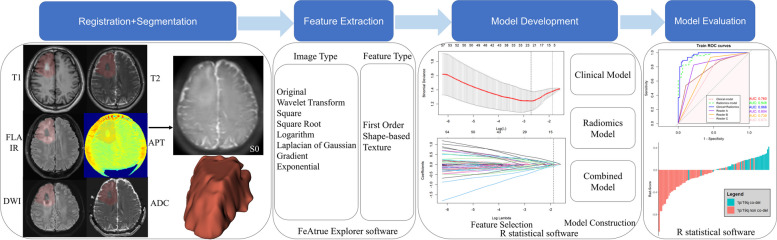
Abstract
Background
1p/19q co-deletion in low-grade gliomas (LGG, World Health Organization grade II and III) is of great significance in clinical decision making. We aim to use radiomics analysis to predict 1p/19q co-deletion in LGG based on amide proton transfer weighted (APTw), diffusion weighted imaging (DWI), and conventional MRI.
Methods
This retrospective study included 90 patients histopathologically diagnosed with LGG. We performed a radiomics analysis by extracting 8454 MRI-based features form APTw, DWI and conventional MR images and applied a least absolute shrinkage and selection operator (LASSO) algorithm to select radiomics signature. A radiomics score (Rad-score) was generated using a linear combination of the values of the selected features weighted for each of the patients. Three neuroradiologists, including one experienced neuroradiologist and two resident physicians, independently evaluated the MR features of LGG and provided predictions on whether the tumor had 1p/19q co-deletion or 1p/19q intact status. A clinical model was then constructed based on the significant variables identified in this analysis. A combined model incorporating both the Rad-score and clinical factors was also constructed. The predictive performance was validated by receiver operating characteristic curve analysis, DeLong analysis and decision curve analysis. P < 0.05 was statistically significant.
Results
The radiomics model and the combined model both exhibited excellent performance on both the training and test sets, achieving areas under the curve (AUCs) of 0.948 and 0.966, as well as 0.909 and 0.896, respectively. These results surpassed the performance of the clinical model, which achieved AUCs of 0.760 and 0.766 on the training and test sets, respectively. After performing Delong analysis, the clinical model did not significantly differ in predictive performance from three neuroradiologists. In the training set, both the radiomic and combined models performed better than all neuroradiologists. In the test set, the models exhibited higher AUCs than the neuroradiologists, with the radiomics model significantly outperforming resident physicians B and C, but not differing significantly from experienced neuroradiologist.
Conclusions
Our results suggest that our algorithm can noninvasively predict the 1p/19q co-deletion status of LGG. The predictive performance of radiomics model was comparable to that of experienced neuroradiologist, significantly outperforming the diagnostic accuracy of resident physicians, thereby offering the potential to facilitate non-invasive 1p/19q co-deletion prediction of LGG.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12880-024-01262-z.
Keywords: Low-grade glioma, 1p/19q Co-deletion, Prediction, Radiomics, Magnetic resonance imaging, Amide proton transfer weighted imaging, Diffusion weighted imaging
Introduction
Low-grade gliomas (LGGs) are invasive neoplasms that arise in the cerebral hemispheres of adults including diffuse low-grade and intermediate-grade gliomas (World Health Organization [WHO] grades II and III) [1]. The fifth edition of the WHO Classification of Tumors of the Central Nervous System divides adult-type gliomas into three subtypes based on molecular markers: (1) astrocytoma, isocitrate dehydrogenase (IDH) -mutant, (2) oligodendroglioma, IDH-mutant, and 1p/19q-codeleted, and (3) glioblastoma, IDH-wildtype [2].
Studies have shown that oligodendroglioma has the best prognosis between these three categories [3]. In addition, studies have illustrated that even small residual tumor has a negative impact on overall survival in 1p/19q intact astrocytoma than on 1p/19q co-deleted oligodendroglioma [4]. Therefore, the noninvasive assessment of the molecular subtype of 1p/19q is particularly valuable in guiding clinical decision making.
The radiological features of 1p/19q co-deleted tumors frequently display calcifications, and they predominantly occur in the frontal lobe, with a tendency to invade the gray matter. These tumors typically exhibit heterogeneous signal intensities on both T1- and T2-weighted MR imaging, often lacking a distinct tumor margin [5, 6].
With artificial intelligence technique applied to MRI, radiogenomics becoming a promising tool for discriminating genotype of gliomas in a non-invasive fashion. The largest amount of literature researches focused on conventional MRI, such as T1-weighted imaging (T1WI), T2 weighted imaging (T2WI) and fluid-attenuated inversion recovery (FLAIR) [7–9]. Advanced MRI techniques such as amide proton transfer weighted (APTw) or diffusion weighted imaging (DWI) remain less studied.
APTw imaging is one of the most developed branch of chemical exchange saturation transfer (CEST) imaging [10, 11]. Previous studies have demonstrated that APTw imaging has important value in detecting molecular biomarkers in gliomas, such as IDH mutation, O6-methylguanine methyltransferase (MGMT), and Lys-27-Met mutations in histone 3 genes (H3K27M) [12–16]. In the study by Su et al. [17], CEST imaging was used to identify 1p/19q co-deletion, and statistically significant indices included direct saturation of water (DSW), semi-solid magnetization transfer contrast (MTC), and MTRasym (2.0 ppm). However, the APT value was not statistically significant. In addition, a growing number of researches have shown that features extracted from DWI have predictive values in predicting of glioma molecular subtypes [18–20].
The aim of this retrospective study was to develop a radiogenomics method to predict 1p/19q co-deletion of LGG based on advanced and conventional MRI features.
