ABSTRACT
Despite optimistic predictions on the eventual end of COVID-19 (Coronavirus Disease 2019), caution is necessary regarding the emergence of new variants to sustain a positive outlook and effectively address any potential future outbreaks. However, ongoing efforts to track COVID-19 variants are concentrated in developed countries and unique social practices and remote habitats of indigenous peoples present additional challenges. By combining small-sized equipment that is easily accessible and inexpensive, we performed SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) whole genome sequencing and measured the sample-to-answer time and accuracy of this portable variant tracking tool. Our portable design determined the variant of SARS-CoV-2 in an infected individual within 9 hours and 15 minutes without external power or internet connection, surpassing the speed of previous portable tools. It took only 16 minutes to complete sequencing run, whole genome assembly, and lineage determination using a single standalone laptop. We then demonstrated the capability to produce 289 SARS-CoV-2 whole genome sequences in a single portable sequencing run, representing a significant improvement over an existing throughput of 96 sequences per run. We verified the accuracy of portable sequencing by comparison with two other independent sequencing methods. We showed that our high-throughput data consistently represented the circulating variants in Los Angeles, United States, when compared with publicly available sequences. Our scheme is designed to be flexible, rapid, and accurate, making it a valuable tool for large-scale surveillance operations in low- and middle-income countries as well as targeted surveys for vulnerable populations in remote locations.
IMPORTANCE
There have been significant efforts to track COVID-19 (Coronavirus Disease 2019) variants, accumulating over 15 million SARS-CoV-2 sequences as of 2023. However, the distribution of global survey data is highly skewed, with nearly half of all countries having inadequate or low levels of genomic surveillance. In addition, indigenous peoples face more severe threats from COVID-19, due to their generally remote residence and unique social practices. Cost-effective portable sequencing tools have been used to investigate Ebola and Zika outbreaks. However, these tools have a sample-to-answer time of around 24 hours and require an internet connection for data transfer to an off-site cloud server. In our study, we rapidly determined COVID-19 variants using only small and inexpensive equipment, with a completion time of 9 hours and 15 minutes. Furthermore, we produced 289 near-full-length SARS-CoV-2 genome sequences from a single portable Nanopore sequencing run, representing a threefold increase in throughput compared with existing Nanopore sequencing methods.
KEYWORDS: COVID variant, portable sequencing, next-generation sequencing
INTRODUCTION
The Omicron variant has garnered significant public concern due to its high levels of immune escape and infectivity since its first appearance in late 2021 (1, 2). This has resulted in a significant reduction in vaccine efficacy compared with earlier viruses (3). It has been demonstrated that the Omicron variant (XBB.1.5) evaded the existing humoral immunity produced by mRNA vaccines or natural infection (4). However, the revising bivalent mRNA vaccine indicated progress in producing neutralizing antibodies against XBB.1.5, emphasizing the importance of updating vaccines in response to currently circulating viruses (4) .
Mutations in the target regions of therapeutic drugs can potentially result in treatment failure. Mutations conferring resistance to FDA-approved antivirals Nirmatrelvir or Remdesivir have been detected with a prevalence as high as 0.4% (5). Continuous surveillance of evolving variants is of utmost importance to ensure the effectiveness of prophylactic vaccines and therapeutic drugs.
Significant efforts have been made to sequence and track novel variants, but the global survey data are highly imbalanced, with around half of all countries having limited or low levels of genomic surveillance (6). As of January 2023, the top 10 countries contributed more than 80% of the global sequences deposited to the Global Initiative on Sharing Avian Influenza Data (GISAID) (7). Addressing geographical disparities in variant tracking should be a priority (8), but this is complicated by challenges faced in low- and middle-income countries, including limited access to high-cost laboratory equipment (9). In response to these challenges, we here provide an affordable and large-scale solution for SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) molecular surveillance in LMICs. Scaling up the local genomic surveillance capacity for the SARS-CoV-2 pandemic will be also essential in preventing and controlling potential future pandemics.
The UN Human Rights Council has urgently called for attention to be given to indigenous peoples facing more severe threats from COVID-19 (Coronavirus Disease 2019) (10). Recent surveys have clearly shown that indigenous peoples experience higher rates of COVID-19 infection and mortality compared with non-indigenous individuals (11, 12). We thus create a portable lab that operates completely independently of electrical and internet infrastructure, making it fully deployable to vulnerable populations in remote locations.
Our portable laboratory’s essential component is the palm-sized MinION sequencer (Oxford Nanopore Technologies). This portable sequencer has been used to investigate Ebola and Zika outbreaks (13, 14) and to perform in-flight sequencing on the Space Station (15). Its advantages include cost-effectiveness and long-read sequencing capabilities. However, its utility in monitoring COVID variants has been hampered by moderate throughput (16–18). Furthermore, to the best of our knowledge, there has been no demonstration of portable sequencing of the SARS-CoV-2 whole genome and its lineage assignment without reliance on an external power source or an internet connection. Herein, we demonstrate rapid determination of COVID-19 variants using only small and inexpensive equipment, with a completion time of 9 hours and 15 minutes. In addition, we produced 289 near-full-length SARS-CoV-2 genome sequences from a single portable Nanopore sequencing run, representing a threefold increase in throughput compared with existing Nanopore sequencing methods. With its portable and cost-efficient design, our system could serve as a valuable resource for tracking COVID-19 variants and supporting public health efforts in LMICs and underserved areas, including indigenous communities.
MATERIALS AND METHODS
COVID-19 positive specimens
We accessed 290 de-identified nasopharyngeal (NP) and/or oropharyngeal (OP) remnant swab specimens that tested positive for COVID-19 using the Roche cobas 6800 SARS-CoV-2 qualitative EUA assay or the Abbott Alinity m SARS-CoV-2 EUA assay at USC Clinical Laboratories, Keck Medicine of USC (Los Angeles, California, USA). This study (HS-20-00326) was approved by the Institutional Review Board of the University of Southern California with a waiver of informed consent. The specimen sequenced by the portable system (TU5327) was collected on 13 December 2022. The cycle threshold (Ct) value of this specimen was 11.75. The other 289 specimens were collected between 22 November 2020 and 5 January 2021. These specimens’ collection date and cycle threshold (Ct) value from the diagnostic testing assays were provided in Table S1.
