ABSTRACT
The alphaproteobacterium Caulobacter crescentus thrives in oligotrophic environments and is able to optimally exploit minimal resources by entertaining an intricate network of gene expression control mechanisms. Numerous transcriptional activators and repressors have been reported to contribute to these processes, but only few studies have focused on regulation at the post-transcriptional level in C. crescentus. Small RNAs (sRNAs) are a prominent class of regulators of bacterial gene expression, and most sRNAs characterized today engage in direct base-pairing interactions to modulate the translation and/or stability of target mRNAs. In many cases, the ubiquitous RNA chaperone, Hfq, contributes to the establishment of RNA-RNA interactions. Although the deletion of the hfq gene is associated with a severe loss of fitness in C. crescentus, the RNA ligands of the chaperone have remained largely unexplored. Here we report on the identification of coding and non-coding transcripts associated with Hfq in C. crescentus and demonstrate Hfq-dependent post-transcriptional regulation in this organism. We show that the Hfq-bound sRNA RusT is transcriptionally controlled by the NtrYX two-component system and induced in response to iron starvation. By combining RusT pulse expression with whole-genome transcriptome analysis, we determine 16 candidate target transcripts that are deregulated, many of which encode outer membrane transporters. We hence suggest RusT to support remodeling of the C. crescentus cell surface when iron supplies are limited.
IMPORTANCE
The conserved RNA-binding protein Hfq contributes significantly to the adaptation of bacteria to different environmental conditions. Hfq not only stabilizes associated sRNAs but also promotes inter-molecular base-pairing interactions with target transcripts. Hfq plays a pivotal role for growth and survival, controlling central metabolism and cell wall synthesis in the oligotroph Caulobacter crescentus. However, direct evidence for Hfq-dependent post-transcriptional regulation and potential oligotrophy in C. crescentus has been lacking. Here, we identified sRNAs and mRNAs associated with Hfq in vivo, and demonstrated the requirement of Hfq for sRNA-mediated regulation, particularly of outer membrane transporters in C. crescentus.
KEYWORDS: Caulobacter, small RNA, Hfq, iron starvation, NtrYX
INTRODUCTION
One essential hallmark of bacteria is their ability to effectively sense and react to fluctuations in the environment. Complex regulatory networks coordinate bacterial stress responses by converting external stimuli into defined changes in gene expression, and RNA-mediated control mechanisms efficiently complement protein-based regulation (1). Especially bacterial regulatory small RNAs (sRNAs) have consistently been linked to environmental responses and can critically impact gene expression control at the post-transcriptional level. Diverse in structure, size, and function, the most prevalent class of sRNAs commonly interact with complementary target messenger RNAs (mRNAs) through the formation of limited base-pairing interactions which results in changes in translation and/or stability of target transcripts (2). While sRNAs have the capacity to positively affect target gene expression, they most commonly act as repressors. When binding in the vicinity of the translation initiation region, sRNAs block ribosome recruitment. As a consequence, the sRNA:mRNA complex is usually rapidly turned over by ribonucleases (RNases) (3, 4). Alternatively, sRNAs can recruit RNases to the base-pairing site to facilitate transcript cleavage and decay without affecting translation initiation (5). Gene activation by sRNAs is based on target mRNA stabilization and stimulation of translation via base-pairing to alleviate self-inhibitory structures within the target, interfere with pre-mature transcription termination, or mask RNase recognition sites (6–9).
Bacterial riboregulators oftentimes require the assistance of RNA chaperones which stabilize sRNAs and facilitate annealing of target transcripts. The three major RNA chaperones currently known to foster sRNA activity in bacteria are the FinO family member ProQ, CsrA/Rsm, and Hfq, which belongs to the Lsm/Sm protein superfamily [reviewed in reference (10)]. Widely conserved in bacterial species, Hfq assembles in a homo-hexameric ring structure offering binding sites for single-stranded RNAs at both faces of the ring, the rim, and the intrinsically unstructured C-terminal domain which is appended to the core (11–13). The C-terminal domain has been resolved in a crystal structure of Caulobacter crescentus Hfq, revealing its ability to interact with positively charged residues on the rim to foster selective annealing of RNA (14).
In comparison to the well-understood standard models of bacterial RNA biology such as the Gram-negative enterobacteria Escherichia coli and Salmonella Typhimurium, studies on the role of Hfq in post-transcriptional control in C. crescentus have only scratched the surface (15). Most of the ~130 sRNA candidates previously identified by transcriptomic studies in Caulobacter still await further characterization, and a function of Hfq for the activity of the four sRNAs studied in more detail as yet has either not been observed (ChvR and GsrN) or not been addressed (CrfA and CcnA) (16–21).
A member of the highly diverse clade of Alphaproteobacteria, C. crescentus inhabits nutrient-poor freshwater environments and has evolved diverse mechanisms to adapt to severe nutrient fluctuations in this niche (22). The oligotrophic lifestyle of C. crescentus is for example reflected in the abundance of more than 60 genes encoding paralogs of outer membrane transporters of the family of TonB-dependent transporters (TBDRs). TBDRs catalyze the specific uptake of nutrients from the dilute environment including substrates like carbohydrates, vitamin B12, and iron-scavenging siderophores (23–27). Iron serves as an essential cofactor in central metabolism and respiration, yet due to its poor solubility in the abundant, ferric form (Fe3+), it is a limiting nutrient in most biological systems (28). On the other hand, free intracellular ferric iron can damage biomolecules through reactive oxygen species, which eventually causes cell death (29). Iron homeostasis is thus finely controlled in bacteria, and oftentimes involves a transcriptional master regulator termed Fur (ferric uptake regulator) which, when in complex with iron, binds so-called Fur boxes predominantly in promoters to repress expression of genes controlling iron uptake (30). In E. coli and related enterobacteria, the Fur regulon includes an Hfq-dependent sRNA, RyhB, which serves as the negative arm of the regulatory circuit and represses the synthesis of non-essential iron-binding proteins to increase the availability of iron (31). While Fur is also the master transcriptional regulator of iron homeostasis in C. crescentus (32), an sRNA modulating gene expression at the post-transcriptional level in response to iron depletion remains to be identified.
Here, we conducted RNA co-immunoprecipitation experiments to catalogue direct ligands of Hfq in C. crescentus. Our results reveal that of all sRNAs expressed in the early stationary phase, only a subset is associated with Hfq in this bacterium, including sRNA CCNA_R0199. We found that the transcript is specifically induced in response to high levels of zinc and iron starvation, and renamed it RusT (RNA upregulated in iron starvation). Forward genetics in combination with biochemical analyses unveiled that transcription of RusT is under the control of NtrYX, a conserved two-component system (TCS) recently implicated in cell envelope regulation in alphaproteobacteria. Moreover, transcriptome analyses revealed that RusT, together with Hfq, post-transcriptionally controls at least 16 transcripts, including seven membrane transporters. In summary, our study presents evidence for Hfq-dependent post-transcriptional regulation through an sRNA in C. crescentus, confirming that the chaperone is not only mediating RNA stability but also functions as an important mediator of RNA-based post-transcriptional control of the membrane proteome in this oligotroph.
RESULTS
Profiling of Hfq-associated RNA in C. crescentus
A previous study reporting on the purification of RNA crosslinked to Hfq via UV illumination identified nine potential sRNA ligands in C. crescentus (15), but we assumed that the Hfq interactome was likely to be more extensive. To close this gap, we performed co-immunoprecipitation (co-IP) of RNA with a functional Flag-tagged derivative of Hfq (Fig. S1). Setting a cutoff at ≥3-fold enrichment in the co-IP sample, we detected 311 transcripts bound by Hfq, including 290 mRNAs, 19 sRNAs, and 2 rRNAs (Table S1; Fig. 1A).
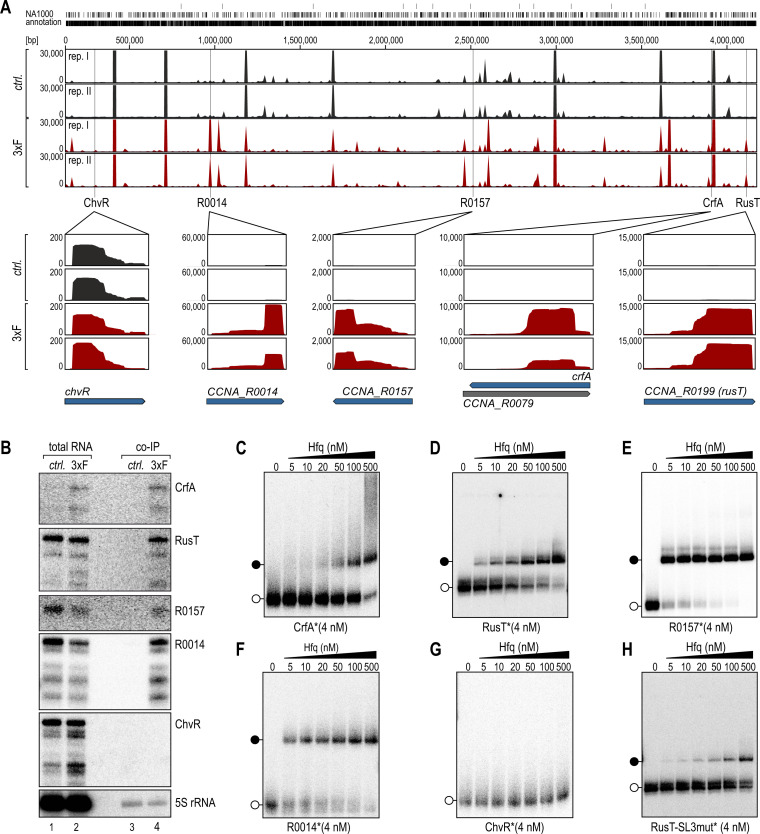
Fig 1.