Materials and methods
Patients
This study was approved by The Ethics Committee of the Zhujiang Hospital of Southern Medical University, and because its nature of retrospective study, the requirement of obtaining informed consent was waived. We retrospectively analyzed all patients from July 2017 to January 2023, with pathologically diagnosed LGG according to the WHO 2016 Classification and completed preoperative 3D APTw imaging evaluation. A total of 95 patients with initial diagnosed as LGG were reviewed (Fig. 1). The inclusion criteria were as follows: (1) LGG with histopathological confirmation and known 1p/19q co-deletion status; (2) LGG with preoperative MRI including APTw, DWI and corresponding apparent diffusion coefficient (ADC) maps; (3) LGG without any previous treatment at initial diagnosis, and (4) patients over 18 years old. Cases with insufficient MRI data (n = 2), MRI data had intense motion artifacts (n = 1), patients with recurrent glioma (n = 2) were excluded from the study, rendering 90 LGGs in the dataset.
Fig. 1.
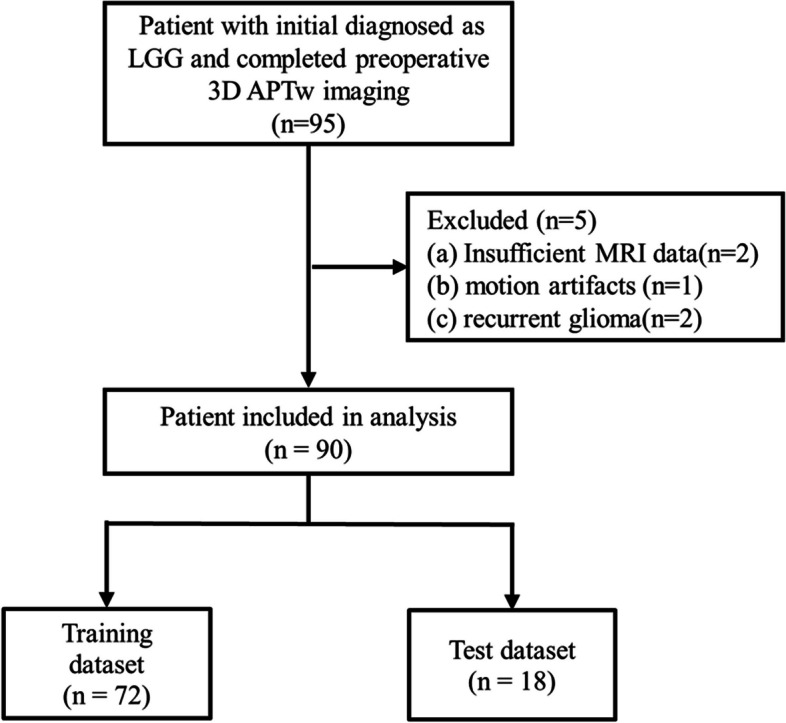
Flowchart of the study population
Evaluation of 1p/19q co-deletion status
1p/19q co-deletion was assessed by a fluorescence in situ hybridization (FISH) locus specific identifier (LSI) probe sets 1p36/1q21 and 19q13/19p13. The assessment was consensus-classified by two pathologists over 6 and 20 years of experience, respectively.
MRI acquisition
All patients were examined on two 3.0 T Philips scanners (Ingenia Elition 3.0 T X and Ingenia 3.0 T; Philips Medical Systems, Best, The Netherlands) with a 20-channel head-neck coil. In addition to conventional anatomic sequences, each MRI consists of 3D APTw, DWI and the corresponding ADC maps. All image processing and reconstruction algorithms were automatically implemented on MRI scanning system. Among them, the APT sequence encompasses two sets of images: one set is APTw images, and the other set is S0 images. Both sets of images possess the identical spatial resolution. The S0 images represent the control signal intensity without saturation, and the imaging is performed at a frequency offset of − 1560 ppm [11]. Detailed information on imaging parameters are available in the Supplementary Table 1, Additional File 1.
APTw and DWI imaging parameters
APTW imaging was implemented with a fat-suppressed, mDIXON 3D turbo spin-echo sequence, with RF saturation powers of 2μT and a saturation duration of 2 s were used [11]. The detailed parameters were as follows: SENSE factor, 1.4; repetition time /echo time (TR/TE) = 5900/8 ms; field of view (FOV) = 212 × 182 mm2; slice thickness = 5.4 mm; matrix = 120 × 102 (reconstructed to 224 × 224); and voxel size = 1.80 × 1.80 × 5.40 mm3 (0.95 × 0.95 × 5.40 mm3, reconstructed). A multi-offset, multi-acquisition APTw imaging acquisition protocol was used [7 offsets = ± 2.7, − 3.5, + 3.5 (3), ± 4.28, − 1560 ppm; value in parentheses is the number of acquisitions, which was considered as one, if not specified]. The total scan time was 4 min 48 s.
DWI was implemented with a 2D single-shot echo-planar imaging sequence. The detailed parameters were as follows: SENSE factor, 2; TR/TE = 3284/200 ms; field of view = 230 × 230 mm2; slice thickness = 4.4 mm; matrix = 152 × 122 (reconstructed to 256 × 256); and voxel size = 1.50 × 1.89 × 4.40 mm3 (0.90 × 0.90 × 4.40 mm3, reconstructed). The ADC maps were calculated using b values of 0 and 1000 s/mm2 images.
