Graphical abstract
Keywords: Ribavirin, Cyclodextrin, Electrospinning, Nanofibers, Chronic wounds, Pseudomonas aeruginosa, Candida albicans
Abstract
Background
Skin is regarded as an essential first line of defense against harmful pathogens and it hosts an ecosystem of microorganisms that create a widely diverse skin microbiome. In chronic wounds, alterations in the host-microbe interactions occur forming polymicrobial biofilms that hinder the process of wound healing. Ribavirin, an antiviral drug, possesses antimicrobial activity, especially against Pseudomonas aeruginosa and Candida albicans, which are known as the main opportunistic pathogens in chronic wounds.
Rationale
In this study, electrospun nanofiber systems loaded with ribavirin were developed as a potential wound dressing for topical application in chronic wounds. Ribavirin was chosen in this study owing to the emerging cases of antimicrobial (antibiotics and antifungal) resistance and the low attempts to discover new antimicrobial agents, which encouraged the repurposing use of current medication as an alternative solution in case of resistance to the available agents. Additionally, the unique mechanism of action of ribavirin, i.e., perturbing the bacterial virulence system without killing or stopping their growth and rendering the pathogens disarmed, might be a promising choice to prevent drug resistance. Cyclodextrin (CD) was utilized to formulate ribavirin as an electrospun nanofibers delivery system to enhance the absorption and accelerate the release of ribavirin for topical use.
Results
The results demonstrated a successful ribavirin nanofibers fabrication that lacked beads and pores on the nanofibrous surfaces. Ribavirin underwent a physical transformation from crystalline to amorphous form, as confirmed by X-ray diffraction analysis. This change occurred due to the molecular dispersion after the electrospinning process. Additionally, the CD enhanced the encapsulation efficiency of ribavirin in the nanofibers as observed from the drug-loading results. Polyvinylpyrrolidone (PVP) and CD increased ribavirin released into the solution and the disintegration of fibrous mats which shrank and eventually dissolved into a gel-like substance as the ribavirin-loaded fibers began to break down from their border toward the midpoint. Cytotoxicity of ribavirin and CD was evaluated against human dermal fibroblasts (HFF-1) and the results showed a relatively safe profile of ribavirin upon 24-hour cell exposure, while CD was safe within 24- and 48-hour.
Conclusion
This study provides valuable insights into the potential application of our nanofibrous system for treating chronic wounds; however, further antimicrobial and in-vivo studies are required to confirm its safety and effectiveness.
1. Introduction
Skin is the body's largest multifunctional organ acting as its primary defense mechanism against physical, chemical, and immunological threats from the external environment. It also hosts an ecosystem of a widely diverse population of microbes. Bacteria, viruses and microeukaryotes such as fungi form complex communities of microorganisms inhabiting various parts of the skin. The skin itself provides a niche of microbial populations that are exposed to variability of ecological factors such as pH, temperature, and humidity, and are associated with microbial diversity (Grice et al., 2009). The relationships between microbes within a host can be categorized into three groups based on their effects on the species involved. These relationships may have a negative, positive, or neutral impact on the host or the microbes. Under normal physiological conditions, the majority of these microbes are commensals (i.e., nonpathogenic) and are maintained in homeostasis. However, microbial imbalance (known as microbial dysbiosis), host immune system status or genetic alterations may lead to the transition of skin microbiota from being commensal to becoming pathogenic (Findley and Grice, 2014). In other words, alterations in the host-microbe interactions can be linked to disease state.
It has been reported that there are at least nineteen phyla on human skin, with four exhibiting the highest abundance of bacterial species: Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes (Schommer and Gallo, 2013, Callewaert et al., 2020). The bacterial distribution and diversity of such species depend on the physiological characteristics of the skin niches; for instance, sebaceous sites are dominated by Propionibacterium (Actinobacteria), while Staphylococcus (Firmicutes) and Corynebacterium (Actinobacteria) thrive in humid environments (Grice et al., 2009, Schommer and Gallo, 2013, Byrd et al., 2018). Fungal communities, unlike bacteria, have a similar composition across the core body predominately by the genus Malassezia (Byrd et al., 2018). Reports of viruses such as human papillomavirus, human polyomaviruses, circoviruses, and bacteriophages indicated the presence of viral flora in healthy and diseased skin (Foulongne et al., 2012, Findley and Grice, 2014).
The process of wound healing is complex and distinctive as it is regulated by numerous growth factors, chemokines, and cytokines and involves the interaction of cells resulting in cell migration and wound closure (Tottoli et al., 2020). Whenever this process is perturbed, chronic wounds occur with a lack of migration and an increased rate of keratinocyte proliferation leading to extracellular matrix remodeling and, in most cases, persistent inflammation (Sachdeva et al., 2022). Some of the most common forms of chronic wounds include diabetic ulcers, venous ulcers, and pressure ulcers (Sen, 2021, Sachdeva et al., 2022). In a typical wound-healing process, commensal microorganisms interact with the skin cells to initiate an intrinsic immune reaction (Harrison et al., 2019, Sachdeva et al., 2022). However, chronic wounds harbor activities of polymicrobial biofilms hindering the process of wound healing. Wolcott et al reported a biofilm formation that was associated with 60 % of chronic wound patients, predominantly caused by Pseudomonas aeruginosa (P. aeruginosa) (Wolcott et al., 2016). Persistent biofilms contribute to a higher prevalence of opportunistic infections and antimicrobial resistance which can further hinder effective treatments (Sachdeva et al., 2022). It has been reported that fungi such as Candida spp. play an important role as an opportunistic pathogen in chronic wounds. Moreover, fungi associated with non-healing wounds contribute to antimicrobial resistance by enabling commensal bacteria to evade the host's immune response turning wounds into reservoirs for multi-resistant species.
Numerous studies suggested novel wound treatments to offer long-term relief and lower healthcare expenses, intending to achieve more effective wound healing. Skin wound therapies can be categorized into conventional or regenerative therapies. In an attempt to overcome functional alterations often associated with conventional therapy, regenerative wound therapy became a rapidly growing field in biomedical research, intending to rejuvenate skin to its original state by repairing damaged cells and tissues without leaving scars.
The emergence of antimicrobial resistance of a wide range of bacterial species encouraged researchers to repurpose the use of currently used and effective agents. One of the medications explored for their antimicrobial activity is ribavirin. It has been reported that ribavirin successfully protected Caenorhabditis elegans and mice from P. aeruginosa infection (Yuan et al., 2022). Ribavirin, an early-stage antiviral drug, has unique antimicrobial activities, especially against P. aeruginosa and Candida albicans (C. albicans). The mechanism by which ribavirin acts as an antimicrobial drug varies according to the type of its targeted microbe. Unlike conventional antibiotics, which are associated with the risk of antibiotic resistance, they act by perturbing the bacterial virulence system without killing or stopping their growth, rendering the pathogens disarmed, which might be a promising mechanism to prevent drug resistance (Mandal et al., 2016, Yuan et al., 2022). Hence, this antiviral drug is a promising agent that can be fabricated and repurposed as anti-pseudomonal drugs.
Methyl-β-cyclodextrin (CD), in itself, does not have any known antimicrobial activity, however, it was reported that it can delay bacterial growth when combined with an antimicrobial drug (Jaiswal et al., 2010, Boczar and Michalska, 2022). The CD might also improve the drug delivery and the stability of nanofiber formulation while maintaining the antimicrobial properties of ribavirin. The CD and its derivatives have robust properties due to the ability to complex with various compounds since the internal hydrophobicity and external hydrophilicity characteristics. Moreover, the biocompatibility of CD polymers increases their use in the field of drug delivery to carry different laded materials to targeted sites and improve the efficacy of encapsulated drugs (Pinho et al., 2014). It has been demonstrated that CD polymer has adhesive properties that can be exploited in wound-dressing manufacturing (Wang et al., 2023a). The formulation of bilirubin/CD adhesive hydrogel has been reported to promote wound healing in diabetic and non-diabetic in vivo, indicating the ability to be used as a wound dressing agent (Yao et al., 2021).
Polyvinylpyrrolidone (PVP) has unique physicochemical properties making it one of the most used polymers in biomedical applications. The pharmaceutical industry is the most recognized field that benefited from the use of PVP polymer since it is chemically stable, nontoxic, biodegradable, biocompatible, temperature-resistant, pH-stable, nonionic, soluble in water and several organic solvents and has an affinity to complex with hydrophilic and hydrophobic compounds (Teodorescu and Bercea, 2015). Dynamic interaction between PVP copolymer hydrogels and skin cells was investigated and results indicated the biocompatibility of this polymer and its ability to form macromolecular network with living cells and ability to support the growth of skin cells (Smith et al., 2006). Moreover, PVP polymer has an important function in bonding strength and increasing product viscosity. Therefore, it has been demonstrated that PVP can be used in different biomedical adhesive applications such as the fabrication of nanofibers for drug delivery, nanofibers for wound healing, and other different applications in the biomedical field (Chen et al., 2012).
Several studies showed the effectiveness of PVP for in vivo applications. For example, it was reported that PVP was used with celecoxib to formulate solid dispersion nanoparticles that have a size lower than 300 nm and the formulation was used in vitro and in vivo (Ha et al., 2014). Their results demonstrate that their formulation can enhance the bioavailability of celecoxib which is hydrophobic. Another study evaluates PVP to form a film with an anti-inflammatory Trans-resveratrol drug and the result shows that this formulation can decrease the inflammation in vivo by approximately 66 % (Riccio et al., 2022). In addition, several other studies demonstrated a lack of toxicity of PVP owing to its biologically inertness (Wessel et al., 1971, Nair, 1998). Furthermore, PVP is an FDA-approved polymer and several marketed products use PVP in their formulation such as Inadine® which is presented as PVP-Iodine that is used as an antiseptic for wound healing (Kurakula and Rao, 2020).