Specific association of RNA with the Hfq protein in C. crescentus. (A) C. crescentus NA1000 carrying a chromosomally encoded 3xFLAG-hfq allele was cultured in complex PYE medium to an OD660 of 1.0, an untagged wild-type strain served as control. Following the co-immunoprecipitation of RNA-Hfq complexes from cell lysates using a monoclonal antibody directed against the FLAG-tag, isolated RNA was converted into cDNA, and subjected to high-throughput sequencing analysis. cDNA reads of the control (ctrl.; black) and 3xFLAG-Hfq co-IP (3xF; red) were mapped to the C. crescentus NA1000 genome (two replicates each, rep. I and rep. II). Examples of enrichment patterns of five sRNAs are shown as enlarged panels below the sequencing tracks. For each plot, a scale indicates the number of mapped reads per position. (B) Northern blot analysis of total RNA samples and upon co-IP of C. crescentus wild-type (ctrl.) and cells expressing 3xFLAG-Hfq (3xF) to determine recovery profiles for five selected sRNAs. 5S rRNA served as loading control. (C–H) Native gel electrophoretic mobility shift assay (EMSA) using in vitro-synthesized, 5′ end-labeled sRNAs (C) CrfA, (D) RusT, (E) R0157, (F) R0014, (G) ChvR, or (H) RusT-SL3mut (all 4 nM) with increasing concentrations of purified C. crescentus Hfq protein as indicated. Open circles indicate free sRNAs, black circles indicate sRNA-Hfq complexes.
The fraction of sRNAs purified from the co-IP sample more than doubled relative to the control (Fig. S2A), and we listed the candidates based on their enrichment (Table 1). We verified the results of the RNA sequencing experiment for four sRNAs with the highest enrichment (CrfA, RusT, R0157, and R0014) using Northern blot analysis (Fig. 1B), and included ChvR as Hfq-independent control sRNA (14, 17). As expected, ChvR was not recovered with Hfq, while CrfA, RusT, R0157, and R0014 were detected in the co-IP sample. However, we observed that expression of CrfA, an sRNA induced during starvation (18), was increased in the total RNA input sample of the 3xFLAG::hfq strain when compared to the control strain (Fig. 1B; Fig. S3), resulting in a potential overestimation of the relative enrichment of this RNA. All other tested sRNAs had comparable expression patterns in both strains (Fig. S3). To further corroborate our results, we performed native gel electrophoretic mobility shift assays (EMSAs) using purified C. crescentus Hfq protein and in vitro transcribed sRNAs. As expected, Hfq formed stable complexes with CrfA, RusT, R0157, and R0014 (Fig. 1C through F), but not with ChvR (Fig. 1G).
TABLE 1.
Hfq-associated sRNAs (≥3-fold enrichment)
| Gene | Enrichment | FDR-adj. P value |
|---|---|---|
| CCNA_R0093 (crfA) | 172.1 | 9.5 E−80 |
| CCNA_R0199 (rusT) | 73.8 | 5.8 E−150 |
| CCNA_R0157 | 63.8 | 1.8 E−111 |
| CCNA_R0014 | 51.7 | 3.4 E−53 |
| CCNA_R0188 | 49.2 | 1.4 E−120 |
| CCNA_R0143 | 34.9 | 8.2 E−32 |
| CCNA_R0097 | 20.7 | 1.9 E−63 |
| CCNA_R0195 | 17.6 | 1.7 E−63 |
| CCNA_R0040 | 11.4 | 1.9 E−40 |
| CCNA_R0133 | 9.1 | 1.0 E−41 |
| CCNA_R0025 | 8.1 | 1.2 E−29 |
| CCNA_R0127 | 7.0 | 5.4 E−44 |
| CCNA_R0098 | 5.7 | 1.0 E−18 |
| CCNA_R0156 | 5.6 | 3.1 E−14 |
| CCNA_R0016 | 5.0 | 1.3 E−15 |
| CCNA_R0184 | 4.3 | 2.3 E−16 |
| CCNA_R0175 | 3.4 | 3.6 E−15 |
| CCNA_R0117 | 3.3 | 4.5 E−11 |
RusT is a conserved sRNA interacting with Hfq
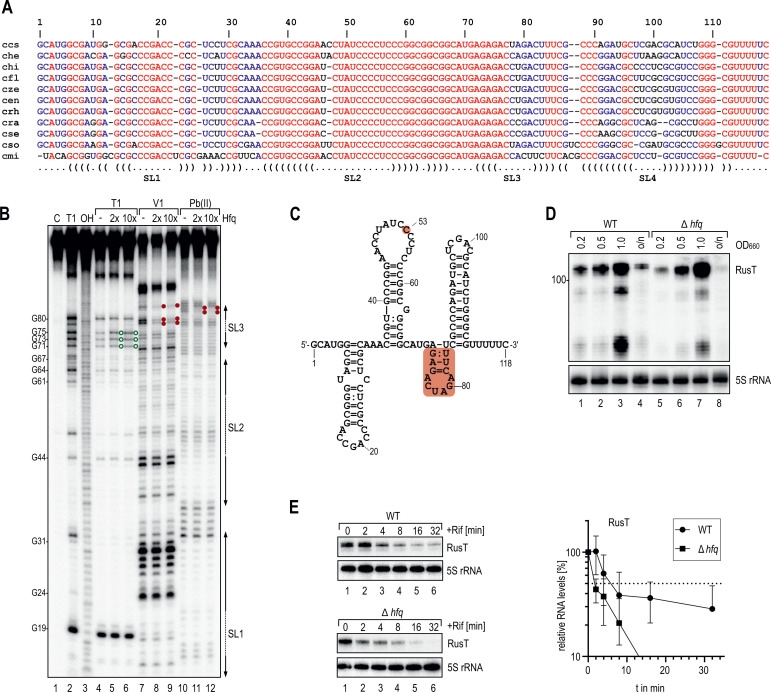
We selected the previously uncharacterized RusT sRNA (annotated as CCNA_R0199) which was strongly enriched in our co-IP experiment (>70-fold; Table 1), to study the functions of an Hfq-associated sRNA in C. crescentus. RusT is transcribed as a 118-nt long RNA from the plus-strand of the intergenic region between CCNA_03820 (encoding an outer membrane lipoprotein carrier protein; lolA) and CCNA_03821 (encoding exodeoxy-ribonuclease III involved in DNA repair; xth). Homologs of rusT with little sequence variation are encoded at this genomic locus in other Caulobacteraceae closely related to C. crescentus (Fig. S4; Fig. 2A). To determine the secondary structure of RusT in the presence and absence of Hfq we employed chemical probing. To this end, we incubated in vitro transcribed, 5′ end-labeled RusT sRNA with increasing concentrations of purified Hfq protein, and subsequently subjected the RNA to partial digestion with RNase T1 (preferentially cleaving after single-stranded guanine residues), RNase V1 (preferentially cleaving double-stranded regions), and lead(II) acetate (preferentially cleaving single-stranded regions). The resulting digestion pattern (Fig. 2B) revealed that RusT contains four stem-loops, of which the last one is followed by a polyU-stretch likely functioning as a Rho-independent termination signal (Fig. 2C). The addition of Hfq caused a change in the cleavage pattern between nt 72 and 87 (corresponding to stem-loop 3), suggesting that the RNA-binding protein alters the secondary structure of the sRNA when occupying this site. Indeed, when we used a mutant of RusT lacking the third stem-loop (RusT-SL3mut) in EMSAs with Hfq, we observed a strong decrease in the binding affinity compared to wild-type RusT (Fig. 1D and H).
Fig 2.
Conservation, structure, and Hfq-dependence of RusT. (A) Non-redundant alignment of the RusT sRNA with orthologous sRNAs of the genus Caulobacter (ccs: C. crescentus NA1000; che: C. henricii CB4; chi: C. hibisci strain KACC 18849; cfl: C. flavus strain RHGG3; cze: C. zeae strain 410 SGCZ15; cen: C. endophyticus strain 774; crh: C. rhizosphaerae strain KCTC 52515; cra: C. radicis strain 695; cse: C. segnis strain TK0059; cso: C. soli strain Ji-3-8; cmi: C. mirabilis strain FWC 38). Colors indicate full (red); partial (blue) or no (black) conservation. The predicted secondary structure of C. crescentus RusT is indicated below the alignment. (B) In vitro structure probing of 5′ end-labeled RusT sRNA (0.4 pmol) with RNase T1 (lanes 4–6), RNase V1 (lanes 7–9), and lead(II) (lanes 10–12) in the presence and absence of purified Hfq protein (0, 0.8 pmol or 4 pmol, respectively). RNase T1 and alkaline (OH) ladders of RusT were used to map cleavage fragments, and positions of mapped G-residues are marked relative to the TSS. Increased cleavage is marked with green open circles, reduced cleavage is marked with red filled circles. (C) Secondary structure of RusT as determined by chemical probing shown in (B). The sRNA region interacting with Hfq (nt 73–84) and residue C53 are highlighted in red. (D) Northern blot analysis of RusT expression in C. crescentus wild-type and an isogenic hfq mutant strain. RNA was collected at the indicated optical density (OD660) or after 24 h of growth (o/n) in PYE medium. 5S rRNA served as loading control. (E) Determination of RusT stability. Northern blot analysis of RNA obtained from wild-type or Δhfq cells grown in minimal M2G medium to exponential phase (OD660 of 0.5). Total RNA samples were collected at indicated time points prior to, and after addition of rifampicin to inhibit transcription. The time point at which 50% of RusT had been decayed (dashed line) was calculated to determine RNA half-life. Error bars indicate the standard deviation of three independent biological replicates.
To test if Hfq protects RusT from degradation, we compared the RusT levels in wild-type and hfq mutant strains (Fig. 2D) and observed equal or even elevated levels (depending on the time point of sampling) of RusT in the absence of Hfq. To determine whether this result was due to either higher transcription rates or increased stability of RusT in the mutant, we first determined the half-life of the sRNA. Both wild-type and hfq mutant cells were grown to exponential phase, and RNA decay was monitored after inhibition of transcription with rifampicin (Fig. 2E). RusT stability decreased more than threefold in the absence of Hfq (Fig. 2E), as frequently observed for Hfq-associated sRNAs in other species (6, 33, 34). Similarly, decay of the Hfq-associated sRNA R0014 was accelerated in the hfq mutant strain, while half-life of the Hfq-independent ChvR sRNA was not affected (Fig. S5A and B). Since RusT stability was decreased in the absence of Hfq, the increase of RusT steady-state levels in the hfq mutant strain was likely a result of increased sRNA transcription. Indeed, when we expressed rusT under control of the unrelated, vanillate-inducible promoter (pPvan-RusT), sRNA levels in the absence of Hfq were no longer augmented (Fig. S5C).