MRI feature evaluation
Three neuroradiologists, labeled as Reader A (an experienced neuroradiologist with 7 years of experience) and, Reader B and C (both being resident physicians with 3 and 1 years of experience in neuroradiology respectively) independently evaluated the MR images while blinded to the pathology results. In cases of disagreement, a consensus was reached. The evaluation encompassed the following aspects: gray matter involvement, calcification, hemorrhage, tumor margin clarity (indistinct vs. sharp), and contrast enhancement. Due to limitations in data availability, contrast-enhanced T1-weighted imaging (T1C) was not incorporated into this study. However, where such data were available, we did analyze whether lesions exhibited enhancement.
For each tumor, the readers were also asked to assess whether they believed it exhibited 1p/19q codeletion or was intact, providing a confidence score ranging from 1 (indicating very unsure) to 5 (indicating very sure). This confidence score was then transformed into a prediction "score" by dividing it by 5 and multiplying the result by 1 if the predicted label was 1p/19q codeleted, or by -1 if the predicted label was 1p/19q intact. This approach allowed for the calculation of an Area Under the Curve (AUC) for the manual classification [21].
Image preprocessing and tumor segmentation
A rigid co-registration was conducted between T1, T2, FLAIR, APT DWI images, and ADC map using SPM12 (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/). The reference volume for coregistration was the unsaturated images (S0 image). Three-dimensional volume of interest (VOI) of whole-tumor were delineated by consensus between two neuroradiologists (reader A and reader B with 7 and 3 years of experience in neuroradiology), blinded to 1p/19q status, using the ITK-SNAP software (http://www.itksnap.org/pmwiki/pmwiki.php). Necrosis, cystic cavities, large vessels, calcification and hemorrhagic components were excluded.
Extraction of radiomic features
All radiomic features were extracted using an open-source software package named FeAture Explorer software (version 0.5.5 https://github.com/salan668/FAE), which was built based on the PyRadiomics package (https://github.com/Radiomics/pyradiomics) [22, 23]. Features were extracted on original image and preprocessed imaging, including wavelet transform, square, square root, logarithm, laplacian of gaussian, gradient and exponential. There are 7 feature types: shape features, first-order features, gray level cooccurrence matrix (GLCM) features, gray level run length matrix (GLRLM) features, gray level size zone matrix (GLSZM) features, neighboring gray tone difference matrix (NGTDM) features, and gray level dependence matrix (GLDM). A total of 8454 features were extracted from the MRI data and 1409 features each from T1WI, T2WI, FLAIR, APTw, DWI and ADC map. A detailed information of the workflow is presented in the Fig. 2.
Fig. 2.
Workflow of the study
Feature selection
The training and test datasets were randomly selected from the dataset at a ratio of 8:2, where the clinical characteristics in the two datasets were balanced. Standardization of radiomic features was performed using z-score intensity normalization and the upsampling was employed to remove the unbalance of the training dataset. Then, a least absolute shrinkage and selection operator (LASSO) algorithm was employed in combination with fivefold cross-validation, which was aimed to identify the best features subset via the one-standard error of the minimum criteria.
Model development
A total of 3 models were built. Demographic factors and MRI features of the training set were compared between patients with 1p/19q codeleted and those with 1p/19q intact status using multivariable logistic regression analysis. The significant variables identified in this analysis were then used to build a clinical model. Radiomic model was established by using the final selected radiomics features, and a radiomics score (Rad-score) was generated using a linear combination of the values of the selected features weighted for each patient. A combined model was established by employing logistic regression analysis with the significant variables previously identified, along with the Rad-score of the patient.
Model evaluation
The receiver operating characteristic (ROC) curves and area under the curve (AUC), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and accuracy were calculated for all models. We used Delong's test to compare the prediction performance of the three models against the individual readers (Reader A, B, and C). A p < 0.05 was considered significant. Decision curve analysis (DCA) was conducted to evaluate the clinical consequences of three models by plotting their net benefits across different threshold probabilities [24].
Statistical analysis
SPSS v.26.0 (IBM SPSS Statistic Version19, Chicago, IL, USA) and R statistical software (v.4.2.2; https://www.r-project.org) were used for statistical analysis. We used independent samples t-test for quantitative data, and Wilcoxon test, chi-square test and Fisher’s exact test for qualitative data. A two-sided p-value of < 0.05 was considered significant.
Results
Clinical characteristics of patients
The characteristics of the patients in both the training and test datasets are outlined in Table 1. Within the training dataset, the variables of IDH1 status, gray matter involvement, calcification, and tumor margin exhibited statistically significant differences between the patients with 1p/19q codeletion and those with intact 1p/19q status. However, upon performing a multiple logistic regression analysis, only calcification and tumor margin clarity were identified as independent predictors in the clinical model. When evaluating the enhancement of lesion, after excluding four cases without T1C, there was no statistically significant difference in the enhancement patterns of tumors between the 1p/19q co-deleted and 1p/19q intact groups among the remaining 86 cases (p = 0.941). Among these cases, 44 (51.2%) cases showed no evidence of enhancement.
Table 1.