SARS-CoV-2 whole genome sequencing with portable laboratory
We listed all the components of our portable laboratory in Table S2. SARS-CoV-2 RNA was extracted from NP/OP swab specimens using the QIAamp Viral RNA Mini Kit (Qiagen) with the following modifications: using a tabletop centrifuge for spin-down steps, adding 85 µL of elution buffer, and increasing the elution incubation time to 5 minutes. Extracted RNA was then used as input for three individual long-range reverse transcription polymerase chain reactions (RT-PCRs) as previously described (19). The master mixes were composed of 25 µL Platinum SuperFi RT-PCR Master Mix (Thermo Fisher Scientific), 2.5 µL of each 10 µM forward and reverse primer, 0.5 µL SuperScript IV RT mix (Thermo Fisher Scientific), and 19.5 µL viral RNA to a total volume of 50 µL. We used PCR primers of 1_LEFT-IF11/Rev-c-IR1 for the first amplicon, covering positions 31 to 10,267, 31.5_LEFT-IF11/67_RIGHT-IR1 for the second amplicon (9,634-20,572), and 67_LEFT-IF11/98_RIGHT-IR1 for the third amplicon (20,173-29,866) (Table S3). Here, 1_LEFT, 67_RIGHT, 67_LEFT, and 98_RIGHT primers were selected from the ARTIC network V3 primers (20) and Rev-c and 31.5_LEFT were designed in-house. Our six RT-PCR primer regions were highly conserved across Omicron variants, with a maximum of one base substitution being observed in each region among the 813 GISAID Omicron variant sequences depicted in Fig. 1C. A volume of 25 µL of mineral oil (GaudiLabs) was added to the top of each reaction. The reactions were PCR cycled using a PocketPCR instrument (GaudiLabs). After 10 minutes of reverse transcription at 50°C, the samples were incubated for 2 minutes at 98°C, followed by 45 cycles of 10 seconds at 98°C, 10 seconds at 55°C, and 5 minutes at 72°C.
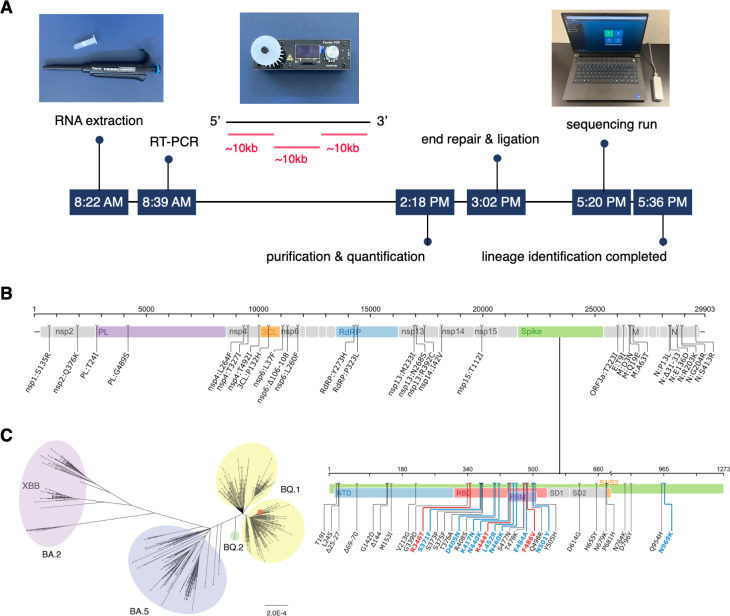
Fig 1.
COVID variant tracking portable laboratory. (A) SARS-CoV-2 whole genome sequencing timeline from sample to result. SARS-CoV-2 RNA was extracted and amplified via one round of three overlapping RT-PCRs (~10,000 base long each). Following purification, quantification, end-repair, and ligation, three amplicons were sequenced by a Nanopore MinION sequencer. It took 9 hours 15 minutes to identify the lineage from a COVID-positive individual’s NP/OP swab specimen. (B) Mutation map of the specimen sequenced by our portable lab (TU5327), compared with the reference sequence Wuhan-Hu-1 (GenBank accession number: MN908947). This map was generated using the Stanford Coronavirus Antiviral & Resistance Database. (C) Phylogenetic tree of specimen TU5327 (red dot) and 813 GISAID sequences from Los Angeles, California, USA, collected in December 2022. Sequences clustered into four lineages, BA.2 (pink), BA.5 (blue), BQ.2 (green), and BQ.1 (yellow).
The PCR products were separated from the mineral oil and then size selected using a SPRI bead clean-up protocol (21). A volume of 35 µL of the SPRI beads was added to each of three amplicons, which were mixed well, incubated for 5 minutes at room temperature, and washed with 70% ethanol two times. The bead pellets were resuspended in 40 µL of Nuclease-free water, which was incubated for 5 minutes at room temperature. The purified and size-selected product was then eluted from the SPRI beads and quantified using a Qubit 4 Fluorometer and Qubit 1× dsDNA High Sensitivity Assay (Thermo Fisher Scientific). Subsequently, the three amplicons were pooled equimolarly, yielding a concentration of 62 ng/µL. A volume of 4.03 µL of this pool was added to 5.70 µL of the pre-made control sample from specimen YT2631 (Table S1) and 37.27 µL of Nuclease-free water, yielding a pooled library of 499.52 ng in 47 µL.
The pooled library was subjected to Oxford Nanopore MinION library preparation using the Ligation Sequencing kit V14 (Oxford Nanopore Technologies) and NEBNext Companion Module (NEB) with modifications. The end-repair and A-tailing steps were performed using the PocketPCR instrument at 20°C for 30 minutes and 65°C for 10 minutes. Pre-made washed AMPure XP Beads (Beckman Coulter) were used for the clean-up step. AMPure XP Beads were washed by pelleting the beads on a magnetic rack, removing the supernatant, washing five times with Nuclease-free water, washing once with 10 mM Tris-Cl (pH 8.5), and finally resuspending the beads in the original supernatant. The ligation incubation time was increased to 20 minutes. Washed AMPure XP Beads were used for the following clean-up with a final incubation time of 20 minutes. The final library was then diluted to a concentration of 129 ng in 12 µL and sequenced by the MinION Mk1B sequencer with the R10.4.1 flow cell. While not used here, an offline version of the sequencing software, MinKNOW, is available upon request from Oxford Nanopore Technologies (13).
High-throughput sequencing library preparation
SARS-CoV-2 RNA was extracted from remnant NP/OP swab specimens using an automated nucleic acid purification system (KingFisher Duo Prime), as previously described (19). A volume of 200 µL of viral media was diluted in 1× PBS buffer to a total volume of 400 µL prior to RNA extraction. Extracted RNA was then used as input for three long-range RT-PCRs using the SuperScript IV One-Step RT-PCR System (Thermo Fisher Scientific). Each reaction mixture contained 6.25 µL of 2× Platinum SuperFi RT-PCR Master Mix, 0.625 µL of each 2 µM forward and reverse primer, 0.125 µL of SuperScript IV RT Mix, and 4.875 µL of RNA to a total volume of 12.5 µL. RT-PCR primers were For-c/Rev-c for the first amplicon (381-10,267), 31.5_LEFT/67_RIGHT for the second (9,634-20,572), and 67_LEFT/98_RIGHT for the third (20,173-29,866) (Table S3). The samples were then incubated at 50°C for 10 minutes, followed by 98°C for 2 minutes, then 35 cycles of 98°C for 10 sec, 55°C for 10 sec, and 72°C for 5 minutes, with a final extension of 72°C for 5 minutes and a 4°C hold until the next step.
