ABSTRACT
Serum polymerase chain reaction (PCR) for the detection of Coxiella burnetii DNA has been suggested for rapid Q fever diagnosis. We evaluated the role of PCR testing in serum in the diagnosis of acute Q fever in an endemic setting. We examined patients suspected of acute Q fever tested for C. burnetii-specific serum real-time PCR in a tertiary hospital between January 2019 toand December 2022. In the first half, PCR orders were consultation-based by infectious diseases specialists, while in the second half, they were guided by serology, positive IgM2, and negative IgG1 and IgG2, indicating early acute infection. Logistic regression analyzed independent predictors for positive PCR. PCR positivity rates were calculated using various clinical criteria in the diagnostic algorithm. Out of 272 patients, 13 (4.8%) tested positive and 130 exhibited serologically suspected early infection. Presentation during April–July and aspartate aminotransferase (AST) > 3× upper normal limit (UNL) were independently associated with positive PCR with an odds ratio (OR) = 15.03 [95% confidence interval (CI), 1.58–142.46], P = 0.018 and OR = 55.44 [95% CI, 6.16–498.69], P < 0.001, respectively. PCR positivity rate was 8.5% in serologically suspected early infection vs 1.4% in other serology, yielding OR = 6.4 [95% CI, 1.4–29.7], P = 0.009. Adding AST > 3× UNL increased OR to 49.5 [95% CI, 5.9–408.7], P ≤ 0.001 reducing required PCR tests for a single acute Q fever case from 11.8 to 3. Elevated AST in serologically suspected early Q fever is proposed to be used in a diagnostic stewardship algorithm integrating PCR in serum in an endemic setting.
IMPORTANCE
Our study suggests in a diagnostic stewardship approach the integration of molecular testing (Coxiella burnetii targeted PCR) for the diagnosis of acute Q fever in a reliable time in the endemic setting. Integrating PCR detecting Coxiella burnetii in serum in routine testing of suspected early acute Q fever based on serology result increased the PCR positivity rate significantly. Adding increased transaminases optimizes PCR utility which is highly requested particularly in endemic areas.
KEYWORDS: Coxiella burnetii, acute Q fever, polymerase chain reaction, diagnosis, endemic
INTRODUCTION
Q fever is a zoonotic infection caused by the obligate intracellular Gram-negative bacteria Coxiella burnetti. Although the infection has been documented worldwide, it has been reported with a wide range of endemicity (1). In 2019, about 1,000 cases of Q fever were reported from European Union with the highest incidence rate reported from Spain (0.7 per 100,000) (2). In the same year, about 210 cases of Q fever were reported in the United States of America (estimated incidence of 0.06 per 100,000) (3). Israel has been endemic for Q fever for many years, with the most recent estimated incidence being around 3 per 100,000 in 2022 and of more than 400 expected new cases in 2023 (4).
The clinical presentation of acute Q fever varies, mostly being asymptomatic (5). Symptomatic patients may present with fever and other non-specific symptoms. A wide spectrum of clinical manifestations has been reported with pneumonia and hepatitis being the most typical (6). Less common manifestations include neurological (7) and cardiac (8, 9) as well as other rare manifestations (1). This wide spectrum of clinical presentation makes acute Q fever to be frequently included in the differential diagnoses in the endemic setting.
The laboratory diagnosis of acute Q fever is based mainly on the detection of phase 2 C.Coxiella burnetii antibodies by serology testing. As the antibodies response takes time to appear and serology-based diagnosis is not straightforward especially when based on a single sample result (particularly in endemic places), the uncertainty about acute Q fever diagnosis in real time leads frequently to additional unnecessary tests. Indeed, in the majority of cases, the diagnosis of acute Q fever is made in retrospect with a repeated serology test documenting seroconversion or a significant increase of IgG 2 antibodies titer. This delay in diagnosis is frequently associated with prolonged hospitalization, unnecessary and costly laboratory and imaging studies, and inappropriate/excessive antimicrobial treatment such as therapy with beta-lactams for community-acquired pneumonia.
