Abstract
Since the Simplified Molecular Input Line Entry System (SMILES) is oriented to the atomic-level representation of molecules and is not friendly in terms of human readability and editable, however, IUPAC is the closest to natural language and is very friendly in terms of human-oriented readability and performing molecular editing, we can manipulate IUPAC to generate corresponding new molecules and produce programming-friendly molecular forms of SMILES. In addition, antiviral drug design, especially analogue-based drug design, is also more appropriate to edit and design directly from the functional group level of IUPAC than from the atomic level of SMILES, since designing analogues involves altering the R group only, which is closer to the knowledge-based molecular design of a chemist. Herein, we present a novel data-driven self-supervised pretraining generative model called “TransAntivirus” to make select-and-replace edits and convert organic molecules into the desired properties for design of antiviral candidate analogues. The results indicated that TransAntivirus is significantly superior to the control models in terms of novelty, validity, uniqueness, and diversity. TransAntivirus showed excellent performance in the design and optimization of nucleoside and non-nucleoside analogues by chemical space analysis and property prediction analysis. Furthermore, to validate the applicability of TransAntivirus in the design of antiviral drugs, we conducted two case studies on the design of nucleoside analogues and non-nucleoside analogues and screened four candidate lead compounds against anticoronavirus disease (COVID-19). Finally, we recommend this framework for accelerating antiviral drug discovery.
Introduction
Greater understanding of viral life cycles, prompted, in particular, by the need to combat the human immunodeficiency virus, has resulted in the discovery and validation of several targets for therapeutic intervention. Consequently, the current antiviral repertoire now includes more than 90 drugs.1 But we still lack effective therapies for several viral infections, and established treatments are not always effective or well-tolerated, highlighting the need for further refinement of antiviral drug design and development. Here, Erik De Clercq describes the rationale behind current and future drug-based strategies for combating viral infections.2 For the clinical use of the antiviral drugs, you can read the literature.3 This review address currently used antiviral drugs, mechanism of action, and antiviral agents reported against COVID-19.4
The global pandemic substantiated by coronavirus disease 2019 (COVID-19) is an unexpected public health crisis that demands the timely development of new therapeutics and viral detection platforms.5 Merck’s Molnupiravir (inducing viral error catastrophe) has once again entered the spotlight in the same way as Remdesivir (a coronavirus RNA polymerase inhibitor), but with a slightly different mechanism of action, as Molnupiravir mainly causes genetic mutations to exert antiviral effects.6 To date, only one oral drug, Paxlovid from Pfizer, has been approved for marketing in China for coronavirus. Paxlovid is a 3CL protease inhibitor and hence differs from Molnupiravir which is an RNA polymerase inhibitor. Molnupiravir binds to RNA polymerase of the progeny coronavirus and incorporates a wrong nucleotide into the newly synthesized RNA molecule, thus masking the survival capability of offspring viruses.
In the recent past, a small amount of research has been committed to identifying nucleoside analogues for the development of novel and efficacious antiviral drugs. As the viruses are prone to hypermutation and breed resistant variants, the development of nucleoside analogue drugs holds great promise. Conclusively, nucleoside analogues substantiate a pharmacological class of compounds with cytotoxic, immunosuppressive, and antiviral properties.
To classify antiviral drugs based on their types of mechanism of action, we consider two types of antiviral analogs: nucleoside analogues and non-nucleoside analogues. There are several nucleoside analogues and non-nucleoside analogues as antiviral drugs.7
The last decade has witnessed successful applications of a diverse array of novel drug development methods including expert manual design, computer-aided drug design (CADD), and quantitative structure–activity relationships (QSARs).8 With the emergence of deep learning and the incorporation of the new paradigm of artificial intelligence (AI) in science, there are overwhelmingly animated fascinating discoveries. Extensive research has primarily focused on the deep generation model and indeed accomplished astonishing results.9 De novo molecular generation models contain several different branches, graph-based model10 and transcriptome-based model,11 antibody-based design method,12 protein–protein interaction-based model,13 molecular substructure tree generative model,14 targeting RNA for small molecule drug design,15 three-dimensional (3D) equivariant diffusion for target-aware molecule generation,16 reinforcement learning (RL) tunes pretrained networks to generate molecules with user-defined properties,17 multitarget RL has also been incorporated to optimize drug similarity and molecular similarity,18 etc. According to the published literature, many variants of the transformer model have obtained enormous success in the natural language processing (NLP) discipline, such as BERT,19 GPT,20 T5,21 DETR,22 ViT,23 NAT,24 and Wav2Vec2.25 The codes of all these models can be found in ref (26). IUPAC names and SMILES strings can be employed as a type of language and can be used as inputs for different models. For instance, LSTM,27 cGAN,28 MolGPT,29 C5T5,30 and smiles-gpt.31
Notably, numerous evidence manifested that conditional and interpretable generative models can generate molecules that successfully meet our needs better than generic molecular generative models on a specific target.32
IUPAC Representation and SMILES Representation
The future of chemistry is language.33
The IUPAC (International Union of Pure and Applied Chemistry) nomenclature is a globally recognized unique naming system that assigns names to chemical compounds. The SMILES (Simplified Molecular Input Line Entry System) is another naming system, which allocates symbolic representation to compounds, known as SMILES strings. IUPAC harmonized chemical names globally and established the nomenclature of organic chemistry that documented instruction on the unambiguous names for all compounds.34 Furthermore, we can clearly observe that the IUPAC name is close to natural language, an abstract representation and easy handling for human, consisting of English words, numbers, etc., while the SMILES string is a combination of chemical elements, low-level representation, hard to read but programming friendly, consisting of atomic symbols, bonds, etc.
Antiviral drug design, especially analogue-based drug design, is also more appropriate to edit and design directly from the functional group level of IUPAC than from the atomic level of SMILES, since designing analogues involves altering the R group only, which is closer to the knowledge-based molecular design of a chemist.
In this study, considering the advantage of the human-based IUPAC name and computer-based SMILES strings, we integrate them to the transformer-based model. We propose a novel transformer-based molecular generative model (TransAntivirus) for the design of antiviral lead compounds. First, we perform pretraining on the constructed training set. Thereafter, the two antiviral tasks as the downstream task will be fine-tuning, and finally, 30 000 molecules will be generated for model evaluation.
Although several models exist to establish a mapping relationship between the IUPAC name and SMILES string, molecular optimization for specific molecular properties with the IUPAC name as input and the SMILES string as output has not been explored yet. Hence, our model is the first exploration of the relationship between the chemical semantics and molecular structure for the molecular design and optimization by editing the IUPAC name and adding different prefixes, especially for the design and optimization of antiviral drug. In addition, the data set for our fine-tuning was collected specifically for antiviral drug design, and although our approach is general, from the perspective of application, the results demonstrate that our model can be used for antiviral drug discovery and generates several potential candidates for anticoronavirus disease (COVID-19) drug design that can go for further experimental validation or provide reference compounds.
Data and Methods
Data Preparation
The SMILES strings and IUPAC name of all molecules were downloaded from PubChem.35 They were filtered using a series of criteria: (i) filtering out the SMILES strings containing disconnected ions or fragments; (ii) compound standardization by RDKit36 for the removal of salts and isotopes as well as charge neutralization. Upon filtration, a total of 106 459 817 molecules were retained. Subsequently, we calculated the essential properties of these molecules including molecular weight (MolWt), partition coefficient (LogP),37 synthetic accessibility score (SAscore),38 rotatable bonds (ROTB), hydrogen bond donor (HBD), hydrogen bond acceptor (HBA), topological polar surface area (TPSA), and quantitative estimate of drug-likeness (QED)39 by RDKit tool. The filtration rules were parametrized as follows.
