ABSTRACT
A member of the Retroviridae, human immunodeficiency virus type 1 (HIV-1), uses the RNA genome packaged into nascent virions to transfer genetic information to its progeny. The genome packaging step is a highly regulated and extremely efficient process as a vast majority of virus particles contain two copies of full-length unspliced HIV-1 RNA that form a dimer. Thus, during virus assembly HIV-1 can identify and selectively encapsidate HIV-1 unspliced RNA from an abundant pool of cellular RNAs and various spliced HIV-1 RNAs. Several “G” features facilitate the packaging of a dimeric RNA genome. The viral polyprotein Gag orchestrates virus assembly and mediates RNA genome packaging. During this process, Gag preferentially binds unpaired guanosines within the highly structured 5′ untranslated region (UTR) of HIV-1 RNA. In addition, the HIV-1 unspliced RNA provides a scaffold that promotes Gag:Gag interactions and virus assembly, thereby ensuring its packaging. Intriguingly, recent studies have shown that the use of different guanosines at the junction of U3 and R as transcription start sites results in HIV-1 unspliced RNA species with 99.9% identical sequences but dramatically distinct 5′ UTR conformations. Consequently, one species of unspliced RNA is preferentially packaged over other nearly identical RNAs. These studies reveal how conformations affect the functions of HIV-1 RNA elements and the complex regulation of HIV-1 replication. In this review, we summarize cis- and trans-acting elements critical for HIV-1 RNA packaging, locations of Gag:RNA interactions that mediate genome encapsidation, and the effects of transcription start sites on the structure and packaging of HIV-1 RNA.
KEYWORDS: HIV, RNA packaging, retrovirus, RNA dimerization, virus assembly, Gag, transcription start site
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1), a pandemic pathogen, causes acquired immunodeficiency syndrome (AIDS) (1). To date, HIV-1 has caused the loss of >40 million lives and currently, there are ~39 million people in the world living with HIV-1. As a retrovirus, the genetic material in HIV-1 viral particles is in the form of RNA. After entering the host cells, HIV-1 uses the RNA genome as a template to synthesize viral DNA, which is then integrated into the host chromosomes to form a provirus (2). The HIV-1 provirus relies on host machinery for gene expression including transcription, RNA processing, and translation. Host RNA polymerase II (Pol II) transcribes the provirus using a single promoter in the 5′ long terminal repeat; the resulting RNA can be unspliced or undergo complex alternative splicing to generate partially and completely spliced HIV1-RNA species (3). All three types of RNA—unspliced, partially spliced, or completely spliced—are exported from the nucleus and translated to produce viral proteins. In addition, unspliced HIV-1 RNA is selectively packaged into assembling virions to serve as the viral genome (4–6).
HIV-1 unspliced RNA is translated to generate the Gag polyprotein and, by a −1 frameshift near the end of the gag gene, the Gag-Pol polyproteins. The Gag polyprotein orchestrates the assembly of infectious virus particles including packaging the unspliced RNA as its genome (4, 7). Although each unspliced RNA contains all the genetic information, all retroviruses, including HIV-1, package two copies of RNA into the virus particle (8). This unique genome packaging feature allows recombination to occur during reverse transcription to generate a hybrid DNA containing portions of genetic information from each copackaged RNA (9). Frequent recombination is required for maintaining genome integrity (10) and assorts mutations in the viral population allowing better-adapted variants to emerge, such as those that escape host immune responses or antiviral treatments (11, 12). Therefore, HIV-1 genome packaging has a strong impact on virus replication and evolution. By imaging viral RNA in individual particles, it was shown that the packaging of the HIV-1 RNA genome is highly efficient and regulated. The vast majority of viral particles (>90%) contain two copies of HIV-1 unspliced RNA (13). This review focuses on how HIV-1 efficiently selects and packages its unspliced RNA. For simplicity, HIV-1 unspliced RNA is referred to as HIV-1 RNA hereafter.
It has long been recognized that Gag needs to select HIV-1 RNA from an abundant pool of cellular RNAs and spliced viral RNAs as the virion genome. However, recent studies revealed that genome packaging is more selective than originally appreciated because not all HIV-1 RNA molecules are created equal. Using neighboring sequences to initiate transcription, HIV-1 can generate multiple species of unspliced RNAs that are 99.9% identical, differing by only a few nucleotides at the 5′ end. However, HIV-1 selectively packages one RNA species that has a single guanosine at the 5′ end (1G RNA) over another species that is 2-nt longer with three guanosines at the 5′ end (3G RNA) (14–17). The mechanism used by HIV-1 to distinguish two 9 kb RNAs with a 2-nt difference and the possible selective advantage of such a strategy will also be discussed in this review.
Viral elements important for HIV-1 RNA genome packaging
Trans-acting element: Gag
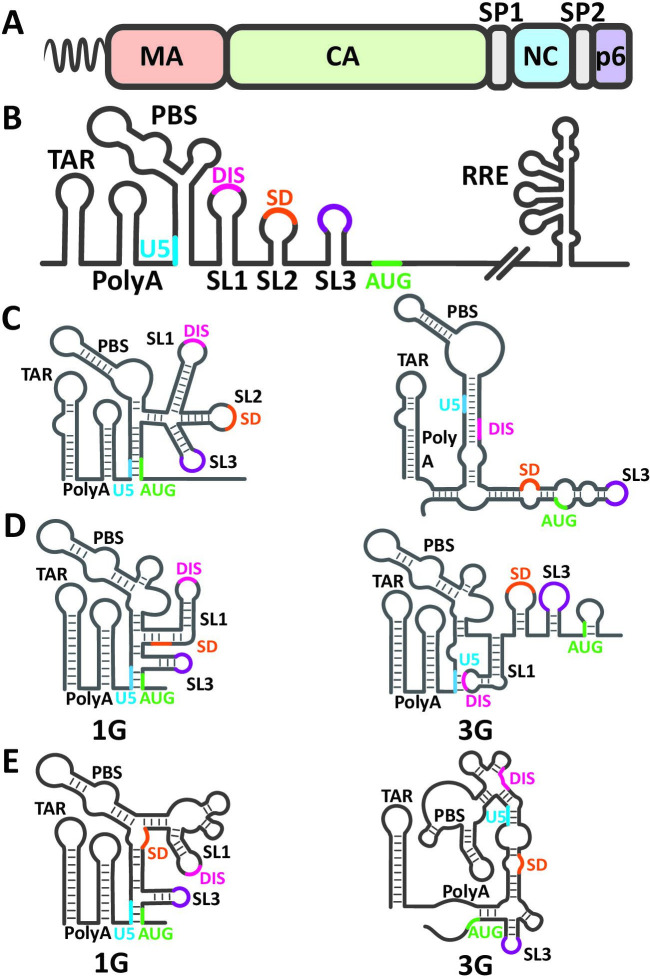
Efficient HIV-1 RNA genome packaging is mediated by trans- and cis-acting elements within Gag and the viral RNA, respectively (2). The Gag polyprotein is composed of several functional domains: matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), and p6 (Fig. 1A). These domains act in concert to assemble virus particles at the plasma membrane and to ensure efficient genome packaging (4). MA is a globular protein that contains a highly basic patch able to interact with negatively charged lipids present within the plasma membrane, most notably phosphatidylinositol 4,5-bisphosphate (PIP2) (18–22). Furthermore, the first glycine residue is modified with the addition of a myristic acid moiety that ensures Gag anchoring to the plasma membrane (23). The CA domain consists of N- and C-terminal domains that contain residues essential for Gag multimerization during assembly and, following Gag polyprotein cleavage, during virion maturation for capsid core formation (24). In tandem with CA-driven Gag oligomerization, CA-SP1 forms a stable 6-helix bundle with inositol hexakisphosphate (IP6) to enhance Gag lattice formation during assembly (25, 26). NC is an RNA-binding domain and acts as a nucleic acid chaperone (27, 28). This domain contains multiple basic residues important for RNA binding and two critical zinc fingers, or zinc knuckles, each with a conserved cysteine-histidine motif, CCHC, that coordinates a Zn2+ molecule (29, 30). The C-terminal domain, p6, contains critical PTAP and YPXL-type binding motifs necessary for interaction with host machinery important for assembled viruses to bud off from the host cells (31–33).
Fig 1.
Cis- and Trans-acting elements important for selective HIV-1 RNA packaging. (A) Schematic of HIV-1 Gag polyprotein containing matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), and p6 domains. Gag is also N-terminally myristoylated as denoted at the N-terminus of MA (squiggle). (B) Structural elements within the 5′ UTR of HIV-1 RNA (left) and necessary for nuclear export (right). Conserved stem loops within the 5′ UTR are highlighted from left to right as the trans-activation response (TAR) element, polyA, primer-binding site (PBS), SL1, SL2, and SL3. Additional features highlighted are a portion of U5 sequences (blue), dimerization initiation signal (DIS; pink), splice donor (SD; orange), exposed guanosines in SL3 (purple), and start codon of Gag (AUG; green). The highly structured rev-response element (RRE) located within the env gene is depicted on the right. Structures of the 5’UTR conformational switch models proposed by (C) Berkhout group, (D) Summers/Telesnitsky groups, and the zip and pack model proposed by (E) Musier-Forsyth/Hu groups. The 5′ UTR structures predicted to favor and inhibit packaging are shown on the left and right, respectively. Highlighted features correspond to labeling in (B).