Over the past years, molecular tests have been used in the diagnosis of Q fever, where polymerase chain reaction (PCR) has been introduced mainly in the diagnosis of the chronic persistent form of the disease as well as for monitoring treatment response (10, 11). In acute Q fever, several studies have focused on the accuracy of PCR (12–14), its role in epidemiologic surveillance (15) or in outbreaks in small cohorts (16, 17). No data are available on the role of PCR in the diagnosis of acute Q fever in an endemic setting.
The objective of this study was to assess the added value of C. burnetii-specific real-time PCR in serum in the diagnosis of hospitalized patients with suspected acute Q fever in an endemic setting. We evaluated the yield of PCR testing in cases with serologically suspected early acute Q fever compared to a restricted testing approach mandating an infectious diseases (ID) specialist approval. In addition, we looked for clinical and laboratory parameters that might improve the integration of serum PCR testing in the diagnostic algorithm of patients with suspected acute Q fever.
MATERIALS AND METHODS
Setting
We retrospectively identified all adult patients (age > 18 years) with suspected acute Q fever who had been tested for C. burnetii PCR in serum between 1 January 2019 and 31 December 2022. The study was conducted at Rambam Health Care Campus (RHCC), a tertiary university-affiliated hospital with 1,000 beds, located in Haifa, the largest city in the north of Israel, a region with a high prevalence of Q fever.
Up to the end of 2020, C. burnetii serum PCR has been in use at RHCC microbiology laboratory for the diagnosis of suspected acute Q fever according to ID specialist approval (consultation-based approach). Since January 2021, routine testing of C. burnetii PCR has been performed for all serology specimens with a result suspected of early infection of acute Q fever in hospitalized patients (serology-based approach). The definition of serologically suspected early infection was the presence of phase 2 IgM (IgM2) and absence of phase 2 and phase 1 IgG (IgG2 and IgG1) as detected in enzyme-linked immunosorbent assay (ELISA), used as the screening test in the hospital laboratory.
Study design
This study retrospectively analyzes data from a prospective database of Q fever. We compared the yield of serum PCR testing to detect acute Q fever in each of the two periods, consultation-based vs serology-based periods.
In addition, we focused on the group of patients with serologically suspected early acute infection. Clinical and laboratory characteristics of patients with positive and negative PCR results within this group were compared to identify independent factors associated with positive PCR.
In a subgroup analysis for patients with serologically suspected early infection who had clinical and serological follow-up, we evaluated the diagnostic accuracy of the PCR in serum in the diagnosis of acute Q fever by calculating the sensitivity, specificity, positive predictive value, and negative predictive value. The reference standard for diagnosis of acute Q fever was defined as a composite of appropriate clinical illness along with serology dynamics compatible with acute Q fever, i.e., seroconversion or fourfold increase in phase 2 IgG within 3–6 weeks. Moreover, we compared the characteristics of patients according to the PCR result as compared to the final diagnosis—true positive (TP), true negative (TN), and false negative (FN) looking for factors that could differentiate between these groups.
Finally, we compared PCR positivity rates applying serological and clinical criteria looking for the optimal situation in which the integration of PCR testing will be beneficial. Based on these results, we suggest a laboratory diagnostic algorithm in which the integration of PCR in serum might be helpful in rapid diagnosis of acute Q fever in the endemic setting.
Microbiology testing
Real-time PCR for the detection of C. burnetii was performed in serum samples targeting IS1111 a 295-bp fragment of the transposase gene of the C. burnetii IS1111a element (18). DNA extraction was performed on the QIAcube instrument (Qiagen, Crawley, UK) up to the end of 2021 and on MagLEAD 12gc extraction system (Percision System Science, Japan) thereafter. Each sample was tested for inhibition by testing globin as an internal control. Cycle threshold (Ct) value cutoff ≤ 35 was reported positive, while the value ≥45 was reported negative. For borderline results (at least one of the duplicates with Ct value 36–44), the test was repeated and reported as weak positive if similar result was achieved (Ct > 35 and Ct < 45).
For serology testing, the screening test that was used in RHCC microbiology laboratory was enzyme-linked immunoassay (ELISA (Serion Diagnostics, Germany) which tests for phase 2 IgM and phase 2 and phase 1 IgG. For any positive IgG result, an indirect immunofluorescence assay (IFA) testing was performed for confirmation and titer determination using an in-house test at the national reference laboratory for rickettsiosis.