Clinical characteristics of the patients
| Clinical factors | Training set (n = 72) | Test set (n = 18) | p value | ||||
|---|---|---|---|---|---|---|---|
| 1p/19q co-del | 1p/19q non co-del | P value | 1p/19q co-del | 1p/19q non co-del | p value | ||
| Number | 26 (36.1) | 46 (63.9) | - | 7 (38.9) | 11(61.1) | - | 0.827 |
| Age(years)a | 42 (34–46) | 43 (33–51) | 0.560 | 52 (41–63) | 43(33–59) | 0.303 | 0.081 |
| Sex | 0.100 | 1.000 | 0.164 | ||||
| Male | 13 (50.0) | 32 (69.6) | 3 (42.9) | 5 (45.5) | |||
| Female | 13 (50.0) | 14 (30.4) | 4 (57.1) | 6 (54.5) | |||
| WHO grades | 0.903 | 1.000 | 0.642 | ||||
| Grade II | 19 (73.1) | 33 (71.7) | 5 (71.4) | 7 (63.6) | |||
| Grade III | 7 (26.9) | 13 (28.3) | 2 (28.6) | 4 (36.4) | |||
| Histology | 0.826 | ||||||
| Oligodendroglioma | 26 | 6 | |||||
| Astrocytoma | 46 | 12 | |||||
| IDH | < 0.001 | 0.316 | 0.642 | ||||
| Mutation | 25(96.2) | 27(58.7) | 6(85.7) | 6(54.5) | |||
| Wildtype | 1(3.8) | 19(41.3) | 1(14.3) | 5(45.5) | |||
| Gray matter involvement | 0.003 | 0.637 | 0.039 | ||||
| Absent | 0(0) | 12(26.1) | 2(28.6) | 5(45.5) | |||
| Present | 26(100) | 34(73.9) | 5(71.4) | 6(54.5) | |||
| Calcification | < 0.001 | 0.141 | 0.743 | ||||
| Absent | 9(34.6) | 36(78.3) | 3(42.9) | 9(81.8) | |||
| Present | 17(65.4) | 10(21.7) | 4(57.1) | 2(18.2) | |||
| Hemorrhage | 1.000 | 1.000 | 0.316 | ||||
| Absent | 23(88.5) | 40(87.0) | 5(71.4) | 9(81.8) | |||
| Present | 3(11.5) | 6(13.0) | 2(28.6) | 2(18.2) | |||
| Tumor margin | 0.009 | 0.316 | 0.820 | ||||
| Indistinct | 23(88.5) | 27 (58.7) | 6(85.7) | 6(54.5) | |||
| Sharp | 3(11.5) | 19(41.3) | 1(14.3) | 5(45.5) | |||
| Contrast enhancement | 0.357 | 0.035 | 0.050 | ||||
| Absent | 13(50.0) | 24(52.2) | 2(28.6) | 5(45.5) | |||
| Present | 12(46.2) | 22(47.8) | 2(28.6) | 6(54.5) | |||
| Not available | 1(3.8) | 0(0) | 3(42.9) | 0(0) | |||
Data are numbers of patients, with percentages in parentheses
aData are medians, with inter-quartile range
Feature selection and development of a radiomics model
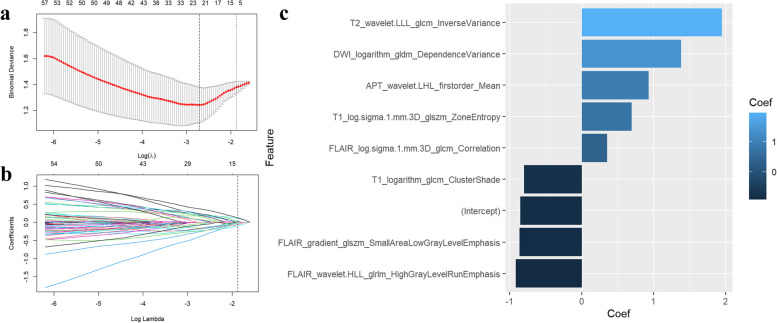
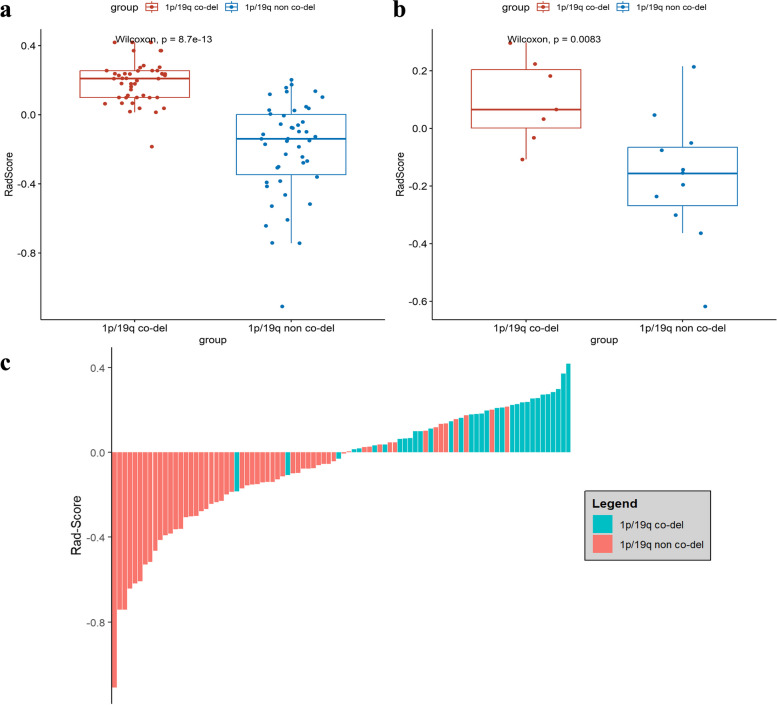
Overall, 8454 radiomic features were extracted from multiparametric MRI sequences (T1, T2, FLAIR, DWI, APTw images, and ADC maps) for each patient. After application of the LASSO regression model, 8 highly relevant radiomic features were selected to construct the radiomic model (Fig. 3, a and b, Table 2) and the coefficients of the features were demonstrated in Fig. 3, c. There was a significant difference in the Rad-score between 1p/19q co-deleted and 1p/19q non co-deleted gliomas in the both training and test sets (Fig. 4 a and b). The bar chart for Rad-score can be found in Fig. 4c.