The electrospinning technique involves applying a high electric potential with two electrodes of opposite polarity to completely evaporate a polymeric solution and form a fibrous structure. The concept behind electrospinning involves the application of a high voltage to a viscous polymer droplet, overcoming its surface tension, converting the droplet to a cone-like shape, and finally, allowing the solvent to evaporate and fibers to form. Electrospinning is a powerful tool that enables the formation of fibers with various diameters. Since fabricating flexible and adaptable formulations are simple, electrospun fibers were utilized in many applications which include the delivery of drugs and the healing of tissue wounds. These fibers are also desirable substrates for drug administration due to their capacity to encapsulate a variety of active substances, including medicines, peptides, proteins, nucleic acids, and cells (Aburayan et al., 2020, Alshaya et al., 2022).
One of the most important parameters that play a key role in the preparation and loading of active pharmaceutical ingredient (API) into electrospun nanofibers is the optimal controlling of preparation conditions such as using a suitable temperature that is safe for the integrity of loaded drug. The high temperature could affect the chemical integrity and properties of the loaded drug. Different techniques can be taken into consideration to offer sustainable and efficient methods for improving the stability of loaded API such as the ultrasound-assisted extraction (UAE) approach which is conducted normally in low temperatures (Ponphaiboon et al., 2023). This method refers to ultrasonic waves or energy that exceeds the human hearing threshold of 20 kHz. The main advantage of applying ultrasonic energy is the protection of heat-sensitive compounds by eliminating the need for high temperatures (Picot-Allain et al., 2021). Therefore, applying this approach can protect the loaded drug from the effect of elevated temperature during the electrospinning process that is created from the use of high voltage and maintain its chemical integrity and properties during the process of electrospinning.
Consequently, in this study, ribavirin-loaded electrospun fiber systems were fabricated using CD and PVP to enhance its release, bioavailability and absorption, and these fibers were evaluated for their potential topical use for chronic wounds applications.
2. Materials and methods
2.1. Materials
Polyvinylpyrrolidone (PVP) that has a molecular weight of 1,300 kDa, CD with a molecular weight of 1.303 kDa, ethyl alcohol (≥99.5 %) and all buffer salts used, i.e., potassium phosphate monobasic, sodium phosphate dibasic, sodium chloride and potassium chloride, were bought from Sigma-Aldrich (St. Louis, MO, USA). Triethanolamine was obtained from Loba Chemie (Mumbai, India) and the ribavirin (Mw = 244.20) was purchased from Biosynthesis Carbosynth (Compton, UK). P. aeruginosa – ATCC 27853, ATCC 1744, and PAO1, C. albicans – ATCC 66027, Staphylococcus aureus (S. aureus)ATCC 29,213 and MRSA—ATCC 43300, Escherichia coli (E. coli) ATCC 25922, and Acinetobacter baumannii (A. baumannii) ATCC 747 reference microbes were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Clinical isolates of P. aeruginosa (7067), E. coli (1060) and A. baumannii (3034) were also used in this study. Mueller–Hinton agar (MHA) and Mueller-Hinton broth (MHB) were purchased from Scharlau (Barcelona, Spain) and afterwards prepared according to the manufacturer's instructions. Water used in experimental preparations was distilled via the Milli-Q® IQ 7005 Purification System (Millipore SAS Molsheim, France).
2.2. Methods
2.2.1. Preparation of the ribavirin-loaded fibers
All fibrous systems were fabricated according to the modified methods of these previous studies Aburayan et al., 2020, Alamer et al., 2023, Alshaya et al., 2022. A solution of PVP and ethanol (8 % w/v) was stirred for one hour at an ambient temperature on a magnetic stirrer. After the solution became homogeneous, 20 % w/v CD was mixed with the mixture for another hour. After the complete dissolution of both materials, 2 % w/v of ribavirin was mixed with the solution and it was left under stirring for an hour. Spraybase® electrospinning instrument (Dublin, Ireland) was used for the fabrication of the fibers. The solution was added into a syringe (5 mL) with a 0.9 mm needle diameter. The syringe stream was controlled by a pump, and the flow rate, collector distance and voltage were adjusted at 0.8 mL/hour, 20 cm and 7–9 kV, respectively. The metallic collector was wrapped with aluminum foil for easy collection of the fibers. The electrospinning was performed at ambient temperature and approximate humidity of 30 to 40 %. A similar electrospinning set-up was used to prepare blank fibers (i.e., ribavirin-free fibers), PVP-only fibers, and ribavirin-loaded PVP-only fibers. All fibrous systems were peeled off easily from the aluminum foil using a surgical tweezer and kept in an air-tight chamber (dry-keeper) for storage.
2.2.2. Morphology assessment of ribavirin-loaded fibers using scanning electron microscopy (SEM)
SEM (JSM-IT500HR SEM, JEOL Inc., Peabody, USA) was used to analyze the morphology of the fibers. A piece of the foil with collected fibers was put on a metal stub and the samples were analyzed without the need to coat the sample with a conductive material similar to the previous studies of Aburayan et al., (2020) and Alshaya et al., (2022). The fiber's diameter was evaluated by the microscope’s software (SEM Operation: 3.010, version 1.010, JEOL TECHNICS Ltd., Tokyo, Japan) by measuring the diameter of a minimum of 30 fibers.
2.2.3. Characterization of the solid-state using X-ray diffraction (XRD)
Rigaku miniflex 300/600 (Tokyo, Japan), with a Cu Kα radiation point (λ) of 1.5148 227 Å was used to characterize the solid state of ribavirin after being electrospun according to a previous study by Alshaya et al., (2022). The specimen was directly positioned on a glass mount and inspected at 40 kV, 15 mA and a scan speed of 5°/min (2θ: 2–60°).
2.2.4. Characterization of the solid-state using Fourier-Transform Infrared Spectroscopy (FTIR)
Thermo smart ATR IS20 Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to conduct FTIR analysis. Ribavirin, CD, PVP, three different physical mixtures (PM), blank and ribavirin-loaded nanofiber systems were placed on the sampling spot one sample at a time in small quantities (around 5 mg). Samples were analyzed at a specific wavenumber range of 4000 to 400 cm−1 based on a modified method of Alkahtani et al., (2021). The spectral resolution was configured at 4 cm−1, and 32 scans were conducted for each sample.
2.2.5. Quantification of ribavirin using high-performance liquid chromatography (HPLC)
HPLC was used to identify and quantify ribavirin. A serial dilution of six ribavirin concentrations (200–6.25 μg/mL) was prepared by dissolving ribavirin in phosphate buffer saline (PBS, pH 6.8). An HPLC set-up of Waters e2695 was utilized that consists of a Waters 600 binary pump, Waters® 717 plus autosampler, and Waters 2489 UV/detector (Waters Technologies Corporation, Milford, MA, USA), which is similar to the HPLC set-up of the previous study of Alamer et al., (2023). Ribavirin was identified according to a modified method of Inoue et al., 2004, D'Avolio et al., 2006, and Haggag et al., 2014 using a C18 column (4.6 mm × 250 mm, 5 μm), while the mobile phase was 1 % v/v triethanolamine (pH 3.9). An isocratic elution with a flow rate of 1 mL/min was used. The column temperature, the volume of injection and the detection wavelength were adjusted at ambient temperature, 10 μL and 254 nm, respectively. Ribavirin was identified at a retention time (Rt) of 4.5 min, which is very close to the reported ribavirin Rt of the previous studies (i.e. Rt 4.1–4.7 min) (Inoue et al., 2004, D'Avolio et al., 2006, Haggag et al., 2014).
2.2.6. Quantification of ribavirin-loaded fibers drug loading (DL) and encapsulation efficiency (EE%)
The DL and EE% of the ribavirin-loaded PVP/CD fibers and ribavirin-loaded PVP-only fibers were quantified by dissolving three pieces of the fiber at a weight of 4 ± 0.2 mg in PBS (pH 6.8), for four hours at room temperature, according to a slightly modified method of Alamer et al., (2023). The samples were measured after the complete dissolving of the ribavirin-loaded fibers and subjected to the previously HPLC-developed method. The EE% and DL were analyzed by the subsequent formulas:
| (1) |
| (2) |
2.2.7. Determination of ribavirin-loaded fibers disintegration rate
The disintegration of blank PVP/CD fibers, PVP-only fibers, ribavirin-loaded PVP/CD fibers, and ribavirin-loaded PVP-only fibers was evaluated using the petri dish method, according to the previous studies of Alamer et al., 2023, Alshaya et al., 2022 and Alkahtani et al., 2021. Briefly, certain weights of each nanofibrous system were positioned inside Petri dishes half-filled with pre-warmed PBS (37 °C and pH 6.8), mimicking the pH of an open wound that ranges from 6.5 to 8.5 (Louisa et al., 2017). The dishes were kept in a trembling incubator (Excella E24 Incubator Shaker Series, New Brunswick Scientific Co., Enfield, CT, USA) until a complete detachment of the fiber mats occurred, which was recorded in seconds.