-
(a)
100 ≤ MolWt ≤ 900
-
(b)
–5 ≤ LogP < 8
-
(c)
SAscore <4
-
(d)
ROTB < 10
-
(e)
HBD < 5
-
(f)
HBA < 10
-
(g)
TPSA < 150
-
(h)
QED ≥ 0.3
Right after the filtration, 79 156 024 molecules were retained. Finally, we sorted out 30 million molecules as the training set, and every molecule contained the IUPAC name, SMILES string, and LogP value.
The fine-tuning data set was divided into two components: nucleoside analogues and non-nucleoside analogues. For nucleoside analogues, we collected SIMILES strings from the literature40 and retreived their IUPAC names from the PubChem database. Afterward, their properties were calculated by RDKit. For non-nucleoside analogues, we took non-nucleoside analogues as our fine-tuned data set and collected SMILES strings from this paper,41 while the postprocessing remained the same as for nucleoside analogues.
In this study, we first applied the Tokenization procedure on the sequence. Consequently, we assigned a character-based SMILES tokenization with tokens in SMILES strings corresponding to individual atoms and bonds. In contrast, we constructed a rule-based IUPAC tokenizer with tokens in the IUPAC names analogues to well-known functional groups and moieties. In order to understand the SMILES and IUPAC tokenizers in depth, the readers are referred to these references.30,34,40,41
For each molecule, a particular property is usually a continuous value; thus, we should convert the continuous value of a property to its discrete values, for example, high, middle, and low. Herein, we choose LogP as our manageable property, where −0.4 and 5.6 as two breakpoints are used to delineate three intervals. For each molecule, a token (one either of <high, middle, low>) will be appended in the front of the sequence.
After sequence tokenization and appending the discretized property value, we encoded all the tokens as the incremental integer sequence from 3, specifically, 0 as padding mark, 1 as start mark, and 2 as the end mark. Notably, the start mark is the encoded integer of the discretized property value in the IUPAC sequence, but in the SMILES sequence, the start mark is 1, and the end mark and padding marks are the same in both the IUPAC sequence and SMILES sequence.
In this section, we introduce our model and the improvement from three aspects. Conclusively, an end-to-end learning-based prediction model is proposed to solve the molecular generation and optimization problem.
Overall, our encoder-decoder transformer implementation closely follows that of its originally proposed platform. We can conclude two critical points from the T5 model. At first, the T5 model is trained with a maximum likelihood objective regardless of the task.21 On the other hand, we note that the choice of text prefix used for a given task is essentially a hyperparameter.
Inspired by the two aforementioned points, we developed our own property-controlled molecular generative model. First, we add a prefix to the head of each input as a property-controlled condition and prepend the resulting sequence with a token indicating the computed property value of the original molecule. To obtain these property value tokens, we discretized the distribution of the property values into three buckets. Octanol–water partition coefficient (LogP); low: (−∞,–0.4), middle: (−0.4,5.6), and high: (5.6,∞). The second end point is to get the output of maximum likelihood objective as IUPAC embedding representation.
Model Architecture
IUPAC-Based T5 Model
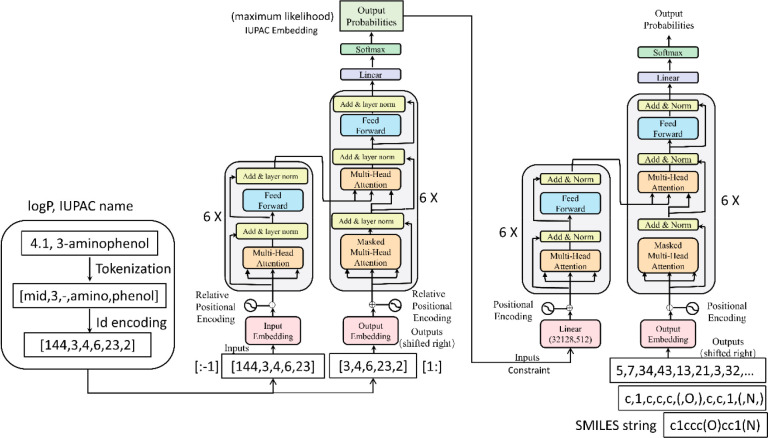
T5 model implementation closely follows its originally (Transformer) proposed form. First, an input sequence of tokens is mapped to a sequence of embedding that is then passed into the encoder. The encoder consists of a stack of “blocks”, each of which comprises two subcomponents: a self-attention layer followed by a small feed-forward network. Layer normalization is applied to the input of each subcomponent. T5 uses a simplified version of layer normalization where the activations are only rescaled and no additive bias is applied. After layer normalization, a residual skip connection adds the input of each subcomponent to its output. Dropout is applied within the feed-forward network, on the skip connection, on the attention weights, and at the input and output of the entire stack. The decoder is similar in structure to the encoder, except that it includes a standard attention mechanism after each self-attention layer that attends to the output of the encoder. The self-attention mechanism in the decoder also uses a form of autoregressive or causal self-attention, which allows the model to attend only to past outputs. The output of the final decoder block is fed into a dense layer with a softmax output whose weights are shared with the input embedding matrix. All attention mechanisms in the Transformer are split up into independent “heads” whose outputs are concatenated before being further processed.
T5 uses a simplified form of position embedding where each “embedding” is simply a scalar that is added to the corresponding logit used for computing the attention weights. To summarize, T5 is roughly equivalent to the original Transformer with the exception of removing the Layer Norm bias, placing the layer normalization outside the residual path, and using a different position embedding scheme.21
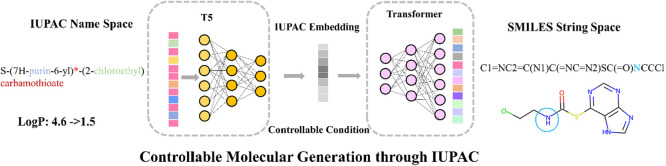
In contrast to the idea implemented in T5, such as C5T5,30 we mainly fed the output (maximum likelihood objective) of T5 into next transformer model, as input to the next step of learning to map brand-new intersequence relationships. Since the whole framework is joined together for training, instead of training the T5 model first and then another Transformer model, the output of T5 (the first part) in our framework is an IUPAC embedding vector considered as a constraint, which is not transformed into an IUPAC sequence but directly used as input vector for the next module, as detailed in Figure 2.
Figure 2.
Efficient and enhanced sampling of molecular chemical space for virtual screening and molecular design using TransAntivirus model.
But there is a question: the vocabulary size is 32 128 in T5, IUPAC: 1491, SMILES: 870; thus, the size of IUPAC’s encoding vocabulary in T5 is different from the next transformer model. To accommodate the size of the next encoded dictionary table, a linear layer was added as an adaptive layer. Therefore, we added a linear layer (32 128, 512) before entering into an Embedding layer of Transformer, and dropped the linear layer (1491, 512) in the original transformer.42
Similar to the IUPAC2Struct model,34 our model also focuses on learning the chemical semantic relationship between the IUPAC name and SMILES. As we know, the IUPAC name and SMILES are two different levels of language coding systems. The former is close to our abstract natural language, which is composed of some chemical terms, numbers, and special symbols. Nevertheless, the latter consists of atomic symbols, bonds, and special symbols. We can first obtain the embedding vector representation of tokens at the natural language semantic level through the IUPAC predictor and then use their contextual information to encode and guide the vector representation of the context relationship of low-level semantic tokens in the SMILES predictor. Therefore, by learning the embedding vector representation space of IUPAC, we can guide and expand the space of SMILES molecule generation, combining randomness and controllability to achieve the ability for molecule generation and optimization under specific conditions.
In the IUPAC2Struct model, only the mapping relationship between the two encoding systems is considered, and the relationship within the sequence is not explicitly modeled. Therefore, for the IUPAC2Struct model, we can only assume that it is a language translation system from IUPAC to SMILES and not a molecular generative model.