The RT-PCR products were then subjected to long-range index PCR as previously described (19). Each specimen was indexed by a unique combination of a 12-base long forward index and a 12-base long reverse index. Index PCR product bands were confirmed via 96-well 1% agarose gel electrophoresis with the E-Gel Electrophoresis System (Thermo Fisher Scientific). Samples were then quantified via Quant-iT PicoGreen dsDNA Assay (Thermo Fisher Scientific) and pooled equimolarly. An open-source liquid handler (Opentrons OT-2) was used to prepare the PCR master mixes, the PicoGreen dsDNA Assay plates, and the pooled libraries.
Nanopore MinION sequencing and data processing
Half of the pooled library was size selected using the SRE XS kit (Pacific Biosciences) and subjected to Oxford Nanopore MinION library preparation using the Ligation Sequencing kit V14 (Oxford Nanopore Technologies) and NEB with modifications: the end-repair and A-tailing steps were carried out at 20°C for 30 minutes and 65°C for 30 minutes. The ligation time and final clean-up incubation time were increased to 20 minutes. Pre-made washed AMPure XP Beads were used for all clean-up steps. The library was then diluted to 129 ng in 12 µL and sequenced via the MinION Mk1b sequencer using the R10.4.1 flow cell (Oxford Nanopore Technologies) for 72 hours.
We used the graphics processing unit (GPU) version of the Guppy basecaller (version 6.4.2) with the AI network model dna_r10.4.1_e8.2_400bps_sup, which operates in Super Accuracy (SUP) mode (22). The fastq output files containing 1.43 million reads were de-multiplexed, and fasta files were generated for each specimen’s three amplicons. After de-multiplexing, 31 raw reads were selected for each amplicon of every specimen, chosen based on their median quality score and read length to construct the consensus sequence. Among reads with a median quality score greater than 30, priority was given to longer reads, starting with the maximum-length read. If the resulting number of reads was less than 31, reads with lower quality scores were also included. The selected 31 reads were then aligned using MUSCLE (23).
Our first error correction step is to perform local realignment of amplicons surrounding gap columns. We selected columns with gap frequencies ranging from 15% to 85% and used MUSCLE (23) to perform the realignment. For both the left and right directions from the gap column, we identified the nearest column where all sequences shared the same nucleotide base, starting our search three columns away from the gap column. If no such column was found within 12 columns of the gap column, the 12th column was selected. Therefore, the span for local alignment ranged from 7 to 25 columns, depending on the proximity of identical base columns. Any gap column within this alignment range was locally realigned only once. The realignment replaced the original alignment if the average pairwise nucleotide substitution distance was equal to or lower than that of the original alignment. Additionally, we identified regions with a pair of gap and non-gap bases in both the forward and reverse orders and realigned these regions.
In the second error correction step, we selected gap bases with a frequency greater than 65% when determining the consensus sequence from the alignment. To correct deletion errors in homopolymer regions with five or more of the same bases, we calculated the distribution of homopolymer lengths in the raw reads and selected the most frequent length, except in the following cases. If the homopolymer length of the consensus sequence was greater than or equal to the most frequent length observed in the raw reads, we increased the homopolymer length by one from this prevalent length, provided that the cumulative proportion of homopolymers extending by one, two, and three bases beyond the prevalent length exceeded 60% of the raw reads with the most frequent length. The cumulative proportion cutoff was set to 55% when the most frequent homopolymer length was over 8. Examples illustrating such corrections were presented in Fig. S1. The resulting three amplicons’ consensus sequences were then assembled to produce each specimen’s near-full-length SARS-CoV-2 whole genome sequence. The obtained SARS-CoV-2 whole genome sequences were then assigned a lineage using Pangolin (version 4.1.3) (24).
Pacbio HiFi sequencing and data processing
The other half of the pooled library was subjected to size selection using the Bluepippin instrument (Sage Science) before undergoing HiFi sequencing on the PacBio Sequel II system for a 30-hour data collection according to the manufacturer instructions at HistoGenetics (NY, USA). Around 6.86 million circular consensus sequence reads were obtained, and the output fasta file was de-multiplexed. A total of 31 raw reads were selected for each specimen’s amplicon, with preference given to reads that were closer to the peak of the read length distribution of the raw reads. The 31 selected reads were aligned using MUSCLE (23), and regions surrounding gap columns were locally realigned as described above. Gap bases were selected when they were the most frequent base during the consensus sequence building procedure. A mixed base was indicated if the frequency of the second most abundant base was greater than 15%.
Control run
For our control run, we used three genomic RNAs of NY1-PV08001 (GenBank: MT370904.1), WI-CDC-03041142-001 (GenBank; MT039887.1), and CA-CDC-03039618-001 (GenBank: MT027062.1), along with two clinical specimens, USA/CA-LAC-USC1 and USA/CA-LAC-USC2 (19), as detailed in the Supplementary Materials. The reference sequences of three genomic RNA strains differed from the sequences in GenBank, with NY1-PV08001 having one mutation, WI-CDC-03041142-001 having two mutations and nine deletions, and CA-CDC-03039618-00 having one mutation. We confirmed these differences through Sanger sequencing (Fig. S2). The reference sequence of USA/CA-LAC-USC2 had two mutations in comparison to the Pacbio HiFi sequencing data. These mutations were verified by the presence of double peaks in the Sanger chromatogram data (Fig. S3).
Sanger sequencing
Index PCR products of NY1-PV08001, WI-CDC-03041142-001, and CA-CDC-03039618-001 were purified by pre-made Washed AMPure XP Beads using a 1.2× bead volume (24 µL beads), two 70% ethanol washes, and elution in 20 µL of Nuclease-free water. A total of 10 µL of purified sample was combined with 5 µL of 5 µM Sanger sequencing primer. Six primers, 91_LEFT-mod, 39_LEFT-mod, 73_LEFT-mod, 36_LEFT, 78_LEFT, and 34_LEFT, were selected from the V3 ARTIC network primers (20) with minor modifications (Table S3). The samples were then shipped to Azenta Life Sciences for Sanger sequencing. The near-full-length sequence of USA/CA-LAC-USC2 was obtained by Sanger sequencing, as previously described (19).