2.2.8. Quantification of ribavirin-loaded fibers release rate
The drug release rate of ribavirin-loaded PVP/CD fibers and ribavirin-loaded PVP fibers were assessed after weighing a certain amount of both fibrous systems and dissolving them into 20 mL PBS (pH 6.8) using custom-made sinkers to keep the fibers at the bottom of the vials, by a modified method of the release studies of Alamer et al., 2023, Alshaya et al., 2022 and Alkahtani et al., 2021. The vials were kept inside a trembling incubator (Excella E24 Incubator Shaker Series, New Brunswick Scientific Co., Enfield, CT, USA) at a shaking rate of 100 RPM and a temperature of 37 degrees Celsius. One mL of samples was obtained from each vial after 1, 3, 5, 10, 15, 30, 60, 90, 120, 180, 240, and 300 min. To maintain the sink conditions, an equivalent volume of fresh PBS was replaced. The determination of the released ribavirin amount was obtained by the previously HPLC-developed method. The cumulative release % was quantified according to the following equation:
| (3) |
2.2.9. Determination of ribavirin and CD in-vitro cytotoxicity
The assessment of in-vitro cellular viability of ribavirin, CD and their combination was performed following 24 and 48-hour cellular exposure to the human fibroblast HFF-1 cells (ATCC- SCRC-1401). The evaluation of the metabolic activity of ribavirin was conducted using the modified method of Ahmed et al., (2023), using the colorimetric MTS assay (cell Titer 96® Aqueous one solution cell proliferation assay, Promega, Southampton, UK). HFF-1 cells were sub-cultured in Dulbecco's modified eagle medium (DMEM) containing the following: streptomycin (100 μg/mL), FBS (10 % (v/v)), and penicillin (100 U/mL), which were brought from Sigma-Aldrich (St. Louis, MO, USA). Following 90–100 % cell confluency, trypsin solution was applied and incubated for several minutes, and then HFF-1 cells were counted after harvested using the trypan blue exclusion test. The collected cells were then seeded into 96-well plates with a seeding density of 1.5 × 104 cells/well. The seeded cells were then incubated in a cell culture incubator overnight at 37 °C and 5 % CO2. 100 µL of diluted concentration of the ribavirin (5,000 to 156.25 µg/mL), CD (20,000 to 156.25 µg/mL) and their combination in a ratio of 20:1CD: ribavirin (20,000/1,000 to 156.25/7.81 µg/mL) were then added to the HFF-1 cells for 24 and 48-hour. Negative and positive controls of cells incubated with DMEM only and Triton X-100 (0.2 %) treated cells, respectively were used as experimental controls. After the designated incubation times (i.e., 24 and 48 h. Thereafter, removal of the medium from each well was performed and then wells were washed gently with PBS followed by adding fresh DMEM (100 μL) in each well. 20 μL of the MTS reagent was then added to each well and cells were incubated in the CO2 incubator for a further 3 h. The absorbance of formazan color produced by living cells was measured at 492 nm using the Cytation 3 absorbance microplate reader (BIOTEK Instruments Inc., Winooski, VT, USA. The relative percentage of living cells was measured according to the following equation of Jarallah et al., (2023):
| Cell Viability % = (S-T)/ (H-T) × 100 | (4) |
where S, T and H are the absorbances of cells treated with ribavirin, Triton X-100 and media only, respectively.
2.2.10. Determination of ribavirin and CD minimum inhibitory concentration (MIC)
The broth microdilution method was applied as outlined by CLSI guidelines to test ribavirin and CD alone and in combination but with few modifications (Wayne, 2015). A serially diluted (2-fold dilutions) ranging concentrations of ribavirin (2,500–19.53 μg/mL), CD (25,000–195.31 μg/mL) and 1:10 combination (2,500/25,000–19.53/195.31 μg/mL) was tested against P. aeruginosa (ATCC 27853, ATCC 1744, PAO1, and the clinical isolate 7067), S. aureus (ATCC 29213), MRSA (ATCC 43300), E. coli (ATCC 25922, and the clinical isolate 1060), A. baumannii (ATCC 747 and the clinical isolate 7067, and the clinical isolate 3034), and C. albicans (ATCC 66027). Then, 100 μL of the microbial inoculums (0.5 McFarland standard) was added to each well with a final cell density of 1.5 × 105 CFU/mL. Wells with microbe without treatment were considered as a positive control and wells with media (MHB) only were considered as a negative control. The 96-well microtiter plates of the bacterial strains were incubated overnight at 37 °C, while the C. albicans plates were incubated at 25 °C and with a continuous shaking speed of 160 RPM. The lowest concentration with no visible growth was considered as the MIC. Microbial growth inhibition was also assessed by measuring UV absorbance at 600 nm with a PowerWave XS2 plate reader (bioMérieux, Marcy L'Etoile, France).
2.2.11. Determination of the antimicrobial activity of ribavirin-loaded fibers
The antimicrobial activity of the ribavirin-loaded PVP/CD fibers and ribavirin-loaded PVP fibers was evaluated, against the same bacterial and C. albicans strains used in the MIC test, using the agar-diffusion method per the modified protocol of He et al., (2020). The plates were inoculated with 100 μL of 1.5 × 108 (CFU)/mL (0.5 McFarland standard) bacteria and fungus inoculums and spread onto the surface of MHA plates. Then, certain amounts of the ribavirin-loaded fibers, which contained 625 μg of the drug, and their corresponding weights for the blank fiber systems were positioned on the surface of the MHA plates. As a test control, 20 μL of ribavirin diluted in MHB was added on filter discs, which is equivalent to a concentration of the ribavirin-loaded fibers. Following incubation at 37 °C overnight for the bacterial strains and at 25 °C for C. albicans, the diameters of the clear zones of microbial inhibitions were measured in millimeters (mm).
2.2.12. Statistical analysis
The results of fibers’ diameter, XRD, FTIR, HPLC calibration curve and linearity, and ribavirin release rate were analyzed and plotted by OriginPro 2021 (OriginLab Corporation, Northampton, MA, USA). All findings in the DL, EE%, drug release, fibers' disintegration, cellular viability and antimicrobial activity were presented as the average ± standard deviation (SD) calculated from three repeated measurements.
3. Results
3.1. Morphology assessment of ribavirin-loaded fibers using SEM
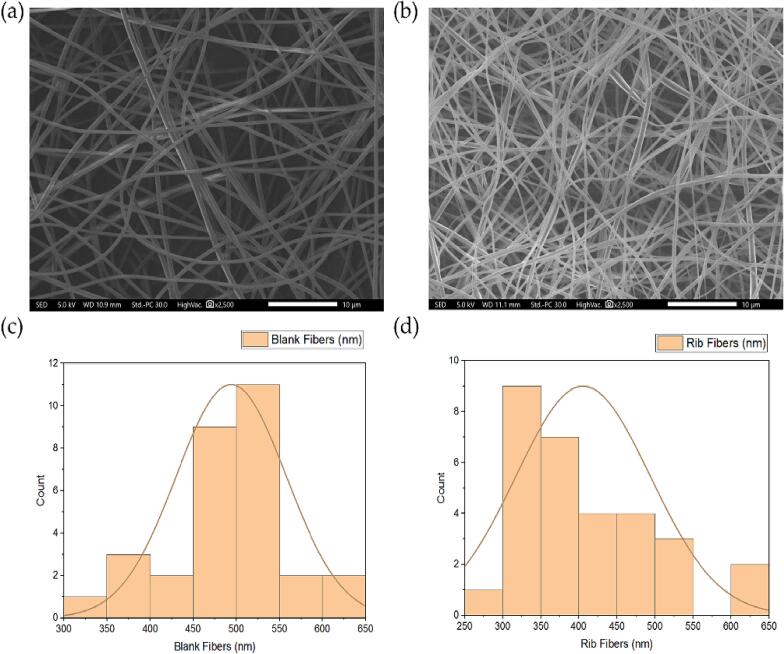
The prepared PVP fibers and ribavirin-loaded PVP fibers in Fig. 1 showed successful fiber fabrication criteria of smooth surface, lacked any beads and pores formation, having mean diameters of approximately 494 ± 64 nm and 406 ± 88 nm, respectively. The fabrication blank CD/PVP fibers showed average fiber diameters of 790 ± 431 nm, while the addition of ribavirin nanofibers increased the fibers' diameter to 1,133 ± 234 nm (Fig. 2).
Fig. 1.
SEM images that represent (a) PVP fibers and (b) ribavirin-loaded PVP fibers. The diameter distribution of (c) PVP fibers and (d) ribavirin-loaded PVP fibers showed their average diameters of 494 ± 64 nm and 406 ± 88 nm, respectively (n = 30).
Fig. 2.
SEM images that represent (a) blank PVP/CD fibers and (b) ribavirin-loaded PVP/CD fibers. The distribution of diameter of (c) blank PVP/CD fibers and (d) ribavirin-loaded PVP/CD fibers showed their average fiber diameters of 790 ± 431 nm and 1,133 ± 234 nm, respectively (n = 30).
3.2. Characterization of the solid-state using XRD
The raw materials' physical state, i.e., ribavirin, CD, PVP and their PM, as well as blank and ribavirin-loaded fibers (fabricated with CD), were all evaluated. As illustrated in Fig. 3, a sequence of intense Bragg reflections was observed in ribavirin raw material, precisely at 2θ: 21°, 23–29°, 36° and 39°. The pattern of CD exhibited two distinctive sequences of Bragg reflections at 2θ: 32° and 46°, while no distinctive peaks were observed in the PVP polymer pattern (i.e., a broad-halo diffraction). All CD distinctive reflections at 32° and 46° were also presented in the PM that contained PVP and CD only, while the characteristic peaks of ribavirin at 21°, 23-29°, 36° and 39° were demonstrated in the PM that contained this drug along with PVP and CD, suggesting its presence in the crystalline form. Both blank and ribavirin-loaded nanofibers showed a broad halo diffraction indicating their presence in the amorphous form.
Fig. 3.
XRD patterns of PVP, CD, ribavirin, blank PM (PVP + CD), ribavirin PM (PVP + Rib + CD), blank and ribavirin-loaded PVP/CD nanofibers demonstrating the molecular dispersion of ribavirin in the ribavirin-loaded fibers. Grey arrows represent the distinctive peaks. PM, physical mixture; PVP, polyvinylpyrrolidone; CD, cyclodextrin; Rib, ribavirin.