In contrast to C5T5, which learns only within one coding language system, IUPAC or SMILES, our model directly learns the mapping relationship between the two language systems, IUPAC and SMILES.
In summary, we used a pair of encoders and decoders to extract the interrelationships within the IUPAC names. Then, we use another pair of encoders and decoders to learn the mapping relationships between the IUPAC representation and SMILES strings. The former can simultaneously capture the contextual semantic relationships inside the chemical language, and the latter can learn the translation relationships and expand the space of SMILES molecule generation.
To prove that it is supposed to lead to improved performance, we compared the TransAntivirus model with the C5T5 model in the Results and Discussion section. We concluded that TransAntivirus presented an optimal performance in terms of the generated molecular distribution, wherein a possible explanation is the introduction of the additional IUPAC embedding space for learning semantic relationships within the SMILES strings through a self-attention mechanism.
End-to-End Learning Models
It is worth mentioning that the TransAntivirus prediction model is integrated into a unified end-to-end neural network learning framework (Figure 1). At first, TransAntivirus is leveraged to learn the IUPAC name internal relationship over a property-controlled deformed transformer model and pretraining and fine-tuned training patterns, respectively. Second, the original transformer prediction model receives the first part of TransAntivirus’s output (maximum Likelihood) to employ further nonlinear transformations. The final predictions are obtained through the decoding of the transformed softmax vector. All parameters Winvolved in TransAntivirus prediction model are simultaneously optimized via a gradient descent with adaptive moment estimation.43 The Figure 2 modified from the original transformer illustrates our proposed framework.
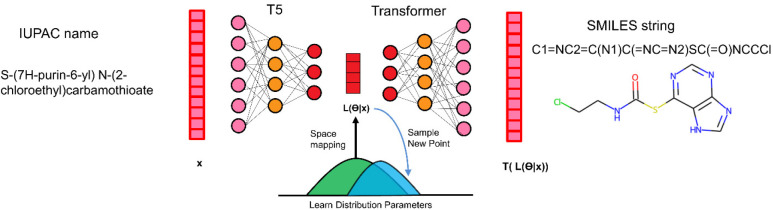
Figure 1.
Framework of TransAntivirus model.
The proposed model is mainly composed of two components.
-
(1)
IUPAC embedding representation: operates directly on IUPAC name that intuitively encodes rich chemical semantics for organic chemists and performs transfer learning with a unified text-to-text transformer to leverage their intuitions about chemical knowledge.
-
(2)
IUPAC2SMILES generator: yields IUPAC-SMILES association relationship with the learned latent representations from first component as input to enable the controllable SMILES string generation and optimization.
Experimental Setup
The implemented experimental code is based on the open-source machine learning framework “Pytorch” (https://pytorch.org). The employed variant transformer models are based on the open-source deep learning platform “Hugging Face library” (https://huggingface.co/t5-base). All experiments were performed on Windows 10 operating system, an Ubuntu 20.04.4 LTS operating system of an Intel(R) Xeon(R) Gold 6226R CPU 32 cores, 2.9 GHz CPU and 128G memory, and a single A40 GPU, 24G memory.
Model Evaluation
In this study, we randomly split a preprocessed data set into two subsets, 30 million molecules for pretraining, and 50 000 molecules as the test data set. The batch size is 64 in both training and evaluation steps, and 10 epochs are executed during each period. The whole pretraining and evaluation processes elapsed 20 days, which is almost 2 days per epoch.
After training and fine-tuning, the model was applied to generate molecules for the assessment of their Fraction of valid (Valid), Uniqueness (Unique@k), Novelty, Frechet ChemNet Distance (FCD),44 Internal diversity (IntDiv),45 QED, LogP, and sample spatial distribution.
Docking Protocol and MM-GBSA Energy Calculation
The three-dimensional (3D) structure of the receptor is subjected to the protein preparation and docking modules of the Schrödinger Release 2022-1 version. The preparation involved the assignment of the hydrogen bonds, bond orders, addition of hydrogen optimization by OPLS4 force field, minimization of the proteins, and deletion of water molecules beyond 5 Å of the Het group in the complex.46 Using the Glide application, a protein receptor grid was generated (allocation of ligand binding site for docking). Additionally, docking of all ligands was carried out using Glide’s Ligand Docking module.47
The Prime MM-GBSA module of the Schrödinger Tool calculates the energy of optimized free receptors, free ligand, and a complex of the ligand with a receptor.48 It also calculates the ligand strain energy by placing the ligand in a solution that was autogenerated by the VSGB 2.0 suite. The prime MMGBSA method computes the relative binding-free energy (ΔG binding) of each ligand molecule by the following equation
| 1 |
where ΔG(solv) is the difference between the GBSA solvation energy of the receptor-inhibitor complex and the sum of the solvation energies for the unliganded receptor and inhibitor. ΔE(MM) is the difference of molecular mechanics energy between the receptor-inhibitor complex and the sum of the molecular mechanics energies of the unliganded receptor and inhibitor. ΔG(SA) is a difference in surface area energies of the complex and the sum of the surface area energies for the unliganded receptor and inhibitor.
Results and Discussion
Herein, we present the training and fine-tuning process and then perform analysis of the model performance, analysis of the property distribution of the generated molecules, chemical space analysis, and property optimization analysis. Furthermore, to demonstrate the application of our model for antiviral drug design, we provide two case studies for targets of two types of antiviral drugs (nucleoside analogues and non-nucleoside analogues). Finally, we perform molecular docking, MM-GBSA energy, and virtual screening of novel lead compounds as candidate antiviral drugs.
Analysis of Model Performance
To evaluate the efficiency of TransAntivirus over existing models, we calculated the same metrics from generated molecules and the original molecules by MOSES platform.45 The experimentally compared results of our proposed model and the other three generative models (LSTM, cGAN, and C5T5) have been summarized in Table 1.
Table 1. Properties of the Generated Compounds for Three Generative Models: Properties Include Numbers of Valid, Unique, and Novelty Compounds; FCD, Fréchet ChemNet Distance.
| Model | Valid | Unique@1000 | Unique@10000 | FCD | IntDiv | Novelty |
|---|---|---|---|---|---|---|
| LSTM | 0.9954 | 0.9944 | 0.9956 | 5.6139 | 0.8558 | 0.9857 |
| cGAN | 0.9926 | 0.9911 | 0.9927 | 4.8349 | 0.8819 | 0.9807 |
| C5T5 | 0.9913 | 0.9925 | 0.9934 | 9.1964 | 0.8939 | 0.9890 |
| TransAntivirus | 0.9998 | 0.9991 | 0.9989 | 10.9471 | 0.8953 | 0.9993 |
Herein, each metric depends on the generated set and reference (training) set. We have rounded each number to four decimal places, where the Valid and Unique@1k and Unique@10k indicate that the molecular generated by TransAntivirus is better than LSTM, cGAN, and C5T5 in terms of the validation and uniqueness. In Table 1, the FCD, IntDiv, and Novelty indicate that TransAntivirus is preferentially better than other models. Relative to C5T5, our model performs the better novelty, FCD, and diversity by introducing the new framework, especially regarding the maximum likelihood of latent variant space and bridging gap between the IUPAC name and SMILES strings. It indicates that TransAntivirus could be improved by learning the inter- and inner-sequences relationship.
Based on the above results, it can be confirmed that TransAntivirus is significantly superior to the control methods in terms of novelty, validity, uniqueness, FCD, and IntDiv.