Iron starvation induces RusT transcription
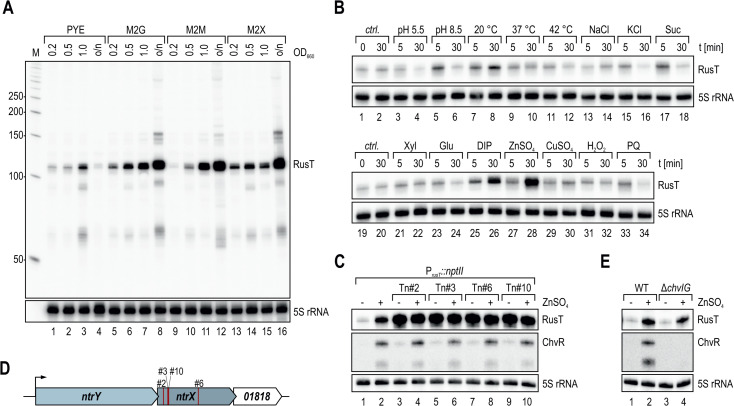
To probe the transcriptional regulation of RusT in C. crescentus, we next determined RusT abundance by Northern blot analysis of total RNA samples collected from cells grown in complex (PYE) and defined media (M2 supplemented with either glucose, maltose, or xylose as sole carbon source), and after stress induction under 16 different conditions (Fig. 3A and B). RusT levels were elevated in cells cultivated in minimal medium and most abundant in samples collected after overnight growth (Fig. 3A). However, when exponentially growing cells were treated with either the chelator 2,2-dipyridyl (DIP) or 75 µM of ZnSO4, respectively (Fig. 3B, lanes 26 and 28), a ~5-fold and ~9-fold increase in RusT levels was observed. Addition of DIP is known to induce iron starvation (31), and similarly, excess of zinc results in perturbation of the iron homeostasis, leading to potential mismetallation of iron-dependent enzymes (35–37). In many bacteria, iron starvation and concomitant de-repression of the Fur regulon also induces sRNAs including RyhB in the enterobacteria or PrrF, NrrF, and FsrA sRNAs of Pseudomonas aeruginosa, Neisseria meningitidis, and Bacillus subtilis, respectively (38–40). However, contrary to the abovementioned examples, we suspected that rusT transcription was not directly repressed by Fur as we were unable to identify the well-defined consensus binding site within the promoter PrusT (32), suggesting that another regulator controls the expression of the sRNA.
Fig 3.
RusT is induced in response to iron starvation. (A) Northern blot analysis of RusT expression in PYE or minimal M2 medium supplemented with glucose (M2G), maltose (M2M), or xylose (M2X) as carbon source. RNA was collected at the indicated optical density (OD660) or after 24 h of growth (o/n). 5S rRNA served as loading control. (B) Total RNA was prepared from cells grown in PYE to OD660 of 0.4 at different time points without (ctrl.) or upon treatment (5 min and 30 min) of the culture with the indicated stresses (pH 5.5: PYE at pH 5.5; pH 8.5: PYE at pH 8.5; 20 °C/37 °C/42°C: growth at alternative temperature; NaCl/KCl/Suc: osmotic stress at 85 mM NaCl/40 mM KCl/150 mM sucrose, respectively; Xyl: addition of xylose [f.c. 0.2%]; Glu: addition of glucose [f.c. 0.2%]; DIP: addition of 2,2-dipyridyl [f.c. 200 µM]; ZnSO4: addition of ZnSO4 [f.c. 75 µM]; CuSO4: addition of CuSO4 [f.c. 30 µM]; H2O2/PQ: oxidative stress at 10 mM hydrogen peroxide or paraquat, respectively). RusT expression in response to stress was determined by Northern blot analysis; 5S rRNA served as loading control. (C) Northern Blot analysis of RusT and ChvR expression in C. crescentus carrying the transcriptional reporter PrusT::nptII, without and selected clones with insertion of a mini-himar1 transposon (Mar2xT7). Total RNA samples were prepared from cells at OD660 of 0.5 in PYE prior to and 30 min after addition of 75 µM ZnSO4. 5S rRNA served as loading control. (D) Mapped positions of transposon insertions in ntrX of the clones analyzed in (C) are indicated in red. (E) Expression of sRNAs RusT and ChvR was analyzed on Northern blots using RNA samples prepared from C. crescentus wild-type and a ΔchvIG-hprK mutant strain grown as described in (C).
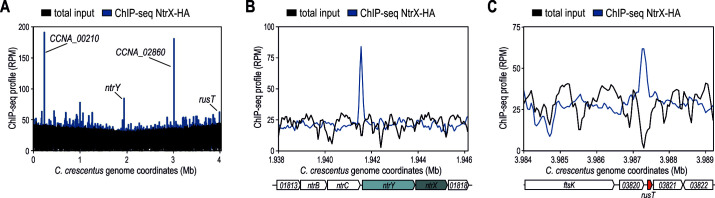
To identify regulators of rusT transcription, we integrated a transcriptional reporter into the Caulobacter chromosome in which expression of the nptII gene (conferring resistance to kanamycin) is driven by PrusT. We capitalized on the low activity of PrusT and the resulting inability of the reporter strain to grow on plates supplemented with high concentrations of kanamycin ( 20 µg/mL; Kan20). Upon mutagenesis with the mini-himar1 transposon (Mar2xT7, conferring resistance to gentamycin), we selected clones growing on Kan20 and gentamycin. When testing RusT levels in response to zinc for 10 individual clones, we identified four mutants in which the sRNA was constitutively expressed independent of the presence of the inducer (Fig. 3C). The himar1 insertion site in all four mutants was mapped to ntrX, encoding the response regulator of the conserved NtrYX TCS (Fig. 3D). Originally, the NtrYX system has been associated with the regulation of nitrogen metabolism including denitrification, nitrogen assimilation, and fixation in diverse bacteria (41–45), but has also been implicated to respond to iron starvation of Paracoccus denitrificans (46). To probe for a direct involvement of NtrX in RusT transcription, we performed chromatin immunoprecipitation coupled to deep sequencing (ChIP-Seq) in a C. crescentus strain harboring an HA-tagged version of ntrX (ntrX::HA; (47)) in lieu of the wild-type version. We detected 23 peaks that were significantly enriched (fold-enrichment ≥2) over the background, including a site upstream of the ntrYX transcriptional start site (TSS) (Fig. 4A and B; Table S2). Potential autoregulation has likewise been suggested for the NtrYX system of Rhodobacter sphaeroides where NtrX also binds its own promoter (45). In addition, the ChIP-Seq analysis revealed efficient occupancy of PrusT by NtrX (Fig. 4C), indeed suggesting that the response regulator is directly involved in transcriptional control of RusT and functioning as a repressor of sRNA expression under non-inducing conditions.
Fig 4.
Determination of NtrX binding sites on the genome of C. crescentus. (A) Genome-wide profiles of reads for NtrX-HA ChIP-Seq (blue) and total input control (black). Selected peaks are assigned, a list of all significantly enriched sites is provided in Table S2. (B, C) Traces of NtrX-HA ChIP-Seq (blue) and total input control (black) upstream of the ntrXY-CCNA_01818 operon (B) or at the rusT promoter (C).
Identification of RusT target genes
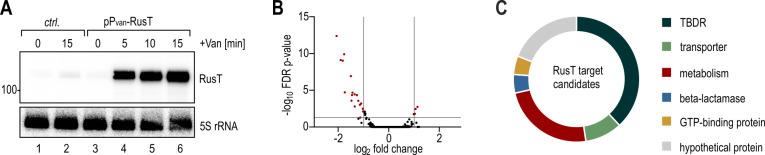
While no Hfq-dependent activity of a trans-acting sRNA is currently known for C. crescentus (48, 49), we were curious to determine the physiological function of RusT. To identify potential direct targets of the sRNA, we globally assessed transcriptome differences in response to pulse-overexpression of RusT. To this end, we cultivated wild-type cells carrying either an empty control vector or the vanillate-inducible pPvan-RusT in M2G (OD660 of 0.8), and isolated total RNA samples before and after addition of vanillate. Under this condition, RusT is only weakly expressed, and competition with the endogenous sRNA is not relevant to the experiment. Compared to the control, RusT levels increased ~13-fold within 15 min in the presence of the inducer (Fig. 5A; Fig. S6). We next converted the total RNA samples into cDNA which we subjected to high-throughput sequencing (HTPS) analysis. In total, 23 transcripts met our cutoff criteria for differentially expressed genes (≥10 reads; ≥2-fold regulation; FDR-adj. P value ≤ 0.05), of which four were upregulated and 19 were downregulated in the presence of RusT (Table 2; Fig. 5B). Not surprisingly, RusT sRNA levels were strongly induced. In addition, also CrfA sRNA increased ~2-fold, a result which is likely an artifact of the pulse-overexpression due to increased Hfq occupancy by RusT upon the addition of vanillate to the culture (Fig. S6). Two additional transcripts were more abundant in response to RusT expression when compared to the control, CCNA_03360 (encoding tagatose-1,6-bisphosphate aldolase; 2-fold upregulation) as well as CCNA_01820 (2.1-fold upregulation), encoding hflX, the second gene in the hfq-hflX operon [the hfq (CCNA_01819) mRNA increased 1.98-fold in our experiment, and was excluded by our cutoff criteria]. The large set of repressed genes included transcripts encoding TBDRs (CCNA_02895, CCNA_03263, CCNA_01042, CCNA_01738, CCNA_00210, CCNA_03248, CCNA_00338, and CCNA_00573), other types of transporters [major facilitator superfamily (MFS) transporter CCNA_02400, porin CCNA_01475 (ompW)], various metabolic factors [CCNA_02380, CCNA_03181, CCNA_03774, and CCNA_02047 (glnA)], a beta-lactamase family protein (CCNA_03294) as well as several hypothetical proteins (CCNA_01043, CCNA_02741, CCNA_02342, and CCNA_02379). When we analyzed the list of potential RusT targets we realized that a significant portion had been previously identified to be down-regulated in response to iron starvation and/or in strains deficient for Fur (Table 2; (50)), corroborating our finding that RusT is activated when iron homeostasis is disturbed (Fig. 3B).
