing and decreased as prescribing of oxycodone increased due to FDA warnings about codeine. Education about the opioid-focused pharmacogenetic test was provided to prescribers (eg, the pain and sickle cell teams) as well as patients and families. Education and electronic health record capability increased provider compliance with genotype-guided postsurgical prescribing of oxycodone, although there was a perceived lack of utility for oxycodone prescribing.
Conclusion
The implementation of pharmacogenetic testing to inform opioid prescribing for children has evolved with accumulating evidence and guidelines, requiring changes in reporting of results and recommendations.
Keywords: analgesics, opioid, cytochrome P450 CYP2D6, pharmacogenetics
Key Points.
Implementing CYP2D6 testing to inform opioid prescribing required a multidisciplinary team that has adapted to advances in knowledge and changing guidelines.
When implementing CYP2D6 testing to inform opioid prescribing, great consideration must be given to the alleles to test, the assay method used to detect duplication, testing turnaround time, and education of providers.
At our institution, the enthusiasm for genotyping CYP2D6 to guide opioid prescribing has decreased as the rate of codeine prescribing has decreased and due to the lack of consensus recommendations on pharmacogenetics-guided oxycodone prescribing.
Oral opioids, including codeine, tramadol, hydrocodone, and oxycodone, are often prescribed to children and infants for analgesia after surgeries, for cancer pain, and occasionally for chronic pain. While they are all metabolized by cytochrome P450 (CYP) isozyme 2D6, there are some important differences. Codeine and tramadol are prodrugs primarily metabolized by CYP2D6 into active metabolites, morphine and (+)-O-desmethyltramadol, respectively.1-4 Unlike codeine and tramadol, hydrocodone and oxycodone have some analgesic properties not involving metabolism by CYP2D6. The Food and Drug Administration (FDA) issued a black box warning in 2013, followed by a contraindication in 2017, regarding codeine and tramadol use in children younger than 12 years of age due to life-threatening and fatal events associated with CYP2D6 metabolizer phenotype.5 After these warnings, codeine use dramatically declined and alternative pain management post tonsillectomy with/without adenoidectomy became necessary.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) published guidelines for opioid prescribing based on CYP2D6 metabolizer phenotype in 2012, with guideline updates in 2014 and 2021.6-8 There are more than 140 CYP2D6 alleles, with complete gene duplication and deletion being possible.9 CYP2D6 metabolizer phenotype is determined by calculation of an activity score based on allele activity values.10,11 For poor metabolizers, CPIC recommends avoiding codeine and tramadol due to poor analgesia.8 For ultrarapid metabolizers, CPIC recommends avoiding codeine and tramadol due to a risk of serious toxicity.12,13 The recommendations for intermediate metabolizers are to prescribe usual doses of codeine or tramadol but, if the patient experiences poor analgesia, to switch to a different medication.
At Cincinnati Children’s Hospital Medical Center (CCHMC), we have approximately 1.5 million patient encounters per year and perform more than 6,000 inpatient surgeries and more than 25,000 outpatient surgeries per year. CCHMC has a perioperative, medical pain, and palliative service for management of pain in inpatients with postsurgical pain, medical/chronic pain, and cancer pain, respectively. Our pain clinic manages outpatients with chronic pain. Codeine is restricted to use only by the pain team, and tramadol use in patients less than 12 years of age requires prior approval from the pain or palliative care team. We have a Genetic Pharmacology Service that facilitates the implementation of pharmacogenetic testing in clinical care across multiple gene-drug pairs for a variety of medications.14 We began performing pharmacogenetic testing in 2004, with CYP2C19 and CYP2D6 tests to guide use of neuropsychiatric medications and CYP2D6 testing to guide codeine use. In this report, we describe the implementation and evolution of CYP2D6 pharmacogenetic testing to inform opioid prescribing and our learnings.