Comparison of Chemical Properties Distribution
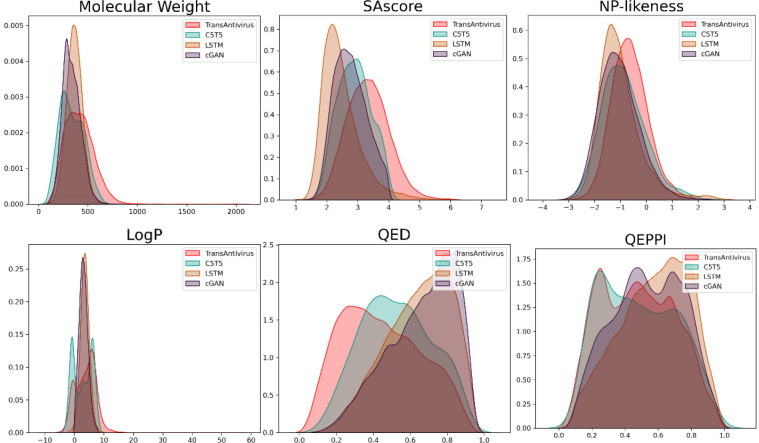
Chemical property distribution is an efficient presentation for visually evaluating the generative model. To gain further insight into the performance of TransAntivirus, we conducted extensive experiments to systematically compare the properties distribution of the molecules generated by LSTM, cGAN, C5T5, and TransAntivirus.
We randomly selected 30 000 compounds from train data set and input them into the MOSES platform.45 After training and generating 30 000 molecules, we calculated these properties including molecular weight, natural products likeness (NP-likeness), LogP, SAscore, QED, and quantitative estimate of protein–protein interaction targeting drug-likeness (QEPPI).13,49
It is noteworthy that both C5T5 and our model are conditional generation models. It means that the properties distribution of generated molecules is decided to be the input sequence and the condition set; instead, the other two models can sample without input. Thus, here, we need to focus on these two conditional models for how to generate the novel molecules. To be fair, we fed the same conditions and same sequences into C5T5 and TransAntivirus, then generated 10 000 compounds, and filtered out the valid molecules.
In Figure 3, the red curve represents the property distribution of molecules generated by the TransAntivirus. Other curves colored by blue, pink, and yellow represent the property distribution of molecules generated C5T5, cGAN, and LSTM, respectively. For MolWt distribution, all the compounds are less than 1000, and the peak of all curves is about 250. Compared to C5T5, the molecules generated by our model have overall larger MolWt. Furthermore, Figure 3 shows a clear difference between the distribution of QED and LogP properties between TransAntivirus and C5T5. The peak of the curve for TransAntivirus shows the biggest value than other models in term of NP-likeness. The compounds generated by TransAntivirus arrive at the peak of distribution in a bigger horizontal coordinate than C5T5 except for QED and LogP in Figure 3. However, we cannot suppose that the distribution curve can indicate which model is better, since each model has its own target and merit.
Figure 3.
Performance Analysis of TransAntivirus model.
Chemical Space Analysis
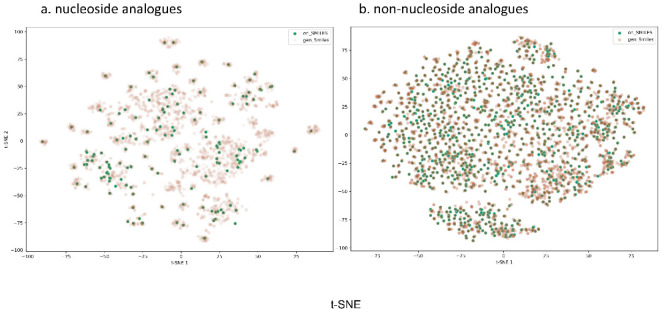
After fine-tuning on both nucleoside analogues and non-nucleoside analogues, we performed calculation of MACCS fingerprint for each molecule. Then, we applied t-SNE (t-distributed stochastic neighborhood embedding) to map the high dimensional features space to two-dimensional (2D) space.50Figures 4a and 4b represent nucleoside analogues and non-nucleoside analogues, respectively. The dense green dots in the scattered plot represent the original data, while the light grayish dots represent the generated sample. The results in Figure 4 demonstrated that the generated data have occupied the larger space than the original data. For the generated data that have widely spread and pervaded to more blank spaces, we assume that it is caused by the introduction of maximum likelihood, the space conversion between the input set (regarded as discrete space), and the output maximum likelihood (regarded as continuous space). Thus, there are several different molecules to be generated for the same input, but they are similar and very close in continuous space for the randomness.
Figure 4.
Chemical space analysis for (a) nucleoside analogues and (b) non-nucleoside analogues.
Property Optimization Analysis
In this section, we compared data space before and after optimization of properties for nucleoside analogues and non-nucleoside analogues, especially in terms of the LogP property space. A detailed account of the prediction results can be accessed in the Supporting Information.
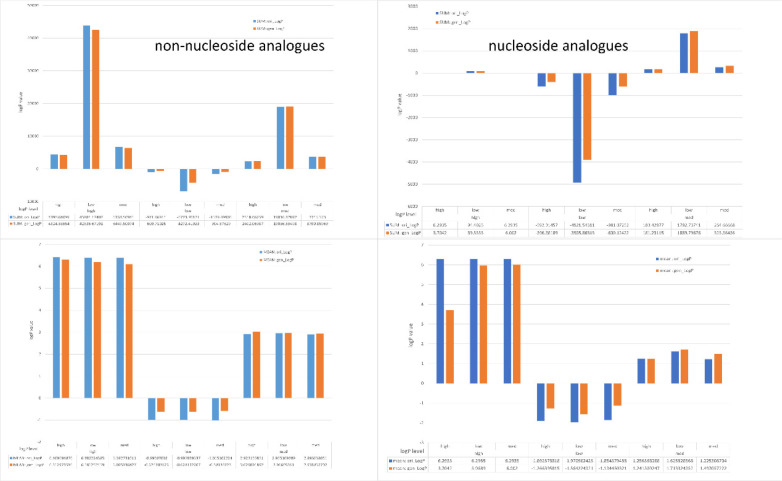
For nucleoside analogues, we collected 208 compounds from the reference article.40 The nucleoside analogues included the parent nucleosides (four distinct nucleotide triphosphates, i.e., adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine (CTP), thymidine (TTP), and uridine (UTP, which replaces TTP in RNA)). In addition, we selected a group of approximately 188 synthetic nucleoside analogues (the synthetic nucleosides). To prepare the input data before fine-tuning, at first, we converted the SMILES strings to IUPAC names and calculated the LogP value for each molecule. Thereafter, the selected nucleoside analogues (total of 170 molecules) were fed to the model and consequently generated 5000 compounds with the target level of LogP property. After calculating the LogP value of the generated molecules and filtering out the molecules by rule like molecular mass >1000, invalid molecules were recognized by the RDKit. Hence, we applied another parameter levenshtein distance >10, wherein a total of 4822 molecules were retained. The results are illustrated in Figure 5. The upper and lower left panels of the bar chart portrayed the sum and mean LogP values for the nucleoside analogues, respectively. Since the input data were divided into three groups including low (LogP value < −0.4), medium (−0.4 > LogP value <5.6), and high (LogP value >5.6), the expected LogP level for each molecule was also categorized as low, medium, and high. Conclusively, our results confirmed that the experimental LogP level (“ori” in bar chart) and the expected LogP level have significant difference. To this end, we argued that our model is reliable enough to predict nearly expected LogP level to the experimental LogP score of the newly generated nucleoside analogues.
Figure 5.
Comparison between the input data and generated data in three levels of LogP for nucleoside analogues and non-nucleoside analogues.