Fig. 3.
LASSO coefficient profiles of the 8454 radiomics features. A coefficient profile plot was generated versus the selected log λ value using fivefold cross-validation (a). Optimal parameter selection in the LASSO model via 1-standard error criterion (b) The coefficients of 8 most relevant radiomics features and intercept (c)
Table 2.
Radiomics feature selection results
| Variables | Radiomics feature name | Variables | Radiomics feature name |
|---|---|---|---|
| A | T1_logarithm_glcm_ClusterShade | E | FLAIR_log.sigma.1.mm.3D_glcm_Correlation |
| B | T1_log.sigma.1.mm.3D_glszm_ZoneEntropy | F | FLAIR_wavelet.HLL_glrlm_HighGrayLevelRunEmphasis |
| C | T2_wavelet.LLL_glcm_InverseVariance | G | DWI_logarithm_gldm_DependenceVariance |
| D | FLAIR_gradient_glszm_SmallAreaLowGrayLevelEmphasis | H | APT_wavelet.LHL_firstorder_Mean |
Fig. 4.
Comparation of Rad-Score between glioma patients with 1p/19q co-deleted and 1p/19q non co-deleted in training (a) and test sets (b). The bar chart of Rad-score (c)
Classification performance and model comparison with neuroradiologist
The performance of the three models classifying the molecular subtypes of LGG in the training and test sets are shown in Table 3. The results of the DeLong test comparing the predictive performance of the three models against neuroradiologists (Reader A, B, and C) in both the training and test sets are summarized in Table 4.
Table 3.
Diagnostic performance by different models
| Model | AUC (95% CI) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| Training set (n = 72) | ||||||
| Clinical model | 0.761 (0.946–0.871) | 73.7 | 77.4 | 53.8 | 89.1 | 76.4 |
| Radiomics model | 0.948 (0.909–0.987) | 83.7 | 88.4 | 89.1 | 82.6 | 85.9 |
| Combined model | 0.966(0.932–0.999) | 87.5 | 90.9 | 91.3 | 87.0 | 89.1 |
| Reader A | 0.804(0.723–0.886) | 82.6 | 78.3 | 79.2 | 81.8 | 80.4 |
| Reader B | 0.739(0.658–0.820) | 93.5 | 54.3 | 67.2 | 89.3 | 73.9 |
| Reader C | 0.674(0.588–0.760) | 89.1 | 45.7 | 62.1 | 80.8 | 67.4 |
| Test set (n = 18) | ||||||
| Clinical model | 0.766 (0.525–1) | 80.0 | 76.9 | 57.1 | 90.9 | 77.8 |
| Radiomics model | 0.909 (0.770–1) | 80.0 | 76.9 | 57.1 | 90.9 | 77.8 |
| Combined model | 0.896 (0.733–1) | 80.0 | 76.9 | 57.1 | 90.9 | 77.8 |
| Reader A | 0.740(0.523–0.957) | 57.1 | 90.9 | 80.0 | 76.9 | 77.8 |
| Reader B | 0.604(0.356–0.852) | 57.1 | 63.6 | 50.0 | 70.0 | 61.1 |
| Reader C | 0.513(0.262–0.764) | 57.1 | 45.4 | 40.0 | 62.5 | 50.0 |
AUC area under the curve, CI confidence interval, PPV positive predictive value, NPV negative predictive value
Table 4.
P values in comparing AUC of Delong test
| Training set | Test set | |||||
|---|---|---|---|---|---|---|
| Clinical model | Radiomics model | Combined model | Clinical model | Radiomics model | Combined model | |
| Rader A | 0.53 | 0.001 | < 0.001 | 0.88 | 0.26 | 0.28 |
| Rader B | 0.76 | < 0.001 | < 0.001 | 0.38 | 0.03 | 0.053 |
| Rader C | 0.23 | < 0.001 | < 0.001 | 0.13 | 0.02 | 0.02 |
After performing the Delong analysis, it was found that there was no statistically significant difference in predictive performance between the clinical model and the three readers (A, B, C). In the training set. Both the radiomic model and the combined model performed significantly better than the three readers. In the test set, the AUC values of the three models were higher than those of the three readers, but there was no statistically significant difference compared to the experienced neuroradiologist (Reader A). On the other hand, the radiomics model significantly outperformed the resident physicians (Reader B and C). The ROC curves for the three models and the three neuroradiologists in both the training and test sets are presented in Fig. 5.
Fig. 5.
ROC curves of the models and signatures in the training (a) and test set (b)
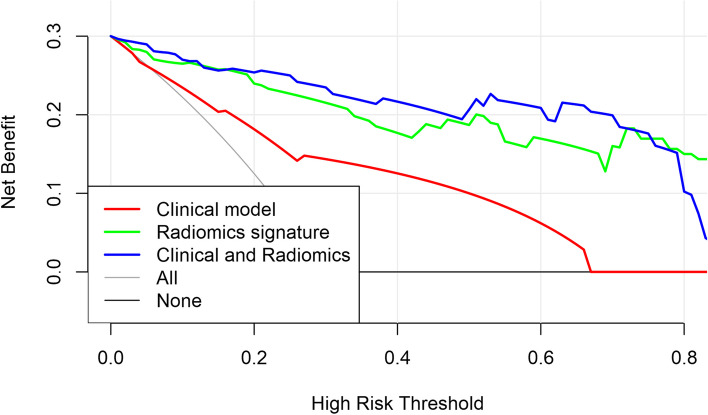
Meanwhile, the decision curves for molecular subtype classification among the three models demonstrate that the radiomic model and combined model have a better overall net benefit compared to the clinical model (Fig. 6).