Among the Gag domains, NC has RNA binding activity and was first examined for its role in specifically packaging viral RNAs. NC serves multiple functions during HIV-1 replication. After polyprotein cleavage and virus maturation, the NC protein with its nucleic acid chaperone activity plays important roles during reverse transcription and integration (27, 34–38). Thus, NC mutations often cause pleiotropic effects in viral replication; here, we focus on the role of NC in genome packaging. By replacing the NC domain with that of another retrovirus, it was shown that such hybrid Gag proteins can gain the ability to package RNA from a different retrovirus (39). For example, replacing the HIV-1 NC with that of murine leukemia virus (MLV), or vice versa, allowed the hybrid Gag to package MLV RNA or HIV-1 RNA, respectively (40, 41). However, replacing the NC domain is insufficient to alter RNA packaging specificity in some other cases (42); for example, a hybrid HIV-1 Gag with mouse mammary tumor virus (MMTV) NC does not preferentially package MMTV RNA (43).
Extensive mutational studies have shown that multiple regions of the HIV-1 NC, including the zinc fingers and the basic residues, are important for selective packaging of HIV-1 RNA (29, 34–36, 44–52). Abolishing the conserved CCHC motifs causes severe genome packaging defects and even replacing these motifs with other zinc-chelating motifs such as CCCC or CCHH can cause packaging defects (30, 49, 52, 53). Interestingly, the two zinc fingers of HIV-1 NC are not functionally equivalent (30); the first or N-terminal zinc finger is more important for genome packaging although both motifs are required for optimal genome packaging (30, 54). Removal or mutating zinc fingers has also been reported to cause mislocalization of Gag within cells (55). In addition to the zinc-chelating CCHC, other residues within the zinc fingers, such as the conserved aromatic residues, also play an important role in HIV-1 RNA packaging specificity (30, 48, 52). Although the two zinc finger motifs are critical for RNA genome packaging, deleting both zinc fingers does not completely abolish RNA packaging, suggesting that other regions of Gag contribute to RNA binding (41). The HIV-1 NC domain contains multiple basic residues flanking and within the two zinc finger motifs. These basic residues are also important in RNA packaging. Abolishing multiple basic residues can cause very severe RNA packaging defects, whereas mutating a single basic residue often causes mild defects (36, 45, 56, 57).
In addition to NC, the MA and CA domains in Gag have also been shown to play critical roles in HIV-1 RNA genome packaging. Mutations in the MA or CA domain often cause assembly defects, making it difficult to study their contributions to RNA packaging (58–63). To probe this question, a complementation system was established to separate the assembly and the RNA-binding functions of Gag using one type of Gag to assemble particles and a second type of Gag to bind and package the RNA genome (64). It was shown that mutations in the RNA-binding Gag that abolish membrane targeting or Gag-Gag multimerization also cause RNA packaging defects (64). The requirement for Gag multimerization has been confirmed by another study analyzing interactions between CA-NC and HIV-1 RNA and showed that mutations that affect Gag multimerization also impact its ability to bind HIV-1 RNA specifically (65). Interestingly, MA can bind RNA (66); however, the MA domain mainly binds host RNA including tRNA in the cytoplasm (67). The RNA binding activity of MA regulates Gag trafficking by preventing nonspecific membrane binding by Gag and ensuring that Gag is targeted to the plasma membrane (68). Using an in vitro binding assay, the p6 domain was suggested to be important in Gag binding to HIV-1 RNA (69). However, other studies found that the p6 domain is dispensable in Gag:RNA-specific binding (70) and RNA genome packaging (64), leaving the role of the p6 domain in RNA genome packaging unsubstantiated. The importance of membrane targeting by the MA domain and the Gag:Gag multimerization of the CA domain in facilitating RNA packaging suggests that HIV-1 Gag multimerizes on the RNA at the plasma membrane to package the RNA genome (64) (Fig. 2).
Fig 2.
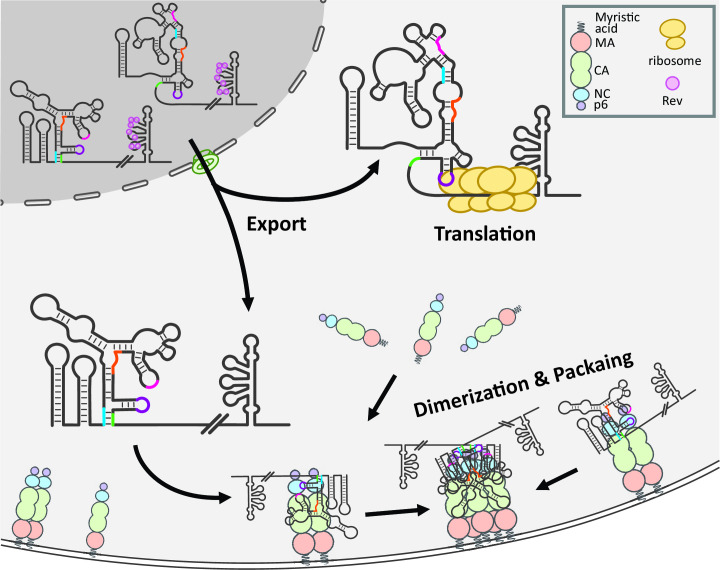
Steps of HIV-1 RNA packaging and virus assembly. Unspliced HIV-1 RNA nuclear export is facilitated by Rev:RRE interactions with host machinery. These RNA species can form distinct 5′ UTR structures that lead to preferential packaging or translation. HIV-1 RNA is held at the plasma membrane by a low number of Gag, anchored by the myristic acid moiety and PIP2-binding site on MA. Dimerization of HIV-1 RNA occurs on the plasma membrane and facilitates the rapid accumulation of Gag proteins.
Cis-acting elements: 5′ UTR and other regions
Analogous to host mRNAs, following transcription, HIV-1 RNA is modified with a 5′ cap and a 3′ polyadenylic acid (polyA) tail (2, 71, 72). HIV-1 RNA contains several cis-acting elements to ensure its nuclear export and efficient genome packaging. The export of intron-containing HIV-1 RNAs is accomplished with the trans-acting viral protein Rev and cis-acting Rev-response element (RRE), which is embedded within the env gene (73, 74). Rev is expressed by completely spliced transcripts exported through the NXF1 pathway used by most host mRNAs. Rev shuttles back into the nucleus where it multimerizes on the RRE, a ~350 nt highly structured RNA region, and recruits host factors necessary for nuclear export through the CRM1-dependent pathway. RRE is present in all unspliced and partially spliced HIV-1 RNAs allowing them to be exported efficiently from the nucleus (75–92). It was previously shown that inactivating mutations in Rev or RRE cause inefficient genome packaging and reduced translation (93, 94). However, it was later demonstrated that the major impact of these mutations is caused by defects in the nuclear export of HIV-1 RNA; indeed, such defects are rescued by replacing RRE with an unrelated cis-acting element, Mason Pfizer monkey virus constitutive transport element (CTE) that allows Rev-independent nuclear export of unspliced HIV-1 RNA (94–96). CTE- and RRE-containing HIV-1 RNAs can be packaged at similar efficiencies (95, 97–99).
The 5′ UTR of the HIV-1 RNA along with sequences that extend into the 5′ end of the gag gene are necessary and sufficient to mediate genome packaging (100). This region can adopt different structural conformations that affect RNA functions. The HIV-1 5′ UTR is highly structured and contains multiple elements important for viral replication, including those essential for Gag recognition and packaging of a dimeric RNA (Fig. 1B) (4). Generally, the 5′ UTR secondary structure is composed of multiple stem loop motifs (Fig. 1B): trans-acting response (TAR) stem loop, polyA stem loop, a stem-loop region containing the primer-binding site (PBS), and a set of three stem loop structures termed SL1, SL2, and SL3. Through numerous studies, several functional roles have been attributed to these elements and are briefly discussed here. The TAR stem loop contains binding sites for the viral protein Tat and host protein cyclinT1. Tat recruits the host super elongation complex, which includes cyclinT1, to abrogate the pausing of RNA polymerase II and allow the elongation of RNA transcription to continue (101–108). The polyA stem contains the polyadenylation signal. HIV-1 RNA has two polyadenylation signals located near the 5′ and 3′ end of the RNA. The 3′ polyadenylation signal is used for cleavage and the addition of the polyA tail to the HIV-1 RNA (2). The PBS stem loop plays an important role in reverse transcription; using sequence complementarity, a host tRNA is annealed to the 18-nt PBS and serves as a primer for the initiation of viral DNA synthesis (109, 110). The SL1, SL2, and SL3 contain multiple Gag/NC- binding sites, although the locations of these sites may vary in different studies (111–116). In addition, the loop of SL1 contains a GC-rich 6-nt palindromic sequence known as the dimerization initiation signal (DIS), which plays a critical role in the selection of co-packaged RNA (117–119). DIS elements in two HIV-1 RNA molecules interact to form a “kissing loop” and initiate RNA dimerization (120–123). SL2 contains the major splice donor site (SD), which is used by HIV-1 to generate partially spliced and completely spliced transcripts (124). Interestingly, although these elements are important in HIV-1 RNA packaging, deleting any one or two stem loops including TAR, polyA, PBS, SL1, Sl2, and SL3 may cause genome packaging defects but does not completely abolish HIV-1 RNA packaging (100, 125–131). These studies demonstrate that the cis-acting sequence important for packaging is not located within a single element; instead, multiple elements with redundant functions may exist to facilitate HIV-1 genome packaging.