Implementation and evolution of the CYP2D6-opioid pharmacogenetics panel
The Genetic Pharmacology Service at CCHMC began with a team composed of a molecular geneticist, clinical pharmacologists, a clinical pharmacy specialist, advanced practice nurses, and an informaticist, neurologist, and psychiatrist champion.14 When the service began, testing was implemented at once for several gene-drug pairs, including CYP2D6 and CYP2C19 for antidepressants, VKORC1 and CYP2C9 for warfarin, TPMT for thiopurines, and CYP2D6 for codeine. Collaboration by a multidisciplinary team was necessary to select the variants to test, estimate dosing adjustments for each medication (a task led by the clinical pharmacologist), write template reports for each combination of metabolizer status and drug (with contributions from the nurses, pharmacist, and physicians), implement decision support and result reporting (a task led by the informaticist), and provide education to clinicians on how to order, view, understand, and use the testing (with contributions from the entire team).
A test focused on codeine based on 3 CYP2D6 alleles was launched in 2004 but was rarely ordered. CCHMC is a member of the Electronic Medical Records and Genomics (eMERGE) Network, which is organized and funded by the National Human Genome Research Institute to implement genomic medicine.15 eMERGE is a national network that combines DNA biorepositories with electronic medical record systems for large-scale, high-throughput genetic research in support of implementing genomic medicine, including site-specific projects.16 As part of a research project conducted with the eMERGE Network, an anesthesiologist champion worked with the Genetic Pharmacology Service team to replace the codeine test with an opioid panel that included more comprehensive CYP2D6 testing and recommendations for 4 opioids (codeine, tramadol, hydrocodone, and oxycodone).17,18 The effort of the anesthesiologist and advanced practice nurse that led the 2013 update was supported by an eMERGE II grant to CCHMC.19 The membership of the Genetic Pharmacology Service team also evolved but always included at least one (but often more than one) physician, pharmacist, pharmacologist, laboratory director, and advanced practice nurse. The service does not support any eMERGE Network research efforts of the team members, but it provides the codirectors a small fee for each completed test, which is built into the cost of the testing.
Evolution of the assay.
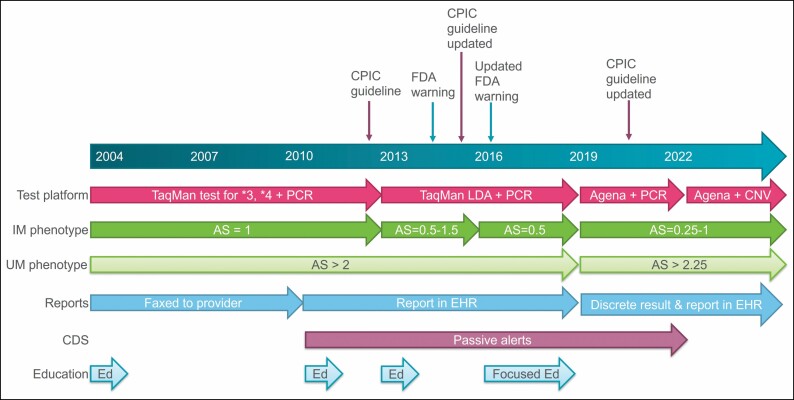
Since our service started before most commercial pharmacogenetic tests were available, there was little consideration of external tests at the onset. In designing the pharmacogenetic test, there were many considerations regarding which alleles to include and what assay to use for testing. When testing was launched in 2004, only CYP2D6*3, *4, and *5 (deletion) and duplication were tested to balance the 2-day turnaround time and cost of testing. These alleles were chosen based on then-available evidence supporting their impact on function and their population frequency; many of the alleles that are now commonly tested had not yet been described or characterized at that time. In 2013, the assay was updated to test for 21 alleles on a TaqMan low-density array (Applied Biosystems, Forest City, CA) and perform long-range polymerase chain reaction (PCR) testing for duplication, which indicates whether a duplication is present but not which allele is duplicated or how many times it is duplicated (Figure 1). Again the evidence was evaluated for alleles that impact the function of CYP2D6, and the testing platform was chosen based on cost and turnaround time. In 2019, the molecular genetics laboratory validated a new assay using