Fig. 6.
Decision curve analysis for the three models
Discussion
In this study, we developed a radiogenomics method that predicts 1p/19q co-deletion status in LGG based on APTw, DWI, conventional sequences, as well as additional MRI features. Our results showed that the radiomics model and the combined model exhibited excellent performance in distinguishing 1p/19q co-deletion status in both the training and test sets, with AUCs of 0.948 and 0.966 for the training set, and 0.909 and 0.896 for the test set, respectively. Furthermore, the predictive performance of our model was comparable to that of experienced neuroradiologist, significantly outperforming the diagnostic accuracy of resident physicians. 1p/19q co-deletion is associated with longer progression-free and overall survival, and better response to radiotherapy and chemotherapy [1, 25, 26]. For the suspected LGG patients, maximal safe surgical resection is advocated as the standard of care [27]. However, recent studies demonstrated that gross total resection was not related with prolonged survival of patients with oligodendroglioma [28, 29]. Therefore, preoperative identification of 1p/19q genotype could help with surgical planning.
There are growing number of studies using machine learning algorithms to predict molecular subtypes such as 1p/19q co-deletion and IDH mutations [7, 21, 30–33]. However, most of these studies only used conventional anatomical MR sequences because of the widespread usage. The 3D APTw and DWI in our datasets enabled us to extract molecular information from the tumor. Recently, two pioneering APTw-based radiomics studies have been undertaken, aiming to distinguish glioblastomas from gliomas and brain metastases, as well as to discriminate treatment response from tumor progression [34, 35]. Our research findings indicate that APTw-based radiomics also holds value in predicting the 1p/19q co-deletion in LGG.
In the training set, our findings indicate statistically significant differences in calcifications, tumor margin clarity, and gray matter involvement between gliomas with 1p/19q co-deletion and those without, which is consistent with prior studies. Notably, among these features, only calcifications and tumor margin clarity independently predict 1p/19q co-deletion. This may be related to the relatively small sample size of our training dataset. In the radiomics model, our results demonstrate that among the three most contributing features two are texture features from T2WI and DWI while the third is a histogram feature from APTw (Fig. 3, c). This finding could partly be explained by the fact that 1p/19q-codeleted glioma frequently had heterogeneous signal intensity on T2WI [6, 36], mixed/restricted diffusion characteristics [37], and concentration of endogenous cellular proteins in tissue, which can be reflected by radiomic features from APTw, varies across different molecular subtypes of gliomas [38].
Of all these models, the radiomics model achieved the best performance in the testing set, which means the radiomics model had strong predictive power for 1p/19q co-deletion. The clinical model demonstrates similar performance in both the training and testing sets with an AUC of 0.760 and 0.766. However, in the training set, the AUC value of the radiomics model is lower than that of the combined model. Contrastingly, in the testing set, the AUC value of the radiomics model is higher than that of the combined model. This observed difference could be attributed to the relatively smaller sample size of the testing set, indicating the need for a larger dataset evaluation to draw more conclusive results.
Our study has several limitations. First, it was based on a single-center, retrospectively collected dataset, multi-center data will be needed to allow external validation. Secondly, other medical images including perfusion-weighted imaging (PWI) and computed tomography (CT) may provide extra functional and calcified information; accordingly, we suggest that further work includes more imaging modalities to explore the performance of radiomic models. Finally, our work only focused on the prediction of the 1p/19q co-deletion genotype, the analysis of other molecular subtype, including IDH1/2, and CDKN2A/B will permit additional comprehensive understanding of the diffuse gliomas.
Conclusions
Radiomic features from APTw, DWI and conventional MRI sequences can preoperatively and non-invasively distinguish the 1p/19q co-deletion genotype in patients with LGG. The predictive performance of radiomics model was comparable to that of experienced neuroradiologist, significantly outperforming the diagnostic accuracy of resident physicians. These findings suggest that our radiogenomics approach has the potential to become a valuable tool in clinical decision making for LGG patients.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- LGG
Low-grade gliomas
- APTw
Amide proton transfer weighted
- DWI
Diffusion weighted imaging
- LASSO
Least absolute shrinkage and selection operator
- Rad-score
Radiomics score
- IDH
Isocitrate dehydrogenase
- T1WI
T1-weighted imaging
- FLAIR
Fluid-attenuated inversion recovery
- T1C
Contrast-enhanced T1-weighted imaging
- CEST
Chemical exchange saturation transfer
- MGMT
Human O6-methylguanine DNA methyltransferase
- H3K27M
Lys-27-Met mutations in histone 3 genes
- DSW
Direct saturation of water
- MTC
Semi-solid magnetization transfer contrast
- MTR
Magnetization transfer ratio
- FISH
Fluorescence in situ hybridization
- LSI
Locus specific identifier
- TR
Repetition time
- TE
Echo time
- VOI
Volume of interest
- ADC
Apparent diffusion coefficient
- GLCM
Gray level co-occurrence matrix
- GLRLM
Gray level run length matrix
- GLSZM
Gray level size zone matrix
- NGTDM
Neighboring gray tone difference matrix
- GLDM
Gray level dependence matrix
- AUC
Area under the curve
- ROC
Receiver operating characteristics
- PPV
Positive predictive value
- NPV
Negative predictive value
- PWI
Perfusion-weighted imaging
- CT
Computed tomography
Authors’ contributions
AM, YQ and XY analyzed data. AM drafted the manuscript text. YQ and ZW substantively revised the manuscript text. YQ, JM, HW, AM, XY, XZ, XL and ML acquired MRI data. All authors read and approved the final manuscript.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province (2022A1515011585).
