Abstract
Background and Aims
Early diagnosis of aortic stenosis (AS) is critical to prevent morbidity and mortality but requires skilled examination with Doppler imaging. This study reports the development and validation of a novel deep learning model that relies on two-dimensional (2D) parasternal long axis videos from transthoracic echocardiography without Doppler imaging to identify severe AS, suitable for point-of-care ultrasonography.
Methods and results
In a training set of 5257 studies (17 570 videos) from 2016 to 2020 [Yale-New Haven Hospital (YNHH), Connecticut], an ensemble of three-dimensional convolutional neural networks was developed to detect severe AS, leveraging self-supervised contrastive pretraining for label-efficient model development. This deep learning model was validated in a temporally distinct set of 2040 consecutive studies from 2021 from YNHH as well as two geographically distinct cohorts of 4226 and 3072 studies, from California and other hospitals in New England, respectively. The deep learning model achieved an area under the receiver operating characteristic curve (AUROC) of 0.978 (95% CI: 0.966, 0.988) for detecting severe AS in the temporally distinct test set, maintaining its diagnostic performance in geographically distinct cohorts [0.952 AUROC (95% CI: 0.941, 0.963) in California and 0.942 AUROC (95% CI: 0.909, 0.966) in New England]. The model was interpretable with saliency maps identifying the aortic valve, mitral annulus, and left atrium as the predictive regions. Among non-severe AS cases, predicted probabilities were associated with worse quantitative metrics of AS suggesting an association with various stages of AS severity.
Conclusion
This study developed and externally validated an automated approach for severe AS detection using single-view 2D echocardiography, with potential utility for point-of-care screening.
Keywords: Deep learning, Echocardiography, Aortic stenosis, Digital health
Structured Graphical Abstract
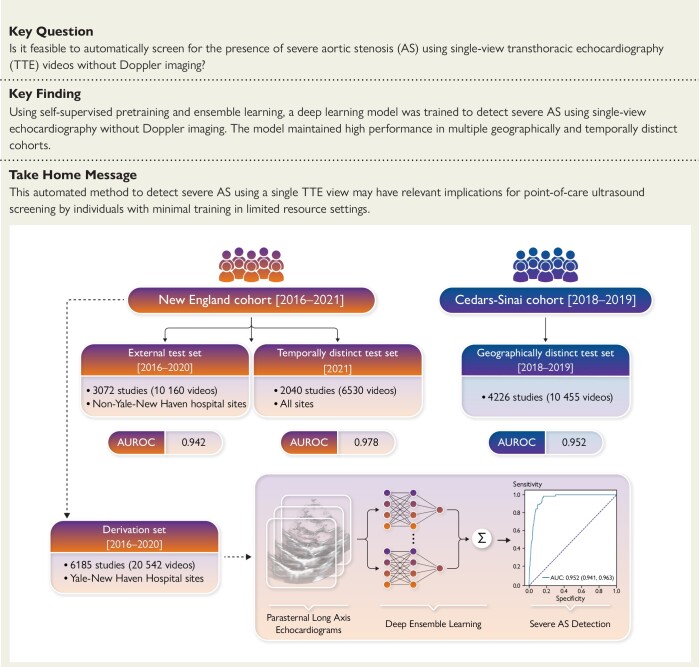
Structured Graphical abstract.
An automated deep learning approach for severe AS detection from single-view echocardiography evaluated across geographically and temporally distinct cohorts. AUROC, area under the receiver operating characteristic curve.
Introduction
Aortic stenosis (AS) is a chronic, progressive disease, and associated with morbidity and mortality.1,2 With advances in both surgical and transcatheter aortic valve replacement,3 there has been an increasing focus on early detection and management.4–6 The non-invasive diagnosis of AS can be made with hemodynamic measurements using Doppler echocardiography,2,7–10 but that requires dedicated equipment and skilled acquisition and interpretation. On the other hand, even though two-dimensional (2D) cardiac ultrasonography is increasingly available with handheld devices that can visualize the heart,11 it has not been validated for the diagnosis or longitudinal monitoring of AS. With an estimated prevalence of 5% among individuals aged 65 years or older,8 there is a growing need for user-friendly screening tools which can be used in everyday practice by people with minimal training to screen for severe AS. This need for timely screening is further supported by evidence suggesting improved outcomes with early intervention, even in the absence of symptomatic disease.5,12
Machine learning offers opportunities to standardize the acquisition and interpretation of medical images.13 Deep learning algorithms have successfully been applied in echocardiograms, where they have shown promise in detecting left ventricular dysfunction,14 and left ventricular hypertrophy.15 With the expanded use of point-of-care ultrasonography,11 developing user-friendly screening algorithms relying on single 2D echocardiographic views would provide an opportunity to improve AS screening by operators with minimal experience through time-efficient protocols. This is often limited by the lack of carefully curated, labeled datasets, as well as efficient ways to utilize the often noisy real-world data for model development.16,17
In the present study, we hypothesized that a deep learning model trained on 2D echocardiographic views of parasternal long-axis (PLAX) videos can reliably predict the presence of severe AS without requiring Doppler input. The approach leverages self-supervised learning (SSL) of PLAX videos along with two other neural network initialization methods to form a diverse ensemble model capable of identifying severe AS from raw 2D echocardiograms. The model is trained based on a dataset from different operators and machines, with its external performance assessed both in geographically and temporally distinct cohorts. Combined with automated view classification, our approach serves as an end-to-end automated solution for deep learning applications in the field of point-of-care echocardiography.
Methods
Study population and data source
New England cohort (Yale-New Haven Health network)
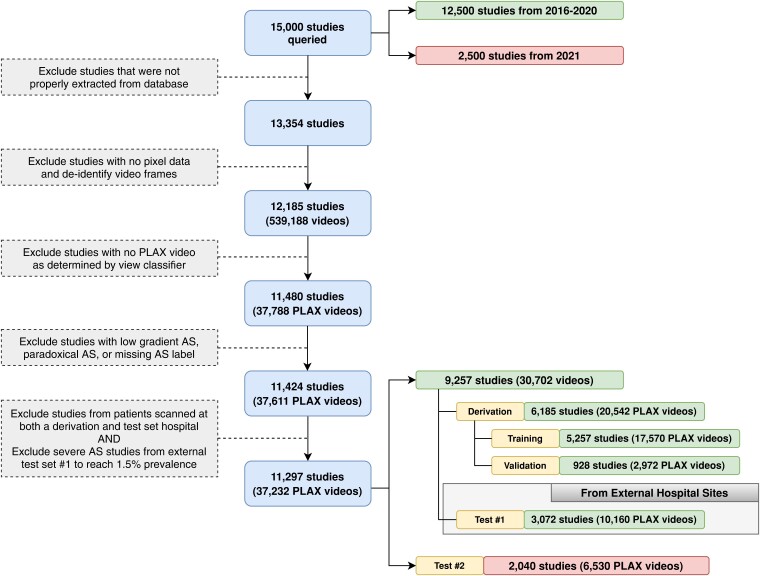
A total of 15 000 studies were queried from all transthoracic echocardiography (TTE) exams performed between 2016 and 2021 across the Yale New Haven Health System (YNHHS, including Connecticut and Rhode Island), and were used for model derivation and testing across different hospitals and time periods. For model development and evaluation across New England sites, 12 500 studies from 2016 to 2020 were randomly queried with AS oversampled to mitigate class imbalance during model training. Specifically, this query sampled normal studies uniformly (including ‘no AS’ and ‘sclerosis without stenosis’), oversampled non-severe AS studies by 5-fold (including ‘mild AS’, ‘mild-moderate AS’, and ‘moderate-severe AS’), and oversampled severe AS by 50-fold. This strategy was designed to ensure that the model encounters sufficient examples of severe AS to learn the signatures of the disorder; however, all test cohorts were ensured to have severe AS prevalence in the range of 1%–1.5%, mirroring the expected prevalence in a general screening population.18 The 12 500 studies were then split at the patient level into a derivation set (consisting of all patients scanned in the Yale-New Haven Hospital (YNHH), Connecticut, USA, including satellite locations) and a geographically distinct, external testing set from New England (consisting of patients scanned at four other hospitals—namely Bridgeport Hospital, Lawrence & Memorial Hospital, and Greenwich Hospital, all in Connecticut, USA—as well as the Westerly hospital in Rhode Island, USA). The remaining 2500 studies of the query were all conducted across the previously mentioned centers a year later in 2021, with no oversampling to serve as a challenging temporally distinct testing set, where severe AS represents ∼1% of all cases. The study population is summarized in Figure 1.