Fig 5.
Pulse-overexpression of RusT reveals target genes of the sRNA. (A) C. crescentus ΔvanAB carrying either an empty control vector (pBV-MCS6; ctrl.) or the expression plasmid pPvan-RusT were grown in M2G to an OD660 of 0.8. Total RNA was prepared from cells collected prior to and at indicated time points after addition of vanillate. Expression of RusT and 5S rRNA (loading control) was determined by Northern blot analysis. (B) Volcano-plot analysis of gene expression changes in response to pulse-induction (15 min) of RusT. Genes with fold changes ≥2 and an FDR-adj. P value of ≤0.05 were considered as significantly different and are highlighted in red. (C) Classification of RusT target candidates based on annotated gene functions.
TABLE 2.
Differentially expressed transcripts in response to RusT pulse-expression (≥2-fold change)
| Gene ID | Gene product | Fold change | FDR-adj. P value |
gfp fusion (relative to TSS) |
Downregulation in low iron conditionsa |
|---|---|---|---|---|---|
| CCNA_02895 | TonB-dependent receptor | −4.2 | 4.1 E−13 | −269 to +60 | Yes |
| CCNA_03263 | TonB-dependent receptor | −3.8 | 7.7 E−10 | −116 to +60 | Yes |
| CCNA_01042 | TonB-dependent receptor | −3.5 | 9.5 E-10 | −225 to +60 | Yes |
| CCNA_02400 | Transporter, major facilitator superfamily | −3.4 | 1.2 E−10 | −32 to +60 | |
| CCNA_01475 | OmpW family outer membrane protein | −3.3 | 1.9 E−05 | −40 to +75 | Yes |
| CCNA_01738 | TonB-dependent outer membrane receptor | −2.8 | 1.2 E−07 | −86 to +60 | |
| CCNA_00210 | TonB-dependent receptor | −2.8 | 2.6 E−4 | −59 to +60 | Yes |
| CCNA_02380 | Aspartate 1-decarboxylase | −2.8 | 4.6 E−05 | −71 to +60 | |
| CCNA_03181 | Alcohol dehydrogenase | −2.7 | 2.1 E−05 | −156 to +60 | |
| CCNA_01043 | Hypothetical protein | −2.6 | 3.6 E−05 | −43 to +60 | |
| CCNA_03248 | TonB-dependent receptor | −2.5 | 1.6 E−03 | −49 to +60 | |
| CCNA_02741 | Conserved hypothetical protein | −2.4 | 4.6 E−05 | −99 to +75 | |
| CCNA_03774 | Citrate lyase beta chain/citryl-CoA lyase subunit | −2.3 | 2.7 E−04 | −29 to +60 | Yes |
| CCNA_00338 | TonB-dependent receptor | −2.2 | 9.2 E−04 | −94 to +60 | |
| CCNA_02047 | Glutamine synthetase GlnA | −2.2 | 0.05 | −424 to +60; −42 to +60 |
|
| CCNA_00573 | TonB-dependent receptor | −2.1 | 6.4 E−04 | No tss | |
| CCNA_02342 | Hypothetical protein with pentapeptide repeats | −2.1 | 6.4 E−04 | −35 to +60 | |
| CCNA_03294 | Beta-lactamase family protein | −2.0 | 3.3 E−03 | −96 to +60 | |
| CCNA_02379 | Hypothetical protein | −2.0 | 7.7 E−04 | −71 to +60 | |
| CCNA_03360 | Tagatose-1,6-bisphosphate aldolase | 2.0 | 0.02 | −282 to +60 | |
| CCNA_01820 | GTP-binding protein | 2.1 | 3.6 E−03 | −179 to +60; −53 to +60 |
|
| CCNA_R0093 | Non-coding RNA CrfA | 2.2 | 1.8 E−03 | ||
| CCNA_R0199 | Non-coding RNA RusT | 72.4 | 5.6 E−50 |
As determined in reference (50).
Reporter fusions confirm post-transcriptional regulation by RusT
To investigate the potential post-transcriptional regulation of the newly identified RusT target candidates, we designed 21 translational fusions to the green fluorescent protein (gfp) under control of the constitutive rsaA promoter (17). Base-pairing sRNAs typically interact with their target transcripts at sites located within the 5′ UTR or the early coding sequence of the mRNA (51). We fused the sequence from the TSS up to the first 15–20 codons of each target gene to the second codon of gfp (Table 2; Fig. S7). For genes organized in operons, we made a fusion to the first gene of the polycistronic mRNA. We omitted target candidate CCNA_00573 from our analysis as we and others were unable to detect the TSS of the corresponding mRNA (17, 52).
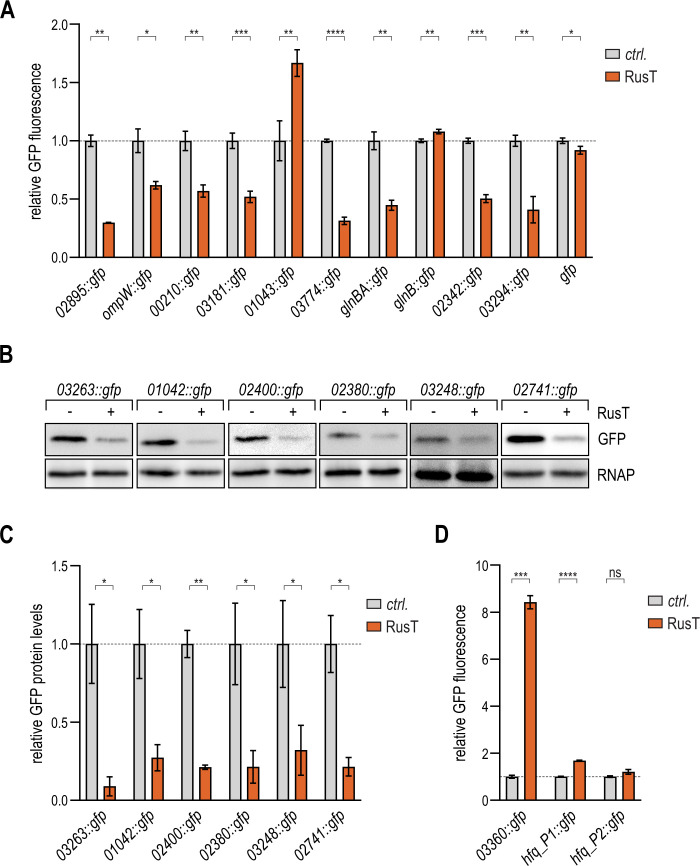
We integrated the gfp reporter fusions in a C. crescentus ΔvanAB ΔrusT mutant carrying either an empty control plasmid, or the construct pPvan-RusT to overexpress RusT. Cells were harvested upon overnight growth in PYE (supplemented with the inducer vanillate), and GFP expression was quantified by fluorescence intensity measurements (Fig. 6A and D). Fluorescence was reduced in the presence of RusT for eight target fusions (02895::gfp, ompW::gfp, 00210::gfp, 03181::gfp, 03774::gfp, glnBA::gfp, 02342::gfp, and 03294::gfp), while sRNA overexpression did not affect GFP production of a gfp control construct. An additional six fusions (03263::gfp, 01042::gfp, 02400::gfp, 02380::gfp, 03248::gfp, and 02741::gfp) were validated as negatively regulated by RusT through immunoblot analysis using an anti-GFP antibody since the fluorescence of these reporters was below the detection limit (Fig. 6B and C). No negative regulation of gfp fluorescence in the presence of RusT was observed for two fusions, glnB::gfp and 01043::gfp. In both cases, however, the two target candidates encode short leader peptides for the glnBA and CCNA_01043-01042 operons, respectively. The longer reporter fusions comprising the second genes of these operonic structures are both regulated by RusT (Fig. 6A through C). We thus assume that target repression requires a regulatory element located downstream of the leader sequence. Reporter fusions 01738::gfp and 00338::gfp were excluded from the analysis as we were unable to detect GFP production for these constructs (not shown).
Fig 6.
Regulation of RusT target reporter fusions. (A–D) Regulation of target reporter fusions by RusT. C. crescentus ΔvanAB ΔrusT cells carrying the indicated gfp reporter fusion in combination with either an empty control vector (pBV-MCS6; ctrl.), or the expression plasmid pPvan-RusT were grown overnight in the presence of vanillate, and GFP expression was quantified either by fluorescence intensity measurements (A, D) or by Western blot analysis of total protein samples using RNA polymerase (RNAP) as loading control [B; quantification in (C)]. For the quantification of each GFP-fusion, expression levels in the presence of the control plasmid were set to 1, and relative changes were determined for cells expressing RusT. GFP levels were calculated from three biological replicates; error bars indicate the standard deviation.
Our transcriptome analysis also revealed a positive effect of RusT expression on two target candidates (Table 2; Fig. 5B), and we confirmed upregulation of the 03360::gfp fusion in the presence of RusT (Fig. 6D). As expression of the hfq-hflX operon is controlled through two promoter elements P1 and P2 (17, 52), we constructed two reporter variants (Fig. S7U and V). An increase in GFP fluorescence upon RusT overexpression was solely observed for the reporter fusion harboring the longer 5′ UTR (hfq_P1::gfp; Fig. 6D). Since input through the second, proximal promoter P2 included in this fusion could not be excluded, we also performed primer extension analysis of the native hfq-hflX transcripts. Pulse expression of RusT resulted in an accumulation of the shorter but not the longer transcript (Fig. S8). Taken together, these results point toward an increase of transcriptional activity from the second, proximal promoter P2 in response to RusT overexpression; however, the exact mechanism of regulation awaits further characterization.