Phylogenetic tree analysis
The phylogenetic trees were produced using MAFFT (version 7.392) (25), FastTree (version 2.1.8) (26), and FigTree (version 1.4.4) (Supplementary Materials).
RESULTS
Portable laboratory for COVID-19 variant tracking
Our portable system can be transported in a single suitcase and occupies minimal space, fitting within a 1.5 × 0.5-meter area (Fig. S4). Assuming extremely resource-limited settings without external power and internet connection, our system consists of a tabletop centrifuge, vortex mixer, open-source miniaturized thermocycler, fluorometer, Nanopore MinION portable sequencer, laptop computer, and battery (Table S2). We used an aluminum block and ice packs for heat-sensitive reagents. We conducted MinION sequencing, super-accurate basecalling, error correction, whole genome assembly, and lineage assignment on a single standalone laptop without the need for external power or an internet connection.
Our compact laboratory, powered by a single portable battery (296 watt-hours) and a laptop (86 watt-hours), successfully completed SARS-CoV-2 whole genome sequencing and lineage identification in 9 hours and 15 minutes, starting from the RNA extraction of a COVID-positive individual’s NP/OP swab specimen (Fig. 1A). To cover the near-full genome, we conducted a single round of three RT-PCRs (19) and sequenced the resulting three amplicons using a Nanopore MinION portable sequencer (Fig. 1A).
Our long-read approach had clear advantages in both sequencing run time and data analysis. Compared with the ARTIC nCoV-2019 protocol that involves 98 amplicons and approximately 6 hours of Nanopore sequencing (16), our protocol employs only three amplicons for whole genome assembly, reducing the sequencing run time to just 9 minutes. The minimal number of amplicons also reduced the downstream data processing time for basecalling, assembly, and lineage identification to just 7 minutes. The accuracy of long-read sequencing was improved by utilizing the latest AI-based basecalling algorithms and improved Nanopore technology (22, 27). To manage the significant computational demands and high power consumption, we employed state-of-the-art GPUs that are specifically designed for low-power laptops. Equipped with an RTX 3070 Ti Laptop GPU, our laptop was capable of promptly running the AI-assisted Guppy basecaller in Super Accuracy (SUP) mode (22). Our benchmark test showed that the laptop GPU’s basecalling rate was 1.4 million reads per second, which was comparable to an RTX 3060 Ti desktop GPU’s rate, 1.5 million reads per second. This optimized design allowed us to complete the computationally intensive basecalling in 4 minutes and 11 seconds. We were then able to complete error corrections, whole genome assembly, and lineage identification in 2 minutes and 45 seconds. Our fast sample-to-answer time holds promise for enabling real-time monitoring of COVID variants.
The specimen sequenced by our portable laboratory (TU5327) was found to have key antibody resistance mutations, including R346T, K444T, and N460K in the spike protein (Fig. 1B) (28, 29). This sequence was classified as the BQ.1 lineage. A phylogenetic tree was generated using this sequence in addition to 813 other GISAID sequences collected in December 2022 from Los Angeles, California, USA (Fig. 1C). The tree showed that this sequence grouped closely with other sequences belonging to the BQ.1 lineage that were present in Los Angeles.
Affordable and high-throughput solution for SARS-CoV-2 whole genome sequencing
Another advance of our long-read sequencing protocol is the capacity to process a high volume of specimens in a single sequencing run, which can facilitate large-scale genomic surveillance in LMICs. We obtained remnant NP/OP specimens that had tested positive for SARS-CoV-2 during diagnostic testing at Keck Medicine of University of Southern California (Table S1). We generated 289 near-full-length SARS-CoV-2 genome sequences from a single portable Nanopore sequencing run. This represents a significant improvement compared with existing Nanopore sequencing methods that demonstrated sequencing approximately 96 samples in a single run (16–18).
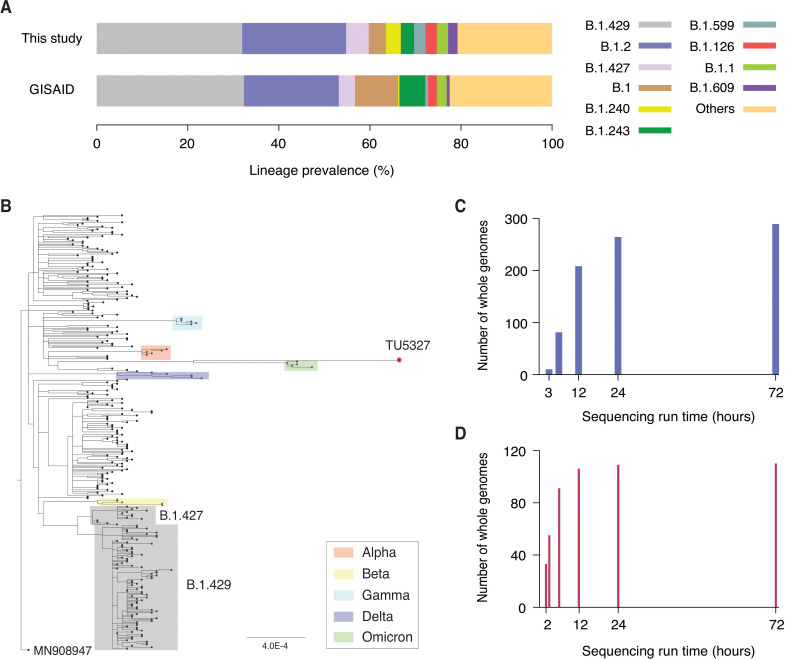
We observed that our high-throughput sequence data yielded a reasonable depiction of the variants present in Los Angeles in December 2020. This provides a promising outlook for the usage of our tool for real-time surveillance. As shown in Fig. 2A, our specimens collected in December 2020 exhibited a high degree of consistency in lineage profile with all 1,918 GISAID sequences obtained from Los Angeles during the same period (spearman correlation of 0.6, P < 0.001). A diverse range of 43 different PANGO lineages was identified (Table S1). The most prevalent lineage, accounting for 31% of the sequences, was B.1.429, which is a member of the Epsilon variant (Fig. 2A). This frequency was consistent with the reported prevalence of B.1.429 in Los Angeles, which was 32%. Moreover, we detected B.1.397, B.1.399, and C.23, which were not previously reported in Los Angeles in December 2020. A phylogenetic tree was created for the 289 whole genome sequences (Fig. 2B). This tree also includes representative sequences of Alpha, Beta, Gamma, Delta, and Omicron variants (30). The tree confirmed that our sequences did not cluster with any of these variants.
Fig 2.