Figure 1.
Inclusion-exclusion flowchart for the New England study population. Exclusion criteria for transthoracic echocardiography (TTE) studies and videos included in this study from the Yale-New Haven Health network. Studies with valid pixel data were de-identified frame by frame, and the parasternal long axis (PLAX) view was determined by an automated view classifier. A sample of 12 500 studies from 2016 to 2020 were split into a derivation set and external test set, which comprised studies from hospital sites not encountered during model development. An independent random sample of 2500 studies from 2021 was used as an additional test set to evaluate robustness to temporal shift
All studies underwent de-identification, view classification, and preprocessing to curate a dataset of PLAX videos for deep-learned severe AS prediction. The full process describing the extraction of the echocardiographic videos, loading of image data, masking of identifying information, conversion to Audio Video Interleave format, and downsampling for further processing and automated view classification is described in the Supplement. After excluding studies that were not properly extracted or contained no pixel data, 12 185 studies with 539 188 videos underwent automated view classification based on a pretrained TTE view classifier.19 We retained videos where the automated view classifier most confidently predicted the presence of a PLAX view. We then excluded cases of low-flow, low-gradient, and paradoxical AS (determined based on the final clinical report) and excluded severe AS cases such that our geographically distinct test cohort reached 1.5% prevalence, in accordance with estimated prevalence in individuals aged 55 years and older.18 After these steps, the final YNHHS dataset consisted of 37 232 videos in 11 297 TTE studies, with 6185 studies from YNHH and satellite centers (with AS oversampled as described above) forming the derivation set. Another 3072 studies from 2016 to 2020 at hospital sites not found in the derivation set formed a geographically distinct testing set, and 2040 studies from 2021 formed a temporally distinct testing set (see Study population & data source, Figure 1).
Cedars-Sinai cohort
For further testing in an additional geographically distinct cohort, all transthoracic echocardiograms performed at the Cedars-Sinai Medical Center (Los Angeles, California, USA) between 1 January 2018 and 31 December 2019 were retrieved. AS severity was determined from finalized TTE reports. After excluding studies with prosthetic aortic valves, 4000 TTEs were sampled at random and combined with 1572 TTEs from this period (not part of the random sample) all with severe AS to create a 5572 study cohort that was enriched for AS. For consistency with the geographically distinct New England 2016–20 and temporally distinct New England 2021 cohort, where the severe AS prevalence of 1%–1.5% mirrors that of a general screening population,18 we downsampled the Cedars-Sinai cohort to a collection of 4226 studies with 1.5% prevalence by removing severe AS studies. To avoid bias, each patient with severe AS contributes no more than one study to this downsampled cohort.
Consent
The study was reviewed by the Yale and Cedars-Sinai Institutional Review Boards, which approved the study protocol and waived the need for informed consent as the study represents secondary analysis of existing data (Yale IRB ID #2000029973).
Echocardiogram interpretation
All studies were performed by trained echocardiographers or cardiologists and reported by board-certified cardiologists with specific training cardiac echocardiography. These reports were a part of routine clinical care, in accordance with the recommendations of the American Society of Echocardiography.20,21 The presence of AS severity was adjudicated based on the original echocardiographic report. Further details on the measurements obtained are presented in the Supplement.
Model training and development
Self-supervised learning
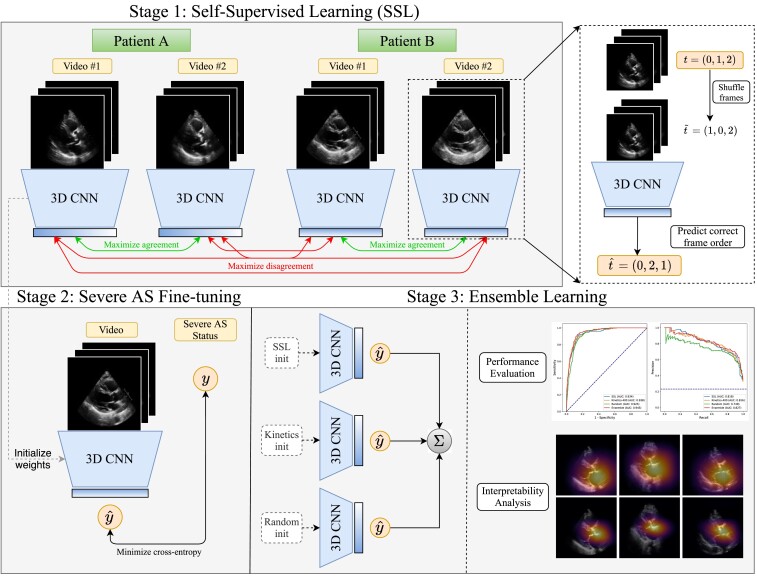
We used our previously described novel approach of self-supervised contrastive pretraining for echocardiogram videos.22 This approach demonstrated that classification tasks could be performed in a more data-efficient manner through ‘in-domain’ pretraining on echocardiograms,22 as opposed to other standard approaches such as random initialization of weights and transfer learning.14,23,24 Briefly, this SSL was performed on the training set videos with a novel combination of: (i) a multi-instance contrastive learning task and (ii) a frame re-ordering pretext task, both explained in detail in the Supplementary data online and summarized in Figure 2. For this, we adopted ‘multi-instance’ contrastive learning, where the model was trained to learn similar representations of different videos from the same patient, which allowed the model to learn the latent space of PLAX-view echocardiographic videos. To additionally encourage the temporal coherence of our model, we included a frame re-ordering ‘pretext’ task to our SSL method, where we randomly permuted the frames of each input echo, then trained the model to predict the original order of frames.25
Figure 2.
Overview of the proposed approach. We first perform self-supervised pretraining on parasternal long axis (PLAX) echocardiogram videos, selecting different PLAX videos from the same patient as ‘positive samples’ for contrastive learning. After this representation learning step, we then use these learned weights as the initialization for a model that is fine-tuned to predict severe aortic stenosis (AS) in a supervised fashion
Deep neural network training for severe AS prediction
The 3D-ResNet1826 architecture described above was also leveraged to detect severe AS. Three different methods were used to initialize the parameters of this network: an SSL initialization, a Kinetics-400 initialization, and a random initialization (see Figure 2 and Supplementary data online). All fine-tuning models were trained on randomly sampled video clips of 16 consecutive frames from training set echocardiograms. Models were trained for a maximum of 30 epochs with early stopping if the validation area under the receiver operating characteristic curve (AUROC) did not improve for five consecutive epochs. Severe AS models were trained on a single NVIDIA RTX 3090 GPU with the Adam optimizer, a learning rate of (except the SSL-pretrained model, which used a learning rate of 0.1) and a batch size of 88 using a sigmoid cross-entropy loss. We additionally used class weights computed with the method provided by scikit-learn27 to accommodate class imbalance in addition to label smoothing28 with α = .1. Learning curves depicting loss throughout training can be found in Supplementary data online, Figure S1.