the MassARRAY System (Agena Bioscience, San Diego, CA) but was still using long-range PCR to detect duplications. This test was chosen for cost-effectiveness and speed, though it did not include some of the other alleles or genes that we wanted (eg, CYP2D6*40, TPMT, NUDT15). In 2022, the laboratory validated a custom panel to add these genes and alleles. In 2022, we also migrated to using the VeriDose CYP2D6 CNV Panel reagent (Agena Bioscience) for CYP2D6 copy number variation (CNV) to help determine which allele is duplicated and to detect hybrid alleles (eg, when CYP2D6 has portions of CYP2D7 inserted). The turnaround time remains at 2 business days. As there is no gold standard for allele coverage in pharmacogenetic testing, the Association for Molecular Pathology has begun categorizing alleles into tier 1 (ie, “must test” alleles) and tier 2 (ie, “nice to include” alleles).9 When the CYP2D6 categorizations were published in 2021, we ensured that we were testing all the tier 1 alleles (our test also includes many, but not all, of the tier 2 alleles). We prefer not to test alleles that CPIC categorizes as “unknown function” so that we do not report a ranged or “possible” phenotype (eg, “possible intermediate metabolizer”). The CPIC and PharmVar websites are the resources we use to determine the activity of each allele, calculate the activity score, and determine the phenotype.20-23 The benefits of in-house testing over commercial testing remain: quick turnaround, reporting of results in the electronic health record (EHR), and ensuring our standards for allele coverage, genotype-to-phenotype translation, and dosing recommendations are met.
Figure 1.
Timeline of the evolution of CYP2D6 testing for opioid guidance at Cincinnati Children’s Hospital Medical Center (including assay selection, interpretation of genotype to phenotype for intermediate and ultrarapid metabolizers, reporting, clinical decision support, and education). CNV indicates copy number variation; AS, activity score; CDS, clinical decision support; CPIC, Clinical Pharmacogenetics Implementation Consortium; FDA, Food and Drug Administration; PCR, long-range polymerase chain reaction; EHR, electronic health record; Ed, provider education; IM, intermediate metabolizer.
Evolution of genotype-to-phenotype interpretation.
We used best available evidence to assign a predicted metabolizer phenotype to a diplotype. In preparation for our 2004 launch, we used the literature’s most common diplotype interpretation without the subsequently published activity scoring system. The interpretation of intermediate, normal, and ultrarapid metabolizers has changed since 2004. With our initial minimal set of alleles, the combination of a functional allele (CYP2D6*1) and no-function allele (CYP2D6*3, *4, or *5) was interpreted as “intermediate metabolizer” (IM). When our test was expanded to include decreased-function alleles in 2013, we used the activity scoring system and assigned the IM designation to individuals with a total activity scores of 0.5 (1 no-function and 1 reduced-function allele), 1.0 (2 reduced-function alleles or 1 no-function and 1 normal-function allele), and 1.5 (1 reduced-function and 1 normal-function allele) (Figure 1). We updated our interpretations in 2016 to be consistent with the 2014 CPIC guideline for codeine,7 which classified patients with total activity scores of 1.0 and 1.5 as normal metabolizers (NMs). In 2019, CPIC published a consensus paper on the standardization of CYP2D6 genotype-to-phenotype translation.21 This led to the inclusion of the activity score of 1.0 in the IM classification and changed the definition of ultrarapid metabolizer (UM), and our interpretation was modified accordingly.
Evolution of results reporting.