For non-nucleoside analogues, we essentially collected the fine-tuning data from the published article.41 We randomly took 1000 molecules and fine-tuned the model for ten epochs. Afterward, the fine-tuned model generated 10 000 molecules. Finally, 8102 compounds were obtained for LogP calculation after the filtration while applying the same rules as in the case of nucleoside analogues. If the LogP value of input molecules is under −0.4, this compound is identified to low level, and if over 5.6, designated as high level; otherwise, it is medium. As illustrated in upper and lower right panels of Figure 5, the obtained results closely resembled the same pattern as established by the nucleoside analogues. Consequently, it indicates that the optimization of properties across one or two level is achieved successfully. For instance, if the input molecule’s LogP is low level, the expected LogP level is med, which is called low → med (regard as a case), and it is successful when the model predicts a molecule with the med level of LogP. For other cases, such as med → high, high → med, med → low, low → high, and high → low, in our experiment, all tests except for two boundaries are achieved from the overall statistics. In Figure 5, we compared the sum and mean for each case.
For the exception of two boundary conditions: high → high and low → low, where the prediction could not achieve the target, it may probably be explained by two reasons. First, it may not be possible to find elements that are able to establish the LogP value of the target compound higher or lower. In other words, comparing to the original compound, replacing new elements may also destroy the group of high or low LogP value. Second, the fraction of higher and lower chemical groups and molecules may probably be very little in the training set and fine-tuning data.
In general, we compared the distribution of LogP and molecular fingerprint space. Although the molecules generated by TransAntivirus are structurally similar to the fine-tuning sets, they showed diversity in terms of other properties such as QED and LogP. For the optimization of properties in nucleoside analogues and non-nucleoside analogues, TransAntivirus has shown good performance, especially on the LogP and NP-likeness.
Case Study
To further evaluate the potential of our proposed model for the design of antiviral drugs, we conducted two case studies on two different targets for coronavirus disease (COVID-19). We fine-tuned the known active molecules for the specific targets by generating models and then generated virtual molecular libraries. Finally, the generated molecules were ranked and evaluated by docking score and MM-GBSA energy score. The case study further confirms the potential of the proposed antiviral drug generative model for the design of antiviral candidate compounds.
A Case for the Design of Antiviral Nucleoside Analogues
In this case study, we collected 170 nucleoside analogues with bioactivity for SARS-Cov-2 RNA-dependent-polymerase, which involved five basic nucleosides (adenosine, guanosine, cytidine, uridine, and thymidine) and 15 nucleoside analogues (Molnupiravir, Remdesivir, 6-Mercaptopurine, 6-Thio-dG, 6-Thiopurine riboside, 8-Azaguanine, Azathiopurine, BCNA,Cloturin, Flufylline, Gemcitabine, GS-441524, Thiamiprine, Thioguanine, and Tubercidin) that have reported in vitro activity against the SARS-CoV-2 Nucleosides. We hypothesize that nucleoside analogues, targeting the SARSCoV-2 RNA-dependent RNA polymerase, contain implicit chemical information that contributes to their activity against the enzyme. Thus, searching the molecular space that optimizes the properties of the anti-SARS-CoV-2 Nucleosides could lead to more inhibitors against the viral enzyme. For the anti-SARS-CoV-2 nucleosides, the model was applied to generate 5000 molecules after fine-tuning, and 4802 molecules were retained after filtering out by the rules.
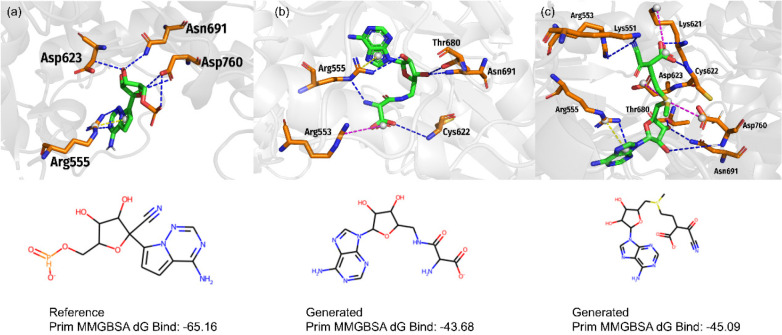
According to our findings, the candidates showed the highest similarity with the reference drug and corroborated high novelty and strong binding affinity for nucleoside analogues. Referring to the structure of Remdesivir, first, we computed the MACCS fingerprint of 4802 compounds and applied Tanimoto Similarity to exclude those candidates who have score less than 0.5. Thereafter, the 1546 retained candidates were used to perform docking and MMGBSA energy computation. The three-dimensional (3D) structure of the SARS-Cov-2 RNA-dependent-polymerase (PDB ID: 7BV2) was retrieved from Protein Data Bank (PDB) and subjected to the protein preparation and docking modules. To this end, based on several scoring and ranking criteria from both docking and MMPBSA energy computation, we shortlisted the top-8 candidate drug-like molecules in Table S1 in the Supporting Information. Furthermore, the top-2 candidate molecules were recommended for further evaluation as the promising candidate hits against the Covid-19 disease.
Since crucial molecular interactions between the potential drug candidates and protein targets are essential to guide structure–activity optimization, the binding interactions of promising antiviral drug candidate hits and SARS-Cov-2 RNA-dependent-polymerase were explored. The molecular interaction pattern of the final two candidate hits is depicted in Figure 6. As previously reported, a traditional hydrogen bond (H-bond) will form when the two atoms (capable of H-bond formation) are oriented in such a position that the angle and distance between the donor and acceptor is less than 135° and 3.0 Å (acceptor to donor heavy atom), respectively.51 The binding interaction of Remdesivir triphosphate (Reference molecule) accomplished several attractive charge interactions between its triphosphate group and the residues Asp623, Arg555, Asn691, and Asp760 of the SARS-Cov-2 RNA-dependent-polymerase(Figure 6a). Compared to Remdesivir triphosphate, the interaction of our newly generated two candidate compounds showed a preferential binding pattern.
Figure 6.
Ligand interaction diagrams of inhibitors into the binding site of SARS-CoV-2 RNA-dependent-RNA-polymerase (RdRp). The inhibitors are (A) Remdesivir triphosphate (RTP), (B) Compound a2, and (C) Compound a1. The binding free energy (kcal/mol) for the protein-inhibitor complexes predicted by the MM/GBSA method.
The docking results of the TransAntivirus generated candidate hit molecule 1 (Compound a2) showed the formation of five hydrogen bonds with SARS-Cov-2 RNA-dependent-polymerase (Figure 6b). The in-depth analyses depicted that the sugar moiety of Compound a2 established three H-bonds with Arg555, Thr680, and Asn691 of SARS-Cov-2 RNA-dependent-polymerase. In parallel, the other two H-bonds were formed between the terminal amino and carboxylic groups of Compound a2 and the Arg555 and Cys622 residues of the SARS-Cov-2 RNA-dependent-polymerase. Moreover, it was also noticed that a π-cation interaction and a salt bridge were formed with the aromatic ring of the nucleoside moiety and the terminal carboxyl group of Compound a2 and the Arg555 and Arg553 residues of the SARS-Cov-2 RNA-dependent-polymerase, respectively (Figure 6b).
Compound a1 in Table S1, which exhibited the highest binding affinity among the hit molecules, obtained a high hydrogen bond occupancy as compared to Remdesivir. Compound a1 formed a total of eight hydrogen bonds with SARS-Cov-2 RNA-dependent-polymerase (Figure 6c). As illustrated in Figure 6c, the sugar moiety of compound a1 formed three H-bonds with Thr680, Asn691, and Asp760 of the SARS-Cov-2 RNA-dependent-polymerase. Furthermore, the terminal nitrile and carboxyl groups of Compound a1 formed two H-bonds each with Lys551 and Arg553 and Lys621 and Cys622 residues of the SARS-Cov-2 RNA-dependent-polymerase. Similar to the Reference molecule, the adenine moiety of Compound a1 also established a H-bond and a π-cation interaction with the Arg555 residue of the SARS-Cov-2 RNA-dependent-polymerase (Figure 6c). In addition, π-alkyl hydrophobic interactions were observed with Asp623 and Lys621. Since the newly generated candidate hits established a strong network of H-bonds with the conserved binding site residues of the SARS-Cov-2 RNA-dependent-polymerase, it is therefore suggested that polar interactions (H-bonds) could improve the binding affinity of the candidate hits.