In addition to the elements described above, the ribosomal frameshift signal was once thought to play a role in RNA packaging (132), but a later study showed that this element does not affect HIV-1 genome encapsidation (133). Similar to cellular mRNAs, RNA modifications have been identified on HIV-1 RNA including N6-methyladenosine (m6A) and acetylation of the N4 position of cytidine (ac4C). The reported effects of HIV-1 RNA modifications are mainly on RNA stability and gene expression (134–138), although the possible role of m6A on genome packaging has also been suggested (139). It was reported that the 5′ cap of HIV-1 RNA is trimethylated (71). The hypermethylated cap is reported to allow HIV-1 RNA to be translated through a pathway distinct from that of cellular RNAs (72); whether the cap modification affects packaging is unclear. It was shown that at the plasma membrane, Gag preferentially binds A-rich host mRNAs. However, the exact contribution of the nucleotide composition on HIV-1 RNA packaging is difficult to assess. HIV-1 and other viruses have evolved to have low 5′-CG-3′ dinucleotide content in their RNA genomes to mimic host mRNAs and avoid the detection of zinc-finger antiviral protein (ZAP) (140). Global alteration of HIV-1 sequences can inadvertently trigger antiviral response (140) and/or alter intricately regulated HIV-1 splicing (141). Thus, whether the nucleotide composition affects HIV-1 genome packaging remains unclear.
DIS sequence and RNA dimerization
HIV-1 RNA dimerization relies heavily on complementary interactions between the DIS elements of two RNA molecules. In vitro experiments also demonstrated that the presence of DIS in SL1 promotes dimerization of RNA segments (117, 142–146). The exact DIS sequence in the SL1 loop region varies and most HIV-1 strains have a 6-nt palindromic DIS. Most subtype B viruses have a GCGCGC palindrome, whereas many subtype A, C, or G viruses have a GTGCAC palindrome (147, 148). Early genetic studies showed that the complementarity of the DIS sequences affects recombination rates. Progeny between two proviruses with the same DIS exhibits high recombination rates, whereas progeny between two proviruses with DIS sequences that cannot form perfect base pairings exhibits low recombination rates (149). These studies suggest that the DIS identity affects the frequency of viral RNA copackaging (150). This was later demonstrated by directly examining viral RNA in particles. HIV-1 RNAs with identical DIS sequences are copackaged at a frequency similar to random assortment, indicating that RNAs generated from different proviruses can dimerize efficiently. However, two RNAs with different DIS sequences that cannot form perfect base-pairing, such as one RNA containing the subtype B DIS and the other RNA containing the subtype C DIS, are copackaged together much less frequently than random distribution. Conversely, two RNAs containing engineered DISs that are complementary to each other, such as one virus containing GGGGGG and the other virus containing CCCCCC, are copackaged together more efficiently than random distribution (13, 98, 151). These observations demonstrated that the two copackaged RNAs interact via the DIS to dimerize and the dimerization step occurs prior to packaging. Although the initial RNA:RNA interactions are mediated by DIS, HIV-1 RNAs in the virion engage in dimeric contacts far beyond DIS interactions and Gag/NC plays an important role in the extended dimerization (152–157). Despite the proven role of the DIS in HIV-1 RNA dimerization, it has been reported that HIV-1 with deletions of the DIS, or SL1 including the DIS, can replicate; furthermore, their virion RNAs are dimeric (147, 150, 158–160). These results suggest that there may be other elements within the RNA that facilitate dimerization or that HIV-1 can employ alternate mechanisms to promote RNA dimerization.
HIV-1 5′ UTR structures and Gag/NC-binding sites
Although the HIV-1 5′ UTR is known to be highly structured, its precise functional conformation(s) has been highly debated. Many predicted 5′ UTR RNA conformations have similar structures with minor variations such as the length of the stems and the locations of the bulges. Most, but not all, structures suggest that to facilitate genome packaging, TAR, polyA, SL1, SL2, and SL3 are formed, with DIS located at the top of SL1, and a region of U5 interacting with the Gag AUG start codon and its flanking sequences. Because HIV-1 RNA serves as the template for translation and the virion genome, it has been suggested that the 5′ UTR can adopt structures to favor translation or packaging (161) (Fig. 1C). In the conformation facilitating packaging, TAR, polyA, PBS, SL1, SL2, and SL3 RNA stem loops are formed with the DIS displayed within the loop of SL1, available for RNA dimerization (Fig. 1C, structure on the left). In the RNA structure facilitating translation, many of the stem loops, including the SL1, are not formed; the DIS sequence forms base-pairing with sequences in the polyA stem and is not available for RNA dimerization (Fig. 1C, structure on the right). Mutagenesis experiments have been performed to test the proposed structures, and it was found that elements within the DIS-exposed conformation are important for RNA packaging, but translation is not dependent on the formation of predicted translation-competent structures (162).
Multiple studies have shown that Gag/NC preferentially binds unpaired guanosine (G) residues within the HIV-1 5′ UTR (111, 163–165). Using a chemical probe reverse-footprinting assay, the NC binding of HIV-1 RNA within virions has been mapped (164). This study identified two primary binding sites, both located in bulges of SL1, two secondary binding sites, one located immediately upstream of SL1 and one located in SL2, and two tertiary binding sites, located within SL3 (164). The primary NC-binding sites described in this virion chemical probing study were also later identified as the major Gag-binding sites in an in vitro binding assay (116, 166). Mutating one or more of these binding sites affects in vitro NC:RNA binding (116, 166–168). The contribution of these Gag/NC-binding sites to HIV-1 RNA packaging was examined using a cell-based assay and mutating the guanosines in the primary binding sites located within SL1 were shown to have the most effects. Importantly, combining mutations at different NC-binding sites causes synergistic defects in RNA packaging (111). These results suggest that there is redundancy in Gag-RNA interactions and multiple Gag polyproteins need to bind to HIV-1 RNA to facilitate packaging.
Based on nuclear magnetic resonance (NMR) spectroscopy studies, a three-way junction structure was recently proposed (115, 169). In this structure, SL1 exists with exposed DIS, but SL2 is not formed, instead its sequences base pair with part of the PBS stem and sequences upstream of SL1 forming the first three-way junction (Fig. 1D). The second three-way junction is composed of SL3 with downstream AUG region interacting with U5. The three-way junction structure model also predicted 17 exposed guanosines as putative NC-binding sites. It is worth noting that many of the predicted exposed guanosines overlap with the previously described NC-binding sites identified using chemical probing of virion RNA (164). To test the three-way junction model, mutants with multiple substitutions of these guanosines were generated and shown to have RNA packaging defects (111, 115, 169). Further studies identified a UUUU:GGAG base-paring region to be critical for RNA packaging, with the two consecutive guanosines as a high-affinity NC-binding site (165). The UUUU and GGAG flank the SL3 shown in Fig. 1B; in addition, the two consecutive guanosines are described as an NC tertiary binding site in virion RNA (164, 170). It was shown that the lability of the base-pairing is important for its function, stabilizing the interactions by substituting the uridines to cytosines abolishes high-affinity NC binding (165).
Using in-gel chemical probing, it was shown that RNA containing the HIV-1 5′ UTR forms multiple conformations (16) (Fig. 1E). Hence, it was also proposed that HIV-1 5′ UTR forms an ensemble of conformations, some suitable for packaging, whereas others are not. RNAs with conformations that exhibit exposed DIS and many Gag-binding sites are likely to be packaged and serve as virion genomes (Fig. 1E, left). By contrast, RNA in other conformations, the DIS and major Gag-binding sites form intramolecular base-pairing and are sequestered, and therefore not accessible to interact with another HIV-1 RNA or allow Gag binding (Fig. 1E, right) (16).
Although each of these studies proposes distinct RNA structures and Gag/NC:RNA interactions, there are at least three points of consensus. First, HIV-1 5′ UTR RNA structures play an important role in genome packaging. Second, the formation of SL1 along with the availability of the DIS and SL3 are two common structural features essential for efficient genome packaging. Third, multiple Gag/NC-binding sites exist in the 5′ UTR and multiple proposed sites are used to mediate RNA packaging.
Despite being efficiently packaged during virus assembly (13), HIV-1 RNA is not required for Gag particle formation. In the absence of HIV-1 RNA, Gag can still assemble into particles and instead packages cellular mRNA (171). The mechanism by which HIV-1 ensures the packaging of viral RNA when it is not required for particle assembly is intriguing. It was shown that when Gag is expressed at levels reflecting those in infected cells, HIV-1 RNA promotes virus assembly and particle production (172). These findings lead to the hypothesis that with many Gag-binding sites, the dimerized HIV-1 RNA allows the binding of multiple Gag proteins and increases the local Gag concentration, which facilitates a switch in Gag conformation to promote Gag-Gag interactions leading to particle assembly (172). By contrast, lacking multiple specific Gag-binding sites, cellular mRNAs are less efficient at increasing a local Gag concentration needed to facilitate the Gag conformation switch and virus assembly. Therefore, compared to Gag binding to cellular mRNA, the Gag-dimeric RNA complex has a significant advantage in initiating virus assembly (172). This property provides a mechanism to ensure high-efficiency RNA genome packaging by HIV-1 (5).