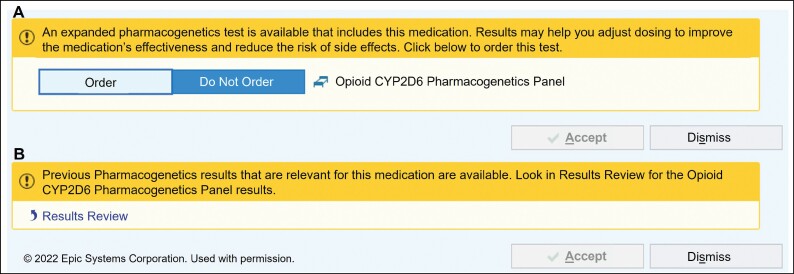
As our service began prior to the implementation of the electronic medical record at our institution, results were delivered to the ordering clinician by fax (Figure 1). When the EHR was introduced in 2010, text reports were included in the results review tab, where all routine clinical laboratory results are reported. Additionally, clinical decision support alerts were deployed in 2010 to inform the ordering clinician that a genetic test applicable to a medication was available (Figure 2). When the alerts were implemented with the introduction of the EHR, there were not many options for customizing the alerts, so they were designed to simply state that a test was available for an ordered medication or that results relevant to the medication ordered were available. If the results were already in the chart, the alert said that they were available in the results review tab; however, it took the clinicians several clicks to view the results. The report available in the results review tab includes recommendations for codeine, tramadol, hydrocodone, and oxycodone that are consistent with the most recent CPIC guidelines.8 The pain team was able to provide feedback about how the reports were structured and what guidance was provided. When CPIC determined that there was insufficient evidence to support making any recommendations for oxycodone based on CYP2D6 metabolizer status,8 the report was updated to reflect this recommendation. For each CYP2D6 phenotype, all 4 medications are included in the report along with the associated recommendations (Table 1). The reason we include oxycodone and hydrocodone in the report is because the clinicians wanted to know the recommendations for alternatives to codeine and tramadol.
Figure 2.
Best practice alerts that appear after an opioid is ordered to prompt ordering the CYP2D6 test (A) or to indicate that results are available (B).
Table 1.
Current Recommendations for Opioid Prescribing Based on CYP2D6 phenotype at Cincinnati Children’s Hospital Medical Center
| CYP2D6 phenotype | Codeine and tramadol recommendations | Oxycodone and hydrocodone recommendations |
|---|---|---|
| PM | Not recommended | 100% standard starting dose |
| IM | When not contraindicated, 100% standard starting dose | 100% standard starting dose |
| NM | When not contraindicated, 100% standard starting dose | 100% standard starting dose |
| UM | Not recommended | Insufficient evidence to make recommendation |
Abbreviations: CYP2D6, cytochrome P450 isozyme 2D6; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
During the period 2016-2018, the postoperative pain team physicians, nurse practitioners, and surgeons were directed through education and EHR alerts to prescribe hydromorphone or morphine instead of oxycodone for CYP2D6 PMs and UMs as part of a research project. During the time period when the education on genotype guidance was provided in addition to passive alerts, 29% of CYP2D6 PM/UMs were prescribed CYP2D6-dependent opioids, compared to 67% during the preceding period, when the results were only provided passively.18 In the genotype-directed group (N = 127), 95% of NM/IMs versus 29% of PM/UMs were prescribed CYP2D6-dependent opioids (P < 0.001). Logistic regression showed that PM/UMs in the genotype-directed group had a lower chance of being prescribed CYP2D6-dependent opioids (effect size, −2.775; SE, 1.566; P = 0.076) than the control group (n = 66). The study was not powered to study the clinical effectiveness of the program. However, we found that oxycodone requirements to maintain analgesia and the risk of opioid-induced respiratory depression (in subjects with higher oxycodone use relative to total opioid use) were different by CYP2D6 genotype.
In 2019, the molecular genetics lab transitioned to a different reporting system that enabled inclusion of CYP2D6 phenotype as a discrete result in the results review tab, with only one click required to view the recommendations. New best practice alerts that included the CYP2D6 phenotype when an opioid was prescribed were created, but dosing recommendations could not be included in the alert due to lack of infrastructure (ie, there was no genomics module in our EHR); however, they are presented in a table in the results review tab, and one goal for the future is to provide dosing recommendations in the alert when infrastructure allows.