RusT function requires the aid of Hfq
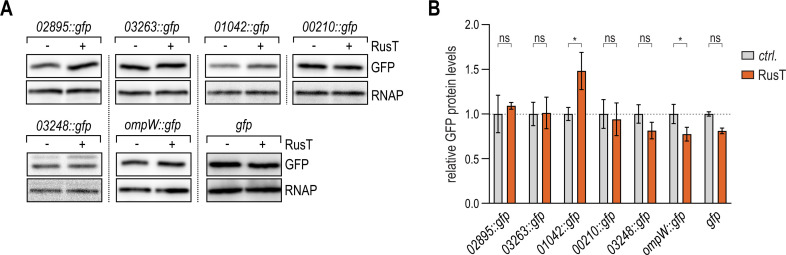
Tight control of outer membrane proteins is critical for cell survival, and many proteins involved in substrate transport across the outer membrane are controlled through the activity of sRNAs in enterobacteria (53, 54). Likewise, TBDRs spanning the outer membrane represented the largest class of confirmed RusT targets in Caulobacter, suggesting that the sRNA is involved in remodeling the cell envelope in response to iron deficiency. Different from many other Hfq-associated sRNAs, RusT was also well expressed in an hfq mutant background (Fig. 2D). We therefore sought to determine whether Hfq was at all required for RusT-dependent regulation. To this end, we selected the top five deregulated candidates encoding TBDRs (02895::gfp, 03263::gfp, 01042::gfp, 00210::gfp, and 03248::gfp) and ompW::gfp for further analysis, and transferred our reporter system into ΔvanAB ΔrusT Δhfq cells. We again determined GFP levels for each of the six fusions in the presence or absence of RusT by immunoblot analysis using antibodies to GFP. Although we confirmed that overexpression from the Pvan promoter resulted in similar sRNA levels when compared to the wild-type background (Fig. S9), RusT-dependent downregulation of the reporter fusions was abrogated in all cases (Fig. 7A and B). Collectively, our results suggest that the binding of Hfq not only increases the stability of RusT but that the sRNA also requires Hfq for the post-transcriptional regulation of target mRNAs.
Fig 7.
Target regulation by RusT is dependent on Hfq. (A, B) Regulation of target reporter fusions by RusT in the absence of Hfq. C. crescentus ΔvanAB ΔrusT Δhfq cells carrying the indicated gfp reporter fusion in combination with either an empty control vector (pBV-MCS6; ctrl.), or the expression plasmid pPvan-RusT were grown overnight in the presence of vanillate. Total protein samples were analyzed on Western blots to determine GFP expression; RNAP served as loading control. (B) Quantification of GFP expression as described in (A); error bars indicate the standard deviation from three biological replicates.
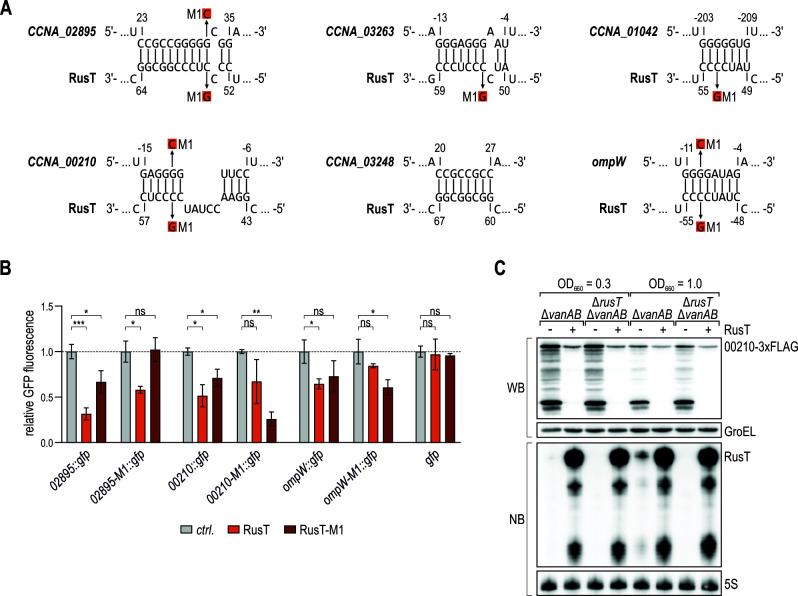
In analogy to regulatory proteins, sRNAs are assembled of modular domains that administrate either structural or functional roles (55). Typically, target recognition sites—referred to as seed sequences—are characterized by a high degree of sequence conservation between homologs, and low structural complexity. We thus assumed that an internal segment of RusT including the conserved sequence of the second stem-loop (nt 45–57; Fig. 2A) was relevant for base-pairing with target mRNAs. To test this hypothesis, we used the RNAhybrid and IntaRNA algorithms (56, 57) to predict interactions between the highly conserved RusT sequence element and the five TBDR-encoding and ompW mRNA portions present in the respective gfp reporter constructs (Fig. 8A). For five of the six target fusions, a conserved cytosine within the loop of the second hairpin of RusT (C53; Fig. 2B) was involved in the interaction. We thus inserted the relevant point mutation in pPvan-RusT (C53G; RusT-M1) which did not affect the sRNA’s expression level (Fig. S9), and tested its effect on the regulation of the two TBDR reporters that were amenable to analysis via fluorescence intensity measurements, 02895::gfp and 00210::gfp, as well as on the ompW::gfp reporter. When compared to wild-type RusT, RusT-M1 displayed reduced GFP repression of the 02895::gfp and 00210::gfp fusions, indicating that the conserved C53 residue of the sRNA was involved in its regulatory activity in both cases (Fig. 8B). To restore target gene regulation, we next introduced compensatory single-point mutations in the respective reporter fusions (Fig. 8A). RusT had reduced capacity to repress 02895-M1::gfp; however, we did not observe the restoration of the regulation in the presence of RusT-M1. In contrast, 00210-M1::gfp and ompW-M1::gfp were specifically repressed by RusT-M1, and only partially amenable to regulation by the native RusT version (Fig. 8B).
Fig 8.
RusT employs a conserved sequence stretch for target recognition. (A) Base-pairing interactions between RusT and target mRNAs encoding transporters CCNA_02895, CCNA_03263, CCNA_01042, CCNA_00210, CCNA_03248, and ompW as predicted using the RNA hybrid and IntaRNA algorithms (56, 57). Nucleotide positions in the mRNA are marked relative to the start codon. The M1 (C53G) mutation in RusT and the corresponding M1 mutations in the mRNAs are indicated. (B) Regulation of target reporter fusions by RusT or RusT-M1. C. crescentus ΔvanAB ΔrusT cells carrying the indicated gfp reporter fusion in combination with either an empty control vector (pBV-MCS6; ctrl.), or the expression plasmid pPvan-RusT or pPvan-RusT-M1 were grown overnight in the presence of vanillate, and GFP expression was determined and quantified by fluorescence intensity measurements as described for Fig. 6. (C) CCNA_00210-3xFLAG protein expression in response to RusT overexpression. C. crescentus ΔvanAB CCNA_00210::3xFLAG cells carrying either an empty control vector [pBV-MCS6; (−)], or the expression plasmid pPvan-RusT (+) were grown to mid-exponential and early stationary phase (OD660 of 0.3 and 1.0, respectively) in the presence of vanillate. Total protein samples were analyzed by Western blot to determine CCNA_00210–3xFLAG levels. Expression of RNA polymerase (RNAP) was probed as loading control. Induction of RusT from pPvan-RusT was confirmed by Northern blot analysis; 5S rRNA served as loading control.
As we had successfully confirmed that regulation of the 00210::gfp target fusion was due to a direct, Hfq-dependent base-pairing interaction with RusT we tested whether the sRNA also affected the levels of the endogenous CCNA_00210 TBDR. We inserted a C-terminal 3xFLAG tag in the chromosomal copy of CCNA_00210, and compared protein levels in the presence and absence of RusT. Caulobacter wild-type and rusT mutant cells carrying either a control plasmid or the RusT overexpression construct pPvan-RusT were grown in the presence of vanillate to mid-exponential and early stationary phase (OD660 of 0.3 and 1.0, respectively), and while the deletion of RusT did not significantly affect CCNA_00210 levels, we observed strongly reduced protein levels in the strain overexpressing RusT (Fig. 8C).
DISCUSSION
Specific binding of Caulobacter Hfq to RNA
Although Hfq was originally identified as a co-factor required for the replication of RNA phage Qβ in E. coli (58), the high degree of conservation in various bacterial species early on suggested an additional, more general physiological function of the RNA binding protein. Indeed, the primary role of Hfq is today seen in its complex contribution to post-transcriptional gene regulation, fostering the interaction between sRNAs and their cognate trans-encoded target mRNAs during stress responses (55). Base-pairing of sRNAs and mRNAs results in either positive or negative regulation of target translation and typically also affects transcript stability. The Hfq-governed networks are best studied in enterobacteria, where more than 100 sRNAs entertain thousands of individual interactions to modulate gene expression (10, 59). Hfq deficiency manifests in pleiotropic phenotypes including reduced growth, increased sensitivity to intrinsic and environmental stresses, alterations in central metabolism and reduced virulence of bacterial pathogens (60, 61). In Caulobacter, hfq null mutants suffer from severe defects in cell morphology and decreased tolerance to antibiotics targeting the cell wall (49), likely reflecting pervasive changes in gene expression and the accumulation of alpha-ketoglutarate which redirects peptidoglycan precursor metabolism. Hundreds of transcripts are deregulated in the absence of Hfq; however, the RNAs directly bound by Hfq are barely known (15, 49). In this study, we used co-immunoprecipitation of RNA with 3xFLAG-tagged Hfq protein in C. crescentus to capture the chaperone’s interactome in early stationary growth. In total, we recovered 311 transcripts including 21 non-coding RNAs with at least threefold enrichment in the co-IP sample when compared to the control (Fig. 1; Fig. S2; Table 1; Table S1). Of note, we also identified an increase in abundance of the two 23S rRNA transcripts (~17.5-fold and 22-fold for CCNA_R0084 and CCNA_R0066, respectively), while 16S and 5S rRNA transcripts were unaffected. As two distinct regions within the 23S transcripts were highly enriched, Hfq seems to interact specifically rather than spuriously with the transcripts. An association with stable house-keeping RNAs is not uncommon, E. coli Hfq interacts with 16S rRNA and r-protein S12 (62, 63), and is involved in the biogenesis of 30S ribosomes (64).