Ensemble learning
Since models were trained on 16-frame video clips, we averaged clip-level predictions to obtain video-level predictions of severe AS at inference time. To form study-level predictions, we averaged predicted severe AS probabilities from all videos acquired during the same study. The final ensemble model was then formed by averaging the study-level output probabilities from the SSL-pretrained model, the Kinetics-400-pretrained model, and the randomly initialized model after fine-tuning each ensemble member to detect severe AS. Since no quality control is applied when selecting PLAX videos for this work, averaging results over multiple videos in the same study has a stabilizing effect that boosts predictive performance.29
Assessing diagnostic performance in the testing sets
We evaluate the model’s performance on both AUROC and the area under the precision-recall curve, with the latter being specifically informative when class imbalance is present.30 We additionally reported metrics that assess performance at specific decision thresholds such as F1 score, positive predictive value (PPV), and negative predictive value (NPV). For these metrics, we proceed with a fixed decision threshold of 0.607, which was selected to maximize F1 score in the validation set of the derivation cohort.
Model explainability
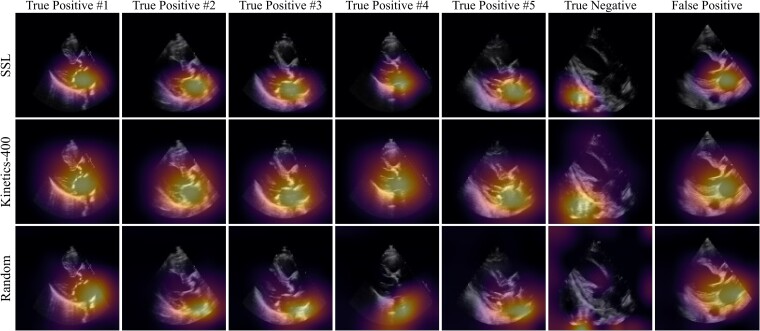
We evaluated the predictive focus of the models using saliency maps. These were generated using the Grad-CAM method31 for obtaining visual explanations from deep neural networks (see Supplementary data online). This method was used to produce a frame-by-frame ‘visual explanation’ of where the model was focusing to make its prediction. To generate a single 2D heatmap for a given echo clip, the pixelwise maximum along the temporal axis was taken to capture the most salient regions for severe AS predictions across all time points. These spatial attention maps were visualized based on the outputs of each ensemble member (the randomly initialized, Kinetics-400-pretrained, and SSL-pretrained AS models) for five true positive examples, a true negative (TN), and a false positive (FP). We used the ‘inferno’ colormap (https://matplotlib.org/stable/tutorials/colors/colormaps.html) for visualization, where pixels closer to bright yellow are highly salient for the model prediction and pixels closer to dark purple or black are negligible.
Statistical analysis
All 95% confidence intervals for model performance metrics were computed by bootstrapping. Specifically, 10 000 stratified bootstrap samples (samples with replacement having positive- and negative-label sample sizes equal to those of the original evaluation set) of the test cohort were drawn, metrics were computed on this set of studies, and nonparametric confidence intervals were constructed with the percentile method.32 Bootstrapping was performed at the study level since the severe AS labels are provided for each echocardiographic study. To assess the calibration of the model at a 1% prevalence of severe AS, a logistic regression model was fitted in a down-sampled version of the New England training set. Calibration was assessed by the Brier score (the average squared distances between the actual and predicted probability), as well as the calibration slope and intercept. The logistic model was then applied in the remaining New England and California testing sets, where the same calibration metrics are reported.33 For analysis of the correlation between model outputs and quantitative measures of AS, categorical variables were summarized as percentages, whereas continuous variables are reported as mean values with standard deviation and visualized using violin plots. Continuous variables between the two groups were compared using the Student’s t-test. Pearson’s r was used to assess the pairwise correlation between continuous variables. Spearman’s rank-order correlation test was used to analyze the relationship between model outputs and AS severity, which was represented ordinally with increasing severity (e.g. 0 = none, 1 = mild-moderate, 2 = severe). The independent association of the model output with various echocardiographic indices of AS severity and diastolic function was assessed using multivariable linear regression modelling. All statistical tests were two-sided with a significance level of 0.05, unless specified otherwise. Analyses were performed using Python (version 3.8.5) and R (version 4.2.3). Reporting of the study methods and results stands consistent with the CONSORT-AI (Consolidated Standards of Reporting Trials–Artificial Intelligence)34 and CODE-EHR (electronic health record) guidelines.35
Results
Study population
In the New England cohort, after removing studies with no pixel data, de-identifying video frames, and using an automated view classifier to determine the PLAX view, our final derivation set consisted of 6185 studies with 20 542 videos (1 294 197 frames) [mean age 70 ± 16 years, n = 2992 (48.4%) women], with mild, moderate, and severe AS in 12.6% (n = 780), 8.0% (n = 495), and 23.1% (n = 1427) of studies, respectively. To evaluate generalization across local hospital sites, we curated a test set of 3072 studies (10 160 videos) from separate New England hospitals in the YNHHS network that were not present in the derivation set, with a prevalence of mild, moderate, and severe AS in 196 (6.4%), 130 (4.2%), and 47 (1.5%) of studies, respectively. The temporally distinct test set consisted of 2040 randomly selected scans with a total of 6530 videos performed between 1 January 2021 and 15 December 2021 across YNHHS (mean age 66 ± 16 years, n = 997 (48.9%) women). These were used for time-dependent model validation, with mild, moderate, and severe AS estimated in 4.1% (n = 83), 2.9% (n = 59), and 1.0% (n = 20) of the studies, respectively. Finally, a set of 4226 studies performed at the Cedars-Sinai Medical Center between 2018 and 2019 (65 studies with severe AS out of 4226, 1.5%) [mean age 69 ± 17 years, n = 1852 (43.8%) women], was also used for further external testing (Figure 1). Further information on patient characteristics is presented in the Methods and Table 1.
Table 1.
Table of baseline demographic and echocardiographic characteristics
| New England (Yale-New Haven Health) | California | ||||
|---|---|---|---|---|---|
| Derivation | Geographically distinct testing #1 | Temporally distinct testing | Geographically distinct testing #2 | ||
| Number of patients | 5749 | 3069 | 2034 | 3923 | |
| Number of echo studies | 6185 | 3072 | 2040 | 4226 | |
| Number of studies per patient, n (%) | 1 | 5367 (93.3) | 3066 (99.9) | 2028 (99.7) | 3677 (93.7) |
| 2 | 336 (5.8) | 3 (0.1) | 6 (0.3) | 201 (5.1) | |
| 3 or more | 46 (0.8) | 0 (0.0) | 0 (0.0) | 45 (1.1) | |
| Study location | YNHH | BH, GH, LMH, WH | YNHH, BH, GH, LMH, WH | CSMC | |
| Year of study | 2016–2020 | 2016–2020 | 2021 | 2018–2019 | |
| Age (years), mean (SD) | 69.9 (15.7) | 66.7 (16.6) | 65.7 (16.4) | 65.2 (17.3) | |
| Gender, n (% a ) | Female | 2992 (48.4) | 1581 (51.5) | 997 (48.9) | 1852 (43.8) |
| Male | 3194 (51.6) | 1491 (48.5) | 1043 (51.1) | 2374 (56.2) | |
| Race & Ethnicity, n (% a ) | Asian | 67 (1.2) | 45 (1.7) | 40 (2.2) | 329 (7.8) |
| Black | 503 (9.0) | 381 (14.3) | 200 (11.1) | 600 (14.2) | |
| Hispanic | 364 (6.5) | 347 (13.0) | 158 (8.8) | 479 (11.3) | |
| Other | 249 (4.4) | 317 (11.9) | 133 (7.4) | 308 (7.3) | |
| Unknown | 221 (3.9) | 23 (0.9) | 63 (3.5) | 111 (2.6) | |
| White | 4575 (81.5) | 1904 (71.3) | 1359 (75.7) | 2399 (56.8) | |
| LVIDd Index (cm/m2), mean (SD) | 2.4 (0.4) | 2.4 (0.4) | 2.4 (0.4) | 2.4 (0.6) | |
| RVSP (mmHg), mean (SD) | 32.5 (13.3) | 32.2 (14.4) | 29.8 (12.0) | 31.6 (14.2) | |
| EF (%), mean (SD) | 59.5 (10.8) | 59.3 (11.4) | 59.1 (10.2) | 57.5 (14.9) | |
| AVA by VTI (cm2), mean (SD) | 1.3 (0.8) | 2.0 (0.9) | 2.1 (0.9) | 1.9 (1.4) | |
| AV mean gradient (mmHg), mean (SD) | 23.2 (18.2) | 8.7 (9.3) | 9.0 (9.4) | 10.9 (11.4) | |
| AV peak velocity (m/s), mean (SD) | 2.4 (1.3) | 1.6 (0.7) | 1.6 (0.6) | 1.6 (0.9) | |
| Patients with severe AS, n (%) | 1213 (21.1) | 47 (1.5) | 20 (1.0) | 65 (1.5) | |
| Studies with severe AS, n (%) | 1427 (23.1) | 47 (1.5) | 20 (1.0) | 65 (1.5) | |
Since splits were made at the study level, distinct studies from n = 9 patients contributed to both the ‘Geographically distinct testing #1’ and ‘Temporally distinct testing set’ cohorts. Similarly, distinct studies from n = 53 patients were including in both the derivation set and the ‘Temporally distinct testing set.’