A Case for the Design of Antiviral Non-Nucleoside Analogues
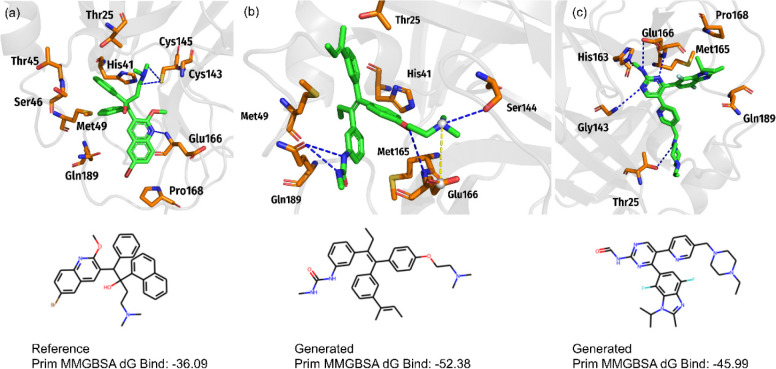
To evaluate the potential of the proposed model for the design of non-nucleoside analogues, we chose the SARS-CoV-2 3CLpro protein. The 3D structure of SARS-CoV-2 3CLpro protein (PDB ID: 6W63)52 was retrieved from PDB. First, we collected 70 molecules as input with bioactivity for SARS-CoV-2 3CLpro. Thereafter, we sampled 10 000 molecules from a fine-tuned model and filtered out some molecules by applying the rules like molecular mass >1000, invalid molecules recognized by the RDKit, levenshtein distance >10, (refer to the input molecules) 8104 molecules were retained. Then, we calculated the Tanimoto Similarity based on MACCS fingerprint with the reference molecule Bedaquiline.53 So, molecules that secured similarity greater than 0.5 were retained; hence, a total of 3706 molecules were shortlisted. Finally, these 3706 molecules and Bedaquiline were evaluated for their SAscore and QED. To this end, only 987 molecules could successfully fulfill the aforementioned criteria. Next, the successful candidate molecules were subjected to molecular docking, Prim MM-GBSA energy calculation, and virtual screening for identification of novel lead. The docking and energy calculation procedure remained the same as those in the first case study.
To this end, based on several scoring and ranking criteria from both docking and MM-GBSA energy computations, we shortlisted the top-18 candidate drug-like molecules in Table S2 in Supporting Information. Furthermore, the top-2 candidate molecules were recommended (through the eyes) for further evaluation as the promising candidate compounds against the Covid-19 disease. Figure 7 shows the selected candidates that exhibit how to interact with the receptor. The binding interaction of Bedaquiline (Reference molecule) accomplished several attractive charge interactions between its group and the residues Cys145 and Glu166 of the SARS-Cov-2 3CLpro (Figure 7a). The hydrophobic interactions of Bedaquiline were established with residues His41, Met49, and Glu166 of 3CLpro. Compared to Bedaquiline, the interaction of our newly generated two candidate compounds showed preferential binding pattern. The docking results of the TransAntivirus generated candidate hit molecule (Compound b7) showed the formation of four hydrogen bonds, four hydrophobic interactions, and a Salt Bridge with SARS-Cov-2 3CLpro (Figure 7b). In Figure 7b, the generated molecule (Compound b7 in Table S2) formed hydrophobic interactions with residues Thr25, His41, Met49, and Met165 and hydrogen bonds established with residues Ser144, Glu166, and Gln189 of SARS-Cov-2 3CLpro. Furthermore, Salt Bridges formed between the group Tertamine of Compound b7 and residue Glu166 of SARS-Cov-2 3CLpro.
Figure 7.
Ligand interaction diagrams of the inhibitors into the binding site of SARS-CoV-2 3CLpro. The inhibitors are (A) bedaquiline, (B) Compound b7, and (C) Compound b18. The binding free energy (kcal/mol) for the protein-inhibitor complexes predicted by the MM/GBSA method.
The generated molecule (Compound b18 in Table S2) showed the formation of five hydrogen bonds with Thr25, Gly143, His163, and Glu166 residues and hydrophobic interactions with the residues Met165, Pro168, and Gln189 of SARS-Cov-2 3CLpro (Figure 7c). The MM-GBSA energy of Compound b18 is −52.38 kcal/mol and more stable than that of the Reference molecule (−36.09 kcal/mol), as shown in Table S2. The same goes for Compound b18 (−45.99 kcal/mol).
Discussion
Overwhelmingly, the transformer-based model has the potential to model long-range dependencies and symmetric molecular structures. We compared the proposed framework to other baseline models. The results show that TransAntivirus performs significantly better than the control methods in terms of novelty, validity, uniqueness, and diversity. TransAntivirus showed excellent performance in the design and optimization of nucleoside and non-nucleoside analogues by chemical space analysis and property prediction analysis. Furthermore, for the applicability of TransAntivirus in the design of antiviral drugs, the two case studies show that the generated antiviral analogues demonstrate similar features and diversity with their inputs. However, we find that the generated compounds could not go beyond the reference molecule and only performed well on specific properties but worse on other properties. It indicates that the sequence-based single modality approach only performs conditional permutations in the dimension of sequence. Although we achieve conditional control generation and optimal generation of molecular properties, the generated molecules are still screened in the subsequent molecular docking using many physics-based methods, so if more information can be fused into the model, it will help to find drug molecules that meet the requirements more quickly, which is what is studied in multiobjective property optimization.
Conclusion
In this study, to generate chemically valid molecules and compare performance with other models, we trained our model and three control models on a large data set. For the optimization of molecular properties, the model was fine-tuned on the small data set, and then we generated molecules and compared them with the input data for similarity. To validate the applicability of TransAntivirus in the design of antiviral drugs, the model was fine-tuned on a small data set consisting of the parent nucleosides, the SARS-CoV-2 Nucleosides, and the Synthetic Nucleosides for the design of nucleoside analogues (non-nucleoside analogues have similar operations). Finally, we performed two case studies and screened four candidate lead compounds against anticoronavirus disease.
Compared to most molecule generation models based on SMILES, using the IUPAC-directed expansion of the SMILES molecule generation space can help optimize the generated molecules more accurately from a chemical semantic perspective. This is mainly achieved through two ways: (1) by adding attribute encoding prefixes at the beginning of IUPAC input sequences, thereby achieving overall changes in specific attributes of the molecule, and (2) by masking a position in the IUPAC token sequence to achieve pointwise optimization, thus achieving molecule generation and optimization at the IUPAC language level.
The significance of our study in the fields of fundamental and translational biology is demonstrated by several observations. It shows that the TransAntivirus was fine-tuned on only a small set of 20 nucleosides, generating several molecules that are similar or identical to natural or synthetic nucleoside analogues. These generated nucleoside analogues have chemical alterations that involved either the ribose or nucleobase moiety. Then, that focused molecular generation could also be directly leveraged to explore the molecular space around antiviral nucleosides and non-nucleoside analogues; specifically, they are active against SARS-CoV-2. Finally, the generative models could aid in the molecular design of nucleosides and non-nucleoside analogues with a wide range of applications from prebiotic chemistry to drug discovery.
It is important to highlight some limitations of our study. First, the metrics for the assessment of generative design models continue to evolve, and it is not feasible to explore all metrics that have been reported in the literature. It is critical that performance assessments of generative models are always taken in the context of the metrics that are applied in the goals of a given project. Second, gold-standard validation sets for generative molecular design problems do not exist. Thus, assessing the similarity between the generated molecules and the reference sets, we recognize that some otherwise biologically significant molecules may be missed.