Binding of Hfq to client RNAs can be mediated via different domains. Six Hfq protomers assemble into a ring-like structure in which four different sites—the distal and proximal surfaces of the ring, the rim as well as the unstructured C-terminal domain—are available for interactions with RNA (65). Each binding site shows distinct preferences for RNA substrates and their differential usage enables Hfq to perform its regulatory functions. The distal surface typically interacts with purine-rich (ARN)n motifs found in mRNA transcripts whereas sRNAs preferentially bind to the proximal face and the rim (66, 67). The flexible C-terminus is also able to contact the rim and has been implicated to restrict non-specific binding to Hfq (14). Initial recognition of sRNAs at the proximal face involves the 3′ uridine-rich stretch downstream of the Rho-independent terminator, a common feature of the 3′ ends of sRNAs (68, 69). In addition, Hfq frequently resides at AU-rich sequences just upstream the terminal hairpin (55). For RusT, our chemical probing experiments have confirmed an Hfq-specific foot-print at this position preceding the terminator (Fig. 2B and C), and deletion of the respective sequence patch in RusT-SL3mut greatly impeded Hfq-binding compared to the wild-type RNA (Fig. 1D and H). We likewise predicted the secondary structures of the 16 additional Hfq-binding sRNAs using RNAfold (70), and found a terminal hairpin structure for 12 transcripts, 10 of which were followed by a U-run (Table S3). When analyzed for the nucleotide composition of their sequences including 15 nt upstream of the sRNA terminators, we in almost all cases detected a lower GC content compared to the full molecule. Collectively, these results point toward a recognition mode by Hfq for at least a subset of sRNAs in Caulobacter that parallels the model established for enterobacterial species (55) However, alternative binding modes are possible, for example, when occupying sites at the distal face and the rim of Hfq.
RusT is a trans-acting sRNA induced in response to iron starvation
Bacteria depend on iron as a critical and oftentimes scarce nutrient which is required as a cofactor in fundamental biological processes (28, 71). While iron-limited environments hence pose a serious constraint to survival, an excess of the metal can be toxic, and tight control of iron uptake, consumption and efflux to maintain metal homeostasis is necessary. In addition to transcription factors such as Fur, sRNAs have been repeatedly associated with the coordination of the cellular response to iron limitation (72).
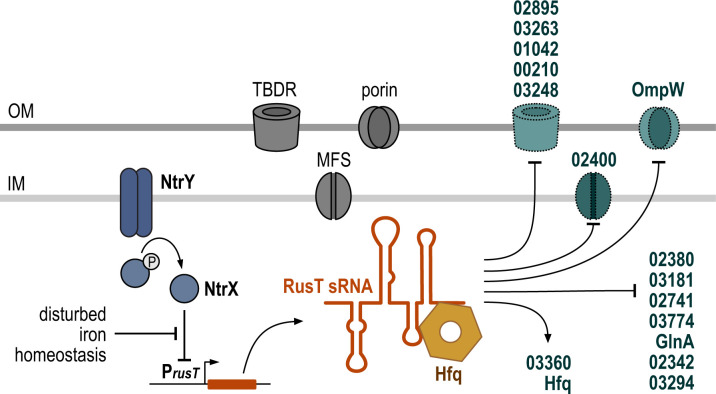
The enterobacterial sRNA RyhB is only transcribed under iron-limited conditions and in conjunction with Hfq reduces the demand by redirecting iron to essential pathways and optimizing iron uptake. While RyhB and its functional analogues are Fur-dependent (73), it is intriguing that no sRNA has been identified in the Fur regulon in Caulobacter. E. coli RyhB functions as the non-coding switch to change the sign of regulation by Fur which acts as a repressor under iron-replete conditions (1). Different from E. coli, Fur has been demonstrated to also function as a direct transcriptional activator for a subset of genes in several organisms, including N. meningitidis (74) and C. crescentus (32). Its ability to act as a dual transcriptional regulator itself might alleviate the need for an sRNA within the direct Fur regulon. Other Fur-independent sRNAs, including FnrS and CyaR in the enterobacteria (72, 75), affect iron homeostasis and we identified the conserved Hfq-associated sRNA RusT (CCNA_R0199) to be synthesized under iron-limiting conditions and to function as an Hfq-dependent post-transcriptional regulator of 16 transcripts in Caulobacter (Fig. 6 and 9). When compared to the wild-type, a rusT deletion strain was not impaired for growth in the presence of DIP or ZnSO4 (Fig. S10), suggesting that the sRNA acts as a modulator rather than a central regulator of the stress response.
The largest functional class of RusT targets encode membrane proteins, such as the porin OmpW, the major facilitator family protein CCNA_02400 as well as the TBDRs CCNA_02895, CCNA_03263, CCNA_01042, CCNA_00210, and CCNA_03248 (Fig. 5C; Table 2). Caulobacter is equipped with an extraordinarily large set of outer membrane proteins, including more than 60 TBDRs which mediate gated, high-affinity uptake of vitamins, carbohydrates, and ferric siderophores (27, 76). The cell needs to constantly tune its envelope profile to optimize nutrient uptake in a given situation while at the same time avoid over-crowding and hence impairment of membrane integrity or fluidity. At the post-transcriptional level, sRNAs play a crucial role in regulation of outer membrane protein synthesis (1), and RusT joins the Caulobacter sRNAs CrfA and ChvR which both have been shown to affect TBDR expression in response to starvation and upon activation of the ChvGI TCS, respectively (17, 18). Except for CCNA_02400 and CCNA_03248, expression of all the aforementioned transporter genes in the RusT regulon has also been identified to be downregulated in response to iron depletion in a previous transcriptome analysis (50), and RusT potentially contributes to this pattern through its regulatory activity at the post-transcriptional level. Similarly, also CCNA_03181 (encoding an alcohol dehydrogenase) and CCNA_03774 (encoding a citrate lyase beta chain/citryl-CoA lyase subunit) are repressed by RusT and in iron-scarce conditions [Fig. 6A and reference (50)].
Transcriptional regulation of RusT
In this study, we demonstrated that RusT is induced in Caulobacter cells treated with the chelator DIP or by high amounts of zinc, two stressors that interfere with the cellular iron homeostasis (Fig. 3B). Transposon insertions in ntrX, encoding the response regulator of the conserved NtrYX TCS, abrogated stress regulation of RusT and resulted in constitutively high expression of the sRNA (Fig. 3C).
In Caulobacter, the ntrYX operon likely originated from gene duplication of the neighboring ntrBC locus; however, different from the latter one it has functionally not been linked to nitrogen metabolism (77). Unconventionally, NtrY is proposed to primarily act as a phosphatase on NtrX, and this activity is inhibited by a Caulobacter-specific periplasmic protein, NtrZ (47). Both functionally and genetically, NtrYX is linked to the ChvGI TCS. The two systems control partially overlapping networks to tune cell envelope integrity, and synergize to mediate growth under distinct environmental conditions (47, 78) involving dedicated sRNAs: ChvR is regulated by ChvI, while RusT is controlled by NtrX (Fig. 3C and D).
Homologs of the NtrYX TCS have been found in many alpha- and betaproteobacteria. While generally annotated to function in nitrogen assimilation, recent studies have reported the involvement of NtrYX in various additional cellular processes including the response to low oxygen tension in Brucella abortus, cell shape, motility, and biofilm formation in Sinorhizobium meliloti as well as cell envelope homeostasis in Rhodobacter sphaeroides (41–45). The NtrYX homolog of P. denitrificans responds to iron starvation, and inactivation of the TCS in this species results in the deregulation of factors involved in metal homeostasis and a subset of TBDRs (46). Our study likewise points toward a functional role of NtrYX in response to perturbations of the iron homeostasis, and a role for the sRNA RusT to aid membrane reorganization under this condition (Fig. 9).
Fig 9.
RusT expression and regulatory activity. Expression of RusT is under control of the NtrYX system and induced in response to disturbance of the iron homeostasis. Together with the RNA chaperone Hfq, RusT interacts with target mRNAs, including a number of transcripts encoding for different types of membrane-standing transporters.
RusT is also one of the Caulobacter sRNA candidates that shows differential regulation over the course of the cell cycle, with a peak of expression in swarmer and predivisional cells (21, 79). Involvement of several cell cycle regulators in the modulation of rusT promoter activity has either been predicted in bioinformatic analyses [two “half site motifs” for CtrA, two DnaA sites, one CcrM recognition motif (79)] or has been confirmed experimentally [ChIP-Seq peak for GcrA (21, 80); differential methylation in mucR1/2 mutant cells (81)].
Iron deprivation has been identified to affect spatiotemporal localization of the division machinery and to interfere with peptidoglycan incorporation in dividing cells, thus blocking the late stages of cytokinesis (82). Hence, dissecting the transcriptional control of rusT might reveal a potential, RNA-mediated link between cell cycle control and iron homeostasis in Caulobacter.
MATERIALS AND METHODS
DNA oligonucleotides and plasmids
Sequences of all oligonucleotides employed in this study are listed in Table S4. All plasmids used in this study are summarized in Table S5, and details on plasmid construction are specified in the Supplemental Data file.
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table S6.
C. crescentus strain NA1000 (KFS-006) is referred to as the wild-type strain and was used for mutant generation. All details on strain construction and growth conditions are provided in the Supplemental Data file.