AV, aortic valve; BH, Bridgeport Hospital; CSMC, Cedars-Sinai Medical Center; EF, ejection fraction; GH, Greenwich Hospital; LAD, left atrium; LMH, Lawrence & Memorial Hospital; LVIDd, left ventricular internal diastolic diameter; RVSP, right ventricular systolic pressure; SD, standard deviation; VTI, velocity time integral; WH, Westerly Hospital; YNHH, Yale-New Haven Hospital.
aPercentages represent valid percentages after excluding cases with missing information.
Performance of a deep learning model for severe AS detection based on PLAX videos
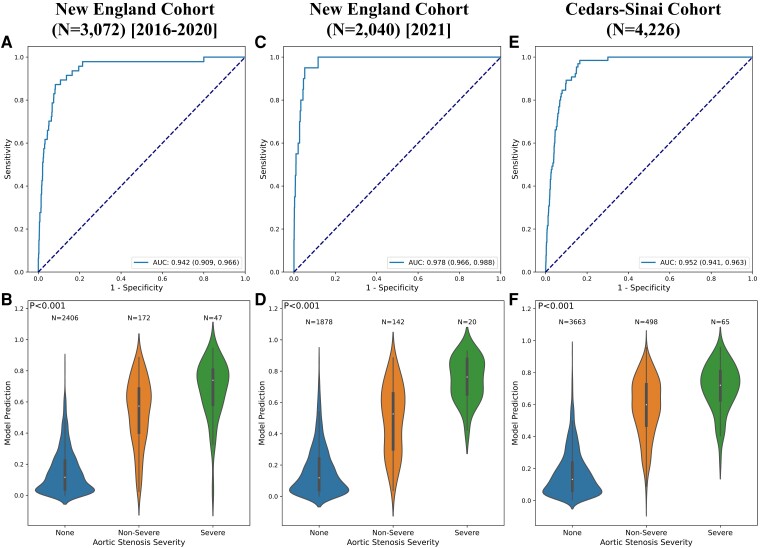
The ensemble model was able to reliably detect the presence of severe AS using single-view, 2D PLAX videos, demonstrating an AUROC of 0.942 (95% CI: 0.909, 0.966) on the geographically distinct testing set of New England hospitals not included in the derivation set. The model also demonstrated consistent performance across time in the same hospital system, maintaining its discriminatory performance with an AUROC of 0.978 (95% CI: 0.966, 0.988) on the temporally distinct testing set from 2021. Finally, in further geographically distinct testing using scans performed at Cedars-Sinai, the model generalized well across institutions, reaching an AUROC of 0.952 (95% CI: 0.941, 0.963). Receiver operating characteristic (ROC) curves and the distribution of model probabilities across disease groups (no AS, mild-moderate AS, severe AS), showing a graded relationship across severity groups, are shown in Figure 3; see Supplementary data online, Table S1 for full detailed results. Furthermore, in sensitivity analyses without averaging predictions from multiple videos in the same study we observed overall consistent results, as summarized in Supplementary data online, Tables S2 and S3. Additional analysis demonstrated that our model continued to exhibit high performance in identifying cases where one or more quantitative measures indicated severe AS, regardless of the reading cardiologist’s interpretation (see Supplementary data online, Table S4).
Figure 3.
Model performance in the external validation sets. Receiver operating characteristic curves (first row) and violin plots showing relationship of model output with aortic stenosis severity (second row) for the external New England cohort (first column), temporally distinct New England cohort (second column), and external Cedars-Sinai cohort (third column)
Due to the extremely low prevalence of severe AS in the external test cohorts, our model’s PPV remained consistent at 0.159 [95% CI: (0.133, 0.188)] in the geographically distinct New England cohort (1.5% prevalence) and 0.159 [95% CI: (0.130, 0.192)] in the temporally distinct New England cohort (<1% prevalence). In auxiliary experiments downsampling the Cedars-Sinai cohort to various levels of severe AS prevalence, PPV varied from 0.155 [95% CI: (0.137, 0.173)] at 1.5% prevalence to 0.556 [95% CI: (0.530, 0.582)] at 10% prevalence, and up to 0.796 [95% CI: (0.780, 0.812)] at the full 25.3% prevalence before downsampling (see Supplementary data online, Table S5).
When fitted in the training set downsampled to a severe AS prevalence of 1.5% (n = 61 cases), the model demonstrated good calibration performance across all levels of risk in both the training (Brier score of 0.012) as well as the three distinct testing sets (Brier scores ranging between 0.008 and 0.013) (see Supplementary data online, Figure S2).
Explainable predictions through saliency maps
We used Gradient-weighted Class Activation Mapping (Grad-CAM) to identify the regions in each video frame that contributed the most to the predicted label. In the examples shown in Figure 4, the first five columns represent the five most confident severe AS predictions, the sixth column represents the most confident ‘normal’ (no severe AS) prediction and the seventh column represents the most confident incorrect severe AS prediction. The saliency maps from our SSL approach demonstrated consistent and specific localization of the activation signal in the pixels corresponding to the aortic valve and annulus (bottom row). For frame-by-frame saliency visualizations for each ensemble member, see Supplementary data online, Videos S1–S5 for each true positive, Supplementary data online, Video S6 for the true negative, and Supplementary data online, Video S7 for the false positive (left = randomly initialized model, middle = Kinetics-400-initialized model, right = SSL-initialized model).
Figure 4.
Saliency map visualization. Spatial attention maps for the self-supervised learning (SSL)-pretrained model (top row), Kinetics-pretrained model (middle row), and randomly initialized model (bottom row) for five true positives (first five columns), a true negative (sixth column), and a false positive (last column). As determined by the Kinetics-pretrained model, the first five columns represent the five most confident severe AS predictions, the sixth column represents the most confident ‘normal’ (no severe AS) prediction, and the seventh column represents the most confident incorrect severe AS prediction. Saliency maps were computed with the GradCAM method and reduced to a single 2D heatmap by maximum intensity projection along the temporal axis
Model identification of features of AS severity
In the temporally distinct testing set from 2021 (reflecting the normal prevalence of severe AS in an echocardiographic cohort), we observed that the predictions of the ensemble model correlated with continuous metrics of AS severity, including the peak aortic valve velocity (r = 0.59, P < .001), trans-valvular mean gradient (r = 0.66, P < .001) and the mean aortic valve area (r = −0.53, P < .001). On the other hand, the model predictions were independent of the left ventricular ejection fraction (LVEF) (r = −0.02, P = .37), a negative control. Of note, a higher model-derived probability of severe AS was associated with higher average E/e’ values [ratio of early diastolic mitral inflow velocity to early diastolic mitral annulus velocity; coef. 0.60 (95%CI: 0.37–0.83), P < 0.001], greater indexed left atrial volumes [mL/m2; coef. 0.09 (0.02–0.17), P = 0.016], and higher maximal tricuspid regurgitation velocities [TRVmax, m/sec; coef. 0.15 (0.05–0.25), P = .003], independent of the peak aortic valve velocity measured in each study (see Supplementary data online, Table S6).