The Genetic Pharmacology Service does not update previously released results when the interpretation changes, but patients could be retested if their first test was prior to the 2013 update, since there is the potential for CYP2D6*1 allele carriers to have an allele that was not tested for previously. The molecular genetics laboratory also does not typically perform a reinterpretation of genetic testing when the assay or genotype-to-phenotype translation changes unless it is requested by the clinician. Therefore, when the CYP2D6 variants tested or genotype-to-phenotype interpretations changed, we did not issue new reports to patients tested prior to the change. Because CYP2D6*1 is the default allele when none of the tested variants are detected, patients with one or more *1 alleles are retested on an updated assay when clinicians request a reinterpretation or order pharmacogenetic testing for a different panel of drugs, such as our psychiatry panel. If the patient had a genotype of CYP2D6*4/*4 on the 2012 test, any subsequent new test order is cancelled in the laboratory because the phenotype is unlikely to change with the addition of more alleles on the test. In the event that there are 2 test results, they are both available in the results review tab. The phenotype from the most recent test will be shown to the clinician in a best practice alert when a medication is ordered. With the implementation of the CNV assay in 2022, it is possible to detect which allele is duplicated and whether there are 3 copies or more than 3 copies of the gene. This is important because a patient who was previously genotyped as a CYP2D6*1/*4 with duplication could be either an IM (if the *4 allele is the one duplicated) or a normal metabolizer (if the *1 allele is the one duplicated). Prior to the implementation of the CNV assay in 2022, this patient’s phenotype would have been reported as “uncertain” and the report would have indicated that the response to these opioids could not be predicted because the duplicated allele could not be determined.
Experience with use of the opioid pharmacogenetic panel
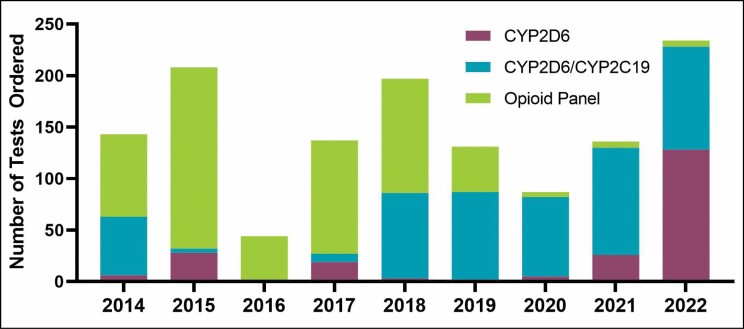
There was very slow uptake of the test for codeine pharmacogenetics prior to 2013; however, since implementation of the expanded opioid panel, more than 1,300 tests that include opioid recommendations have been ordered (Figure 3). Growth in testing was due to clinician awareness of the expanded test rather than direct marketing to patients. As with other laboratory tests, clinicians inform patients about the rationale for planned testing and patients or their parents have the option to refuse such testing. Clinician orders gradually decreased because the acute pain team perceived a lack of utility for oxycodone prescribing, ineffective clinical decision support, and lack of insurance coverage. The clinician perceptions were shared with the director of the perioperative pain service through personal communication, not a rigorous survey. Yet, recent analysis of retrospective data demonstrated an association of CYP2D6 phenotype and oxycodone requirements after surgery, affirming that alerts for genotype-guided dosing affected prescribing after surgery.18 When codeine use was first restricted by FDA, the hematologists would order CYP2D6 testing for patients with sickle cell disease because codeine was the only opioid that could be prescribed electronically or by phone and didn’t require a prescription that the patient would have to physically pick up.24 When the ability to prescribe any opioid electronically was available, the perceived need for testing in this population fell off sharply, and hematologists prescribed other opioids, most often oxycodone. The number of codeine prescriptions for any indication at our institution fell by 90% after the FDA restriction, from more than 5,500 prescriptions in 2012 to 550 in 2017.
Figure 3.