Hfq co-IP and transcriptome analysis using RNA-seq
Details on the Hfq co-IP and the transcriptome analysis upon RusT overexpression using RNA-seq are provided in the Supplemental Data file.
RNA isolation and Northern blot analysis
Total RNA from bacterial samples was prepared using the Hot Phenol method as described previously (17). For Northern blot analysis, 5–10 µg of total RNA were separated on 6–8% polyacrylamide (7 M urea) gels and electroblotted. Membranes were hybridized with gene-specific, 5′ end-labeled DNA-oligonucleotides at 42°C in Roti-Hybri-Quick hybridization solution (Roth), and washed in three subsequent steps with SSC wash buffers (5×/1×/0.5× SSC) supplemented with 0.1% SDS.
In vitro RNA biochemistry
Details on the preparation of in vitro synthesized RNA and associated experiments are provided in the Supplemental Data file.
Transposon screen
For genome-wide transposon mutagenesis of C. crescentus carrying a chromosomal PrusT::nptII fusion (integration of pKF776-1), the mini-himar1 transposon conferring resistance to gentamycin (pMAR2xT7; (83)) was introduced via conjugation from the DAP-auxotroph E. coli WM3064 donor (84). Mutants with increased tolerance for kanamycin were selected on PYE plates supplemented with gentamycin (1 µg/mL) and kanamycin (20 µg/mL). Insertion sites were mapped by semi-random PCR as described previously (83) using transposon-specific primers KFO-2067 and KFO-2068 (nested), arbitrary primers KFO-0899 or KFO-0901 and KFO-0898 (nested) and sequencing primer KFO-2071.
Chromatin immunoprecipitation coupled to deep sequencing (ChIP-Seq)
All details on ChIP-Seq with the C. crescentus CB15 ntrX::ntrX-HA strain are provided in the Supplemental Data file.
Protein sample analysis
Whole-cell protein samples were prepared as described previously (17). Fusion proteins to GFP or the 3xFLAG epitope tag were detected using a monoclonal anti-FLAG antibody (1:1,000; mouse; Sigma #F1804) or an anti-GFP antibody (1:1,000; mouse, Roche #11814460001), respectively. RNA polymerase (1:10,000; rabbit; BioLegend #663205) or GroEL (1:20,000; rabbit; Merck; #G6532) served as a loading controls.
Fluorescence intensity measurements
GFP expression of translational reporter fusions was determined from cells cultivated overnight in PYE supplemented with the appropriate antibiotics and supplements. Upon collection of samples by centrifugation (3 min; 9,000 rpm; 4°C), the cells were washed once in 1× phosphate buffer, and resuspended in phosphate buffer. Background fluorescence was determined by a control sample not expressing GFP. Fluorescence intensity in the presence of the sRNA control plasmid was set to 1, and relative expression was calculated from three biological replicates (error bars represent standard deviation). Statistical significance was determined using unpaired t tests with Welch correction. The P value is indicated as follows—ns for P > 0.05, * for P ≤ 0.05, ** for P ≤ 0.01, *** for P ≤ 0.001, and **** for P ≤ 0.0001.
ACKNOWLEDGMENTS
The authors are grateful to Sean Crosson and Aretha Fiebig (Michigan State University) for sharing strain FC3561 and to Andreas Brachmann (LMU Munich, Faculty of Biology, Genomics Service Unit) and Helmut Blum (LMU Munich, Gene Center, Functional Genome Analysis) for help with RNA sequencing.
Work in the Fröhlich and Papenfort laboratories is supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany´s Excellence Strategy—EXC 2051—Project number 390713860. This project was supported by DFG grant FR 3502/2-1 (to K.S.F.), the Vallee Foundation and European Research Council grant ArtRNA/CoG-101088027 (to K.P.), and Swiss National Science Foundation (SNSF) project grant 310030_219565 (to P.H.V).
Contributor Information
Kathrin S. Fröhlich, Email: kathrin.froehlich@uni-jena.de.
Paul Babitzke, The Pennsylvania State University, University Park, Pennsylvania, USA.
DATA AVAILABILITY
Sequencing data have been deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) repository (85) and are available via the GEO accession numbers GSE148206 (Hfq co-IP), GSE148208 (RusT pulse overexpression), and GSE247928 (ChIP-seq).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.03153-23.
Hfq-associated transcripts.
NtrX-HA ChIP-seq analysis.
Figures S1 to S10, Tables S3 to S6, and supporting methodology.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Fröhlich KS, Gottesman S. 2018. Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol Spectr 6. doi: 10.1128/microbiolspec.RWR-0022-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 17:6069–6075. doi: 10.1093/emboj/17.20.6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prévost K, Desnoyers G, Jacques J-F, Lavoie F, Massé E. 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25:385–396. doi: 10.1101/gad.2001711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16:840–846. doi: 10.1038/nsmb.1631 [DOI] [PubMed] [Google Scholar]
- 6. Fröhlich KS, Papenfort K, Fekete A, Vogel J. 2013. A small RNA activates CFA synthase by Isoform-specific mRNA stabilization. EMBO J 32:2963–2979. doi: 10.1038/emboj.2013.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 95:12462–12467. doi: 10.1073/pnas.95.21.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153:426–437. doi: 10.1016/j.cell.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. 2016. sRNA-mediated control of transcription termination in E. coli. Cell 167:111–121. doi: 10.1016/j.cell.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmqvist E, Vogel J. 2018. RNA-binding proteins in bacteria. Nat Rev Microbiol 16:601–615. doi: 10.1038/s41579-018-0049-5 [DOI] [PubMed] [Google Scholar]
- 11. Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. 2014. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife 3:e05375. doi: 10.7554/eLife.05375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11:1206–1214. doi: 10.1038/nsmb858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santiago-Frangos A., Kavita K, Schu DJ, Gottesman S, Woodson SA. 2016. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc Natl Acad Sci U S A 113:E6089–E6096. doi: 10.1073/pnas.1613053113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santiago-Frangos A, Fröhlich KS, Jeliazkov JR, Małecka EM, Marino G, Gray JJ, Luisi BF, Woodson SA, Hardwick SW. 2019. Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc Natl Acad Sci U S A 116:10978–10987. doi: 10.1073/pnas.1814428116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Assis NG, Ribeiro RA, da Silva LG, Vicente AM, Hug I, Marques MV. 2019. Identification of Hfq-binding RNAs in Caulobacter crescentus. RNA Biol 16:719–726. doi: 10.1080/15476286.2019.1593091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beroual W, Prévost K, Lalaouna D, Ben Zaina N, Valette O, Denis Y, Djendli M, Brasseur G, Brilli M, Robledo Garrido M, Jimenez-Zurdo J-I, Massé E, Biondi EG. 2022. The noncoding RNA CcnA modulates the master cell cycle regulators CtrA and GcrA in Caulobacter crescentus. PLoS Biol 20:e3001528. doi: 10.1371/journal.pbio.3001528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fröhlich KS, Förstner KU, Gitai Z. 2018. Post-transcriptional gene regulation by an Hfq-independent small RNA in Caulobacter crescentus. Nucleic Acids Res 46:10969–10982. doi: 10.1093/nar/gky765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Landt SG, Lesley JA, Britos L, Shapiro L. 2010. CrfA, a small noncoding RNA regulator of adaptation to carbon starvation in Caulobacter crescentus. J Bacteriol 192:4763–4775. doi: 10.1128/JB.00343-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schrader JM, Li GW, Childers WS, Perez AM, Weissman JS, Shapiro L, McAdams HH. 2016. Dynamic translation regulation in Caulobacter cell cycle control. Proc Natl Acad Sci U S A 113:E6859–E6867. doi: 10.1073/pnas.1614795113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tien M, Fiebig A, Crosson S. 2018. Gene network analysis identifies a central post-transcriptional regulator of cellular stress survival. Elife 7:e33684. doi: 10.7554/eLife.33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Govers SK, Jacobs-Wagner C. 2020. Caulobacter crescentus: model system extraordinaire. Curr Biol 30:R1151–R1158. doi: 10.1016/j.cub.2020.07.033 [DOI] [PubMed] [Google Scholar]
- 23. Balhesteros H, Shipelskiy Y, Long NJ, Majumdar A, Katz BB, Santos NM, Leaden L, Newton SM, Marques MV, Klebba PE. 2017. TonB-dependent heme/hemoglobin utilization by Caulobacter crescentus HutA. J Bacteriol 199:e00723-16. doi: 10.1128/JB.00723-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eisenbeis S, Lohmiller S, Valdebenito M, Leicht S, Braun V. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J Bacteriol 190:5230–5238. doi: 10.1128/JB.00194-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menikpurage IP, Barraza D, Meléndez AB, Strebe S, Mera PE. 2019. The B12 receptor BtuB alters the membrane integrity of Caulobacter crescentus. Microbiology (Reading) 165:311–323. doi: 10.1099/mic.0.000753 [DOI] [PubMed] [Google Scholar]
- 26. Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol 187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. doi: 10.1073/pnas.061029298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 29. Keyer K, Imlay JA. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A 93:13635–13640. doi: 10.1073/pnas.93.24.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee JW, Helmann JD. 2007. Functional specialization within the fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7 [DOI] [PubMed] [Google Scholar]