Dynamics of HIV-1 RNA packaging: when, where, and how
The interactions between HIV-1 RNA and Gag are interwoven: HIV-1 RNA is translated to generate Gag/Gag-Pol polyproteins and Gag packages HIV-1 unspliced RNA as the viral genome during assembly. Where the initial Gag-RNA interaction occurs has been a long-standing question. Translation of HIV-1 RNA occurs within the cytoplasm where both Gag and RNA are present, providing ample opportunity for Gag:RNA interactions. It was once proposed that Gag packages the RNA from which it is translated (packaging in cis or cis-packaging) (173). This hypothesis was not supported by later studies, which showed that in cells expressing two proviruses, HIV-1 RNAs that do not encode functional Gag can be packaged as efficiently as those that encode functional Gag (174); thus, Gag does not preferentially package in cis.
Imaging studies reported that most of the cytoplasmic HIV-1 Gag is monomeric and uses diffusion as a main transport mechanism (175, 176). By employing biochemical assays, HIV-1 RNA and Gag have been shown to interact in the cytoplasm (177). Using crosslinking-immunoprecipitation sequencing (CLIP-seq) analyses, it was shown that cytoplasmic Gag can bind HIV-1 RNA; however, most of the cytoplasmic RNA-associated Gag appears to be bound to host RNAs (67). By contrast, at the plasma membrane, Gag is predominantly bound to HIV-1 RNA. Thus, compared to the membrane-bound Gag, cytoplasmic Gag has far less selectivity toward binding to HIV-1 RNA (67). Because Gag is known to target and assemble at the plasma membrane, it was often suggested that the cytoplasmic interaction with Gag facilitates HIV-1 RNA trafficking to the plasma membrane. However, imaging studies showed that HIV-1 RNA can travel to the plasma membrane without Gag, indicating that Gag:RNA interactions are not required to localize HIV-1 RNA at the plasma membrane (178, 179). Therefore, although Gag: HIV-1 RNA can interact in the cytoplasm, whether RNA packaging is a direct result of such interactions is unknown. Further studies will be needed to clarify the initial point of Gag:HIV-1 RNA interaction that leads to specific genome packaging. It was also reported that Gag can traffic to the nucleus and bind to RNA, leading to the hypothesis that Gag retrieves HIV-1 RNA for genome packaging (180). Deletion of a majority of MA, including the regions proposed to contain a nuclear localization signal (NLS) and nuclear export signal (NES), does not disrupt HIV-1 RNA packaging, suggesting that Gag nuclear trafficking is not essential for RNA packaging (181, 182). The biological significance of Gag nuclear trafficking, or how frequently it occurs, awaits further studies.
A major location for Gag:HIV-1 interactions that lead to genome packaging is the plasma membrane (Fig. 2). After exiting the nucleus, HIV-1 RNA mainly travels through the cytoplasm by diffusion; the presence of Gag does not affect the mode of transport or the speed of HIV-1 RNA movement (178). However, Gag does affect the time HIV-1 RNA stays on the plasma membrane (183). In the absence of Gag, HIV-1 RNA can travel to the plasma membrane but only remains there briefly. In the presence of Gag, even mutant Gag containing multimerization defects, HIV-1 RNA extends its stay at the plasma membrane. By contrast, the Gag lacking the NC domain cannot retain HIV-1 RNA at the plasma membrane. Thus, the initial Gag:RNA interactions that allow HIV-1 RNA to be retained at the plasma membrane require the ability of Gag to bind RNA, but not Gag multimerization (183). Because the presence of Gag changes the behavior of most HIV-1 RNA, these findings indicate that most of the HIV-1 RNAs at the plasma membrane are associated with Gag.
HIV-1 RNA packaging and virus assembly processes have been studied using live-cell imaging techniques by fluorescently tagging both HIV-1 RNA and a portion of the Gag. In these studies, the RNA signal is often observed first, then Gag signals can be seen to colocalize with RNA with increasing intensity over time, suggesting that Gag assembles on the RNA scaffold (178, 179, 183, 184). In these studies, a mixture of tagged and untagged Gag was used to make morphologically normal viral particles (185). In addition, each tagged Gag only had one fluorescent protein and could not be detected at a single-molecule level, whereas each HIV-1 RNA was tagged with multiple fluorescent proteins and could be detected at a single RNA level. For these technical reasons, the appearance of RNA signal without a Gag signal should not be interpreted as Gag-free HIV-1 RNAs.
Most viral particles contain two HIV-1 RNA molecules; however, the addition of a second, complementary, DIS in the same RNA allows the formation of a self-dimer and leads to the packaging of only one copy of self-dimerized RNA (151). Therefore, HIV-1 regulates its genome encapsidation by packaging a dimeric RNA. It was suggested that during MLV replication, RNA dimerization occurs in the nucleus (186, 187). To probe this possibility, a cell-fusion approach was employed using two cell lines each harboring a provirus and conditions that facilitated cytoplasmic but not nuclear fusion (98). These experiments demonstrated that most of the RNA dimerization and partner selection process does not occur in the nucleus, but instead, in the cytoplasm or at the plasma membrane. A live-cell imaging approach was also used to examine the location of HIV-1 RNA dimerization (184). In this approach, two different HIV-1 RNAs were tagged with distinct fluorescent proteins so that dimerized RNA was visualized by colocalization of the two fluorescent RNA signals. In addition, HIV-1 Gag was tagged with a third fluorescent protein, and the process of RNA dimerization and virus assembly on the plasma membrane was studied. These experiments showed that two different RNA signals often merge at the plasma membrane, followed by an increase in Gag signals consistent with particle assembly (184). HIV-1 RNA molecules also interact in the absence of Gag, but these complexes quickly fall apart, indicating that Gag is required to stabilize the RNA dimer (184). These findings indicate that HIV-1 RNAs dimerize not in the cytoplasm but on the plasma membrane and stabilization of the RNA dimer requires the Gag protein. Dimerization often occurs at an early stage of the virus assembly process. Furthermore, the dimerization process is likely mediated by the interactions of two RNA-Gag complexes, rather than two RNAs (184).
HIV-1 RNA can undergo two distinct biological processes: translation and packaging. The relationship between translation and packaging was explored using a live-cell imaging assay that can distinguish between HIV-1 RNA molecules that are being actively translated from those that are not (188). It was found that both types of HIV-1 RNA can reach the plasma membrane. However, HIV-1 RNA molecules that are being actively translated do not stay near the plasma membrane, whereas nontranslating HIV-1 RNA molecules are retained on the plasma membrane to allow Gag accumulation. These results indicate that the nontranslating RNA is selected as a substrate to facilitate Gag multimerization and virus assembly (188). It should be noted that this study distinguishes between whether an RNA is or is not being actively translated but does not address whether a previously translated RNA can serve as a virion genome.
The impact of transcription start sites on HIV-1 RNA packaging
HIV-1 has three consecutive guanosines at the junction of U3 and R. Most of the time, host RNA Pol II initiates at one of the three guanosines to generate unspliced RNAs with three, two, or one guanosine, referred to as 3G, 2G, and 1G RNA, respectively (14). It was reported that 3G RNA is more abundant in infected cells; however, 1G RNA is selected over 3G RNA and is enriched in virions (14), which was later confirmed by other groups (15–17, 111). These findings revealed that the 5′ context of the HIV-1 RNA can affect its functions. All the RNA elements present in 1G RNA are also contained in 3G RNA; thus, it is intriguing how HIV-1 can distinguish between two 9 kb RNAs that only differ by 2-nt. The functions of many RNA elements in the HIV-1 5′ UTR are structure-dependent. Therefore, the current prevailing hypothesis is that the 5′ UTR of 1G and 3G RNA folds into different conformations: the 1G RNA, but not 3G RNA, folds into conformation(s) that facilitates packaging.
The precise conformations of the 5′ UTR that render 3G and 1G RNA non-packageable and packageable, respectively, are still being debated. The guanosine(s) at the very 5′ end of the HIV-1 RNA, along with the 5′ cap, can form base-pairing with downstream sequences and become part of the TAR stem (Fig. 1D and E). The U5:DIS alternate base-pairing model proposes that the DIS can form base-pairing with U5 sequences, making it unavailable for RNA dimerization and preventing genome packaging (Fig. 1D). This alternative structure is more stable in 3G RNA than in 1G RNA (15). In addition, binding studies using in vitro transcribed RNA containing the HIV-1 5′ UTR showed that the host factor eIF4E binds 3G-like short RNA better than 1G-like short RNA (189). Thus, this model also proposes that the cap of 1G RNA is sequestered, preventing RNA from being translated. Therefore, 1G RNA is used as the viral genome, whereas 3G RNA is used as the mRNA (189).
By contrast, the “zip and pack” model proposes that the polyA stem is a keystone structural element and that its formation promotes the downstream 5′ UTR sequences to fold into conformations with exposed DIS and major Gag-binding sites so that the RNA can be packaged (16). In the 1G RNA, the cap and the first guanosine base pair with two cytosines at the end of TAR, allowing stable polyA stem formation and the subsequent folding of the 5′ UTR into packageable conformers. Conversely, the additional guanosines present in 3G RNA can extend beyond base-pairing with TAR and invade the polyA stem, destabilizing the structure. Thus, 3G RNA tends to have a destabilized polyA stem; consequently, the downstream 5′ UTR folds into structures in which the DIS and the major Gag-binding sites are sequestered, disfavoring RNA packaging (Fig. 2). This model is supported by mutational analyses: mutants with a 2-nt insertion between the TAR and polyA stems can package 3G RNA efficiently because the insertion prevents the very 5′ guanosines in the 3G RNA from destabilizing the polyA stem (16).