Availability of data and materials
The datasets generated or analyzed in the current study are not publicly available because of patient privacy protection, but are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was carried out in accordance with the Declaration of Helsinki. We confirm that all methods were carried out in accordance with relevant guide-lines and regulations. The Ethics Committee of the Zhujiang Hospital of Southern Medical University approved this retrospective single institution study and waived the requirement for informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cancer Genome Atlas Research N. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubbink HJ, Atmodimedjo PN, Kros JM, French PJ, Sanson M, Idbaih A, Wesseling P, Enting R, Spliet W, Tijssen C, et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC brain tumor group 26951 phase III trial. Neuro Oncol. 2016;18(1523-5866 (Electronic)):388–400. doi: 10.1093/neuonc/nov182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wijnenga MMJ, French PJ, Dubbink HJ, Dinjens WNM, Atmodimedjo PN, Kros JM, Smits M, Gahrmann R, Rutten GJ, Verheul JB, et al. The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol. 2018;20(1):103–112. doi: 10.1093/neuonc/nox176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi T, Ohno M, Matsushita Y, Takahashi M, Miyakita Y, Kitagawa Y, Kondo E, Tsushita N, Satomi K, Yoshida A, et al. Radiological characteristics based on isocitrate dehydrogenase mutations and 1p/19q codeletion in grade II and III gliomas. Brain Tumor Pathol. 2018;35(3):148–158. doi: 10.1007/s10014-018-0321-4. [DOI] [PubMed] [Google Scholar]
- 6.Smits M. Imaging of oligodendroglioma. Br J Radiol. 2016;89(1060):20150857. doi: 10.1259/bjr.20150857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su MY, Cha S, Filippi CG, Bota D, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39(7):1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kempen EJ, Post M, Mannil M, Kusters B, ter Laan M, Meijer FJA, Henssen DJHA. Accuracy of machine learning algorithms for the classification of molecular features of gliomas on MRI: a systematic literature review and meta-analysis. Cancers. 2021;13(11):2606. doi: 10.3390/cancers13112606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams S, Layard Horsfall H, Funnell JP, Hanrahan JG, Khan DZ, Muirhead W, Stoyanov D, Marcus HJ. Artificial intelligence in brain tumour surgery-an emerging paradigm. Cancers (Basel) 2021;13(19):5010. doi: 10.3390/cancers13195010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Zaiss M, Knutsson L, Sun PZ, Ahn SS, Aime S, Bachert P, Blakeley JO, Cai K, Chappell MA, et al. Review and consensus recommendations on clinical APT-weighted imaging approaches at 3T: application to brain tumors. Magn Reson Med. 2022;88(2):546–574. doi: 10.1002/mrm.29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang S, Rui Q, Wang Y, Heo HY, Zou T, Yu H, Zhang Y, Wang X, Du Y, Wen X, et al. Discriminating MGMT promoter methylation status in patients with glioblastoma employing amide proton transfer-weighted MRI metrics. Eur Radiol. 2018;28(5):2115–2123. doi: 10.1007/s00330-017-5182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Zou T, Eberhart CG, Villalobos MAV, Heo HY, Zhang Y, Wang Y, Wang X, Yu H, Du Y, et al. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI. Magn Reson Med. 2017;78(3):1100–1109. doi: 10.1002/mrm.26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuo Z, Qu L, Zhang P, Duan Y, Cheng D, Xu X, Sun T, Ding J, Xie C, Liu X, et al. Prediction of H3K27M-mutant brainstem glioma by amide proton transfer-weighted imaging and its derived radiomics. Eur J Nucl Med Mol Imaging. 2021;48(13):4426–36. [DOI] [PubMed]
- 15.Paech D, Windschuh J, Oberhollenzer J, Dreher C, Sahm F, Meissner JE, Goerke S, Schuenke P, Zaiss M, Regnery S, et al. Assessing the predictability of IDH mutation and MGMT methylation status in glioma patients using relaxation-compensated multipool CEST MRI at 7.0 T. Neuro-Oncology. 2018;20(1523-5866 (Electronic)):1661–1671. doi: 10.1093/neuonc/noy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancini L, Casagranda S, Gautier G, Peter P, Lopez B, Thorne L, McEvoy A, Miserocchi A, Samandouras G, Kitchen N, et al. CEST MRI provides amide/amine surrogate biomarkers for treatment-naïve glioma sub-typing. Eur J Nucl Med Mol Imaging. 2022;49(7):2377–91. [DOI] [PMC free article] [PubMed]
- 17.Su C, Xu S, Lin D, He H, Chen Z, Damen FC, Ke C, Lv X, Cai K. Multi-parametric Z-spectral MRI may have a good performance for glioma stratification in clinical patients. Eur Radiol. 2021;32(1):101–111. doi: 10.1007/s00330-021-08175-3. [DOI] [PubMed] [Google Scholar]
- 18.Sun C, Fan L, Wang W, Wang W, Liu L, Duan W, Pei D, Zhan Y, Zhao H, Sun T, et al. Radiomics and qualitative features from multiparametric MRI predict molecular subtypes in patients with lower-grade glioma. Front Oncol. 2021;11:756828. doi: 10.3389/fonc.2021.756828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MK, Park JAO, Jo Y, Park SY, Kim SJ, Kim HS. Advanced imaging parameters improve the prediction of diffuse lower-grade gliomas subtype, IDH mutant with no 1p19q codeletion: added value to the T2/FLAIR mismatch sign. European Radiol. 2020;30(1432-1084 (Electronic)):844–854. doi: 10.1007/s00330-019-06395-2. [DOI] [PubMed] [Google Scholar]