Large-scale SARS-CoV-2 whole genome sequencing. (A) Lineage profile of 241 specimens that were collected in December 2020 (top) and 1,918 GISAID sequences collected in Los Angeles at the same period (bottom). (B) Phylogenetic tree of 289 SARS-CoV-2 whole genome sequences along with the sequence obtained by our portable laboratory (TU5327) and representative sequences of Alpha (red), Beta (yellow), Gamma (light blue), Delta (blue), and Omicron (green) variants. (C) Number of SARS-CoV-2 whole genome sequences produced as a function of portable Nanopore sequencing run time for 289 specimens. A total of 208 whole genome sequences were obtained within the first 12 hours of sequencing run. (D) Number of SARS-CoV-2 whole genome sequences obtained as a function of Nanopore sequencing run time for 110 specimens. A total of 55 whole genome sequences were produced within the first 3 hours of sequencing run.
We used an open-source liquid handler (Opentrons) and inexpensive MinION sequencer (Oxford Nanopore Technologies) to reduce the overall equipment cost. Moreover, our high-volume sequencing analysis was efficiently managed with a single laptop, making it more cost-effective than cloud computing. The sequencing run time can be shortened depending on the number of samples processed. Within the first 12 hours of the portable sequencing run, we obtained over 200 SARS-CoV-2 whole genome sequences (Fig. 2C). We were able to produce more than 80 sequences within the first 6 hours (Fig. 2C). When sequencing a smaller number of samples, we were able to produce 55 whole genome sequences within the first 3 hours (Fig. 2D). This flexibility makes it suitable for on-demand scheduling and optimized regional allocation of molecular surveillance networks in LMICs.
Error-free sequencing
We verified our error-free sequencing by conducting a series of comparative analyses. First, we sequenced three genomic RNA control strains (NY1-PV08001, WI-CDC-03041142-001, and CA-CDC-03039618-001) and two clinical specimens (USA/CA-LAC-USC1 and USA/CA-LAC-USC2) (19). We verified that our data processing pipeline produced five whole genome sequences that precisely matched their reference sequences (Fig. S2 and S3). Second, we conducted an independent PacBio HiFi sequencing run to verify the accuracy of our 289 Nanopore sequences. The accuracy of PacBio HiFi sequencing had been demonstrated through both control strain sequencing and Sanger reference sequencing (19). We confirmed that our pipeline was free of errors by verifying that the 289 whole genome sequences were identical to those produced by HiFi reference sequencing. Finally, we conducted an additional independent sequencing run, in which 110 out of 289 specimens were re-sequenced. This run also produced 110 whole genome sequences that were identical to the HiFi reference sequences.
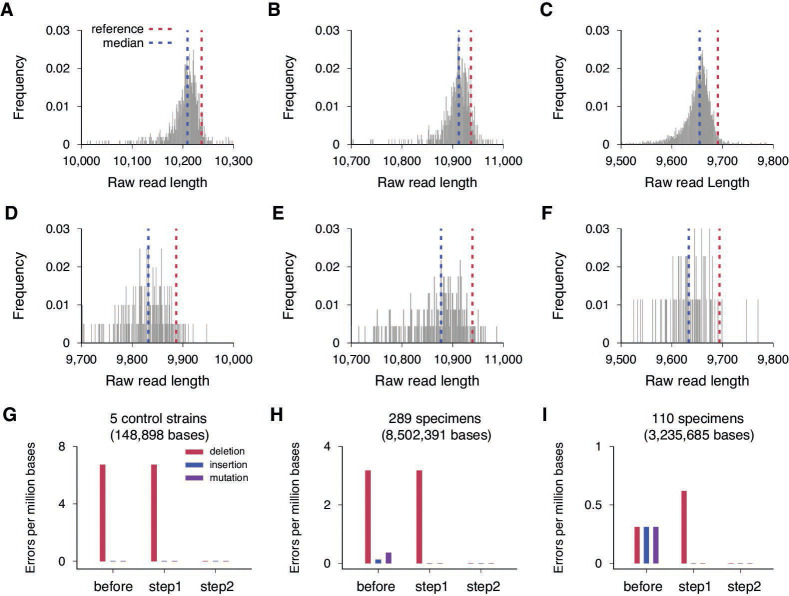
We found that this high level of accuracy was attained by coupling a refined basecalling that leverages recently improved nanopores and advanced AI algorithms (22, 27), with our additional error-correction steps. We plotted the length distribution of raw reads for the three amplicons of the WI-CDC-03041142-001 control strain (Fig. 3A through C). The first amplicon of the reference sequence was 10,237 base-pairs long, but the median length of the raw reads was 10,209, which was 28 bases shorter than the reference (Fig. 3A). The short-sequence bias was consistently observed in all three amplicons (Fig. 3A through C). This bias caused deletion errors in the consensus sequence of the aligned raw reads, resulting in a deletion rate of 6.7 deletions per million bases across the five control sequences (one deletion out of 148,898 bases) (Fig. 3G). We did not observe any insertion or mutation errors in the control run.
Fig 3.
Short sequence bias and error corrections. (A) Length distribution of raw reads for the first amplicon of the WI-CDC-03041142-001 control strain (GenBank: MT039887.1) with R10.4.1 flow cell and dna_r10.4.1_e8.2_400bps_sup model. The median length of the raw reads was 10,209 (blue line), which was 28 bases shorter than the reference sequence length, 10,237 (red line). (B) Length distribution of raw reads for WI-CDC-03041142-00l’s second amplicon with R10.4.1 flow cell and dna_r10.4.1_e8.2_400bps_sup model. The median was 10,912 (blue line), which was 24 bases shorter than the reference sequence length, 10,236 (red line). (C) Length distribution of raw reads for WI-CDC-03041142-00l’s third amplicon with R10.4.1 flow cell and dna_r10.4.1_e8.2_400bps_sup model. The median was 9,655 (blue line), and the reference sequence length was 9,691 (red line). (D) Length distribution of raw reads for the first amplicon of the USA-WA1/2020 control strain (GenBank: MN985325.1) with R9.4.1 and dna_r9.4.1_e8.1_sup model. The median was 9,832 (blue line), which was 55 bases shorter than the reference sequence length, 9,887 (red line). (E) Length distribution of raw reads for USA-WA1/2020’s second amplicon with R9.4.1 and dna_r9.4.1_e8.1_sup model. The median was 10,877 (blue line), which was 62 bases shorter than the reference sequence length, 10,939 (red line). (F) Length distribution of raw reads for USA-WA1/2020’s third amplicon with R9.4.1 and dna_r9.4.1_e8.1_sup model. The median was 9,633.5 (blue line), and the reference sequence length was 9,694 (red line). (G) Error rates of consensus sequences for the five control strains: three genomic RNA control strains [NY1-PV08001 (GenBank: MT370904.1), WI-CDC-03041142-001 (GenBank; MT039887.1), and CA-CDC-03039618-001 (GenBank: MT027062.1) and two clinical specimens [USA/CA-LAC-USC1 (GISAID: EPI_ISL_569664) and USA/CA-LAC-USC2 (GISAID: EPI_ISL_569665). Deletions (red), insertions (blue), and mutations (purple) were counted across 148,898 nucleotide bases of the five consensus sequences before error correction (before), after local realignment (step 1), and after gap and homopolymer length corrections (step 2). (H) Error rates of consensus sequences for the 289 specimens. In a total of 8,502,391 bases, 27 deletions, 1 insertion, and 3 mutations were detected before error correction (before). After the local-realignment step (step 1), the insertion and mutations were eliminated. The deletions were removed after step 2. (I) Error rates of consensus sequences for the 110 re-sequenced specimens. In a total of 3,235,685 bases, 1 deletion, 1 insertion, and 1 mutation were observed before error correction (before), but these were removed after error correction steps 1 and 2.