Statistical analysis
Data were analyzed using SPSS version 26. Patients' characteristics were summarized by standard descriptive summaries using medians with interquartile range for continuous variables and counts and percentages for categorical variables. The χ2 test was used to assess the correlation between potential predictors and serum PCR for Q fever. Binary logistic regression was used for multivariate analysis. Variables with P < 0.05 in the bivariate analysis were included in the binary logistic regression. The association was considered significant if its coefficient in the binary logistic regression equation remained significant at P < 0.05. All statistic tests are two-tailed, P value 0.05 was considered statistically significant [95% confidence interval (CI)] for all tests.
RESULTS
Between January 2019 and December 2022, 4,388 sera were tested for Q fever serology at RHCC microbiology laboratory. Overall serology tests during the study period classified according to results and proportion of samples tested by PCR and their positivity rates are presented in Table 1. The most prevalent result was IgM−/IgG− which was observed in 83%, while serology of possible early infection, IgM+/IgG− was observed in 5.3%. PCR was tested in 272 (6.2%) of overall sera of patients with suspected acute Q fever; 96 during the consultation-based period and 176 during serology-based period. Positive PCR results in serum were detected in 4.8% (13/272) in the overall, comprising 2% (2/96) in the consultation-based period and 6.3% (11/176) in the serology-based period, P = 0.14. The demographic data, comorbidities, and clinical presentation of patients were similar in both periods, except for lower C-reactive protein (CRP) levels in the consultation-based period with a median of 5.4 (mg/dL) compared to 8.3 in the serology-based period (P = 0.04; see Table S1 at https://figshare.com/articles/figure/Supplemental_material-_The_integration_of_Coxiella_burnetii_PCR_testing_in_serum_into_the_diagnostic_algori/25303375/1). No difference was observed in the PCR positivity rates between the two extraction systems used during the study period in which PCR positivity rates were 9/188 (4.8%) and 4/88 (4.8%) with the QIAcube instrument and the MagLEAD system, respectively.
TABLE 1.
The overall serology profile results and their proportion from those tested by PCR and corresponding positivity rates
| Serology profile | Overall serology result N (%) |
Samples tested by PCR N (%) |
PCR positive Nn (%) |
|---|---|---|---|
| IgM− IgG− | 3,631 (83) | 110 (3) | 2 (1.8) |
| IgM+ IgG− | 249 (5.6) | 130 (52) | 11 (8.4) |
| IgM− IgG+ | 341 (7.7) | 27 (8) | 0 (0) |
| IgM+ IgG+ | 167 (3.8) | 6 (3.6) | 0 (0) |
| Total | 4,388 | 272 (6.2) | 13 (4.8) |
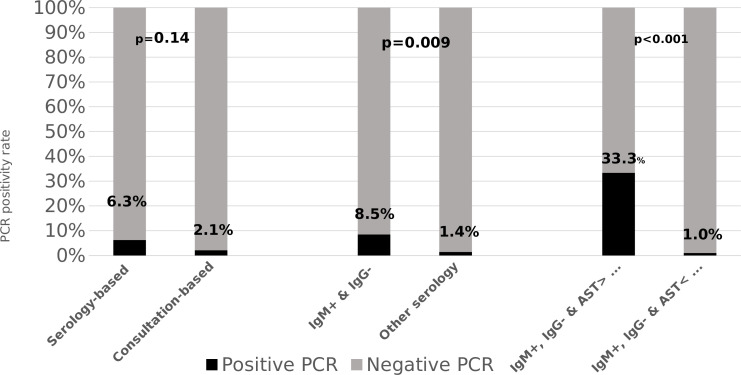