In further sensitivity analysis, we stratified cases without AS or mild/moderate AS based on the predictions of our model as TNs or FPs. Compared to TNs, FP cases had significantly higher peak aortic velocities [FP: 3.4 (25th–75th percentile: 2.9–3.7) m/sec; TN: 1.6 (1.3–2.3) m/sec, P < .001], trans-valvular mean gradients [FP: 26.0 (25th–75th percentile: 20.5–31.8) mmHg; TN: 5.0 (3.8–9.0) m/sec, P < .001], and mean aortic valve area [FP: 1.04 (25th–75th percentile: 0.86–1.28) cm2; TN: 1.99 (1.49–2.67) cm2, P < .001], but no significant difference in the LVEF [FP: 65.4% (55.0%–67.8%); TN: 60.0% (55.0%–65.0%), P = .19] (Figure 5).
Figure 5.
Comparison between model predictions and echocardiographic left ventricular and aortic valve assessment among patients without severe aortic stenosis. Violin plots demonstrating the distribution of LVEF (left ventricular ejection fraction, (A) peak aortic valve velocity (B), mean aortic valve gradient (C) and mean aortic valve area (D) for patients without severe AS, stratified based on the predicted class based on the final ensemble model. These results are based on the temporally distinct cohort of patients scanned in 2021, without oversampling for severe aortic stenosis cases
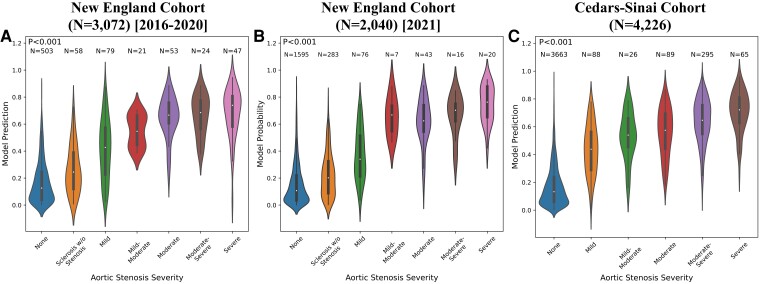
Further, we observed that predicted model probabilities significantly correlated with fine-grained cardiologist-determined AS severity (P < .001 for the New England 2016–20, New England 2021, and Cedars-Sinai cohorts independently). Though the model was only trained to discriminate severe AS from all other designations, its predicted probabilities identify a gradient of AS severity in aggregate. For example, in the Cedars-Sinai test cohort, our model predicted a mean ± standard deviation severe AS probability of 0.171 ± 0.143 for normal studies (N = 3663), 0.436 ± 0.180 for mild AS (N = 88), 0.531 ± 0.168 for mild-moderate AS (N = 26), 0.560 ± 0.173 for moderate AS (N = 89), 0.638 ± 0.163 for moderate-severe AS (N = 295), and 0.708 ± 0.145 (N = 65) for severe AS (Figure 6).
Figure 6.
Correlation of model probabilities with fine-grained AS severity. Violin plots depicting the distribution of predicted model probabilities stratified by cardiologist-determined AS severity for the New England 2016–2020 (A), New England 2021 (B), and Cedars-Sinai (C) cohorts
Model generalization to paradoxical low-flow, low-gradient AS
Though paradoxical low-flow, low-gradient AS cases were removed from our cohorts for training and evaluation purposes, a post-hoc analysis revealed that our model was able to discriminate low-flow, low-gradient AS from normal cases. On the N = 44 low-flow, low-gradient studies originally excluded from the New England cohorts (2016–21 pooled together), our model produced a mean ± standard deviation predicted severe AS probability of 0.697 ± 0.135, compared with 0.157 ± 0.153 for normal studies (see Supplementary data online, Figure S3). In fact, if interpreting low-flow, low-gradient cases as severe AS, our model achieves a PPV of 1.000, sensitivity of 0.727, and an F1 score of 0.842.
Discussion
We have developed and validated an automated algorithm that can efficiently screen for and detect the presence of severe AS based on a single-view 2D TTE video. The algorithm demonstrates excellent performance (AUROCs ranging from 0.92 to 0.98), with high sensitivity (85%) at high specificity (96%), maintaining its robustness and discriminatory performance across several geographically and temporally distinct cohorts with varying prevalence of severe AS (Structured Graphical Abstract). We also leverage a novel self-supervised step leveraging multi-instance contrastive learning, which allowed our algorithm to learn key representations that define each patient’s unique phenotype through contrastive pre-training, independent of the expected technical variation in image acquisition, including differences in probe orientation, beam angulation and depth. Visualization of saliency maps introduces explainability to our algorithms and confirms the key areas of the PLAX view, including the aortic valve, mitral annulus, and left atrium, that contributed the most to our predictions. Furthermore, features learned by the model generalize to lower severity cases as well as cases of low-flow, low-gradient AS, highlighting the potential value of our model in the longitudinal monitoring of AS, a disease with a well-defined, progressive course.30 Our approach has the potential to expand the use of echocardiographic screening for suspected AS, shifting the burden away from dedicated echocardiographic laboratories to point-of-care screening in primary care offices or low-resource settings. It may also enable operators with minimal echocardiographic experience to screen for the condition by obtaining simple two-dimension PLAX views without the need for comprehensive Doppler assessment, which can then be reserved for confirmatory assessment. Given the low prevalence of severe AS in a general screening population, however, our model would likely be best suited for ruling out severe AS, given its >99% NPV.
In recent years, several artificial intelligence applications have been described in the field of echocardiography,36 ranging from automated classification of echocardiographic views,37 video-based beat-to-beat assessment of left ventricular systolic dysfunction,14 detection of left ventricular hypertrophy and its various subtypes,15 diastolic dysfunction,38 to expert-level prenatal detection of complex congenital heart disease.39 Of note, machine learning methods further enable individuals without prior ultrasonography experience to obtain diagnostic TTE studies for limited diagnostic use.40 Despite this and even though the diagnosis and grading of AS remains dependent on echocardiography,2,20 most artificial intelligence solutions for timely AS screening have focused on alternative data types, such as audio files of cardiac auscultation,41 12-lead electrocardiograms,42–44 cardio-mechanical signals using non-invasive wearable inertial sensors,45 as well as chest radiographs.46 For 12-lead electrocardiograms, AUROCs were consistently <0.90,42–44 whereas for alternative data types, analyses were limited to small datasets without external validation.41,45 Other studies have explored the value of structured data derived from comprehensive TTE studies in defining phenotypes with varying disease trajectories.47 More recently, the focus has shifted to AI-assisted AS detection through automated TTE interpretation. In a recent study, investigators employed a form of SSL to automate the detection of AS, with their method, however, discarding temporal information by only including the first frame of each video loop while also relying on the acquisition of images from several different views.48 The approach that relies on ultrasonography is also safer than the alternative screening strategies, such as those using chest computed tomography and aortic valve calcium scoring,47,49 which expose patients to radiation.