The error profiles of the portable sequencer have been significantly improved compared with previous versions (22, 27). We directly compared the performance of the version we used (R10.4.1 and AI network model: dna_r10.4.1_e8.2_400bps_sup) with the previous version (R9.4.1 and AI network model: dna_r9.4.1_e8.1_sup). In a control run that sequenced the genomic RNA of USA-WA1/2020 using the previous version, the median length of raw reads was 55, 62, and 60.5 bases shorter than the first, second, and third amplicons of the reference sequence, respectively (Fig. 3D through F). This deviation was approximately two times greater than that of the new version. As a result, the previous version showed a much higher error rate than the most recent kit we used: 29.3 deletions per million bases, 6.2 insertions per million bases, and 1.5 mutations per million bases.
This improved raw read accuracy allowed us to eliminate all errors in the Nanopore sequencing data in two steps. First, we locally realigned the raw sequencing reads near gap columns (Fig. S5). Second, we corrected deletion errors by choosing the gap base if its frequency was more than 65%, instead of 50%. In addition, we addressed homopolymer errors by determining the length of each homopolymer based on the distribution of raw reads’ homopolymer lengths (Fig. S1). The sole deletion observed in our control run was eliminated in our second step (Fig. 3G), resulting in five whole genomes that fully matched their reference sequences. Compared with the HiFi sequencing data, our Nanopore sequencing data showed 27 deletions, 1 insertion, and 3 base substitutions in a total of 8,502,391 bases, resulting in an error rate of 3.2 deletions per million bases, 0.12 insertions per million bases, and 0.35 mutations per million bases (Fig. 3H). All the insertion and mutation errors were corrected in the first re-alignment step, and all the deletion errors were removed in the second step, producing 289 SARS-CoV-2 whole genomes that matched those produced by the HiFi reference sequencing (Fig. 3H). From the 110 specimens re-sequenced independently, 1 deletion, 1 insertion, and 1 mutation were observed in a total of 3,235,685 bases. Our two-step error correction approach again successfully eliminated all these errors (Fig. 3I).
DISCUSSION
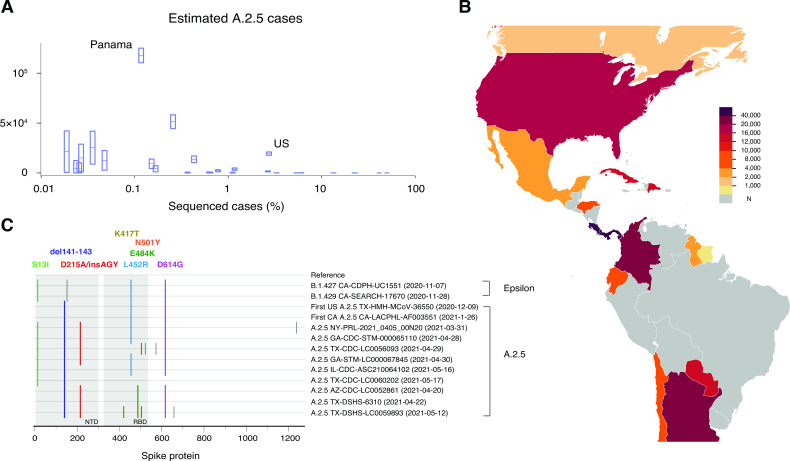
Geographical disparities in COVID-19 variant tracking leave emerging variants at risk of remaining undetected and spreading widely in regions with limited whole genome sequencing capacity. We identified one such instance: the spread of the S clade A.2.5 lineage. This lineage was primarily found in countries with limited SARS-CoV-2 whole genome sequencing (Fig. 4A; Table S4). The A.2.5 lineage was prevalent in several countries in the Americas, including Panama (86%) and Costa Rica (37%) (7), where less than 1% of COVID-19 cases had been sequenced between January and June 2021 (Fig. 4B) (31, 32). This lineage showed evidence of convergent evolution, which is characterized by the rapid accumulation of shared amino acid substitutions in the spike protein, including concerning mutations with immune evasion signatures, S-L452R, S-E484K, and S-N501Y (Fig. 4C) (33, 34).
Fig 4.
A.2.5 lineage and geographic disparities in SARS-CoV-2 genomic surveillance. (A) Sequencing efforts which were limited in countries with a high abundance of the A.2.5 lineage. Around 1.3% of global cases (1,235,031 out of 92,006,675) was sequenced between January and June 2021. To estimate the number of A.2.5 cases, we multiplied the confirmed cases of each country between January and June 2021 (30, 31) by the proportion of A.2.5 sequences out of the total sequenced cases deposited to GISAID in the same period (7). Except for the United States, countries with over 10,000 A.2.5 cases had sequenced less than 1% of their total cases (S4 Table). (B) Estimated A.2.5 cases of each country in North and South America between January and June 2021. Here “N” indicates that either no A.2.5 lineage sequences were reported or no cases were sequenced. (C) A.2.5 sequences showed several mutations of concern in the receptor binding domain of the spike protein, including L452R (sky-blue), E484K (dark-green), and N501Y (orange) in reference to Wuhan-Hu-1 (MN908947). Three amino acid deletions (del141-143) and an amino acid substitution with three amino acid additions (D215A/InsAGY) within the N-terminal domain of the spike protein were also frequently observed among the A.2.5 lineage sequences. The A.2.5 lineage shared S13I, L452R, and D614G with the California variants, B.1.427 (Epsilon) and B.1.429 (Epsilon). Gray ticks denoted W152C, A570D, H655Y, and C1235F. The highlighter plot was generated using the Highlighter tool, which is available at HIV Databases at Los Alamos National Laboratory.