Number of tests that include CYP2D6 ordered per year. The CYP2D6 test results include no therapeutic recommendations. The opioid panel test results include recommendations for 4 opioids. The CYP2D6/CYP2C19 panel includes recommendations on prescribing of voriconazole and opioids.
In 2017, the clinical pharmacy specialist on the bone marrow transplant service requested a combination test report that included CYP2C19 data for voriconazole dosing and CYP2D6 data for opioid selection. Since then, this combination test has been part of the standard order set for all patients receiving a bone marrow transplant.
Because our clinicians prefer ordering drug panels relevant to their specialties, it is possible that CYP2D6 test results are in patients’ EHRs without therapeutic recommendations specific to opioids. If a clinician notices the CYP2D6 test result and wants the opioid recommendations, they can order a reinterpretation and the laboratory will add the report, along with recommendations, to the patient’s chart without the need for a new sample to be tested. In response to clinicians’ requests, we recently began offering CYP2D6 as well as other pharmacogenes without drug specific therapeutic recommendations. Uptake is low for gene-only tests.
Education.
Education about pharmacogenetic testing in general was made available on the CCHMC website. Codeine- and opioid-focused CYP2D6 testing education was initiated in preparation for research studies using pharmacogenetics to inform pain management drug selection.25 Therefore, presentations were given to otolaryngology, orthopedic surgery, and same-day surgery faculty and staff. The research-focused education quickly expanded to presentations with the clinical pain team at faculty meetings. The clinical pharmacy specialists were made aware of the pharmacogenetic tests via periodic presentations at their monthly meetings. Printable handouts about pharmacogenetic testing and about specific results are available on the website for parents and patients. Additional parent education about results occurred when the advanced practice nurse returned results as part of eMERGE-related pharmacogenetic studies.17,18
Cost and reimbursement issues.
When pharmacogenetic testing was first implemented at CCHMC, the cost of testing was balanced with the turnaround time and evidence-based determination of the most impactful variants in the patient population. The cost billed to patients was approximately $300 for CYP2D6 testing that included alleles *3 and *4 CNV. When the panel was expanded in 2013, the price was increased over the initial price at implementation to reflect improved coverage of the gene, and the price has continued to increase to keep up with inflation of the cost of testing. If the test is ordered during an inpatient stay, the cost is included in the bundle payment from insurance and the hospital ensures that the molecular genetics lab is reimbursed for the test. When testing is performed at an outpatient visit, insurance coverage is variable, as some insurance companies consider it experimental.26 Recent changes in insurance coverage have likely improved reimbursement, but many insurers still will not cover CYP2D6 testing. During our education sessions with clinicians, cost and insurance coverage issues are addressed and clinicians are provided with instructions on how to check an individual patient’s insurance coverage with the Current Procedural Terminology codes that will be used for billing.
Lessons learned.
CYP2D6 is a complicated locus to genotype, with many alleles having been discovered and characterized after our first implementation. When beginning implementation, the decision of which alleles to test and how to test for duplication of alleles is a necessary consideration that has to be balanced with cost and turnaround time. The interpretation of genotype to phenotype has changed several times over the last 18 years as more information about the impact of alleles and the effect on opioid pharmacokinetics accumulated. This required our service to update our therapeutic recommendations several times to keep pace with the updated guidelines.
The use of the test changed as clinician champions (doctors, nurses, and pharmacists) became aware of the test and understood how to use it through education with direct instruction. It was common to incorporate testing into the standard order sets for patients they knew were likely to receive opioids as part of their treatment course (those having very painful surgeries, bone marrow transplant, or cancer treatment). For future efforts in implementing pharmacogenetic tests, we will engage clinical champions in the divisions expected to utilize the testing most frequently to allow for education to reach all members of the divisions and to achieve faster uptake and appropriate utilization.
Planned updates for the future.