Additional studies demonstrated that these two features—multiple transcription start sites and preferential packaging of one RNA species—are not unique to HIV-1 but are also present in other primate lentiviruses (190). Isolates of simian immunodeficiency virus (SIV), closely related to the ancestral viruses that caused zoonotic infection of humans, exhibit similar utilization of multiple transcription start sites and selective packaging indicating that HIV-1 retains these conserved characteristics from ancestral viruses (190). In addition, HIV-2 also uses multiple transcription start sites and preferentially packages a specific RNA. Features in viral replication often provide advantages in replication fitness. To probe whether the transcription start site usage provides HIV-1 with a replication advantage, HIV-1 mutants were generated that either predominantly expressed 3G RNA with very little 1G RNA or predominantly expressed 1G RNA with no 3G RNA (17). It was found that both mutants can undergo multiple rounds of replication, indicating neither 3G nor 1G RNA, two major HIV-1 transcripts, are strictly required for spreading infection (17). Another study with similar mutants also confirmed these observations (191). However, neither mutant virus can replicate as well as the wild-type virus (17). Thus, HIV-1 expresses different unspliced transcripts with specified functions to optimize replication fitness (17).
SUMMARY AND PROSPECTIVES
HIV-1 must package its RNA genome to generate infectious viruses to spread to new hosts. How HIV-1 identifies and packages two copies of unspliced viral RNA from a vast pool of other cellular RNAs has been a long-standing question in the field. Studies suggest that the cis-acting elements important for genome encapsidation reside in HIV-1 5′ UTR and their functions are structure-dependent. The HIV-1 5′ UTR forms complex structures, some of which have exposed DIS and multiple Gag-binding sites that facilitate genome packaging. Gag interacts with HIV-1 RNA at the plasma membrane and Gag:RNA complexes initiate dimerization using the DIS sequences, followed by accumulation of Gag on the RNA dimer complex to assemble HIV-1 particles. The selection of HIV-1 RNA dimer is based on Gag-binding sites on the 5′ UTR and the ability of the dimeric HIV-1 RNA to serve as a preferred scaffold to promote Gag assembly. However, many aspects of this process are not well defined or understood such as the HIV-1 RNA structure(s) that promote packaging, the properties of HIV-1 RNA that make it a suitable scaffold to promote virus assembly, how two RNA:Gag complexes merge and dimerize, and whether RNA modifications can affect the packaging of HIV-1 RNA. Lastly, as a critical step of viral replication, whether strategies can be developed to interfere with genome packaging, thereby abolishing the production of infectious particles remains an open question. Future studies on these topics and others will advance our understanding of HIV-1 RNA genome packaging and explore the development of novel antiviral strategies.
ACKNOWLEDGMENTS
We thank E. O. Freed and V.K. Pathak for their helpful discussions and critical reading of the manuscript and K. Delviks-Frankenberry for suggesting the title of this review.
This work is supported by Intramural Research, National Cancer Institute, National Institutes of Health (NIH); Supplemental funds from the Center for Cancer Research to WSH, and Innovation funds from the Office of AIDS Research, NIH to WSH.
Biographies

Alice Duchon is a Staff Scientist in the HIV Dynamics and Replication Program at the National Cancer Institute, USA. She received her Bachelor of Science degree in Biochemistry and Molecular Biology from Dickinson College and Ph.D. in Biochemistry from The Ohio State University under Dr. Karin Musier-Forsyth’s guidance. Over the last 12 years, she has focused on studying HIV-1 virus assembly. Her current research utilizes live-cell microscopy techniques to elucidate the mechanisms by which efficient HIV-1 RNA packaging and viral particle assembly are regulated.

Wei-Shau Hu received her Ph.D. in Genetics from the University of California, Davis. She studied the mechanisms of retroviral replication as a postdoctoral fellow at the University of Wisconsin under Dr. Howard Temin's guidance. Dr. Hu joined the faculty of West Virginia University as an Assistant Professor and was promoted to Associate Professor with tenure. She later joined the National Cancer Institute as Senior Investigator and Head of the Viral Recombination Section in the HIV Drug Resistance Program (renamed the HIV Dynamics and Replication Program in 2015). For the past two decades, her research has been focused on the mechanism used by HIV to transfer genetic information to progeny. Her lab uses multidisciplinary approaches to study mechanisms of HIV replication including RNA genome packaging, virus assembly, and reverse transcription.
Contributor Information
Alice Duchon, Email: Alice.Duchon@nih.gov.
Wei-Shau Hu, Email: Wei-Shau.Hu@nih.gov.
Vinayaka R. Prasad, Albert Einstein College of Medicine, Bronx, New York, USA
REFERENCES
- 1. Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ott M, Freed EO. 2023. Human immunodeficiency viruses: replication, p 558–617. In Fields Virology. Vol. 3. [Google Scholar]
- 3. Emery A, Swanstrom R. 2021. HIV-1: to splice or not to splice, that is the question. Viruses 13:181. doi: 10.3390/v13020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rein A. 2019. RNA packaging in HIV. Trends Microbiol 27:715–723. doi: 10.1016/j.tim.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanson HM, Willkomm NA, Yang H, Mansky LM. 2022. Human retrovirus genomic RNA packaging. Viruses 14:1094. doi: 10.3390/v14051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lerner G, Weaver N, Anokhin B, Spearman P. 2022. Advances in HIV-1 assembly. Viruses 14:478. doi: 10.3390/v14030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goff SP, Roth MJ. 2023. Retroviridae. Fields Virology 3:465–526. [Google Scholar]
- 9. Hu WS, Temin HM. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A 87:1556–1560. doi: 10.1073/pnas.87.4.1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rawson JMO, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS. 2018. Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic Acids Res 46:10535–10545. doi: 10.1093/nar/gky910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Streeck H, Li B, Poon AFY, Schneidewind A, Gladden AD, Power KA, Daskalakis D, Bazner S, Zuniga R, Brander C, Rosenberg ES, Frost SDW, Altfeld M, Allen TM. 2008. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med 205:1789–1796. doi: 10.1084/jem.20080281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kellam P, Larder BA. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J Virol 69:669–674. doi: 10.1128/JVI.69.2.669-674.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U.S.A. 106:13535–13540. doi: 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Masuda T, Sato Y, Huang YL, Koi S, Takahata T, Hasegawa A, Kawai G, Kannagi M. 2015. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep 5. doi: 10.1038/srep17680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, Russo E, Swanson C, Shuey A, Telesnitsky A, Summers MF. 2016. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A 113:13378–13383. doi: 10.1073/pnas.1616627113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nikolaitchik OA, Liu S, Kitzrow JP, Liu Y, Rawson JMO, Shakya S, Cheng Z, Pathak VK, Hu WS, Musier-Forsyth K. 2021. Selective packaging of HIV-1 RNA genome is guided by the stability of 5' untranslated region polyA stem. Proc Natl Acad Sci U S A 118:e2114494118. doi: 10.1073/pnas.2114494118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nikolaitchik OA, Islam S, Kitzrow JP, Duchon A, Cheng Z, Liu Y, Rawson JMO, Shao W, Nikolaitchik M, Kearney MF, Maldarelli F, Musier-Forsyth K, Pathak VK, Hu WS. 2023. HIV-1 USURPS transcription start site heterogeneity of host RNA polymerase II to maximize replication fitness. Proc Natl Acad Sci U S A 120:e2305103120. doi: 10.1073/pnas.2305103120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A 101:14889–14894. doi: 10.1073/pnas.0405596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ono A. 2009. HIV-1 assembly at the plasma membrane: Gag trafficking and localization. Future Virol 4:241–257. doi: 10.2217/fvl.09.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ono A. 2010. HIV-1 assembly at the plasma membrane. Vaccine 28:B55–B59. doi: 10.1016/j.vaccine.2009.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphospnate is essential for efficient Gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A 107:1600–1605. doi: 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang C, Ndassa Y, Summers MF. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol 9:537–543. doi: 10.1038/nsb806 [DOI] [PubMed] [Google Scholar]
- 24. Campbell EM, Hope TJ. 2015. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13:471–483. doi: 10.1038/nrmicro3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dick RA, Zadrozny KK, Xu C, Schur FKM, Lyddon TD, Ricana CL, Wagner JM, Perilla JR, Ganser-Pornillos BK, Johnson MC, Pornillos O, Vogt VM. 2018. Inositol phosphates are assembly co-factors for HIV-1. Nature 560:509–512. doi: 10.1038/s41586-018-0396-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mallery DL, Faysal KMR, Kleinpeter A, Wilson MSC, Vaysburd M, Fletcher AJ, Novikova M, Böcking T, Freed EO, Saiardi A, James LC. 2019. Cellular IP(6) levels limit HIV production while viruses that cannot efficiently package IP(6) are attenuated for infection and replication. Cell Reports 29:3983–3996. doi: 10.1016/j.celrep.2019.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80:217–286. doi: 10.1016/S0079-6603(05)80006-6 [DOI] [PubMed] [Google Scholar]
- 28. Rein A. 2010. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol 7:700–705. doi: 10.4161/rna.7.6.13685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dorfman T, Luban J, Goff SP, Haseltine WA, Göttlinger HG. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol 67:6159–6169. doi: 10.1128/JVI.67.10.6159-6169.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol 67:4027–4036. doi: 10.1128/JVI.67.7.4027-4036.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fossen T, Wray V, Bruns K, Rachmat J, Henklein P, Tessmer U, Maczurek A, Klinger P, Schubert U. 2005. Solution structure of the human immunodeficiency virus type 1 p6 protein. J Biol Chem 280:42515–42527. doi: 10.1074/jbc.M507375200 [DOI] [PubMed] [Google Scholar]