- 20.Xiong J, Tan W, Wen J, Pan J, Wang Y, Zhang J, Geng D. Combination of diffusion tensor imaging and conventional MRI correlates with isocitrate dehydrogenase 1/2 mutations but not 1p/19q genotyping in oligodendroglial tumours. Eur Radiol. 2015;26(6):1705–1715. doi: 10.1007/s00330-015-4025-4. [DOI] [PubMed] [Google Scholar]
- 21.van der Voort SR, Incekara F, Wijnenga MMJ, Kapas G, Gardeniers M, Schouten JW, Starmans MPA, Nandoe Tewarie R, Lycklama GJ, French PJ, et al. Predicting the 1p/19q codeletion status of presumed low-grade glioma with an externally validated machine learning algorithm. Clin Cancer Res. 2019;25(24):7455–7462. doi: 10.1158/1078-0432.CCR-19-1127. [DOI] [PubMed] [Google Scholar]
- 22.Song Y, Zhang J, Zhang YD, Hou Y, Yan X, Wang YD, Zhou MX, Yao YF, Yang G. FeAture Explorer (FAE): a tool for developing and comparing radiomics models. Plos One. 2020;15(8):e0237587. doi: 10.1371/journal.pone.0237587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts H. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(1538-7445 (Electronic)):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–74. [DOI] [PMC free article] [PubMed]
- 25.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 27.Buckner J, Giannini C, Eckel-Passow J, Lachance D, Parney I, Laack N, Jenkins R. Management of diffuse low-grade gliomas in adults - use of molecular diagnostics. Nat Rev Neurol. 2017;13(1759-4766 (Electronic)):340–351. doi: 10.1038/nrneurol.2017.54. [DOI] [PubMed] [Google Scholar]
- 28.Alattar AA, Brandel MG, Hirshman BR, Dong X, Carroll KT, Ali MA, Carter BS, Chen CC. Oligodendroglioma resection: a Surveillance, Epidemiology, and End Results (SEER) analysis. J Neurosurg. 2018;128(1933-0693 (Electronic)):1076–1083. doi: 10.3171/2016.11.JNS161974. [DOI] [PubMed] [Google Scholar]
- 29.Hou Z, Zhang K, Liu X, Fang S, Li L, Wang Y, Jiang T. Molecular subtype impacts surgical resection in low-grade gliomas: a Chinese Glioma genome atlas database analysis. Cancer Lett. 2021;522:14–21. doi: 10.1016/j.canlet.2021.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Xie Z, Zang Y, Zhang S, Gu D, Zhou M, Gevaert O, Wei J, Li C, Chen H, et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J Neurooncol. 2018;140(2):297–306. doi: 10.1007/s11060-018-2953-y. [DOI] [PubMed] [Google Scholar]
- 31.He J, Ren J, Niu G, Liu A, Wu Q, Xie S, Ma X, Li B, Wang P, Shen J, et al. Multiparametric MR radiomics in brain glioma: models comparation to predict biomarker status. BMC Med Imaging. 2022;22(1):137. doi: 10.1186/s12880-022-00865-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaragori T, Oster J, Roch V, Hossu G, Chawki MB, Grignon R, Pouget C, Gauchotte G, Rech F, Blonski M, et al. (18)F-FDOPA PET for the noninvasive prediction of glioma molecular parameters: a radiomics study. J Nucl Med. 2022;63(1):147–157. doi: 10.2967/jnumed.120.261545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Ma Z, Pei D, Duan W, Guo Y, Liu Z, Guan F, Wang Z, Xing A, Guo Z, et al. Improving noninvasive classification of molecular subtypes of adult gliomas with diffusion-weighted MR imaging: an externally validated machine learning algorithm. J Magn Reson Imaging. 2023;58(4):1234–42. doi: 10.1002/jmri.28630. [DOI] [PubMed] [Google Scholar]
- 34.Jiang S, Guo P, Heo HY, Zhang Y, Wu J, Jin Y, Laterra J, Eberhart CG, Lim M, Blakeley JO. Radiomics analysis of amide proton transfer-weighted and structural MR images for treatment response assessment in malignant gliomas. NMR in Biomedicine. 2022;36(1):e4824. doi: 10.1002/nbm.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartoretti E, Sartoretti T, Wyss M, Reischauer C, van Smoorenburg L, Binkert CA, Sartoretti-Schefer S, Mannil M. Amide proton transfer weighted (APTw) imaging based radiomics allows for the differentiation of gliomas from metastases. Sci Rep. 2021;11(1):5506. doi: 10.1038/s41598-021-85168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batchala PP, Muttikkal TJE, Donahue JH, Patrie JT, Schiff D, Fadul CE, Mrachek EK, Lopes MB, Jain R, Patel SH. Neuroimaging-based classification algorithm for predicting 1p/19q-codeletion status in IDH-mutant lower grade gliomas. AJNR Am J Neuroradiol. 2019;40(3):426–432. doi: 10.3174/ajnr.A5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park YW, Han K, Ahn SS, Bae S, Choi YS, Chang JH, Kim SH, Kang SG, Lee SK. Prediction of IDH1-Mutation and 1p/19q-Codeletion Status Using Preoperative MR Imaging Phenotypes in Lower Grade Gliomas. AJNR Am J Neuroradiol. 2018;39(1):37–42. doi: 10.3174/ajnr.A5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Payen JF, Wilson DA, Traystman RJ, Van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9(1078-8956 (Print)):1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed in the current study are not publicly available because of patient privacy protection, but are available from the corresponding author upon reasonable request.