Long-read sequencing is planned for future testing, which will improve the accuracy of allele-calling (ie, determining which star allele is represented by the combination of variants) and genotype-to-phenotype interpretation.27 We are working to develop a comprehensive panel that includes the CYP2D6-guided opioid recommendations with several of the other gene-drug pairs we have implemented. The clinical pharmacy specialist in oncology will incorporate this comprehensive test into the standard order set for patients with acute lymphoblastic leukemia. We will work with the teams serving the populations we believe can benefit from CYP2D6 testing to incorporate the testing into routine care in the future, as well as perform cost-benefit analyses of the testing.
Conclusion
The evidence, FDA recommendations, CPIC guidelines, and implementation of CYP2D6 gene-drug pairs have evolved over the last 18 years, which has required the Genetic Pharmacology Service team at CCHMC to update its testing, interpretation, and reporting several times. The use of the testing has also varied considerably over the years as clinician interest and prescribing trends have changed.
Acknowledgments
We thank Carole Jayne, BA, for providing the number of tests ordered.
Contributor Information
Laura B Ramsey, Department of Pediatrics, Division of Clinical Pharmacology and Division of Research in Patient Services, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
Cynthia A Prows, Division of Human Genetics and Division of Patient Services, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Vidya Chidambaran, Department of Anesthesiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
Senthilkumar Sadhasivam, Department of Anesthesiology and Perioperative Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA.
Charles T Quinn, Department of Pediatrics and Division of Hematology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
Ashley Teusink-Cross, Division of Pharmacy, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Sonya Tang Girdwood, Department of Pediatrics, Division of Clinical Pharmacology; and Division of Hospital Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
D Brian Dawson, Department of Pediatrics and Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Alexander A Vinks, Department of Pediatrics, Division of Clinical Pharmacology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
Tracy A Glauser, Department of Pediatrics, Division of Neurology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; College of Medicine, University of Cincinnati, Cincinnati, OH, USA.
Disclosures
The research that required implementation of CYP2D6 testing was funded by the National Human Genome Research Institute through grants U01HG006828 and U01HG008666, the National Institute of Arthritis and Musculoskeletal and Skin Diseases through grant R01AR075857, and the National Institute of Child Health and Human Development through grants R01HD089458, R21HD094311, and R01HD096800. Dr. Sadhasivam’s salary during the work described and manuscript preparation were partially supported by the following National Institutes of Health principal investigator (PI) or multiple principal investigator (MPI) grants: R01HD089458 (PI, Sadhasivam); R21HD094311 (PI, Sadhasivam); R01HD096800 (PI, Sadhasivam); R44DA055407, R44DA056280, and R41DA053877 (MPI, Sadhasivam); R01DA054513 (MPI, Chelly and Sadhasivam); and U01TR003719 (PI, Sadhasivam). Dr. Sadhasivam, Dr. Prows, and Dr. Chidambaran are inventors on US patents 9944985, 10662476, 16/946401, and 16/850537, and Dr. Sadhasivam is an inventor on US patents 16/946399 and 10878939. Dr. Sadhasivam is the founder and chief medical officer of OpalGenix, Inc. The other authors have declared no potential conflicts of interest.
Previous affiliations
At the time of the work performed, Dr. Sadhasivam was affiliated with Department of Anesthesia, Cincinnati Children’s Hospital Medical Center.