There are several directions for future studies, which we can exclude from our study. As our proposed TransAntivirus is a general approach for bimodal molecular generation and property optimization, it would be interesting to apply it to other domains and problems, for instance, learning of relationship between SMARTS and SMILES. Moreover, multiobjective optimization of molecular properties is a very challenging problem. Nevertheless, TransAntivirus can be easily extended to multiobjective molecular optimization by encoding it as a prefix to the top of the molecular sequence.
Finally, multiobjective and multimodal research is the current critical direction for molecular generation. Incorporating more dimensional data and meeting the requirements of more objectives are crucial for the development of such an AI model; for example, a reinforcement learning method can be combined with TransAntivirus, and a prompt-based approach can be used to fine-tune.
Acknowledgments
This work was supported by Yonsei University graduate school “Integrative Biotechnology & Translational Medicine” and the Establishment and demonstration of biomaterial data platform, P0014714.
Data Availability Statement
The implemented code and experimental data set are available online at https://github.com/AspirinCode/TransAntivirus.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.3c00536.
Author Contributions
J.M., J.W. conceived the idea. J.M. wrote the article, developed the codes, and prepared the figures. A.Z. aided to write the docking analysis part and drew some figures, editing the language expression. K.N. provided the financial support. H.J. performed the first case study. K.H.C. provided some advice on writing skills. All the authors discussed the results and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- De Clercq E.; Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016, 29, 695–747. 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discovery. 2002, 1, 13–25. 10.1038/nrd703. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Recent highlights in the development of new antiviral drugs. Curr. Opin. Microbiol. 2005, 8, 552–560. 10.1016/j.mib.2005.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; De Clercq E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004, 30, 115–133. 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]; De Clercq E. Antiviral drugs: current state of the art. J. Clin. Virol. 2001, 22, 73–89. 10.1016/S1386-6532(01)00167-6. [DOI] [PubMed] [Google Scholar]
- Kausar S.; Said Khan F.; Ishaq Mujeeb Ur Rehman M.; Akram M.; Riaz M.; Rasool G.; Hamid Khan A.; Saleem I.; Shamim S.; Malik A.. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol Pharmacol.2021, 35, 10.1177/20587384211002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y. C.; Lu R. M.; Su S. C.; Chiang P. Y.; Ko S. H.; Ke F. Y.; Liang K. H.; Hsieh T. Y.; Wu H. C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed Sci. 2022, 29, 1. 10.1186/s12929-021-00784-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabinger F.; Stiller C.; Schmitzova J.; Dienemann C.; Kokic G.; Hillen H. S.; Hobartner C.; Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct Mol. Biol. 2021, 28, 740–746. 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfour H. H. Jr Antiviral drugs. N. Engl. J. Med. 1999, 340, 1255–1268. 10.1056/NEJM199904223401608. [DOI] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X. M.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; Li C.; Li Y.; Bai F.; Wang H.; Cheng X.; Cen X.; Hu S.; Yang X.; Wang J.; Liu X.; Xiao G.; Jiang H.; Rao Z.; Zhang L.; Xu Y.; Yang H.; Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Sci. 2020, 368, 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sun L.; Mao J.; Zhao Y.; Quan C.; Zhong M.; Fan S. Coarse-grained molecular dynamics simulation of interactions between cyclic lipopeptide Bacillomycin D and cell membranes. Mol. Simul. 2018, 44, 364. 10.1080/08927022.2017.1384632. [DOI] [Google Scholar]; Mao J. S.; Akhtar J.; Zhang X.; Sun L.; Guan S. H.; Li X. Y.; Chen G. M.; Liu J. X.; Jeon H. N.; Kim M. S.; No K. T.; Wang G. Y. Comprehensive strategies of machine-learning-based quantitative structure-activity relationship models. iscience. 2021, 24, 103052. 10.1016/j.isci.2021.103052. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abdolmaleki A.; Ghasemi J.; Ghasemi F. Computer aided drug design for multi-target drug design: SAR/QSAR, molecular docking and pharmacophore methods. Curr. Drug Targets. 2017, 18, 556–575. 10.2174/1389450117666160101120822. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Gong Y.; Liu Y.; Song B.; Zou Q.. Molecular design in drug discovery: a comprehensive review of deep generative models. Briefings Bioinf. 2021, 22. 10.1093/bib/bbab344. [DOI] [PubMed] [Google Scholar]; Martinelli D. D. Generative machine learning for de novo drug discovery: A systematic review. Comput. Biol. Med. 2022, 145, 105403. 10.1016/j.compbiomed.2022.105403. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Gong Y.; Liu Y.; Song B.; Zou Q.. Molecular design in drug discovery: a comprehensive review of deep generative models. Briefings Bioinf. 2021, 22. 10.1093/bib/bbab344. [DOI] [PubMed] [Google Scholar]; De Cao N.; Kipf T.. MolGAN: An implicit generative model for small molecular graphs. arXiv 2018. 10.48550/arXiv.1805.11973 [DOI] [Google Scholar]
- Méndez-Lucio O.; Baillif B.; Clevert D.-A.; Rouquié D.; Wichard J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 2020, 11, 10. 10.1038/s41467-019-13807-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hie B. L.; Shanker V. R.; Xu D.; Bruun T. U.; Weidenbacher P. A.; Tang S.; Wu W.; Pak J. E.; Kim P. S.. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 2023. 10.1038/s41587-023-01763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Chu Y.; Mao J.; Jeon H. N.; Jin H.; Zeb A.; Jang Y.; Cho K. H.; Song T.; No K. T.. De novo molecular design with deep molecular generative models for PPI inhibitors. Briefings Bioinf. 2022, 23. 10.1093/bib/bbac285. [DOI] [PubMed] [Google Scholar]
- Wang S.; Song T.; Zhang S.; Jiang M.; Wei Z.; Li Z.. Molecular substructure tree generative model for de novo drug design. Briefings Bioinf. 2022, 23. 10.1093/bib/bbab592. [DOI] [PubMed] [Google Scholar]
- Childs-Disney J. L.; Yang X.; Gibaut Q. M.; Tong Y.; Batey R. T.; Disney M. D. Targeting RNA structures with small molecules. Nat. Rev. Drug Discovery. 2022, 21, 736–762. 10.1038/s41573-022-00521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.; Qian W. W.; Peng X.; Su Y.; Peng J.; Ma J.. 3D Equivariant Diffusion for Target-Aware Molecule Generation and Affinity Prediction. arXiv 2023. 10.48550/arXiv.2303.03543 [DOI] [Google Scholar]
- Olivecrona M.; Blaschke T.; Engkvist O.; Chen H. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 2017, 9, 1–14. 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Ye K.; van Vlijmen H. W.; Emmerich M. T.; IJzerman A. P.; van Westen G. J. DrugEx v2: de novo design of drug molecules by Pareto-based multi-objective reinforcement learning in polypharmacology. J. Cheminf. 2021, 13, 85. 10.1186/s13321-021-00561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J.; Chang M. W.; Lee K.; Toutanova K.. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In NAACL ;Association for Computational Linguistics, 2019: pp 4171–4186. 10.18653/v1/N19-1423. [DOI] [Google Scholar]
- Radford A.; Narasimhan K.; Salimans T.; Sutskever I.. Improving language understanding by generative pre-training. Technical report, OpenAI, 2018.