In this context, our work represents an advance both in the clinical and methodological space. First, we describe a method that can efficiently screen for a condition associated with significant morbidity and mortality,44 with increasing prevalence in the setting of an aging population.50 Our method can potentially shift the initial burden away from trained echocardiographers and specialized core laboratories as part of a more cost-effective screening and diagnostic cascade that can detect the condition at its earliest stages, particularly by ruling out the presence of severe AS thanks to its high NPV.11,40 Furthermore, it provides an additional layer of information in any setting where point-of-care ultrasound is used, where more detailed echocardiographic assessment may fall outside the scope of the original study indication or may be limited by the available equipment, time, or skills of the provider. In this regard, major strengths of our model include its reliance on a single echocardiographic view that can be obtained by individuals with limited experience and focused training,40 and its ability to process temporal information through analysis of videos rather than isolated frames. By adjusting the optimal threshold for a positive screen depending on the specific patient population and clinical setting, potential applications of the method range from the screening of asymptomatic individuals in the community, to the rapid assessment of patients in emergency settings, as well as the screening of lower severity or low-flow low-gradient cases where Doppler-based measurements may result in misclassification of disease severity. The importance of this is further supported by expanding evidence regarding the possible benefits of timely intervention in AS across asymptomatic,5 low-risk severe,51 or even moderate severity cases.52 The overarching goal is to develop screening tools that can be deployed cost-effectively, gatekeeping access to comprehensive TTE assessment, which can be used as a confirmatory test to establish the suspected diagnosis. The current model further offers the ability to retrospectively interrogate databases for potentially missed disease and/or prospectively guide the need for aortic valve interrogation with Doppler in limited studies obtained for alternative indications.
Second, our work describes an end-to-end framework to boost artificial intelligence applications in echocardiography. We present an algorithm that automatically detects echocardiographic views, then performs self-supervised representation learning of PLAX videos with a multi-instance, contrastive learning approach. This novel approach further enables our algorithm to learn key representations of a patient’s cardiac phenotype that generalize and remain consistent across different clips and variations of the same echocardiographic views. By optimizing the detection of an echocardiographic fingerprint for each patient, this important pretraining step has the potential to boost AI-based echocardiographic assessment across a range of conditions. Furthermore, unlike previous approaches,48 our method benefits from multi-instance contrastive learning, which learns key representations using different videos from the same patient, a method that has been shown to improve predictive performance in the classification of dermatology images.53
Further to detecting severe AS, our algorithm learns features of aortic valvular pathology that generalize across different stages of the condition, including various severity stages as well as the diagnostically challenging low-flow, low-gradient AS phenotype.54 When restricting our analysis to patients without severe AS, the model’s predictions strongly correlated with Doppler-derived, quantitative features of stenosis severity. This is in accordance with the known natural history of AS, a progressive, degenerative condition, the hallmarks of which are aortic valve calcification, restricted mobility, functional stenosis and eventual ventricular decompensation.18 As such, our algorithm’s predictions also carry significant value as quantitative predictors of the stage of AV severity and could theoretically be used to monitor the rate of AS progression.
On this note, saliency maps demonstrate that the model focuses on the aortic valve, possibly learning features such as aortic valve calcification and restricted leaflet mobility,20 as well as the mitral annulus and left atrium. These findings are further supported by a cross-sectional correlation of the model output with echocardiographic markers of elevated filling pressures, left atrial dilation, and elevated pulmonary pressures.55 These observations align with the known hemodynamic effects of worsening AS, characterized by worsening diastolic dysfunction, often defined based on changes in mitral annular tissue velocities and left atrial structure.56,57 Notably, prior studies suggest that among patients with AS and normal LVEF, annular tissue Doppler and echocardiographic markers of left atrial mechanics may better reflect the hemodynamic consequences of an increasing afterload burden on the left ventricle than traditional echocardiographic markers of AS severity.58,59
Limitations of our study include the lack of prospective validation of our findings. To this end, we are working on deploying this method in prospective cohorts of patients referred for routine TTE assessment to understand its real-world implications as a screening tool. Second, our model is limited to using PLAX views, which often represent the first step of TTE or point-of-care ultrasound protocols in cardiovascular assessment. Though there is no technical restriction to expanding these methods to alternative views, increasing the complexity of the screening protocol is likely to negatively impact its adoption in busy clinical settings. Future work will incorporate multiple TTE views into the proposed AS detection framework. Finally, this study used data from formal TTEs, which generally produce higher-quality images than machines in point-of-care ultrasound settings. Though videos were downsampled for model development, further validation is needed to ensure robustness across acquisition technologies.
Conclusion
In summary, we propose an efficient method to screen for severe AS using single-view (PLAX) TTE videos without the need for Doppler signals. More importantly, we describe an end-to-end approach for deploying artificial intelligence solutions in echocardiography, starting from automated view classification to self-supervised representation learning to accurate and explainable detection of severe AS. Our findings have significant implications for point-of-care ultrasound screening of AS as part of routine clinic visits and in limited resource settings and for individuals with minimal training.
Supplementary data
Supplementary data are available at European Heart Journal online.
Supplementary Material
Contributor Information
Gregory Holste, Department of Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, USA; Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
Evangelos K Oikonomou, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
Bobak J Mortazavi, Department of Computer Science & Engineering, Texas A&M University, College Station, TX, USA; Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, 195 C hurch St 5th Floor, New Haven, CT, USA.
Andreas Coppi, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA; Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, 195 C hurch St 5th Floor, New Haven, CT, USA.
Kamil F Faridi, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
Edward J Miller, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
John K Forrest, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
Robert L McNamara, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA.
Lucila Ohno-Machado, Section of Biomedical Informatics and Data Science, Yale School of Medicine, New Haven, CT, USA.
Neal Yuan, Department of Medicine, University of California San Francisco, San Francisco, CA, USA; Division of Cardiology, San Francisco Veterans Affairs Medical Center, San Francisco, CA, USA.
Aakriti Gupta, Department of Cardiology, Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
David Ouyang, Department of Cardiology, Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Division of Artificial Intelligence in Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Harlan M Krumholz, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA; Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, 195 C hurch St 5th Floor, New Haven, CT, USA; Department of Health Policy and Management, Yale School of Public Health, New Haven, CT, USA.
Zhangyang Wang, Department of Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, USA.
Rohan Khera, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520-8056, USA; Center for Outcomes Research and Evaluation, Yale-New Haven Hospital, 195 C hurch St 5th Floor, New Haven, CT, USA; Section of Biomedical Informatics and Data Science, Yale School of Medicine, New Haven, CT, USA; Section of Health Informatics, Department of Biostatistics, Yale School of Public Health, 60 College St, New Haven, CT, USA.
Declarations
Disclosure of Interest
E.K.O. is a co-inventor of the U.S. Provisional Patent Application 63/177,117, a co-founder of Evidence2Health, and reports a consultancy and stock option agreement with Caristo Diagnostics Ltd (Oxford, U.K.), all unrelated to the current work. B.J.M. reported receiving grants from the National Institute of Biomedical Imaging and Bioengineering, National Heart, Lung, and Blood Institute, US Food and Drug Administration, and the US Department of Defense Advanced Research Projects Agency outside the submitted work. In addition, B.J.M. has a pending patent on predictive models using electronic health records (US20180315507A1). A.H.R. is supported in part by CNPq (310679/2016-8 and 465518/2014-1) and FAPEMIG (PPM-00428-17 and RED-00081-16) and is funded by Kjell och Märta Beijer Foundation. J.K.F. has received grant support/research contracts and consultant fees/honoraria/Speakers Bureau fees from Edwards Lifesciences and Medtronic. H.M.K. works under contract with the Centers for Medicare & Medicaid Services to support quality measurement programs, was a recipient of a research grant from Johnson & Johnson, through Yale University, to support clinical trial data sharing; was a recipient of a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; receives payment from the Arnold & Porter Law Firm for work related to the Sanofi clopidogrel litigation, from the Martin Baughman Law Firm for work related to the Cook Celect IVC filter litigation, and from the Siegfried and Jensen Law Firm for work related to Vioxx litigation; chairs a Cardiac Scientific Advisory Board for UnitedHealth; was a member of the IBM Watson Health Life Sciences Board; is a member of the Advisory Board for Element Science, the Advisory Board for Facebook, and the Physician Advisory Board for Aetna; and is the co-founder of Hugo Health, a personal health information platform, and co-founder of Refactor Health, a healthcare AI-augmented data management company. D.O. is supported by NIH K99 HL157421-01 and has provisional patents in AI and echocardiography. R.K. is an Associate Editor at JAMA. He receives support from the National Heart, Lung, and Blood Institute of the National Institutes of Health (under award K23HL153775) and the Doris Duke Charitable Foundation (under award, 2022060). R.K. further receives research support, through Yale, from Bristol-Myers Squibb and Novo Nordisk. He is also a coinventor of U.S. Pending Patent Applications 63/177,117, 63/428,569, 63/346,610, and 63/484,426, unrelated to the current work. He is also a founder of Evidence2Health, a precision health platform to improve evidence-based cardiovascular care. The remaining authors have no competing interests to disclose.