The cumulative incidence rate in Panama was 8.95% as of June 2021, which was considerably higher than the global rate of 2.26%. During this period of high incidence in Panama, only 0.12% of positive cases had their sequence data shared, which limited the ability to link the severity to the A.2.5 lineage. Importantly, we found the prevalence of A.2.5 was extremely high in Chiriquí, Panama, where only a limited number of sequences were reported (11 A.2.5 sequences out of 16 total sequences in July 2020 and 29 out of 32 in March 2021) (7). This region is home to various indigenous peoples collectively known as Guaymi (or Ngabe), who not only reside in Chiriquí but have communities spread from neighboring indigenous areas in Panama to Costa Rica. Furthermore, of the initial 20 A.2.5 sequences collected in Costa Rica and deposited to GISAID before March 2020, 50% originated from the border region with Panama (7). Despite the potential association between the indigenous people and the A.2.5 outbreak in Panama and Costa Rica, the scarcity of available sequencing data hampered further investigation. This highlights the significance of establishing a more effective real-time surveillance system.
It has been reported that 58% of LMICs sequenced less than 0.5% of their COVID-19 cases (8). The sequencing throughput presented in this study is sufficient to cover 0.5% of the approximately 58,000 COVID-19 cases, making it a potentially valuable tool to enhance LMICs’ genomic surveillance capacity. Our system also offered the flexibility to adjust sample-to-answer time and throughput, providing an ideal solution for on-demand scheduling and strategic allocation of genomic surveillance resources across different regions.
Our portable laboratory system can perform SARS-CoV-2 whole genome sequencing and lineage identification in 9 hours and 15 minutes. However, approximately 5 hours and 40 minutes is spent on RT-PCR alone. We can potentially integrate ultrafast PCR technologies, such as photonic thermocycling, which can significantly reduce our sample-to-answer time (35–37). Currently, our portable system requires many different components which limits its streamlined operations. These components could be combined into a lab-on-a-chip, as demonstrated in microbial genomics (38) and COVID-19 diagnostics (39).
Another limitation of our study was that the maximum cycle threshold value for the remnant NP/OP specimens we processed, which had been stored for more than 6 months, was 24.6. However, our long-read amplification protocol successfully generated SARS-CoV-2 whole genome sequences from remnant NP/OP specimens with cycle threshold values between 25 and 30, which were stored for less than a few months (19). Therefore, it is important to monitor the sequencing success rate depending on the remnant specimen’s storage conditions.
In summary, we developed a portable tool for rapid on-site COVID-19 variant tracking. This tool can facilitate timely public health responses and reduce the disease burden in remote communities. In addition, our high-throughput solution can accurately produce almost 300 whole genome sequences in a single run, which can narrow down the molecular surveillance data gap between high-income countries and low- and middle-income counties. Our portable tool also has the potential to be adapted for the genomic analysis of various pathogens, which could help diminish global health disparities.
ACKNOWLEDGMENTS
We thank Dr. Jane Emerson for providing the study specimens. We acknowledge all laboratories and authors who shared global SARS-CoV-2 whole genome sequences by depositing to GISAID.
We thank BEI Resources (NIAID, NIH) for providing genomic RNAs of NY1-PV08001, WI-CDC-03041142–001, and CA-CDC-03039618–001.
This study was supported by NIH grant R01-AI095066.
S.Y.P., G.F. and H.Y.L. designed the sequencing workflow and conducted sequencing experiments. S.Y.P. and H.Y.L. formulated the bioinformatics pipeline and analyzed the sequencing data. P.W. provided the study specimens. All authors participated in the writing of the manuscript.
Contributor Information
Ha Youn Lee, Email: hayoun@usc.edu.
Alexander Mellmann, Westfalische Wilhelms-Universitat Munster, Münster, Germany.
DATA AVAILABILITY
The sequences reported in this study are available in the GISAID database (www.gisaid.org) (Accession ID: EPI_ISL_17067675–EPI_ISL_17067953 and EPI_ISL_17072870–EPI_ISL_17072880).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.01558-23.
All supplemental materials.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KK-H, Yuen TT-T, Yoon C, To KK-W, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen K-Y, Ho DD. 2022. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602:676–681. doi: 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
- 2. Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, Liu C, Mentzer AJ, Supasa P, Duyvesteyn HME, et al. 2022. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 185:2422–2433. doi: 10.1016/j.cell.2022.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lau JJ, Cheng SMS, Leung K, Lee CK, Hachim A, Tsang LCH, Yam KWH, Chaothai S, Kwan KKH, Chai ZYH, Lo THK, Mori M, Wu C, Valkenburg SA, Amarasinghe GK, Lau EHY, Hui DSC, Leung GM, Peiris M, Wu JT. 2023. Real-world COVID-19 vaccine effectiveness against the Omicron BA.2 variant in a SARS-CoV-2 infection-naive population. Nat Med 29:348–357. doi: 10.1038/s41591-023-02219-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uraki R, Ito M, Kiso M, Yamayoshi S, Iwatsuki-Horimoto K, Furusawa Y, Sakai-Tagawa Y, Imai M, Koga M, Yamamoto S, Adachi E, Saito M, Tsutsumi T, Otani A, Kikuchi T, Yotsuyanagi H, Halfmann PJ, Pekosz A, Kawaoka Y. 2023. Antiviral and bivalent vaccine efficacy against an Omicron XBB.1.5 isolate. Lancet Infect Dis 23:402–403. doi: 10.1016/S1473-3099(23)00070-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. SARS-CoV-2 resistance mutations - 3CLpro inhibitors. 2023. Available from: https://covdb.stanford.edu/drms/3clpro/
- 6. Chen Z, Azman AS, Chen X, Zou J, Tian Y, Sun R, Xu X, Wu Y, Lu W, Ge S, Zhao Z, Yang J, Leung DT, Domman DB, Yu H. 2022. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet 54:499–507. doi: 10.1038/s41588-022-01033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 1:33–46. doi: 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brito AF, Semenova E, Dudas G, Hassler GW, Kalinich CC, Kraemer MUG, Ho J, Tegally H, Githinji G, Agoti CN, et al. 2022. Global disparities in SARS-CoV-2 genomic surveillance. Nat Commun 13:7003. doi: 10.1038/s41467-022-33713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okeke IN, Ihekweazu C. 2021. The importance of molecular diagnostics for infectious diseases in low-resource settings. Nat Rev Microbiol 19:547–548. doi: 10.1038/s41579-021-00598-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. 2021. Indigenous peoples have been disproportionately affected by COVID-19 – senior United Nations oficial tells human rights council. Available from: https://www.ohchr.org/en/press-releases/2021/09/indigenous-peoples-have-been-disproportionately-affected-covid-19-senior