Overall, 130/272 patients had a serology result of phase 2 IgM+, IgG−, consistent with suspected early infection of acute Q fever; 22 in the consultation-based period and 108 from the serology-based period. PCR positivity rate at this group of patients with suspected early infection was 8.5% (11/130) compared to 1.4% (2/142), for patients with any other serology result, with an odds ratio (OR) [95% CI)] of 6.4 [1.4–29.7], P = 0.009. Comparison of the characteristics of patients with positive and negative PCR results in this group is presented in Table 2. The basic demographic characteristics including age, gender, and ethnicity were similar. Presentation between April toand July was observed in 10 cases (91%) of the PCR positive group compared to 51 (43%) in the PCR negative group, P = 0.002. Although there was a statistically significant difference in leukocyte count on presentation, values in both groups fell within the normal range. The median CRP was increased in both groups,; 9.1 mg/dL in the PCR positive and 5.0 mg/dL in the PCR negative group, with P = 0.05. Significantly higher levels of liver enzymes were observed in the PCR positive group compared to PCR negative group. Notably, the proportion of patients with aspartate aminotransferase (AST) and alanine transaminase (ALT) above threefold the upper normal limit (>3× xUNL) was significantly higher in the PCR positive group. In multivariate logistic regression analysis including presentation between April toand July, AST > 3× xUNL and CRP, it was independently observed that presentation between April toand July and AST > 3× xUNL were associated with positive PCR with OR [95% CI] of 15.03 [1.58–142.46], P = 0.018 and 55.44 [6.16–498.69], P =<≤ 0.001, respectively. Data of Ct were available in 12/13 positive PCR cases with a mean of 35.8 ± 6.0 [range: 24–42]. In five out of 12 samples (42%), PCR was positive (the Ct value was <35), while in seven (58%), the PCR result was weak positive (Ct between 36 and 44). The association between the duration of fever and Ct values is illustrated in Fig. S1 at https://figshare.com/articles/figure/Supplemental_material-_The_integration_of_Coxiella_burnetii_PCR_testing_in_serum_into_the_diagnostic_algori/25303375/1, demonstrating that all cases were detected during the first two2 weeks of fever with a trend of increasing Ct values with increasing time since fever onset.
TABLE 2.
Comparison between positive and negative PCR results in patients with serologically suspected early Q fever infection (with IgM2+/IgG−)
| Variable | Positive PCR N = 11 |
Negative PCR N = 119 |
Univariate analysis P value |
Multivariate analysis OR [95% CI], P value |
|---|---|---|---|---|
| Age | 47 [26, 67] | 47 [32, 66] | 1.00 | |
| Male | 6 (54.5) | 68 (57.1) | 1.00 | |
| Jewish | 10 (91) | 87 (73) | 0.28 | |
| Rural residence | 0 (0) | 30 (25.2) | 0.06 | |
| Presentation between April and July | 10 (91) | 51 (43) | 0.002 | 15.03 [1.58–142.46], 0.018 |
| Charlson comorbidities index | 6 [0, 6] | 4 [2, 8] | 0.87 | |
| Malignancy | 1 (9) | 12 (10) | 1.00 | |
| Cerebral vascular accident (CVA) | 1 (9) | 11 (9.2) | 1.00 | |
| Congestive heart failure | 2 (18) | 14 (11.7) | 0.61 | |
| Ischemic heart disease | 2 (18) | 8 (6.7) | 0.20 | |
| Diabetes | 1 (9) | 12 (10) | 1.00 | |
| Chronic kidney disease | 2 (18) | 21 (17.6) | 1.00 | |
| Valvular heart disease | 2 (18) | 7 (5.8) | 0.56 | |
| Vascular aneurysm | 2 (18) | 1 (0.8) | 0.24 | |
| White blood cell count (103/µL) | 6.7 [4.6, 7.2] | 8.0 [5.6, 11.2] | 0.01 | |
| Hemoglobin (g/dL) | 12.4 [11.5, 14] | 12 [10.1, 13.2] | 0.52 | |