- Raffel C.; Shazeer N.; Roberts A.; Lee K.; Narang S.; Matena M.; Zhou Y.; Li W.; Liu P. J.. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 2020, 21, Article 140. [Google Scholar]
- Carion N.; Massa F.; Synnaeve G.; Usunier N.; Kirillov A.; Zagoruyko S.. End-to-end object detection with transformers. In European conference on computer vision ;Springer, 2020; pp 213–229. [Google Scholar]
- Dosovitskiy A.; Beyer L.; Kolesnikov A.; Weissenborn D.; Zhai X.; Unterthiner T.; Dehghani M.; Minderer M.; Heigold G.; Gelly S.; Uszkoreit J.; Houlsby N.. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. ICLR 2021. [Google Scholar]
- Hassani A.; Walton S.; Li J.; Li S.; Shi H.. Neighborhood Attention Transformer. arXiv 2022. 10.48550/arXiv.2204.07143 [DOI] [Google Scholar]
- Baevski A.; Zhou Y.; Mohamed A.; Auli M. wav2vec 2.0: A framework for self-supervised learning of speech representations. Advances in Neural Information Processing Systems 2020, 33, 12449–12460. [Google Scholar]
- Wolf T.; Debut L.; Sanh V.; Chaumond J.; Delangue C.; Moi A.; Cistac P.; Rault T.; Louf R.; Funtowicz M.; Davison J.; Shleifer S.; Von Platen P.; Ma C.; Jernite Y.; Plu J.; Xu C.; Le Scao T.; Gugger S.; Drame M.; Lhoest Q.; Rush A.. Transformers: State-of-the-Art Natural Language Processing ;Association for Computational Linguistics, 2020; pp 38–45. 10.18653/v1/2020.emnlp-demos.6. [DOI] [Google Scholar]
- Ertl P.; Lewis R.; Martin E.; Polyakov V. In In silico generation of novel, drug-like chemical matter using the LSTM neural network. arXiv 2017. 10.48550/arXiv.1712.07449 [DOI] [Google Scholar]
- Prykhodko O.; Johansson S. V.; Kotsias P.-C.; Arús-Pous J.; Bjerrum E. J.; Engkvist O.; Chen H. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminf. 2019, 11, 1–13. 10.1186/s13321-019-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagal V.; Aggarwal R.; Vinod P. K.; Priyakumar U. D. MolGPT: Molecular Generation Using a Transformer-Decoder Model. J. Chem. Inf. Model. 2022, 62, 2064–2076. 10.1021/acs.jcim.1c00600. [DOI] [PubMed] [Google Scholar]
- Rothchild D.; Tamkin A.; Yu J. H.; Misra U.; Gonzalez J.. C5T5: Controllable Generation of Organic Molecules with Transformers. arXiv 2021. 10.48550/arXiv.2108.10307 [DOI] [Google Scholar]
- Adilov S.Generative Pre-Training from Molecules ChemRxiv. 2021. 10.26434/chemrxiv-2021-5fwjd [DOI] [Google Scholar]
- Kang S.; Cho K. Conditional Molecular Design with Deep Generative Models. J. Chem. Inf. Model. 2019, 59, 43–52. 10.1021/acs.jcim.8b00263. [DOI] [PubMed] [Google Scholar]
- White A. D. The future of chemistry is language. Nat. Rev. Chem. 2023, 10.1038/s41570-023-00502-0. [DOI] [PubMed] [Google Scholar]
- Krasnov L.; Khokhlov I.; Fedorov M. V.; Sosnin S.. Transformer-based artificial neural networks for the conversion between chemical notations. Sci. Rep. 2021, 11. 10.1038/s41598-021-94082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Thiessen P. A.; Bolton E. E.; Chen J.; Fu G.; Gindulyte A.; Han L.; He J.; He S.; Shoemaker B. A.; Wang J.; Yu B.; Zhang J.; Bryant S. H. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–1213. 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RDKit: Open-source cheminformatics; http://www.rdkit.org.
- Wildman S. A.; Crippen G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Model. 1999, 39, 868–873. 10.1021/ci990307l. [DOI] [Google Scholar]
- Ertl P.; Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 2009, 1, 1–11. 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton G.; Paolini G.; Besnard J.; Muresan S.; Hopkins A. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]; Article
- Dablain D.; Siwo G.; Chawla N.. Generative AI Design and Exploration of Nucleoside Analogs ChemRxiv. 2021. 10.26434/chemrxiv-2021-l5pr9 [DOI] [Google Scholar]
- Hu F.; Wang L.; Hu Y.; Wang D.; Wang W.; Jiang J.; Li N.; Yin P. A novel framework integrating AI model and enzymological experiments promotes identification of SARS-CoV-2 3CL protease inhibitors and activity-based probe. Briefings Bioinf. 2021, 22, bbab301. 10.1093/bib/bbab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Mao J.; Wang M.; Le X.; Wang Y.. Explore drug-like space with deep generative models. Methods 2023.21052. 10.1016/j.ymeth.2023.01.004 [DOI] [PubMed] [Google Scholar]; Vaswani A.; Shazeer N.; Parmar N.; Uszkoreit J.; Jones L.; Gomez A. N.; Kaiser Ł.; Polosukhin I.. Attention is all you need. Advances in neural information processing systems. 2017, 30. [Google Scholar]
- Kingma D. P.; Ba J.. Adam: A method for stochastic optimization. arXiv 2014. 10.48550/arXiv.1412.6980 [DOI] [Google Scholar]
- Preuer K.; Renz P.; Unterthiner T.; Hochreiter S.; Klambauer G. Frechet ChemNet Distance: A Metric for Generative Models for Molecules in Drug Discovery. J. Chem. Inf. Model. 2018, 58, 1736–1741. 10.1021/acs.jcim.8b00234. [DOI] [PubMed] [Google Scholar]
- Polykovskiy D.; Zhebrak A.; Sanchez-Lengeling B.; Golovanov S.; Tatanov O.; Belyaev S.; Kurbanov R.; Artamonov A.; Aladinskiy V.; Veselov M.; et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 2020, 11, 565644. 10.3389/fphar.2020.565644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavi Sastry G.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Du J.; Sun H.; Xi L.; Li J.; Yang Y.; Liu H.; Yao X. Molecular modeling study of checkpoint kinase 1 inhibitors by multiple docking strategies and prime/MM–GBSA calculation. J. Comput. Chem. 2011, 32, 2800–2809. 10.1002/jcc.21859. [DOI] [PubMed] [Google Scholar]
- Kosugi T.; Ohue M.. Quantitative Estimate of Protein-Protein Interaction Targeting Drug-likeness. 2021 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB)2021, 135–142, Proceedings Paper. 10.1109/CIBCB49929.2021.9562931. [DOI]; Kosugi T.; Ohue M.. Quantitative Estimate Index for Early-Stage Screening of Compounds Targeting Protein-Protein Interactions. Int. J. Mol. Sci. 2021, 22, 10925. 10.3390/ijms222010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maaten L.; Hinton G.. Visualizing data using t-SNE. JMLR 2008, 9. [Google Scholar]
- Uengwetwanit T.; Chutiwitoonchai N.; Wichapong K.; Karoonuthaisiri N. Identification of novel SARS-CoV-2 RNA dependent RNA polymerase (RdRp) inhibitors: From in silico screening to experimentally validated inhibitory activity. Comput. Struct. Biotechnol. J. 2022, 20, 882–890. 10.1016/j.csbj.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecar A.A taxonomically-driven approach to development of potent, broad-spectrum inhibitors of coronavirus main protease including SARS-CoV-2 (COVID-19). Be Publ. 2020. 10.2210/pdb6W63/pdb.2020. [DOI] [Google Scholar]
- Ghahremanpour M. M.; Tirado-Rives J.; Deshmukh M.; Ippolito J. A.; Zhang C.-H.; Cabeza de Vaca I.; Liosi M.-E.; Anderson K. S.; Jorgensen W. L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020, 11, 2526–2533. 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The implemented code and experimental data set are available online at https://github.com/AspirinCode/TransAntivirus.