3.3. Characterization of the solid-state using Fourier-Transform Infrared Spectroscopy (FTIR)
The FTIR spectrum of the pure PVP polymer, CD, ribavirin, and their PM displayed distinct peaks for each raw material as exhibited in Fig. 4A. A broad band of O–H stretching was observed between 3600 and 3000 cm−1 for PVP. Furthermore, the peaks at 1660, 1463, and 1293 cm−1 represented the stretching vibrations of the following C = O, C–H (in the aliphatic compound), and C–N (in aromatic amine), respectively. The FTIR spectrum of the pure ribavirin showed stretching at the function group region representing O–H stretching between 3500 and 3050 cm−1, amine group stretching (–N–H 3450 cm−1), carbonyl group stretching at 1653 cm−1, and the C–N stretching was observed at 1157 cm−1. Other absorption bands at 1086 and 1040 cm−1 of the aliphatic ether group were observed within the ribavirin spectrum. The IR spectrum of the pure CD showed the O–H stretching between 3550 and 3000 cm−1 and absorption bands at 2923 cm−1 that correspond to –CH2– stretching, 1651 cm−1 that correspond to the carbonyl group stretching, 1459, 1436, 1423 cm−1 that indicate for the –CH– band vibrations and characteristic C–O/C–C vibrations at 1288 cm−1.
Fig. 4.
FTIR transmissions at a wavenumber range 4000 to 400 cm−1 of (A) PVP, CD, ribavirin, PVP/CD PM, ribavirin/PVP PM, ribavirin/PVP/CD PM, and (B) blank PVP nanofibers, PVP/CD nanofibers, ribavirin-loaded PVP nanofibers and ribavirin-loaded PVP/CD nanofibers. PM, physical mixture; PVP, polyvinylpyrrolidone; CD, cyclodextrin; Rib, ribavirin.
Three PMs were analyzed, PVP/CD, ribavirin/PVP, and ribavirin/PVP/CD (Fig. 4A). The O–H stretching between 3600 and 3000 cm−1, and the carbonyl group stretching at ∼ 1652 cm−1 were presented in all three PM spectra. Three distinctive peaks of CD were presented clearly in the PVP/CD and ribavirin/PVP/CD PMs compared to the ribavirin/PVP spectrum, indicating the presence of CD within those PMs. Characteristic peaks of ribavirin at ∼ 1070 and 1030 cm−1 (i.e., aliphatic ether group) were shown in the PM that contained ribavirin (ribavirin/PVP, and ribavirin/PVP/CD). Fig. 4B exhibited the FTIR spectra of all prepared nanofiber groups, which observed similar distinctive peaks to the PMs with no noticeable shifts in those peaks.
3.4. Quantification of ribavirin using HPLC
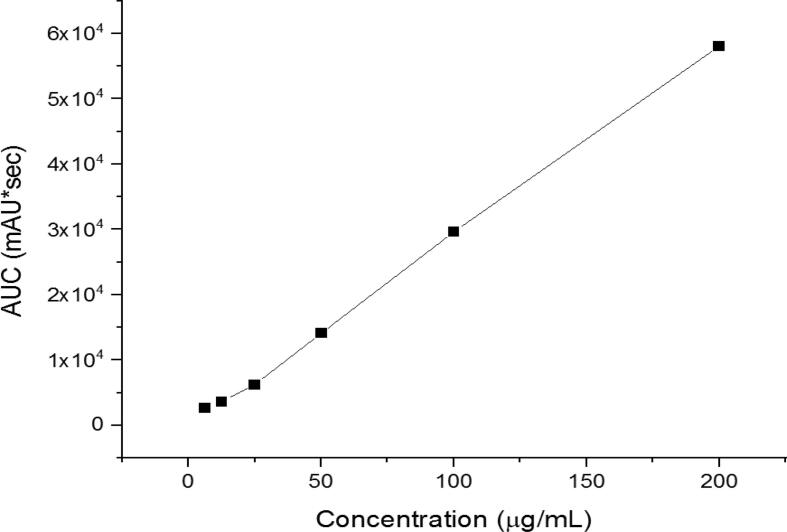
The development and application of an HPLC for quantifying ribavirin-loaded fibers demonstrated its precision and reliability in drug separation and identification. The calibration curve yielded favorable outcomes as shown in Fig. 5. The HPLC analysis exhibited a successful separation of ribavirin revealing a separation of the drug in around 4.5 min (i.e., Rt). The regression equation was y = 291.11x − 55.552 and the coefficient of determination (R2) was 0.9991, indicating an excellent linear correlation and effective separation of the drug achieved by this developed method.
Fig. 5.
HPLC calibration curve of ribavirin-loaded nanofibers showed excellent linearity (R2 = 0.9991).
3.5. Quantification of ribavirin-loaded fibers drug loading (DL) and encapsulation efficiency (EE%)
In the analysis of the DL and EE% for the two different formulations; ribavirin-loaded PVP fibers and ribavirin-loaded PVP/CD fibers, the results showed that the EE% and DL were 102 ± 12 % and 204 ± 24 μg/mg and 117 ± 15 % and 78 ± 10 μg/mg respectively.
3.6. Determination of ribavirin-loaded fibers disintegration rate
As seen in Fig. 6, the desired disintegration time was obtained at all tested nanofiber systems, including the blanks, which dissolved quite quickly (i.e., PBS pH 6.8). Blank PVP/CD fibers, ribavirin-loaded PVP/CD fibers and ribavirin-loaded PVP fibers showed a complete disintegration in 2 ± 1 s, while the blank PVP fibers exhibited a complete disintegration in 1 ± 1 s.
Fig. 6.
The disintegration of PVP/CD fibers (PVP + CD), ribavirin-loaded fibers (PVP + Rib + CD), blank PVP fibers (blank fibers) and ribavirin-loaded fibers (Rib fibers) showing an ultra-rapid disintegration of all systems in ≤ 3 s. PVP, polyvinylpyrrolidone; CD, cyclodextrin; Rib, ribavirin.
3.7. Quantification of ribavirin-loaded fibers release rate
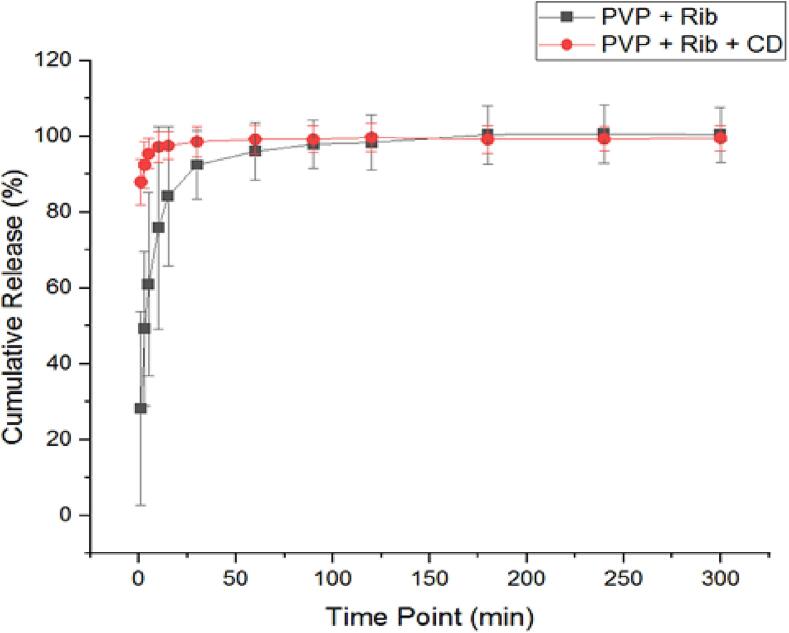
As shown in Fig. 7, ribavirin-loaded fibers demonstrated a burst release of ribavirin from the PVP only and PVP/CD fibers in the first 10 min and at 1 min, respectively. This was followed by > 85 % and > 97.5 % cumulative release of ribavirin after 15 min, and a complete drug release after 180 min and 60 min, respectively.
Fig. 7.
The release profile of the ribavirin demonstrated a rapid release in the initial 10 min and at 1 min from ribavirin-loaded fibers (PVP + Rib) and ribavirin-loaded PVP/CD fibers (PVP + Rib + CD), respectively. Results are shown as the average ± SD (n = 3). PVP, polyvinylpyrrolidone; CD, cyclodextrin; Rib, ribavirin.
3.8. Determination of ribavirin and CD in-vitro cytotoxicity
As shown in Fig. 8, ribavirin was exposed to HFF-1 cells in the current study for two time points which are 24 and 48 h. The MTS results following 24-hour incubation time demonstrated that a high level of cellular metabolic activity was observed in all assessed ribavirin concentrations, except the highest concentration (i.e., 5,000 µg/mL) which reduced the cellular viability to less than 60 %. However, the application of ribavirin for 48 h showed a substantial decrease in the HFF-1 cell viability when the concentration was increased. Cell viability was shown to be below 50 %, at all ribavirin concentrations more than 312.5 µg/mL, while it was found to be high (>60 %) at the lowest applied concentration, i.e., 156 μg/mL.
Fig. 8.
Cell viability of (A) ribavirin, (B) CD, and (C) ribavirin and CD combined in a ratio of 1:20, after being applied at different concentrations upon 24- and 48-hour exposure to HFF-1 cells. Results are shown as average ± SD (n = 3).