| Platelets (103/µL) | 181 [173, 273] | 221 [146, 328] | 0.97 | |
| Aspartate AST (U/L) | 191 [112, 283] | 45 [30, 86] | 0.001 | |
| AST > 3x UNL (U/L) | 10 (91) | 20 (17.2) | <0.001 | 55.44 [6.16–498.69], <0.001 |
| ALT (U/L) | 254 [111, 499] | 50 [19, 127] | 0.01 | |
| ALT > 3x UNL (U/L) | 7 (63.6) | 24 (21) | 0.005 | |
| Alkaline phosphatase (U/L) | 136 [95, 214] | 90 [67, 165] | 0.18 | |
| GGT (U/L) | 134 [97, 546] | 68 [31, 219] | 0.01 | |
| Bilirubin (µmol/L) | 0.3 [0.3, 0.7] | 0.6 [0.4, 0.8] | 0.20 | |
| Creatinine (mg/dL) | 0.7 [0.5, 0.9] | 0.7 [0.6, 0.9] | 0.87 | |
| Sodium (µmol/L) | 137 [136, 139] | 137 [135, 139] | 0.95 | |
| CRP reactive protein (mg/dL), normal < 0.5 (N = 126) | 9.1 [6.2, 12.3] | 5.0 [1.5, 12.0] | 0.05 | 1.55 [0.67–3.55], 0.30 |
| Length of hospital stay (days) | 4.1 [3.5, 5.0] | 6.0 [3.0, 11.7] | 0.029 |
In the subgroup analysis of the patients with serologically suspected early infection; 41/130 (31%) patients had clinical and serology follow-up in whom the PCR result was compared to the final diagnosis. Eighteen patients were diagnosed with acute Q fever; 11 had positive PCR (TP = 11) and 7 patients had negative PCR (FN = 7). Twenty-three patients had other diagnoses (TN = 23), and no patient with positive PCR was diagnosed with other than acute Q fever (FP = 0). The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were 61%, 100%, 100%, and 77%, respectively. Characteristics of these three groups (TP, TN, and FN) appear in Table S2 at https://figshare.com/articles/figure/Supplemental_material-_The_integration_of_Coxiella_burnetii_PCR_testing_in_serum_into_the_diagnostic_algori/25303375/1. Of notice, no difference in duration of symptoms was observed with a median of 9 days in all three groups, while the only difference was in elevated transaminases, mainly AST which was higher in TP group compared to both TN and FN groups. Indeed, reassessment of the PCR positivity rate by adding the AST > 3× xUNL in serologically suspected early infection increased the OR to 49.5 [95% CI, 5.9–408.7], P= <≤ 0.001. In this scenario, the number of PCR tests that was needed to be performed would be reduced from 130 to 30 in our cohort, translated to 77% of minimizing of total tests that were performed and leading to a decrease in the number of tests needed to perform in order to diagnose a one case of acute Q fever from 11.8 (130/11) to 3 (30/10). Comparison of PCR positivity rates according to periods, serology, and elevated AST is illustrated in Fig. 1. Paired serology tests were found in 74 patients of whom 28 had final diagnosis of acute Q fever, 13 who had positive PCR, and 15 with negative PCR. For acute Q fever negative PCR group, the serology profile result of first test was as follows: one had IgM−/IgG−, seven had IgM+/IgG−, and seven had IgM+/IgG+. None had IgM−/IgG+.
Fig 1.
Positivity rates of PCR across various clinical scenarios.
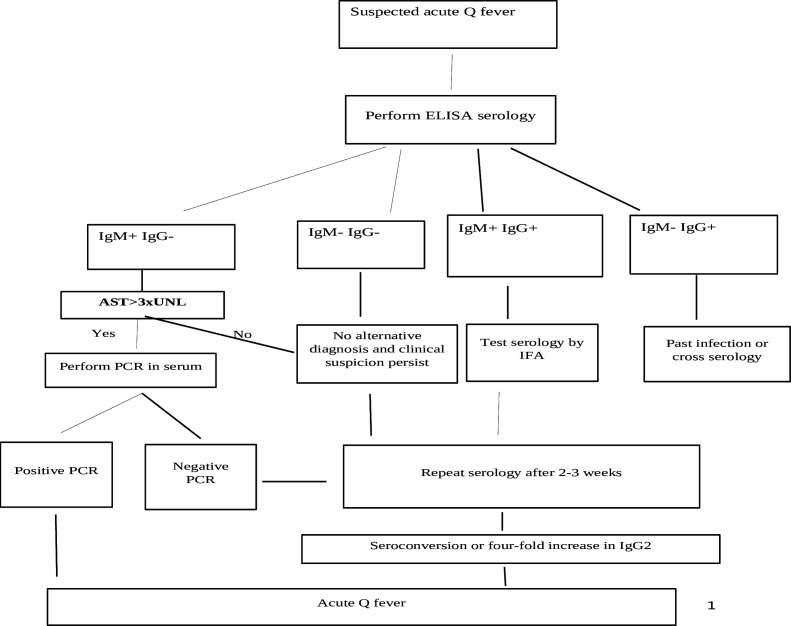
Based on the results, we suggest a diagnostic algorithm that incorporates AST > 3× xUNL into the PCR testing protocol for Q fever in patients with suspected acute Q fever, identified through serology results of IgM+, IgG− particularly in the endemic setting as presented in Fig. 2.