Data Availability
The data are not available for public sharing, given the restrictions in our institutional review board approval. Deidentified test set data may be available to researchers under a data use agreement after the study has been published in a peer-reviewed journal.
Funding
The study was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (under award K23HL153775 to R.K.).
Ethical Approval
The study was reviewed by the Yale and Cedars-Sinai Institutional Review Boards, which approved the study protocol and waived the need for informed consent as the study represents secondary analysis of existing data (Yale IRB ID #2000029973).
Pre-registered Clinical Trial Number
Not applicable.
Code Availability
The code repository for this work can be found at https://github.com/CarDS-Yale/echo-severe-AS.
References
- 1. Marc E, Piotr D, Bernard P, Olaf W, Cécile L, Jean-Luc M, et al. Contemporary management of severe symptomatic aortic stenosis. J Am Coll Cardiol 2021;78:2131–43. 10.1016/j.jacc.2021.09.864 [DOI] [PubMed] [Google Scholar]
- 2. Otto CM, Prendergast B. Aortic-valve stenosis—from patients at risk to severe valve obstruction. N Engl J Med 2014;371:744–56. 10.1056/NEJMra1313875 [DOI] [PubMed] [Google Scholar]
- 3. Smith CR, Leon MB, Mack MJ, Miller DC, Moses JW, Svensson LG, et al. Transcatheter versus surgical aortic-valve replacement in high-risk patients. N Engl J Med 2011;364:2187–98. 10.1056/NEJMoa1103510 [DOI] [PubMed] [Google Scholar]
- 4. Reardon MJ, Van Mieghem NM, Popma JJ, Kleiman NS, Søndergaard L, Mumtaz M, et al. Surgical or transcatheter aortic-valve replacement in intermediate-risk patients. N Engl J Med 2017;376:1321–31. 10.1056/NEJMoa1700456 [DOI] [PubMed] [Google Scholar]
- 5. Kang D-H, Park S-J, Lee S-A, Lee S, Kim D-H, Kim H-K, et al. Early surgery or conservative care for asymptomatic aortic stenosis. N Engl J Med 2020;382:111–9. 10.1056/NEJMoa1912846 [DOI] [PubMed] [Google Scholar]
- 6. The Early Valve Replacement in Severe Asymptomatic Aortic Stenosis Study . https://clinicaltrials.gov/ct2/show/NCT04204915 (June 2, 2022)
- 7. Otto CM, Nishimura RA, Bonow RO, Carabello BA, Erwin JP III, Gentile F, et al. 2020 ACC/AHA guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2021;143:e72–e227. 10.1161/CIR.0000000000000932 [DOI] [PubMed] [Google Scholar]
- 8. Baumgartner H, Falk V, Bax JJ, De Bonis M, Hamm C, Holm PJ, et al. 2017 ESC/EACTS guidelines for the management of valvular heart disease. Eur Heart J 2017;38:2739–91. 10.1093/eurheartj/ehx391 [DOI] [PubMed] [Google Scholar]
- 9. Vahanian A, Beyersdorf F, Praz F, Milojevic M, Baldus S, Bauersachs J, et al. 2021 ESC/EACTS Guidelines for the management of valvular heart disease: developed by the Task Force for the management of valvular heart disease of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS). Eur Heart J 2022;43:561–632. 10.1093/eurheartj/ehab395 [DOI] [PubMed] [Google Scholar]
- 10. Siontis GCM, Overtchouk P, Cahill TJ, Modine T, Prendergast B, Praz F, et al. Transcatheter aortic valve implantation vs. surgical aortic valve replacement for treatment of symptomatic severe aortic stenosis: an updated meta-analysis. Eur Heart J 2019;40:3143–53. 10.1093/eurheartj/ehz275 [DOI] [PubMed] [Google Scholar]
- 11. Narula J, Chandrashekhar Y, Braunwald E. Time to add a fifth pillar to bedside physical examination: inspection, palpation, percussion, auscultation, and insonation. JAMA Cardiol 2018;3:346–50. 10.1001/jamacardio.2018.0001 [DOI] [PubMed] [Google Scholar]
- 12. Windecker S, Okuno T, Unbehaun A, Mack M, Kapadia S, Falk V. Which patients with aortic stenosis should be referred to surgery rather than transcatheter aortic valve implantation? Eur Heart J 2022;43:2729–50. 10.1093/eurheartj/ehac105 [DOI] [PubMed] [Google Scholar]
- 13. Dey D, Slomka PJ, Leeson P, Comaniciu D, Shrestha S, Sengupta PP, et al. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol 2019;73:1317–35. 10.1016/j.jacc.2018.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouyang D, He B, Ghorbani A, Yuan N, Ebinger J, Langlotz CP, et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020;580:252–6. 10.1038/s41586-020-2145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duffy G, Cheng PP, Yuan N, He B, Kwan AC, Shun-Shin MJ, et al. High-throughput precision phenotyping of left ventricular hypertrophy with cardiovascular deep learning. JAMA Cardiol 2022;7:386–95. 10.1001/jamacardio.2021.6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newgard CD, Lewis RJ. Missing data: how to best account for what is not known. JAMA 2015;314:940–1. 10.1001/jama.2015.10516 [DOI] [PubMed] [Google Scholar]
- 17. Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, et al. Preparing medical imaging data for machine learning. Radiology 2020;295:4–15. 10.1148/radiol.2020192224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strange GA, Stewart S, Curzen N, Ray S, Kendall S, Braidley P, et al. Uncovering the treatable burden of severe aortic stenosis in the UK. Open Heart 2022;9:e001783. 10.1136/openhrt-2021-001783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 2018;138:1623–35. 10.1161/CIRCULATIONAHA.118.034338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baumgartner H, Hung J, Bermejo J, Chambers JB, Evangelista A, Griffin BP, et al. Echocardiographic assessment of valve stenosis: EAE/ASE recommendations for clinical practice. J Am Soc Echocardiogr 2009; 22:1–23; quiz 101–102. 10.1016/j.echo.2008.11.029 [DOI] [PubMed] [Google Scholar]
- 21. Mitchell C, Rahko PS, Blauwet LA, Canaday B, Finstuen JA, Foster MC, et al. Guidelines for performing a comprehensive transthoracic echocardiographic examination in adults: recommendations from the American Society of Echocardiography. J Am Soc Echocardiogr 2019;32:1–64. 10.1016/j.echo.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 22. Holste G, Oikonomou EK, Mortazavi B, Wang Z, Khera R. Self-supervised learning of echocardiogram videos enables data-efficient clinical diagnosis. arXiv [cs.CV]. 2022. [DOI] [PMC free article] [PubMed]
- 23. Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv [cs.CV]. 2017.
- 24. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402–10. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 25. Jiao J, Droste R, Drukker L, Papageorghiou AT, Noble JA. Self-supervised representation learning for ultrasound video. Proc IEEE Int Symp Biomed Imaging 2020; 2020:1847–50. 10.1109/ISBI45749.2020.9098666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. Proc. IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016:2818–26. [Google Scholar]
- 27. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- 28. When does label smoothing help? Adv Neural Inf Process Syst 2019. [Google Scholar]
- 29. Dietterich TG. Ensemble methods in machine learning. In: Multiple Classifier Systems. Springer; 2000. p 1–15. [Google Scholar]
- 30. Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432. 10.1371/journal.pone.0118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. arXiv [cs.CV]. 2016.