On the other hand, the CD exhibited a better safety profile than ribavirin, with > 60 % cell viability after 24- and 48-hour HFF-1 exposure. Interestingly, the cell viability % was higher after 48 h compared to 24 h after being incubated with these cell lines, especially at the concentrations 2,500 and 1,250 μg/mL, but this variation is irrelevant to this current study. The cytotoxicity of both ribavirin and CD in combination at a ratio of 1:20, respectively, which mimics the ratio upon the fabrication of ribavirin-loaded PVP/CD nanofibers (i.e., 8:20:1 PVP: CD: ribavirin), was also tested. It seems that the cell viability of this combination follows the trend of both compounds, i.e., ribavirin and CD, in being more toxic after 48 h of HFF-1 cell exposure (ribavirin) and being less toxic (>60 % cell viability) at a concentration of 5,000 μg/mL (CD).
3.9. Determination of ribavirin and CD minimum inhibitory concentration (MIC)
The antimicrobial activity of ribavirin and CD was initially tested using the MIC assay, with the results presented in Table 1. Ribavirin alone had MIC values between 156.25 and 625 μg/mL. However, the clinical isolate of P. aeruginosa required a higher inhibitory concentration, i.e., 1250 μg/mL. No inhibition was found against the other bacterial strains at all tested concentrations (i.e., ≤ 2500 μg/mL). In addition, no direct synergistic effects were observed against the tested pathogens at equivalent concentrations of ribavirin in combination with CD (Table 1). In most previous studies, the effect of ribavirin was only observed at concentrations higher than 500 μg/mL at attainable therapeutic levels (Snell, 2001). Additionally, the combination with CD did not exhibit any synergistic effect.
Table 1.
The MICs of ribavirin and CD alone or in combination against P. aeruginosa (ATCC 27853, ATCC 1744, PAO1, and the clinical isolate 7067), C. albicans (ATCC 66027), S. aureus (ATCC 29213, MRSA—ATCC 43300), E.coli (ATCC 25,922 and clinical isolate 1060) and A. baumannii (ATCC 747 and clinical isolate 3034). Strains treated with ribavirin at concentrations (2,500–19.53 μg/mL), CD (25,000–195.31 μg/mL) and combination 1:10 at (2,500/25,000––19.53/195.31 μg/mL). The results are shown as average ± SD (n = 3).
| Microorganisms |
MIC (μg/mL) |
||
|---|---|---|---|
| Ribavirin | CD | Combination 1:10 | |
| P. aeruginosa – (ATCC 27853) | 156.25 ± 0.02 | >25,000 ± 0.00 | 156.25 ± 0.01 |
| P. aeruginosa – (ATCC 1744) | 312.5 ± 0.01 | >25,000 ± 0.00 | 312.5 ± 0.01 |
| P. aeruginosa – (PAO1) | 625 ± 0.02 | >25,000 ± 0.00 | 625 ± 0.01 |
| P. aeruginosa – (Clinical isolate, 7067) | 1250 ± 0.01 | >25,000 ± 0.00 | 1250 ± 0.00 |
| C. albicans – (ATCC 66027) | 156.25 ± 0.02 | >25,000 ± 0.00 | 156.25 ± 0.01 |
| S. aureus – (ATCC 29213) | >2500 ± 0.03 | >25,000 ± 0.07 | >2500 ± 0.01 |
| MRSA – (ATCC 43300) | >2500 ± 0.03 | >25,000 ± 0.06 | >2500 ± 0.03 |
| E. coli – (ATCC 25922) | >2500 ± 0.08 | >25,000 ± 0.06 | >2500 ± 0.01 |
| E. coli – (Clinical isolate, 1060) | >2500 ± 0.04 | >25,000 ± 0.09 | >2500 ± 0.05 |
| A. baumannii – (ATCC 747) | >2500 ± 0.10 | >25,000 ± 0.04 | >2500 ± 0.25 |
| A. baumannii – (Clinical isolate, 3034) | >2500 ± 0.14 | >25,000 ± 0.07 | >2500 ± 0.15 |
3.10. Determination of the antimicrobial activity of ribavirin-loaded fibers
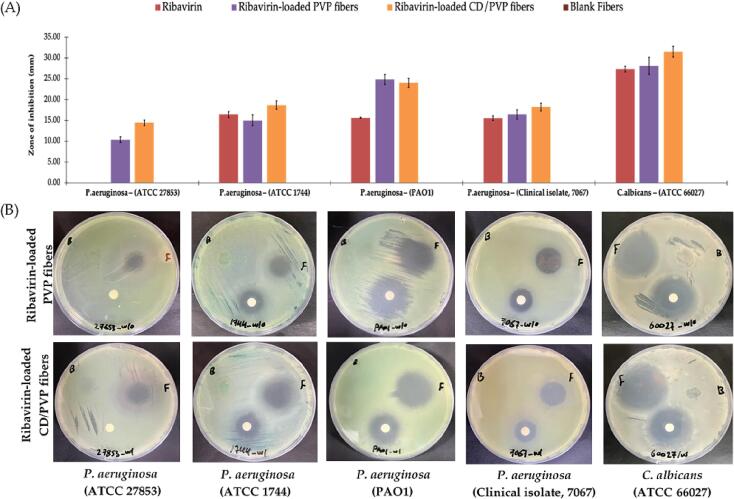
In this current study, novel skin antimicrobial wound dressings of ribavirin-loaded PVP/CD fibers were successfully prepared. The effectiveness of this fabricated nanofiber system compared to the ribavirin-loaded PVP nanofibers, ribavirin disc (positive control) and blank fibers (negative control) was tested against P. aeruginosa (ATCC 27853, ATCC 1744, PAO1, and the clinical isolate 7067), and C. albicans (ATCC 66027), at a similar dose (625 μg) through the disc diffusion assay. As shown in Fig. 9, both nanofiber systems were able to inhibit the growth of all strains of P. aeruginosa, and C. albicans, suggesting that the drug efficacy was retained after being electrospun, which is consistent with another antimicrobial-loaded PVP nanofiber system against different microbes (Aburayan et al., 2020). There is no clear advantage between the antimicrobial effect of ribavirin-loaded PVP/CD nanofibers and ribavirin-loaded PVP nanofibers, as the slightly larger diameter could contribute to the drug-loading variation within the batch. The combination with CD did not exhibit any synergistic effect.
Fig. 9.
The zone of inhibition assay showing (A) the antimicrobial activity of ribavirin, ribavirin-loaded PVP nanofibers, ribavirin-loaded PVP/CD nanofibers and blank nanofibers at dose equivalent to 625 μg against P. aeruginosa (ATCC 27853, ATCC 1744, PAO1, and the clinical isolate 7067) and C. albicans, and (B) MHA plate images that were taken after an overnight incubation against all test microbes. Data shown are the diameters of the zones of inhibition measured in millimeters (mm). The results are shown as average ± SD (n = 3). PVP, polyvinylpyrrolidone; CD, cyclodextrin; Rib, ribavirin.
Interestingly, ribavirin only (disc) did not show any effect against the P. aeruginosa ATCC 27853 compared to both ribavirin-loaded nanofiber systems, despite showing an effect in the MIC assay (MIC = 156.25 ± 0.02). It is worth mentioning that this was repeated twice, each was in triplicates.
4. Discussion
The morphology of the fabricated PVP nanofibers was consistent with previously reported PVP nanofibers encapsulated by different active compounds including halicin, melittin, and imeglimin (Aburayan et al., 2020, Tawfik et al., 2020, Tawfik et al., 2021a, Tawfik et al., 2021b, Alshaya et al., 2022, Alamer et al., 2023). Moreover, both nanofibrous systems exhibited successful fiber fabrication criteria as mentioned in 3.1. The enhanced fiber diameter of the blank and ribavirin-loaded PVP/CD fibers was expected, as the used concentration for the CD in the spinning solution was 20 % (w/v) led to a higher spinning solution concentration, and thus, a larger fiber diameter. This was consistent with (Williams et al., 2018).
The XRD analysis was conducted to demonstrate the molecular dispersion of ribavirin after being electrospun. Ribavirin XRD diffractogram in its solid form (Fig. 3) showed a pattern of Bragg reflections similar to reported results by Li et al., (2023). The absence of reflections suggests that the polymer is in amorphous form, which is in agreement with (Aburayan et al., 2020) Aburayan et al., 2020, Alkahtani et al., 2021. The diffractogram of the PM of both PVP and CD (representing the blank fibers) confirmed the presence of the same two distinctive sequences of CD, suggesting the conformity of an intact CD within the polymer mixture. Similarly, the diffractogram of the PM of ribavirin, CD and PVP (representing the ribavirin-loaded fibers) demonstrated a mix of the characteristic peaks of the drug and CD suggesting their presence in the crystalline form (Fig. 3). The blank and ribavirin-loaded fibers shared a similar XRD pattern to the PVP polymer, demonstrating the absence of highly distinguishable reflections and only appearing as broad haloes. The XRD results indicated that both blank and ribavirin-loaded nanofiber systems are in an amorphous state which is cotenant with Zhao et al., (2021). Moreover, the molecular dispersion of ribavirin from the crystalline form to the amorphous state after being processed by electrospinning is in agreement with other previous studies (Aburayan et al., 2020, Alkahtani et al., 2021). This alteration may arise from the instant solvent evaporation, which hindered the molecular organization into a crystalline lattice. This subsequently promotes a disordered alignment of molecules within the fibrous mat (Bukhary et al., 2018).