Fig 2.
Proposed algorithm for diagnosis of acute Q fever in an endemic setting.
DISCUSSION
In the present study, we found that in the endemic setting, including suspected early acute Q fever serology (IgM+, IgG−) in decision-making process for serum PCR testing for hospitalized patients with clinically compatible Q fever yielded a higher positivity rate compared to an approach based solely on clinical suspicion and ID consultation. This finding highlights the challenge in making the diagnosis of acute Q fever on a clinical basis, especially in an endemic setting. Consequently, prioritizing PCR testing should be supported by additional well-defined criteria such as serology results and elevated AST as proposed in our diagnostic algorithm.
The high specificity and PPV endorse the use of serum PCR in serologically suspected cases. This finding is supported by similar results from previous studies demonstrating the high specificity of PCR in serum for diagnosis of acute Q fever ranging between 90%- and 100% (12–15). FromOn the other side, the relatively low sensitivity and NPV in our study as had been reported from other studies (12, 14), should be considered when interpreting a negative PCR result. Based on our results, negative PCR should be followed by a repeated serology test to exclude acute Q fever.
In settings of a high demand for Q fever testing, such as during an outbreak or in an endemic area, starting with a screening serology test such as IgM2 is reasonable as has been proposed previously (19). The low specificity of IgM that cross-react with other infectious and non-infectious etiologies alongside the expected increased costs and efforts needed for the implementation of routine use of both IFA and PCR in endemic setting necessitates a diagnostic stewardship approach as we proposed in our algorithm. The addition of increased AST > 3× xUNL, decreases the number needed to test from 11.8 to 3 in our cohort. Although not addressed in our study, such an approach seems to improve the cost-effectiveness of the integration of PCR in serum, but this should be further evaluated. This is particularly important in the endemic setting where making the diagnosis on a clinical basis is challenging, often leading to its consideration in the differential diagnosis of variable clinical syndromes as mentioned previously. This approach is further supported by the high demand for serology tests in the endemic setting as presented in Table 1 in which an average of 100 tests per month were ordered in our hospital. Patients with acute Q fever might have a serology profile of either IgM−/IgG−, IgM+/IgG−, or IgM+/IgG+, depending on the time point in which serology was taken in relation to illness course and factors related to variable host immune reaction, making a single sample serology result highly challenging for interpretation. Therefore, our proposed algorithm suggests either repeating the serology test after a while in order to show seroconversion/significant antibodies rise or testing for PCR where we expect to have a high yield of this test (in IgM+/IgG− with elevated AST as supported by our study results).
It is important to emphasize that our suggested algorithm is derived from hospitalized patients,; thus, it is suggested to be applied in a similar setting. Typically, patients are hospitalized usually one1 week or more after the onsetstart of a non-resolving fever. In addition, abnormal laboratory results, mainly elevated transaminases, may contribute to selection bias for hospitalization and even prompt serology testing for Q fever. In contrast to previous studies (14, 20), we found that elevated AST, rather than duration of illness, was significantly associated with positive PCR. Here to emphasize, that the vast majority of sera that were tested by PCR in our study were with IgG negative (88%) as it is already known that PCR yield falls dramatically at the time IgG is detected (17),; thus, duration of symptoms was relatively short and homogenous in the whole cohort. Still, the association between elevated AST and positive PCR might be related to other factors. One possible explanation for this finding might be that bacteremia of C. burnetii are more prevalent in cases involving hepatitis compared to those without hepatitis, such as isolated pneumonia where the infection may be contained in the respiratory tract and lung. Such differences might be related to different routes of acquiring the infection as had been suggested in an animal model study (21). Whether elevated AST could be used as a criterion for testing also in the absence of IgM2 (IgM−/IgG−) should be further evaluated in future studies.