- 11. Kelsey P. B, Carla L. B, Megan L. T, Ellen M. P. 2022. COVID-19 cases, hospitalizations, and deaths among American Indian or Alaska Native persons — Alaska, 2020–2021. Available from: https://www.cdc.gov/mmwr/volumes/71/wr/mm7122a2.htm [DOI] [PMC free article] [PubMed]
- 12. Sansone NMS, Boschiero MN, Ortega MM, Ribeiro IA, Peixoto AO, Mendes RT, Marson FAL. 2022. Severe acute respiratory syndrome by SARS-CoV-2 infection or other etiologic agents among Brazilian indigenous population: an observational study from the first year of coronavirus disease (COVID)-19 pandemic. Lancet Reg Health Am 8:100177. doi: 10.1016/j.lana.2021.100177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, et al. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232. doi: 10.1038/nature16996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faria NR, Quick J, Claro IM, Thézé J, de Jesus JG, Giovanetti M, Kraemer MUG, Hill SC, Black A, da Costa AC, et al. 2017. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546:406–410. doi: 10.1038/nature22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castro-Wallace SL, Chiu CY, John KK, Stahl SE, Rubins KH, McIntyre ABR, Dworkin JP, Lupisella ML, Smith DJ, Botkin DJ, Stephenson TA, Juul S, Turner DJ, Izquierdo F, Federman S, Stryke D, Somasekar S, Alexander N, Yu G, Mason CE, Burton AS. 2017. Nanopore DNA sequencing and genome assembly on the international space station. Sci Rep 7:18022. doi: 10.1038/s41598-017-18364-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Josh Q. 2019. nCoV-2019 sequencing protocol V.1. Available from: https://www.protocols.io/view/ncov-2019-sequencing-protocol-bp2l6n26rgqe/v1
- 17. Baker DJ, Aydin A, Le-Viet T, Kay GL, Rudder S, de Oliveira Martins L, Tedim AP, Kolyva A, Diaz M, Alikhan N-F, et al. 2021. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med 13:21. doi: 10.1186/s13073-021-00839-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker A, Houwaart T, Finzer P, Ehlkes L, Tyshaieva A, Damagnez M, Strelow D, Duplessis A, Nicolai J, Wienemann T, et al. 2022. Characterization of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection clusters based on integrated genomic surveillance, outbreak analysis and contact tracing in an urban setting. Clin Infect Dis 74:1039–1046. doi: 10.1093/cid/ciab588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park SY, Faraci G, Ward PM, Emerson JF, Lee HY. 2021. High-precision and cost-efficient sequencing for real-time COVID-19 surveillance. Sci Rep 11:13669. doi: 10.1038/s41598-021-93145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, Choi JH, Lapointe H, Kamelian K, Smith AD, Prystajecky N, Goodfellow I, Wilson SJ, Harrigan R, Snutch TP, Loman NJ, Quick J. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. Genomics. doi: 10.1101/2020.09.04.283077 [DOI]
- 21. Oxford nanopore Technologies. 2023. SPRI size selection protocol. Available from: https://community.nanoporetech.com/extraction_methods/spri-size-selection
- 22. Oxford Nanopore Technologies. 2023. How basecalling workds. Available from: https://nanoporetech.com/how-it-works/basecalling
- 23. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oxford Nanopore Technologies . 2023. Nanopore sequencing accuracy. Available from: https://nanoporetech.com/platform/accuracy
- 28. Cao Y, Jian F, Wang J, Yu Y, Song W, Yisimayi A, Wang J, An R, Chen X, Zhang N, et al. 2023. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614:521–529. doi: 10.1038/s41586-022-05644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Groenheit R, Galanis I, Sondén K, Sperk M, Movert E, Bacchus P, Efimova T, Petersson L, Rapp M, Sahlén V, Söderholm S, Valentin Asin K, Zanetti S, Lind Karlberg M, Bråve A, Klingström J, Blom K. 2023. Rapid emergence of Omicron sublineages expressing spike protein R346T. Lancet Reg Health Eur 24:100564. doi: 10.1016/j.lanepe.2022.100564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. World Health Organization . 2020. Tracking SARS-CoV-2 variants. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 31. World Health Organization . 2021. Weekly epidemiological update - 5 January 2021. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update---5-january-2021
- 32.World Health Organization. 2021. Weekly epidemiological update - 15 June 2021. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---15-june-2021
- 33. Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, et al. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361. doi: 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, Ngare I, Kimura I, Uriu K, Kosugi Y, et al. 2021. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host & Microbe 29:1124–1136. doi: 10.1016/j.chom.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roche PJR, Beitel LK, Khan R, Lumbroso R, Najih M, Cheung MC-K, Thiemann J, Veerasubramanian V, Trifiro M, Chodavarapu VP, Kirk AG. 2012. Demonstration of a plasmonic thermocycler for the amplification of human androgen receptor DNA. Analyst 137:4475–4481. doi: 10.1039/c2an35692a [DOI] [PubMed] [Google Scholar]
- 36. Cheong J, Yu H, Lee CY, Lee J-U, Choi H-J, Lee J-H, Lee H, Cheon J. 2020. Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat Biomed Eng 4:1159–1167. doi: 10.1038/s41551-020-00654-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blumenfeld NR, Bolene MAE, Jaspan M, Ayers AG, Zarrandikoetxea S, Freudman J, Shah N, Tolwani AM, Hu Y, Chern TL, Rogot J, Behnam V, Sekhar A, Liu X, Onalir B, Kasumi R, Sanogo A, Human K, Murakami K, Totapally GS, Fasciano M, Sia SK. 2022. Multiplexed reverse-transcriptase quantitative polymerase chain reaction using plasmonic nanoparticles for point-of-care COVID-19 diagnosis. Nat Nanotechnol 17:984–992. doi: 10.1038/s41565-022-01175-4 [DOI] [PubMed] [Google Scholar]
- 38. Kim S, De Jonghe J, Kulesa AB, Feldman D, Vatanen T, Bhattacharyya RP, Berdy B, Gomez J, Nolan J, Epstein S, Blainey PC. 2017. High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nat Commun 8:13919. doi: 10.1038/ncomms13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibani MM, Toumazou C, Sohbati M, Sahoo R, Karvela M, Hon T-K, De Mateo S, Burdett A, Leung KYF, Barnett J, et al. 2020. Assessing a novel, lab-free, point-of-care test for SARS-CoV-2 (CovidNudge): a diagnostic accuracy study. Lancet Microbe 1:e300–e307. doi: 10.1016/S2666-5247(20)30121-X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All supplemental materials.
Data Availability Statement
The sequences reported in this study are available in the GISAID database (www.gisaid.org) (Accession ID: EPI_ISL_17067675–EPI_ISL_17067953 and EPI_ISL_17072870–EPI_ISL_17072880).