- 31. Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. da Silva Neto JF, Braz VS, Italiani VCS, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res 37:4812–4825. doi: 10.1093/nar/gkp509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heidrich N, Bauriedl S, Barquist L, Li L, Schoen C, Vogel J. 2017. The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res 45:6147–6167. doi: 10.1093/nar/gkx168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schu DJ, Zhang A, Gottesman S, Storz G. 2015. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34:2557–2573. doi: 10.15252/embj.201591569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imlay JA. 2014. The mismetallation of enzymes during oxidative stress. J Biol Chem 289:28121–28128. doi: 10.1074/jbc.R114.588814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foster AW, Young TR, Chivers PT, Robinson NJ. 2022. Protein metalation in biology. Curr Opin Chem Biol 66:102095. doi: 10.1016/j.cbpa.2021.102095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z, Wang P, Wang H, Yu ZH, Au-Yeung HY, Hirayama T, Sun H, Yan A. 2019. Zinc excess increases cellular demand for iron and decreases tolerance to copper in Escherichia coli J Biol Chem 294:16978–16991. doi: 10.1074/jbc.RA119.010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 101:9792–9797. doi: 10.1073/pnas.0403423101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. 2008. The Bacillus subtilis iron-sparing response is mediated by a fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105:11927–11932. doi: 10.1073/pnas.0711752105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis . J Bacteriol 189:3686–3694. doi: 10.1128/JB.01890-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Calatrava-Morales N, Nogales J, Ameztoy K, van Steenbergen B, Soto MJ. 2017. The NtrY/NtrX system of Sinorhizobium meliloti GR4 regulates motility, EPS I production, and nitrogen metabolism but is dispensable for symbiotic nitrogen fixation. Mol Plant Microbe Interact 30:566–577. doi: 10.1094/MPMI-01-17-0021-R [DOI] [PubMed] [Google Scholar]
- 42. Wang D, Xue H, Wang Y, Yin R, Xie F, Luo L. 2013. The Sinorhizobium meliloti ntrX gene is involved in succinoglycan production, motility, and symbiotic nodulation on alfalfa. Appl Environ Microbiol 79:7150–7159. doi: 10.1128/AEM.02225-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carrica M del C, Fernandez I, Martí MA, Paris G, Goldbaum FA. 2012. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol Microbiol 85:39–50. doi: 10.1111/j.1365-2958.2012.08095.x [DOI] [PubMed] [Google Scholar]
- 44. Carrica MDC, Fernandez I, Sieira R, Paris G, Goldbaum FA. 2013. The two-component systems PrrBA and NtrYX co-ordinately regulate the adaptation of Brucella abortus to an oxygen-limited environment. Mol Microbiol 88:222–233. doi: 10.1111/mmi.12181 [DOI] [PubMed] [Google Scholar]
- 45. Lemmer KC, Alberge F, Myers KS, Dohnalkova AC, Schaub RE, Lenz JD, Imam S, Dillard JP, Noguera DR, Donohue TJ. 2020. The NtrYX two-component system regulates the bacterial cell envelope. mBio 11:11. doi: 10.1128/mBio.00957-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olaya-Abril A, Luque-Almagro VM, Hidalgo-Carrillo J, Chicano-Gálvez E, Urbano FJ, Moreno-Vivián C, Richardson DJ, Roldán MD. 2022. The NtrYX two-component system of Paracoccus denitrificans is required for the maintenance of cellular iron homeostasis and for a complete denitrification under iron-limited conditions. Int J Mol Sci 23:9172. doi: 10.3390/ijms23169172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stein BJ, Fiebig A, Crosson S. 2021. The ChvG-ChvI and NtrY-NtrX two-component systems coordinately regulate growth of Caulobacter crescentus. J Bacteriol 203:e0019921. doi: 10.1128/JB.00199-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fröhlich KS, Velasco Gomariz M. 2021. RNA-controlled regulation in Caulobacter crescentus. Curr Opin Microbiol 60:1–7. doi: 10.1016/j.mib.2021.01.002 [DOI] [PubMed] [Google Scholar]
- 49. Irnov I, Wang Z, Jannetty ND, Bustamante JA, Rhee KY, Jacobs-Wagner C. 2017. Crosstalk between the tricarboxylic acid cycle and peptidoglycan synthesis in Caulobacter crescentus through the homeostatic control of alpha-ketoglutarate. PLoS Genet 13:e1006978. doi: 10.1371/journal.pgen.1006978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. da Silva Neto JF, Lourenço RF, Marques MV. 2013. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. doi: 10.1186/1471-2164-14-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. 2008. Small RNA binding to 5' mRNA coding region inhibits translational initiation. Mol Cell 32:827–837. doi: 10.1016/j.molcel.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 52. Schrader JM, Zhou B, Li G-W, Lasker K, Childers WS, Williams B, Long T, Crosson S, McAdams HH, Weissman JS, Shapiro L. 2014. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet 10:e1004463. doi: 10.1371/journal.pgen.1004463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guillier M, Gottesman S, Storz G. 2006. Modulating the outer membrane with small RNAs. Genes Dev 20:2338–2348. doi: 10.1101/gad.1457506 [DOI] [PubMed] [Google Scholar]
- 54. Vogel J, Papenfort K. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9:605–611. doi: 10.1016/j.mib.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 55. Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. doi: 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mann M, Wright PR, Backofen R. 2017. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res 45:W435–W439. doi: 10.1093/nar/gkx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517. doi: 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Franze de Fernandez MT, Eoyang L, August JT. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588–590. doi: 10.1038/219588a0 [DOI] [PubMed] [Google Scholar]
- 59. Hör J, Matera G, Vogel J, Gottesman S, Storz G. 2020. Trans-acting small RNAs and their effects on gene expression in Escherichia coli and Salmonella enterica. EcoSal Plus 9. doi: 10.1128/ecosalplus.esp-0030-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33. doi: 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 61. Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. 2006. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol 71:1–11. doi: 10.1101/sqb.2006.71.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de Haseth PL, Uhlenbeck OC. 1980. Interaction of Escherichia coli host factor protein with Q beta ribonucleic acid. Biochemistry 19:6146–6151. doi: 10.1021/bi00567a030 [DOI] [PubMed] [Google Scholar]
- 63. Strader MB, Hervey WJ 4th, Costantino N, Fujigaki S, Chen CY, Akal-Strader A, Ihunnah CA, Makusky AJ, Court DL, Markey SP, Kowalak JA. 2013. A coordinated proteomic approach for identifying proteins that interact with the E. coli ribosomal protein S12. J Proteome Res 12:1289–1299. doi: 10.1021/pr3009435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrade JM, Dos Santos RF, Chelysheva I, Ignatova Z, Arraiano CM. 2018. The RNA-binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J 37:e97631. doi: 10.15252/embj.201797631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weichenrieder O. 2014. RNA binding by Hfq and ring-forming (L)Sm proteins: a trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol 11:537–549. doi: 10.4161/rna.29144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Link TM, Valentin-Hansen P, Brennan RG. 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 106:19292–19297. doi: 10.1073/pnas.0908744106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. 2014. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res 42:2736–2749. doi: 10.1093/nar/gkt1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Otaka H, Ishikawa H, Morita T, Aiba H. 2011. Polyu tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci U S A 108:13059–13064. doi: 10.1073/pnas.1107050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sauer E, Weichenrieder O. 2011. Structural basis for RNA 3'-end recognition by Hfq. Proc Natl Acad Sci U S A 108:13065–13070. doi: 10.1073/pnas.1103420108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gruber AR, Bernhart SH, Lorenz R. 2015. The ViennaRNA web services. Methods Mol Biol 1269:307–326. doi: 10.1007/978-1-4939-2291-8_19 [DOI] [PubMed] [Google Scholar]
- 71. Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. 2008. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13:1205–1218. doi: 10.1007/s00775-008-0404-5 [DOI] [PubMed] [Google Scholar]
- 72. Charbonnier M, González-Espinoza G, Kehl-Fie TE, Lalaouna D. 2022. Battle for metals: regulatory RNAs at the front line. Front Cell Infect Microbiol 12:952948. doi: 10.3389/fcimb.2022.952948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chareyre S, Mandin P. 2018. Bacterial iron homeostasis regulation by sRNAs. Microbiol Spectr 6. doi: 10.1128/microbiolspec.RWR-0010-2017 [DOI] [PubMed] [Google Scholar]
- 74. Delany I, Rappuoli R, Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol 52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x [DOI] [PubMed] [Google Scholar]
- 75. Sy BM, Tree JJ. 2022. The small RNA CyaR activates translation of the outer membrane haem receptor chuA in enterohemorrhagic Escherichia coli. Front Microbiol 13:821196. doi: 10.3389/fmicb.2022.821196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Braun V, Endriss F. 2007. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20:219–231. doi: 10.1007/s10534-006-9072-5 [DOI] [PubMed] [Google Scholar]
- 77. Capra EJ, Perchuk BS, Skerker JM, Laub MT. 2012. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150:222–232. doi: 10.1016/j.cell.2012.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Quintero-Yanes A, Mayard A, Hallez R. 2022. The two-component system ChvGI maintains cell envelope homeostasis in Caulobacter crescentus. PLoS Genet 18:e1010465. doi: 10.1371/journal.pgen.1010465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Beroual W, Brilli M, Biondi EG. 2018. Non-coding RNAs potentially controlling cell cycle in the model Caulobacter crescentus: a bioinformatic approach. Front Genet 9:164. doi: 10.3389/fgene.2018.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Murray SM, Panis G, Fumeaux C, Viollier PH, Howard M. 2013. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol 11:e1001749. doi: 10.1371/journal.pbio.1001749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ardissone S, Redder P, Russo G, Frandi A, Fumeaux C, Patrignani A, Schlapbach R, Falquet L, Viollier PH. 2016. Cell cycle constraints and environmental control of local DNA hypomethylation in alpha-proteobacteria. PLoS Genet 12:e1006499. doi: 10.1371/journal.pgen.1006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Santos TMA, Lammers MG, Zhou M, Sparks IL, Rajendran M, Fang D, De Jesus CLY, Carneiro GFR, Cui Q, Weibel DB. 2018. Small molecule chelators reveal that iron starvation inhibits late stages of bacterial cytokinesis. ACS Chem Biol 13:235–246. doi: 10.1021/acschembio.7b00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. doi: 10.1073/pnas.1834303100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Edgar R, Domrachev M, Lash AE. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hfq-associated transcripts.
NtrX-HA ChIP-seq analysis.
Figures S1 to S10, Tables S3 to S6, and supporting methodology.
Data Availability Statement
Sequencing data have been deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) repository (85) and are available via the GEO accession numbers GSE148206 (Hfq co-IP), GSE148208 (RusT pulse overexpression), and GSE247928 (ChIP-seq).