FTIR spectroscopic analysis was conducted to evaluate the compatibility of ribavirin and CD with the polymer, specifically focusing on intermolecular interactions between the drug and polymer, to verify the stability of the resulting formulation. FTIR spectrum of PVP showed a broad band of O–H stretching between 3600 and 3000 cm−1 which can be explained by the hygroscopic nature of PVP (Fig. 4A). Additionally, stretching vibrations at the functional groups of the C = O, C–H, and C–N were observed which agree with previous reports (Aburayan et al., 2020, Alshaya et al., 2022). FTIR analysis of ribavirin in its solid form indicated stretching of the hydroxyl, aliphatic aromatic amine, and carbonyl groups which aligns with Rizwana et al., (2018). The stretching of –CH2– (2923 cm−1), C = O (1651 cm−1) and C–O/C–C (1288 cm−1) in the FTIR spectrum of CD (Fig. 4A) are by (Ribeiro et al., 2008, Nyairo et al., 2017). Ribavirin distinctive peaks were shown within the drug containing PM (i.e., ribavirin/PVP and ribavirin/PVP/CD), but with different intensities suggesting different degrees of interaction. However, for the FTIR spectra of all prepared nanofiber systems (Fig. 4B), there was a clear variation between the spectra that contained CD within their fibers which showed a strong reduction to the complete disappearance of the distinctive peaks of ribavirin at the fingerprint region (∼1070 and 1030 cm−1, the aliphatic ether group). This might indicate the strong ribavirin–CD interactions and possibly the inclusion complexation of the drug within the fibrous system that was explained in the previous study of Ribeiro et al., (2008). Additionally, these aliphatic ether group peaks that were observed in the ribavirin-loaded PVP-only fibers were slightly shifted compared to the blank PVP fibers, which could suggest the intermolecular interaction between ribavirin and PVP explained in the previous study (Alkahtani et al., 2021).
The DL and EE% analyses indicate that the actual amount of ribavirin in the nanofibers exceeds the theoretical amount, which could be due to variations in the preparation process. Overall, these results facilitate the effective formulation of this nanofibrous system. The addition of CD increased the EE% of ribavirin in the nanofibers, which suggests that CD could play a role in enhancing the integration or protection of ribavirin within the nanofiber matrix. While the EE% is enhanced with the addition of CD, the overall DL is reduced. This could mean that while a higher percentage of the ribavirin is successfully encapsulated, the total amount of ribavirin in the fiber was lower owing to the higher ratio of CD that was used in the preparation of the ribavirin-loaded PVP/CD fibers. Another reason is that the presence of CD takes up space or interacts with ribavirin in a way that reduces its overall concentration. The addition of CD seems to enhance the encapsulation of ribavirin, which aligns with the hypothesis that encapsulating the drug in the fibers can enhance drug delivery and protect it from degradation. However, the reduced DL in the presence of CD suggests a potential trade-off. Yet, in general, CDs have been investigated for their potential to enhance drug delivery and stability of electrospun fibers. CDs are known to form inclusion complexes with drugs, increasing their solubility and stability (Celebioglu and Uyar, 2020, Topuz et al., 2021). The observed enhancement in EE% with CD might be due to this complex formation, leading to better encapsulation.
When using a topical dosage form, the disintegration time is a crucial qualitative characteristic. The electrospun fibers' ultra-rapid disintegration (i.e., ≤ 3 s) is consistent with earlier studies using comparable polymeric systems, i.e., PVP fibers (Chuangchote et al., 2009, Yu et al., 2018). Another important criterion to assess is the release rate of the drug-loaded fibers. A substantial release rate of ribavirin from the PVP/CD fibers, especially at ≤ 30-minute time point was expected due to the addition of CD which is known to enhance drug solubility, absorption and bioavailability. Another reason is due to the utilization of a hydrophilic polymer, i.e., PVP, which has the property of speeding up the fibrous mats' dissolution and disintegration, releasing the loaded medications more rapidly. This result is in agreement with (Sriyanti et al., 2018, Moydeen et al., 2019, Aburayan et al., 2020). Furthermore, the quick release of the medications was made possible by the increased contact area between the fibers and the dissolving medium as a result of their high surface-to-volume ratio (Alkahtani et al., 2021). Moreover, ribavirin's molecular dispersion during electrospinning is another element that facilitated the release of the drug (Yu et al., 2018). Using a hygroscopic polymer, such as PVP, the rate of the encapsulated drug being released will increase owing to the disintegration of fibrous mats. This is considered one of the various ways to enhance the solubility and dissolution rate of drugs. A high contact area with the dissolving medium can be ensured by the high surface-area-to-volume ratio, which will improve drug release. In addition, there may be no need to overcome any lattice energy barriers that solid-state molecules cause when dissolving due to the drug's amorphous nature (Yu et al., 2018). The fibrous mat shrank and eventually dissolved into a gel-like substance as the ribavirin-loaded fibers began to break down from the edges to the center. Consequently, it is possible to see these drug-loaded fibrous systems as appropriate delivery methods for ribavirin topical administration.
Before administering any medication for therapeutic purposes or as a first step toward biomedical application, it is imperative to evaluate its in vitro cytotoxicity to determine its safety profile and prevent any adverse effects on living tissues. The cytotoxicity profile for both compounds showed that ribavirin is considered safe throughout a 24-hour application at a concentration of ≤ 2,500 µg/mL, while CD is considered safe upon 24- and 48-hour application at a concentration of ≤ 5,000 µg/mL. Since medications provided in a topical dosage form can increase their bioavailability by avoiding first-pass metabolism and entering the bloodstream directly, the results suggest that greater care should be used when administering drugs in this manner. Further evaluation should be performed to confirm these observations and to explore their mechanism. Further in-vitro and in-vivo experiments on the safety of ribavirin-loaded nanofibers should be considered.
In a recent study by Yuan et al, a combination of dimetridazole and ribavirin had no significant effect on the growth of P. aeruginosa PAO1 at concentrations lower than 244.2 μg/mL (Yuan et al., 2022). These results suggest that dimetridazole and ribavirin at routinely used concentrations had poor antibacterial activity against P. aeruginosa PAO1 but might inhibit the quorum sensing (QS) system. Therefore, they further studied the anti-virulence activities of P. aeruginosa, which concluded that ribavirin can significantly inhibit the QS-activated regulatory genes, such as rhIR, lasR, and pqsR, as well as their downstream genes in P. aeruginosa. Moreover, P. aeruginosa PAO1 could express virulence factors, specifically QS, which ribavirin targets to disarm the bacterium (Hernández-Jiménez et al., 2013). The same holds for clinical isolate 7067, which showed a higher MIC of 1250 μg/mL.
A study done by Bonaventura et al tested several antivirals in clinical isolates of P. aeruginosa and reported that ribavirin was the sole medication that could effectively reduce biofilm biomass at lower concentrations (Di Bonaventura et al., 2022) (Di Bonaventura et al., 2022). In comparison, higher concentrations were required to achieve the same effect on planktonic cells (>1024 μg/mL). Furthermore, they found that ribavirin significantly increased the expression of P. aeruginosa clinical isolates virulence genes, including mexA and mexC acting as an efflux pump system, and the most toxic virulence factor exotoxin A – toxA gene. These results indicate that the production of QS system-controlled proteases, pyocyanin, and biofilm formation was significantly reduced in P. aeruginosa in the presence of ribavirin (Di Bonaventura et al., 2022, Yuan et al., 2022).
In another study, ribavirin was shown to antagonize AphB, a LysR-type transcriptional regulator (LTTR) in Vibrio cholerae, which regulates the expression of genes encoding cholera toxin, leading to the inhibition of cholera toxin production and intestinal colonization in animal models (Mandal et al., 2016). Additionally, the same study reported that the suppression of the functions of the Salmonella Typhi LTTR hydrogen peroxide resistance gene (Hrg) by ribavirin led to the inhibition of intracellular survival and systemic infection (Jurado-Martín et al., 2021). It has been found that ribavirin can interfere with pathways that prevent drug cell invasion or adhesion. These findings support the idea that the newly developed wound dressing could potentially treat wound pathogens that express virulence mechanisms, nevertheless, further research is necessary to evaluate the ability of the ribavirin-loaded fiber systems against different bacterial virulence factors.
Ribavirin has also shown a potent antifungal activity both as a free drug and when combined with C, with an MIC of ≤ 156.25 μg/mL. Zhang et al reported a high antifungal activity of ribavirin when used alone or in combination with fluconazole (Zhang et al., 2020). The combination exhibited synergistic effects for all fluconazole-resistant C. albicans strains, with a significant decrease in the MIC from > 512 μg/mL to 0.25–1 μg/mL. Interestingly, fluconazole-susceptible C. albicans strains did not improve against the combination, and the MIC values were between 0.56 and 1.25 μg/mL (Zhang et al., 2020). Another study by Wang et al. tested ribavirin alone and in combination with caspofungin (Wang et al., 2023b). The MIC assay showed that ribavirin and caspofungin had synergistic antifungal effects against fluconazole-susceptible and fluconazole-resistant C. albicans. In the combination group, ribavirin reduced the MIC of caspofungin from 0.5 to 1 μg/mL to 0.0625–0.25 μg/mL. Both studies indicated that this inhibition is associated with the mechanisms of action contributing to the synergistic impact of the antifungal medications targeting C. albicans virulence factors. These include inhibiting the biofilm formation, inhibiting the hyphal growth, activating the metacaspase enzymes, and the accumulation of the reactive oxygen species (Zhang et al., 2020, Talapko et al., 2021 Wang et al., 2023b). Furthermore, in a different study that screened about 100 Candida sp. strains against ribavirin, the MIC of ribavirin was reported as 3.02 to 0.37 μg/mL (Yousfi et al., 2019). These findings provide new insights into using ribavirin as an antifungal agent against skin infections.