- 32. Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A 88:3195–3199. doi: 10.1073/pnas.88.8.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6(Gag) is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol 69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. 1998. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem 273:33781–33786. doi: 10.1074/jbc.273.50.33781 [DOI] [PubMed] [Google Scholar]
- 35. Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. 2013. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication. Virus Res 171:263–277. doi: 10.1016/j.virusres.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu H, Mitra M, Naufer MN, McCauley MJ, Gorelick RJ, Rouzina I, Musier-Forsyth K, Williams MC. 2014. Differential contribution of basic residues to HIV-1 nucleocapsid protein’s nucleic acid chaperone function and retroviral replication. Nucleic Acids Res 42:2525–2537. doi: 10.1093/nar/gkt1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouhand A, Pasi M, Catala M, Zargarian L, Belfetmi A, Barraud P, Mauffret O, Tisné C. 2020. Overview of the nucleic-acid binding properties of the HIV-1 nucleocapsid protein in its different maturation states. Viruses 12:1109. doi: 10.3390/v12101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 353:41–51. doi: 10.1016/j.virol.2006.05.014 [DOI] [PubMed] [Google Scholar]
- 39. Dupraz P, Spahr PF. 1992. Specificity of rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol 66:4662–4670. doi: 10.1128/JVI.66.8.4662-4670.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berkowitz RD, Ohagen A, Höglund S, Goff SP. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol 69:6445–6456. doi: 10.1128/JVI.69.10.6445-6456.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Barklis E. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol 69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaye JF, Lever AM. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the P2 domain of gag in RNA encapsidation. J Virol 72:5877–5885. doi: 10.1128/JVI.72.7.5877-5885.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poon DTK, Li G, Aldovini A. 1998. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J Virol 72:1983–1993. doi: 10.1128/JVI.72.3.1983-1993.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375:592–610. doi: 10.1016/j.virol.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 45. Poon DTK, Wu J, Aldovini A. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid P7 protein involved in RNA packaging and infectivity. J Virol 70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paillart JC, Berthoux L, Ottmann M, Darlix JL, Marquet R, Ehresmann B, Ehresmann C. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol 70:8348–8354. doi: 10.1128/JVI.70.12.8348-8354.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92–104. doi: 10.1006/viro.1999.9629 [DOI] [PubMed] [Google Scholar]
- 48. Gorelick RJ, Nigida SM Jr, Bess JW Jr, Arthur LO, Henderson LE, Rein A. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol 64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwartz MD, Fiore D, Panganiban AT. 1997. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J Virol 71:9295–9305. doi: 10.1128/JVI.71.12.9295-9305.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol 64:1920–1926. doi: 10.1128/JVI.64.5.1920-1926.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y, Barklis E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol 71:6765–6776. doi: 10.1128/JVI.71.9.6765-6776.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc- coordinating sequences. Virology 256:92–104. doi: 10.1006/viro.1999.9629 [DOI] [PubMed] [Google Scholar]
- 53. Mark-Danieli M, Laham N, Kenan-Eichler M, Castiel A, Melamed D, Landau M, Bouvier NM, Evans MJ, Bacharach E. 2005. Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the Gag protein and enhance packaging and infectivity. J Virol 79:7756–7767. doi: 10.1128/JVI.79.12.7756-7767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dannull J, Surovoy A, Jung G, Moelling K. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grigorov B, Décimo D, Smagulova F, Péchoux C, Mougel M, Muriaux D, Darlix J-L. 2007. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 4:54. doi: 10.1186/1742-4690-4-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fu W, Hu WS. 2003. Functional replacement of nucleocapsid flanking regions by heterologous counterparts with divergent primary sequences: effects of chimeric nucleocapsid on the retroviral replication cycle. J Virol 77:754–761. doi: 10.1128/jvi.77.1.754-761.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cimarelli A, Sandin S, Höglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol 74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Göttlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:5781–5785. doi: 10.1073/pnas.86.15.5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol 68:5311–5320. doi: 10.1128/JVI.68.8.5311-5320.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 87:523–527. doi: 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ono A, Freed EO. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol 73:4136–4144. doi: 10.1128/JVI.73.5.4136-4144.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yuan X, Yu X, Lee TH, Essex M. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol 67:6387–6394. doi: 10.1128/JVI.67.11.6387-6394.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/jvi.77.9.5439-5450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duchon A, Santos S, Chen J, Brown M, Nikolaitchik OA, Tai S, Chao JA, Freed EO, Pathak VK, Hu WS. 2021. Plasma membrane anchoring and Gag: Gag multimerization on viral RNA are critical properties of HIV-1 Gag required to mediate efficient genome packaging. mBio 12:e0325421. doi: 10.1128/mbio.03254-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lei X, Gonçalves-Carneiro D, Zang TM, Bieniasz PD. 2023. Initiation of HIV-1 Gag lattice assembly is required for recognition of the viral genome packaging signal. Elife 12:e83548. doi: 10.7554/eLife.83548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alfadhli A, McNett H, Tsagli S, Bächinger HP, Peyton DH, Barklis E. 2011. HIV-1 matrix protein binding to RNA. J Mol Biol 410:653–666. doi: 10.1016/j.jmb.2011.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 Gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thornhill D, Olety B, Ono A. 2019. Relationships between MA-RNA binding in cells and suppression of HIV-1 Gag mislocalization to intracellular membranes. J Virol 93:e00756-19. doi: 10.1128/JVI.00756-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dubois N, Khoo KK, Ghossein S, Seissler T, Wolff P, McKinstry WJ, Mak J, Paillart J-C, Marquet R, Bernacchi S. 2018. The C-terminal P6 domain of the HIV-1 Pr55Gag precursor is required for specific binding to the genomic RNA. RNA Biol 15:923–936. doi: 10.1080/15476286.2018.1481696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sarni S, Biswas B, Liu S, Olson ED, Kitzrow JP, Rein A, Wysocki VH, Musier-Forsyth K. 2020. HIV-1 gag protein with or without P6 specifically dimerizes on the viral RNA packaging signal. J Biol Chem 295:14391–14401. doi: 10.1074/jbc.RA120.014835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yedavalli VSRK, Jeang K-T. 2010. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A 107:14787–14792. doi: 10.1073/pnas.1009490107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Singh G, Seufzer B, Song Z, Zucko D, Heng X, Boris-Lawrie K. 2022. HIV-1 hypermethylated guanosine cap licenses specialized translation unaffected by mTOR. Proc Natl Acad Sci U S A 119:e2105153118. doi: 10.1073/pnas.2105153118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pfeifer K, Weiler BE, Ugarkovic D, Bachmann M, Schröder HC, Müller WE. 1991. Evidence for a direct interaction of Rev protein with nuclear envelope mRNA–translocation system. Eur J Biochem 199:53–64. doi: 10.1111/j.1432-1033.1991.tb16091.x [DOI] [PubMed] [Google Scholar]
- 74. Behrens RT, Sherer NM. 2023. Retroviral hijacking of host transport pathways for genome nuclear export. mBio 14:e0007023. doi: 10.1128/mbio.00070-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fritz CC, Green MR. 1996. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol 6:848–854. doi: 10.1016/s0960-9822(02)00608-5 [DOI] [PubMed] [Google Scholar]
- 76. Fritz CC, Zapp ML, Green MR. 1995. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 376:530–533. doi: 10.1038/376530a0 [DOI] [PubMed] [Google Scholar]
- 77. Madore SJ, Tiley LS, Malim MH, Cullen BR. 1994. Sequence requirements for Rev multimerization in vivo. Virology 202:186–194. doi: 10.1006/viro.1994.1334 [DOI] [PubMed] [Google Scholar]
- 78. Yi R, Bogerd HP, Cullen BR. 2002. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J Virol 76:2036–2042. doi: 10.1128/jvi.76.5.2036-2042.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stade K, Ford CS, Guthrie C, Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0 [DOI] [PubMed] [Google Scholar]
- 80. Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W. 1986. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 321:412–417. doi: 10.1038/321412a0 [DOI] [PubMed] [Google Scholar]
- 81. Malim MH, Böhnlein S, Hauber J, Cullen BR. 1989. Functional dissection of the HIV-1 Rev trans-activator-derivation of a trans-dominant repressor of Rev function. Cell 58:205–214. doi: 10.1016/0092-8674(89)90416-9 [DOI] [PubMed] [Google Scholar]
- 82. Malim MH, Cullen BR. 1991. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241–248. doi: 10.1016/0092-8674(91)90158-u [DOI] [PubMed] [Google Scholar]
- 83. Malim MH, Hauber J, Fenrick R, Cullen BR. 1988. Immunodeficiency virus Rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181–183. doi: 10.1038/335181a0 [DOI] [PubMed] [Google Scholar]
- 84. Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. 1989. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254–257. doi: 10.1038/338254a0 [DOI] [PubMed] [Google Scholar]
- 85. Meyer BE, Malim MH. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev 8:1538–1547. doi: 10.1101/gad.8.13.1538 [DOI] [PubMed] [Google Scholar]