Our study has limitations. First, being a single and tertiary center study, might affected the type of patients included in our cohort, although the clinical characteristics of our patients did not have major differences compared to cohorts of acute Q fever that were reported from other non-tertiary hospitals in Israel (22, 23). Second, data that were collected retrospectively limiting our ability to evaluate the overall clinical impact of making a diagnosis with positive PCR in acute Q fever. A previous small study had reported the shorter time to diagnosis in acute Q fever when diagnosis was made by PCR compared to serology (4 days compared to 17 days, respectively) (16). The shorter length of hospital stays in the positive PCR cases compared to negative PCR cases in our study might be attributed to the rapid definite diagnosis achieved by PCR. Third, including only hospitalized patients probably represents more severe cases of acute Q fever with longer duration of symptoms with the expected detection of IgM2 antibodies already upon presentation. For ambulatory milder cases of acute Q fever, a different diagnostic algorithm might be needed, focusing on the seronegative cases (with a corresponded shorter course of illness). Here to highlight that in the present study, we did not check PCR routinely in the group of cases with totally negative results (IgM−, IgG−), in which the number of positive PCR cases might be higher than that in the group of suspected early infection (IgM+, IgG−). As the totally sero-negative group consistituted 83% of the ordered tests for Q fever serology at our hospital, routine testing of PCR in this group is unreliable. Further studies should address clinical and laboratory criteria that could assist in directing the use of PCR in this large group of cases. Finally, as it had been learned from other aspects of Q fever research, data coming from a restricted geographic area should be re-assessed in other areas in order to provide external validation. This is especially true knowing the variability in clinical manifestations of the diseases in different countries (1).
ACKNOWLEDGMENTS
We would like to thank Mr. Tomer Karny from the Information and Computing systems department at Rambam Health Care Campus for his assistance in data extraction and processing. We would like to thank Mrs. Daniel Haber for her research technical assistance.
Contributor Information
Nesrin Ghanem-Zoubi, Email: n_ghanem@rambam.health.gov.il.
Daniel J. Diekema, Maine Medical Center, Portland, Maine, USA
ETHICS APPROVAL
The study was approved by the Institutional Review Board (RMB-19-0729). Informed consent was waived due to the retrospective design of the study.
REFERENCES
- 1. Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. doi: 10.1128/CMR.12.4.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. European Centre for Disease Prevention and Control . 2021. Q fever - annual epidemiological report for 2019. Available from: https://www.ecdc.europa.eu/en/publications-data/q-fever-annual-epidemiological-report-2019 [PubMed]
- 3. CDC . 2019. Q fever: epidemiology and statistics. Available from: https://www.cdc.gov/qfever/stats/index.html
- 4. Israeli Ministry of Health . 2023. Weekly epidemiological reports. Available from: https://www.gov.il/he/departments/dynamiccollectors/weekly-epidemiological-report?skip=0
- 5. Raoult D, Marrie TJ, Mege JL. 2005. Natural history and pathophysiology of Q fever. Lancet Infect Dis 5:219–226. doi: 10.1016/S1473-3099(05)70052-9 [DOI] [PubMed] [Google Scholar]
- 6. Terheggen U, Leggat PA. 2007. Clinical manifestations of Q fever in adults and children. Travel Med Infect Dis 5:159–164. doi: 10.1016/j.tmaid.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 7. Bernit E, Pouget J, Janbon F, Dutronc H, Martinez P, Brouqui P, Raoult D. 2002. Neurological involvement in acute Q fever: a report of 29 cases and review of the literature. Arch Intern Med 162:693–700. doi: 10.1001/archinte.162.6.693 [DOI] [PubMed] [Google Scholar]
- 8. Fournier P-E, Etienne J, Harle J-R, Habib G, Raoult D. 2001. Myocarditis, a rare but severe manifestation of Q fever: report of 8 cases and review of the literature. Clin Infect Dis 32:1440–1447. doi: 10.1086/320159 [DOI] [PubMed] [Google Scholar]