References
- 1. Stamer UM, Musshoff F, Kobilay M, Madea B, Hoeft A, Stuber F. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther. 2007;82(1):41-47. doi: 10.1038/SJ.CLPT.6100152 [DOI] [PubMed] [Google Scholar]
- 2. Lötsch J, Rohrbacher M, Schmidt H, Doehring A, Brockmöller J, Geisslinger G. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain. 2009;144(1-2):119-124. doi: 10.1016/J.PAIN.2009.03.023 [DOI] [PubMed] [Google Scholar]
- 3. Poulsen L, Arendt-Nielsen L, Brøsen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther. 1996;60(6):636-644. doi: 10.1016/S0009-9236(96)90211-8 [DOI] [PubMed] [Google Scholar]
- 4. Kirchheiner J, Keulen JTHA, Bauer S, Roots I, Brockmöller J. Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol. 2008;28(1):78-83. doi: 10.1097/JCP.0B013E318160F827 [DOI] [PubMed] [Google Scholar]
- 5. Ramsey LB, Ong HH, Schildcrout JS, et al. Prescribing prevalence of medications with potential genotype-guided dosing in pediatric patients. JAMA Netw Open. 2020;3(12):e2029411. doi: 10.1001/JAMANETWORKOPEN.2020.29411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91(2):321-326. doi: 10.1038/clpt.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376-382. doi: 10.1038/clpt.2013.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6, OPRM1, and COMT genotype and select opioid therapy. Clin Pharmacol Ther. Published online January 2, 2021. doi: 10.1002/cpt.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical CYP2D6 genotyping allele selection: a joint consensus recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23(9):1047-1064. doi: 10.1016/J.JMOLDX.2021.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaedigk A, Simon S, Pearce R, Bradford L, Kennedy M, Leeder J. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234-242. doi: 10.1038/sj.clpt.6100406 [DOI] [PubMed] [Google Scholar]
- 11. Gaedigk A, Dinh J, Jeong H, Prasad B, Leeder J. Ten years’ experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J Pers Med. 2018;8(2):15. doi: 10.3390/jpm8020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orliaguet G, Hamza J, Couloigner V, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics. 2015;135(3):e753-e755. doi: 10.1542/PEDS.2014-2673 [DOI] [PubMed] [Google Scholar]
- 13. Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351(27):26-27. doi: 10.1056/NEJMOA041888 [DOI] [PubMed] [Google Scholar]
- 14. Ramsey LB, Prows CA, Zhang K, et al. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin Pharmacol Ther. 2019;105(1):49-52. doi: 10.1002/cpt.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761-771. doi: 10.1038/GIM.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. eMERGE Network. Home page. Accessed October 5, 2022. https://emerge-network.org/
- 17. Myers MF, Zhang X, McLaughlin B, et al. Prior opioid exposure influences parents’ sharing of their children’s CYP2D6 research results. Pharmacogenomics. 2017;18(13):1199-1213. doi: 10.2217/PGS-2017-0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Merchant S, Prows CA, Yang F, et al. Association of CYP2D6 genotype predicted phenotypes with oxycodone requirements and side effects in children undergoing surgery. Ann Transl Med. 2022;10(23):1262. doi: 10.21037/atm-2022-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96(4):482-489. doi: 10.1038/CLPT.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nofziger C, Turner AJ, Sangkuhl K, et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther. 2020;107(1):154-170. doi: 10.1002/cpt.1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2020;13(1):116-124. doi: 10.1111/cts.12692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin Pharmacol Ther. 2020;107(1):171-175. doi: 10.1002/CPT.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther. 2018;103(3):399-401. doi: 10.1002/cpt.910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gammal RS, Caudle KE, Quinn CT, et al. The case for pharmacogenetics-guided prescribing of codeine in children. Clin Pharmacol Ther. 2019;105(6):1300-1302. doi: 10.1002/cpt.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohrer Vitek CR, Abul-Husn NS, Connolly JJ, et al. Healthcare provider education to support integration of pharmacogenomics in practice: the eMERGE Network experience. Pharmacogenomics. 2017;18(10):1013-1025. doi: 10.2217/PGS-2017-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers SL, Keeling NJ, Giri J, et al. PARC report: a health-systems focus on reimbursement and patient access to pharmacogenomics testing. Pharmacogenomics. 2020;21(11):785-796. doi: 10.2217/PGS-2019-0192 [DOI] [PubMed] [Google Scholar]
- 27. Qiao W, Yang Y, Sebra R, et al. Long-read single molecule real-time full gene sequencing of cytochrome P450-2D6. Hum Mutat. 2016;37(3):315-323. doi: 10.1002/humu.22936 [DOI] [PMC free article] [PubMed] [Google Scholar]