- 32. Wilcox RR. Applying Contemporary Statistical Techniques: Elsevier; 2003. [Google Scholar]
- 33. Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW, Topic group. ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative. Calibration: the Achilles heel of predictive analytics. BMC Med 2019;17:230. 10.1186/s12916-019-1466-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu X, Rivera SC, Moher D, Calvert MJ, Denniston AK; SPIRIT-A; Working Group CONSORT-AI . Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. BMJ 2020;370:m3164. 10.1136/bmj.m3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotecha D, Asselbergs FW, Achenbach S, Anker SD, Atar D, Baigent C, et al. CODE-EHR best practice framework for the use of structured electronic healthcare records in clinical research. BMJ 2022;378:e069048. 10.1136/bmj-2021-069048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghorbani A, Ouyang D, Abid A, He B, Chen JH, Harrington RA, et al. Deep learning interpretation of echocardiograms. NPJ Digit Med 2020;3:10. 10.1038/s41746-019-0216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madani A, Arnaout R, Mofrad M, Arnaout R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med 2018;1:6. 10.1038/s41746-017-0013-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chiou Y-A, Hung C-L, Lin S-F. AI-assisted echocardiographic prescreening of heart failure with preserved ejection fraction on the basis of intrabeat dynamics. JACC Cardiovasc Imaging 2021;14:2091–104. 10.1016/j.jcmg.2021.05.005 [DOI] [PubMed] [Google Scholar]
- 39. Arnaout R, Curran L, Zhao Y, Levine JC, Chinn E, Moon-Grady AJ. An ensemble of neural networks provides expert-level prenatal detection of complex congenital heart disease. Nat Med 2021;27:882–91. 10.1038/s41591-021-01342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Narang A, Bae R, Hong H, Thomas Y, Surette S, Cadieu C, et al. Utility of a deep-learning algorithm to guide novices to acquire echocardiograms for limited diagnostic use. JAMA Cardiol 2021;6:624–32. 10.1001/jamacardio.2021.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Voigt I, Boeckmann M, Bruder O, Wolf A, Schmitz T, Wieneke H. A deep neural network using audio files for detection of aortic stenosis. Clin Cardiol 2022;45:657–63. 10.1002/clc.23826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kwon J-M, Lee SY, Jeon K-H, Lee Y, Kim K-H, Park J, et al. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J Am Heart Assoc 2020;9:e014717. 10.1161/JAHA.119.014717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cohen-Shelly M, Attia ZI, Friedman PA, Ito S, Essayagh BA, Ko W-Y, et al. Electrocardiogram screening for aortic valve stenosis using artificial intelligence. Eur Heart J 2021;42:2885–96. 10.1093/eurheartj/ehab153 [DOI] [PubMed] [Google Scholar]
- 44. Hata E, Seo C, Nakayama M, Iwasaki K, Ohkawauchi T, Ohya J. Classification of aortic stenosis using ECG by deep learning and its analysis using grad-CAM. Annu Int Conf IEEE Eng Med Biol Soc 2020;2020:1548–51. 10.1109/EMBC44109.2020.9175151 [DOI] [PubMed] [Google Scholar]
- 45. Yang C, Ojha BD, Aranoff ND, Green P, Tavassolian N. Classification of aortic stenosis using conventional machine learning and deep learning methods based on multi-dimensional cardio-mechanical signals. Sci Rep 2020;10:17521. 10.1038/s41598-020-74519-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ueda D, Yamamoto A, Ehara S, Iwata S, Abo K, Walston SL, et al. Artificial intelligence-based detection of aortic stenosis from chest radiographs. Eur Heart J Digit Health 2022;3:20–8. 10.1093/ehjdh/ztab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sengupta PP, Shrestha S, Kagiyama N, Hamirani Y, Kulkarni H, Yanamala N, et al. A machine-learning framework to identify distinct phenotypes of aortic stenosis severity. JACC Cardiovasc Imaging 2021;14:1707–20. 10.1016/j.jcmg.2021.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang Z, Long G, Wessler B, Hughes MC. A new semi-supervised learning benchmark for classifying view and diagnosing aortic stenosis from echocardiograms. In: Jung K, Yeung S, Sendak M, Sjoding M, Ranganath R, eds. Proceedings of the 6th Machine Learning for Healthcare Conference PMLR, 06–07 Aug 2021, 614–47.
- 49. Pawade T, Clavel M-A, Tribouilloy C, Dreyfus J, Mathieu T, Tastet L, et al. Computed tomography aortic valve calcium scoring in patients with aortic stenosis. Circ Cardiovasc Imaging 2018;11:e007146. 10.1161/CIRCIMAGING.117.007146 [DOI] [PubMed] [Google Scholar]
- 50. Bonow RO, Greenland P. Population-wide trends in aortic stenosis incidence and outcomes. Circulation 2015;131:969–71. 10.1161/CIRCULATIONAHA.115.014846 [DOI] [PubMed] [Google Scholar]
- 51. Forrest JK, Deeb GM, Yakubov SJ, Rovin JD, Mumtaz M, Gada H, et al. 2-year outcomes after transcatheter versus surgical aortic valve replacement in low-risk patients. J Am Coll Cardiol 2022;79:882–96. 10.1016/j.jacc.2021.11.062 [DOI] [PubMed] [Google Scholar]
- 52. Généreux P, Bax JJ, Makkar R. The PROGRESS trial: a prospective, randomized, controlled trial to assess the management of moderate aortic stenosis by clinical surveillance or transcatheter aortic valve replacement. 2021.https://clinicaltrials.gov/study/NCT04889872. Date accessed 18 July 2023. [Google Scholar]
- 53. Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, et al. Big self-supervised models advance medical image classification. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV) IEEE; 2021.
- 54. Asami M, Windecker S, Praz F, Lanz J, Hunziker L, Rothenbühler M, et al. Transcatheter aortic valve replacement in patients with concomitant mitral stenosis. Eur Heart J 2019;40:1342–51. 10.1093/eurheartj/ehy834 [DOI] [PubMed] [Google Scholar]
- 55. Nagueh SF, Smiseth OA, Appleton CP, Byrd BF, Dokainish H, Edvardsen T, et al. Recommendations for the evaluation of left ventricular diastolic function by echocardiography: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur J Echocardiogr 2016;17:1321–60. 10.1016/j.echo.2016.01.011 [DOI] [PubMed] [Google Scholar]
- 56. Stassen J, Ewe SH, Butcher SC, Amanullah MR, Mertens BJ, Hirasawa K, et al. Prognostic implications of left ventricular diastolic dysfunction in moderate aortic stenosis. Heart 2022;108:1401–7. 10.1136/heartjnl-2022-320886 [DOI] [PubMed] [Google Scholar]
- 57. Ong G, Pibarot P, Redfors B, Weissman NJ, Jaber WA, Makkar RR, et al. Diastolic function and clinical outcomes after transcatheter aortic valve replacement: PARTNER 2 SAPIEN 3 registry. J Am Coll Cardiol 2020;76:2940–51. 10.1016/j.jacc.2020.10.032 [DOI] [PubMed] [Google Scholar]
- 58. Poh K-K, Chan MY-Y, Yang H, Yong Q-W, Chan Y-H, Ling LH. Prognostication of valvular aortic stenosis using tissue Doppler echocardiography: underappreciated importance of late diastolic mitral annular velocity. J Am Soc Echocardiogr 2008;21:475–81. 10.1016/j.echo.2007.08.031 [DOI] [PubMed] [Google Scholar]
- 59. Marques-Alves P, Marinho AV, Teixeira R, Baptista R, Castro G, Martins R, et al. Going beyond classic echo in aortic stenosis: left atrial mechanics, a new marker of severity. BMC Cardiovasc Disord 2019;19:215. 10.1186/s12872-019-1204-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are not available for public sharing, given the restrictions in our institutional review board approval. Deidentified test set data may be available to researchers under a data use agreement after the study has been published in a peer-reviewed journal.