Several previous studies demonstrated the antimicrobial ability of the electrospun fibers loaded with different natural and synthetic antimicrobial agents. One recent study by Aung et al., (2024) has successfully prepared and assessed electrospun nanofibers using shellac fiber for loading Senna alata (SA) leaf extract. Their findings showed a diameter of ∼ 300 nm, a high EE for SA of 96 % and the antibacterial effect of these nanofibers against S. aureus, P. aeruginosa, and E. coli. Another study by Krongrawa et al., (2023) utilized shellac fiber by electrospinning to load Kaempferia parviflora (KP) extract. The results exhibited that the diameter of the prepared nanofiber was ∼ 570 nm and the KP was dispersed through the nanofiber, with a sustained drug release rate within 10 h. Additionally, the nanofibers had an antibacterial activity against S. aureus.
Lastly, we suggest that the reason for this lack of antibacterial efficacy of ribavirin discs might be due to an experimental error that would need further investigation. Another interesting result is that there is a substantial variation between both ribavirin-loaded nanofiber systems and ribavirin discs against P. aeruginosa PAO1 strain. This observation may be attributed to the virulence factors expressed by this strain that could have affected the antibacterial activity of the pure ribavirin compared to the molecularly dispersed (electrospun) drug, which can also be an experimental error and would need further investigation. Despite the above issues, the ribavirin-loaded nanofiber systems seemed effective and showed promising results, yet, we do not claim the superiority of both systems to the ribavirin disc. Accordingly, further in-vivo investigation is needed to assess the efficacy of ribavirin as an antimicrobial wound dressing against a suitable animal model.
5. Conclusion
To conclude, this study demonstrated a successful ribavirin-loaded nanofibers fabrication criteria (i.e., lack of the surface pores and beads formation), with average diameters of 494 ± 64 nm and 406 nm ± 88 nm for the PVP fibers and ribavirin-loaded PVP fibers, respectively. The addition of CD enhanced the fibrous diameter to 790 ± 431 nm for the blank PVP/CD fibers and 1,133 ± 234 nm for the ribavirin-loaded PVP/CD fibers, while it did not impact the successful fibers fabrication criteria. The EE% and DL were measured as 102 ± 12 % and 204 ± 24 μg/mg, respectively for the ribavirin-loaded PVP fibers and 117 % ± 15 and 78 ± 10 μg/mg, respectively for the ribavirin-loaded PVP/CD fibers. The drug release demonstrated a burst release of ribavirin from ribavirin-loaded PVP fibers and ribavirin-loaded PVP/CD fibers in the first 10 min and at 1 min, respectively, followed by a complete release after 180 min and 60 min. The cytotoxicity test against HFF-1 cell lines demonstrated a safe ribavirin concentration at 2,500 μg/mL after 24-hour cell exposure but not 48 h, while the CD was considered safe to be used up to 5,000 μg/mL after 24- and 48-hours cell exposure. Both ribavirin-loaded nanofiber systems exhibited a potent antimicrobial efficacy against all tested P. aeruginosa strains, as well as, the C. albicans. This study introduces an innovative method for delivering ribavirin in a nanofibrous system for topical applications, which offers a rapid delivery and quick release of the loaded drug. Further experiments are required for the assessment and validation of the fabricated ribavirin nanofibers in a relevant in-vivo model.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Funding
This study was funded by the Health Sector laboratories’ resources at King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia.
CRediT authorship contribution statement
Khulud A. Alsulami: Formal analysis, Investigation, Project administration, Writing – original draft. Abrar A. Bakr: Formal analysis, Investigation, Methodology, Writing – original draft. Abdullah A. Alshehri: Methodology, Supervision, Writing – review & editing. Alhassan H. Aodah: Methodology, Supervision, Writing – review & editing. Fahad A. Almughem: Formal analysis, Visualization, Writing – review & editing. Ali A. Alamer: Investigation, Methodology. Lujain A. Alharbi: Investigation. Deema S. Alsuwayeh: Investigation. Abdulrahman A. Halwani: Investigation, Writing – original draft. Abdullah A. Alamoudi: Methodology, Supervision, Writing – review & editing. Haya A. Alfassam: Conceptualization, Methodology, Supervision. Essam A. Tawfik: Conceptualization, Resources, Supervision, Visualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Haya A. Alfassam, Email: halfassam@kacst.gov.sa.
Essam A. Tawfik, Email: etawfik@kacst.gov.sa.
References
- Aburayan W.S., Booq R.Y., BinSaleh N.S., et al. The delivery of the novel drug ‘halicin’ using electrospun fibers for the treatment of pressure ulcer against pathogenic bacteria. Pharmaceutics. 2020;12:1189. doi: 10.3390/pharmaceutics12121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T.A., Almehmady A.M., Alharbi W.S., et al. Incorporation of perillyl alcohol into lipid-based nanocarriers enhances the antiproliferative activity in malignant glioma cells. Biomedicines. 2023;11:2771. doi: 10.3390/biomedicines11102771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamer A.A., Alsaleh N.B., Aodah A.H., et al. Development of imeglimin electrospun nanofibers as a potential buccal antidiabetic therapeutic approach. Pharmaceutics. 2023;15:1208. doi: 10.3390/pharmaceutics15041208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkahtani M.E., Aodah A.H., Abu Asab O.A., et al. Fabrication and characterization of fast-dissolving films containing escitalopram/quetiapine for the treatment of major depressive disorder. Pharmaceutics. 2021;13:891. doi: 10.3390/pharmaceutics13060891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshaya H.A., Alfahad A.J., Alsulaihem F.M., et al. Fast-dissolving nifedipine and atorvastatin calcium electrospun nanofibers as a potential buccal delivery system. Pharmaceutics. 2022;14:358. doi: 10.3390/pharmaceutics14020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung W.W., Krongrawa W., Limmatvapirat S., et al. Fabrication and optimization of electrospun shellac fibers loaded with Senna alata leaf extract. Polymers. 2024;16:183. doi: 10.3390/polym16020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczar D., Michalska K. Cyclodextrin inclusion complexes with antibiotics and antibacterial agents as drug-delivery systems—A pharmaceutical perspective. Pharmaceutics. 2022;14:1389. doi: 10.3390/pharmaceutics14071389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhary H., Williams G.R., Orlu M. Electrospun fixed dose formulations of amlodipine besylate and valsartan. Int. J. Pharm. 2018;549:446–455. doi: 10.1016/j.ijpharm.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- Callewaert C., Ravard Helffer K., Lebaron P. Skin microbiome and its interplay with the environment. Am. J. Clin. Dermatol. 2020;21:4–11. doi: 10.1007/s40257-020-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celebioglu A., Uyar T. Development of ferulic acid/cyclodextrin inclusion complex nanofibers for fast-dissolving drug delivery system. Int. J. Pharm. 2020;584 doi: 10.1016/j.ijpharm.2020.119395. [DOI] [PubMed] [Google Scholar]
- Chen M., Qu H., Zhu J., et al. Magnetic electrospun fluorescent polyvinylpyrrolidone nanocomposite fibers. Polymer. 2012;53:4501–4511. doi: 10.1016/j.polymer.2012.07.046. [DOI] [Google Scholar]
- Chuangchote S., Sagawa T., Yoshikawa S. Electrospinning of poly(vinyl pyrrolidone): effects of solvents on electrospinnability for the fabrication of poly(p-phenylene vinylene) and TiO2 nanofibers. J. Appl. Polym. Sci. 2009;114:2777–2791. doi: 10.1002/app.30637. [DOI] [Google Scholar]
- D'Avolio A., Ibañez A., Sciandra M., et al. Validation of liquid/liquid extraction method coupled with HPLC-UV for measurement of ribavirin plasma levels in HCV-positive patients. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006;835:127–130. doi: 10.1016/j.jchromb.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Di Bonaventura G., Lupetti V., De Fabritiis S., et al. Giving drugs a second chance: antibacterial and antibiofilm effects of ciclopirox and ribavirin against cystic fibrosis Pseudomonas aeruginosa strains. Int. J. Mol. Sci. 2022;23:5029. doi: 10.3390/ijms23095029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K., Grice E.A. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. 2014;10:e1004436. doi: 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V., Sauvage V., Hebert C., et al. Human skin Microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Kong H.H., Conlan S., et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha E.S., Choo G.H., Baek I.H., Kim M.S. Formulation, characterization, and in vivo evaluation of celecoxib-PVP solid dispersion nanoparticles using supercritical antisolvent process. Molecules. 2014;19:20325–20339. doi: 10.3390/molecules191220325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggag R.S., Belal S.F., Hewala I.I., El Rouby O.A. Stability-indicating HPLC-DAD determination of ribavirin in capsules and plasma. J. Chromatogr. Sci. 2014;52:493–500. doi: 10.1093/chromsci/bmt067. [DOI] [PubMed] [Google Scholar]