- 9. Badarni K, Blich M, Atiya-Nasagi Y, Ghanem-Zoabi N. 2022. Acute Q fever with atrioventricular block, Israel. Emerg Infect Dis 28:1886–1889. doi: 10.3201/eid2809.212565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kampschreur LM, Wegdam-Blans MCA, Wever PC, Renders NHM, Delsing CE, Sprong T, van Kasteren MEE, Bijlmer H, Notermans D, Oosterheert JJ, Stals FS, Nabuurs-Franssen MH, Bleeker-Rovers CP, Dutch Q Fever Consensus Group . 2015. Chronic Q fever diagnosis—consensus guideline versus expert opinion. Emerg Infect Dis 21:1183–1188. doi: 10.3201/eid2107.130955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diagnosis and management of Q fever — United States. 2013. Available from: https://www.cdc.gov/mmwr/preview/mmwrhtml/rr6203a1.htm
- 12. Fournier PE, Raoult D. 2003. Comparison of PCR and serology assays for early diagnosis of acute Q fever. J Clin Microbiol 41:5094–5098. doi: 10.1128/JCM.41.11.5094-5098.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pradeep J, Stephen S, Ambroise S, Gunasekaran D. 2017. Diagnosis of acute Q fever by detection of Coxiella burnetii DNA using real-time PCR, employing a commercial genesig easy kit. J Clin Diagn Res 11:DC10–DC13. doi: 10.7860/JCDR/2017/31005.10606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bae M, Jin CE, Park JH, Kim MJ, Chong YP, Lee S-O, Choi S-H, Kim YS, Woo JH, Shin Y, Kim S-H. 2019. Diagnostic usefulness of molecular detection of Coxiella burnetii from blood of patients with suspected acute Q fever. Medicine (Baltimore) 98:e15724. doi: 10.1097/MD.0000000000015724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Angelakis E, Mediannikov O, Socolovschi C, Mouffok N, Bassene H, Tall A, Niangaly H, Doumbo O, Znazen A, Sarih M, Sokhna C, Raoult D. 2014. Coxiella burnetii-positive PCR in febrile patients in rural and urban Africa. Int J Infect Dis 28:107–110. doi: 10.1016/j.ijid.2014.05.029 [DOI] [PubMed] [Google Scholar]
- 16. Turra M, Chang G, Whybrow D, Higgins G, Qiao M. 2006. Diagnosis of acute Q fever by PCR on sera during a recent outbreak in rural South Australia. Ann N Y Acad Sci 1078:566–569. doi: 10.1196/annals.1374.112 [DOI] [PubMed] [Google Scholar]
- 17. Schneeberger PM, Hermans MHA, van Hannen EJ, Schellekens JJA, Leenders A, Wever PC. 2010. Real-time PCR with serum samples is indispensable for early diagnosis of acute Q fever. Clin Vaccine Immunol 17:286–290. doi: 10.1128/CVI.00454-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, Appel B. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol 6:6. doi: 10.1186/1471-2180-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jager MM, Weers-Pothoff G, Hermans MHA, Meekelenkamp JCE, Schellekens JJA, Renders NHM, Leenders A, Schneeberger PM, Wever PC. 2011. Evaluation of a diagnostic algorithm for acute Q fever in an outbreak setting. Clin Vaccine Immunol 18:963–968. doi: 10.1128/CVI.00009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boden K, Wagner-Wiening C, Seidel T, Baier M, Bischof W, Straube E, Kimmig P. 2010. Diagnosis of acute Q fever with emphasis on enzyme-linked immunosorbent assay and nested polymerase chain reaction regarding the time of serum collection. Diagn Microbiol Infect Dis 68:110–116. doi: 10.1016/j.diagmicrobio.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 21. Marrie TJ, Stein A, Janigan D, Raoult D. 1996. Route of infection determines the clinical manifestations of acute Q fever. J Infect Dis 173:484–487. doi: 10.1093/infdis/173.2.484 [DOI] [PubMed] [Google Scholar]
- 22. Reisfeld S, Hasadia Mhamed S, Stein M, Chowers M. 2019. Epidemiological, clinical and laboratory characteristics of acute Q fever in an endemic area in Israel, 2006–2016. Epidemiol Infect 147:e131. doi: 10.1017/S0950268818003576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn T, Babushkin F, Geller K, Alexander H, Paikin S, Lellouche J, Atiya-Nasagi Y, Cohen R. 2021. Epidemiological, clinical and laboratory features of acute Q fever in a cohort of hospitalized patients in a regional hospital, Israel, 2012-2018. PLoS Negl Trop Dis 15:e0009573. doi: 10.1371/journal.pntd.0009573 [DOI] [PMC free article] [PubMed] [Google Scholar]