- 86. Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen BR. 1992. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A 89:758–762. doi: 10.1073/pnas.89.2.758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim SY, Byrn R, Groopman J, Baltimore D. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol 63:3708–3713. doi: 10.1128/JVI.63.9.3708-3713.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. 1991. Characterization of HIV-1 Rev protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 19:1577–1583. doi: 10.1093/nar/19.7.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Daly TJ, Cook KS, Gray GS, Maione TE, Rusche JR. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342:816–819. doi: 10.1038/342816a0 [DOI] [PubMed] [Google Scholar]
- 90. Kjems J, Frankel AD, Sharp PA. 1991. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell 67:169–178. doi: 10.1016/0092-8674(91)90580-r [DOI] [PubMed] [Google Scholar]
- 91. Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci U S A 86:1495–1499. doi: 10.1073/pnas.86.5.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rosen CA, Terwilliger E, Dayton A, Sodroski JG, Haseltine WA. 1988. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci U S A 85:2071–2075. doi: 10.1073/pnas.85.7.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. 2007. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog 3:e54. doi: 10.1371/journal.ppat.0030054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J 23:2632–2640. doi: 10.1038/sj.emboj.7600270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjöld ML. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A 91:1256–1260. doi: 10.1073/pnas.91.4.1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Helga-Maria C, Hammarskjöld ML, Rekosh D. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J Virol 73:4127–4135. doi: 10.1128/JVI.73.5.4127-4135.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wodrich H, Schambach A, Kräusslich HG. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28:901–910. doi: 10.1093/nar/28.4.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Moore MD, Nikolaitchik OA, Chen J, Hammarskjöld ML, Rekosh D, Hu WS. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog 5:e1000627. doi: 10.1371/journal.ppat.1000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. 2010. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol 84:6598–6604. doi: 10.1128/JVI.02264-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu Y, Nikolaitchik OA, Rahman SA, Chen J, Pathak VK, Hu WS. 2017. HIV-1 sequence necessary and sufficient to package non-viral RNAs into HIV-1 particles. J Mol Biol 429:2542–2555. doi: 10.1016/j.jmb.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gaynor RB. 1995. Regulation of HIV-1 gene expression by the transactivator protein tat. Curr Top Microbiol Immunol 193:51–77. doi: 10.1007/978-3-642-78929-8_3 [DOI] [PubMed] [Google Scholar]
- 102. Jones KA, Peterlin BM. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem 63:717–743. doi: 10.1146/annurev.bi.63.070194.003441 [DOI] [PubMed] [Google Scholar]
- 103. Hauber J, Cullen BR. 1988. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol 62:673–679. doi: 10.1128/JVI.62.3.673-679.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Parada CA, Roeder RG. 1996. Enhanced processivity of RNA polymerase II triggered by tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375–378. doi: 10.1038/384375a0 [DOI] [PubMed] [Google Scholar]
- 105. Rounseville MP, Kumar A. 1992. Binding of a host cell nuclear protein to the stem region of human immunodeficiency virus type 1 trans-activation-responsive RNA. J Virol 66:1688–1694. doi: 10.1128/JVI.66.3.1688-1694.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Molecular Cell 38:428–438. doi: 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38:439–451. doi: 10.1016/j.molcel.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schulze-Gahmen U, Hurley JH. 2018. Structural mechanism for HIV-1 TAR loop recognition by tat and the super elongation complex. Proc Natl Acad Sci U S A 115:12973–12978. doi: 10.1073/pnas.1806438115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, Ivanoff L, Petteway SR Jr, Pearson ML, Lautenberger JA, Papas TS, Ghrayeb J, Chang NT, Gallo RC, Wong-Staal F. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277–284. doi: 10.1038/313277a0 [DOI] [PubMed] [Google Scholar]
- 110. Rhim H, Park J, Morrow CD. 1991. Deletions in the tRNA(Lys) primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J Virol 65:4555–4564. doi: 10.1128/JVI.65.9.4555-4564.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nikolaitchik OA, Somoulay X, Rawson JMO, Yoo JA, Pathak VK, Hu WS. 2020. Unpaired guanosines in the 5= untranslated region of HIV-1 RNA act synergistically to mediate genome packaging. J Virol 94:e00439-20. doi: 10.1128/JVI.00439-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. 1998. Structure of the HIV-1 nucleocapsid protein bound to the Sl3 Ψ-RNA recognition element. Science 279:384–388. doi: 10.1126/science.279.5349.384 [DOI] [PubMed] [Google Scholar]
- 113. Ding P, Kharytonchyk S, Waller A, Mbaekwe U, Basappa S, Kuo N, Frank HM, Quasney C, Kidane A, Swanson C, Van V, Sarkar M, Cannistraci E, Chaudhary R, Flores H, Telesnitsky A, Summers MF. 2020. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc Natl Acad Sci U S A 117:17737–17746. doi: 10.1073/pnas.2008519117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLOS Biol. 6:e96. doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. 2015. RNA structure. structure of the HIV-1 RNA packaging signal. Science 348:917–921. doi: 10.1126/science.aaa9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Abd El-Wahab EW, Smyth RP, Mailler E, Bernacchi S, Vivet-Boudou V, Hijnen M, Jossinet F, Mak J, Paillart J-C, Marquet R. 2014. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun 5:4304. doi: 10.1038/ncomms5304 [DOI] [PubMed] [Google Scholar]
- 117. Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barré-Sinoussi F. 1990. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol 216:689–699. doi: 10.1016/0022-2836(90)90392-Y [DOI] [PubMed] [Google Scholar]
- 118. Awang G, Sen D. 1993. Mode of dimerization of HIV-1 genomic RNA. Biochemistry 32:11453–11457. doi: 10.1021/bi00093a024 [DOI] [PubMed] [Google Scholar]
- 119. Laughrea M, Jetté L. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464–13474. doi: 10.1021/bi00249a035 [DOI] [PubMed] [Google Scholar]
- 120. Clever JL, Wong ML, Parslow TG. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol 70:5902–5908. doi: 10.1128/JVI.70.9.5902-5908.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. 2006. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res 34:343–352. doi: 10.1093/nar/gkj427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. 1996. “A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA”. Proc Natl Acad Sci U S A 93:5572–5577. doi: 10.1073/pnas.93.11.5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. 1997. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J Mol Biol 270:36–49. doi: 10.1006/jmbi.1997.1096 [DOI] [PubMed] [Google Scholar]
- 124. Purcell DFJ, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67:6365–6378. doi: 10.1128/JVI.67.11.6365-6378.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Das AT, Harwig A, Vrolijk MM, Berkhout B. 2007. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol 81:7742–7748. doi: 10.1128/JVI.00392-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Luban J, Goff SP. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol 68:3784–3793. doi: 10.1128/JVI.68.6.3784-3793.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lu K, Heng X, Summers MF. 2011. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410:609–633. doi: 10.1016/j.jmb.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. McBride MS, Panganiban AT. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol 70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. McBride MS, Panganiban AT. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol 71:2050–2058. doi: 10.1128/JVI.71.3.2050-2058.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Clever JL, Parslow TG. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol 71:3407–3414. doi: 10.1128/JVI.71.5.3407-3414.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rong L, Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. 2003. Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1. Virology 314:221–228. doi: 10.1016/s0042-6822(03)00405-7 [DOI] [PubMed] [Google Scholar]
- 132. Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, Le Grice SFJ, Arts EJ. 2013. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 13:181–192. doi: 10.1016/j.chom.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Nikolaitchik OA, Hu WS. 2014. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. J Virol 88:4040–4046. doi: 10.1128/JVI.03745-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kennedy EM, Bogerd HP, Kornepati AVR, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, Cullen BR. 2016. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host & Microbe 19:675–685. doi: 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. doi: 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5:e15528. doi: 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26:217–227. doi: 10.1016/j.chom.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR. 2020. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28:306–312. doi: 10.1016/j.chom.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Pereira-Montecinos C, Toro-Ascuy D, Ananías-Sáez C, Gaete-Argel A, Rojas-Fuentes C, Riquelme-Barrios S, Rojas-Araya B, García-de-Gracia F, Aguilera-Cortés P, Chnaiderman J, Acevedo ML, Valiente-Echeverría F, Soto-Rifo R. 2022. Epitranscriptomic regulation of HIV-1 full-length RNA packaging. Nucleic Acids Res 50:2302–2318. doi: 10.1093/nar/gkac062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. doi: 10.1038/nature24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Takata MA, Soll SJ, Emery A, Blanco-Melo D, Swanstrom R, Bieniasz PD. 2018. Global synonymous mutagenesis identifies cis-acting RNA elements that regulate HIV-1 splicing and replication. PLoS Pathog 14:e1006824. doi: 10.1371/journal.ppat.1006824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U S A 91:4945–4949. doi: 10.1073/pnas.91.11.4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. 2004. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol 336:369–379. doi: 10.1016/j.jmb.2003.12.010 [DOI] [PubMed] [Google Scholar]
- 144. Sakuragi JI, Panganiban AT. 1997. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol 71:3250–3254. doi: 10.1128/JVI.71.4.3250-3254.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Laughrea M, Jetté L. 1997. HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5' splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry 36:9501–9508. doi: 10.1021/bi970862l [DOI] [PubMed] [Google Scholar]