- Harrison O.J., Linehan J.L., Shih H.-Y., et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363:eaat6280. doi: 10.1126/science.aat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Chen M., Dou Y., et al. Electrospun silver Nanoparticles-embedded feather keratin/poly(vinyl alcohol)/poly(ethylene oxide) antibacterial composite nanofibers. Polymers. 2020;12:305. doi: 10.3390/polym12020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Jiménez E., del Campo R., Toledano V., et al. Biofilm vs. planktonic bacterial mode of growth: which do human macrophages prefer? Biochem. Biophys. Res. Commun. 2013;441:947–952. doi: 10.1016/j.bbrc.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Homma M., Matsuzaki Y., et al. Liquid chromatography assay for routine monitoring of cellular ribavirin levels in blood. Antimicrob. Agents Chemother. 2004;48:3813–3816. doi: 10.1128/aac.48.10.3813-3816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Duffy B., Jaiswal A.K., et al. Enhancement of the antibacterial properties of silver nanoparticles using β-cyclodextrin as a capping agent. Int. J. Antimicrob. Agents. 2010;36:280–283. doi: 10.1016/j.ijantimicag.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Jarallah S.J., Aldossary A.M., Tawfik E.A., et al. GL67 lipid-based liposomal formulation for efficient siRNA delivery into human lung cancer cells. Saudi Pharm J. 2023;31:1139–1148. doi: 10.1016/j.jsps.2023.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado-Martín I., Sainz-Mejías M., McClean S. Pseudomonas aeruginosa: an audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021;22:3128. doi: 10.3390/ijms22063128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krongrawa W., Limmatvapirat S., Vollrath M.K., et al. Fabrication, optimization, and characterization of antibacterial electrospun shellac fibers loaded with Kaempferia parviflora Extract. Pharmaceutics. 2023;15:123. doi: 10.3390/pharmaceutics15010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakula M., Rao G. Pharmaceutical assessment of polyvinylpyrrolidone (PVP): as excipient from conventional to controlled delivery systems with a spotlight on COVID-19 inhibition. J. Drug Deliv. Sci. Technol. 2020;60 doi: 10.1016/j.jddst.2020.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chen S., Hu H., et al. Crystallization selectivity of ribavirin solution and amorphous phase. Molecules. 2023;28:6320. doi: 10.3390/molecules28176320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louisa R.B., Charne N.M., Richard J.S., et al. The pH of wounds during healing and infection: a descriptive literature review. Wound Practice Res. 2017:25. [Google Scholar]
- Mandal R.S., Ta A., Sinha R., et al. Ribavirin suppresses bacterial virulence by targeting LysR-type transcriptional regulators. Sci. Rep. 2016;6:39454. doi: 10.1038/srep39454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moydeen A.M., Padusha M.S.A., Thamer B.M., et al. Single-nozzle core-shell electrospun nanofibers of PVP/Dextran as drug delivery system. Fibers Polym. 2019;20:2078–2089. doi: 10.1007/s12221-019-9187-2. [DOI] [Google Scholar]
- Nair B. Final report on the safety assessment of polyvinylpyrrolidone (PVP) Int. J. Toxicol. 1998;17:95–130. doi: 10.1177/109158189801700408. [DOI] [Google Scholar]
- Nyairo W.N., Eker Y.R., Kowenje C., et al. Efficient removal of lead(II) ions from aqueous solutions using methyl-β-cyclodextrin modified graphene oxide. Water Air Soil Pollut. 2017;228:406. doi: 10.1007/s11270-017-3589-9. [DOI] [Google Scholar]
- Picot-Allain C., Mahomoodally M.F., Ak G., Zengin G. Conventional versus green extraction techniques — a comparative perspective. Curr. Opin. Food Sci. 2021;40:144–156. doi: 10.1016/j.cofs.2021.02.009. [DOI] [Google Scholar]
- Pinho E., Grootveld M., Soares G., Henriques M. Cyclodextrin-based hydrogels toward improved wound dressings. Crit. Rev. Biotechnol. 2014;34:328–337. doi: 10.3109/07388551.2013.794413. [DOI] [PubMed] [Google Scholar]
- Ponphaiboon J., Krongrawa W., Aung W.W., et al. Advances in natural product extraction techniques, electrospun fiber fabrication, and the integration of experimental design: a comprehensive review. Molecules. 2023;28:5163. doi: 10.3390/molecules28135163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A., Figueiras A., Santos D., Veiga F. Preparation and solid-state characterization of inclusion complexes formed between miconazole and methyl-β-cyclodextrin. AAPS PharmSciTech. 2008;9:1102–1109. doi: 10.1208/s12249-008-9143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio B.V.F., Nascimento A.L.C.S.D., Meneguin A.B., et al. Solid dispersions incorporated into PVP films for the controlled release of trans-resveratrol: development, physicochemical and in vitro characterizations and in vivo cutaneous anti-inflammatory evaluation. Pharmaceutics. 2022;14:1149. doi: 10.3390/pharmaceutics14061149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwana B., Fathima, Muthu S., Prasana J.C., et al. Spectroscopic (FT-IR, FT-raman) investigation, topology (ESP, ELF, LOL) analyses, charge transfer excitation and molecular docking (dengue, HCV) studies on ribavirin. Chem. Data Collect. 2018;17–18:236–250. doi: 10.1016/j.cdc.2018.09.003. [DOI] [Google Scholar]
- Sachdeva C., Satyamoorthy K., Murali T.S. Microbial interplay in skin and chronic wounds. Current Clin. Microbiol. Reports. 2022;9:21–31. doi: 10.1007/s40588-022-00180-4. [DOI] [Google Scholar]
- Schommer N.N., Gallo R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21:660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen C.K. Human wound and its burden: updated 2020 compendium of estimates. Adv. Wound Care. 2021;10:281–292. doi: 10.1089/wound.2021.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.E., Rimmer S., MacNeil S. Examination of the effects of poly(N-vinylpyrrolidinone) hydrogels in direct and indirect contact with cells. Biomaterials. 2006;27:2806–2812. doi: 10.1016/j.biomaterials.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Snell N.J.C. Ribavirin - current status of a broad spectrum antiviral agent. Expert Opin. Pharmacother. 2001;2:1317–1324. doi: 10.1517/14656566.2.8.1317. [DOI] [PubMed] [Google Scholar]
- Sriyanti I., Edikresnha D., Rahma A., et al. Mangosteen pericarp extract embedded in electrospun PVP nanofiber mats: physicochemical properties and release mechanism of α-mangostin. Int. J. Nanomed. 2018;4927–4941 doi: 10.2147/IJN.S167670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talapko J., Juzbašić M., Matijević T., et al. Candida albicans—The virulence factors and clinical manifestations of infection. J. Fungi. 2021;7:79. doi: 10.3390/jof7020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik E.A., Craig D.Q.M., Barker S.A. Dual drug-loaded coaxial nanofibers for the treatment of corneal abrasion. Int. J. Pharm. 2020;581 doi: 10.1016/j.ijpharm.2020.119296. [DOI] [PubMed] [Google Scholar]
- Tawfik E.A., Alshamsan A., Abul Kalam M., et al. In vitro and in vivo biological assessment of dual drug-loaded coaxial nanofibers for the treatment of corneal abrasion. Int. J. Pharm. 2021;604 doi: 10.1016/j.ijpharm.2021.120732. [DOI] [PubMed] [Google Scholar]
- Tawfik E.A., Scarpa M., Abdelhakim H.E., et al. A potential alternative orodispersible formulation to Prednisolone sodium phosphate orally disintegrating tablets. Pharmaceutics. 2021;13:120. doi: 10.3390/pharmaceutics13010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodorescu M., Bercea M. Poly(vinylpyrrolidone) – a versatile polymer for biomedical and beyond medical applications. Polym.-Plast. Technol. Eng. 2015;54:923–943. doi: 10.1080/03602559.2014.979506. [DOI] [Google Scholar]
- Topuz F., Kilic M.E., Durgun E., Szekely G. Fast-dissolving antibacterial nanofibers of cyclodextrin/antibiotic inclusion complexes for oral drug delivery. J. Colloid Interface Sci. 2021;585:184–194. doi: 10.1016/j.jcis.2020.11.072. [DOI] [PubMed] [Google Scholar]
- Tottoli E.M., Dorati R., Genta I., et al. Skin wound healing process and new emerging technologies for skin wound care and regeneration. Pharmaceutics. 2020;12:735. doi: 10.3390/pharmaceutics12080735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., He L., Ding L., et al. Fabrication of cyclodextrin-based hydrogels for wound healing: progress, limitations, and prospects. Chem. Mater. 2023;35(15):5723–5743. doi: 10.1021/acs.chemmater.3c00926. [DOI] [Google Scholar]
- Wang Y., Yan H., Li J., et al. Antifungal activity and potential mechanism of action of caspofungin in combination with ribavirin against Candida albicans. Int. J. Antimicrob. Agents. 2023;61 doi: 10.1016/j.ijantimicag.2023.106709. [DOI] [PubMed] [Google Scholar]
- Wayne P. 10th ed. Clinical and Laboratory Standards Institute CLSI; 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- Wessel W., Schoog M., Winkler E. Polyvinylpyrrolidone (PVP), its diagnostic, therapeutic and technical application and consequences thereof. Arzneimittelforschung. 1971;21:1468–1482. [PubMed] [Google Scholar]
- Williams G.R., Raimi-Abraham B.T., Luo C. UCL Press; 2018. Nanofibres in drug delivery. [Google Scholar]
- Wolcott R.D., Hanson J.D., Rees E.J., et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016;24:163–174. doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- Yao Q., Shi Y., Xia X., et al. Bioadhesive hydrogel comprising bilirubin/β-cyclodextrin inclusion complexes promote diabetic wound healing. Pharm. Biol. 2021;59:1137–1147. doi: 10.1080/13880209.2021.1964543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousfi H., Cassagne C., Ranque S., et al. Repurposing of ribavirin as an adjunct therapy against invasive <i>Candida</i> strains in an <i>In Vitro</i> study. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00263-19. 10.1128/aac.00263-00219. doi:10.1128/aac.00263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.-G., Li J.-J., Williams G.R., Zhao M. Electrospun amorphous solid dispersions of poorly water-soluble drugs: a review. J. Control. Release. 2018;292:91–110. doi: 10.1016/j.jconrel.2018.08.016. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Yang X., Zeng Q., et al. Repurposing dimetridazole and ribavirin to disarm Pseudomonas aeruginosa virulence by targeting the quorum sensing system. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.978502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yan H., Lu M., et al. Antifungal activity of ribavirin used alone or in combination with fluconazole against Candida albicans is mediated by reduced virulence. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Zhao L., Orlu M., Williams G.R. Electrospun fixed dose combination fibers for the treatment of cardiovascular disease. Int. J. Pharm. 2021;599 doi: 10.1016/j.ijpharm.2021.120426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.