- 146. Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. 1997. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J Mol Biol 270:36–49. doi: 10.1006/jmbi.1997.1096 [DOI] [PubMed] [Google Scholar]
- 147. St. Louis DC, Gotte D, Sanders-Buell E, Ritchey DW, Salminen MO, Carr JK, McCutchan FE. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol 72:3991–3998. doi: 10.1128/JVI.72.5.3991-3998.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Laughrea M, Shen N, Jetté L, Wainberg MA. 1999. Variant effects of non-native kissing-loop hairpin palindromes on HIV replication and HIV RNA dimerization: role of stem-loop B in HIV replication and HIV RNA dimerization. Biochemistry 38:226–234. doi: 10.1021/bi981728j [DOI] [PubMed] [Google Scholar]
- 149. Chin MPS, Rhodes TD, Chen J, Fu W, Hu WS. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci U S A 102:9002–9007. doi: 10.1073/pnas.0502522102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol 81:4002–4011. doi: 10.1128/JVI.02589-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SHS, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS. 2013. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog 9:e1003249. doi: 10.1371/journal.ppat.1003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ye L, Gribling-Burrer A-S, Bohn P, Kibe A, Börtlein C, Ambi UB, Ahmad S, Olguin-Nava M, Smith M, Caliskan N, von Kleist M, Smyth RP. 2022. Short- and long-range interactions in the HIV-1 5' UTR regulate genome dimerization and packaging. Nat Struct Mol Biol 29:306–319. doi: 10.1038/s41594-022-00746-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sakuragi JI, Sakuragi S, Shioda T. 2007. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J Virol 81:7985–7992. doi: 10.1128/JVI.00429-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mujeeb A, Ulyanov NB, Georgantis S, Smirnov I, Chung J, Parslow TG, James TL. 2007. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stemloop of HIV-1: sequence effects and mechanism of RNA refolding. Nucleic Acids Res 35:2026–2034. doi: 10.1093/nar/gkm097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Muriaux D, De Rocquigny H, Roques BP, Paoletti J. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem 271:33686–33692. doi: 10.1074/jbc.271.52.33686 [DOI] [PubMed] [Google Scholar]
- 156. Fu W, Gorelick RJ, Rein A. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol 68:5013–5018. doi: 10.1128/JVI.68.8.5013-5018.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. 1996. “HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro”. Proc Natl Acad Sci U S A 93:7577–7581. doi: 10.1073/pnas.93.15.7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hill MK, Shehu-Xhilaga M, Campbell SM, Poumbourios P, Crowe SM, Mak J. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J Virol 77:8329–8335. doi: 10.1128/jvi.77.15.8329-8335.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Berkhout B, van Wamel JL. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol 70:6723–6732. doi: 10.1128/JVI.70.10.6723-6732.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Clever JL, Parslow TG. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol 71:3407–3414. doi: 10.1128/JVI.71.5.3407-3414.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Abbink T.E.M, Berkhout B. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem 278:11601–11611. doi: 10.1074/jbc.M210291200 [DOI] [PubMed] [Google Scholar]
- 162. Abbink Truus E M, Ooms M, Haasnoot PCJ, Berkhout B. 2005. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 44:9058–9066. doi: 10.1021/bi0502588 [DOI] [PubMed] [Google Scholar]
- 163. De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388. doi: 10.1126/science.279.5349.384 [DOI] [PubMed] [Google Scholar]
- 164. Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ding P, Kharytonchyk S, Waller A, Mbaekwe U, Basappa S, Kuo N, Frank HM, Quasney C, Kidane A, Swanson C, Van V, Sarkar M, Cannistraci E, Chaudhary R, Flores H, Telesnitsky A, Summers MF. 2020. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc Natl Acad Sci U S A 117:17737–17746. doi: 10.1073/pnas.2008519117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Smyth RP, Despons L, Huili G, Bernacchi S, Hijnen M, Mak J, Jossinet F, Weixi L, Paillart J-C, von Kleist M, Marquet R. 2015. Mutational interference mapping experiment (MIME) for studying RNA structure and function. Nat Methods 12:866–872. doi: 10.1038/nmeth.3490 [DOI] [PubMed] [Google Scholar]
- 167. Rye-McCurdy T, Olson ED, Liu S, Binkley C, Reyes JP, Thompson BR, Flanagan JM, Parent LJ, Musier-Forsyth K. 2016. Functional equivalence of retroviral MA domains in facilitating psi RNA binding specificity by Gag. Viruses 8:256. doi: 10.3390/v8090256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Comas-Garcia M, Kroupa T, Datta SAK, Harvin DP, Hu WS, Rein A. 2018. Efficient support of virus-like particle assembly by the HIV-1 packaging signal. Elife 7:e38438. doi: 10.7554/eLife.38438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Keane SC, Van V, Frank HM, Sciandra CA, McCowin S, Santos J, Heng X, Summers MF. 2016. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci U S A 113:13033–13038. doi: 10.1073/pnas.1614785113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess Jr JW, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. doi: 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Rulli SJ, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81:6623–6631. doi: 10.1128/JVI.02833-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Dilley KA, Nikolaitchik OA, Galli A, Burdick RC, Levine L, Li K, Rein A, Pathak VK, Hu WS. 2017. Interactions between HIV-1 Gag and viral RNA genome enhance virion assembly. J Virol 91:e02319-16. doi: 10.1128/JVI.02319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Poon DTK, Chertova EN, Ott DE. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55Gag: functional linkage between translation and RNA packaging. Virology 293:368–378. doi: 10.1006/viro.2001.1283 [DOI] [PubMed] [Google Scholar]
- 174. Nikolaitchik O, Rhodes TD, Ott D, Hu WS. 2006. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J Virol 80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hendrix J, Baumgärtel V, Schrimpf W, Ivanchenko S, Digman MA, Gratton E, Kräusslich H-G, Müller B, Lamb DC. 2015. Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J Cell Biol 210:629–646. doi: 10.1083/jcb.201504006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Fogarty KH, Berk S, Grigsby IF, Chen Y, Mansky LM, Mueller JD. 2014. Interrelationship between cytoplasmic retroviral Gag concentration and Gag–membrane association. J Mol Biol 426:1611–1624. doi: 10.1016/j.jmb.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog 6:e1001200. doi: 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Chen J, Grunwald D, Sardo L, Galli A, Plisov S, Nikolaitchik OA, Chen D, Lockett S, Larson DR, Pathak VK, Hu WS. 2014. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc Natl Acad Sci U S A 111:E5205–13. doi: 10.1073/pnas.1413169111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Jouvenet N, Simon SM, Bieniasz PD. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A 106:19114–19119. doi: 10.1073/pnas.0907364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Tuffy KM, Maldonado RJK, Chang J, Rosenfeld P, Cochrane A, Parent LJ. 2020. HIV-1 Gag forms ribonucleoprotein complexes with unspliced viral RNA at transcription sites. Viruses 12:1281. doi: 10.3390/v12111281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Baluyot MF, Grosse SA, Lyddon TD, Janaka SK, Johnson MC. 2012. CRM1-dependent trafficking of retroviral Gag proteins revisited. J Virol 86:4696–4700. doi: 10.1128/JVI.07199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Grewe B, Hoffmann B, Ohs I, Blissenbach M, Brandt S, Tippler B, Grunwald T, Überla K. 2012. Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with Gag. J Virol 86:2990–3002. doi: 10.1128/JVI.06874-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Sardo L, Hatch SC, Chen J, Nikolaitchik O, Burdick RC, Chen D, Westlake CJ, Lockett S, Pathak VK, Hu WS. 2015. Dynamics of HIV-1 RNA near the plasma membrane during virus assembly. J Virol 89:10832–10840. doi: 10.1128/JVI.01146-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Chen J, Rahman SA, Nikolaitchik OA, Grunwald D, Sardo L, Burdick RC, Plisov S, Liang E, Tai S, Pathak VK, Hu WS. 2016. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc Natl Acad Sci U S A 113:E201–8. doi: 10.1073/pnas.1518572113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Larson DR, Johnson MC, Webb WW, Vogt VM. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci U S A 102:15453–15458. doi: 10.1073/pnas.0504812102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Kharytonchyk SA, Kireyeva AI, Osipovich AB, Fomin IK. 2005. Evidence for preferential copackaging of moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site. Retrovirology 2:3. doi: 10.1186/1742-4690-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Flynn JA, Telesnitsky A. 2006. Two distinct moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology 344:391–400. doi: 10.1016/j.virol.2005.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chen J, Liu Y, Wu B, Nikolaitchik OA, Mohan PR, Chen J, Pathak VK, Hu WS. 2020. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc Natl Acad Sci U S A 117:6145–6155. doi: 10.1073/pnas.1917590117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Brown JD, Kharytonchyk S, Chaudry I, Iyer AS, Carter H, Becker G, Desai Y, Glang L, Choi SH, Singh K, Lopresti MW, Orellana M, Rodriguez T, Oboh U, Hijji J, Ghinger FG, Stewart K, Francis D, Edwards B, Chen P, Case DA, Telesnitsky A, Summers MF. 2020. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 368:413–417. doi: 10.1126/science.aaz7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Rawson JMO, Nikolaitchik OA, Shakya S, Keele BF, Pathak VK, Hu WS. 2022. Transcription start site heterogeneity and preferential packaging of specific full-length RNA species are conserved features of primate lentiviruses. Microbiol Spectr 10:e0105322. doi: 10.1128/spectrum.01053-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kharytonchyk S, Burnett C, Gc K, Telesnitsky A. 2023. Transcription start site heterogeneity and its role in RNA fate determination distinguish HIV-1 from other retroviruses and are mediated by core promoter elements. J Virol 97:e0081823. doi: 10.1128/jvi.00818-23 [DOI] [PMC free article] [PubMed] [Google Scholar]